Abstract
RNA structures contain many bulges and loops that are expected to be sites for inter- and intra-molecular interactions. Nucleotides in the bulge are expected to influence the structure and recognition of RNA. The same stability is assigned to all trinucleotide bulged RNA in the current secondary structure prediction models. In this study thermal denaturation experiments were performed on four trinucleotide bulged RNA, in the context of HIV-1 TAR RNA, to determine whether the bulge sequence affects RNA stability and its divalent ion interactions. Cytosine-rich bulged RNA were more stable than uracil-rich bulged RNA in 1 M KCl. Interactions of divalent ions were more favorable with uracil-rich bulged RNA by ∼2 kcal/mol over cytosine-rich bulged RNA. The UCU-TAR RNA (wild type) is stabilized by 1.7 kcal/mol in 9.5 mM Ca2+ as compared with 1 M KCl, whereas no additional gain in stability is measured for CCC-TAR RNA. These results have implications for base substitution experiments traditionally employed to identify metal ion binding sites. To our knowledge, this is the first systematic study to quantify the effect of small sequence changes on RNA stability upon interactions with divalent ions.
Keywords: RNA thermodynamics, metal–RNA interactions, divalent ion interactions with bulged RNA, bulge stabilities
INTRODUCTION
RNA structures are made up of helical regions that are interspersed with a large number of nonhelical regions, such as loops and bulges, which are crucial for creating the diversity seen in the three-dimensional structures of RNA (Turner 1992; Hermann and Patel 2000). Bulges are one of the most common nonhelical structural features in RNA and the size of the bulge varies from one to several nucleotides. In an asymmetric bulge, additional nucleotides on one strand have no partner on the corresponding strand. The unpaired nucleotides of the bulge can either be stacked in the helix or looped out, causing them to be stacked or unstacked outside the helix. Bulges are often sites for intramolecular interactions and intermolecular recognition; bulge nucleotides can serve as recognition sites for sequence-specific interactions and their presence distorts the surrounding helices, creating additional features for specific recognition by target molecules (White and Draper 1987; Wu and Uhlenbeck 1987; Roy et al. 1990; Zacharias and Hagerman 1995b; Hermann and Patel 2000). Bulge nucleotides are being introduced into antisense RNA for various applications, including targeted chemical cleavage of RNA (Huesken et al. 1996; Madder et al. 2003).
Asymmetric bulge regions are expected to be sites for interactions with positively charged ions, thus neutralizing the charges on phosphates that are in close proximity due to distortion of the RNA backbone (Hermann and Patel 2000). Metal ion binding sites are often identified by RNA crystallography when crystals are grown in high concentrations of monovalent and divalent ions (Cate and Doudna 1996; Correll et al. 1997; Basu et al. 1998). Biophysical studies provide additional data on sites of interaction and binding constants for metal–RNA interactions (Aboul-ela et al. 1995; Zacharias and Hagerman 1995a; Edwards and Sigurdsson 2003; Casiano-Negroni et al. 2007). However, the extent to which divalent ions stabilize RNA structures, including bulges, is not known.
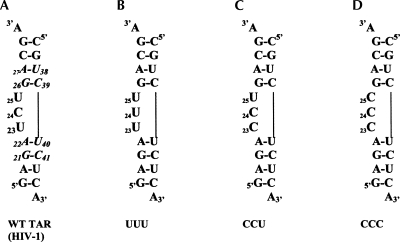
TAR RNA is a hairpin formed between position +1 and +59 (approximately) of HIV-1 RNA (Coffin et al. 1997; Frankel and Young 1998) and contains a UCU bulge. The bulge region interacts with the Tat protein, in complex with cyclin T1, to increase the rate of transcription by nearly 100-fold (Coffin et al. 1997; Frankel and Young 1998). Hence, TAR RNA has been extensively studied by biochemical and biophysical methods (Muesing et al. 1987; Dingwall et al. 1989; Cordingley et al. 1990; Weeks et al. 1990; Calnan et al. 1991; Puglisi et al. 1992; Tao and Frankel 1992; Aboul-ela et al. 1995, 1996; Zacharias and Hagerman 1995a; Ippolito and Steitz 1998; Edwards and Sigurdsson 2003; Casiano-Negroni et al. 2007). The trinucleotide (UCU) bulge is at position 23–25 (HIV-1 TAR RNA numbering system is being used here) (Fig. 1A); the two adjoining base pairs on each of the upper and lower helical stem are conserved (Fig. 1A, shown in italics). The crystal structure of HIV-1 TAR RNA shows four Ca2+ binding sites that stabilize an alternate conformation of TAR RNA that is not expected to bind to Tat protein (Ippolito and Steitz 1998). Transient electric birefringence experiments on TAR RNA show that Mg2+ ions straighten the bend in TAR RNA by nearly 50% and do not interfere with the binding of arginine (Zacharias and Hagerman 1995a). Residual dipolar coupling NMR experiments show that binding of arginine changes the interhelical angle and arrests interhelical motion; Na+ and Mg2+ stabilize TAR RNA by different modes of interactions (Pitt et al. 2004; Casiano-Negroni et al. 2007). In EPR studies, Na+ and Ca2+ caused similar changes in mobility, while Mg2+ affected RNA differently at one of the bulge positions (Edwards and Sigurdsson 2003). Two bulge residues are expected to be part of the “hinge” region for RNA motions in TAR RNA (Musselman et al. 2007). Thus, TAR RNA is an ideal candidate to study the role of bulge nucleotides in RNA stability and to quantify the magnitude of RNA–metal interactions.
FIGURE 1.
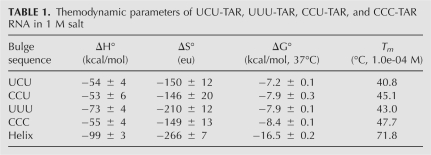
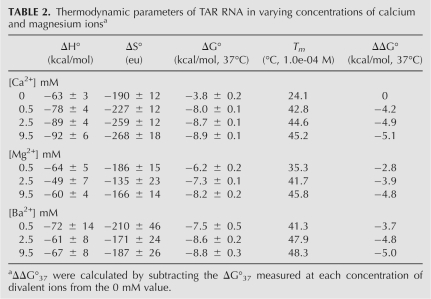
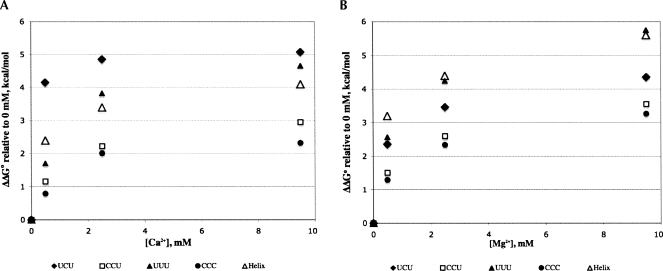
TAR RNA construct and modifications used in this study. (A) Wild-type TAR RNA construct from HIV-1 containing the UCU bulge at positions 23–25 and the conserved base pairs A27–C38, C26–C39, A22–U40, and G21–C41 are shown in italics; (B) UUU-TAR, where C24 has been changed to U24; (C) CCU-TAR, where U23 changed to C23; and (D) CCC-TAR, where U23 and U25 have been changed to cytosine.
RESULTS
RNA with the UCU bulge is the least stable in 1 M salt
The thermodynamic properties of four trinucleotide-bulged RNAs, with pyrimidine nucleotides in the bulge of HIV-1 TAR RNA (Fig. 1), and a core helix, were measured in a buffer containing 1 M KCl. The UCU-TAR RNA is 1.2 kcal/mol less stable than CCC-TAR RNA (Table 1). The stability of CCU-TAR and UUU-TAR RNA is identical and in-between the stability of UCU-TAR and CCC-TAR RNA. The core helix was significantly destabilized by the interruption caused by the bulge (Table 1).
TABLE 1.
Themodynamic parameters of UCU-TAR, UUU-TAR, CCU-TAR, and CCC-TAR RNA in 1 M salt
TAR RNA is stabilized by a low concentration of calcium ions
Thermal denaturation experiments were performed on the wild-type TAR RNA construct (UCU-TAR RNA) in buffers containing varying concentrations of calcium, magnesium, and barium ions (Table 2) following the protocols presented in the Materials and Methods section. As expected, UCU-TAR RNA is relatively unstable in a 10 mM KCl buffer, with a ΔG°37 value of −3.8 kcal/mol. Addition of just 0.5 mM Ca2+ increases RNA stability by an additional 4 kcal/mol (Fig. 2A, filled diamonds); a further increase in calcium concentration, up to 9.5 mM, added 0.9 kcal/mol to RNA stability. In low concentrations of divalent ions (0–4 mM range), Ca2+ stabilizes UCU-TAR RNA more than Mg2+. As the concentration of metal ions is increased (>5 mM), the stability of TAR RNA in Ca2+ and Mg2+ is within 1 kcal/mol (Fig. 2, cf. A and B). Interactions of UCU-TAR RNA with Ba2+ ions were similar to those with Ca2+ (Table 2).
TABLE 2.
Thermodynamic parameters of TAR RNA in varying concentrations of calcium and magnesium ionsa
FIGURE 2.
Plot of ΔΔG°37 values in varying concentrations of calcium (A) and magnesium ions (B) relative to no divalent ion (0 mM) buffer. The ΔΔG°37 values were calculated by subtracting the ΔG°37 value in a given concentration of divalent ion from the value in 0 mM buffer.
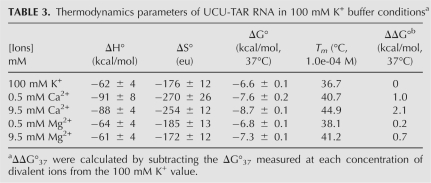
A few metal titration experiments were also performed in a high-salt buffer (Table 3). Even under these conditions, UCU-TAR RNA was 0.4 kcal/mol more stable in 0.5 mM Ca2+ and 1.5 kcal/mol more stable in 9.5 mM Ca2+ as compared with 1 M K+ (and 9.5 mM Mg2+). Thus, high concentrations of monovalent ions competed more effectively with Mg2+ interactions with UCU-TAR RNA than with Ca2+ interactions.
TABLE 3.
Thermodynamics parameters of UCU-TAR RNA in 100 mM K+ buffer conditionsa
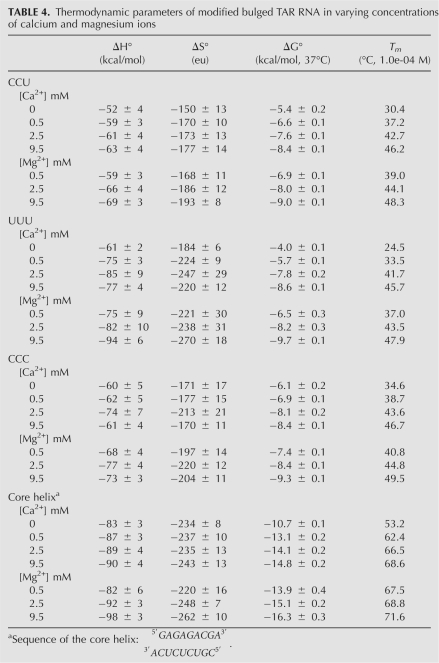
Thermodynamic properties of modified bulge nucleotides and the core helix
Table 4 shows the thermodynamic parameters for CCU-TAR RNA, UUU-TAR RNA, CCC-TAR RNA, and the core helix in various concentrations of divalent ions. A small change in the bulge sequence changed the RNA stability in low-salt conditions, with a ΔG°37 of −6.1 kcal/mol for CCC-TAR RNA and ΔG°37 of −3.8 kcal/mol for UCU-TAR RNA.
TABLE 4.
Thermodynamic parameters of modified bulged TAR RNA in varying concentrations of calcium and magnesium ions
As RNA with different bulge sequences have different ΔG°37 values in the absence of divalent ions, changes in RNA stability (ΔΔG° relative to 0 mM condition) were compared in various concentrations of Ca2+ (Fig. 2A) and Mg2+ (Fig. 2B) ions rather than the measured ΔG°37 values. Interactions with Mg2+ or Ca2+ ions stabilized the cytosine-rich bulged RNA by nearly 2–3 kcal/mol (Fig. 2, open rectangles represent CCU, filled circles represent CCC). The uracil-rich bulged RNA were stabilized by interactions with divalent ions: a ΔΔG°37 of 5.7 kcal/mol for UUU-TAR in 9.5 mM Mg2+ (Fig. 2B, filled triangles) and a ΔΔG°37 of 5.1 kcal/mol for UCU-TAR in 9.5 mM Ca2+ (Fig. 2A, filled diamonds). Interactions with Mg2+ ions were more favorable for the core helix than with Ca2+ (Fig. 2, open triangles).
Contributions of monovalent and divalent ions to RNA stability
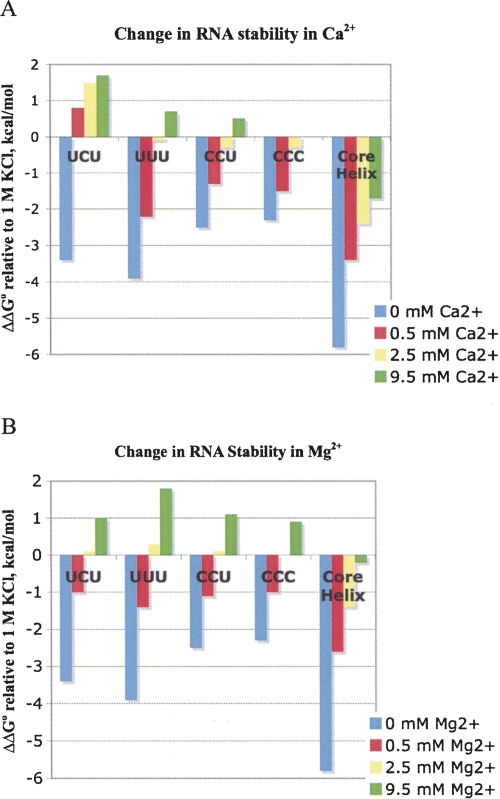
RNA thermodynamic data are usually collected in 1 M salt condition to obtain ΔGo 37 values for RNA under conditions where charge–charge interactions are satisfied. Hence, a comparative analysis was performed between ΔG°37 values obtained in 1 M KCl and in various concentrations of divalent ions (ΔΔG° relative to 1 M KCl) (Fig. 3). Based on the polyelectrolyte condensation theory (Bloomfield et al. 2000), TAR RNA in 9.5 mM divalent ions is expected to be less stable than in 1 M monovalent ions if the interactions between ions and RNA are purely electrostatic. Among the four bulge modifications examined here, only UCU-TAR RNA shows a 1.7 kcal/mol gain in stability in Ca2+ (Fig. 3A, green bars); all other constructs show favorable interactions with Mg2+, with UUU–TAR interactions being the most favorable (ΔΔG°37 of 1.8 kcal/mol). This is ∼0.8 kcal/mol more favorable than the other bulged RNA sequences (Fig. 3B, green bars). The core helix is maximally stabilized in 1 M KCl; interactions with Mg2+ and Ca2+ ions do not increase core helix stability (Fig. 3A,B). Cytosine-rich bulged RNA behaved similarly in both divalent ions.
FIGURE 3.
Plot of ΔΔG°37 values relative to 1 M K+ ions. The ΔΔG°37 values were calculated by taking the difference in the ΔG°37 in a given concentration of calcium (A) or magnesium ions (B) and 1 M KCl. The positive ΔΔG°37 value indicates that the RNA is more stable in divalent ions relative to 1 M KCl.
DISCUSSION
Bulged RNA play a significant role in RNA biochemistry by serving as recognition sites for target molecules and are expected to be sites for interactions with metal ions due to the close proximity of phosphates in the bulge. In this study we performed a thermodynamic analysis of trinucleotide bulged RNA, in the context of a well-studied HIV-1 TAR RNA, to demonstrate that under 1 M salt conditions, the order of stability is: UCU-TAR < UUU-TAR = CCU-TAR < CCC-TAR, with the difference between UCU-TAR and CCC-TAR being 1.2 kcal/mol, and the average stability of the trinucleotide bulged RNA being −7.9 kcal/mol. Thus, RNA with the bulge is additionally destabilized by ∼1 kcal/mol over current prediction models.
The bulged region of TAR RNA was chosen as a model system to quantify the contribution of divalent ions to bulged RNA stability under various ionic conditions and to determine if all bulged sequences would give similar thermodynamic signatures. Four different bulged RNA, with pyrimidine nucleotides in the bulge, were examined here to show that the identity of the bulge nucleotides affects the stability of trinucleotide bulged RNA, with a clear difference between uracil-rich and cytosine-rich bulged RNA (Fig. 2; Tables 2, 4). In 1 M KCl (Table 1) and low-salt buffers (Tables 2, 4), cytosine-rich trinucleotide-bulged RNAs were more stable. Addition of millimolar concentration of Mg2+ and Ca2+ ions increases stability for all RNA as expected (Figs. 2), but to varying degrees. In general, cytosine-rich bulge sequences gained ∼3 kcal/mol, whereas uracil-rich bulge sequences gained ∼5 kcal/mol in stability upon interactions with divalent ions (over the low-salt conditions). The average stability of pyrimidine-containing bulged RNA was −8.6 kcal/mol in 9.5 mM Ca2+ and −9.1 kcal/mol in 9.5 mM Mg2+. The ability of bulged nucleotides to interact with each other and with the neighboring base pairs can effect the local electrostatic environment, hydration, conformations, and ion binding (Auffinger and Westhof 2000; Draper 2004); all of these differences are reflected in the measured ΔG°37 values reported here. In the case of TAR RNA, various conformations have indeed been seen under various ionic conditions (Aboul-ela et al. 1995, 1996; Edwards and Sigurdsson 2003; Pitt et al. 2004; Casiano-Negroni et al. 2007; Zhang et al. 2007).
As shown in Figure 3, changing U23 to C23 significantly alters the Ca2+ interactions but does not alter Mg2+ interactions. Changing C24 to U24 significantly alters both the Ca2+ and Mg2+ interactions. Changing both uracil 23 and 25 to cytosine primarily alters the Ca2+ interactions and not Mg2+ interactions. These results suggest that divalent interactions with U23 and C24 are sufficiently different to lead to measurable differences in ΔGo 37 values. NMR data from Al-Hashimi and coworkers showed differences in interactions for U23 between Na+ and Mg2+ ions (Pitt et al. 2004; Casiano-Negroni et al. 2007). EPR data from Sigurdsson and coworkers showed Na+ and Ca2+ interactions were similar and influence U23; Mg2+ additionally influences mobility at position U25 (EPR data was not collected for cytosine in the bulge) (Edwards and Sigurdsson 2003). Thermodynamic analyses presented here suggest that both U23 and C24 play a role in divalent ion interactions and could potentially play a role in allowing bulged RNA to sample various stable conformations. Our results show that single nucleotide substitution or modification can lead to differences in divalent ion interactions. Thus, single-base substitution or base-modification experiments should be interpreted with caution in identifying metal ion binding sites, especially when these sites involve nonhelical, dynamic regions in RNA structure. A simple thermodynamic profile of the native and modified RNA (under experimentally relevant conditions) may serve as a diagnostic for changes in RNA structure.
In this study we show that bulged RNA have varying stability in both low salt and 1 M KCl condition, and their interactions with divalent ions can be quantified. Inclusion of the divalent ion interactions of RNA under biochemically relevant conditions is likely to improve RNA secondary structure prediction and in vitro experimental design, especially as large RNA contain multiple ion interaction sites.
MATERIALS AND METHODS
RNA design and purification
To study the stability of trinucleotide bulge sequences and their interactions with metal ions, a TAR-RNA construct (UCU-TAR, WT TAR) (Fig. 1) containing the U23C24U25 bulge (numbering system of HIV-1 TAR RNA) and the two conserved base pairs on each helical stem was designed. To understand the effect of the UCU sequence on TAR RNA stability and its interactions with divalent ions, single modifications in pyrimidine sequence in the bulge region were designed by changing one bulge nucleotide at a time—U23 to C23 modification (CCU-TAR) and C24 to U24 (UUU-TAR). One construct containing a double modification, U23 to C23 and U25 to C25 (CCC-TAR), was also designed to study any combined effect of positions 23 and 25 on RNA stability and ion interactions. A core helix sequence,
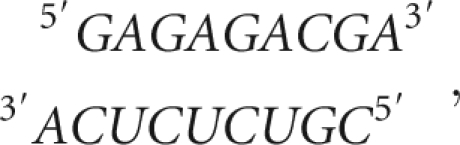 |
was designed by eliminating the bulge sequence (RNA designed by base pairing the bulge sequence forms a structure that is too stable for these experiments.)
All RNA constructs were designed to be non-self-complementary and were ordered from Dharmacon, Inc. A hairpin RNA was not used for several reasons: the loop region is expected to interact with divalent ions, most bulges are present in helices, base substitution experiments are easier and cheaper to design in a duplex RNA, and data analysis for duplexes is more robust. RNA were purified with a thin-layer chromatography using 20 cm × 20 cm, 500-mm thick silica plates with a running buffer of 6:3:1 (v/v/v) 1-propanol, 30% ammonium hydroxide, and water. The samples were spin filtered to remove any excess silica and dried using SPD1010 SpeedVac System (Savant); samples were deprotected in a 100-mM acetic acid buffer adjusted to pH 3.8 with TEMED. The samples were desalted using C18 Sep-Pak column (Waters). The loading buffer was 5 mM sodium bicarbonate at pH 6. Elution was performed twice using 2 mL of 30% acetonitrile, and once using 2 mL of 100% acetonitrile. The fractions containing RNA were dried. The concentrations of RNA were determined using extinction coefficient derived from pairwise values for nearest neighbors.
Thermal denaturation experiments and data analysis
All thermodynamic data were collected on a Cary 100 Bio UV-Visible Spectrophotometer fitted with 6 × 6 Peltier unit. Buffer conditions used were similar to those used for biochemical experiments. Samples were prepared with equimolar amounts of each RNA strand. All samples were run in a buffer containing 10 mM cacodylic acid, 10 mM KCl, and 0.5 mM EDTA at pH 6 (0 mM buffer). For divalent metal ion titration experiments, appropriate amounts of calcium chloride or magnesium chloride were added to the 0 mM buffer. The concentrations of divalent ions were established by complexometric titrations or atomic absorption spectroscopy (with the detection limit in low- to submicromolar range, respectively). The buffers were labeled based on the net concentration of divalent ions. For example, 10 mM cacodylic acid, 10 mM KCl, 0.5 mM EDTA, and 10 mM CaCl2 buffer was labeled as 9.5 mM Ca2+ buffer. Stability of the UCU-TAR RNA was also measured in buffer containing various concentrations of BaCl2. Thermodynamic parameters for each construct were also measured in 1 M KCl (added to the 0 mM buffer), as it provides a reference for maximum stability in the absence of divalent-specific interactions. A few samples of TAR RNA were tested for their interactions with divalent metal–RNA interactions in a buffer containing 100 mM K+. Thermodynamic data were collected using a nine-step dilution scheme to generate an ∼50-fold concentration range for thermal denaturation experiments. The change in RNA absorbance was monitored at a wavelength of 280 nm.
The melt curves generated by thermal denaturation experiments were analyzed using the two-state model (Xia et al. 1998; Matthews et al. 2004). Linear sloping baselines and temperature-independent enthalpy and entropy values were assumed to generate thermodynamic parameters using the Meltwin program to fit individual melt curves (McDowell and Turner 1996). Thermodynamic parameters generated by individual curve fit were compared with those generated by T −1 m versus log(CT/4) plot for non-self-complementary RNA using equations 1 and 2 (where, Tm is the melting temperature, CT is the total concentration of RNA, and R is the gas constant).
Thermodynamic data reported is from the van't Hoff analysis [T −1 m versus log(CT/4) plots].
ACKNOWLEDGMENTS
This work was funded through a National Science Foundation Grant, MCB-0621509 (N.G.), Venture Grants, and through divisional and college-wide funding. NSF-REU grant CHE 024412 (PI Dr. Krugh) funded I.C.-O. research in the Turner laboratory. I.C.-O. and D.B. were both funded by the Boettcher Foundation of Colorado for their undergraduate education at the Colorado College. We are grateful to Dr. Douglas Turner for providing an intellectually stimulating environment for N.G.'s sabbatical and I.C.-O.'s summer research. We thank Dr. Harold Jones for editorial assistance and the Department of Chemistry and Biochemistry at the Colorado College for prioritizing undergraduate research.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1004108.
REFERENCES
- Aboul-ela F., Karn J., Varani G. The structure of the human immunodeficiency virus type-I TAR RNA reveals principles of RNA recognition by Tat protein. J. Mol. Biol. 1995;253:313–332. doi: 10.1006/jmbi.1995.0555. [DOI] [PubMed] [Google Scholar]
- Aboul-ela F., Karn J., Varani G. Structure of HIV-1 TAR RNA in the absence of ligands reveals a novel conformation of the trinucleotide bulge. Nucleic Acids Res. 1996;24:3974–3981. doi: 10.1093/nar/24.20.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffinger P., Westhof E. Water and ion binding around RNA and DNA (C,G) oligomers. J. Mol. Biol. 2000;300:1113–1131. doi: 10.1006/jmbi.2000.3894. [DOI] [PubMed] [Google Scholar]
- Basu S., Rambo R.P., Strauss-Soukup J., Cate J.H., Ferré-D'Amaré A.R., Strobel S.A., Doudna J.A. A specific monovalent metal ion integral to the AA platform of the RNA tetraloop receptor. Nat. Struct. Biol. 1998;5:986–992. doi: 10.1038/2960. [DOI] [PubMed] [Google Scholar]
- Bloomfield V.A., Crothers D.M., Tinoco I. University Science Books; Sausalito, CA: 2000. Nucleic acids: Structure, properties, and function, Chap. 11. [Google Scholar]
- Calnan B.J., Tidor B., Biancalana S., Hudson D., Frankel A.D. Arginine-mediated RNA recognition: The arginine fork. Science. 1991;252:1167–1171. doi: 10.1126/science.252.5009.1167. [DOI] [PubMed] [Google Scholar]
- Casiano-Negroni A., Sun X., Al-Hashimi H.M. Probing Na+-induced changes in the HIV-1 TAR conformational dynamics using NMR residual dipolar couplings: New insights into the role of counterions and electrostatic interactions in adaptive recognition. Biochemistry. 2007;46:6525–6535. doi: 10.1021/bi700335n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cate J.H., Doudna J.A. Metal-binding sites in the major groove of a large ribozyme domain. Structure. 1996;4:1221–1229. doi: 10.1016/s0969-2126(96)00129-3. [DOI] [PubMed] [Google Scholar]
- Coffin J.M., Hughes S.H., Varmus H.E. Retroviruses. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. [PubMed] [Google Scholar]
- Cordingley M.J., LaFemina R.L., Callahan P.L., Condra J.H., Sardana V.V., Graham D.J., Nguyen T.M., LeGrow K., Gotlib L., Schlabach A., et al. Sequence specific interaction of Tat protein and Tat peptides with the transactivation-responsive sequence element of human immunodeficiency virus type 1 in vitro. Proc. Natl. Acad. Sci. 1990;87:8985–8989. doi: 10.1073/pnas.87.22.8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correll C.C., Freeborn B., Moore P.B., Steitz T.A. Metals, motifs, and recognition in the crystal structure of a 5S rRNA domain. Cell. 1997;91:705–712. doi: 10.1016/s0092-8674(00)80457-2. [DOI] [PubMed] [Google Scholar]
- Dingwall C., Ernberg I., Gait M.J., Green S.M., Heapy S., Karn J., Lowe A.D., Singh M., Skinner M.A., Valerio R. Human immunodeficiency virus 1 Tat protein binds trans-activation-responsive region (TAR) RNA in vitro. Proc. Natl. Acad. Sci. 1989;86:6925–6929. doi: 10.1073/pnas.86.18.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper D.E. A guide to ions and RNA structure. RNA. 2004;10:335–343. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards T.E., Sigurdsson S.T. EPR spectroscopic analysis of TAR RNA-metal interactions. Biochem. Biophys. Res. Commun. 2003;303:721–725. doi: 10.1016/s0006-291x(03)00411-x. [DOI] [PubMed] [Google Scholar]
- Frankel A.D., Young J.A.T. HIV-1: Fifteen proteins and an RNA. Annu. Rev. Biochem. 1998;67:1–25. doi: 10.1146/annurev.biochem.67.1.1. [DOI] [PubMed] [Google Scholar]
- Hermann T., Patel D.J. RNA bulges as architectural and recognition motifs. Structure. 2000;8:R47–R54. doi: 10.1016/s0969-2126(00)00110-6. [DOI] [PubMed] [Google Scholar]
- Huesken D., Goodall G., Blommers M.J.J., Jahnke W., Hall J., Haener R., Moser H.E. Creating RNA bulges: Cleavage of RNA in RNA/DNA duplexes by metal ion catalysis. Biochemistry. 1996;35:16591–16600. doi: 10.1021/bi961700c. [DOI] [PubMed] [Google Scholar]
- Ippolito J.A., Steitz T.A. A 1.3 Å resolution crystal structure of the HIV-1 trans-activation response region RNA stem reveals a metal ion-dependent bulge conformation. Proc. Natl. Acad. Sci. 1998;95:9819–9824. doi: 10.1073/pnas.95.17.9819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madder A., Ehrl R., Stroemberg R. Stabilization of RNA bulges by oligonucleotide complements containing an adenosine analogue. ChemBioChem. 2003;4:1194–1200. doi: 10.1002/cbic.200300531. [DOI] [PubMed] [Google Scholar]
- Matthews D.H., Disney M., Childs J., Schroeder S., Zuker M., Turner D.H. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell J.A., Turner D.H. Investigation of the structural basis for thermodynamic stabilities of tandem GU mismatches: Solution structure of (rGAGGUCUC)2 by two-dimensional NMR and simulated annealing. Biochemistry. 1996;35:14077–14089. doi: 10.1021/bi9615710. [DOI] [PubMed] [Google Scholar]
- Muesing M.A., Smith D.H., Capon D.J. Regulation of mRNA accumulation by a human immunodeficiency virus trans-activator protein. Cell. 1987;48:691–701. doi: 10.1016/0092-8674(87)90247-9. [DOI] [PubMed] [Google Scholar]
- Musselman C., Al-Hashimi H.M., Andricioaei I. iRED analysis of TAR RNA reveals motional coupling, long-range correlations, and a dynamical hinge. Biophys. J. 2007;93:411–422. doi: 10.1529/biophysj.107.104620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt S.W., Majumdar A., Serganov A., Patel D.J., Al-Hashimi H.M. Argininamide binding arrests global motions in HIV-1 TAR RNA: Comparison with Mg2+-induced conformational stabilization. J. Mol. Biol. 2004;338:7–16. doi: 10.1016/j.jmb.2004.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi J.D., Tan R., Calnan B.J., Frankel A.D., Williamson J.R. Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science. 1992;257:76–80. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]
- Roy S., Delling U., Chen C.H. A bulge structure in HIV-1 TAR RNA is required for Tat binding and Tat mediated transactivation. Genes & Dev. 1990;4:1365–1373. doi: 10.1101/gad.4.8.1365. [DOI] [PubMed] [Google Scholar]
- Tao J., Frankel A.D. Specific binding of arginine to TAR RNA. Proc. Natl. Acad. Sci. 1992;89:2723–2726. doi: 10.1073/pnas.89.7.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D.H. Bulges in nucleic acids. Curr. Opin. Struct. Biol. 1992;2:334–337. [Google Scholar]
- Weeks K.M., Ampe C., Schultz S.C., Steitz T.A., Crothers D.M. Fragments of the HIV-1 Tat protein specifically bind TAR RNA. Science. 1990;249:1281–1285. doi: 10.1126/science.2205002. [DOI] [PubMed] [Google Scholar]
- White S.A., Draper D.E. Single base bulges in small RNA hairpins can enhance intercalation of ethidium. Nucleic Acids Res. 1987;15:4049–4064. doi: 10.1093/nar/15.10.4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.-N., Uhlenbeck O.C. Role of a bulged A residue in a specific RNA protein interaction. Biochemistry. 1987;26:8221–8227. doi: 10.1021/bi00399a030. [DOI] [PubMed] [Google Scholar]
- Xia T.B., SantaLucia J., Bruckard M.E., Kierzek R., Schroeder S.J., Zuker M., Turner D.H. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson–Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- Zacharias M., Hagerman P.J. The bend in RNA created by the trans-activation response element bulge of human immunodeficiency virus is straightened by arginine and by Tat-derived peptide. Proc. Natl. Acad. Sci. 1995a;92:6052–6056. doi: 10.1073/pnas.92.13.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias M., Hagerman P.J. Bulge-induced bends in RNA; quantification by transient electric birefringence. J. Mol. Biol. 1995b;247:486–500. doi: 10.1006/jmbi.1995.0155. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Stelzer A.C., Fisher C.K., Al-Hashimi H.M. Visualizing spatially correlated dynamics that directs RNA conformational transitions. Nature. 2007;450:1263–1267. doi: 10.1038/nature06389. [DOI] [PubMed] [Google Scholar]