Abstract
The EBNA1 protein of Epstein-Barr virus (EBV) is essential for EBV latent infection in ensuring the replication and stable segregation of the EBV genomes and in activating the transcription of other EBV latency genes. We have tested the ability of four host proteins (Brd2, Brd4, DEK, and MeCP2) implicated in the segregation of papillomavirus and Kaposi's sarcoma-associated herpesvirus to support EBNA1-mediated segregation of EBV-based plasmids in Saccharomyces cerevisiae. We found that Brd4 enabled EBNA1-mediated segregation while Brd2 and MeCP2 had a general stimulatory effect on plasmid maintenance. EBNA1 interacted with Brd4 in both yeast and human cells through N-terminal sequences previously shown to mediate transcriptional activation but not segregation. In keeping with this interaction site, silencing of Brd4 in human cells decreased transcriptional activation by EBNA1 but not the mitotic chromosome attachment of EBNA1 that is required for segregation. In addition, Brd4 was found to be preferentially localized to the FR enhancer element regulated by EBNA1, over other EBV sequences, in latently EBV-infected cells. The results indicate that EBNA1 can functionally interact with Brd4 in native and heterologous systems and that this interaction facilitates transcriptional activation by EBNA1 from the FR element.
As part of their life cycle, gammaherpesviruses and papillomaviruses establish persistent infections in proliferating cells in which their double-stranded circular DNA genomes are maintained at a constant copy number. Maintenance of copy number involves the doubling of the population of viral genomes each cell cycle and a segregation mechanism to ensure equal delivery of the genomes to the daughter cells during cell division. The mechanism of mitotic segregation is conserved in the gammaherpesviruses and papillomaviruses in that, in all cases, the viral genomes are tethered to the host mitotic chromosomes through the viral origin DNA binding protein, which binds directly to the viral segregation element and interacts with one or more host chromosomal proteins.
The mechanism of papillomavirus genome segregation has been studied most extensively using the bovine papillomavirus (BPV). The BPV E2 protein tethers the viral genomes to host chromosomes through interactions with multiple E2 recognition sites in the minichromosome maintenance element (MME) (22, 30, 38, 47). E2 binds the MME through its DNA binding and dimerization domain and interacts with mitotic chromosomes through the domain responsible for transcriptional activation (6, 47). The mechanism by which the E2 transactivation domain contacts mitotic chromosomes to mediate segregation has been the subject of several studies, and considerable evidence has implicated an interaction with the bromodomain protein Brd4 in this process. Brd4 was identified as a binding partner of E2 that colocalized with E2 on host mitotic chromosomes and was shown to enable E2 to maintain plasmids containing the MME in budding yeast (Saccharomyces cerevisiae) (9, 34, 62). Interference with the E2-Brd4 interaction through point mutation of the E2 transactivation domain was found to decrease the chromosome binding of E2 (7), while overexpression of the E2 binding domain of Brd4 promoted the release of both BPV and human papillomavirus (HPV) genomes from mitotic chromosomes (1, 63).
The E2-Brd4 interaction also occurs in interphase (34), and recent studies point to a role for this interaction in the transcriptional activation function of E2 (23, 33, 42). In fact, the E2s from all HPV strains bind Brd4 and require this interaction for transcriptional activation, while only a subset of these E2s colocalize with Brd4 in mitosis and require the Brd4 interaction for mitotic chromosome attachment (33, 44). In addition, Brd4 binding has recently been shown to be important for the transcriptional repression role of HPV E2s (60), further indicating that Brd4 plays multiple roles in the papillomavirus life cycle. Finally, while Brd4 may contribute to the segregation of some papillomaviruses, another E2 binding partner, the ChlR1 DNA helicase, also contributes to this process. Parish et al. (37) showed that a BPV point mutant that binds Brd4 but not ChlR1 fails to associate with mitotic chromosomes and that silencing of ChlR1 (but not silencing of Brd4) reduced E2 binding to mitotic chromosomes.
The segregation mechanisms of three gammaherpesviruses have been studied to various degrees: namely, the two human viruses Epstein-Barr virus (EBV) and Kaposi's sarcoma-associated herpesvirus (KSHV), and the monkey virus herpesvirus saimiri (HVS). The segregation of KSHV is mediated by the viral LANA protein, which binds to the terminal repeat sequences of the viral genome and, like E2, can also activate and repress transcription (2, 3). The association of LANA with mitotic chromosomes has been reported to involve two regions of the protein, the extreme N terminus and the extreme C terminus (4, 29, 39, 53), each of which can interact with chromosome-associated host proteins. The N-terminal LANA sequence binds histones H2A and H2B and the methyl CpG binding protein MeCP2, while the C-terminal domain can bind Brd4, Brd2 (also called Ring3), and DEK (5, 29, 36, 54, 64). Both DEK and MeCP2 have been shown to enable LANA to associate with mouse chromosomes (29), while Brd4 has been found to colocalize with LANA and KSHV genomes on mitotic chromosomes (64). However, it is not yet known how important each of these protein interactions is for mitotic chromosome tethering and segregation, as opposed to playing roles in the transcriptional activation or repression functions of LANA.
HVS is closely related to KSHV, and the HVS episomes also persist in proliferating cells through association with mitotic chromosomes. This association is mediated by ORF73, a LANA homologue, which binds to the HVS terminal repeats and interacts with mitotic chromosomes through its C terminus (10, 11, 52). The similarities between LANA and ORF73 prompted Griffiths and Whitehouse to investigate the ability of MeCP2 and DEK to mediate HVS episome maintenance. They found that MeCP2 (but not DEK) interacted with ORF73 and that silencing of MeCP2 resulted in loss of HVS-based plasmids from human cells (17). It remains to be determined if plasmids were lost due to segregation failure or other effects.
Finally, many studies from several different laboratories have shown that EBV episomes persist in proliferating cells by tethering to mitotic chromosomes (reviewed in reference 16). Attachment to the host chromosomes occurs through the viral EBNA1 protein, which binds to the family of repeats (FR) element of the latent origin of replication, oriP (24). Like E2 and LANA, EBNA1 has roles in the replication of the viral episomes and transcriptional activation of viral genes in addition to its role in mitotic segregation (reviewed in reference 16) (Fig. 1). Two regions of EBNA1 have been identified as having affinity for mitotic chromosomes when excised from EBNA1, the N-terminal 89 amino acids and an internal Gly-Arg-rich region between amino acids 325 and 376 (325-376 sequence) (20, 32). However, mutational analyses of EBNA1 indicate that the 325-376 sequence is the more important region for mitotic chromosome attachment and segregation function (45, 57, 58). The 325-376 region is also important for the transcriptional activation function of EBNA1 as is a sequence between residues 61 and 83 (61-83 sequence) (28, 58). Deletion of either sequence abrogates transcriptional activation, and the requirement for the 61-83 sequence distinguishes the transcriptional activation role of EBNA1 from the other EBNA1 functions.
FIG. 1.
EBNA1 domains and mutants. Schematic representation of the wild-type EBNA1 protein is shown on top along with some of the functional elements, nuclear localization signal (NLS), and amino acid numbers. A version of the EBNA1 protein lacking most of the glycine-alanine repeat sequence was used in these studies (EBNA1) and retains all EBNA1-associated activities. EBNA1 deletion mutants are also shown along with a summary of their previously determined activities in DNA replication (Repl), segregation (Segr), and transcriptional activation (Trans) from the work of Shire et al. (45), Ceccarelli and Frappier (12), and Wu et al. (58), where + denotes complete activity, +/− denotes partial activity, − denotes no activity, and ND means “not determined”.
The EBNA1 325-376 region has been shown to mediate an interaction with one cellular protein that is associated with mitotic chromosomes, termed EBP2 (EBNA1 binding protein 2) (45). A functional role for EBP2 in EBNA1-mediated segregation was initially shown using a budding yeast system in which plasmids containing a yeast origin of replication (ARS element) and the EBV FR segregation element were tested for their ability to be stably maintained in yeast (27). While EBNA1 did not support the maintenance of these plasmids on its own, expression of human EBP2 (hEBP2) enabled plasmid maintenance in an EBNA1- and FR-dependent manner. Further analyses showed that this effect required the EBNA1 325-376 region as well as the EBNA1 binding region of EBP2, that EBP2 enabled EBNA1 to attach to the yeast mitotic chromosomes, and that chromosome attachment by EBP2 was required for it to support segregation (25, 27). EBP2 was subsequently shown to be important for EBNA1 attachment to human mitotic chromosomes, as silencing of EBP2 in human cells resulted in greatly decreased association of EBNA1 and oriP plasmids with metaphase chromosomes (26). EBP2 is currently the only host protein identified as playing a role in EBNA1-mediated segregation, although one group has suggested that a direct interaction of EBNA1 with DNA might also contribute to chromosome attachment (43). However, given the findings that multiple cellular proteins appear to contribute to the segregation of BPV and KSHV, it seems likely that additional cellular proteins will be involved in EBV segregation, either acting in conjunction with EBP2 or providing alternative mechanisms.
The purpose of this study was to test the possibility that Brd2, Brd4, MeCP2, and DEK, which have been implicated in the segregation process of at least one of the viruses BPV, KSHV, and HVS, may also contribute to EBV segregation. Since plasmid segregation systems in budding yeast have previously identified host factors important for both BPV and EBV segregation, we used this approach to test functional effects of these proteins on EBNA1-mediated segregation. This led to the identification of an interaction between EBNA1 and Brd4, which was verified in human cells and shown to contribute to transcriptional activation by EBNA1.
MATERIALS AND METHODS
Plasmid constructs for yeast plasmid loss assays.
Plasmid loss assays were conducted using the centromeric plasmid pRS314 as a positive control (46) and the replicating plasmid YRp7, which lacks a centromere, as a negative control (48). Both plasmids contain a tryptophan (Trp) selectable marker. The FR element of EBV was inserted in YRp7 to generate YRp7FR as described in the work of Kapoor et al. (27). EBNA1 proteins were expressed from p416MET25 (containing a URA3 selection marker), and these constructs are described in the work of Kapoor et al. (27) and Wu et al. (58). hEBP2, DEK, Brd2, and MeCP2 were expressed from the phosphoglycerate kinase (PGK) promoter of the pR425/PGK plasmid, containing a LEU2 selectable marker (31). The construction of pR425/PGK.hEBP2 was previously described (27). The cDNA clones for human Brd2, DEK, and MeCP2 were purchased from the Mammalian Gene Collection (MGC) IMAGE consortium distribution network. These were PCR amplified with primers containing BglII sites and inserted in the BglII site of p425/PGK. Mouse Brd4 was expressed from pADNS (also containing a LEU2 selectable marker), which was kindly supplied by Alison McBride and is described in the work of Brannon et al. (9).
Yeast plasmid loss assay.
The plasmid loss assay was performed as described in the work of Kapoor et al. (27) except that Saccharomyces cerevisiae strain YPH499 (MATa ura3-52 lys2-801 1de2-101 trp1Δ63 his3Δ200 leu2Δ1) was used (46). Briefly, YPH499 was transformed to Ura prototrophy with p416MET25 expressing EBNA1, an EBNA1 mutant, or no protein; transformed to Leu prototrophy with pADNS, pADNS.mBrd4, p425/PGK, p425/PGK.hEBP2, p425/PGK.DEK, p425/PGK.Brd2, or p425/PGK.MeCP2; and to Trp prototrophy with either YRp7, YRp7.FR, or pRS314. Cells were grown in appropriate selectable medium (synthetic complete [SC] medium lacking uracil, leucine, and tryptophan when all three plasmids were present [SC-Ura,Leu,Trp]) to mid-log phase and then diluted into medium that did not select for the segregation plasmid (SC-Ura,Leu). After 28 to 30 doublings in nonselective medium, cultures were spotted in 10-fold serial dilutions on selective and nonselective plates with respect to the segregation plasmid. In parallel, equal amounts of culture were spread on SC-Ura,Leu,Trp and SC-Ura,Leu plates, and the resulting colonies were counted. Plasmid stability was calculated as the number of colonies on SC-Ura,Leu,Trp plates over those on SC-Ura,Leu plates. Standard two-tailed t tests were performed to determine the statistical significance between specific samples.
The expression of EBNA1, hEBP2, DEK, MeCp2, Brd2, and Brd4 in the yeast cells was verified by Western blotting approximately 2 × 107 cells transformed with these expression plasmids. Blots were probed with OT1x monoclonal antibody against EBNA1 (a gift from Jaap Middeldorp), rabbit anti-Brd4 (Abcam ab46199), goat anti-Brd2 (Abcam ab3718), rabbit anti-MeCP2 (Abcam ab2828), or rabbit anti-DEK (Santa Cruz sc-30213). Actin was also probed using a monoclonal antibody (Abcam ab8224) as a loading control. Specific antibodies were detected using secondary anti-mouse, anti-rabbit, or anti-goat antibodies conjugated to horseradish peroxidase (all from Santa Cruz) and chemiluminescence reagents (Perkin-Elmer).
Coimmunoprecipitation assays.
Coimmunoprecipitations of Brd4 with EBNA1 and EBNA1 mutants were performed in 293T cells cotransfected with expression plasmids for Brd4 and EBNA1. For these experiments the cDNA for full-length human Brd4 was excised with BamHI and NotI from pGEX-6P-1 (62) (Addgene plasmid 14447; www.addgene.org) and cloned between the BamHI and NotI sites of pcDNA3. EBNA1 and EBNA1 mutants were expressed from pc3DNA3-based plasmids containing EBV oriP (pc3oriPEBNA1) and have been previously described (45, 58). 293T cells at 50% confluence in 10-cm dishes were transfected with 2 μg of pcDNA3.hBRD4 and either 0.5 μg (pc3oriPEBNA1, pc3oriPΔ325-376, and pc3oriPΔ61-83), 4.0 μg (pc3oriPΔ8-67Δ325-376 and pc3oriP452-641), or 6.0 μg (pc3oriPΔ8-67) of EBNA1-expressing constructs using Lipofectamine 2000 (Invitrogen). The amount of EBNA1-expressing plasmids was adjusted in order to obtain similar expression levels of the different EBNA1 proteins. Three days posttransfection, cells were lysed in modified RIPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.5% NP-40, 0.5 mM EDTA, 0.5% sodium deoxycholate, protease inhibitor cocktail [Roche], and 1 mM phenylmethylsulfonyl fluoride [PMSF]) on ice for 30 min. Lysates were incubated with 10 units/ml of DNase I (Fermentas) for 30 min at room temperature, clarified by centrifugation, and preincubated with 50 μl/ml of protein A agarose beads (Santa Cruz; catalog no. sc-2001) for at least 2 h while rotating at 4°C. One milligram of precleared lysate was diluted to a final protein concentration of 3.0 mg/ml with modified RIPA buffer and then incubated overnight at 4°C with the protein A agarose beads with or without the coupled Brd4 antibody. In each case, protein A agarose beads were preincubated with 1 mg/ml of bovine serum albumin prior to addition to the lysate. After incubation with the lysate, the beads were harvested by centrifugation and washed in cold modified RIPA buffer. Proteins were eluted from the beads in 2× sodium dodecyl sulfate (SDS) loading buffer, separated by SDS-polyacrylamide gel electrophoresis, transferred to nitrocellulose, and probed with anti-Brd4 or EBNA1 OT1x antibodies. Input samples in the Western blot contain 50 μg of each lysate (5% of that used in the immunoprecipitations [IPs]).
Brd4 esiRNA.
Endoribonuclease-prepared short interfering RNAs (esiRNAs) were prepared for the long isoform of human Brd4 (henceforth referred to as esiBrd4) in Lawrence Pelletier's laboratory (Samuel Lunenfeld Research Institute) as previously described (61). A 392-bp region spanning nucleotides 153 to 508 of Brd4 (chosen by DEQOR analysis; http://cluster-1.mpi-cbg.de/Deqor/deqor.html) was amplified by PCR using the following primers, each of which contains the T7 promoter sequence (underlined) at the 5′ end: esiBrd4 Fwd, 5′ TCACTATAGGGAGAGCAACCCTAACAAGCCCAAGA 3′, and esiBrd4 Rev, 5′ TCACTATAGGGAGACCGGTTTCTTCTGTGGGTAGC 3′. The PCR product was further amplified using T7 promoter primers, and an aliquot was in vitro transcribed with T7 polymerase overnight at 37°C for 12 to 16 h. The single-stranded RNA transcripts were annealed, and then the double-stranded RNA was digested into 18- to 25-bp oligomers with RNase III to generate esiBrd4 and purified with Q-Sepharose and isopropanol precipitation.
Transcriptional activation assays.
For transfection assays involving Brd4 silencing, 1 × 105 CNE2Z nasopharyngeal carcinoma cells (49) were grown in one well of a six-well plate to 30% confluence and then transfected with two rounds of 200 ng esiBrd4 or siRNA against green fluorescent protein (GFP) (GCAAGCUGACCCUGAAGUUCAU) using Lipofectamine 2000 on subsequent days. At 48 h after the first round of transfection, cells were further transfected with 1.0 μg of pFRTKCAT reporter plasmid, 0.5 μg of plasmid CMVPLAP expressing secreted alkaline phosphatase (SEAP) (56), and 5 ng of pc3oriPEBNA1 or pc3oriP using Fugene HD (Roche), and 48 h later, cell lysates were assayed for chloramphenicol acetyltransferase (CAT) activity as previously described (12, 58). Briefly, 25 μg of each lysate was incubated with acetyl coenzyme A (Sigma) and [14C]chloramphenicol for various times, and reaction products were separated by thin-layer chromatography and quantified by PhosphorImager analysis. The amount of acetylated product produced at each time point was used to determine the acetylation rate for each lysate. Levels of SEAP in the same cells were determined by diluting 10 μl of cell medium to 100 μl with water and then incubating the medium with 100 μl of 1-mg/ml 4-nitrophenyl phosphate in 10 mM diethanolamine (pH 9.5)-0.5 mM MgCl2 at 37°C for 5 to 10 min until a yellow color developed (56). SEAP reaction product was measured at 405 nm using a Spectra Max M2 plate reader. Changes in CAT activity were normalized to changes in SEAP to correct for any general effects of the treatments on gene expression. Standard two-tailed t tests were performed to determine statistical significance differences between esiBrd4- and siRNA against GFP-treated samples.
For the transactivation assays involving Brd4 overexpression, 4 × 105 CNE2Z cells were plated in a 6-cm dish. The next day, cells were cotransfected with 1.0 μg of pFRTKCAT reporter, 0.5 μg pCMVPLAP, 5 ng of pc3oriPEBNA1 or pc3oriP, and 3 μg of pcDNA3.hBrd4 or pcDNA3. At 48 h posttransfection, cells were harvested and CAT and SEAP levels were determined as described above.
ChIP assays.
Chromatin IP (ChIP) assays were performed on EBV-positive Raji Burkitt's lymphoma cells. Raji cells were fixed with 1% paraformaldehyde in phosphate-buffered saline for 15 min at room temperature and then lysed in hypotonic buffer (10 mM HEPES [pH 7.9], 10 mM KCl, 1 mM EDTA, 10% glycerol, 1 mM PMSF, and protease inhibitor cocktail [p8430; Sigma]), by Dounce homogenization with 10 strokes of pestle B. Nuclei were collected by centrifugation and incubated in RIPA buffer (50 mM Tris-Cl [pH 8.0], 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM PMSF, and protease inhibitors). Chromatin was sheared by sonication to an average DNA length of 500 to 1,000 bp using a Branson 450 sonifier. Fifty micrograms of sheared DNA was incubated with 2.0 μg of rabbit immunoglobulin G (IgG; Santa Cruz), anti-EBNA1 R4 rabbit antibody (18), or a rabbit antibody against Brd4 at 4°C overnight. Antibody complexes were recovered by incubation with 50 μl of 50% (vol/vol) salmon sperm DNA-protein A agarose (Upstate Biochemicals) with rocking for at least 1 h at 4°C. After reversal of the cross-links, quantitative real-time PCR was performed using 1/50 of the ChIP DNA and Platinum SYBR Green qPCR superMix-UDG (Invitrogen) in a Rotorgene quantitative PCR system (Corbett Research). Quantitative real-time PCR was also performed on samples directly after the shearing step (input samples) using 1/2,500 of each sample, and values obtained for ChIP samples were normalized to input samples with the same primer sets. The primers for amplification of the BZLF1 promoter region and DS element are as described in the work of Deng et al. (14), while primers for the FR region correspond to oligonucleotides SC3F and SC3B in the work of Schepers et al. (41).
Biochemical fractionation assay for EBNA1 binding to human chromosomes.
HeLa cells expressing EBNA1 were generated by transfecting HeLa cells with pc3oriPEBNA1 and selecting for the plasmid in 0.4 mg/ml Geneticin-G-418 (Invitrogen) for 4 days. Selection was then removed, and cells at 90% confluence in 10-cm dishes were transfected with 1.2 μg of esiBrd4 or siRNA against GFP using Lipofectamine 2000 (2 μl per 100 ng RNA). Three days posttransfection, cells were blocked in mitosis and fractionated as described in the work of Kapoor et al. (26). Briefly, cells were treated with colcemid (0.1 μg/ml medium; Invitrogen) for 15 h and then harvested by mitotic shake-off. Equal cell numbers (∼1.5 × 106 cells) were lysed in 100 μl of buffer A (20 mM Tris-HCl [pH 7.5], 75 mM KCl, 30 mM MgCl2, 0.1% NP-40, 1 mM dithiothreitol, 0.5 mM EDTA,1 mM PMSF, 1 mM benzamidine, and protease inhibitor cocktail [Roche Diagnostics]), and 50 μl of the cell lysate was removed (W sample). The remaining 50 μl of lysate was separated into soluble (S) and pellet (P) fractions by centrifugation. The pellet was resuspended in a final volume of 50 μl of buffer A, and equal volumes of each sample were analyzed by Western blotting using antibodies against EBNA1 as described above. Western blots for Brd4 were also performed on these fractions from siGFP-treated cells.
RESULTS
Effects of candidate proteins on plasmid loss in yeast.
Yeast plasmid loss assays have been used previously to identify the abilities of Brd4 to enable plasmid segregation by BPV E2 and of EBP2 to enable EBNA1-mediated segregation (9, 27). We used this approach to determine if DEK, MeCP2, Brd2, and Brd4 could also facilitate EBNA1-mediated segregation. All of the segregation test plasmids used in this assay contained a yeast ARS element, which serves as the origin of replication for the plasmids. The positive-control plasmid (pRS314) also contains a centromere element, which ensures the stable segregation of the plasmid and hence its persistence in the absence of selection, while the negative-control plasmid (YRp7) lacks any segregation element and is rapidly lost in the absence of selection. EBNA1-mediated segregation was tested using YRp7FR, which contains the EBV segregation element, FR. As shown previously, EBNA1 expression alone does not enable the maintenance of YRp7FR in the absence of selection but coexpression of EBNA1 with EBP2 results in greatly increased plasmid maintenance (Fig. 2A). In contrast, EBP2 expression does not increase the maintenance of pRS314 or YRp7, indicating that the effect is EBNA1 dependent.
FIG. 2.
Effects of candidate proteins on plasmid maintenance in yeast. Plasmid loss assays were conducted for the indicated segregation test plasmid in the presence (+) of expression plasmids for EBNA1 and hEBP2 (A), DEK (B), Brd2 (C), and MeCP2 (D) or equivalent plasmids not expressing protein (−). After 24 h of growth without selection for the segregation plasmid, 10-fold serial dilutions of the cultures were spotted on plates that were nonselective [NS (−UL)] or selective [S (−UTL)] for the segregation plasmids. Cultures were also spread on selective and nonselective plates such that the number of colonies could be counted, and the percentage of cells retaining the segregation plasmid was determined. The histograms show the average percentages of colonies retaining the segregation plasmid from multiple experiments along with standard deviations, where N is the number of experiments performed. Western blots of total yeast extracts are shown on the right, confirming EBNA1 and candidate protein expression. Numbers at right of blots are molecular masses in kilodaltons.
When the effect of DEK expression was tested in the same assay, it was not observed to increase plasmid maintenance with or without EBNA1; in fact it had a slight inhibitory effect on the maintenance of YRp7FR in the presence of EBNA1 (Fig. 2B). When Brd2 and MeCP2 expression was tested in the plasmid loss assays, both proteins were found to enable the maintenance of YRp7FR in the presence of EBNA1 (Fig. 2C and D). However, both proteins also increased the persistence of YRp7FR in the absence of EBNA1, indicating that the effect was independent of EBNA1. In addition, both Brd2 and MeCP2 enabled the maintenance of YRp7 and further increased the maintenance of the CEN/ARS plasmid, indicating that they have a general stimulatory effect on the plasmid maintenance.
We also tested the effect of Brd4 expression on the maintenance of the three plasmids in yeast. We found that Brd4 enabled the maintenance of YRp7FR in the presence of EBNA1 but not in the absence of EBNA1 (Fig. 3A) and that the maintenance of YRp7FR by EBNA1 and Brd4 is not statistically different from the maintenance of the CEN/ARS plasmid (P = 0.17). In addition, Brd4 had no significant effect on the maintenance of YRp7 or the CEN/ARS plasmid. Therefore, like EBP2, Brd4 facilitates plasmid persistence in an EBNA1-dependent manner, suggesting that it can enable EBNA1-mediated plasmid segregation.
FIG. 3.
Brd4 enables EBNA1-mediated plasmid maintenance in yeast. Plasmid loss assays in yeast were conducted as described for Fig. 2 except that Brd4 expression plasmids were included where indicated. Western blots of whole-yeast lysates on the right confirmed protein expression. In panel B, the indicated EBNA1 mutants were tested in the plasmid loss assay in place of wild-type (WT) EBNA1 and their similar expression levels were confirmed by the Western blot on the right.
Identification of EBNA1 sequences that mediate the Brd4 interaction.
The ability of Brd4 to promote EBNA1-dependent plasmid maintenance in yeast suggests that EBNA1 interacts with Brd4 in this system, either directly or indirectly. We initially explored the EBNA1 sequences involved in the Brd4 interaction by repeating the plasmid loss assays with YRp7FR in the presence of Brd4 and one of three EBNA1 mutants which are expressed at similar levels, the Δ325-376, Δ61-83, and Δ8-67 mutants (Fig. 3B). We have previously shown that EBNA1 lacking amino acids 325 to 376 (Δ325-376) has a severe defect in segregating EBV-based plasmids in human cells as well as in transcriptional activation (45, 58) (Fig. 1) and is unable to bind or function with EBP2 to maintain YRp7FR in yeast (27). It is also known that deletion of the EBNA1 61-83 sequence (Δ61-63) abrogates transcriptional activation but has no detectable effect on the replication or segregation functions of EBNA1 in human cells, while deletion of residues 8 to 67 (Δ8-67) has a partial effect on both EBNA1-mediated transcription and segregation (58) (Fig. 1). Examination of the ability of these EBNA1 mutants to function with Brd4 in plasmid segregation in yeast revealed that deletion of amino acids 325 to 376 had only a small effect on YRp7FR plasmid maintenance (not statistically significant according to two-tailed t test, where P = 0.088), while Brd4-mediated plasmid maintenance was abrogated by deletion of the EBNA1 8-67 or 61-83 sequence (Fig. 3B). This suggests that EBNA1 interacts with Brd4 through its N-terminal region and not through residues 325 to 376.
We further examined the interaction between EBNA1 and Brd4 in human cells by cotransfecting 293T cells with plasmids expressing EBNA1 or EBNA1 mutants and human Brd4. Brd4 was immunoprecipitated from the cells, and recovered proteins were Western blotted for Brd4 and EBNA1 (using an antibody that recognizes all the EBNA1 mutants) (Fig. 4, bottom panel). We consistently found that a small proportion of EBNA1 and EBNA1Δ325-376 coimmunoprecipitated with Brd4 antibody but not in the absence of specific antibody. A Brd4 interaction was not detected with EBNA1Δ61-83 or with the EBNA1 C-terminal fragment 452 to 641, consistent with the yeast plasmid loss assays. Interestingly, deletion of EBNA1 residues 8 to 67 increased Brd4 binding (both on its own and when combined with the 325-376 deletion) even though this deletion slightly decreased the level of EBNA1 protein produced (Fig. 4, top panel). This suggests that deletion of amino acids 8 to 67 makes the Brd4-interacting sequence more accessible to Brd4, as might occur if the binding site is adjacent to the 8-67 deletion, as suggested by the loss of binding with the Δ61-83 mutant. Both the yeast plasmid loss assay and the coimmunoprecipitation experiments in human cells point to the importance of the EBNA1 61-83 sequences and the lack of importance of the 325-376 sequence for Brd4 interactions. While the effects of the 8-67 deletion are opposite in the two systems, this might indicate that EBNA1-Brd4 interactions must be transient or weak to function in the yeast segregation system.
FIG. 4.
Coimmunoprecipitation of EBNA1 and EBNA1 mutants with Brd4 in human cells. 293T cells were cotransfected with a plasmid expressing human Brd4 and a plasmid expressing EBNA1 or the indicated EBNA1 mutants. Brd4 was immunoprecipitated from the cell lysates with Brd4 antibody (B lanes, bottom panel). Recovered proteins were separated by SDS-polyacrylamide gel electrophoresis and immunoblotted for EBNA1 and Brd4 (bottom panel). Western blots of equal amounts of starting lysates (1/20 the amount used in IPs) are also shown (top panel), as are negative-control IPs lacking Brd4 antibody (C lanes, bottom panel). Numbers at left are molecular masses in kilodaltons.
Effect of Brd4 on EBNA1-mediated transcriptional activation.
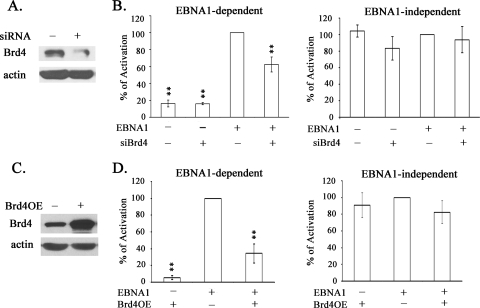
The finding that the EBNA1-Brd4 interaction requires EBNA1 amino acids 61 to 83, which are essential for the transcription function of EBNA1 but dispensable for segregation, suggests that this interaction could be relevant for transcriptional activation by EBNA1. This prompted us to ask whether downregulation of Brd4 affects transcriptional activation by EBNA1. For these studies we used the EBV-negative, nasopharyngeal carcinoma cell line CNE2Z, as this cell type is known to be associated with latent EBV infection. CNE2Z cells were transfected with either siRNA against GFP (negative control) or esiRNA against Brd4, and downregulation of Brd4 was verified by Western blotting (Fig. 5A). Cells were then transfected with a reporter construct in which CAT expression is controlled by EBNA1 binding to the upstream FR element, a SEAP reporter plasmid that is not regulated by EBNA1, and a plasmid containing or lacking an EBNA1 expression cassette. We consistently found that downregulation of Brd4 decreased the ability of EBNA1 to activate the CAT reporter (P = 0.0026 relative to EBNA1 with GFP siRNA treatment) but had no significant effect on the expression of SEAP with or without EBNA1 (P = 0.96 for EBNA1-expressing samples with siRNA treatment for Brd4 versus GFP). The results of the CAT reporter assays after normalization to the SEAP reporter results are shown in Fig. 5B (EBNA1 dependent), as are the results for the SEAP reporter itself (EBNA1 independent). The results indicate that Brd4 contributes to transcriptional activation by EBNA without affecting EBNA1-independent transcription.
FIG. 5.
Effect of Brd4 silencing on transcriptional activation by EBNA1. (A) CNE2Z cells were treated with esiRNA against Brd4 or siRNA against GFP, and downregulation of Brd4 was confirmed by Western blotting equal amounts of cell lysates. (B) After siRNA or esiRNA treatment, cells were transfected with a CAT reporter plasmid under EBNA1 control, a constitutively expressed SEAP reporter plasmid, and an expression plasmid for EBNA1 or empty plasmid control. Chloramphenicol acetylation rates were determined from equal amounts of cell lysate as a measure of CAT activity, and this was normalized to the level of expressed SEAP (EBNA1 dependent). The level of SEAP expressed in each sample is shown on the right (EBNA1 independent). All activities are shown relative to EBNA1 with siGFP samples. Averages and standard deviations are shown from three separate experiments performed in duplicate. Statistically significant differences (compared to EBNA1 activation without silencing Brd4) with 0.001 < P < 0.005 are indicated by double asterisks. (C) A plasmid expressing Brd4 (+) or nothing (−) was cotransfected with EBNA1 expression, CAT reporter, and SEAP reporter plasmids, and Brd4 expression levels were determined by Western blotting. (D) CAT activity was compared in samples with and without Brd4 overexpression (Brd4OE) and normalized to SEAP (EBNA1 dependent). Effects on SEAP activity are also shown (EBNA1 independent).
We also examined the effect of overexpression of Brd4 on EBNA1-dependent and -independent transcription, using the same FR-CAT and SEAP reporter constructs as described above, by including a Brd4 expression plasmid (or empty plasmid) in the transfections (Fig. 5C). Brd4 overexpression inhibited transcriptional activation by EBNA1 approximately threefold (Fig. 5D, left panel) but had no significant effect on the EBNA1-independent SEAP reporter (Fig. 5D, right panel). These results further support a role for Brd4 in EBNA1-mediated transcriptional activation.
Localization of Brd4 to the EBV FR element.
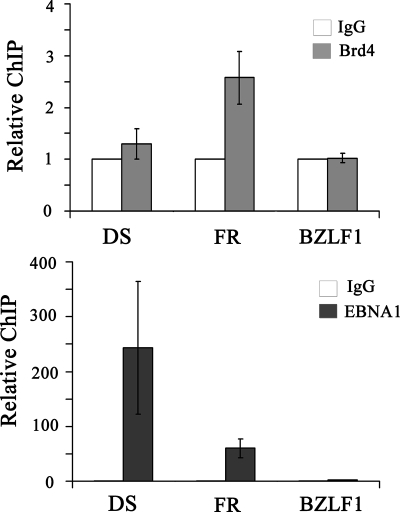
EBV oriP is comprised of two elements: the DS element, which serves to initiate DNA replication, and the FR element, which serves as both the transcriptional enhancer and the segregation element. Both elements are constitutively bound by EBNA1 (40). The finding that Brd4 levels affect transcriptional activation by EBNA1 could be either an indirect effect or a direct effect of Brd4 interaction with EBNA1 on the FR element. To distinguish between these possibilities, we performed ChIP experiments in Raji Burkitt's lymphoma cells, which are latently infected with EBV, to test for localization of endogenous Brd4 to the DS and FR elements and to the EBV BZLF1 gene promoter region located more than 40 kb away from oriP. Antibodies against Brd4 or EBNA1 were used to immunoprecipitate these proteins from sheared Raji DNA, and the levels of the recovered DS, FR, and BZLF1 fragments were compared by quantitative real-time PCR. As expected, EBNA1 was found at both the DS and FR elements (with better recovery of the DS element, as has been observed previously [14, 41]) but not on the BZLF1 fragment (Fig. 6). In contrast, Brd4 antibody pulled down the FR fragment twofold more efficiently than the DS or BZLF1 fragment, indicating that Brd4 is preferentially associated with the FR element.
FIG. 6.
Brd4 is preferentially localized to the FR element of the EBV genome. ChIP experiments were performed on EBV-positive Raji cells using antibodies against Brd4, EBNA1, or nonspecific rabbit IgG as a negative control. Recovered DNA fragments were quantified by real-time PCR using primer sets for the oriP DS and FR elements and the BZLF1 promoter region. The amplification signals were normalized to those from the same cell lysates prior to IP using the same primer pairs. Signals from Brd4 and EBNA1 antibody samples are expressed relative to the control IgG sample, which was set to 1. The results shown are from three independent experiments with PCR quantification performed in triplicate for each experiment. For Brd4 samples, only the FR fragment result is statistically significantly different from the IgG control, with a P value of 0.000007.
Effect of Brd4 silencing on EBNA1 attachment to human mitotic chromosomes.
Since Brd4 can localize with EBNA1 at the FR element in human cells and can support EBNA1-mediated segregation in yeast, we also tested the importance of Brd4 for EBNA1 attachment to human mitotic chromosomes. HeLa cells are known to support EBNA1-mediated segregation, and hence, HeLa cells expressing EBNA1 were used for these studies. We first examined the localization of Brd4 and EBNA1 in mitosis, by fractionating lysates from nocodazole-blocked cells into chromosomal pellet and soluble fractions as previously described (26) and analyzing equal cell equivalents of the fractions by Western blotting (Fig. 7A). Brd4 is known to exist as long and short isoforms generated by alternative splicing (59), and these fractionated somewhat differently, with the larger species distributed in the soluble and pellet fractions and the smaller species distributed predominantly in the chromosomal pellet along with EBNA1. HeLa cells expressing EBNA1 were then transfected with esiRNA against Brd4 or siRNA against GFP, and after the downregulation of Brd4 was confirmed (Fig. 7B), cells were blocked in mitosis and then fractionated as described above. Equal cell equivalents of the soluble and pellet fractions were then analyzed for EBNA1 by Western blotting (Fig. 7C). Although this method previously showed that silencing of hEBP2 resulted in the release of EBNA1 from the pellet into the soluble fraction (26), we did not see any effect of Brd4 silencing on the fractionation of EBNA1, suggesting that Brd4 is unlikely to function by regulating the global attachment of EBNA1 to chromosomes.
FIG. 7.
Brd4 silencing does not noticeably affect EBNA1 attachment to mitotic chromosomes. HeLa cells expressing EBNA1 were transfected with siRNA against GFP (negative control) or esiRNA against Brd4. (A) Negative-control cells were blocked in mitosis, and whole-cell lysates (W) were generated and separated into soluble (S) and chromosomal pellet (P) fractions by centrifugation. Equal cell equivalents of each fraction were then analyzed by Western blotting using Brd4 or EBNA1 antibody. (B) Downregulation of Brd4 by esiRNA was confirmed by Western blotting of whole-cell lysates, compared to siRNA treatment against GFP. (C) Cells from panel B were blocked in mitosis, and whole-cell lysates (W) were separated into soluble (S) and chromosomal pellet (P) fractions. Equal cell equivalents of each fraction were then analyzed by Western blotting using EBNA1 antibody. The same blot was also probed for NAP1, a protein known to be in the soluble fraction in mitosis, as a control for proper fractionation of soluble proteins.
DISCUSSION
The reconstitution of virus-based segregation in yeast has proven to be a useful way of identifying mammalian proteins that function in this process. This approach was initially described for EBV, where segregation mediated by the EBNA1 protein was shown to require hEBP2 (27). EBP2 enabled the attachment of EBNA1 to yeast mitotic chromosomes (25), and this role was verified in human cells, where silencing of EBP2 led to loss of EBNA1 from mitotic chromosomes (26). A similar yeast system was used to study segregation by the BPV E2 protein and showed that mouse Brd4 enabled E2 to maintain plasmids containing multiple E2 binding sites in yeast (9). E2 was also shown to interact with Brd4 in mammalian cells, and interference with this interaction led to loss of E2 from mitotic chromosomes (1, 7, 34, 62, 63).
We have used the same yeast system that identified EBP2 as a segregation factor for EBV to test the ability of other mammalian proteins to facilitate EBNA1-mediated segregation. In particular, we were interested in determining whether proteins implicated in the segregation of BPV, KSHV, and HVS contributed to EBV segregation. Brd4, Brd2, MeCP2, and DEK have all been suggested to contribute to KSHV segregation (5, 29, 36, 54, 64), and Brd4 and MeCP2 have also been implicated in the segregation of papillomavirus (1, 7, 34, 62, 63) and HVS (17), respectively. Therefore, we tested the ability of these proteins to enable the persistence of plasmids containing the EBV segregation element, FR, in yeast with and without EBNA1. While DEK did not contribute to plasmid maintenance, we found that Brd2 and MeCP2 both greatly increased plasmid maintenance. However, these effects did not require EBNA1 or the FR element, and even the maintenance of the CEN/ARS plasmid was improved. This indicates that Brd2 and MeCP2 can have a general effect on plasmid stability in yeast but do not mediate the segregation function of EBNA1. Since both Brd2 and MeCP2 interact with chromatin, they may be functioning to tether plasmid DNA directly to host chromatin, thereby facilitating plasmid maintenance nonspecifically. Alternatively, the expression of these proteins in yeast might have increased the replication efficiency of the test plasmids or altered the cellular environment to favor persistence. However, we did not observe any difference in the doubling rate of yeast cells expressing Brd2 or MeCP2, suggesting that their expression did not cause major changes to the cell cycle. Brd2 was previously tested for effects on E2-mediated plasmid segregation in yeast, and no effect was observed (9). Discrepancies in the Brd2 effects on plasmid maintenance could be due to differences in the expression levels of Brd2 or technical differences in the assay used (Brannon et al. used a sectoring assay). Our results with Brd2 and MeCP2 raise the possibility that these proteins might contribute to the persistence of multiple viral plasmids in mammalian cells in a manner independent of an interaction with the viral segregation protein.
Using the yeast plasmid loss assay, we also found that Brd4 greatly increased the maintenance of the FR-containing plasmid only in the presence of EBNA1 and did not increase the maintenance of the same plasmid lacking the FR or the CEN/ARS plasmid. Therefore, Brd4 enabled EBNA1 to segregate FR plasmids in yeast. The efficiency of FR plasmid maintenance with EBNA1 and Brd4 was similar to that observed with EBNA1 and human EBP2 and for the CEN/ARS plasmid in the same yeast strain. These results are also similar to the findings of Brannon et al. (9), who found that Brd4 enabled E2-mediated segregation in yeast. We have previously shown that EBP2 enables EBNA1 to segregate FR-containing plasmids in yeast by facilitating the attachment of EBNA1 to mitotic chromosomes (25). Like EBP2, Brd4 is known to be associated with mitotic chromosomes (15), as is the yeast homologue of Brd4 (Bdf1) (13), and therefore, it seems likely that the effect of Brd4 on EBNA1-mediated segregation in yeast also involves tethering of EBNA1 to the yeast chromosomes. The fact that EBNA1 is not able to segregate FR-containing plasmids in yeast without mammalian Brd4 (or EBP2) indicates that the yeast Bdf1 was not able to mediate this process. Bdf1 lacks the C-terminal domain of Brd4 that is responsible for E2 binding and required for E2-mediated segregation in yeast (1, 9, 62). Therefore, its inability to function with EBNA1 suggests that the EBNA1 interaction, like the E2 interaction, involves the Brd4 C-terminal domain.
We used EBNA1 mutants in the plasmid loss assay to assess which EBNA1 sequences mediated the interaction with Brd4. EBNA1 residues 325 to 376 are the only EBNA1 region found to be required for segregation in human cells, while the 61-83 sequence is the only EBNA1 region that abrogates transcriptional activation without affecting other EBNA1 functions (45, 58). Therefore, we were particularly interested in assessing the roles of these sequences in the Brd4-mediated segregation assay. We found that the 61-83 deletion abrogated Brd4-mediated segregation while the 325-376 deletion had only a small effect, indicating that the interaction with Brd4 occurred predominantly through transcriptional activation sequences. This result was also supported by coimmunoprecipitation experiments in human cells, where EBNA1Δ325-376 bound EBNA1 to the same degree as did wild-type EBNA1, but no binding was detected with EBNA1Δ61-83. One major difference in these two assays was seen with EBNA1Δ8-67, which is partially defective in both transcriptional activation and segregation in human cells (58). This mutant did not support Brd4-mediated segregation in yeast but was found to bind Brd4 better than wild-type EBNA1 in human cells. Since the 8-67 deletion did not increase the expression level of EBNA1, the increased binding to Brd4 indicates that removal of residues 8 to 67 allows greater access or more stable binding of Brd4 to EBNA1 sequence 61-83. However, this did not translate into more efficient segregation in the yeast system, and we suggest two possible reasons for this discrepancy. First, Brd4 binding may not be the only requirement for segregation in yeast and sequence 8-67 might provide an important role in segregation that is independent of Brd4 binding. Second, constitutive or unusually strong binding of EBNA1 to Brd4 may be detrimental to plasmid maintenance, especially if the Brd4-EBNA1 interaction usually occurs transiently. For example, since Brd4 is known to associate with chromatin throughout the cell cycle, it could be that tethering of EBNA1 to chromatin in interphase through Brd4 interferes with the replication of the plasmids.
The finding that EBNA1 interacted with Brd4 through sequences important for transcriptional activation but not for segregation suggested that the significance of the EBNA1-Brd4 interaction may be in transcription. This was further supported by the finding that downregulation of Brd4 in human cells significantly decreased the ability of EBNA1 to activate transcription from the FR enhancer element, without affecting EBNA1-independent transcription. The degree of this effect (approximately twofold) is in keeping with effects reported for silencing of other histone-associated, transcription coactivator proteins that are recruited to transcriptional elements through interactions with other proteins (21, 35, 51). We also observed that Brd4 overexpression specifically inhibited EBNA1-mediated transcription, further supporting a role for Brd4 in this process. The fact that overexpressing and silencing Brd4 both inhibited EBNA1-mediated transcription suggests that Brd4 mediates interactions between EBNA1 and other proteins involved in transcription and that alterations of the ratio of EBNA1 to Brd4 disrupt this complex. In keeping with this model, Brd4 is known to be associated with the transcriptional complexes Mediator and P-TEFb (59), and overexpression of Brd4 has been shown to inhibit transcription by the human immunodeficiency virus Tat protein by disrupting the interaction of Tat with P-TEFb (8).
A direct role for Brd4 in EBNA1-mediated transcription was further suggested by the finding that Brd4 was preferentially localized to the FR element over the DS element or BZLF1 gene. Since the FR governs both transcriptional activation and segregation, the interaction of Brd4 with this element could be relevant for either process. The lack of association of Brd4 with the DS element suggests that Brd4 is not directly involved in DNA replication from oriP, which occurs from this element. Both the FR and DS are constitutively occupied by EBNA1 (19), suggesting that recruitment of Brd4 does not occur through every type of EBNA1 interaction but rather requires a particular number or arrangement of EBNA1 dimers or flanking DNA sequences.
Our finding that Brd4 contributes to transcriptional activation by EBNA1 is reminiscent of recent results for the E2-Brd4 interaction. Although initially identified as an interaction involved in segregation, the E2-Brd4 interaction now appears to be more important for the transcriptional function of E2. For example, silencing of Brd4 was found to affect transcriptional activation by BPV E2 but not plasmid replication or mitotic chromosome attachment (37, 42). Segregation and transcriptional activation by E2 have been difficult to separate because the same domain of E2 mediates both processes; however, point mutants have been identified that disrupt Brd4 binding without disrupting mitotic chromosome binding and, conversely, that bind Brd4 but fail to attach to mitotic chromosomes or maintain BPV plasmids (33, 37). In addition, the E2s from all HPV strains appear to require the Brd4 interaction for transcriptional activation, while E2s from some strains do not colocalize with Brd4 on mitotic chromosomes (33). In EBNA1 two different transcriptional domains, 325-376 and 61-83, have been identified, and both are required for transcriptional activation, suggesting that they fulfill distinct roles in this process (12, 28, 58). A few host proteins have been shown to interact with the 325-376 region that might mediate its transcriptional effects (18, 50, 55), but to this point, protein interactions with the 61-83 region that are likely to be important for transcription have not been identified. We suggest that an interaction of the 61-83 sequence with Brd4, either directly or indirectly, is at least partially responsible for its contribution to transcriptional activation.
Although Brd4 was able to function with EBNA1 in segregation in the yeast system, this does not necessarily mean that Brd4 contributes to EBNA1-mediated segregation in human cells. In fact the EBNA1 residues involved in the Brd4 interaction do not correspond to those important for attachment to human mitotic chromosomes. In addition, we did not observe any decrease in EBNA1 attachment to chromosomes upon Brd4 silencing, nor was there obvious colocalization of EBNA1 and Brd4 on mitotic chromosome spreads (data not shown). Therefore, taken as a whole, the data suggest that the principal function of the EBNA1-Brd4 interaction is in transcription and that the ability of Brd4 to rescue EBNA1-mediated segregation in yeast reflects an artificial means of tethering EBNA1 to yeast chromosomes. Indeed it has been previously shown that artificial tethering of EBNA1 to human mitotic chromosomes is sufficient to confer segregation (20). However, we would like to stress that, although the functional significance of the EBNA1-Brd4 interaction in human cells may not be in segregation, the yeast-based segregation system successfully identified an interaction between EBNA1 and Brd4 that was indeed functional in human cells, as well as identifying the EBNA1 region that mediated the interaction. This shows the utility of the yeast-based system for identifying functional interactions with chromatin-associated proteins, even if the significance of the interactions in human cells is not in segregation. However, like any reconstituted system, it is important to verify/determine the significance of the interactions observed in the system in human cells.
Although we did not see any effect of downregulation of Brd4 on EBNA1 binding to human mitotic chromosomes, we cannot conclusively rule out the possibility that this interaction contributes to segregation. For example, since Brd4 was not completely silenced, it is possible that the residual Brd4 was sufficient to function with EBNA1 in segregation or that the contribution of Brd4 to segregation is functionally redundant with other protein interactions. Finally, it is possible that EBNA1 may use different mechanisms to attach to mitotic chromosomes in different human cell types. EBP2 has been shown to be important for EBNA1 attachment to mitotic chromosomes in several epithelial cell lines, including HeLa cells, but perhaps Brd4 mediates mitotic chromosome interactions in other cell types (e.g., B lymphocytes).
In summary, we have identified an interaction between EBNA1 and Brd4 that can function in plasmid segregation in a heterologous system and appears to be important for transcriptional activation by EBNA1 in human cells. Brd4 has now been shown to functionally interact with EBNA1, LANA, and E2 proteins from several papillomavirus strains. The fact that these proteins lack obvious sequence homology and yet all have mechanisms for interacting with Brd4 further stresses the functional importance of these interactions in one or more of the conserved functions of these proteins. Therefore, Brd4 is emerging as a general mediator of viral persistence.
Acknowledgments
We gratefully acknowledge Laurence Pelletier, Andrea Tagliaferro, and Steffen Lawo for assistance with generation of the Brd4 esiRNA; Alison McBride for pADNS-Brd4; and Alan Cochrane for the SEAP reporter plasmid. We also thank Brigitte Lavoie for YPH499 and many insightful comments throughout the course of the yeast assays.
This work was funded in part by grant number 12477 from the Canadian Institutes of Health Research and by a grant from the National Cancer Institute of Canada, which receives funds from the Canadian Cancer Society. L.F. is a tier 1 Canada Research Chair in Molecular Virology.
Footnotes
Published ahead of print on 15 October 2008.
REFERENCES
- 1.Abbate, E. A., C. Voitenleitner, and M. R. Botchan. 2006. Structure of the papillomavirus DNA-tethering complex E2:Brd4 and a peptide that ablates HPV chromosomal association. Mol. Cell 24877-889. [DOI] [PubMed] [Google Scholar]
- 2.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284641-644. [DOI] [PubMed] [Google Scholar]
- 3.Ballestas, M. E., and K. M. Kaye. 2001. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J. Virol. 753250-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbera, A. J., M. E. Ballestas, and K. M. Kaye. 2004. The Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 N terminus is essential for chromosome association, DNA replication, and episome persistence. J. Virol. 78294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbera, A. J., J. V. Chodaparambil, B. Kelley-Clarke, V. Joukov, J. C. Walter, K. Luger, and K. M. Kaye. 2006. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science 311856-861. [DOI] [PubMed] [Google Scholar]
- 6.Bastien, N., and A. A. McBride. 2000. Interaction of the papillomavirus E2 protein with mitotic chromosomes. Virology 270124-134. [DOI] [PubMed] [Google Scholar]
- 7.Baxter, M. K., M. G. McPhillips, K. Ozato, and A. A. McBride. 2005. The mitotic chromosome binding activity of the papillomavirus E2 protein correlates with interaction with the cellular chromosomal protein, Brd4. J. Virol. 794806-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bisgrove, D. A., T. Mahmoudi, P. Henklein, and E. Verdin. 2007. Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc. Natl. Acad. Sci. USA 10413690-13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brannon, A. R., J. A. Maresca, J. D. Boeke, M. A. Basrai, and A. A. McBride. 2005. Reconstitution of papillomavirus E2-mediated plasmid maintenance in Saccharomyces cerevisiae by the Brd4 bromodomain protein. Proc. Natl. Acad. Sci. USA 1022998-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calderwood, M., R. E. White, R. A. Griffiths, and A. Whitehouse. 2005. Open reading frame 73 is required for herpesvirus saimiri A11-S4 episomal persistence. J. Gen. Virol. 862703-2708. [DOI] [PubMed] [Google Scholar]
- 11.Calderwood, M. A., K. T. Hall, D. A. Matthews, and A. Whitehouse. 2004. The herpesvirus saimiri ORF73 gene product interacts with host-cell mitotic chromosomes and self-associates via its C terminus. J. Gen. Virol. 85147-153. [DOI] [PubMed] [Google Scholar]
- 12.Ceccarelli, D. F. J., and L. Frappier. 2000. Functional analyses of the EBNA1 origin DNA binding protein of Epstein-Barr virus. J. Virol. 744939-4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chua, P., and G. S. Roeder. 1995. Bdf1, a yeast chromosomal protein required for sporulation. Mol. Cell. Biol. 153685-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng, Z., C. Atanasiu, K. Zhao, R. Marmorstein, J. I. Sbodio, N. W. Chi, and P. M. Lieberman. 2005. Inhibition of Epstein-Barr virus OriP function by tankyrase, a telomere-associated poly-ADP ribose polymerase that binds and modifies EBNA1. J. Virol. 794640-4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dey, A., J. Ellenberg, A. Farina, A. E. Coleman, T. Maruyama, S. Sciortino, J. Lippincott-Schwartz, and K. Ozato. 2000. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G2-to-M transition. Mol. Cell. Biol. 206537-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frappier, L. EBNA1 in viral DNA replication and persistence. In E. S. Robertson (ed.), Epstein-Barr virus latency and transformation, in press. Horizon Scientific Press, Norwich, United Kingdom.
- 17.Griffiths, R., and A. Whitehouse. 2007. Herpesvirus saimiri episomal persistence is maintained via interaction between open reading frame 73 and the cellular chromosome-associated protein MeCP2. J. Virol. 814021-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holowaty, M. N., M. Zeghouf, H. Wu, J. Tellam, V. Athanasopoulos, J. Greenblatt, and L. Frappier. 2003. Protein profiling with Epstein-Barr nuclear antigen 1 reveals an interaction with the herpesvirus-associated ubiquitin-specific protease HAUSP/USP7. J. Biol. Chem. 27829987-29994. [DOI] [PubMed] [Google Scholar]
- 19.Hsieh, D.-J., S. M. Camiolo, and J. L. Yates. 1993. Constitutive binding of EBNA1 protein to the Epstein-Barr virus replication origin, oriP, with distortion of DNA structure during latent infection. EMBO J. 124933-4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hung, S. C., M.-S. Kang, and E. Kieff. 2001. Maintenance of Epstein-Barr virus (EBV) oriP-based episomes requires EBV-encoded nuclear antigen-1 chromosome-binding domains, which can be replaced by high-mobility group-I or histone H1. Proc. Natl. Acad. Sci. USA 981865-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ichijo, T., G. P. Chrousos, and T. Kino. 2008. Activated glucocorticoid receptor interacts with the INHAT component Set/TAF-Ibeta and releases it from a glucocorticoid-responsive gene promoter, relieving repression: implications for the pathogenesis of glucocorticoid resistance in acute undifferentiated leukemia with Set-Can translocation. Mol. Cell. Endocrinol. 28319-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ilves, I., S. Kivi, and M. Ustav. 1999. Long-term episomal maintenance of bovine papillomavirus type 1 plasmids is determined by attachment to host chromosomes, which is mediated by the viral E2 protein and its binding sites. J. Virol. 734404-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ilves, I., K. Maemets, T. Silla, K. Janikson, and M. Ustav. 2006. Brd4 is involved in multiple processes of the bovine papillomavirus type 1 life cycle. J. Virol. 803660-3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanda, T., M. Otter, and G. M. Wahl. 2001. Coupling of mitotic chromosome tethering and replication competence in Epstein-Barr virus-based plasmids. Mol. Cell. Biol. 213576-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kapoor, P., and L. Frappier. 2003. EBNA1 partitions Epstein-Barr virus plasmids in yeast by attaching to human EBNA1-binding protein 2 on mitotic chromosomes. J. Virol. 776946-6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kapoor, P., B. D. Lavoie, and L. Frappier. 2005. EBP2 plays a key role in Epstein-Barr virus mitotic segregation and is regulated by aurora family kinases. Mol. Cell. Biol. 254934-4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kapoor, P., K. Shire, and L. Frappier. 2001. Reconstitution of Epstein-Barr virus-based plasmid partitioning in budding yeast. EMBO J. 20222-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kennedy, G., and B. Sugden. 2003. EBNA-1, a bifunctional transcriptional activator. Mol. Cell. Biol. 236901-6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krithivas, A., M. Fujimuro, M. Weidner, D. B. Young, and S. D. Hayward. 2002. Protein interactions targeting the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus to cell chromosomes. J. Virol. 7611596-11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lehman, C. W., and M. R. Botchan. 1998. Segregation of viral plasmids depends on tethering to chromosomes and is regulated by phosphorylation. Proc. Natl. Acad. Sci. USA 954338-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marcus, S. L., K. S. Miyata, R. A. Rachubinski, and J. P. Capone. 1995. Transactivation by PPAR/RXR heterodimers in yeast is potentiated by exogenous fatty acid via a pathway requiring intact peroxisomes. Gene Expr. 4227-239. [PMC free article] [PubMed] [Google Scholar]
- 32.Marechal, V., A. Dehee, R. Chikhi-Brachet, T. Piolot, M. Coppey-Moisan, and J. Nicolas. 1999. Mapping EBNA1 domains involved in binding to metaphase chromosomes. J. Virol. 734385-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McPhillips, M. G., J. G. Oliveira, J. E. Spindler, R. Mitra, and A. A. McBride. 2006. Brd4 is required for E2-mediated transcriptional activation but not genome partitioning of all papillomaviruses. J. Virol. 809530-9543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McPhillips, M. G., K. Ozato, and A. A. McBride. 2005. Interaction of bovine papillomavirus E2 protein with Brd4 stabilizes its association with chromatin. J. Virol. 798920-8932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murano, K., M. Okuwaki, M. Hisaoka, and K. Nagata. 2008. Transcription regulation of the rRNA gene by a multifunctional nucleolar protein, B23/nucleophosmin, through its histone chaperone activity. Mol. Cell. Biol. 283114-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ottinger, M., T. Christalla, K. Nathan, M. M. Brinkmann, A. Viejo-Borbolla, and T. F. Schulz. 2006. Kaposi's sarcoma-associated herpesvirus LANA-1 interacts with the short variant of BRD4 and releases cells from a BRD4- and BRD2/RING3-induced G1 cell cycle arrest. J. Virol. 8010772-10786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parish, J. L., A. M. Bean, R. B. Park, and E. J. Androphy. 2006. ChlR1 is required for loading papillomavirus E2 onto mitotic chromosomes and viral genome maintenance. Mol. Cell 24867-876. [DOI] [PubMed] [Google Scholar]
- 38.Piirsoo, M., E. Ustav, T. Mandel, A. Stenlund, and M. Ustav. 1996. Cis and trans requirements for stable episomal maintenance of the BPV-1 replicator. EMBO J. 151-11. [PMC free article] [PubMed] [Google Scholar]
- 39.Piolot, T., M. Tramier, M. Coppey, J. C. Nicolas, and V. Marechal. 2001. Close but distinct regions of human herpesvirus 8 latency-associated nuclear antigen 1 are responsible for nuclear targeting and binding to human mitotic chromosomes. J. Virol. 753948-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ritzi, M., K. Tillack, J. Gerhardt, E. Ott, S. Humme, E. Kremmer, W. Hammerschmidt, and A. Schepers. 2003. Complex protein-DNA dynamics at the latent origin of DNA replication of Epstein-Barr virus. J. Cell Sci. 1163971-3984. [DOI] [PubMed] [Google Scholar]
- 41.Schepers, A., M. Ritzi, K. Bousset, E. Kremmer, J. L. Yates, J. Harwood, J. F. X. Diffley, and W. Hammerschmidt. 2001. Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein-Barr virus. EMBO J. 204588-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schweiger, M. R., J. You, and P. M. Howley. 2006. Bromodomain protein 4 mediates the papillomavirus E2 transcriptional activation function. J. Virol. 804276-4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sears, J., M. Ujihara, S. Wong, C. Ott, J. Middeldorp, and A. Aiyar. 2004. The amino terminus of Epstein-Barr virus (EBV) nuclear antigen 1 contains AT hooks that facilitate the replication and partitioning of latent EBV genomes by tethering them to cellular chromosomes. J. Virol. 7811487-11505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Senechal, H., G. G. Poirier, B. Coulombe, L. A. Laimins, and J. Archambault. 2007. Amino acid substitutions that specifically impair the transcriptional activity of papillomavirus E2 affect binding to the long isoform of Brd4. Virology 35810-17. [DOI] [PubMed] [Google Scholar]
- 45.Shire, K., D. F. J. Ceccarelli, T. M. Avolio-Hunter, and L. Frappier. 1999. EBP2, a human protein that interacts with sequences of the Epstein-Barr nuclear antigen 1 important for plasmid maintenance. J. Virol. 732587-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 12219-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skiadopoulos, M. H., and A. A. McBride. 1998. Bovine papillomavirus type 1 genomes and the E2 transactivator protein are closely associated with mitotic chromatin. J. Virol. 722079-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stinchcomb, D. T., K. Struhl, and R. W. Davis. 1979. Isolation and characterization of a yeast chromosomal replicator. Nature 28239-43. [DOI] [PubMed] [Google Scholar]
- 49.Sun, Y., G. Hegamyer, Y. J. Cheng, A. Hildesheim, J. Y. Chen, I. H. Chen, Y. Cao, K. T. Yao, and N. H. Colburn. 1992. An infrequent point mutation of the p53 gene in human nasopharyngeal carcinoma. Proc. Natl. Acad. Sci. USA 896516-6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Scoy, S., I. Watakabe, A. R. Krainer, and J. Hearing. 2000. Human p32: a coactivator for Epstein-Barr virus nuclear antigen-1-mediated transcriptional activation and possible role in viral latent cycle DNA replication. Virology 275145-157. [DOI] [PubMed] [Google Scholar]
- 51.Vardabasso, C., L. Manganaro, M. Lusic, A. Marcello, and M. Giacca. 2008. The histone chaperone protein nucleosome assembly protein-1 (hNAP-1) binds HIV-1 Tat and promotes viral transcription. Retrovirology 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Verma, S. C., and E. S. Robertson. 2003. ORF73 of herpesvirus saimiri strain C488 tethers the viral genome to metaphase chromosomes and binds to cis-acting DNA sequences in the terminal repeats. J. Virol. 7712494-12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Viejo-Borbolla, A., E. Kati, J. A. Sheldon, K. Nathan, K. Mattsson, L. Szekely, and T. F. Schulz. 2003. A domain in the C-terminal region of latency-associated nuclear antigen 1 of Kaposi's sarcoma-associated herpesvirus affects transcriptional activation and binding to nuclear heterochromatin. J. Virol. 777093-7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Viejo-Borbolla, A., M. Ottinger, E. Bruning, A. Burger, R. Konig, E. Kati, J. A. Sheldon, and T. F. Schulz. 2005. Brd2/RING3 interacts with a chromatin-binding domain in the Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 (LANA-1) that is required for multiple functions of LANA-1. J. Virol. 7913618-13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, Y., J. E. Finan, J. M. Middeldorp, and S. D. Hayward. 1997. P32/TAP, a cellular protein that interacts with EBNA-1 of Epstein-Barr virus. Virology 23618-29. [DOI] [PubMed] [Google Scholar]
- 56.Woolaway, K., K. Asai, A. Emili, and A. Cochrane. 2007. hnRNP E1 and E2 have distinct roles in modulating HIV-1 gene expression. Retrovirology 428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu, H., D. F. J. Ceccarelli, and L. Frappier. 2000. The DNA segregation mechanism of the Epstein-Barr virus EBNA1 protein. EMBO Rep. 1140-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu, H., P. Kapoor, and L. Frappier. 2002. Separation of the DNA replication, segregation and transcriptional activation functions of Epstein-Barr nuclear antigen 1. J. Virol. 762480-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu, S. Y., and C. M. Chiang. 2007. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J. Biol. Chem. 28213141-13145. [DOI] [PubMed] [Google Scholar]
- 60.Wu, S. Y., A. Y. Lee, S. Y. Hou, J. K. Kemper, H. Erdjument-Bromage, P. Tempst, and C. M. Chiang. 2006. Brd4 links chromatin targeting to HPV transcriptional silencing. Genes Dev. 202383-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang, D., F. Buchholz, Z. Huang, A. Goga, C. Y. Chen, F. M. Brodsky, and J. M. Bishop. 2002. Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proc. Natl. Acad. Sci. USA 999942-9947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.You, J., J. L. Croyle, A. Nishimura, K. Ozato, and P. M. Howley. 2004. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell 117349-360. [DOI] [PubMed] [Google Scholar]
- 63.You, J., M. R. Schweiger, and P. M. Howley. 2005. Inhibition of E2 binding to Brd4 enhances viral genome loss and phenotypic reversion of bovine papillomavirus-transformed cells. J. Virol. 7914956-14961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.You, J., V. Srinivasan, G. V. Denis, W. J. Harrington, Jr., M. E. Ballestas, K. M. Kaye, and P. M. Howley. 2006. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen interacts with bromodomain protein Brd4 on host mitotic chromosomes. J. Virol. 808909-8919. [DOI] [PMC free article] [PubMed] [Google Scholar]