Abstract
Structural polymorphism is increasingly recognised as a major form of human genome variation, and is particularly prevalent on the Y chromosome. Assay of the Amelogenin Y gene (AMELY) on Yp is widely used in DNA-based sex testing, and sometimes reveals males who have interstitial deletions. In a collection of 45 deletion males from 12 populations, we used a combination of STS (sequence-tagged site) mapping, and binary-marker and Y-STR (short tandem repeat) haplotyping to understand the structural basis of this variation. 41/45 males carry indistinguishable deletions, 3.0-3.8Mb in size. Breakpoint mapping strongly implicates a mechanism of non-allelic homologous recombination between the proximal major array of TSPY-gene-containing repeats, and a single distal copy of TSPY; this is supported by estimation of TSPY copy number in deleted and non-deleted males. The remaining four males carry three distinct non-recurrent deletions (2.5-4.0Mb) which may be due to non-homologous mechanisms. Haplotyping shows that TSPY-mediated deletions have arisen seven times independently in the sample. One instance, represented by 30 chromosomes mostly of Indian origin within haplogroup J2e1*/M241, has a time-to-most-recent-common-ancestor of ∼7700 ± 1300 years. In addition to AMELY, deletion males all lack the genes PRKY and TBL1Y, and the rarer deletion classes also lack PCDH11Y. The persistence and expansion of deletion lineages, together with direct phenotypic evidence, suggests that absence of these genes has no major deleterious effects.
Introduction
Recent studies have revealed a previously unexpected degree of segmental duplication (1), structural polymorphism and copy number variation (reviewed in ref. (2) in the human genome. Among human chromosomes, the Y bears a particularly large proportion of segmental duplications (3), and has long been known to exhibit a high degree of cytogenetically visible structural polymorphism, including heterochromatin and euchromatin length variation (4, 5), inversions (6, 7), and neutral translocations with autosomes (8). Unlike other human chromosomes, the Y has no requirement for pairing along its entire length with a homologue, and this may explain its structurally variable nature. A recent phylogenetically based molecular survey has emphasised the dynamic nature of the chromosome (9), but, other than the well-established variation in copy number at the TSPY tandem array (10) and 3.6-Mb inversion polymorphism (11, 12), discovered no structural variation on Yp.
One locus on Yp that is, in effect, routinely assayed on a large scale for presence-absence polymorphism is the Amelogenin Y gene (AMELY), since it forms part of the DNA-based sex test routinely employed in prenatal diagnosis (13), forensic typing (14), archaeological analysis (15, 16), and paternity testing. The test relies on the simultaneous PCR amplification of differently sized X- and Y-specific fragments from the XY-homologous amelogenin gene pair (14), lying in Xp22.3 and Yp11.1; however, some males have been reported in whom AMELY fails to amplify, and who, on DNA evidence alone, might therefore be misinterpreted as females. Initially, two such males were identified in a sample of 24 Sri Lankans (17), who lacked AMELY through a deletion on the short arm of the Y chromosome. The widespread use of the amelogenin PCR assay provides strong ascertainment of males with apparent interstitial AMELY deletions, and 65 of these have subsequently been reported (18-30). Although limited haplotyping and physical mapping has been undertaken in some of these studies, no systematic analysis of the origin and extent of these structural variants in a substantial collection of chromosomes has been carried out.
The availability of the near-complete sequence of a Y chromosome (3) allows the investigation of the mutational mechanism(s) underlying deletions: in the original cases described (17), evidence from Southern blotting analysis of pulsed-field gels was used to propose a mechanism of non-allelic homologous recombination (NAHR) between copies of the 20.4-kb repeat units containing TSPY genes on Yp. We can now ask whether this mechanism is supported by breakpoint mapping, and whether it is responsible for recurrent deletions in an analogous way to the repeat-mediated deletions of the AZFa (31-33), AZFb (34) and AZFc (35) regions on Yq, associated with male infertility. Aside from their forensic relevance, AMELY deletion males are of phenotypic interest because they lack the Amelogenin Y gene itself, and may also lack other nearby genes on Yp. A study of such males could throw light on the selective influence that the absence of any such genes might have.
Here we describe a collection of 45 males from 12 different populations carrying AMELY deletions, and use a combination of sequence-tagged-site (STS) deletion mapping, binary-marker and Y-STR haplotyping, and TSPY copy number estimation to ask how many independent deletion events are represented in this sample. Based on the Y chromosome reference database sequence (3) we also estimate the sizes of the deletions and the number of genes deleted, and propose molecular mechanisms for their generation.
Results
A collection of 45 DNA samples from males showing amplification of only the X-specific amelogenin PCR product was assembled (Figure 1). Two of these males, m632 and m640, were the first deletion individuals to be described (17), and 30 others have also been previously reported (19, 22, 23, 25, 26, 28, 29). The remaining thirteen (see Figure 1) were discovered when the amelogenin sex test was applied either as part of an autosomal STR multiplex for paternity testing or DNA profiling, or as part of a Y-specific STR multiplex in population studies.
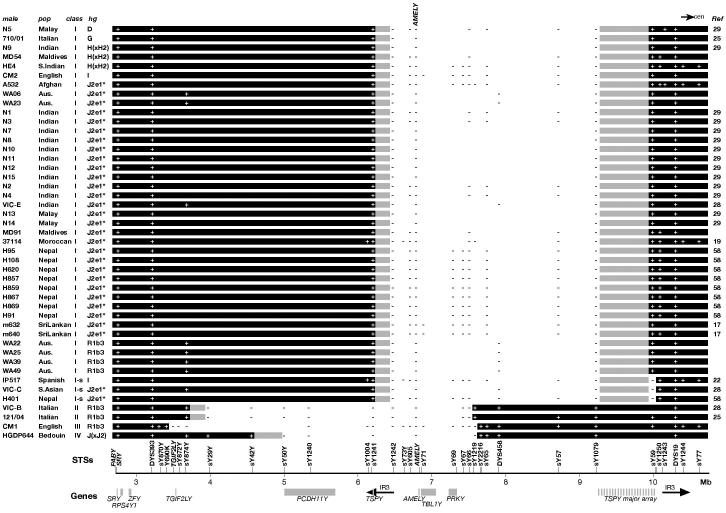
Figure 1. Schematic representation of deletion mapping data.
Black bars indicate regions of Yp observed or inferred to be present, and grey bars indicate regions of uncertainty within deletion breakpoint intervals. ‘+’: marker present; ‘−‘: marker absent; ‘I-s’: Class I deletions also lacking sY59; ‘Aus.’: Australian. Genes are shown below. The scale in Mb shows distance from the Yp telomere, according to the March 2006 (Build 36.1) genome assembly. Haplogroups are given for completeness (see text and Figure 3a), and J2e1* indicates J2e1*/M241. ‘Ref’ – reference; those without numbers are from this study.
Deletion mapping
To estimate the sizes of the deletions, to ask what genes are co-deleted with AMELY, and to throw light on the underlying molecular mechanisms, we undertook deletion mapping using sequence-tagged sites (STSs) from Yp. FISH-based mapping approaches were precluded by the unavailability of cellular material. Many published STSs were initially developed to aid in clone contig construction (3, 36, 37), and therefore do not necessarily represent single-copy Y-specific sequences; they often detect homologs (containing identical primer binding sites) on the X chromosome, on autosomes, or elsewhere on the Y chromosome, making them useless for deletion mapping purposes in genomic DNA. STSs chosen around AMELY were therefore first verified as single-copy by BLAST searching, then tested for Y-specificity in normal male and female DNA samples before use. Within regions of high XY homology, novel STSs were designed near to existing ones, targeting Y-specific bases in XY sequence alignments; these are given the original STS names, with the suffix ‘Y’ (e.g. sY872Y).
Mapping using a total of 33 Y-specific markers distinguished five apparent different deletion classes (referred to here as Class I, I-sY59, II, III and IV) among the 45 chromosomes (Figure 1), suggesting at least five underlying events. The Class I deletion is by far the commonest, being found in 38 chromosomes (84%). It has its proximal breakpoint within the TSPY major array, as indicated by absence of the STS sY1079, lying just distal to the array, and the presence of sY59, lying at the proximal end. Its distal breakpoint lies in a 298-kb interval corresponding to the distal IR3 inverted repeat element, as shown by absence of the proximal marker sY1242 and presence of the distal sY1241. Further refinement of the distal breakpoint is rendered impractical by the presence of the second, highly homologous (99.75%; (3) copy of IR3, lying proximal to the deleted segment on Yp. The distal IR3 element contains a copy of the 20.4-kb TSPY repeat in the same orientation at the major TSPY array, with overall sequence similarity (considering the most distal copy in the array) of 96%, and a longest block of sequence identity of 796bp. The most parsimonious mechanism for this deletion is therefore NAHR between TSPY repeat copies, as originally proposed by Santos et al. (17). Based on the reference sequence, the extent of deleted DNA is between 3.0 and 3.8 Mb, depending on the position of the breakpoint in the major array. Individual deletion sizes are likely to vary further, due to differences in TSPY copy number.
The second apparent category of deletion (‘Class I-sY59’), carried by three individuals, is related to the first. The distal breakpoint is the same as in Class I, but, proximally, sY59 is absent in addition to sY1079. Inspection of the sequence around sY59 shows that it lies just inside the proximal end of the TSPY array, between the penultimate and last copies. This might imply that the recombination event giving rise to these deletions occurred between the TSPY copy in the distal IR3 element and the most proximal copy of TSPY within the major array, to leave only a single copy of the repeat unit; however, when we screened a global collection of 188 AMELY+ males with sY59 we found the marker to be absent in two cases, belonging to haplogroups I and E(xE3b3). Presence/absence polymorphism of sY59 presumably arises through recombination within the proximal end of the TSPY array, and so its absence in some AMELY-deletion males is uninformative about the distal breakpoint. Deletions lacking sY59 should be classified simply as Class I, and therefore a total of 41/45 chromosomes belong to this class (91%).
The Class II deletion occurs in two individuals, and the remaining two classes (III and IV) are found as singletons (Figure 1). These three deletion classes are respectively 3.5-3.9Mb, 3.9-4.0Mb, and 2.5-3.1Mb in size, with uncertainty introduced by marker spacings at the breakpoints. To address the mechanism of deletion in these classes, we searched the breakpoint intervals in each class using the program Reputer (38) for perfect direct repeats that might be involved in sponsoring deletions. This revealed no repeats larger than the ‘minimum efficient processing segment’ length for homologous recombination of ∼200bp (39), implying that the mechanism in all three classes may be non-homologous end-joining. However, there are many smaller perfect repeats forming parts of isolated SINEs, including one of 139bp in the Class IV breakpoint intervals, and these may have been involved in the deletion events; studies of some multi-megabase non-recurrent deletions on 17p11.2 have mapped breakpoints to a sequence identity block as small as 21bp within a SINE (40).
Haplotyping
Having identified four distinct deletion classes, we can ask if these reflect only four deletion events, or if some are recurrent. To do this we exploit the availability of a well resolved Y-chromosomal phylogeny based on binary markers (41, 42): if two deletion chromosomes within Class I or II belong to two different haplogroups within the phylogeny, the deletions are likely to have arisen independently. On the basis of binary marker typing (Figure 1, 2a), Class I deletions are found in six different haplogroups showing that this TSPY-mediated deletion event is recurrent. Haplogroups D and G are each represented once, haplogroup I twice, haplogroup H(xH2) three times, and haplogroup R1b3 four times. The remaining 30 Class I chromosomes all belong to a single well-defined haplogroup, J2e1*/M241 (Figure 1). Class II deletions are present in two chromosomes, but both belong to hgR1b3, so these could represent identity by descent. The Class III deletion belongs to hgR1b3, and the Class IV deletion to hgJ(xJ2). The four deletion classes are therefore due to a minimum of nine events, and include a common founder AMELY Class I deletion lineage within haplogroup J2e1*/M241 - a finding consistent with previous studies that have identified deletions in this haplogroup (29, 30).
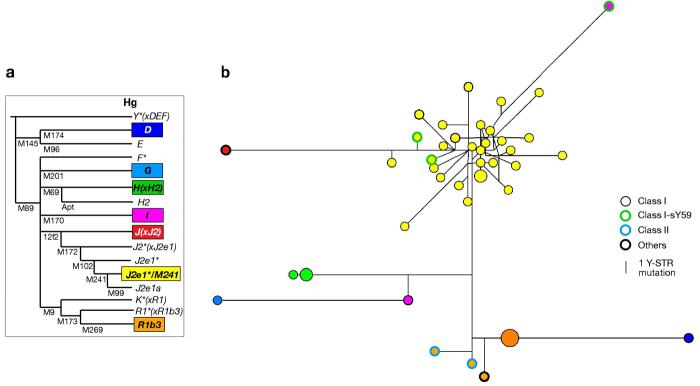
Figure 2. Haplogroups and Y-STR haplotypes of AMELY deletion chromosomes.
a) Haplogroups (Hg) found in deletion chromosomes, indicated by coloured boxes. The phylogeny shows key binary markers typed to define haplogroups.
b) Median-joining network of Y-STR haplotypes. Circles represent haplotypes, with area proportional to frequency, and coloured by haplogroup according to (a); different classes of deletions are indicated by circle outlines, as in the key below the phylogeny. Lines between the circles indicate mutational differences between Y-STR haplotypes, with lengths proportional to the number of inferred mutational steps. The 20 Y-STRs used for construction were DYS19, DYS388, DYS389I, DYS389II-I, DYS390, DYS391, DYS392, DYS393, DYS434, DYS435, DYS436, DYS437, DYS438, DYS439, DYS447, DYS448, DYS460, DYS461, DYS462, and YGATA-H4.1. The partial allele (26.2) at DYS447 in male N5 was considered as allele 26 for network construction.
Y-STR analysis provides further haplotyping resolution, and clustering of haplotypes within a haplogroup can support the idea of common ancestry. We therefore determined 26-locus Y-STR haplotypes (26) for the deletion chromosomes (Supplementary Table 2). None of the 26 STRs typed fall within the deleted intervals of these chromosomes, though we note that DYS458, included in the widely used commercial forensic profiling kit Y-filer (Applied Biosystems), is absent in Class I deletions (Figure 1; (29)). Figure 2b shows a median joining network connecting the haplotypes, in which three clusters are evident. All four Class I deletion chromosomes belonging to hgR1b3 share a single haplotype, and the three Class I chromosomes within hgH(xH2) carry very closely related haplotypes, suggesting identity-by-descent and a single deletion event within each of these sets. The major cluster is formed by the hgJ2e1*/M241 chromosomes, and applying the rho statistic within Network yields a TMRCA (time-to-most-recent-common-ancestor) of ∼7700 ± 1300 years. The two deletion chromosomes within hgI have very different haplotypes, suggesting probable independence of origin; overall, then, Class I deletions appear to have arisen on seven independent occasions in the set of 45 AMELY-deleted males. The two Class II deletion chromosomes both lie within hgR1b3, and the nature of the deletion they share, together with a shared Italian origin, strongly suggests common ancestry; their Y-STR haplotypes differ by only four mutational steps in the network, which is consistent with this. Considering all chromosomes, the 45 cases can be explained by a total of ten independent deletion events in four different classes.
TSPY copy number
If the Class I deletions were indeed caused by TSPY-mediated NAHR, we might expect the copy number of TSPY repeats in deletion chromosomes to be reduced with respect to related non-deleted examples, the degree of reduction depending on the position of the breakpoint within the TSPY array. To investigate this, we carried out quantitative PCR to estimate TSPY copy number in the nine Nepalese Class I deletion chromosomes. For comparison, we also analysed the four available chromosomes from the same population and haplogroup (J2e1*/M241) that were not deleted for AMELY. Copy number estimation in other chromosomes was not possible due to scarcity or poor quality of genomic DNA.
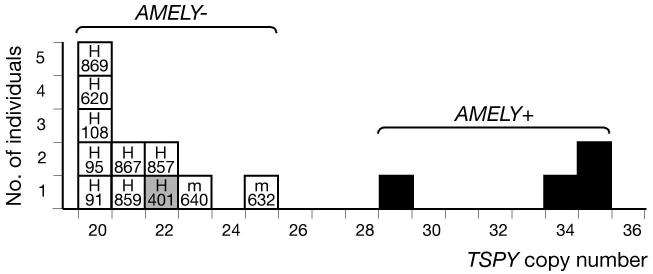
Figure 3 shows TSPY copy number in deleted and non-deleted chromosomes within hgJ2e1*/M241, and includes two deletion cases, m632 and m640, for which TSPY copy number could be estimated from the size of the hybridising XbaI restriction fragment in pulsed-field gel analysis (17). The four non-deleted chromosomes have copy numbers ranging from 29-35 (mean 33), while the 11 deleted chromosomes have copy numbers ranging from 20-25 (mean 21). This difference is significant (P = 0.003, Mann-Whitney U test) and supports the TSPY-mediated recombination mechanism for deletions. Its simplest interpretation is that the founding AMELY deletion was accompanied by the loss of ∼12 TSPY repeat units. Notably, H401, the only Class I sY59-deleted chromosome in this analysis, has a copy number of 22, rather than the single copy expected if absence of both sY59 and AMELY were due to a single deletion event. This confirms that absence of sY59 is an independent event due to mutation within the TSPY array.
Figure 3. TSPY copy number in AMELY-deleted and non-deleted chromosomes within hgJ2e1*/M241.
For deletion chromosomes, names of samples are given within the squares. Grey shading indicates the Class I chromosome lacking sY59.
Discussion
Attempts to understand the nature and dynamics of Y-chromosomal rearrangements through combined physical mapping and haplotyping have been ongoing for a decade (43), but the current availability of the sequence (3), a robust phylogeny (41), and fine resolution of haplotypes (42) now permits detailed characterisation to be carried out. Our analysis of Y chromosomes ascertained by deletion of AMELY shows that recurrent 3.0-3.8-Mb deletions on Yp exist and persist within human populations.
Mechanisms and structural aspects
Deletion mapping and haplotyping analysis confirms that NAHR between TSPY copies is responsible for the deletion in the first two males described (17), and demonstrates that Class I deletions such as this recur, with seven independent instances observed. In principle the mutation rate can be calculated given an estimate of the number of generations encompassed within the phylogeny relating the sampled chromosomes (9, 44). Such an estimate, 52,000 generations, has been calculated for a set of 47 chromosomes representing the major branches of the phylogeny (9). For our Himalayan (26, 45), Southern Indian (46) and Spanish (22) samples, a total of 2471 chromosomes were analysed and three independent Class I deletion events observed. Time elapsed within the phylogeny relating these chromosomes is difficult to estimate; the sample size is large, but many chromosomes are closely related, and the deepest-rooting haplogroups (A and B) are absent, so an estimate of 100,000 generations is likely to be generous, in which case a conservative minimum mutation rate would be of the order of 10−5 per generation. The ease of detection of deletions through the amelogenin sex-test and the propensity to report deletions among the forensic community has led to a strong ascertainment bias, and the inclusion in our collection of three examples of non-recurrent and rare deletions, possibly caused by non-homologous mechanisms.
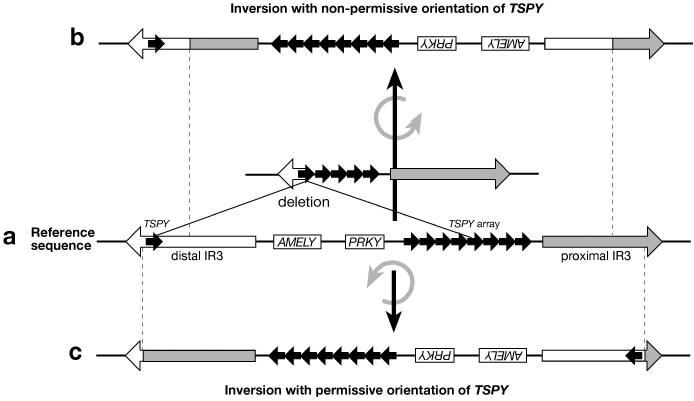
Deletion mapping was undertaken in the knowledge that a 3.6-Mb segment of Yp exists in two different orientations (11, 12) due to a inversion polymorphism sponsored by the IR3 repeats (3). In the reference sequence, the orientations of the two TSPY loci are the same, so the arrangement is permissive for recombination and resulting Class I deletion (Figure 4a). However, because the single copy of TSPY lies distally in the distal IR3 element, most inversion events acting on chromosomes resembling the reference sequence are likely to preserve the distal copy's orientation while inverting the TSPY major array, resulting in a structure that is non-permissive for deletion (Figure 4b). Only inversions with breakpoints towards the outer ends of the IR3 elements will preserve the permissive orientation (Figure 4c). We can therefore conclude either that the seven progenitors of the Class I deletions possessed the same orientation of the inversion as the reference sequence, or that the inversion breakpoints are clustered towards the peripheries of the IR3 element pair. Inversion is recurrent, with at least 12 events observed among 47 Y chromosomes across the Y phylogeny (9), although one specific breakpoint has been shown to be monophyletic, and therefore a probable unique event (Hurles et al., in preparation). Notably, the reference sequence orientation is seen in Class-I-containing haplogroups H and D, as well as R1b3, while the opposite orientation is seen in hgI and J2e (9). Further work is required to understand the molecular details of these inversions.
Figure 4. Schematic illustration of the influence of IR3-mediated inversion on TSPY-mediated deletion.
Some structures are permissive for the deletion; others are not. Diagram not to scale.
As with the other examples of repeat-mediated deletions on the Y chromosome (9, 47), we expect the reciprocal TSPY-mediated duplications to occur. Males carrying such duplications will have a duplicated copy of the Y-STR DYS458, which may be identified as a ‘double-allele’ when Y-STR profiling kits are applied. Two DYS458-duplication cases exist in the HGDP-CEPH diversity panel (HGDP388 and HGDP1153; data not shown), but semi-quantitative analysis of AMELX/Y signals provide no evidence of a duplication of AMELY in these particular cases.
Genes and selection
PRKY, TBL1Y and AMELY all belong to the so-called ‘X-degenerate’ class of Y-chromosomal genes, which were once identical in sequence to their X-chromosomal homologues, but have degenerated in sequence since recombination was suppressed between the regions of the proto-sex-chromosomes in which they resided. However, this degeneration has not been ongoing in recent human evolution, since sequence comparisons of chimpanzee and human orthologues of X-degenerate genes show clear evidence of purifying selection in the human lineage (48). The genes commonly lost in AMELY deletions are, therefore, likely to be of functional significance.
The expansion of deletion lineages such as that in J2e1*/M241, and direct evidence demonstrating heritability in some cases, including 121/04, 710/01 (25), 37114 (19), VIC-B (28) and CM2, indicates that there cannot be a strong general selective disadvantage or negative effect on fertility. However, there may be more subtle effects resulting from the absence of genes on Yp. Because most of our samples were ascertained in population studies, direct phenotypic information is unavailable, so inferences in most cases must be made indirectly.
AMELY itself encodes a protein expressed in developing tooth-buds (49), and has an X-homologue (50), mutation of which causes X-linked amelogenesis imperfecta (51). However, AMELY is expressed at only 10% of the level of AMELX, and examination of two cases of AMELY deletion revealed apparently normal teeth (25), suggesting little or no effect of the deletion on enamel formation. In Class I deletions PRKY, a ubiquitously expressed serine-threonine protein kinase, and TBL1Y (Transducin [beta]-like 1 protein Y), which is expressed in prostate and foetal brain (3) are also absent. The function of these genes is unknown, and again, examination of the phenotypes of AMELY deletion males has revealed no obvious phenotypic consequences of their absence (25). However, both have X-homologues, which may give clues to their functions. PRKX has been proposed to regulate tubulogenesis in the kidney (52), and TBL1X plays an essential role in specific nuclear receptor-mediated gene activation events (53), its deletion being implicated in late-onset sensorineural deafness (54). If the X and Y copies of these genes indeed share identical or similar functions, then males with deletions would be haploinsufficient for their products; interestingly, 45,X Turner Syndrome females, who are haploinsufficient for all XY-homologous genes, show both an increased incidence of renal abnormalities and late-onset sensorineural deafness (55). Careful monitoring of these phenotypes in AMELY deletion males would be worthwhile.
Class I deletions are caused by TSPY-mediated recombination, and the deletion event itself is accompanied by a reduction in the copy number of the repeat containing the TSPY gene. Maintenance of a minimum copy number through selection is suggested by the evolutionary conservation of multiple copies of the gene on the Y chromosomes of other mammals (56-59), and the limited degree of copy number polymorphism observed in two studies of human Y chromosomes (9, 60) (respectively, range: 18-47 or 23-64 copies; median 29 or 32 copies; sample size 89 or 47). In the one case we have been able to examine, within haplogroup J2e1*/M241, the reduction in number results in retention of at least 20 copies of TSPY. It would be of interest to determine copy numbers in further examples of independent Class I deletions.
In addition to the genes discussed above, the non-Class I deletions all lack PCDH11Y (Protocadherin 11, Y-linked), expressed in brain. This gene lies in the ‘X-transposed’ segment of the Y chromosome (3) which has arisen since our divergence from the human-chimpanzee common ancestor, and therefore represents a human-specific genetic novelty. Protocadherins are important in cell-cell interactions within the central nervous system, and differences in structure and expression of the X and Y copies, and accelerated evolution within the human lineage (61), have led to the nomination of PCDH11X/Y as candidates for sex-specific differences in human brain phenotypes (62). Unfortunately information on cognitive ability in the males lacking PCDH11Y is unavailable. The single Class III case also lacks the testis-expressed TGIF2LY (Transforming Growth Factor-Beta-Induced Factor 2-Like, Y-Linked (63)), about which little is known; this male is a singleton, with no phenotypic information, so we cannot exclude the possibility that absence of TGIF2LY has a deleterious effect on spermatogenesis.
Deletion of the AMELY region has one selectively beneficial effect: the descendants of deleted males are protected against sex-reversal caused by the commonest class of XY-translocation. The deleted segment in all of the males encompasses an XY-homologous region, around the PRKY gene, that (in one orientation (64)) sponsors XY-translocations which cause XX maleness (65).
Forensic implications
The geographical origins of males carrying the common hgJ2e1*/M241 Class I deletion are mainly in the Indian subcontinent; nine were from Nepal, ten from a Malaysian population of Indian origin (29), two from Sri Lanka (17), and two from the Maldives Islands in the Indian Ocean, though there are geographical outliers in Afghanistan, Morocco and Australia. Interestingly, a second cluster of Class I deletions, in hgH(xH2), also appears to have an Indian origin. The relatively high frequency of AMELY deletion chromosomes in the Indian subcontinent (∼2%) and around the Indian Ocean seems likely to be a result of genetic drift. However, it has practical implications for the massive forensic identification effort following the South Asian tsunami disaster of 2004, since it is in this part of the world that deletions are most likely to introduce confusion through the misassignment of sex based on DNA evidence. In cases where other evidence is lacking or ambiguous, confirmation using an additional test, such as presence/absence of SRY (17), is necessary.
Materials and Methods
DNA samples
Male HGDP644 is from the CEPH-HGDP diversity panel (66), and was identified as AMELY-deleted during profiling of the panel using the Identifiler autosomal STR kit (Applied Biosystems). Other DNA samples were from collections of the authors, and obtained with appropriate informed consent. CM2 was ascertained through a positive paternity test, demonstrating that he was fertile, and HE4 was ascertained during a study of 100 males from Southern India (46). Some samples were subjected to whole genome amplification (67) using the Genomiphi kit (GE Healthcare) before analysis.
Deletion mapping
Y-specific STSs (primer sequences available from the literature (3, 36, 37)) were amplified by PCR and analysed by agarose gel electrophoresis. Some new or adapted STSs were designed to amplify Y-specific sequences in blocks of XY-homology (Supplementary Table 1), by targeting the 3′ ends of primers at Y-specific bases identified in sequence alignments. An STS was considered to be deleted when absent in the presence of a larger Y-specific control amplicon coamplified in the same PCR reaction. The control sequence generally used was an amplicon from the SRY gene region (715 bp, primers 5′-AAT CGG GTA ACA TTG GCT ACA-3′, and 5′-AGG CTT AAA AGT TAA TAG GCC A-3′). PCR was carried out in a Tetrad Thermocycler (MJR) using 5-10 ng template DNA, the buffer of Jeffreys et al. (68) and 0.1u Taq DNA polymerase (ABgene). PCR conditions were generally: 94°C 30s, 60°C 30s, 70°C 30s, for 33 cycles, though annealing temperature was varied to ensure Y-specificity in some cases. Not all STSs were mapped in all samples, because the amount of DNA was limiting. STS positions were obtained from the UCSC Genome Browser (http://genome.ucsc.edu/), using Build 36.1 of the reference sequence.
Estimation of TSPY copy number
Primers were designed to amplify a 78-bp region of the TSPY repeat and amplification efficiency was checked with a DNA dilution series using a region of the diploid PMP22 gene as a control (69). Four TSPY and four PMP22 replicates were set up for each sample in 96-well plates, each in a total volume of 25μl, containing 1 × SYBR Green master mix (Applied Biosystems), 25ng template DNA, 300nM primer TSPYqF (5′-GAG TCC CCT GAC AGA TCC TAT GTA A-3′) and primer TSPYqR (5′-CTT CAG GTG GCT TCA TCC TCT T-3′). Cycling was performed on an Applied Biosystems 7500 quantitative PCR machine using cycling conditions: 95°C for 10 minutes followed by 40 rounds of 95°C for 15 seconds, 62°C for 1 minute. All DNAs analysed had not been subject to whole genome amplification.
Difference in threshold cycle (ΔCt) was calculated for each individual by subtracting mean PMP22 Ct from mean TSPY Ct. Using one individual as a reference, values were converted to ΔΔCt, from which relative copy number was calculated (70). Absolute copy numbers of TSPY genes were estimated with reference to DNA samples in which copy number was known, from size measurement of the hybridising XbaI fragment in pulsed-field gel analysis (71). Comparison of PCR-based and pulsed-field methods suggest that the former is accurate to ±1 TSPY repeat unit.
Y chromosome haplotyping
Binary markers were typed in a hierarchical fashion using the SNaPshot minisequencing protocol (Applied Biosystems) on an ABI3100 capillary electrophoresis apparatus (Applied Biosystems). Amplification primers and SNaPshot primers were based on ones published previously (72, 73), with additional primers based on published sequences (41).
Twenty-six Y-STRs (DYS19, DYS385a/b, DYS388, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS425, DYS426, DYS434, DYS435, DYS436, DYS437, DYS438, DYS439, DYS447, DYS448, DYS460, DYS461, DYS462, YCAII-sY59/b, and Y-GATA-H4.1) were typed in a 20-plex (74) and an additional 14-plex (26) that incorporates the amelogenin sex-test (14). PCR products were resolved on an ABI3100 capillary electrophoresis apparatus (Applied Biosystems), and analysed using GeneMapper software (Applied Biosystems). Allele nomenclature was as described (26).
Reputer analysis
Perfect direct repeats within deletion breakpoint intervals were identified using the Reputer program (38), at http://bibiserv.techfak.uni-bielefeld.de/reputer/.
Y-STR network construction and dating
A weighted median-joining network (75) was constructed from Y-STR haplotypes using Network 4.0 (http://www.fluxus-engineering.com/sharenet.htm). The network contains all the most parsimonious trees relating the haplotypes, and allows unobserved haplotypes to represent nodes (branch points). Weighting (76) was used to remove some reticulations (closed structures) within the network by taking into account the range of different mutation rates of the Y-STRs, reflected indirectly by their allele length variances among all deletion chromosomes. Thus, fast-mutating loci (high variance) are given lower weight in making links between haplotypes than are slow-mutating loci (low variance). TMRCA of the hgJ2e1*/M241 cluster was estimated within Network from the rho statistic, using a 25-year generation time and a mean per-locus, per-generation mutation rate of 6.9 × 10−4 (77).
Supplementary Material
Acknowledgements
We thank all DNA donors, and Aleksander Bagdonavicius (Path Centre Forensic Biology, WA, Australia), for provision of DNA samples. M.A.J. is a Wellcome Trust Senior Fellow in Basic Biomedical Science (grant number 057559); S.M.A., A.C.L., Y.X. and C.T.S. were supported by the Wellcome Trust. E.J.P. was supported by the Arts and Humanities Research Council and the EC Sixth Framework Programme under Contract no. ERAS-CT-2003-980409, within the framework of the European Science Foundation EUROCORES programme “The Origin of Man, Language and Languages”. T.K. and P.dK. were supported by the NWO (Netherlands Organisation for Scientific Research) project 231-70-001 within the same EUROCORES programme.
References
- 1.Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Adams MD, Myers EW, Li PW, Eichler EE. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 2.Freeman JL, Perry GH, Feuk L, Redon R, McCarroll SA, Altshuler DM, Aburatani H, Jones KW, Tyler-Smith C, Hurles ME, et al. Copy number variation: new insights in genome diversity. Genome Res. 2006;16:949–961. doi: 10.1101/gr.3677206. [DOI] [PubMed] [Google Scholar]
- 3.Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova R, Ali J, Bieri T, et al. The male-specific region of the human Y chromosome: a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 4.Bobrow M, Pearson PL, Pike MC, El-Alfi OS. Length variation in the quinacrine binding segment of human Y chromosomes of different sizes. Cytogenet. 1971;10:190–198. doi: 10.1159/000130138. [DOI] [PubMed] [Google Scholar]
- 5.Verma RS, Dosik H, Scharf T, Lubs HA. Length heteromorphisms of fluorescent (f) and non-fluorescent (nf) segments of human Y chromosome: classification, frequencies, and incidence in normal Caucasians. J. Med. Genet. 1978;15:277–281. doi: 10.1136/jmg.15.4.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verma RS, Rodriguez J, Dosik H. The clinical significance of pericentric inversion of the human Y chromosome: a rare “third” type of heteromorphism. J. Hered. 1982;73:236–238. doi: 10.1093/oxfordjournals.jhered.a109627. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein R, Wadee A, Rosendorff J, Wessels A, Jenkins T. Inverted Y chromosome polymorphism in the Gujerati Muslim Indian population of South Africa. Hum. Genet. 1986;74:223–229. doi: 10.1007/BF00282538. [DOI] [PubMed] [Google Scholar]
- 8.Schmid M, Haaf T, Solleder E, Schempp W, Leipoldt M, Heilbronner H. Satellited Y chromosomes: structure, origin and clinical significance. Hum. Genet. 1984;67:72–85. doi: 10.1007/BF00270562. [DOI] [PubMed] [Google Scholar]
- 9.Repping S, van Daalen SK, Brown LG, Korver CM, Lange J, Marszalek JD, Pyntikova T, van der Veen F, Skaletsky H, Page DC, et al. High mutation rates have driven extensive structural polymorphism among human Y chromosomes. Nat. Genet. 2006;38:463–467. doi: 10.1038/ng1754. [DOI] [PubMed] [Google Scholar]
- 10.Tyler-Smith C, Taylor L, Müller U. Structure of a hypervariable tandemly repeated DNA sequence on the short arm of the human Y chromosome. J. Mol. Biol. 1988;203:837–848. doi: 10.1016/0022-2836(88)90110-6. [DOI] [PubMed] [Google Scholar]
- 11.Affara NA, Ferguson-Smith MA, Tolmie J, Kwok K, Mitchell M, Jamieson D, Cooke A, Florentin L. Variable transfer of Y-specific sequences in XX males. Nucl. Acids Res. 1986;14:5375–5387. doi: 10.1093/nar/14.13.5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Page DC. Sex reversal: deletion mapping the male-determining function of the human Y chromosome. Cold Spring Harb. Symp. Quant. Biol. 1986;51:229–235. doi: 10.1101/sqb.1986.051.01.028. [DOI] [PubMed] [Google Scholar]
- 13.Zhu B, Sun QW, Lu YC, Sun MM, Wang LJ, Huang XH. Prenatal fetal sex diagnosis by detecting amelogenin gene in maternal plasma. Prenat. Diagn. 2005;25:577–581. doi: 10.1002/pd.1192. [DOI] [PubMed] [Google Scholar]
- 14.Sullivan KM, Mannucci A, Kimpton CP, Gill P. A rapid and quantitative DNA sex test - fluorescence-based PCR analysis of X-Y homologous gene amelogenin. Biotechniques. 1993;15:636. [PubMed] [Google Scholar]
- 15.Stone AC, Milner GR, Pääbo S, Stoneking M. Sex determination of ancient human skeletons using DNA. Am. J. Phys. Anthropol. 1996;99:231–238. doi: 10.1002/(SICI)1096-8644(199602)99:2<231::AID-AJPA1>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 16.Faerman M, Kahila G, Smith P, Greenblatt C, Stager L, Filon D, Oppenheim A. DNA analysis reveals the sex of infanticide victims. Nature. 1997;385:212–213. doi: 10.1038/385212a0. [DOI] [PubMed] [Google Scholar]
- 17.Santos FR, Pandya A, Tyler-Smith C. Reliability of DNA-based sex tests. Nat. Genet. 1998;18:103. doi: 10.1038/ng0298-103. [DOI] [PubMed] [Google Scholar]
- 18.Roffey PE, Eckhoff CI, Kuhl JL. A rare mutation in the amelogenin gene and its potential investigative ramifications. J. Forensic Sci. 2000;45:1016–1019. [PubMed] [Google Scholar]
- 19.Henke J, Henke L, Chatthopadhyay P, Kayser M, Dülmer M, Cleef S, Pöche H, Felske-Zech H. Application of Y-chromosomal STR haplotypes to forensic genetics. Croat. Med. J. 2001;42:292–297. [PubMed] [Google Scholar]
- 20.Thangaraj K, Reddy AG, Singh L. Is the amelogenin gene reliable for gender identification in forensic casework and prenatal diagnosis? Int. J. Legal Med. 2002;116:121–123. doi: 10.1007/s00414-001-0262-y. [DOI] [PubMed] [Google Scholar]
- 21.Steinlechner M, Berger B, Niederstatter H, Parson W. Rare failures in the amelogenin sex test. Int. J. Legal Med. 2002;116:117–120. doi: 10.1007/s00414-001-0264-9. [DOI] [PubMed] [Google Scholar]
- 22.Bosch E, Lee AC, Calafell F, Arroyo E, Henneman P, de Knijff P, Jobling MA. High resolution Y chromosome typing: 19 STRs amplified in three multiplex reactions. Forens. Sci. Int. 2002;125:42–51. doi: 10.1016/s0379-0738(01)00627-2. [DOI] [PubMed] [Google Scholar]
- 23.Chang YM, Burgoyne LA, Both K. Higher failures of amelogenin sex test in an Indian population group. J. Forensic Sci. 2003;48:1309–1313. [PubMed] [Google Scholar]
- 24.Michael A, Brauner P. Erroneous gender identification by the amelogenin sex test. J. Forensic Sci. 2004;49:258–259. [PubMed] [Google Scholar]
- 25.Lattanzi W, Di Giacomo MC, Lenato GM, Chimienti G, Voglino G, Resta N, Pepe G, Guanti G. A large interstitial deletion encompassing the amelogenin gene on the short arm of the Y chromosome. Hum. Genet. 2005;116:395–401. doi: 10.1007/s00439-004-1238-z. [DOI] [PubMed] [Google Scholar]
- 26.Parkin EJ, Kraayenbrink T, van Driem GL, Tshering K, de Knijff P, Jobling MA. 26-locus Y-STR typing in a Bhutanese population sample. Forens. Sci. Int. 2006;161:1–7. doi: 10.1016/j.forsciint.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 27.Kashyap VK, Sahoo S, Sitalaximi T, Trivedi R. Deletions in the Y-derived amelogenin gene fragment in the Indian population. BMC Med. Genet. 2006;7:37. doi: 10.1186/1471-2350-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitchell RJ, Kreskas M, Baxter E, Buffalino L, Van Oorschot RA. An investigation of sequence deletions of amelogenin (AMELY), a Y-chromosome locus commonly used for gender determination. Ann. Hum. Biol. 2006;33:227–240. doi: 10.1080/03014460600594620. [DOI] [PubMed] [Google Scholar]
- 29.Chang YM, Perumal R, Keat PY, Yong RY, Kuehn DL, Burgoyne L. A distinct Y-STR haplotype for Amelogenin negative males characterized by a large Y(p)11.2 (DYS458-MSY1-AMEL-Y) deletion. Forens. Sci. Int. 2006 doi: 10.1016/j.forsciint.2006.04.013. in press. [DOI] [PubMed] [Google Scholar]
- 30.Cadenas AM, Regueiro M, Gayden T, Singh N, Zhivotovsky LA, Underhill PA, Herrera RJ. Male amelogenin dropouts: phylogenetic context, origins and implications. Forens. Sci. Int. 2006 doi: 10.1016/j.forsciint.2006.05.002. in press. [DOI] [PubMed] [Google Scholar]
- 31.Sun C, Skaletsky H, Rozen S, Gromoll J, Nieschlag E, Oates R, Page DC. Deletion of azoospermia factor a (AZFa) region of human Y chromosome caused by recombination between HERV15 proviruses. Hum. Mol. Genet. 2000;9:2291–2296. doi: 10.1093/oxfordjournals.hmg.a018920. [DOI] [PubMed] [Google Scholar]
- 32.Kamp C, Hirschmann P, Voss H, Huellen K, Vogt PH. Two long homologous retroviral sequence blocks in proximal Yq11 cause AZFa microdeletions as a result of intrachromosomal recombination events. Hum. Mol. Genet. 2000;9:2563–2572. doi: 10.1093/hmg/9.17.2563. [DOI] [PubMed] [Google Scholar]
- 33.Blanco P, Shlumukova M, Sargent CA, Jobling MA, Affara N, Hurles ME. Divergent outcomes of intra-chromosomal recombination on the human Y chromosome: male infertility and recurrent polymorphism. J. Med. Genet. 2000;37:752–758. doi: 10.1136/jmg.37.10.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Repping S, Skaletsky H, Lange J, Silber S, Van Der Veen F, Oates RD, Page DC, Rozen S. Recombination between palindromes P5 and P1 on the human Y chromosome causes massive deletions and spermatogenic failure. Am. J. Hum. Genet. 2002;71:906–922. doi: 10.1086/342928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuroda-Kawaguchi T, Skaletsky H, Brown LG, Minx PJ, Cordum HS, Waterston RH, Wilson RK, Silber S, Oates R, Rozen S, et al. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nat. Genet. 2001;29:279–286. doi: 10.1038/ng757. [DOI] [PubMed] [Google Scholar]
- 36.Vollrath D, Foote S, Hilton A, Brown LG, Beer-Romero P, Bogan JS, Page DC. The human Y chromosome - a 43-interval map based on naturally-occurring deletions. Science. 1992;258:52–59. doi: 10.1126/science.1439769. [DOI] [PubMed] [Google Scholar]
- 37.Tilford CA, Kuroda-Kawaguchi T, Skaletsky H, Rozen S, Brown LG, Rosenberg M, McPherson JD, Wylie K, Sekhon M, Kucaba TA, et al. A physical map of the human Y chromosome. Nature. 2001;409:943–945. doi: 10.1038/35057170. [DOI] [PubMed] [Google Scholar]
- 38.Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucl. Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lukacsovich T, Waldman AS. Suppression of intrachromosomal gene conversion in mammalian cells by small degrees of sequence divergence. Genetics. 1999;151:1559–1568. doi: 10.1093/genetics/151.4.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaw CJ, Lupski JR. Non-recurrent 17p11.2 deletions are generated by homologous and non-homologous mechanisms. Hum. Genet. 2005;116:1–7. doi: 10.1007/s00439-004-1204-9. [DOI] [PubMed] [Google Scholar]
- 41.Y Chromosome Consortium A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res. 2002;12:339–348. doi: 10.1101/gr.217602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jobling MA, Tyler-Smith C. The human Y chromosome: an evolutionary marker comes of age. Nat. Rev. Genet. 2003;4:598–612. doi: 10.1038/nrg1124. [DOI] [PubMed] [Google Scholar]
- 43.Jobling MA, Samara V, Pandya A, Fretwell N, Bernasconi B, Mitchell RJ, Gerelsaikhan T, Dashnyam B, Sajantila A, Salo PJ, et al. Recurrent duplication and deletion polymorphisms on the long arm of the Y chromosome in normal males. Hum. Mol. Genet. 1996;5:1767–1775. doi: 10.1093/hmg/5.11.1767. [DOI] [PubMed] [Google Scholar]
- 44.Bosch E, Hurles ME, Navarro A, Jobling MA. Dynamics of a human interparalog gene conversion hotspot. Genome Res. 2004;14:835–844. doi: 10.1101/gr.2177404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parkin EJ, Kraayenbrink T, Opgenort JR, van Driem GL, Tuladhar NM, de Knijff P, Jobling MA. Diversity of 26-locus Y-STR haplotypes in a Nepalese population sample: Isolation and drift in the Himalayas. Forens. Sci. Int. 2006 doi: 10.1016/j.forsciint.2006.05.007. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee AC, Kamalam A, Adams SM, Jobling MA. Molecular evidence for absence of Y-linkage of the Hairy Ears trait. Eur. J. Hum. Genet. 2004;12:1077–1079. doi: 10.1038/sj.ejhg.5201271. [DOI] [PubMed] [Google Scholar]
- 47.Bosch E, Jobling MA. Duplications of the AZFa region of the human Y chromosome are mediated by homologous recombination between HERVs and are compatible with male fertility. Hum. Mol. Genet. 2003;12:341–347. doi: 10.1093/hmg/ddg031. [DOI] [PubMed] [Google Scholar]
- 48.Hughes JF, Skaletsky H, Pyntikova T, Minx PJ, Graves T, Rozen S, Wilson RK, Page DC. Conservation of Y-linked genes during human evolution revealed by comparative sequencing in chimpanzee. Nature. 2005;437:100–103. doi: 10.1038/nature04101. [DOI] [PubMed] [Google Scholar]
- 49.Salido EC, Yen PH, Koprivnikar K, Yu LC, Shapiro LJ. The human enamel protein gene amelogenin is expressed from both the X and the Y chromosomes. Am. J. Hum. Genet. 1992;50:303–316. [PMC free article] [PubMed] [Google Scholar]
- 50.Lau EC, Mohandas TK, Shapiro LJ, Slavkin HC, Snead ML. Human and mouse amelogenin gene loci are on the sex chromosomes. Genomics. 1989;6:162–168. doi: 10.1016/0888-7543(89)90295-4. [DOI] [PubMed] [Google Scholar]
- 51.Lagerstrom M, Dahl N, Nakahori Y, Nakagome Y, Backman B, Landegren U, Pettersson U. A deletion in the amelogenin gene (AMG) causes X-linked amelogenesis imperfecta (AIH1) Genomics. 1991;10:971–975. doi: 10.1016/0888-7543(91)90187-j. [DOI] [PubMed] [Google Scholar]
- 52.Li X, Li HP, Amsler K, Hyink D, Wilson PD, Burrow CR. PRKX, a phylogenetically and functionally distinct cAMP-dependent protein kinase, activates renal epithelial cell migration and morphogenesis. Proc. Natl. Acad. Sci. USA. 2002;99:9260–9265. doi: 10.1073/pnas.132051799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 2004;116:511–526. doi: 10.1016/s0092-8674(04)00133-3. [DOI] [PubMed] [Google Scholar]
- 54.Bassi MT, Ramesar RS, Caciotti B, Winship IM, De Grandi A, Riboni M, Townes PL, Beighton P, Ballabio A, Borsani G. X-linked late-onset sensorineural deafness caused by a deletion involving OA1 and a novel gene containing WD-40 repeats. Am. J. Hum. Genet. 1999;64:1604–1616. doi: 10.1086/302408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elsheikh M, Dunger DB, Conway GS, Wass JA. Turner's syndrome in adulthood. Endocr. Rev. 2002;23:120–140. doi: 10.1210/edrv.23.1.0457. [DOI] [PubMed] [Google Scholar]
- 56.Guttenbach M, Müller U, Schmid M. A human moderately repeated Y-specific DNA sequence is evolutionarily conserved in the Y chromosome of the great apes. Genomics. 1992;13:363–367. doi: 10.1016/0888-7543(92)90254-p. [DOI] [PubMed] [Google Scholar]
- 57.Jakubiczka S, Schnieders F, Schmidtke J. A bovine homologue of the human TSPY gene. Genomics. 1993;17:732–735. doi: 10.1006/geno.1993.1394. [DOI] [PubMed] [Google Scholar]
- 58.Raudsepp T, Santani A, Wallner B, Kata SR, Ren C, Zhang HB, Womack JE, Skow LC, Chowdhary BP. A detailed physical map of the horse Y chromosome. Proc. Natl. Acad. Sci. USA. 2004;101:9321–9326. doi: 10.1073/pnas.0403011101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murphy WJ, Pearks Wilkerson AJ, Raudsepp T, Agarwala R, Schaffer AA, Stanyon R, Chowdhary BP. Novel gene acquisition on carnivore Y chromosomes. PLoS Genet. 2006;2:e43. doi: 10.1371/journal.pgen.0020043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mathias N, Bayés M, Tyler-Smith C. Highly informative compound haplotypes for the human Y chromosome. Hum. Mol. Genet. 1994;3:115–123. doi: 10.1093/hmg/3.1.115. [DOI] [PubMed] [Google Scholar]
- 61.Williams NA, Close JP, Giouzeli M, Crow TJ. Accelerated evolution of Protocadherin11X/Y: a candidate gene-pair for cerebral asymmetry and language. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2006;141:623–633. doi: 10.1002/ajmg.b.30357. [DOI] [PubMed] [Google Scholar]
- 62.Blanco P, Sargent CA, Boucher C, Mitchell M, Affara NA. Conservation of PCDHX in mammals; expression of human X/Y genes predominantly in brain. Mamm. Genome. 2000;11:906–914. doi: 10.1007/s003350010177. [DOI] [PubMed] [Google Scholar]
- 63.Blanco-Arias P, Sargent CA, Affara NA. The human-specific Yp11.2/Xq21.3 homology block encodes a potentially functional testis-specific TGIF-like retroposon. Mamm. Genome. 2002;13:463–468. doi: 10.1007/s00335-002-3010-9. [DOI] [PubMed] [Google Scholar]
- 64.Jobling MA, Williams G, Schiebel K, Pandya A, McElreavey K, Salas L, Rappold GA, Affara NA, Tyler-Smith C. A selective difference between human Y-chromosomal DNA haplotypes. Curr. Biol. 1998;8:1391–1394. doi: 10.1016/s0960-9822(98)00020-7. [DOI] [PubMed] [Google Scholar]
- 65.Schiebel K, Winkelmann M, Mertz A, Xu X, Page DC, Weil D, Petit C, Rappold GA. Abnormal XY interchange between a novel isolated protein kinase gene, PRKY, and its homologue, PRKX, accounts for one third of all (Y+)XX males and (Y−)XY females. Hum. Mol. Genet. 1997;6:1985–1989. doi: 10.1093/hmg/6.11.1985. [DOI] [PubMed] [Google Scholar]
- 66.Cann HM, de Toma C, Cazes L, Legrand MF, Morel V, Piouffre L, Bodmer J, Bodmer WF, Bonne-Tamir B, Cambon-Thomsen A, et al. A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- 67.Dean FB, Hosono S, Fang LH, Wu XH, Faruqi AF, Bray-Ward P, Sun ZY, Zong QL, Du YF, Du J, et al. Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl. Acad. Sci. USA. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jeffreys AJ, Neumann R, Wilson V. Repeat unit sequence variation in minisatellites: a novel source of DNA polymorphism for studying variation and mutation by single molecule analysis. Cell. 1990;60:473–485. doi: 10.1016/0092-8674(90)90598-9. [DOI] [PubMed] [Google Scholar]
- 69.McCarroll SA, Hadnott TN, Perry GH, Sabeti PC, Zody MC, Barrett JC, Dallaire S, Gabriel SB, Lee C, Daly MJ, et al. Common deletion polymorphisms in the human genome. Nat. Genet. 2006;38:9–11. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 70.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔ CT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 71.Oakey R, Tyler-Smith C. Y chromosome DNA haplotyping suggests that most European and Asian men are descended from one of two males. Genomics. 1990;7:325–330. doi: 10.1016/0888-7543(90)90165-q. [DOI] [PubMed] [Google Scholar]
- 72.Paracchini S, Arredi B, Chalk R, Tyler-Smith C. Hierarchical high-throughput SNP genotyping of the human Y chromosome using MALDI-TOF mass spectrometry. Nucl. Acids Res. 2002;30:e27. doi: 10.1093/nar/30.6.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bosch E, Calafell F, González-Neira A, Flaiz C, Mateu E, Scheil H-G, Huckenbeck W, Efremovska L, Mikerezi I, Xirotiris N, et al. Male and female lineages in the Balkans show a homogeneous landscape over linguistic barriers, except for the isolated Aromuns. Ann. Hum. Genet. 2006;70:459–487. doi: 10.1111/j.1469-1809.2005.00251.x. [DOI] [PubMed] [Google Scholar]
- 74.Butler JM, Schoske R, Vallone PM, Kline MC, Redd AJ, Hammer MF. A novel multiplex for simultaneous amplification of 20 Y chromosome STR markers. Forens. Sci. Int. 2002;129:10–24. doi: 10.1016/s0379-0738(02)00195-0. [DOI] [PubMed] [Google Scholar]
- 75.Bandelt H-J, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 76.Qamar R, Ayub Q, Mohyuddin A, Helgason A, Mazhar K, Mansoor A, Zerjal T, Tyler-Smith C, Mehdi SQ. Y-chromosomal DNA variation in Pakistan. Am. J. Hum. Genet. 2002;70:1107–1124. doi: 10.1086/339929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhivotovsky LA, Underhill PA, Cinnioglu C, Kayser M, Morar B, Kivisild T, Scozzari R, Cruciani F, Destro-Bisol G, Spedini G, et al. The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am. J. Hum. Genet. 2004;74:50–61. doi: 10.1086/380911. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.