Abstract
Cryptosporidium parvum and Cryptosporidium hominis are two related species of apicomplexan protozoa responsible for the majority of human cases of cryptosporidiosis. In spite of their considerable public health impact, little is known about the population structures of these species. In this study, a battery of C. parvum and C. hominis isolates from seven countries was genotyped using a nine-locus DNA subtyping scheme. To assess the existence of geographical partitions, the multilocus genotype data were mined using a cluster analysis based on the nearest-neighbor principle. Within each country, the population genetic structures were explored by combining diversity statistical tests, linkage disequilibrium, and eBURST analysis. For both parasite species, a quasi-complete phylogenetic segregation was observed among the countries. Cluster analysis accurately identified recently introduced isolates. Rather than conforming to a strict paradigm of either a clonal or a panmictic population structure, data are consistent with a flexible reproductive strategy characterized by the cooccurrence of both propagation patterns. The relative contribution of each pattern appears to vary between the regions, perhaps dependent on the prevailing ecological determinants of transmission.
Cryptosporidium parvum and Cryptosporidium hominis are two species of ubiquitous protozoan parasites responsible for the vast majority of cases of intestinal cryptosporidiosis in humans (42). Cryptosporidium parvum infects humans and animals and is considered zoonotic. In contrast, C. hominis is thought to lack significant animal reservoirs and appears to be transmitted primarily among humans. Transmission is typically through the ingestion of water contaminated with oocysts or through contact with feces. During most of their life cycles, C. parvum and C. hominis are haploid and reside in the intestinal cells of the host, where they undergo consecutive rounds of asexual multiplication. Thereafter, the differentiation and fusion of gametes lead to a transient diploid stage, followed by meiotic division. Meiotic recombination between genetically heterogeneous C. parvum genotypes has been documented in experimental infections (34), but the relative contribution of this process to the diversification of C. parvum and C. hominis in nature is not well understood.
Ascertaining the population genetic structures of C. parvum and C. hominis is relevant to understanding the pathobiology of cryptosporidiosis and to tracking sources of infection.
Cryptosporidium parasite populations have been studied using PCR-based amplification of single or multiple genetic markers in Italy (3, 4), Denmark (9), Scotland (16, 23, 24), India (12), Israel and Turkey (35), England and Wales (20), France and Haiti (27), India, Kenya, the United States, and Peru (13), and Belgium (14). Some studies have used a gene encoding the sporozoite surface glycoprotein GP45/15 (also dubbed GP60) (5, 32). This marker is appealing because of its extensive sequence polymorphism and functional relevance. However, genotypes based on a single locus are limited in their abilities to capture the structure of a population, which is better explored using multilocus sequence typing schemes. A multilocus approach was used to explore the population genetic structures of C. parvum and C. hominis in Scotland (23, 24), Haiti (27), India, Kenya, the United States, and Peru (13).
The aim of the present study was to expand our understanding of the population structures of C. parvum and C. hominis to a global scale. Therefore, a large battery of archived C. parvum and C. hominis DNA samples from seven countries was genotyped at nine polymorphic loci, and the multilocus genotype (MLG) data were mined by using cluster analysis, diversity statistical tests, and measures of linkage disequilibrium.
MATERIALS AND METHODS
Parasite samples.
DNA from 289 human C. hominis isolates from Uganda (UG), the United States, and the United Kingdom and from 217 human and bovine C. parvum isolates from Uganda, New Zealand, Turkey, Israel, Serbia, and the United States were initially included. DNA purification methods varied from country to country as described below. Archived DNA was stored at −20°C.
Israel.
A total of 62 DNA samples from C. parvum-positive fecal specimens collected between March 2004 and April 2005 from 7- to 13-day-old calves on 14 farms (n = 60), from a horse (n = 1), and from a goat kid (n = 1) were used. The collection sites, oocysts purification, and DNA extraction method were previously described (35).
New Zealand.
Twenty-six isolates from humans (n = 19) and calves (n = 7) were used. The isolates originated from spontaneously reported cases diagnosed between August and November 2006 at medical or veterinary diagnostic laboratories in the Waikato region of the North Island (n = 13) and on the South Island (n = 13). To purify the oocysts, approximately 5 g of feces were suspended in water to a volume of 7 ml and strained with gauze. A volume of 3 ml of diethyl ether was added to extract the fat. Oocysts were captured with 50 μl of immunomagnetic beads (Dynabeads; Dynal, Norway). Purified oocysts were suspended in 100 μl Tris-EDTA buffer and subjected to three cycles of freeze-thawing. DNA was extracted from the oocyst lysate by using a commercial column DNA purification kit (HiPure template preparation kit; Roche Diagnostics, Indianapolis, IN) as previously described (35) and the DNA eluted in 50 μl of distilled water.
Serbia.
Cryptosporidium parvum-positive fecal specimens (n = 50) were collected between April 2003 and September 2006 from calves aged between 8 and 30 days raised on 10 farms located 18 to 45 km from the city of Belgrade. Oocysts were purified by sedimentation on sucrose gradients and lysed with three cycles of freeze-thawing. The DNA was purified with DNA purification kits (QIAamp DNA mini kit), as previously reported (25).
Turkey.
Cryptosporidium parvum oocysts were isolated between September 2001 and May 2005 from 15 fecal specimens from calves on 14 farms in the Kars region in northeastern Turkey. Oocyst purification and DNA extraction methods were previously described (35).
Uganda.
Cryptosporidium-positive fecal specimens (n = 237 [62 C. parvum, 175 C. hominis]) were collected from children under 5 years of age diagnosed with persistent diarrhea at the diarrhea treatment ward of Mulago Hospital in Kampala, Uganda. The specimens were collected from November 1999 through January 2001 and from November 2002 through May 2003 during cross-sectional surveys that have been previously described (37, 38). Most specimens originated from patients residing in Kampala and its surroundings. DNA was extracted using the FastDNA SPIN kit for soil (Qbiogene Inc., Carlsbad, CA) as previously described (2).
United Kingdom.
A total of 143 C. hominis DNA samples were selected from an archive composed of over 14,000 samples extracted from human feces submitted between 2000 and 2005 to the United Kingdom Cryptosporidium Reference Unit by diagnostic laboratories throughout England and Wales. Of these, 113 samples were from sporadic cases in immunocompetent patients in the northwest regions of England and Wales. These cases were previously included in a case-control study which ran from November 2000 to February 2002 (21). An additional 21 C. hominis samples from sporadic cases were chosen from the national collection to include human immunodeficiency virus (HIV)-positive cases and increase the geographical and age distribution. Nine samples were from cases involved in two outbreaks linked to water supplies. An outbreak was defined as two or more linked cases of cryptosporidiosis. Twenty-six sporadic cases had a history of having been outside the United Kingdom in the 2 weeks before the onset of illness, of which eight had been in Pakistan. Other places were defined as Mediterranean countries (14), other European destinations (1), Africa (1), New Zealand (1), and one unknown location. Oocysts were separated from fecal material by flotation on a saturated salt solution, then incubated at 100°C for 60 min, and digested with proteinase K, and the DNA was extracted using DNA extraction kits (8). Of all the samples from the United Kingdom, 105 were reidentified as C. hominis by using the LIB13 marker (33).
United States.
Isolates from the United States included those positive for C. parvum (n = 10) and C. hominis (n = 11). The C. parvum isolates were the reference isolate IOWA (1), three laboratory isolates used in human volunteer studies (28, 29), a bovine isolate from the state of Connecticut, as well as isolates from a spontaneous infection and HIV-positive individuals enrolled in a clinical trial (41). The C. hominis isolates were obtained from individuals enrolled in the same clinical trial, from two unrelated HIV-positive individuals, and from two HIV-negative individuals.
Genotyping.
The LIB13 species-specific marker (33) was used to discriminate between C. parvum and C. hominis and rule out Cryptosporidium meleagridis, Cryptosporidium muris, and Cryptosporidium andersoni. Isolates from Uganda were typed using the COWP restriction fragment length polymorphism (31). The United Kingdom isolates had been previously identified at the COWP locus by PCR-restriction fragment length polymorphism and were additionally tested by using the LIB13 marker. Nine mini- and microsatellite markers were used to subtype C. parvum and C. hominis as previously described (35, 36). Markers are located on chromosome I (MSA, MSB), chromosome II (MSC), chromosome III (MSK), chromosome IV (MSE), chromosome V (MS9), chromosome VI (MSG), chromosome VII (1887), and chromosome VIII (TP14). Minisatellite MS9 and microsatellite TP14 were developed by Mallon et al. (23) and adapted as previously described (36). PCR amplifications were carried out in a real-time PCR machine (LightCycler; Roche Applied Science) using Sybr green I master mix (Roche Diagnostics) as described previously (36). Amplicons were fractionated on 15% native polyacrylamide gels and the bands visually scored in comparison to a set of representative alleles and to a 100-bp ladder. The allele-scoring method was validated by sizing selected markers on a capillary electrophoresis instrument (CEQ 8000; Beckman Coulter, Fullerton, CA). Alleles were numbered with a three-digit code indicating their size in base pairs. All genotyping work was performed at Tufts University.
Data analysis.
Only isolates that amplified the LIB13 or COWP marker and all nine loci were included in the analysis. The MLGs of isolates which included double-banded loci were defined by omitting the large bands. This definition was arbitrary and necessary because the species are haploid. No genotypes with three or more bands were observed. The possibility that some MLGs generated by the arbitrary omission of the large bands could have biased the inferences was assessed by repeating the analyses after omitting double-banded isolates from the data set (see below). To eliminate possible bias introduced by resampling substructured populations, only one representative isolate per MLG-farm combination was left for bovine C. parvum, as previously described (16). This resulted in the removal of 65 C. parvum isolates. This procedure was not applied to human C. parvum and C. hominis isolates as residence information was not available. However, most isolates from the United Kingdom originated from sporadic cases of cryptosporidiosis in patients residing at different addresses. Finally, the C. parvum and C. hominis MLG data were compiled in categorical matrixes composed of 11 columns, with isolates characterized by a unique code, nine alleles, and an MLG identifier.
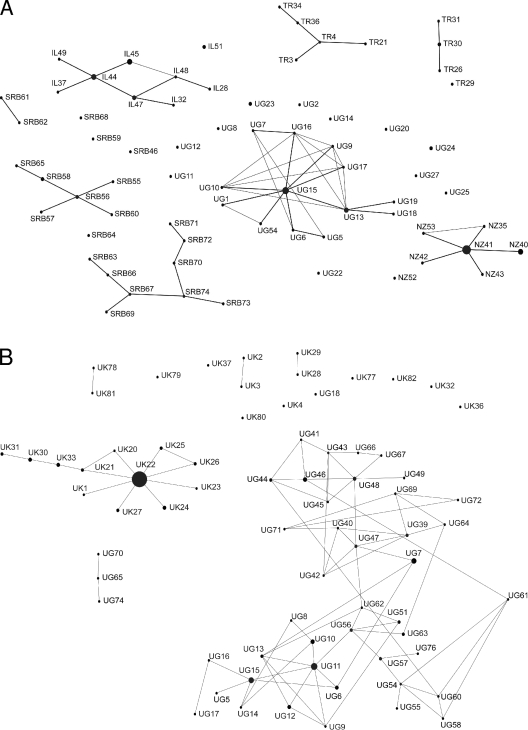
The nine-locus subtyping scheme used in this study defines a genetic distance matrix of only nine possible values. Therefore, dendrograms were not used due to the occurrence of ties and the potential for severe distortions. Instead, the presence of geographical partitions was assessed using a cluster analysis based on the nearest-neighbor principle (7). The pairwise Hamming distances (17), expressed as the number of loci by which the MLGs differ, were used as the genetic distances. For each C. parvum or C. hominis MLG, its nearest neighbors, defined as the MLGs at the shortest Hamming distance, were retrieved manually. Within each country, the measure of clustering was the proportion of MLGs having all the nearest neighbors in the same country. According to this criterion, single-locus variants (i.e., the MLGs that vary at a single locus) are nearest neighbors. Therefore, the degree of clustering within the countries could be assessed partly by a simple inspection of the single-locus variant networks generated using eBURST software (see below). As will be seen, the data did not require any statistical testing to be interpreted.
A combination of methods was used to explore the C. parvum and C. hominis population structures within each country. The genetic diversity of both species was assessed using MLG rank abundance plots (22, 40), with MLG abundances scaled as a percentage. In addition, the MLG richness of C. parvum and C. hominis was compared statistically by means of analytical rarefaction (16, 19). Rarefaction is a statistical method for estimating the number of taxa expected to be present in a random sample of any size taken from a given collection. The approach is useful to compare levels of taxonomic richness among environments, as the taxonomic richness in a sample is dependent on its size and fluctuates stochastically due to sampling variation (16). Rarefaction was performed using the individual-based rarefaction option in PAST software (http://folk.uio.no/ohammer/past/ [downloaded in January 2005]). To assess whether the arbitrary omission of the large bands could have biased the shapes of the rarefaction curves, the analysis was repeated after omitting the double-banded isolates.
Linkage disequilibrium across all loci was assessed using the standardized index of association (sIa) proposed by Habould and Hudson (18). This index, a derivation of the Maynard Smith index of association, fluctuates stochastically around zero in complete panmixia but departs from zero as linkage disequilibrium increases. The index and its probability under a null model of complete panmixia were calculated using LIAN software (http://adenine.biz.fh-weihenstephan.de/cgi-bin/lian/lian.cgi.pl [accessed July 2007]) with 1,000 allele randomizations. Linkage disequilibrium statistics were recalculated after omission of the double-banded isolates from Uganda but were not calculated for the isolates from the United States, Turkey, and Israel, due to the small sample sizes. Finally, the population structures were explored by visual assessment of networks of single-locus variants constructed using the eBURST software (10). Single-locus variant eBURST networks are diagrams composed of MLGs depicted as dots, which are linked to their single-locus variants by lines. A dot's diameter is proportional to the relative frequency of the MLG, so that the largest dots correspond to the most-abundant MLGs and the smallest dots represent singletons.
RESULTS
Genotypes.
A total of 516 isolates were identified as C. parvum (n = 227) or C. hominis (n = 290) and genotyped. Out of 4,644 (516 isolates for each of the nine loci) PCRs, 230 (5%) failed to generate visible amplicons. The proportion of failing reactions was significantly greater in C. hominis (7.0%) than in C. parvum (2.2%) (two-tailed Fisher's exact test; P < 0.001). The same difference was observed between C. parvum and C. hominis isolates from Uganda, a country that provided human samples from both species which were processed in the same manner. Therefore, primer site polymorphisms or a lower C. hominis oocyst output, rather than different DNA extraction techniques or host species, may have contributed to this difference.
Among all PCRs, 30 C. parvum (1.4%) and 42 C. hominis (1.6%) isolates showed double bands. The sizes of the majority of the C. parvum (21/27 [78%]) and C. hominis (33/38 [87%]) bands which were found in double-banded reactions were also found individually in other isolates, suggesting that most double-banded reactions reflected mixed infections rather than PCR artifacts. The proportion of double-banded isolates was significantly greater in Ugandan C. hominis isolates, of which 18.2% had at least one double-banded locus, than in C. hominis isolates from the United Kingdom, of which only two (2%) were double banded (two-tailed Fisher's exact test; P = 0.00) (Table 1). The differences between the proportions of double-banded C. parvum isolates from various countries were also significant. However, there was no difference between the proportion of double-banded C. parvum and C. hominis isolates from Uganda (two-tailed Fisher's exact test; P = 0.24). After removing C. parvum replicates and incomplete C. parvum and C. hominis MLGs, the final data set was composed of 123 C. parvum and 190 C. hominis isolates (see the supplemental material).
TABLE 1.
C. parvum and C. hominis linkage disequilibrium and double-banded genotype statistics according to country of origina
| Isolate | No. of isolates with complete MLGs | sIa (P value) | No. (%) of double-banded isolatesb | Reference for published samples |
|---|---|---|---|---|
| UG C. hominis | 85 | 0.06 (<0.001) | 32 (18) | |
| UG C. parvum | 36 | 0.16 (<0.001) | 7 (11) | |
| UK C. hominis | 95 | 0.38 (<0.001) | 2 (2) | 21 |
| NZ C.parvum | 24 | 0.03 (<0.1) | 0 | |
| IL C. parvum | 20 | ND | 1 (2) | 35 |
| SRB C. parvum | 23 | 0.016 (<0.1) | 7 (13) | 25 |
| TR C. parvum | 11 | ND | 5 (33) | 35 |
Cg. parvum and C. hominis isolates from the United States were excluded. ND, not determined. IL, Israel; NZ, New Zealand; SRB, Serbia; TR, Turkey; UG, Uganda; UK, United Kingdom.
All typed isolates, including incomplete MLGs, were used to calculate the proportion of double-banded isolates.
C. hominis and C. parvum geographic clustering.
Sixty-nine C. parvum and 73 C. hominis MLGs were included in the cluster analysis (see the supplemental material). No MLGs are shared between the two parasite species or between countries, in either C. parvum or C. hominis. Remarkably, with the exception of one C. parvum MLG from Uganda (UG27), which differs from its nearest neighbor at four loci, and one from Serbia (SRB46), which has equidistant nearest neighbors in Serbia and Israel, all the MLGs have their nearest neighbors within their own countries, indicating that more alleles are shared within each country than between the countries. A marked phylogenetic segregation is also evident in the single-locus variant networks, all of which are composed of MLGs from only one country (Fig. 1). Significantly, the C. hominis isolates UK79, UK78, UK81, UK28, and UK29 are not linked to the large C. hominis cluster from the United Kingdom. Retrospective analysis of the patients' data indicated that all these MLGs originated from patients that had traveled to the United Kingdom from Pakistan less than 2 weeks prior to the isolation of the parasites. Together, the clustering of these isolates and the similar travel histories strongly suggested that these isolates were introduced. In contrast to other countries, C. hominis and C. parvum MLGs from the United States did not cluster, which is consistent with the disparate geographic and temporal origins of these samples, and are therefore not shown in Fig. 1.
FIG. 1.
Single-locus variant eBURST networks for C. parvum (A) and C. hominis (B). Each MLG is represented by a dot and designated by a unique identifier. Dot diameters are proportional to the number of isolates. For example, the biggest dot represents C. hominis UK22, which was the most-abundant MLG, found in 59 isolates, and the smallest dots represent singletons. Single-locus variants are connected by lines. IL, Israel; NZ, New Zealand; SRB, Serbia; TR, Turkey; UG, Uganda; UK, United Kingdom.
Population genetic structure.
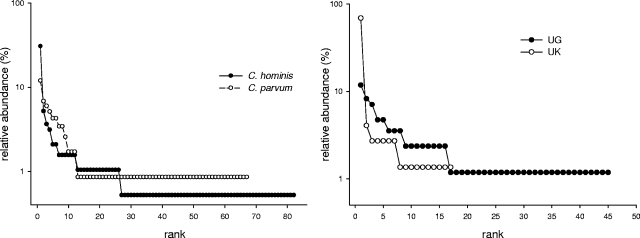
Figure 2A shows the global MLG rank abundance curves for C. hominis and C. parvum. The plot reveals the presence of a small number of highly abundant MLGs and a large number of singletons in both species, consistent with previous data from Scotland (23). In C. hominis, 59 out of 190 (31%) isolates belong to the most abundant MLG (UK22) (see the supplemental material), whereas the most abundant C. parvum MLG (NZ41) accounts for 14/123 (11%) isolates. A comparison between C. hominis rank abundance plots from the United Kingdom and Uganda (Fig. 2B) revealed a more even distribution in Uganda, where the most abundant MLG (UG11) accounts for only 10/85 (12%) of the isolates, compared with 51/74 (69%) in the United Kingdom (isolates from patients reporting travel excluded).
FIG. 2.
C. parvum and C. hominis MLG rank abundance plots. The relative abundance of each MLG is shown on the y axis as a percentage of all isolates. For each curve, the least abundant MLGs are singletons. (Left) Global analysis shows similar rank abundances for both species. (Right) Comparison of Uganda and United Kingdom C. hominis isolates, showing a more-even MLG distribution in Ugandan isolates.
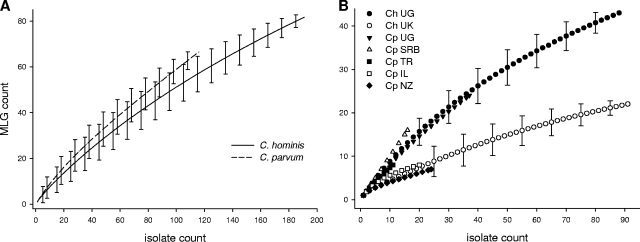
Rarefaction curves were used to compare C. parvum and C. hominis MLG richness levels among countries (Fig. 3). The MLG richness levels in the C. hominis and C. parvum samples from Uganda and C. parvum from Turkey and Serbia, countries with a large proportion of isolates with double-banded loci, are very similar, as indicated by similar rarefaction curves. In contrast, MLG richness in the samples from the United Kingdom, Israel, and New Zealand, countries where only a few double-banded genotypes were observed, is smaller, as indicated by lower curves approaching the asymptote. Rarefaction curves drawn without double-banded C. parvum and C. hominis isolates from Uganda and C. parvum isolates from Serbia and Turkey displayed very similar and overlapping curves (not shown), indicating that the arbitrary omission of large bands did not bias this result. As a substantial number of C. parvum and C. hominis isolates was collected from humans in Uganda as part of the same surveys, it was possible to compare MLG richness levels between C. parvum and C. hominis by using samples originating from the same environment. Rarefied from a sample size of 85 to the C. parvum sample size of 36, C. hominis MLG richness in Uganda has a value of 24.2, which is the same as the MLG richness of 24 observed for C. parvum from the same country.
FIG. 3.
C. parvum and C. hominis analytical rarefaction curves. (A) Global rarefaction; (B) rarefaction by country. Error bars indicate the 95% confidence intervals. Ch, C. hominis; Cp, C. parvum; IL, Israel; NZ, New Zealand; SRB, Serbia; TR, Turkey; UG, Uganda; UK, United Kingdom.
In addition to showing a clear phylogenetic segregation by country, as discussed above in the context of the cluster analysis, the eBURST diagrams (Fig. 1) also reveal differences between countries with respect to the parasites' population structures. The diagram of C. hominis from the United Kingdom has a star-like topology, with the most abundant MLG (UK22) connected to the greatest number of single-locus variants. This topology is reminiscent of eBURST networks of computer-simulated populations diversifying at a high mutation-to-recombination ratio (39). In samples drawn from such populations, most links between the single-locus variants mirror true clonal descent relations. Conversely, the eBURST diagram for C. hominis from Uganda is straggly and shows long chaining of single-locus variants, a feature typical of computer-simulated populations diversifying at a low mutation-to-recombination ratio, in which a significant proportion of links arise from recombination rather than true clonal descent (39). The eBURST diagram for C. parvum from Uganda is also straggly and shows MLG chaining. The exclusion of double-banded isolates from Uganda did not significantly affect the topologies of the eBURST diagrams (not shown), indicating that the omission of the large bands for the definition of the MLGs did not bias these results. Interestingly, the MLGs with the greatest number of single-locus variants (UK22, UG11, UG15, IL-44, NZ41) were also the most abundant in all the countries where abundant MLGs were observed (Fig. 1; see the supplemental material). This pattern argues against a recent epidemic expansion of the abundant MLGs, as in such cases, all the MLGs were equally likely to expand epidemically and prevail in the sample. Instead, this pattern is consistent with a population originating from founders, as proposed for bacterial species by Feil and Spratt (11). According to this structure, abundant genotypes show a greater degree of diversification than do rare genotypes, simply because they undergo more multiplications than rare genotypes.
In support of the differences between the eBURST network topologies and MLG richness levels between Uganda and the United Kingdom, there is strong statistical evidence of allele linkage disequilibrium in C. hominis isolates from the United Kingdom. Conversely, although “statistically significant,” the sIa for C. hominis isolates from Uganda is close to zero (Table 1). Interestingly, the proportion of double-banded isolates of C. hominis is also significantly greater in Uganda than in the United Kingdom, indicating a higher rate of mixed infections in Uganda, but the sIa did not change much after eliminating the double-banded isolates (not shown). To further rule out an epidemic structure or a bias due to the presence of imported, thus reproductively isolated, MLGs in C. hominis isolates from the United Kingdom, linkage disequilibrium analysis was repeated by using one isolate per MLG, without the isolates from patients reporting travel. As expected for a basic clonal population structure (30), linkage disequilibrium persisted (sIa = 0.1786; P < 0.01).
The population structures of C. parvum are less clear, as the sample sizes from Israel, Serbia, and Turkey are relatively small and no C. parvum samples from the United Kingdom were available. Although there is statistical evidence for allele linkage disequilibrium in C. parvum from Uganda, the sIa is only 0.16 and the eBURST diagram is straggly and shows chaining of MLGs. On the other hand, there is no statistical evidence for linkage disequilibrium in C. parvum from New Zealand and Serbia (Table 1). However, the lack of linkage disequilibrium in these countries should not be viewed as evidence for complete panmixia, as it could be due to an underpowered statistical test due to the small sample size, or the presence of only four polymorphic loci in New Zealand C. parvum (see the supplemental material). Indeed, the eBURST network for New Zealand C. parvum is star-like, with the most abundant MLG (NZ41) in the center, reminiscent of a clonal population structure. In contrast, the diagrams from Serbia and Turkey are straggly, consistent with the greater proportion of double-banded C. parvum isolates observed in these countries.
DISCUSSION
In this study, a battery of C. parvum and C. hominis isolates collected from seven countries between 2000 and 2006 was genotyped, enabling inferences on the parasites' global population structures to be made. Our main aims were to assess whether C. parvum and C. hominis populations are geographically partitioned and, if so, to compare the structures of the different populations.
The results of the cluster analysis and the eBURST diagrams show a quasi-complete phylogenetic segregation among countries in both parasite species. Gene flow was therefore not sufficient to erase genetic divergence among these geographically separated populations, which basically remained allopatric. This result is noteworthy considering that C. parvum and C. hominis are cosmopolitan pathogens that may easily travel between countries as asymptomatic infections. Potentially, the ecology of cryptosporidiosis in different countries may have selected for phenotypes which are best adapted to each environment. This hypothesis would predict that imported parasites would be unlikely to spread if environmental factors or transmission patterns are unfavorable. This view is supported by the finding of a number of clustered C. hominis isolates from the United Kingdom, which were apparently introduced from Pakistan and did not cluster with the large network of MLGs from the United Kingdom. Analogous observations were reported in a recent analysis of Scottish and Peruvian isolates (26) and are in accordance with the results of a study using sequence analysis of the GP60 locus (6). The diversity between local and imported MLGs may also explain the putative anthroponotic C. parvum reported by other authors (20, 24), as it is possible that “anthroponotic” isolates were in fact recently imported. Geographic segregation, however, was not reflected in the C. parvum sample from New Zealand, as no partition between North and South Island was observed and the isolates clustered in a single eBURST network (Fig. 1). This interesting result suggests that intense gene flow, as might occur through animal and human movements, may mix the genetic repertoire of C. parvum even in the presence of significant geographical barriers, in this case the Cook Strait. To further evaluate this model, we performed the same cluster analysis with previously published C. parvum MLG data from Scotland and found, as reported by Mallon and coworkers using different analytical methods (24), no phylogenetic partition between the noncontiguous regions of Aberdeenshire and Dumfrieshire (not shown). The lack of partitioning in the Scottish data may be explained by the fact that the cattle population in Aberdeenshire experiences high movement due to the import of cattle from elsewhere in the United Kingdom, therefore preventing the establishment of settled and epidemiologically isolated herds (15). In contrast to these findings, C. parvum samples collected from individual farms in Israel and Turkey showed a marked segregation of the MLGs by farm and a microepidemic structure within the farms (35). Collectively, these results indicate that both micro- and macroecological factors impact C. parvum population structures.
Because C. hominis is thought to propagate primarily in humans but C. parvum has a wider host range, we were interested in assessing whether the differences in host range affected the population structures of these parasites. Cryptosporidium parasites have been defined as being clonal or panmictic based on the results of linkage statistical tests emulating null models of random allele association (23-24, 27). However, these tests are limited in their ability to describe populations propagating both clonally and by recombination, as in such case the test statistic will be below or above the null-model rejection threshold in function of the contribution of each propagation pattern to the population structure (30). Therefore, in this study, we explored the population structures by using a combination of complementary analytical approaches. In contrast to reports of local genetic homogeneity in C. hominis (20, 23, 27), no evidence of any difference between the genetic diversities of C. parvum and C. hominis was found in Uganda, a country that provided samples of both species. Furthermore, the eBURST topology and linkage disequilibrium statistics in Ugandan C. parvum and C. hominis isolates are consistent with similar population structures in both species. Conversely, the coherent differences between C. hominis isolates from Uganda and those from the United Kingdom with respect to MLG richness, eBURST toplogy, and sIa suggest a greater rate of recombination in C. hominis from Uganda than in that from the United Kingdom. The sample from Uganda showed a greater MLG richness and steeper rarefaction curve, as would be expected in a population that, in addition to accumulating variation by mutation, diversifies by recombination. Consistent with this result, there is a greater proportion of double-banded isolates in Uganda, suggesting that many infections were caused by genetically heterogeneous parasites, which enhances the opportunity for recombination. On the other hand, the results of MLG diversity analysis, eBURST network topology, and linkage disequilibrium in the United Kingdom converge in indicating a greater contribution of clonal propagation of C. hominis in this country, and perhaps also of C. parvum in New Zealand. Therefore, rather than being species specific, C. parvum and C. hominis populations appear to be shaped by local or host-related factors. We postulate that frequent transmission from environmental sources increases the probability of coinfections with genetically heterogeneous parasites, favoring recombination. In countries where the sanitary conditions are better and HIV less prevalent (as in the United Kingdom), coinfections with heterogeneous parasites originating from environmental sources may be less frequent, and clonal propagation may prevail.
In summary, the results presented here indicate that although C. parvum and C. hominis are present worldwide, gene flow does not seem sufficient to erase genetic divergence among geographically separated populations, supporting a model of allopatric diversification. As a consequence, multilocus genotyping would enable in many cases the tracking of parasites to their country of origin. Rather than conforming to a strict paradigm of either clonal or panmictic species, the data are consistent with the cooccurrence of both propagation pathways. The relative contribution of each pathway appears to vary according to the prevailing ecological determinants of transmission, indicative of a high adaptive plasticity in C. parvum and C. hominis.
Supplementary Material
Acknowledgments
Our thanks to Niichiro Abe, M. Özkan Arslan, Hidayet Metin Erdoğan, Alex Markovics, and Cynthia L. Winkworth for providing Cryptosporidium samples and to Anne Magurran for advice on statistical analysis. Kristin Elwin and colleagues at the UK Cryptosporidium Reference Unit prepared the Cryptosporidium DNA from the United Kingdom.
Funding from the National Institutes of Allergy and Infectious Diseases (AI052781) is gratefully acknowledged.
Footnotes
Published ahead of print on 3 October 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abrahamsen, M. S., T. J. Templeton, S. Enomoto, J. E. Abrahante, G. Zhu, C. A. Lancto, M. Deng, C. Liu, G. Widmer, S. Tzipori, G. A. Buck, P. Xu, A. T. Bankier, P. H. Dear, B. A. Konfortov, H. F. Spriggs, L. Iyer, V. Anantharaman, L. Aravind, and V. Kapur. 2004. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304:441-445. [DOI] [PubMed] [Google Scholar]
- 2.Akiyoshi, D. E., X. Feng, M. A. Buckholt, G. Widmer, and S. Tzipori. 2002. Genetic analysis of a Cryptosporidium parvum human genotype 1 isolate passaged through different host species. Infect. Immun. 70:5670-5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cacciò, S., W. Homan, R. Camilli, G. Traldi, T. Kortbeek, and E. Pozio. 2000. A microsatellite marker reveals population heterogeneity within human and animal genotypes of Cryptosporidium parvum. Parasitology 120(Pt. 3):237-244. [DOI] [PubMed] [Google Scholar]
- 4.Cacciò, S., F. Spano, and E. Pozio. 2001. Large sequence variation at two microsatellite loci among zoonotic (genotype C) isolates of Cryptosporidium parvum. Int. J. Parasitol. 31:1082-1086. [DOI] [PubMed] [Google Scholar]
- 5.Cevallos, A. M., N. Bhat, R. Verdon, D. H. Hamer, B. Stein, S. Tzipori, M. E. Pereira, G. T. Keusch, and H. D. Ward. 2000. Mediation of Cryptosporidium parvum infection in vitro by mucin-like glycoproteins defined by a neutralizing monoclonal antibody. Infect. Immun. 68:5167-5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chalmers, R. M., S. J. Hadfield, C. J. Jackson, K. Elwin, L. Xiao, and P. Hunter. 2008. Geographic linkage and variation in Cryptosporidium hominis. Emerg. Infect. Dis. 14:496-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cover, T. M., and P. E. Hart. 1967. Nearest neighbor pattern classification. IEEE Trans. Inf. Theory 13:21-27. [Google Scholar]
- 8.Elwin, K., R. M. Chalmers, R. Roberts, E. C. Guy, and D. P. Casemore. 2001. Modification of a rapid method for the identification of gene-specific polymorphisms in Cryptosporidium parvum and its application to clinical and epidemiological investigations. Appl. Environ. Microbiol. 67:5581-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enemark, H. L., P. Ahrens, C. D. Juel, E. Petersen, R. F. Petersen, J. S. Andersen, P. Lind, and S. M. Thamsborg. 2002. Molecular characterization of Danish Cryptosporidium parvum isolates. Parasitology 125:331-341. [DOI] [PubMed] [Google Scholar]
- 10.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feil, E. J., and B. G. Spratt. 2001. Recombination and the population structures of bacterial pathogens. Annu. Rev. Microbiol. 55:561-590. [DOI] [PubMed] [Google Scholar]
- 12.Gatei, W., P. Das, P. Dutta, A. Sen, V. Cama, A. A. Lal, and L. Xiao. 2007. Multilocus sequence typing and genetic structure of Cryptosporidium hominis from children in Kolkata, India. Infect. Genet. Evol. 7:197-205. [DOI] [PubMed] [Google Scholar]
- 13.Gatei, W., C. A. Hart, R. H. Gilman, P. Das, V. Cama, and L. Xiao. 2006. Development of a multilocus sequence typing tool for Cryptosporidium hominis. J. Eukaryot. Microbiol. 53(Suppl. 1):S43-S48. [DOI] [PubMed] [Google Scholar]
- 14.Geurden, T., D. Berkvens, C. Martens, S. Casaert, J. Vercruysse, and E. Claerebout. 2007. Molecular epidemiology with subtype analysis of Cryptosporidium in calves in Belgium. Parasitology 134:1981-1987. [DOI] [PubMed] [Google Scholar]
- 15.Gilbert, M., A. Mitchell, D. Bourn, J. Mawdsley, R. Clifton-Hadley, and W. Wint. 2005. Cattle movements and bovine tuberculosis in Great Britain. Nature 435:491-496. [DOI] [PubMed] [Google Scholar]
- 16.Grinberg, A., N. Lopez-Villalobos, W. Pomroy, G. Widmer, H. Smith, and A. Tait. 2008. Host-shaped segregation of the Cryptosporidium parvum multilocus genotype repertoire. Epidemiol. Infect. 136:273-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamming, R. W. 1950. Error detecting and error correcting codes. Bell Syst. Tech. J. 29:147-160. [Google Scholar]
- 18.Haubold, B., and R. R. Hudson. 2000. LIAN 3.0: detecting linkage disequilibrium in multilocus data. Bioinformatics 16:847-848. [DOI] [PubMed] [Google Scholar]
- 19.Hughes, J. B., J. J. Hellmann, T. H. Ricketts, and B. J. Bohannan. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunter, P. R., S. J. Hadfield, D. Wilkinson, I. R. Lake, F. C. Harrison, and R. M. Chalmers. 2007. Subtypes of Cryptosporidium parvum in humans and disease risk. Emerg. Infect. Dis. 13:82-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hunter, P. R., S. Hughes, S. Woodhouse, Q. Syed, N. Q. Verlander, R. M. Chalmers, K. Morgan, G. Nichols, N. Beeching, and K. Osborn. 2004. Sporadic cryptosporidiosis case-control study with genotyping. Emerg. Infect. Dis. 10:1241-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Magurran, A. E. 2004. Measuring biolocial diversity. Blackwell Publishing, Oxford, United Kingdom.
- 23.Mallon, M., A. MacLeod, J. Wastling, H. Smith, B. Reilly, and A. Tait. 2003. Population structures and the role of genetic exchange in the zoonotic pathogen Cryptosporidium parvum. J. Mol. Evol. 56:407-417. [DOI] [PubMed] [Google Scholar]
- 24.Mallon, M. E., A. MacLeod, J. M. Wastling, H. Smith, and A. Tait. 2003. Multilocus genotyping of Cryptosporidium parvum type 2: population genetics and sub-structuring. Infect. Genet. Evol. 3:207-218. [DOI] [PubMed] [Google Scholar]
- 25.Misic, Z., and N. Abe. 2007. Subtype analysis of Cryptosporidium parvum isolates from calves on farms around Belgrade, Serbia and Montenegro, using the 60 kDa glycoprotein gene sequences. Parasitology 134(Pt. 3):351-358. [DOI] [PubMed] [Google Scholar]
- 26.Morrison, L. J., M. E. Mallon, H. V. Smith, A. Macleod, L. Xiao, and A. Tait. 2008. The population structure of the Cryptosporidium parvum population in Scotland: a complex picture. Infect. Genet. Evol. 8:121-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ngouanesavanh, T., K. Guyot, G. Certad, Y. L. Fichoux, C. Chartier, R. I. Verdier, J. C. Cailliez, D. Camus, E. Dei-Cas, and A. L. Banuls. 2006. Cryptosporidium population genetics: evidence of clonality in isolates from France and Haiti. J. Eukaryot. Microbiol. 53:S33-S36. [DOI] [PubMed] [Google Scholar]
- 28.Okhuysen, P. C., C. L. Chappell, J. H. Crabb, C. R. Sterling, and H. L. DuPont. 1999. Virulence of three distinct Cryptosporidium parvum isolates for healthy adults. J. Infect. Dis. 180:1275-1281. [DOI] [PubMed] [Google Scholar]
- 29.Okhuysen, P. C., S. M. Rich, C. L. Chappell, K. A. Grimes, G. Widmer, X. Feng, and S. Tzipori. 2002. Infectivity of a Cryptosporidium parvum isolate of cervine origin for healthy adults and interferon-gamma knockout mice. J. Infect. Dis. 185:1320-1325. [DOI] [PubMed] [Google Scholar]
- 30.Smith, J. M., N. H. Smith, M. O'Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 90:4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spano, F., L. Putignani, J. McLauchlin, D. P. Casemore, and A. Crisanti. 1997. PCR-RFLP analysis of the Cryptosporidium oocyst wall protein (COWP) gene discriminates between C. wrairi and C. parvum, and between C. parvum isolates of human and animal origin. FEMS Microbiol. Lett. 150:209-217. [DOI] [PubMed] [Google Scholar]
- 32.Strong, W. B., J. Gut, and R. G. Nelson. 2000. Cloning and sequence analysis of a highly polymorphic Cryptosporidium parvum gene encoding a 60-kilodalton glycoprotein and characterization of its 15- and 45-kilodalton zoite surface antigen products. Infect. Immun. 68:4117-4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanriverdi, S., M. O. Arslan, D. E. Akiyoshi, S. Tzipori, and G. Widmer. 2003. Identification of genotypically mixed Cryptosporidium parvum populations in humans and calves. Mol. Biochem. Parasitol. 130:13-22. [DOI] [PubMed] [Google Scholar]
- 34.Tanriverdi, S., J. C. Blain, B. Deng, M. T. Ferdig, and G. Widmer. 2007. Genetic crosses in the apicomplexan parasite Cryptosporidium parvum define recombination parameters. Mol. Microbiol. 63:1432-1439. [DOI] [PubMed] [Google Scholar]
- 35.Tanriverdi, S., A. Markovics, M. O. Arslan, A. Itik, V. Shkap, and G. Widmer. 2006. Emergence of distinct genotypes of Cryptosporidium parvum in structured host populations. Appl. Environ. Microbiol. 72:2507-2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanriverdi, S., and G. Widmer. 2006. Differential evolution of repetitive sequences in Cryptosporidium parvum and Cryptosporidium hominis. Infect. Genet. Evol. 6:113-122. [DOI] [PubMed] [Google Scholar]
- 37.Tumwine, J. K., A. Kekitiinwa, S. Bakeera-Kitaka, G. Ndeezi, R. Downing, X. Feng, D. E. Akiyoshi, and S. Tzipori. 2005. Cryptosporidiosis and microsporidiosis in Ugandan children with persistent diarrhea with and without concurrent infection with the human immunodeficiency virus. Am. J. Trop. Med. Hyg. 73:921-925. [PubMed] [Google Scholar]
- 38.Tumwine, J. K., A. Kekitiinwa, N. Nabukeera, D. E. Akiyoshi, S. M. Rich, G. Widmer, X. Feng, and S. Tzipori. 2003. Cryptosporidium parvum in children with diarrhea in Mulago Hospital, Kampala, Uganda. Am. J. Trop. Med. Hyg. 68:710-715. [PubMed] [Google Scholar]
- 39.Turner, K. M., W. P. Hanage, C. Fraser, T. R. Connor, and B. G. Spratt. 2007. Assessing the reliability of eBURST using simulated populations with known ancestry. BMC Microbiol. 7:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whittaker, R. H. 1965. Dominance and diversity in land plant communities: numerical relations of species express the importance of competition in community function and evolution. Science 147:250-260. [DOI] [PubMed] [Google Scholar]
- 41.Widmer, G., S. Tzipori, C. J. Fichtenbaum, and J. K. Griffiths. 1998. Genotypic and phenotypic characterization of Cryptosporidium parvum isolates from people with AIDS. J. Infect. Dis. 178:834-840. [DOI] [PubMed] [Google Scholar]
- 42.Xiao, L., R. Fayer, U. Ryan, and S. J. Upton. 2004. Cryptosporidium taxonomy: recent advances and implications for public health. Clin. Microbiol. Rev. 17:72-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.