Abstract
The signal-mediated and spatially controlled assembly and dynamics of actin are crucial for maintaining shape, motility, and tip growth of eukaryotic cells. We report that a novel Armadillo repeat protein in Arabidopsis thaliana, ARMADILLO REPEAT ONLY1 (ARO1), is of fundamental importance for polar growth and F-actin organization in tip-growing pollen tubes. ARO1 is specifically expressed in the vegetative cell of pollen as well as in the egg cell. ARO1-GFP (for green fluorescent protein) fusion proteins accumulate most notably in pollen tube tips and partially colocalize with F-actin in the shank of pollen tubes. ARO1 knockout results in a highly disorganized actin cytoskeleton, growth depolarization, and ultimately tube growth arrest. Tip-localized ARO1-GFP is spatially shifted toward the future site of tip growth, indicating a role of ARO1 in the signaling network controlling tip growth and regulating actin organization. After the pollen tube discharges its contents into the receptive synergid, ARO1-GFP colocalizes with emerging F-actin structures near the site of sperm cell fusion, suggesting additional participation in the mechanism of sperm cell tracking toward the female gametes. The variable localization of ARO1 in the cytoplasm, the nucleus, and at the plasma membrane, however, indicates a multifunctional role like that of β-catenin/Armadillo and the p120 catenins.
INTRODUCTION
Establishing and maintaining proper cell polarity are key aspects of life. Polarization and asymmetrical growth in eukaryotic cells require elaborate regulation of a variety of cellular processes, including spatially restricted signaling, reorganization of the cytoskeleton, and polarized membrane trafficking. In plants and fungi, there are several different examples of cells exhibiting polarized growth, including pollen tubes, root hairs, fern/moss protonemata, algal rhizoids, and fungal hyphae (Hepler et al., 2001). As the pollen tube is the fastest growing plant cell, it is widely used as a model system to study tip growth. Once germinated on the surface of the stigma, pollen grains generate an elongating tube that navigates through the female tissues toward the ovule to deliver its two sperm cells for double fertilization (Weterings and Russell, 2004). Similar to axonal growth cones, pollen tubes require a signaling network to recognize and interpret extracellular cues that determine the site and the direction of tip growth (Palanivelu and Preuss, 2000; Geitmann and Palanivelu, 2007).
In recent years, crucial molecular players of the pollen tube tip growth machinery have been identified. A tip-focused calcium gradient, elaborate F-actin dynamics, tip-directed vesicle trafficking, and localized exocytosis are generally accepted as essential intracellular events (reviewed in Cole and Fowler, 2006; Chebli and Geitmann, 2007; Wilsen and Hepler, 2007; Cheung and Wu, 2008). The actin filament dynamics, the equilibrium between globular actin and F-actin pools, and the highly ordered architecture of the actin cytoskeleton are known to be controlled by several classes of actin binding proteins (Ren and Xiang, 2007). Among these, formins and actin-depolymerizing factors are promising key effectors, according to their localization and to misexpression phenotypes (Chen et al., 2003; Cheung and Wu, 2004).
Rho-GTPases are important regulators of the actin cytoskeleton, for example, in budding yeast or in the leading edge of motile mammalian cells, such as moving growth cones or lamellipodia (Etienne-Manneville and Hall, 2002). Likewise, the spatially restricted signaling of tip-localized plant-specific Rops (for Rho of plants) is pivotal for actin dynamics in growing pollen tubes (Gu et al., 2003; Kost, 2008). The recent discoveries of Rop-activating Rho-GEFs (for guanine nucleotide exchange factors) and of the interaction between a Rho-GEF and pollen tube receptor–like kinases suggest a link between extracellular cues and downstream effects in mediating tip growth (Berken et al., 2005; Kaothien et al., 2005; Gu et al., 2006). Likewise, not much is known about effectors and downstream signaling cascades of Rop. In mammalian cells, Rho-GTPases are known to interact with a plethora of distinct target proteins to achieve their various functions (Hall, 2005). In Arabidopsis thaliana, two CRIB (for Cdc42/Rac-interactive binding) motif–containing proteins, RIC3 (for Rop-interactive CRIB motif–containing protein3) and RIC4, were found to be direct targets of ROP1, operating in two counteracting pathways (Gu et al., 2005). While RIC3 is thought to regulate the accumulation of tip-localized Ca2+ and was found to induce localized exocytosis, RIC4, in a yet unknown way, promotes the assembly of F-actin in the pollen tube tip, which is required for the accumulation of exocytotic vesicles to the region near the cortex of the tip (Lee et al., 2008). Ca2+-sensitive actin binding proteins such as profilin or gelsolin may be potential targets of the RIC3 pathway.
The first Armadillo (ARM) repeat protein linked to cytoskeletal regulation in multicellular organisms was β-catenin, a component of adherens junctions in animals (Hatzfeld, 1999; reviewed in Coates, 2003). Besides its important role in Wnt/wingless signaling, β-catenin acts as a dynamic adapter protein linking cadherin transmembrane cell-to-cell adhesion receptors to α-catenin, which in turn interacts with actin. Furthermore, the p120-catenin (p120ctn) subfamily of ARM proteins has a role in regulating adherens junction and desmosome formation by their capacity to cluster cadherins. Recently, members of the p120ctn subfamily were found to have a major function in regulating cell adhesion and motility through their interaction with Rho GTPases and Rho-GEFs (reviewed in Anastasiadis, 2007; Hatzfeld, 2007).
Here, we describe a novel family of ancient plant-specific ARM repeat proteins, which have a characteristic overall structure but no additional protein motifs and therefore were termed ARMADILLO REPEAT ONLY (ARO) proteins. We show that the protein encoded by Arabidopsis ARO1 is expressed exclusively in the egg cell and in the vegetative cell of the male gametophyte. The knockout phenotype of ARO1 and its spatially regulated accumulation in the pollen tube tip indicate that it is of fundamental importance for polar tip growth and for F-actin organization, and it probably also plays a role during the double fertilization process of flowering plants. However, its cytoplasmic and nuclear localization suggests that ARO1 is a multifunctional protein with a function in signaling and transcriptional regulation in addition to its effects on F-actin organization, similar to animal β-catenin and p120ctn.
RESULTS
ARO1 Is a Member of a Small and Phylogenetically Ancient Protein Family in Arabidopsis
A previous analysis of ESTs derived from isolated wheat (Triticum aestivum) egg cells led to the discovery of a novel transcript encoding an ARM repeat–containing protein (EC-123; GenBank accession number EU662204), which was found to be expressed exclusively in egg cells and anthers (Sprunck et al., 2005). BLAST searches revealed significant similarity to hypothetical ARM repeat–containing proteins from rice (Oryza sativa; Os08g0548500) and Arabidopsis (At4g34940). Besides the ARM repeats, the proteins do not contain any recognizable protein motif or any similarity to known proteins; therefore, they were termed ARO proteins. We found At4g34940 to be a member of a small Arabidopsis protein family, designated ARO1 to ARO4. All four proteins are encoded by a single exon, with a length of between 615 (ARO4) and 670 (ARO3) amino acid residues and a predicted molecular mass of 67.8 to 73 kD, respectively. While ARO2 and ARO3 are very similar to ARO1, ARO4 represents the most divergent member of the protein family (see Supplemental Figure 1A and Supplemental Methods online). However, all four proteins share a unique primary structure comprising two conserved ARM repeat–containing domains (ARD1 and ARD2), which are separated by a highly divergent spacer region. A domain of 143 to 153 amino acids is located at the N terminus, and a short stretch of 55 to 59 amino acids follows ARD2 at the C terminus (see Supplemental Figure 1A online). Based solely on sequence homology, only up to three bona fide ARM repeats were recognized in each of the four ARO proteins in Arabidopsis. Nevertheless, when we combined secondary structure and Pfam predictions, the presence of at least six complete and three incomplete putative ARM repeats was indicated (see Supplemental Figures 1B and 2 online). Despite the low sequence conservation of some of the putative ARM repeats, they possess a typical three-helix composition and are predicted to form an elongated superhelical structure similar to that of importin-α (see Supplemental Figure 1C online).
We also found a small family of four ARO-like hypothetical proteins (Os ARO-like; Os AROl-1 to -4) to be present in the rice genome. Furthermore, we identified ARO-like proteins in the genomes of Zea mays (maize), Medicago truncatula (barrel clover), Vitis vinifera (grape), Lotus japonicus (lotus), Populus trichocarpa (black cottonwood), and the moss Physcomitrella patens as well as in the EST data collections of Picea (spruce) and Pinus (pine) species, while we could not find any ARO-like proteins in Chlamydomonas, yeast, or animal. The unrooted phylogenetic tree generated with ARO-like proteins reveals that monocotyledonous, dicotyledonous, gymnosperm, and moss ARO-like proteins form individual subclades (see Supplemental Figure 3 and Supplemental Data Set 1 online), with the exception of Arabidopsis ARO4. When we compared the predicted secondary structure and putative phosphorylation sites of Arabidopsis ARO1 with those of its closest relatives from other plant species, we found the overall architecture as well as several predicted phosphorylation sites to be conserved (see Supplemental Figure 2 online).
ARO1 Is Expressed Exclusively in Egg Cells and Pollen
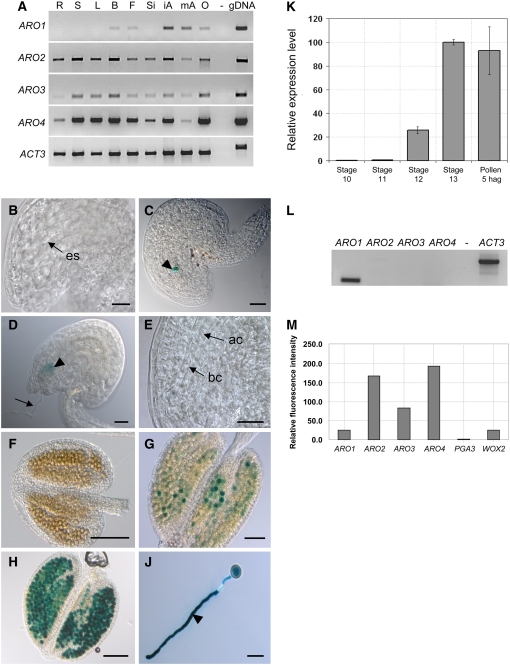
RT-PCR expression studies revealed that Arabidopsis ARO1 is not expressed in vegetative tissues but only in tissues containing the male and female gametophytes. Transcripts of ARO1 were detected in buds and flowers as well as in dissected anthers and ovaries (Figure 1A). By contrast, the other members of the gene family are expressed in all tissues tested. To provide a more detailed view of ARO1 expression during female and male gametophyte development, homozygous transgenic lines were generated, expressing β-glucuronidase (GUS) under the control of the ARO1 promoter. GUS activity was not detected during early stages of ovule development (Figure 1B) but was restricted to mature egg cells of female gametophytes (Figure 1C). While weak GUS activity was still detectable in zygotes (Figure 1D), it was absent in two-celled proembryos (Figure 1E). During anther development, GUS activity was first detected at stage 11 of flower development (Smyth et al., 1990), when pollen reached the trinucleate stage (Figure 1G). Strong GUS activity was observed in mature pollen grains (Figure 1H) as well as in in vitro germinated pollen tubes (Figure 1J). This was confirmed by quantitative RT-PCR, when relative expression levels of ARO1 were determined using Eukaryotic Transcription Initiation Factor4G (eIF4) as a reference housekeeping gene (Figure 1K). During anther maturation (floral stages 10 to 13), ARO1 transcript levels increased significantly, and the highest transcript levels were found in mature pollen and in pollen tubes, 5 h after in vitro germination. RT-PCR analysis using mature pollen but excluding anther tissues revealed that ARO1 is the only member of the gene family expressed in pollen (Figure 1L). By contrast, all four AROs appear to be expressed in the egg cell, as indicated by Affymetrix GeneChip experiments using fluorescently labeled amplified cRNA from isolated Arabidopsis egg cells (Figure 1M). ARO2 to ARO4 seem to be expressed more strongly than ARO1 and WUSCHEL-related homeobox2 (WOX2), a gene that was recently shown to be expressed in egg cells by in situ hybridization (Haecker et al., 2004).
Figure 1.
Expression of Arabidopsis ARO1 to ARO4.
(A) RT-PCR using mRNA isolated from root (R), stem (S), leaf (L), bud (B), flower (F), silique (Si), immature anther (iA), mature anther (mA), and ovary (O) and gene-specific primers for ARO1 to ARO4 and Actin3 (ACT3).
(B) to (J) ARO1 promoter activity in the female and male gametophytes of homozygous ARO1pro:GUS plants.
(B) Immature ovule with developing embryo sac (es) lacking GUS activity.
(C) Mature ovule at 2 d after emasculation. GUS staining is visible in the egg cell (arrowhead).
(D) and (E) ARO1pro:GUS ovules, pollinated with wild-type pollen.
(D) At 18 h after in vitro germination, GUS activity is still visible in the zygote (arrowhead). Arrow points at the pollen tube.
(E) Apical (ac) and basal (bc) cells of two-celled proembryo, lacking GUS signals.
(F) Immature anther (floral stage 10).
(G) Anther at floral stage 11, when GUS activity is first detectable.
(H) Anther at floral stage 13, with mature pollen showing strong GUS activity.
(J) Pollen tube (arrowhead) after 6 h of in vitro germination.
(K) Relative expression levels of ARO1 during anther development and in vitro pollen germination, estimated by quantitative RT-PCR. Highest transcript levels are present in mature pollen and in pollen tubes at 5 h after in vitro germination (hag).
(L) RT-PCR using mRNA from mature pollen and gene-specific primers for ARO1 to ARO4 and ACT3.
(M) Expression of ARO1 to ARO4 in egg cells. Relative signal intensities (P < 0.05) of one ATH1 GeneChip experiment using fluorescently labeled cRNA of isolated egg cells for hybridization. WOX2 and pollen-specific PGA3 were considered as positive and negative controls, respectively.
Bars = 20 μm in (A) to (D), 100 μm in (E) and (G), 50 μm in (F), and 25 μm in (H). Flower stages are according to Smyth et al. (1990).
Polar Tip Growth and Penetration into the Stigma Are Impaired in aro1-3 Pollen Tubes
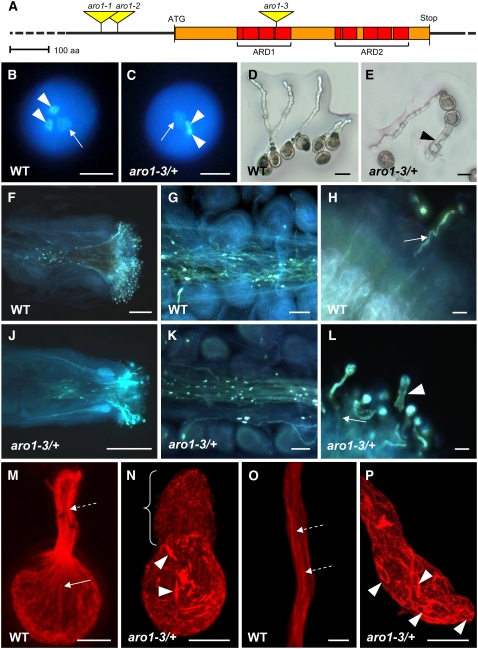
Three independent SALK T-DNA insertion lines were considered for functional analysis of ARO1 (Figure 2A). The mutant alleles were named aro1-1 (insertion at −360 bp upstream of the start codon), aro1-2 (insertion at −327 bp upstream of the start codon), and aro1-3 (insertion at +791 bp, corresponding to amino acid position 263).
Figure 2.
Phenotypic Analysis of aro1-3/+ Pollen and Pollen Tubes.
(A) Genomic structure of ARO1. Positions of ARM repeats are indicated as red boxes, forming the ARM repeat domains 1 and 2 (ARD1 and ARD2). T-DNA insertions in aro1-1, aro1-2, and aro1-3 are indicated. The scale bar = 100 amino acids (aa).
(B) to (L) Phenotypes of aro1-3/+ in comparison with the wild type.
(B) and (C) Epifluorescence microscopy after DAPI staining of wild-type (B) and aro1-3/+ (C) mature pollen. Vegetative nucleus (arrow) and two sperm cell nuclei (arrowheads) are visible.
(D) and (E) Bright-field images of wild-type pollen tubes grown on semisolid medium (D) compared with the aro1-3 pollen tube phenotype ([E], arrowhead).
(F) to (L) Epifluorescence microscopy after aniline blue staining of pollen tubes growing through pistils 24 h after hand pollination with self pollen.
(F) and (J) Pollen tubes of aro1-3/+ (J) appear to grow through the pistil, similar to wild-type pollen tubes (F).
(G) and (K) Close-ups of transmitting tracts. Only aro1-3/+ pollen tubes of wild-type morphology can enter the female tissues (K), as do wild-type pollen tubes (G).
(H) and (L) Close-ups of papillae. Wild-type pollen tubes and aro1-3/+ pollen tubes of wild-type morphology grow into the stigmatic cells and throughout the style (arrows), while pollen tubes with the aro1-3 phenotype remain on the surface of the stigmatic cells (arrowhead in [L]).
(M) to (P) Actin cytoskeleton of in vitro germinated wild-type and aro1-3 pollen, stained with rhodamine–phalloidin and analyzed by CLSM.
(M) and (O) In the wild type, long and polarized actin bundles are visible in the pollen grain (arrow in [M]; projection of 21 optical sections, 0.36 μm each) and pollen tube (dashed arrows in [M] and [O]; single optical section of 0.41 μm).
(N) and (P) aro1-3 pollen exhibit unorganized short actin bundles inside the pollen grain (arrowheads in [N]; projection of 47 optical sections, 0.41 μm each) and pollen tube ([P]; projection of 27 optical sections; 0.36 μm each; bracket in [N]).
Bars = 5 μm in (O), 10 μm in (B), (C), (M), (N), and (P), 20 μm in (D), (E), (H), and (L), 50 μm in (G) and (K), and 200 μm in (F) and (J).
The homozygous offspring of the T-DNA insertion lines aro1-1 and aro1-2 did not show any phenotype in either vegetative development or reproduction. By contrast, no homozygous plant was obtained among the aro1-3 progeny (n = 178). The mutant allele segregated in a ratio of 1:1.02 (+/+:aro1-3/+), indicating a gametophytic effect. Reciprocal crosses of heterozygous aro1-3/+ with the wild type revealed that aro1-3 was only transmitted by the female gametophyte and never by the male (see Supplemental Table 1 online), suggesting a functional role of ARO1 in either pollen development or postpollination events.
Cytological analysis of 4′,6-diamidino-2-phenylindole (DAPI)–stained mature pollen grains from heterozygous aro1-3/+ plants did not show any distinctive feature compared with wild-type pollen (Figures 2B and 2C). The size and shape of these pollen grains were normal, and both the vegetative nucleus and the two sperm cell nuclei were present (Figure 2C). However, ∼50% of pollen from aro1-3/+ plants germinated in vitro displayed a severe phenotype. The pollen tubes either burst immediately after in vitro germination or were short and plump with large vacuole-like structures (Figure 2E). Statistical evaluation revealed that 51.6% of aro1-3/+ pollen tubes appeared phenotypically similar to wild-type controls (long, with uniform diameter), while 28% of these tubes had a short and plump phenotype (short, with variably increased diameter; referred to hereafter as aro1-3 pollen tubes). The diameter of aro1-3 pollen tubes ranged from 15 to 35 μm, with an average of 23 μm, while wild-type pollen tubes were 5 to 7 μm in diameter. Another 20.4% of aro1-3 pollen grains burst immediately after germination (Table 1). Measurement of tube lengths after 3 and 6 h of in vitro germination revealed that the growth rate of aro1-3 pollen tubes was significantly retarded. At 6 h after in vitro germination, the average length of wild-type pollen tubes was 85.65 μm (ranging from 4.1 to 192.9 μm), while aro1-3 pollen tubes were between 6.98 and 47.09 μm long, with an average length of 19.39 μm (Table 2). Successful complementation verified the causal relationship between the observed pollen tube phenotype and the aro1-3 allele and demonstrated that the ARO1-GFP (for green fluorescent protein) fusion protein that we used is functional in planta (see Supplemental Figure 4 online).
Table 1.
Pollen Tube Phenotypes of Wild-Type (+/+) and aro1-3/+ Pollen, Observed 5 h after in Vitro Germination
| Pollen | Normal | Short and Plump | Burst Pollen |
|---|---|---|---|
| aro1-3/+ | 51.6% (n = 1608) | 28.0% (n = 874) | 20.4% (n = 634) |
| +/+ | 98.8% (n = 3956) | 0.01% (n = 3) | 1.1% (n = 44) |
Pollen grains of wild-type and heterozygous aro1-3/+ plants were germinated on the two halves of a Petri dish. Average germination rates were 69% for wild-type and 68% for aro1-3/+ pollen (four independent experiments). Pollen tubes of at least 4 μm in length were scored as germinated. Five hours after germination, phenotypes of germinated pollen were scored microscopically.
Table 2.
Average Length of Wild-Type (+/+) and aro1-3/+ Pollen Tubes Measured after 3 and 6 h of in Vitro Germination
| Tube Length (μm)a
|
||
|---|---|---|
| lPollen | 3 hag | 6 hag |
| +/+ | 37.52 ± 21.43 | 85.65 ± 44.54 |
| aro1-3/+; normal phenotype | 34.09 ± 21.95 | 89.30 ± 38.29 |
| aro1-3/+; short and plump phenotype | 16.90 ± 9.15 | 19.39 ± 10.14 |
Pollen grains of wild-type and heterozygous aro1-3/+ plants were germinated on the two halves of a Petri dish. After the indicated incubation times (hours after germination [hag]), pollen tubes were observed and photographed with a Zeiss Axioskop microscope equipped with a Zeiss AxioVision camera.
Pollen tube length was estimated using the Zeiss AxioVision software.
In vivo germination of pollen from aro1-3/+ plants was analyzed by aniline blue staining of pistils at 24 h after limited hand pollination. Pollen tubes of aro1-3/+ plants were visible in the transmitting tract of pistils, as were pollen tubes of the wild-type controls (Figures 2F and 2J). Closer examination revealed that only pollen tubes with wild-type morphology were able to grow through the style toward the ovules (Figures 2G and 2K). Phenotypic short and plump pollen tubes were only found on the surface of the papillae, which indicates that aro1-3 pollen are in principle able to adhere, hydrate, and germinate in vivo. However, mutant pollen tubes failed to enter the cell wall of stigmatic cells (Figure 2L, arrowhead) and were often observed to form protuberances at the tube tip. As the aro1-3 allele is not transmitted by the male parent (see Supplemental Table 1 online), we conclude that aro1-3/+ plants will become fertilized exclusively by pollen with a wild-type genetic background because aro1-3 pollen tubes are impaired in growing into the stigma.
aro1-3 Pollen Tubes Reveal a Disorganized Actin Cytoskeleton
To examine the cause for the aro1-3 pollen tube morphology, we fixed in vitro germinated pollen tubes, stained them with rhodamine–phalloidin, and analyzed the actin cytoskeleton using confocal laser scanning microscopy (CLSM). Wild-type pollen grains contained thick actin cables, which were strictly oriented toward the germination pore (Figure 2M, solid arrow). In the shank of wild-type pollen tubes, actin cables were oriented in parallel bundles along the growth axis of the tube (Figures 2M and 2O, dashed arrows). Strikingly, the actin cytoskeleton of aro1-3 pollen tubes was severely disorganized. Short fragments of randomly oriented actin bundles were visible in the pollen grains of germinated pollen (Figure 2N, arrowheads). The bulk of very short aro1-3 pollen tubes revealed a disorganized meshwork of short actin filaments throughout the tube (Figure 2N, bracket). In the rare case of longer aro1-3 pollen tubes, short bundles of randomly oriented and occasionally intertwining actin filaments were visible in the shank of the pollen tube and in the tip (Figure 2P, arrowheads). Growth retardation and loss of polarized tip growth of aro1-3 pollen tubes thus coincide with the loss of long and parallel actin bundles along the growth axis of the zygote, together with an accumulation of short and disordered actin filaments at the tip of the pollen tubes.
Subcellular Localization of the ARO1-GFP Fusion Protein in the Female and Male Gametophytes of Arabidopsis
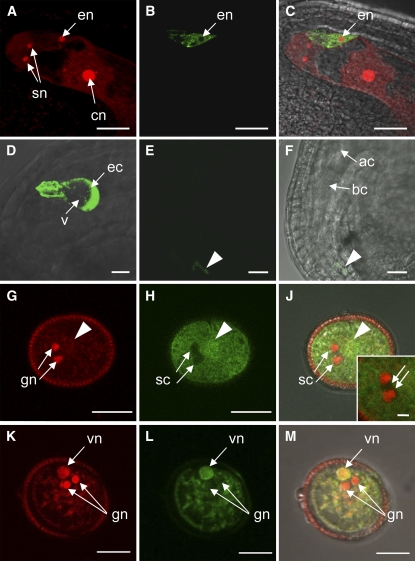
We generated transgenic lines expressing the ARO1-GFP fusion protein under the control of the ARO1 promoter (ARO1pro:ARO1-GFP) and evaluated homozygous progeny for the time frame of GFP expression and subcellular localization of the fusion protein. In the female gametophyte, expression of the ARO1-GFP fusion protein was restricted to mature egg cells (Figures 3A to 3C), consistent with results obtained from ARO1pro:GUS lines. While the GFP fluorescence was found in the cytoplasm of egg cells, we never detected any fluorescence in the egg nucleus (Figure 3B, en) or in the vacuole (Figure 3D, v). After the first asymmetric cell division of the zygote, ARO1-GFP was no longer detectable (Figures 3E and 3F).
Figure 3.
Localization of ARO1-GFP in Gametophytes of Homozygous ARO1pro:ARO1-GFP Lines Using CLSM.
(A) to (C) ARO1-GFP is localized in the egg cell of the mature embryo sac but is excluded from the nucleus (en).
(A) Draq5 staining of the synergid nuclei (sn), the egg cell nucleus (en), and the central cell nucleus (cn) (projection of nine optical sections, 0.34 μm each).
(B) ARO1-GFP in the egg cell (single optical section).
(C) Merged image of (A) and (B).
(D) Close-up view of a mature egg cell (ec). ARO1-GFP is excluded from the large vacuole (v) (projection of 75 optical sections, 0.36 μm each).
(E) and (F) Ovule at 24 h after pollination (hap). After the first zygotic division, ARO1-GFP is no longer detectable (same confocal settings used as in [D]). Remnants of pollen tube–derived fluorescence are visible in the micropylar region (arrowhead). (F) shows a bright-field image with fluorescent signal overlay of (E). ac, apical cell; bc, basal cell.
(G) to (M) Pollen grains stained with propidium iodide.
(G) The two generative nuclei (gn) are stained strongly, and the vegetative nucleus (arrowhead) is not in focus.
(H) ARO1-GFP is not present at the position of the sperm cells (sc; arrows; inset in [J]).
(J) Merged image of (G) and (H).
(K) to (M) The vegetative nucleus (vn) and the two generative nuclei (gn) are visible, but ARO1-GFP is only present in the vegetative nucleus (L), visible as yellow staining in the merged image (M).
(G) to (J) show single 0.5-μm optical sections, and (K) to (M) show single 0.4-μm optical sections. Bars = 20 μm in (A) to (C), (E), and (F), 10 μm in (D) and (G) to (M), and 2 μm in the inset in (J).
Analysis of pollen grains revealed that ARO1-GFP is located in the cytoplasm of the vegetative cell (Figures 3H and 3L). Furthermore, intense fluorescence was detected in the nucleus of the vegetative cell (Figures 3K to 3M, vn), while nuclei and cytoplasm of sperm cells completely lacked GFP fluorescence (Figures 3G to 3M). Both cytoplasmic and nuclear localization of ARO1-GFP were also observed in the majority of transiently transformed epidermal onion cells expressing ARO1-GFP under the control of the maize Ubiquitin (UBI) promoter (see Supplemental Figure 5B online). However, 10% of the transiently transformed cells showed GFP fluorescence in the cytoplasm but not in the nucleus (see Supplemental Figure 5D online). Furthermore, the ARO1-GFP fusion protein appeared to be located exclusively at the plasma membrane in 17% of all analyzed cells (see Supplemental Figure 5F online). This variation in subcellular localization was not observed in Arabidopsis eggs or in pollen expressing ARO1pro:ARO1-GFP, nor was it seen in transiently transformed epidermal spring onion cells expressing an ARO2-GFP fusion protein or the GFP control (see Supplemental Figures 5H and 5K online).
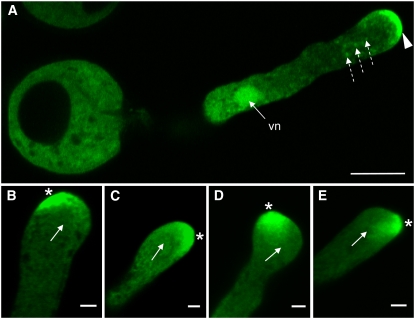
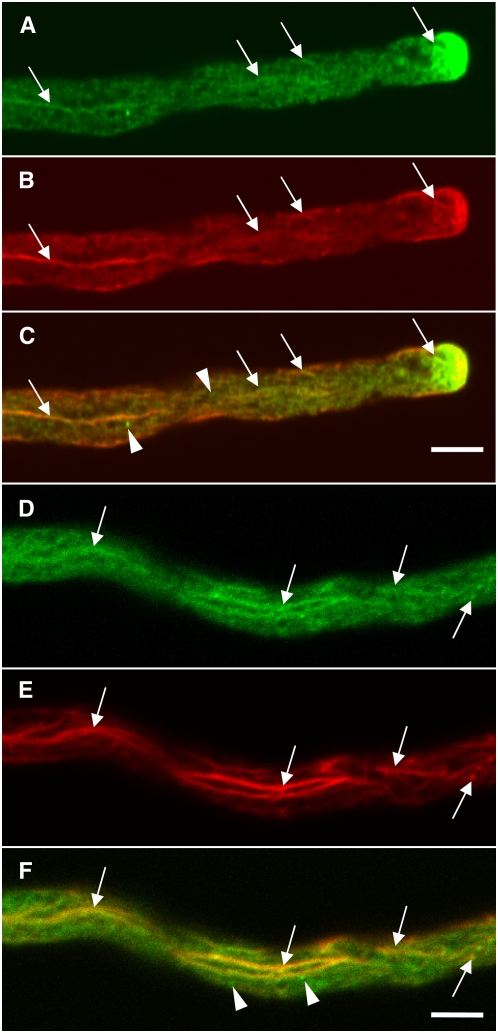
Slightly grainy fluorescence of ARO1-GFP was visible throughout the cytoplasm of in vitro germinated pollen, together with several fluorescing spots (Figure 4A; dashed arrows). Some filament-like fluorescent structures were usually visible, especially in the central part of pollen tubes, parallel to the axis of growth (see Supplemental Figures 6A and 6B online). As in ungerminated pollen, ARO1-GFP was present in the vegetative nucleus (Figure 4A, vn), while sperm cells were free of any fluorescence (see Supplemental Figures 6A and 6B and Supplemental Movie 1 online). Strikingly, ARO1-GFP appeared to accumulate at the very tip of the germinated pollen tube (Figure 4A, arrowhead). This accumulation was present in all pollen tubes analyzed, although the fluorescence intensity was found to vary, and filled 0.7 to 5 μm of the tip. Moreover, a change in the growth direction of pollen tube tips was always accompanied by a relocation of fluorescence toward the future growth site of the tips (Figures 4B to 4E).
Figure 4.
Relocation of ARO1-GFP in the Tip of in Vitro Germinated ARO1pro:ARO1-GFP Pollen Tubes during Growth Reorientation, Analyzed by CLSM.
(A) ARO1-GFP is distributed throughout the cytoplasm of the pollen tube but clearly accumulates at the very tip (arrowhead) of straight-growing pollen tubes. The fusion protein is also present in the vegetative nucleus (vn), and some fluorescent spots are visible in the cytoplasm (dashed arrows). Sperm cells are not in focus.
(B) to (E) Lopsided fluorescence accumulating at the future growth site of the pollen tube tip (indicated by asterisks). Arrows indicate previous growth direction of pollen tubes.
(A) and (B) show single 0.5-μm optical sections, and (C) to (E) show projections of three 0.34-μm optical sections. Bars = 10 μm in (A) and 2 μm in (B) to (E).
ARO1 Partially Colocalizes with Actin Filaments, but Its Accumulation in the Pollen Tube Tip Is BFA-Sensitive
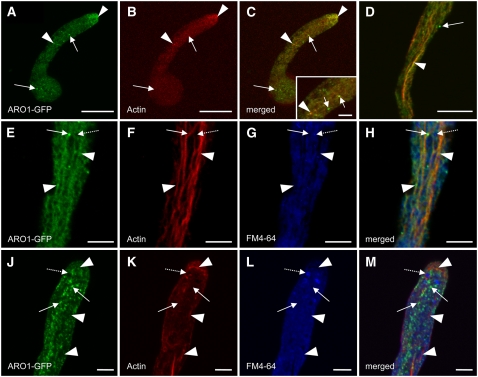
We examined the colocalization of ARO1-GFP and actin by CLSM, after the actin cytoskeleton of in vitro germinated ARO1pro:ARO1-GFP pollen tubes was fixed and stained. Fluorescence of ARO1-GFP was weaker but still visible after fixation and colocalized along longitudinal bundles of actin filaments in the shank of the pollen tube, with some shorter actin filaments in the apical region of the tube tip (Figure 5, arrows). However, the subcellular distribution of ARO1-GFP did not completely cover the labeled actin filaments but appeared somewhat discontinuous, and several fluorescing spots did not show any colocalization with actin (Figures 5C and 5F, arrowheads). Also, without fixation we observed only some filament-like green fluorescent structures occurring particularly in the central part of pollen tubes (see Supplemental Figures 6A and 6B online).
Figure 5.
ARO1-GFP Shows Partial Colocalization with F-Actin.
Colocalization studies of ARO1-GFP and F-actin in ARO1pro:ARO1-GFP pollen tubes (single optical sections). Filament-like fluorescence of ARO1-GFP is visible in the shank of the tube (arrows) ([A] and [D]), corresponding to actin bundles stained with rhodamine–phalloidin ([B] and [E]). Merged images of (A) and (B) and of (E) and (F) are shown in (C) and (F), respectively. Arrowheads indicate GFP signal only. Bars = 5 μm.
We verified the partial colocalization of ARO1-GFP and actin filaments by treating in vitro germinated ARO1pro:ARO1-GFP pollen tubes with latrunculin B (LatB), previously shown to be a potent inhibitor of actin polymerization in pollen tubes (Gibbon et al., 1999). Effective depolymerization of F-actin resulted in scattered weak red fluorescence visible in rhodamine–phalloidin-stained pollen tubes (Figure 6B). The slightly enriched staining in the apical region presumably represents G-actin or patches of fragmented F-actin. The disruption of actin filaments was accompanied by the disappearance of the filament-like ARO1-GFP signal in the shank of pollen tubes (Figure 6A), contrary to control pollen tubes treated with the solvent DMSO only (Figure 6D). The green fluorescence in the apex of LatB-treated pollen tubes appeared weaker and more dispersed compared with that of untreated pollen tubes (Figures 4A and 5A), indicating a correlation between the apical dynamic F-actin and the spatial restriction of ARO1 within the tip. Colocalization of ARO1-GFP with actin remaining after LatB treatment was visible in the apex and in some spots in the shank of the tube (Figure 6C, arrowheads). However, most of the green fluorescent spots in the cytoplasm did not colocalize with actin (Figure 6C, arrows).
Figure 6.
ARO1-GFP Colocalizes with F-Actin but Not with Endocytotic Membrane Compartments, as Shown in Drug-Treated ARO1pro:ARO1-GFP Pollen Tubes.
(A) to (C) LatB causes weak and dispersed staining of ARO-GFP (A) and actin (B). A merged image of (A) and (B) is shown in (C). Punctiform GFP signals (arrows) do not colocalize with actin. Arrowheads point at colocalization of ARO1-GFP and actin remaining after LatB treatment. A magnification of colocalizing (arrowhead) and noncolocalizing (arrows) signals is shown in the inset in (C).
(D) Control pollen tube treated with DMSO (the LatB solvent) only. Colocalization of ARO1-GFP and F-actin is visible at orange areas of the pollen tube shank (arrowhead).
(E) to (H) FM4-64FX–treated pollen tube. ARO1-GFP colocalizes with actin bundles in the pollen tube shank (arrowheads). Punctiform fluorescence of ARO1-GFP (arrows) does not colocalize with endocytotic membrane compartments labeled by FM4-64FX (dashed arrows).
(J) to (M) BFA-treated pollen tube stained with FM4-64FX. ARO1-GFP no longer decorates thick actin bundles (arrowheads) but appears as fluorescent bodies (arrows) that do not colocalize with FM4-64FX–labeled BFA-induced membrane compartments (dashed arrows).
Bars = 20 μm in (A) to (D) and 5 μm in (E) to (M) and the inset in (C).
We investigated a putative plasma membrane– and vesicle-associated localization of ARO1-GFP by the use of the styril dye FM4-64, an established tracer of endocytotic membrane traffic in plant cells (Bolte et al., 2004), and brefeldin A (BFA), an efficient inhibitor of vesicular secretion in plant cells (Satiat-Jeunemaitre et al., 1996). In vitro germinated ARO1pro:ARO1-GFP pollen tubes were incubated for 30 min with the fixable derivative FM4-64FX, to allow the visualization of parts of the endomembrane system (blue staining) together with actin filaments (red staining) (Figures 6E to 6H). Again, the filament- and speckle-like fluorescing signals of ARO1-GFP localized along some actin filaments (Figures 6E to 6H, arrowheads). However, there was no colocalization of FM4-64–labeled membrane compartments (Figures 6G and 6H, dashed arrows) and the fluorescing spots of ARO1-GFP (Figures 6E and 6H, solid arrows). Interestingly, the localization of ARO1-GFP changed significantly upon BFA treatment (Figures 6J to 6M). The tip-focused accumulation of ARO1-GFP disappeared and the filament-like distribution of ARO1-GFP changed into a punctiform fluorescence of variable intensity and size, visible throughout the pollen tube (Figure 6J). Like a string of pearls, ARO1-GFP was visible throughout the pollen tubes but colocalized rather infrequently with thick actin cables (Figures 6K and 6M, arrowheads), which were not significantly altered upon BFA treatment. However, the punctiform fluorescence of ARO1-GFP never colocalized with subapical FM4-64–stained BFA-induced membrane aggregations (Figures 6L and 6M, dashed arrows).
During Fertilization, Male-Derived ARO1 Colocalizes with Emerging Actin Filaments Near the Site of Gamete Fusion
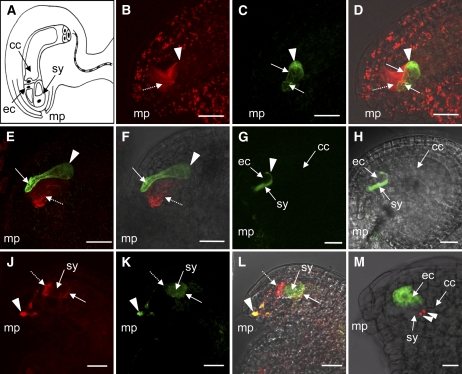
To investigate the localization of ARO1 and actin in the egg apparatus before fertilization, we stained the actin cytoskeleton of emasculated wild-type and ARO1-GFP–expressing Arabidopsis ovules with rhodamine–phalloidin. The synergids exhibited strong staining of actin filaments with a polar orientation along their longitudinal axis (Figures 7B and 7E, dashed arrows). By contrast, egg cells only exhibited weak actin staining, essentially at the apical cortex (Figure 7B, arrowhead). Besides being a matter of detection sensitivity, mature Arabidopsis egg cells may thus either lack extensive actin filaments or could be predominantly occupied by dynamic F-actin.
Figure 7.
Localization of ARO1-GFP in the Female Gametophyte before and during Fertilization.
(A) Schematic representation of an Arabidopsis ovule. The lateral view presents the egg cell (ec) and one of the two synergids (sy) near the micropyle (mp). The large central cell (cc) fills most of the embryo sac.
(B) to (D) An ARO1pro:ARO1-GFP–expressing ovule, stained for actin at 2 d after emasculation (projection of 24 optical sections, 0.34 μm each).
(B) The synergid exhibits prominent axially oriented actin bundles (dashed arrow), while actin staining in the egg cell is scarce and visible mainly at the cortex (arrowhead).
(C) Cytoplasmic ARO1-GFP is predominantly present at the chalazal pole of the mature egg cell (arrowhead). Fluorescence is also seen in transvacuolar strands (arrows).
(E) and (F) ARO1pro:ARO1-GFP ovule at 1 h after hand pollination (projection of 55 optical sections, 0.40 μm each).
(E) Actin staining reveals prominent axial actin bundles (dashed arrow) in the synergids. ARO1-GFP is enriched at the micropylar region of the egg cell (arrow). The arrowhead marks the chalazal pole of the egg cell.
(F) Merged image of (E) and a bright-field image.
(G) Distribution of pollen-derived ARO1-GFP after pollen tube discharge into a wild-type ovule (single 0.40-μm optical section). The fluorescence fills the degenerating synergid (sy) and spreads into the gap (arrowhead) between the egg cell (ec) and the central cell (cc).
(H) Merged image of (G) and a bright-field image.
(J) to (L) Actin organization at 7 h after hand pollination with ARO1pro:ARO1-GFP pollen (projection of 17 optical sections, 0.30 μm each).
(J) Actin filaments in the lumen of the degenerating synergid (sy) disintegrate and aggregate as a dense patch at the micropylar pole (dashed arrow). A narrow band of actin is visible at the chalazal cortex (solid arrow).
(K) ARO1-GFP fills the lumen of the degenerating synergid (sy), and fluorescence is also visible in the pollen tube (arrowhead).
(L) Merged image of (J) and (K) with a bright-field image. Colocalization of actin and ARO1-GFP appears in the pollen tube (arrowhead) and at the chalazal cortex of the degenerated synergid (arrow).
(M) ARO1pro:ARO1-GFP–expressing ovule pollinated with the sperm cell marker line HTR10-mRFP1. The two red fluorescent sperm cells (arrowheads) migrate toward the chalazal pole of the degenerating synergid (sy). The fluorescence of the ARO1-GFP fusion is visible exclusively in the egg cell (ec).
Bars = 20 μm.
To analyze the distribution of ARO1-GFP during the fertilization process, we performed controlled hand pollination of emasculated wild-type pistils using ARO1-GFP–expressing pollen and vice versa. In ARO1-GFP–labeled egg cells, the fluorescence appeared to alter soon after pollination but before fertilization took place. One hour after hand pollination, we observed a stronger punctiform fluorescence of ARO1-GFP, shifting toward the transvacuolar strands and to the micropylar region of the egg cell (Figures 7E and 7F). However, we could not detect ancillary actin filaments in these regions.
We detected the first pollen tubes arriving at the ovules ∼5 h after pollination, which is consistent with previous observations (Ingouff et al., 2007). The typical distribution of male-derived ARO1-GFP in wild-type ovules, after the pollen tube has released its contents into the receptive synergid, is shown in Figures 7G and 7H (live imaging of ARO1-GFP, without fixation and staining, is shown in Supplemental Movie 2 online). The fluorescence is visible throughout the degenerating synergid and spreads into the gap between the egg and the central cell, forming a fluorescent hook covering the chalazal end of the egg cell (Figure 7G, arrowhead). The nuclei of both the egg and the central cell are positioned in this area, the suggested fusion site of male and female gametes (Weterings and Russell, 2004).
In another set of experiments, we performed crosses between ARO1-GFP–expressing pistils and pollen where the sperm cells were labeled with HTR10-mRFP1, a male gamete–specific histone variant fused to monomeric red fluorescent protein 1 (mRFP1) (Ingouff et al., 2007). Indeed, we observed the sperm cells to migrate into the chalazal end of the degenerating synergid (Figure 7M, arrowheads), toward the predicted fusion site. After staining actin filaments of wild-type ovules pollinated with ARO1-GFP pollen at the time of pollen tube discharge and sperm cell migration, we observed significant cytoskeletal modifications in the degenerating synergid (Figures 7J to 7L, sy). The prominent actin filaments, visible in the lumen of synergids before pollen tube arrival, disappeared, whereas fluorescent F-actin aggregated in the micropylar region (Figure 7J, dashed arrow). In addition, we detected a narrow band of F-actin at the chalazal end of the degenerating synergid (Figure 7J, solid arrow), the area where sperm cells migrate toward their fusion sites. Pollen tube-derived ARO1-GFP filled the lumen of the receptive synergid (Figure 7K, sy). Fluorescence of ARO1-GFP was also visible in the shank of the pollen tube, at areas of strong F-actin staining (Figures 7J to 7L, arrowheads). In the egg apparatus, there was no colocalization of the prominent F-actin aggregates at the micropylar region with ARO1-GFP (Figure 7L, dashed arrow). However, ARO1-GFP colocalized with the narrow F-actin band at the chalazal end of the degenerating synergid (Figure 7L, arrow).
DISCUSSION
AROs Represent a Novel Plant-Specific and Ancient ARM Repeat Protein Subfamily
Here, we describe one member of a small family of ARM repeat–containing proteins that is specifically expressed in the egg cells and anthers of Arabidopsis. ARM repeat domains are composed of tandem imperfect repeats of ∼40 amino acids and were first described in the Drosophila melanogaster segment polarity protein Armadillo (Riggleman et al., 1989; Huber et al., 1997), the homolog of mammalian β-catenin. In animals, ARM repeat proteins are known to mediate protein–protein interactions in diverse cellular processes, such as cell junction assembly, cytoskeletal regulation, intracellular signaling, nuclear transport, and transcriptional activation (Hatzfeld, 1999, 2007; Willert and Jones, 2006). A superfamily of >100 proteins with a range of 2 to 32 detected tandem ARM repeats was previously identified in the Arabidopsis genome. On the basis of homologies among each other, homologies to proteins from other organisms, and the presence of additional protein motifs, these proteins were divided into several subfamilies (Coates, 2003; Mudgil et al., 2004). Until now, the functions of only a few plant ARM repeat proteins have been elucidated, and all of them contain other protein motifs in addition to the ARM repeats. These proteins have been associated with functions such as the regulation of self-incompatibility (Stone et al., 2003; Liu et al., 2007), gibberellin and abscisic acid signaling (Amador et al., 2001; Kim et al., 2004), trichome development (Downes et al., 2003), cell death and activation of defense mechanisms (Zeng et al., 2004; González-Lamothe et al., 2006; Yang et al., 2006), as well as the promotion of lateral root development (Coates et al., 2006) and root hair tip growth (Yang et al., 2007; Sakai et al., 2008).
A group of 28 ARM repeat proteins of Arabidopsis appear to lack additional known protein motifs (Mudgil et al., 2004). This group includes the small protein family of four members described here, which we named ARO1 to ARO4, with ARO1 being most similar to the protein encoded by the wheat egg cell cDNA clone Ta ARO1, previously reported by our group (Sprunck et al., 2005). We found that all identified plant AROs share a typical structure composed of two conserved ARM repeat domains separated by a highly divergent spacer of varying size. The N-terminal domain likely constitutes a protein motif of yet unknown function, due to its grade of sequence conservation and predicted secondary structure. Although lacking significant sequence similarity, the number of ARM repeats and the overall structure of plant AROs resemble those of the Armadillo-related p120ctn protein family (Choi and Weis, 2004) involved in regulating intercellular junctions established by calcium-dependent cadherins. AROs are plant-specific and appear to be highly conserved among the plant kingdom, as we found similar predicted proteins or putative open reading frames in other angiosperms, in gymnosperms, and in Physcomitrella patens but not in yeast, animal, or Chlamydomonas. Given that the moss P. patens is separated from flowering plants by >400 million years, we conclude that ARO proteins constitute a phylogenetically ancient plant protein family.
The Gametophyte-Specific ARO1 Exhibits a Dual Subcellular Localization in Both the Cytoplasm and the Vegetative Nucleus of Pollen but Is Excluded from the Egg Cell Nucleus
Molecules of >50 kD generally require nuclear localization sequences (NLS) or nuclear export sequences to pass through the nuclear pore complex of eukaryotic cells (Poon and Jans, 2005). However, ARO1 does not contain a recognizable NLS, and passive diffusion of ARO1-GFP into nuclei of vegetative cells of pollen and of transiently transformed epidermal spring onion cells is unfeasible due to its large size (101.74 kD). Still, several ARM repeat proteins have been reported to be targeted to the nucleus in an NLS-independent manner: β-catenin lacks a classical NLS but is imported by competing with importin-β for docking to components of the nuclear pore complex (Fagotto et al., 1998). Likewise, protein-sorting studies with deletion mutants of the plant ARM repeat proteins PHOR1 (Amador et al., 2001) and ARABIDILLO1 (Coates et al., 2006) indicated that the ARM repeats mediate their nuclear targeting. Thus, nuclear targeting of ARO1 might be mediated by its ARM repeats as well.
Surprisingly, a small but reproducible portion of spring onion epidermal cells transiently transformed with UBIpro:ARO1-GFP exhibited either an almost exclusive cytoplasmic localization of the fusion protein or fluoresced exclusively at the plasma membrane. Moreover, we observed that ARO1-GFP was completely excluded from egg cell nuclei of plants expressing ARO1pro:ARO1-GFP. This variable subcellular localization implies a multifunctional role for ARO1, as is the case, for example, for β-catenin/ARM and plakophilins, which show a dynamic nucleocytoplasmic shuttling (reviewed in Hatzfeld, 1999, 2007). The dual roles of β-catenin in cytoskeletal function and as a regulator of gene expression are regulated by phosphorylation, which is in turn dependent upon the Wnt signaling cascade, induced by extracellular glycoproteins known as Wnts (Willert and Jones, 2006). In resting cells, β-catenin is found mostly at the cell membrane (Tolwinski and Wieschaus, 2001). In plants, the phytohormone gibberellin was reported to induce the migration of PHOR1 into the nucleus (Amador et al., 2001). Given that we found several putative phosphorylation sites of ARO1 to be conserved among ARO1-like proteins of other plant species (see Supplemental Figure 2 online), it is conceivable that changes in intracellular localization of ARO1 are partly mediated by exogenous signals or the physiological state of the individual cell and regulated by phosphorylation.
In Pollen Tubes, ARO1 Is Essential for Tip Growth and F-Actin Organization
A knockdown of ARO1 does not impair egg cells but affects the germination and tip-directed growth of mutant pollen tubes, which is likely due to the fact that ARO1 is the only member of the ARO gene family in Arabidopsis that is expressed in the male gametophyte. Another allele of aro1 (seth4) was previously identified as one of nine male-specific progamic phase Ds transposon mutants (Lalanne et al., 2004). However, so far, there is no additional information about the function of SETH4 in the male gametophyte.
The dynamic organization of the actin cytoskeleton and a tip-localized exocytosis play central roles during pollen germination and tube growth. During hydration, pollen grains are transformed from nonpolar cells to highly polarized cells and arrays of F-actin bundles polarize toward the site of pollen tube emergence (Heslop-Harrison and Heslop-Harrison, 1992). In the shank of growing pollen tubes, axially oriented actin cables are, together with myosins, fundamental for directional cytoplasmic streaming and the transport of organelles and secretory vesicles toward the apex (Hepler et al., 2001), and the inhibition of actin polymerization by LatB and cytochalasins not only hampers pollen germination but effectively blocks tube growth (Gibbon et al., 1999). The observations that aro1-3 pollen tubes exhibit a severely disorganized actin cytoskeleton (Figure 2) and that the ARO1-GFP fusion proteins colocalize, at least in part, along actin filaments (Figures 5 and 6) point to the possibility that ARO1 is directly involved in the organization of actin filaments and that the observed growth depolarization and growth arrest of aro1-3 pollen tubes (Figure 2) is a consequence of unorganized F-actin and thus the breakdown of polar vesicle transport. However, the colocalization of ARO1-GFP with actin cables in the shank of the pollen tube was only partial and the direct binding of ARO1 to actin monomers can be ruled out, as we did not find direct interaction in yeast two-hybrid assays (see Supplemental Figure 7 online). Particularly, the tip-directed accumulation of ARO1-GFP, the spatial shift of tip-localized ARO1-GFP during growth reorientation (Figure 4), and the dramatic effect of BFA on the subcellular localization of ARO1-GFP (Figure 6) suggest a close connection between ARO1 and the tip growth machinery of Arabidopsis pollen tubes. Similar to the regulation of local networks of fine and dynamic cortical F-actin assembled at the leading edge of motile mammalian cells, spatially regulated Rho signaling was found to be crucial for pollen tube tip growth (Gu et al., 2003; Kost, 2008). Active ROP1 is localized to defined regions in the apical region of the pollen tube plasma membrane, and its spatial distribution was shown to control the direction of tube growth by regulating tip-localized F-actin dynamics (Gu et al., 2005; Hwang et al., 2005), thereby coordinating vesicle targeting and exocytosis (Lee et al., 2008). Besides the close correlation of growth direction and spatial distribution of ARO1-GFP within the tube tip, we observed the disordered meshwork of short actin filaments to extend into the very tip of aro1-3 pollen tubes. The apex of wild-type pollen tubes is usually occupied by highly dynamic F-actin that is difficult to visualize with conventional preservation methods (Lovy-Wheeler et al., 2005), suggesting that a loss of functional ARO1 provokes a loss of tip-localized dynamic F-actin. The observed growth depolarization of aro1-3 pollen tubes, therefore, might just as well be the consequence of a failure to target polarity markers such as ROPs to their proper location. Hence, our results here indicate the possibility that the function of ARO1 is upstream of, or parallel to, ROP signaling.
Interestingly, some animal ARM repeat proteins are reported to be directly involved in Rho GTPase–mediated signaling. The non-cadherin-associated cytoplasmic pool of p120-catenin regulates RhoA signaling as well as Rac and Cdc42 activity, leading to actin reorganization and an increase in cell motility (reviewed in Anastasiadis, 2007). By contrast, Diaphanous-related formins (DRFs) act as effectors of Rho GTPases, as they bind to activated Rho GTPases, in part via their ARM repeats. Once activated, the DRFs nucleate and elongate unbranched actin filaments at the site of nascent focal adhesions, filopodia, and at endosomes (reviewed in Faix and Grosse, 2006). The identification of interaction partners of ARO1 will thus help elucidate its mechanistic role during tip growth.
ARO1 Accumulation in the Pollen Tube Tip Is Dependent on the Secretory Pathway
In animal and plant cells, BFA inactivates Golgi-associated ARF-GEFs and impairs the formation of coated vesicles, cargo selection and packaging, as well as the maintenance of the Golgi structure (D'Souza-Schorey and Chavrier, 2006; Richter et al., 2007). The loss of tip-accumulating ARO1-GFP after BFA treatment was accompanied by the appearance of numerous small fluorescent bodies throughout the pollen tube, which we can exclude as being endocytotic vesicles as they did not colocalize with FM4-64–labeled BFA-induced compartments (Figure 6), which are known to appear in the subapical region of BFA-treated pollen tubes (Geitmann et al., 1996). Our results suggest that cytoplasmic ARO1 is, at least in part, associated with membranes of the secretory machinery and thereby is transported together with secretory vesicles toward the pollen tube tip. Passive transport of ARO1 via association with a yet unidentified membrane-localized component of the tip growth machinery is conceivable, as β-catenin, for example, is known to bind to the cytoplasmic tail of the transmembrane receptor E-cadherin early in the biosynthetic pathway. Both proteins are then sorted, as a complex, for directional delivery to the adherens junction plasma membrane domains (Bryant and Stow, 2004, and references cited therein). The observations that ARO1-GFP localizes, in part, at the plasma membrane of transiently transformed cells (see Supplemental Figure 5 online) and that it accumulates in a BFA-sensitive way at the very tip of pollen tubes (Figure 6) suggest that ARO1 is targeted to the apical plasma membrane region in a similar way.
What Is the Role of ARO1 during Double Fertilization?
Previous reports have shown that a dramatic reorganization of F-actin takes place in the embryo sac during pollen tube arrival and discharge. The released sperm cells appear to migrate along emerging F-actin structures (actin coronas), which extend from the degenerating synergid cell toward the membrane fusion sites adjacent to the egg and central cell nucleus (reviewed in Weterings and Russell, 2004). Consistent with previous studies on other plant species, we found prominent actin filaments/bundles in Arabidopsis synergids before pollination that appeared to disintegrate rapidly after pollen tube arrival. Strikingly, pollen-derived ARO1-GFP colocalized with a narrow F-actin band at the chalazal membrane of the degenerating synergid cell (Figure 7), which strongly resembles previously described actin coronas. Despite their important role in tracking the sperm cells to the female gametes (Huang and Sheridan, 1998; Fu et al., 2000, Ye et al., 2002), it is not known how the reorganization of actin filaments into linear corona tracks is regulated. It is tempting to speculate that pollen-derived ARO1 is involved in a process that establishes local dynamic F-actin structures at the chalazal junction between the receptive synergid and the egg cell. However, limited access and the functional redundancy of the ARO protein family in egg cells complicate the analysis of ARO1 function in the embryo sac. Processes in the egg cell/zygote that are likely to rely on spatial-restricted patches of cortical dynamic F-actin are, for example, the formation of a membrane protrusion of the egg cell during sperm cell fusion or the extensive elongation of the zygote. Interestingly, the nun orchid (Phaius tankervilliae) egg cell appears to form a small extension at the site where the sperm cell fuses (Ye et al., 2002), resembling the actin-driven formation of microvilli that participate in sperm–egg binding as well as sperm entry in animal oocytes (Sun and Schatten, 2006). The lack of an aberrant phenotype in aro1-3 egg cells/zygotes indicates that an egg cell–specific knockout of all AROs will probably be required to address this important biological question. However, the most important step toward understanding the mechanistic role of ARO1 during pollen tube tip growth and double fertilization will be the identification of its direct binding partners.
METHODS
All oligonucleotide primers used in this work are specified in the Supplemental Methods and Supplemental Data Set online.
Plant Material and Growth Conditions
Arabidopsis thaliana (ecotype Columbia-0 [Col-0]) plants were grown on soil under a short photoperiod (9 h of light, 22°C, 70% humidity) for 3 to 4 weeks, followed by a long photoperiod (16 h of light, 8500 lux, 22°C, 70% humidity). ARO1 T-DNA insertion lines aro1-1 (SALK_040310), aro1-2 (SALK_147528), and aro1-3 (SALK_033785) were generated by the Salk Institute Genomic Analysis Laboratory (Alonso et al., 2003).
Analysis of T-DNA Insertion Lines
Genomic DNA was isolated using the innuPREP Plant DNA kit (Analytik Jena). T-DNA integrations were identified by PCR using gene-specific primers for aro1-1 (LP1-1 and RP1-1), aro1-2 (LP1-2 and RP1-2), and aro1-3 (LP1-3 and RP1-3) as well as a combination of the T-DNA primer LBa1 with gene-specific primers RP1-1, LP1-2, and LP1-3.
Expression Analysis
Up to 5 mg of frozen tissue was used for mRNA isolation using the Dynabeads mRNA DIRECT Micro kit (Dynal Biotech). Mature pollen and germinated pollen tubes used for mRNA isolation were obtained by dipping freshly dehiscent anthers onto semisolid pollen germination medium as described below and collected either 10 min after rehydration or 5 h after in vitro germination in 5 μL of phosphate buffer. Before RT-PCR, mRNA was treated with 1 unit of RNase-free DNase I (MBI Fermentas) according to the manufacturer's instructions. First-strand cDNA synthesis was performed using oligo(dT)18 primers and RevertAid Moloney Murine Leukemia Virus reverse transcriptase, adding 40 units of RiboLock ribonuclease inhibitor (all enzymes were from MBI Fermentas). The quality and amount of generated cDNAs were checked by PCR using the Actin3 primers Act3fw and Act3rev. Transcripts of ARO1 to ARO4 were amplified using primer pairs ARO1fw/ARO1rev, ARO2fw/ARO2rev, ARO3fw/ARO3rev, and ARO4fw/ARO4rev, respectively (see Supplemental Methods and Supplemental Data Set online). Experiments were repeated at least twice, using two biological replicates. Two microliters of each cDNA was used as template. For quantitative RT-PCR, 1 μL of each of individual cDNA population was used with primer pairs for ARO1 (ARO1fw/ARO1rev) and for the reference gene eIF4G (eIF4fw and eIF4rev). Quantitative PCR was performed in triplicate, using each biological replicate, the SYBR Advantage qPCR Premix (Clontech), and the Mastercycler ep realplex S (Eppendorf). Egg cells were isolated from excised mature Arabidopsis (Col-0) ovaries 2 d after 11 flowers had been emasculated (Smyth et al., 1990). The isolation procedure was performed according to Kranz and Lörz (1993), except that the ovules were dissected in a 730-mOsm enzyme solution using fine hypodermic needles, and the isolated egg cells were washed twice in a droplet of 730 mOsm of mannitol before being collected and frozen in liquid nitrogen. In total, 30 isolated egg cells were used for isolation of mRNA with the Dynabeads mRNA DIRECT Micro kit following the manufacturer's instructions, but scaled down to a volume of 50 μL. The TargetAmp 2-Round Aminoallyl-aRNA Amplification Kit 1.0 (Epicentre) was used for linear amplification of RNA, following the manufacturer's guidelines. Subsequent labeling of cRNA, hybridization of Arabidopsis ATH1 GeneChip arrays, and standard data evaluation were performed by the Center of Excellence for Biochips in Regensburg, Germany.
Generation of Constructs and Transformation of Arabidopsis
A 723-bp genomic fragment (+6 to −717 upstream of the predicted start codon of ARO1) was amplified using ACCUZYME DNA polymerase (Bioline) and the primer pair ARO1pfw/ARO1prev (see Supplemental Methods and Supplemental Data Set online) and cloned into pCR-Blunt II-TOPO (Invitrogen). The binary vector ARO1pro:GUS was created by cloning the BglII-digested promoter fragment upstream of GUS-INT (Vancanneyt et al., 1990), using the vector pMG2002 (M. Gahrtz, unpublished data), after the maize (Zea mays) UBI promoter was cut out by BamHI/BglII. Cloning of the GFP fusion proteins is described in the Supplemental Methods and Supplemental Data Set online. Binary plasmids were delivered into Agrobacterium tumefaciens strain GV3101 (pMP90RK), which was then used for transformation of Arabidopsis Col-0 by floral dip, according to Clough and Bent (1998). Three days after germination, BASTA-resistant seedlings were selected by spraying three times with 200 mg/L BASTA (Bayer Crop Science) supplemented with 0.1% Tween.
In Vitro Pollen Germination and Drug Treatments
Pollen grains were placed on small Petri dishes (4 cm) by dipping freshly dehiscent anthers onto pollen germination medium (PGM) solidified with 0.5% purified agar–agar (Merck), according to Li et al. (1999). Plates were placed in a damp box and incubated at 22°C and 8000 to 9000 lux for 3 to 6 h. Germination and growth rates were evaluated by digital bright-field microscopy of the germinated pollen directly on the solidified medium without a cover slip, using a Zeiss Axioskop microscope equipped with a Zeiss AxioVision camera and Zeiss AxioVision software (release 4.6). For CLSM using the Zeiss Axiovert 200M inverted microscope, a piece of semisolid PGM with germinated pollen was carefully placed upside down onto a cover slip that had been fixed to the bottom of a metal slide provided with an opening (diameter of 20 mm) in the center. For drug treatments, 2.5 mM LatB (Sigma-Aldrich) was dissolved in 100% DMSO and diluted to a final concentration of 20 nM in liquid PGM. BFA (AppliChem) was dissolved in 5 mg/mL methanol and diluted to a final concentration of 5 μg/mL in liquid PGM. After 3 h of in vitro germination, 500 μL of BFA- or LatB-containing PGM was added to the pollen germinated on semisolid PGM, and plates were incubated for another 2 h as described. As controls, corresponding solvents were added to liquid PGM in similar amounts and added to pollen tubes as indicated. Changes in growth rates during drug treatment were not estimated. FM4-64FX (Invitrogen) was dissolved to 1 mM in 100% DMSO, diluted to a final concentration of 4 μM in liquid PGM, and added to germinating pollen 30 min prior to actin staining.
Staining Procedures
GUS staining was performed according to Vielle-Calzada et al. (2000). Before clearing, tissues were washed in 100 mM sodium phosphate buffer (pH 7.0). Ovaries were dissected on a glass slide in a drop of Hoyer's or lactic acid clearing solution, according to Liu and Meinke (1998) and Vielle-Calzada et al. (2000), respectively. Actin labeling of in vitro germinated pollen was performed as described by Monteiro et al. (2005). For actin staining of ovules, pistils were cut open lengthwise using a hypodermic needle (0.4 × 20 mm). To improve penetration of solutions, pistils were incubated under vacuum. For aniline blue staining, pollinated pistils were fixed overnight in ethanol:acetic acid (9:1) at 4°C. After an ethanol series (70, 50, and 30%; 5 min each), pistils were rinsed in 100 mM sodium phosphate buffer (pH 7.0) and softened with 10% chloral hydrate for 5 min at 65°C. After rinsing in sodium phosphate buffer, pistils were incubated in 5 M NaOH for 10 min at 65°C. Samples were mounted on glass slides in 0.1% aniline blue (Fluka) in sodium phosphate buffer. To visualize nuclei, ovules and dehiscent anthers were stained with 10 μg/mL DAPI (Sigma-Aldrich), 10 μM Draq5 (Biostatus Limited), or 15 μg/mL propidium iodide (Invitrogen) for 10 to 30 min on a glass slide before microscopy.
Microscopy
Bright-field and fluorescent specimens were observed with a Zeiss Axioskop FL microscope equipped with an epifluorescence UV light filter set and differential interference contrast optics. Filter set 02 (365 nm excitation, LP 420 nm) was used for DAPI staining, and filter set 09 (450 to 490 nm excitation, LP 515 nm) was used for GFP samples. Images were captured and processed using the Zeiss AxioVision camera and software (release 4.6) and ImageJ 1.38x (http://rsb.info.nih.gov/ij/). A Zeiss Axiovert 200M inverted microscope equipped with a confocal laser scanning module (LSM 510 META) was used for CLSM. For detection of GFP, 488 nm excitation and a BP 505-530 filter were used. For other fluorophores, 543 nm excitation and filter LP 560 (rhodamine–phalloidin) or LP 650 (Draq5, propidium iodide) were used. Simultaneous detection of FM4-64FX and rhodamine–phalloidin was achieved by recording emission maxima for each 10 nm ranging from 552 to 798 nm and subsequent linear unmixing. Capture and processing of images were done using the AxioCam HRc camera, the Zeiss LSM 510 META software, and the Zeiss LSM image browser version 3.5.0.359.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ARO1 (At4g34940), ARO2 (At5g66200), ARO3 (At4g36030), ARO4 (At3g26600), eIF4G (At3g60240), PGA3 (At3g07830), WOX2 (At5g59340), ACT3 (At3g53750), ACT4 (At5g59370), ACT7 (At5g09810), Os AROl-1 (Os08g0548500), Os AROl-2 (Os09g0536200), Os AROl-3 (Os03g0244700), Os AROl-4 (Os10g0147900), Pp AROl-1 (XP_001775927), Pp AROl-2 (XP_001774233), Pp AROl-3 (XP_001784239), Pp AROl-4 (XP_001766369), Pt AROl-1 (FJ189520), and Ta ARO1 (EU662204). Sequence data available in the Third Party Annotation Section of the GenBank/EMBL/DDBJ databases are as follows: Lj AROl-1 (BK006563), Mt AROl-1 (BK006558), Mt AROl-2 (BK006562), Pu AROl-1 (BK006559), Vv AROl-1 (BK006560), Vv AROl-2 (BK006561), Zm AROl-1 (BK006566), Zm_AROl-2 (BK006564), Zm AROl-3 (BK006567), and Zm AROl-4 (BK006565). The putative open reading frame of Pi AROl-1 was translated from assembled ESTs (TC39011) identified in the spruce TIGR Gene Indices database.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Arabidopsis ARO Proteins.
Supplemental Figure 2. Conserved Secondary Structure and Phosphorylation Sites of ARO1-Like Proteins.
Supplemental Figure 3. Phylogenetic Relationship of Members of the ARO Protein Family.
Supplemental Figure 4. Complementation of the aro1-3 Phenotype by At ARO1-GFP.
Supplemental Figure 5. ARO1-GFP Distribution in Spring Onion Epidermal Cells.
Supplemental Figure 6. CLSM Live Imaging of At ARO1-GFP before and after LatB Treatment of in Vitro Germinated Pollen Tubes.
Supplemental Figure 7. Direct Yeast Two-Hybrid Interaction Assay of At ARO1 with Different Actin Monomers.
Supplemental Table 1. Gametophytic Transmission Efficiency of the aro1-3 Allele.
Supplemental Movie 1. Subcellular Localization of ARO1-GFP in an in Vitro Germinated Pollen Tube.
Supplemental Movie 2. Localization of Pollen-Derived ARO1-GFP after Pollen Tube Discharge into the Receptive Synergid.
Supplemental Methods.
Supplemental References.
Supplemental Data Set.
Supplementary Material
Acknowledgments
We thank Lucija Soljic for sharing unpublished data. We are grateful to Rudolph Reimers, Hartmut Quader, and Guido Grossmann for their support in CLSM and to Svenja Rademacher and Nadia Graciele-Krohn for their help during the final phase of the experiments. We thank Fred Berger for providing the marker line HTR10-mRFP1. We acknowledge Rainer Merkl for assistance with alignments and the phylogenetic tree. The German Research Council provided financial support (Grant DR 334/5-2).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Stefanie Sprunck (stefanie.sprunck@biologie.uni-regensburg.de).
Online version contains Web-only data.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Amador, V., Monte, E., García-Martínez, J., and Prat, S. (2001). Gibberellins signal nuclear import of PHOR1, a photoperiod-responsive protein with homology to Drosophila armadillo. Cell 106 343–354. [DOI] [PubMed] [Google Scholar]
- Anastasiadis, P.Z. (2007). p120-ctn: A nexus for contextual signaling via Rho GTPases. Biochim. Biophys. Acta 1773 34–46. [DOI] [PubMed] [Google Scholar]
- Berken, A., Thomas, C., and Wittinghofer, A. (2005). A new family of RhoGEFs activates the Rop molecular switch in plants. Nature 436 1176–1180. [DOI] [PubMed] [Google Scholar]
- Bolte, S., Talbot, C., Boutte, Y., Catrice, O., Read, N.D., and Satiat-Jeunemaitre, B. (2004). FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J. Microsc. 214 159–173. [DOI] [PubMed] [Google Scholar]
- Bryant, D.M., and Stow, J.L. (2004). The ins and outs of E-cadherin trafficking. Trends Cell Biol. 14 427–434. [DOI] [PubMed] [Google Scholar]
- Chebli, Y., and Geitmann, A. (2007). Mechanical principles governing pollen tube growth. Functional Plant Science and Biotechnology 1 232–245. [Google Scholar]
- Chen, C.Y.-H., Cheung, A.Y., and Wu, H.-M. (2003). Actin-depolymerizing factor mediates Rac/Rop GTPase-regulated pollen tube growth. Plant Cell 15 237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, A.Y., and Wu, H.-M. (2004). Overexpression of an Arabidopsis formin stimulates supernumerary actin cable formation from pollen tube cell membrane. Plant Cell 16 257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, A.Y., and Wu, H.-M. (2008). Structural and signaling networks for the polar cell growth machinery in pollen tubes. Annu. Rev. Plant Biol. 59 547–572. [DOI] [PubMed] [Google Scholar]
- Choi, H.-J., and Weis, W.I. (2004). Structure of the armadillo repeat domain of Plakophilin 1. J. Mol. Biol. 346 367–376. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Coates, J.C. (2003). Armadillo repeat proteins: Beyond the animal kingdom. Trends Cell Biol. 13 463–471. [DOI] [PubMed] [Google Scholar]
- Coates, J.C., Laplaze, L., and Haselhoff, J. (2006). Armadillo-related proteins promote lateral root development in Arabidopsis. Proc. Natl. Acad. Sci. USA 103 1621–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, R.A., and Fowler, J.E. (2006). Polarized growth: Maintaining focus at the tip. Curr. Opin. Cell Biol. 9 579–588. [DOI] [PubMed] [Google Scholar]
- Downes, B.P., Stupar, R.M., Gingerich, D.J., and Vierstra, R.D. (2003). The HECT ubiquitin-protein ligase (UPL) family in Arabidopsis: UPL3 has a specific role in trichome development. Plant J. 35 729–742. [DOI] [PubMed] [Google Scholar]
- D'Souza-Schorey, C., and Chavrier, P. (2006). ARF proteins: Roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 7 347–358. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville, S., and Hall, A. (2002). Rho GTPases in cell biology. Nature 420 629–635. [DOI] [PubMed] [Google Scholar]
- Fagotto, F., Glück, U., and Gumbiner, B.M. (1998). Nuclear localization signal-independent and importin/karyopherin-independent nuclear import of β-catenin. Curr. Biol. 8 181–190. [DOI] [PubMed] [Google Scholar]
- Faix, J., and Grosse, R. (2006). Staying in shape with formins. Dev. Cell 10 693–706. [DOI] [PubMed] [Google Scholar]
- Fu, Y., Yuan, M., Huang, B.-H., Yang, H.Y., Zee, S.Z., and O'Brien, T.P. (2000). Changes in actin organization in the living egg apparatus of Torenia fournieri during fertilization. Sex. Plant Reprod. 12 315–322. [Google Scholar]
- Geitmann, A., and Palanivelu, R. (2007). Fertilization requires communication: Signal generation and perception during pollen tube guidance. Floriculture and Ornamental Biotechnology 1 77–89. [Google Scholar]
- Geitmann, A., Wojciechowicz, K., and Cresti, M. (1996). Inhibition of intracellular pectin transport in pollen tubes by monensin, brefeldin A and cytochalasin D. Bot. Acta 109 373–381. [Google Scholar]
- Gibbon, B.C., Kovar, D.R., and Staiger, C.J. (1999). Latrunculin B has different effects on pollen germination and tube growth. Plant Cell 11 2349–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Lamothe, R., Tsitsigiannis, D.I., Ludwig, A.A., Panicot, M., Shirasu, K., and Jones, J.D.G. (2006). The U-box protein CMPG1 is required for efficient activation of defense mechanisms triggered by multiple resistance genes in tobacco and tomato. Plant Cell 18 1067–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Y., Fu, Y., Dowd, D., Li, S., Vernoud, V., Gilroy, S., and Yang, Z. (2005). A Rho family GTPase controls actin dynamics and tip growth via two counteracting downstream pathways in pollen tubes. J. Cell Biol. 169 127–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Y., Li, S., Lord, E.M., and Yang, Z. (2006). Members of a novel class of Arabidopsis Rho guanine nucleotide exchange factors control Rho GTPase-dependent polar growth. Plant Cell 18 366–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Y., Vernoud, V., Fu, Y., and Yang, Z. (2003). ROP GTPase regulation of pollen tube growth through the dynamics of tip-localized F-actin. J. Exp. Bot. 54 93–101. [DOI] [PubMed] [Google Scholar]
- Haecker, A., Gross-Hardt, R., Geiges, B., Sarkar, A., Breuninger, H., Herrmann, M., and Laux, T. (2004). Expression dynamics of WOX genes marks the cell fate decisions during embryonic patterning in Arabidopsis thaliana. Development 131 657–668. [DOI] [PubMed] [Google Scholar]
- Hall, A. (2005). Rho GTPases and the control of cell behaviour. Biochem. Soc. Trans. 33 891–895. [DOI] [PubMed] [Google Scholar]
- Hatzfeld, M. (1999). The Armadillo family of structural proteins. Int. Rev. Cytol. 186 179–224. [DOI] [PubMed] [Google Scholar]
- Hatzfeld, M. (2007). Plakophilins: Multifunctional proteins or just regulators of desmosomal adhesion? Biochim. Biophys. Acta 1773 69–77. [DOI] [PubMed] [Google Scholar]
- Hepler, P.K., Vidali, L., and Cheung, A.Y. (2001). Polarized cell growth in higher plants. Annu. Rev. Cell Dev. Biol. 17 159–187. [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison, Y., and Heslop-Harrison, J. (1992). Germination of monocolpate angiosperm pollen: Evolution of the actin cytoskeleton and wall during hydration, activation and tube emergence. Ann. Bot. (Lond.) 69 385–394. [Google Scholar]
- Huang, B.-Q., and Sheridan, W.F. (1998). Actin coronas in normal and indeterminate gametophyte1 embryo sacs of maize. Sex. Plant Reprod. 11 257–264. [Google Scholar]
- Huber, A.H., Nelson, W.J., and Weis, W.I. (1997). Three-dimensional structure of the armadillo repeat region of β-catenin. Cell 90 871–882. [DOI] [PubMed] [Google Scholar]
- Hwang, J.-U., Gu, Y., Lee, Y.-L., and Yang, Z. (2005). Oscillatory ROP GTPase activation leads the oscillatory polarized growth of pollen tubes. Mol. Biol. Cell 16 5385–5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingouff, M., Hamamura, Y., Gourgues, M., Higashiyama, T., and Berger, F. (2007). Distinct dynamics of HISTONE3 variants between the two fertilization products in plants. Curr. Biol. 17 1032–1037. [DOI] [PubMed] [Google Scholar]
- Kaothien, P., Ok, S.H., Shuai, B., Wengier, D., Cotter, R., Kelley, D., Kiriakopolos, S., Muschietti, J., and McCormick, S. (2005). Kinase partner protein interacts with the LePRK1 and LePRK2 receptor kinases and plays a role in polarized pollen tube growth. Plant J. 42 492–503. [DOI] [PubMed] [Google Scholar]
- Kim, S., Choi, H., Ryu, H., Park, J.H., Kim, M.D., and Kim, S.Y. (2004). ARIA, an Arabidopsis arm repeat protein interacting with a transcriptional regulator of abscisic acid-responsive gene expression, is a novel abscisic acid signalling component. Plant Physiol. 136 3639–3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost, B. (2008). Spatial control of Rho (Rac-Rop) signalling in trip growing plant cells. Trends Cell Biol. 18 119–127. [DOI] [PubMed] [Google Scholar]
- Kranz, E., and Lörz, H. (1993). In vitro fertilization with isolated single gametes results in zygotic embryogenesis and fertile maize plants. Plant Cell 5 739–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalanne, E., Michaelidis, C., Moore, J.M., Gagliano, W., Johnson, A., Patel, R., Howden, R., Vielle-Calzada, J.-P., Grossniklaus, U., and Twell, D. (2004). Analysis of transposon insertion mutants highlights the diversity of mechanisms underlying male progamic development in Arabidopsis. Genetics 167 1975–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y.J., Szumlanski, A., Nielsen, E., and Yang, Z. (2008). Rho-GTPase-dependent filamentous actin dynamics coordinate vesicle targeting and exocytosis during tip growth. J. Cell Biol. 181 1155–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Lin, Y., Heath, R.M., Zhu, M.X., and Yang, Z. (1999). Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to tip-localized calcium influx. Plant Cell 11 1731–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C.M., and Meinke, D.W. (1998). The titan mutants of Arabidopsis are disrupted in mitosis and cell cycle control during seed development. Plant J. 16 21–31. [DOI] [PubMed] [Google Scholar]
- Liu, P., Sherman-Broyles, S., Nasrallah, M.E., and Nasrallah, J.B. (2007). A cryptic modifier causing transient self-incompatibility in Arabidopsis thaliana. Curr. Biol. 17 734–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovy-Wheeler, A., Wilsen, K.L., Baskin, T.I., and Hepler, P.K. (2005). Enhanced fixation reveals the apical cortical fringe of actin filaments as a consistent feature of the pollen tube. Planta 221 95–104. [DOI] [PubMed] [Google Scholar]
- Monteiro, D., Liu, Q., Lisboa, S., Scherer, G.E.F., Quader, H., and Malhó, R. (2005). Phosphoinositides and phosphatidic acid regulate pollen tube growth and reorientation through modulation of [Ca2+]c and membrane secretion. J. Exp. Bot. 56 1665–1674. [DOI] [PubMed] [Google Scholar]
- Mudgil, Y., Shiu, S., Stone, S.L., Salt, J.N., and Goring, D.R. (2004). A large complement of the predicted Arabidopsis ARM repeat proteins are members of the U-box E3 ubiquitin ligase family. Plant Physiol. 134 59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanivelu, R., and Preuss, D. (2000). Pollen tube targeting and axon guidance: Parallels in tip growth mechanisms. Trends Cell Biol. 10 517–524. [DOI] [PubMed] [Google Scholar]
- Poon, I.K.H., and Jans, D.A. (2005). Regulation of nuclear transport: Central role in development and transformation? Traffic 6 173–186. [DOI] [PubMed] [Google Scholar]
- Ren, H., and Xiang, Y. (2007). The function of actin-binding proteins in pollen tube growth. Protoplasma 230 171–182. [DOI] [PubMed] [Google Scholar]
- Richter, S., Geldner, N., Schrader, J., Wolters, H., Stierhof, Y.D., Rios, G., Koncz, C., Robinson, D.G., and Jürgens, G. (2007). Functional diversification of closely related ARF-GEFs in protein secretion and recycling. Nature 448 488–492. [DOI] [PubMed] [Google Scholar]
- Riggleman, B., Wieschaus, E., and Schedl, P. (1989). Molecular analysis of the armadillo locus: Uniformly distributed transcripts and a protein with novel internal repeats are associated with a Drosophila segment polarity gene. Genes Dev. 3 96–113. [DOI] [PubMed] [Google Scholar]
- Sakai, T., et al. (2008). Armadillo repeat-containing kinesins and a NIMA-related kinase are required for epidermal-cell morphogenesis in Arabidopsis. Plant J. 53 157–171. [DOI] [PubMed] [Google Scholar]
- Satiat-Jeunemaitre, B., Cole, L., Bourett, T., Howard, R., and Hawes, C. (1996). Brefeldin A effects in plant and fungal cells: Something new about vesicle trafficking? J. Microsc. 181 162–177. [DOI] [PubMed] [Google Scholar]
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprunck, S., Baumann, U., Edwards, K., Langridge, P., and Dresselhaus, T. (2005). The transcript composition of the egg cell changes significantly following fertilization in wheat (Triticum aestivum L.). Plant J. 41 660–672. [DOI] [PubMed] [Google Scholar]
- Stone, S.L., Anderson, E.M., Mullen, R.T., and Goring, D.R. (2003). ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant Cell 15 885–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Q.-Y., and Schatten, H. (2006). Regulation of dynamic events by microfilaments during oocyte maturation and fertilization. Reproduction 131 193–205. [DOI] [PubMed] [Google Scholar]
- Tolwinski, N.S., and Wieschaus, E. (2001). Armadillo nuclear import is regulated by cytoplasmic anchor Axin and nuclear anchor dTCF/Pan. Development 128 2107–2117. [DOI] [PubMed] [Google Scholar]
- Vancanneyt, G., Schmidt, R., O'Connor-Sanchez, A., Willmitzer, L., and Rocha-Sosa, M. (1990). Construction of an intron-containing marker gene: Splicing of the intron in transgenic plants and its use in monitoring early events in Agrobacterium-mediated plant transformation. Mol. Gen. Genet. 220 245–250. [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada, J.-P., Baskar, R., and Grossniklaus, U. (2000). Delayed activation of the paternal genome during seed development. Nature 404 91–94. [DOI] [PubMed] [Google Scholar]
- Weterings, K., and Russell, S.D. (2004). Experimental analysis of the fertilization process. Plant Cell 16 107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willert, K., and Jones, K.A. (2006). Wnt signalling: Is the party in the nucleus? Genes Dev. 20 1394–1404. [DOI] [PubMed] [Google Scholar]
- Wilsen, K.L., and Hepler, P. (2007). Sperm delivery in flowering plants: The control of pollen tube growth. Bioscience 57 835–844. [Google Scholar]
- Yang, C.W., González-Lamothe, R., Ewan, R.A., Rowland, O., Yoshioka, H., Shenton, M., Ye, H., O'Donell, E., Jones, J.D., and Sadanandom, A. (2006). The E3 ubiquitin ligase activity of Arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell 18 1084–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, G., Gao, P., Zhang, H., Huang, S., and Zheng, Z.-L. (2007). A mutation in MRH2 kinesin enhances the root hair tip growth defect caused by constitutively activated ROP2 small GTPase in Arabidopsis. PLoS ONE 10 e1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, X.-L., Yeung, E.C., and Zee, S.-Y. (2002). Sperm movement during double fertilization of a flowering plant, Phaius tankervilliae. Planta 215 60–66. [DOI] [PubMed] [Google Scholar]
- Zeng, L.-R., Qu, S., Bordeos, A., Yang, C., Baraoidan, M., Yan, H., Xie, Q., Nahm, B.H., Leung, H., and Wang, G.-L. (2004). Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/Armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16 2795–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.