Abstract
The fecundity of RNA viruses can be very high. Thus, it is often assumed that viruses have large populations, and RNA virus evolution has been mostly explained using purely deterministic models. However, population bottlenecks during the virus life cycle could result in effective population numbers being much smaller than reported censuses, and random genetic drift could be important in virus evolution. A step at which population bottlenecks may be severe is host-to-host transmission. We report here an estimate of the size of the population that starts a new infection when Cucumber mosaic virus (CMV) is transmitted by the aphid Aphis gossypii, based on the segregation of two CMV genotypes in plants infected by aphids that acquired the virus from plants infected by both genotypes. Results show very small effective numbers of founders, between one and two, both in experiments in which the three-partite genome of CMV was aphid transmitted and in experiments in which a fourth RNA, CMV satellite RNA, was also transmitted. These numbers are very similar to those published for Potato virus Y, which has a monopartite genome and is transmitted by aphids according to a different mechanism than CMV. Thus, the number of genomic segments seems not to be a major determinant of the effective number of founders. Also, our results suggest that the occurrence of severe bottlenecks during horizontal transmission is general for viruses nonpersistently transmitted by aphids, indicating that random genetic drift should be considered when modeling virus evolution.
Molecular analyses of viral genomes showed, in the late 1970s, that RNA virus populations were intrinsically heterogeneous, and this led to a renewed interest in virus evolution. Since then, the evolution of RNA viruses has been mostly explained by purely deterministic models. Because the number of virus particles in the infected host may be very high (e.g., up to 1011 to 1012 Tobacco mosaic virus particles in an infected tobacco leaf [24, 34] or 107 to 108 cells infected by Human immunodeficiency virus type 1 [29, 49]), it was assumed that viruses have big populations and that selection is the major driver of virus evolution (12, 13). However, the relevant evolutionary parameter is not the total number of individuals in the population, i.e., the census size of the population, but the effective size of the population, which can be grossly assimilated to the number of individuals that pass their genes to the next generation. At small effective population numbers (Ne), random processes (i.e., genetic drift) will predominate over deterministic ones (i.e., selection) (11). Expansions and contractions of population size during the virus life cycle, i.e., the occurrence of population bottlenecks, would reduce Ne even if the population census recovered to the size before the bottleneck. Population bottlenecks could occur when initiating a new deme, for instance, in different organs within a host, in different host individuals, or in different host populations. These bottlenecks result in a type of genetic drift named the founder effect, as the new deme is initiated from a small number of genotypes randomly sampled from the mother deme, which leads to genetic structuring of the population with low within-deme diversities and high between-deme diversities. Thus, to understand virus evolution it is important to identify at what steps of the virus life cycle its population passes through a bottleneck and to estimate the effective size of the population during that bottleneck.
Population bottlenecks have been described during the colonization of different organs within an infected host for both animal and plant viruses, and their sizes have been estimated (20, 21, 22, 28, 29, 41, 50), indicating that the within-host Ne might be much smaller than the census, and genetic drift could be important in within-host virus evolution. Perhaps the occurrence of population bottlenecks during horizontal transmission between hosts would be more relevant for virus evolution (51). Founder effects might explain the random changes in population structure described after mechanical or aphid transmission of different plant viruses to new hosts (1, 2, 5, 27). Indeed, experiments designed to unveil the existence of a bottleneck during aphid transmission of Cucumber mosaic virus (CMV) to new host plants have shown founder effects indicative of severe bottlenecks (3).
Recently, the number of particles of the RNA virus Potato virus Y (PVY) transmitted by aphids was estimated and was found to be very small, 0.5 to 3.2 (35). It is important to know how general these figures can be. A possible cause of variation could be associated with the experimental approach; in the experiments reported by Moury et al. (35), aphids acquired PVY from a solution rather than from leaves of infected plants. Also, the genomic structure of the virus, mono- or multipartite, could influence the size of the transmission-associated bottleneck: it is known that monopartite viruses, such as PVY, are more infectious and are more efficiently transmitted by their vectors than multipartite viruses, such as CMV (25, 44, 52, 54), which require that a set of particles encapsidating the complete genome enters a single host cell for infection to occur. Hence, it could be that a multipartite structure would also affect the size of the transmission bottleneck.
We provide here an estimate of the size of the population bottleneck of CMV during transmission by Aphis gossypii (Glover) in tomato plants. The number of founders starting an infection in a new plant was estimated according to a model based on the proportion of plants infected with only one CMV genotype when aphids acquired the virus from plants infected by a combination of two genotypes. This model was based on one initially developed to estimate the size of the population bottlenecks during systemic colonization of plants by viruses (50). The results show that, as for PVY (35), the effective number of founders is very small, and they suggest that it is not affected by the mono- or multipartite structure, or by the number of genomic segments, of the viral genome.
MATERIALS AND METHODS
Virus isolates and aphids.
Three CMV genotypes were used for all experiments and were derived from T7 RNA polymerase (New England Biolabs, Ipswich, MA) transcripts of biologically active cDNA clones (47) of Fny-CMV, belonging to subgroup IA of CMV isolates, and LS-CMV, belonging to subgroup II (48). The three genotypes were Fny-CMV (hereafter named F1F2F3) and two reassortant genotypes in which either the RNA1 or the RNA2 of LS-CMV was substituted for that of Fny-CMV (genotypes L1F2F3 and F1L2F3, respectively). RNA transcripts were used to inoculate tobacco (Nicotiana tabacum L. cv. Xanthi) plants for virus multiplication. CMV virions were purified from infected tobacco leaves as described in reference 33, and viral RNA was extracted by virion disruption with phenol and sodium dodecyl sulfate.
CMV satellite RNA (CMV-satRNA) was derived from a biologically active cDNA clone of CMV-satRNA 89/20.1 (4), which has a nonnecrogenic phenotype in tomato when supported by either Fny-CMV or LS-CMV. Transcripts were inoculated at 30 μg/ml with virion RNA of satellite-free CMV at 100 μg/ml in 0.1 M Na2HPO4.
A clonal culture of A. gossypii was maintained and multiplied on melon plants (Cucumis melo L. cv. Piel de sapo) in a growth chamber at 23°C (day) and at 18°C (night) with a 16:8 h (light:dark) photoperiod. The aphid colony was started from a single virginiparous aptera collected in Aguadulce, Almeria, Spain, in 1998 from a melon plant.
Transmission test.
Transmission tests were performed essentially as described previously (18). Eight-day-old tomato plants (Solanum lycopersicum L. cv. Rutgers) were mechanically inoculated with the various CMV genotypes, singly or in mixtures, by applying 10 μl of viral RNA at 100 μg/ml in 0.1 M Na2HPO4 into the fully expanded cotyledons. Fifteen days postinoculation, systemically infected leaves from these plants were used as source leaves for virus acquisition by aphids. Young (7- to 8-day-old) apterous aphid females were collected from melon plants, starved for 1 to 1.5 h, and then gently placed by using a paintbrush onto a single source leaf. Aphids were monitored for 1 to 10 min under a magnifying lens until they started to probe on the source leaf (this was assessed by looking at the position of the antennae). Then, aphids were individually transferred to fully expanded cotyledons of 8-day-old tomato plants, on which they were held for 12 h and then sprayed with imidacloprid (Confidor 20 SL). Fifty or 60 test plants were aphid inoculated for each source plant. Virus infection of test plants was analyzed in inoculated leaves 5 days postinoculation. Four or eight replicate experiments were done for each singly or doubly infected plant, respectively.
Analyses of CMV RNA in infected plants.
CMV accumulation in source leaves was quantified as viral RNA accumulation by dot blot hybridization as described previously (14). Total nucleic acid extracts from leaves were spotted onto nylon membranes and analyzed by dot blot hybridization with a 32P-labeled RNA probe complementary to nucleotides (nt) 1933 to 2215 of Fny-CMV RNA3 (GenBank accession no. D10538). This probe hybridizes with the 3′ untranslated region of the three genomic, and subgenomic, RNAs of Fny-CMV (43). To estimate the relative accumulation of Fny-CMV and LS-CMV RNAs 1 or 2 in mixed infected source leaves, total nucleic acid extracts were hybridized with specific 5′-32P-labeled oligonucleotide probes. The genotype-specific oligonucleotide probes used for these analyses were as follows: 5′AGACCGCTAAGCACGAGCAACACATTCGGCGATTAAATCGCCG3′, complementary to nucleotides 3040 to 3082 of Fny-CMV RNA1; 5′CCTTCCGCGTTCAGACTAACGGAATACAAGTAG3′, complementary to nucleotides 3052 to 3085 of LS-CMV RNA1; 5′TCCGCCACGTTCACATGGCGGCATGACCCTGTCAG3′, complementary to nucleotides 2610 to 2645 of Fny-CMV RNA2; and 5′GGACGGAGAGCGAACGACATCAGGAAACCAATCCACGGG3′, complementary to nucleotides 2595 to 2635 of LS-CMV RNA2 (nucleotide numbering as in GenBank accession no. NC002034 and NC002035 for RNA1 or RNA2, respectively, of Fny-CMV, and accession numbers AF416899 and AF416900 for RNA1 and RNA2, respectively, of LS-CMV). In each blot assay, internal standards for each CMV genotype were included as a twofold dilution series of purified RNA (2 to 0.015 μg) in nucleic acid extracts from noninoculated tomato plants. Different amounts of nucleic acid extracts from each sample to be analyzed were blotted to ensure that the hybridization signal was in the linear portion of the RNA concentration-hybridization signal curve. RNA hybridization signal was detected using a Typhoon 9400 scanner (GE Healthcare, Chalfont St. Giles, United Kingdom) after exposure of the Eu2+ store phosphor screens to the labeled samples. CMV quantification was done by densitometry analysis using ImageQuant 5.2 (Molecular Dynamics, GE Healthcare).
To detect CMV infection, or Fny-CMV or LS-CMV RNAs 1 or 2, in test plants, the same probes described above were used in dot blot hybridization analyses. CMV sat-RNA was detected using a probe complementary to the complete sequence of the B2-satRNA (GenBank accession no. M16587). All hybridizations were done at 65°C overnight in 6% SSC (1% SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 5% Denhardt mixture, 0.1% sodium dodecyl sulfate, and yeast tRNA at 250 μg/ml.
In many test plants doubly infected with Fny-CMV and LS-CMV RNA1, LS-CMV RNA1 accumulation was too low to be detected by dot blot hybridization. Thus, in this experiment, Fny-CMV and LS-CMV RNA1 were detected by reverse transcription-PCR. Primers for detection of Fny-CMV RNA1 were 5′ACCACACAATGTGTTTAG3′, complementary to nucleotides 3136 to 3153, and 5′GAATGTGTTGCTCGTGCTTA3′, identical to nt 3056 to 3075. Primers for detection of LS-CMV RNA1 were 5′TAGTTTAAAGCAAACTACC3′, complementary to nt 3130 to 3148, and 5′AGGTGGGGACCTAATCGCTA3′, identical to nt 3034 to 3053.
All statistical analyses were done as described in reference 53 and performed using the Statgraphics package. As the variables analyzed in this work did not follow a normal distribution and showed heterogeneity of variances, analyses were done using nonparametric tests. Comparisons of virus infectivity and transmissibility were done using the Wilcoxon signed rank test. Differences in viral accumulation were analyzed with Kruskal-Wallis tests.
RESULTS
Rationale and model.
To estimate the effective number of founders, Nef, that start a CMV infection when transmitted by the aphid vector A. gossypii, we followed a modification of the model proposed by Sacristán et al. (50). In this model Nef is estimated from the segregation of two alleles, A and B, in demes originated from a mother deme. In the mother deme the frequency of alleles A and B is psA and psB, respectively, so that psA + psB = 1. From this mother deme, alleles A and B are drawn without replacement Nef times, to start daughter demes with a genetic composition given by the binomial 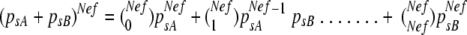 . The probability that in a daughter deme only allele A is present, POB, is given by the first term of the above expansion, resulting in equation 1.
. The probability that in a daughter deme only allele A is present, POB, is given by the first term of the above expansion, resulting in equation 1.
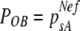 |
(1) |
The effective founder number should lie between that estimated from equation 1 and that estimated from the symmetrical expression of equation 2, as described previously (50).
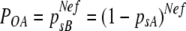 |
(2) |
In our system, mother demes are tomato plants used as sources for aphid transmission. Source plants were infected by two CMV genotypes, so that either at RNA1 or at RNA2 two different alleles, i.e., the RNA1 or -2 derived from Fny-CMV and from LS-CMV (alleles F and L, respectively) are present within the Fny-CMV genetic context at frequencies depending on their relative accumulation. Plants double-infected with RNA3 from both CMV genotypes were not used, as CMV coat protein (CP), encoded by RNA3, is the determinant for aphid transmission (10, 31, 38). POA and POB were estimated from the proportion of daughter demes (i.e., tomato test plants inoculated by aphids) in which only one of the two alleles, F or L, was present. Since new demes are started by aphid transmission, psA and psB in the equations above should represent the probability that allele A, or B, is transmitted from the mother deme, which depends on the relative accumulation of alleles A and B in doubly infected plants (AcA and AcB) and on the transmissibility of the CMV genotypes that carry allele A or allele B, TrA and TrB, so that psA = AcATrA and psB = AcBTrB, and equations 1 and 2 above can be written as shown in equations 3 and 4:
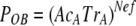 |
(3) |
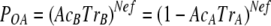 |
(4) |
Note that in our system AcATrAand AcBTrB, but not AcA and AcB, add to 1 in the source plants for aphid transmission.
Accumulation and transmissibility of CMV genotypes in tomato plants.
Tomato plants to be used as source plants for aphid transmission experiments were inoculated at the fully expanded cotyledons by gently rubbing a suspension of viral RNA. The inoculum was 1,000 ng of viral RNA per plant. At this inoculum dose infectivity of the three genotypes, F1F2F3, L1F2F3, and F1L2F3, was the same, as estimated by single-lesion infectivity experiments in Chenopodium quinoa Wild, which showed that the number of necrotic local lesions per leaf did not differ significantly for the three CMV genotypes at 1,000 ng/leaf (mean values and standard errors of four replicates were 126 ± 4.4, 128 ± 19.6, and 124 ± 2.3 for F1F2F3, L1F2F3, and F1L2F3, respectively; P > 0.80 for all comparisons based on a Wilcoxon's signed ranks test). The accumulation and transmissibility of the three CMV genotypes were analyzed in two different experiments: in experiment 1, F1F2F3 and L1F2F3 were compared. In experiment 2, F1F2F3 and F1L2F3 were compared. In a third experiment (experiment 3), F1F2F3 and F1L2F3 supporting CMV-satRNA were compared. In each experiment four replicate plants were inoculated with each CMV genotype, and eight replicate plants were inoculated with both genotypes, at 1,000 ng RNA/plant in each of these experiments.
As shown in Table 1, CMV accumulation in infected plants was significantly different between experiments (compare data for F1F2F3 in experiments 1 and 2). The accumulation of L1F2F3 was significantly lower than that of F1F2F3 (P < 0.00001, Kruskal-Wallis test), but the accumulation of F1F2F3 and F1L2F3 was similar in both experiment 2 and experiment 3 (P > 0.29). In all three experiments, CMV accumulation in mixed infection plants was not different from that in plants inoculated with the reassortant genotype.
TABLE 1.
Transmissibility of CMV isolates in tomato plants
| Expt no. and treatmenta | Accumulation of CMVb | Frequency of transmissionc | Transmissibilityd |
|---|---|---|---|
| Expt 1 | |||
| F1F2F3 | 205.53 ± 4.88 | 0.47 ± 0.038 (97/207) | 0.228 ± 0.018 |
| L1F2F3 | 121.01 ± 5.86 | 0.12 ± 0.006 (25/209) | 0.099 ± 0.011 |
| F1F2F3 + L1F2F3 | 116.12 ± 6.73 | 0.34 ± 0.046 (146/425) | |
| Expt 2 | |||
| F1F2F3 | 94.98 ± 17.37 | 0.31 ± 0.054 (65/212) | 0.384 ± 0.079 |
| F1L2F3 | 73.15 ± 20.40 | 0.21 ± 0.031 (44/215) | 0.373 ± 0.077 |
| F1F2F3 + F1L2F3 | 75.49 ± 15.02 | 0.23 ± 0.029 (103/448) | |
| Expt 3 | |||
| F1F2F3 + sat | 48.91 ± 9.60 | 0.11 ± 0.027 (21/188) | 0.242 ± 0.061 |
| F1L2F3 + sat | 26.93 ± 6.17 | 0.07 ± 0.014 (13/197) | 0.285 ± 0.086 |
| F1F2F3 + F1L2F3 + sat | 41.46 ± 5.86 | 0.14 ± 0.016 (61/440) |
For each treatment, source plants were infected with the indicated CMV genotypes.
Accumulation of CMV, expressed in micrograms of total viral RNA per gram (fresh weight) of leaf. Data are means ± standard errors of four replicates for singly infected plants or of eight replicates for doubly infected plants.
Frequency of plants infected with CMV. Data are means ± standard errors of four replicates for singly infected plants or of eight replicates for doubly infected plants. Between parentheses are indicated the number of infected plants over the total number of inoculated plants of all replicates pooled together.
Transmissibility is expressed as the percentage of infected plants per unit virus accumulation in source leaf. Data are means ± standard errors of four replicates.
Transmission frequency was estimated from the success of infection in 50 to 60 test plants for each source plant. Transmissibility, then, was estimated as the ratio of transmission frequency to virus accumulation. As shown in Table 1, the transmissibility of F1F2F3 and F1L2F3 did not differ in either experiment 2 or 3 (P > 0.67), but the transmissibility of L1F2F3 was significantly lower than that of F1F2F3 (P = 0.030, Wilcoxon's signed ranks test).
Estimation of the number of founders for aphid-inoculated plants.
To estimate psA and psB it is necessary to quantify the relative accumulation of alleles A and B in the source leaves. Despite the care that was taken to inoculate double-infected source plants with similar amounts of both CMV genotypes, i.e., with equal frequencies of alleles F and L, as estimated from infectivity experiments in C. quinoa, in the systemically infected leaves used as a source for virus acquisition the F allele accumulated always to higher levels than the L allele (Table 2). Accumulation of F1 was 45 times higher than accumulation of L1 (Table 2, experiment 1), while accumulation of F2 was 3 times higher than accumulation of L2 (Table 2, experiments 2 and 3). The probability of transmission of each allele depends on the product of its accumulation and the transmissibility of the genotype carrying this allele. These numbers (shown in the second column of Table 2), transformed to add to 1, are the transmission probabilities of each allele and are shown in the third column of Table 2. We will consider as allele A in equations 3 and 4 the most abundant allele, i.e., the F allele.
TABLE 2.
Accumulation and frequency of each allele in doubly infected source plants
| Expt no., treatmenta | Accumulation of each allele (Aci)b
|
Accumulation × transmissibility (AciTri)c
|
Estimated probability of transmissiond
|
|||
|---|---|---|---|---|---|---|
| F allele | L allele | F allele | L allele | F allele | L allele | |
| Expt 1, F1F2F3 + L1F2F3 | 7.68 ± 2.08 | 0.17 ± 0.06 | 1.752 | 0.016 | 0.99 | 0.01 |
| Expt 2, F1F2F3 + F1L2F3 | 5.58 ± 2.57 | 1.86 ± 0.32 | 2.143 | 0.695 | 0.75 | 0.25 |
| Expt 3, F1F2F3 + F1L2F3 + sat | 4.75 ± 1.20 | 1.57 ± 0.24 | 1.151 | 0.447 | 0.72 | 0.28 |
For each experiment, the treatment (i.e., genotypes that infect source plants) is indicated.
Accumulation of each allele, F or L, at either RNA1 or RNA2, expressed in micrograms of RNA per gram (fresh weight) of leaf. Data are means ± standard errors from eight doubly infected plants.
Data are the products of the accumulation of each allele times the transmissibility of the CMV carrying that allele, as shown in Table 1.
Data are AciTri values made to add to 1.
POA and POB in equations 3 and 4 were estimated from the number of test plants infected only with allele L, or only with allele F, respectively, after aphid inoculation from source plants doubly infected with both alleles. For this, only the inoculated leaves of test plants were analyzed. The segregation of alleles F and L in test plants from experiments 1 to 3 is shown in Table 3. In experiments 1 and 2 about 70% of plants were infected with allele F only, 10% with allele L only, and 20% with both alleles. The results from experiment 3 were similar, with 61% of plants infected with allele F only, 23% with allele L only, and 16% with both alleles. Experiment 3 was designed with the aim to determine if a fourth “genomic” RNA, the satRNA, would affect the effective number of founders. Out of 61 test plants infected with CMV, 38 plants (62%) were also infected with CMV-satRNA. The segregation of F and L alleles (68% F, 8% L, and 24% both) in this set of plants is the estimate of POA and POB for this experiment.
TABLE 3.
Segregation of F and L alleles at CMV RNA1 and RNA2 in aphid transmissions from double-infected source plants
| Treatmenta | Frequency of plants infected withb:
|
||
|---|---|---|---|
| F allele only | L allele only | F allele + L allele | |
| F1F2F3 + L1F2F3 | 0.70 ± 0.08 (103/146) | 0.10 ± 0.03 (14/146) | 0.20 ± 0.05 (29/146) |
| F1F2F3 + F1L2F3 | 0.71 ± 0.07 (73/103) | 0.09 ± 0.05 (9/103) | 0.20 ± 0.06 (21/103) |
| F1F2F3 + F1L2F3 + sat | 0.61 ± 0.09 (37/61) | 0.23 ± 0.04 (14/61) | 0.16 ± 0.08 (10/61) |
| 0.68 ± 0.08 (26/38)c | 0.08 ± 0.04 (3/38)c | 0.24 ± 0.05 (9/38)c | |
In each treatment group, source plants were infected with the indicated CMV genotypes.
Frequencies of plants infected only with the F or L alleles or with both alleles. Data are means ± standard errors from eight doubly infected source plants. Between parentheses are indicated the number of infected plants/number of plants treated for the eight replicates pooled together.
Frequencies of plants infected only with the F or L alleles or with both alleles, of those also infected with CMV-satRNA. Data are means ± standard errors from eight doubly infected source plants. Between parentheses are indicated the number of infected plants/number of plants treated for the eight replicates pooled together.
Once AcA, AcB, TrA, TrB, POA, and POB were estimated, the effective number of founders, Nef, could be obtained from equations 3 and 4. Table 4 shows that the upper and lower threshold estimates from experiments 2 and 3 are very similar and indicate an effective founder number between 1 and 2. The upper and lower threshold estimates from experiment 1 define a much larger range for Nef, which includes the range derived from experiments 1 and 2.
TABLE 4.
Estimated effective sizes of founder populations
| Treatmenta | POBb | POAb | psAc | psBc | Effective founder sized |
|---|---|---|---|---|---|
| F1F2F3 + L1F2F3 | 0.70 | 0.10 | 0.99 | 0.01 | 0.50-35.48 |
| F1F2F3 + F1L2F3 | 0.71 | 0.09 | 0.75 | 0.25 | 1.19-1.73 |
| F1F2F3 + F1L2F3 + sat | 0.68 | 0.08 | 0.72 | 0.28 | 1.17-1.98 |
In each treatment group, source plants were infected with the indicated CMV genotypes.
Frequency of plants infected only by the F allele (POB) or only by the L allele (POA). Values are from Table 3.
Estimated probability of transmission of the F allele (psA) or of the L allele (psB) from the source leaf. Values are from Table 2.
DISCUSSION
Population bottlenecks during horizontal transmission of plant viruses by aphids have been postulated to occur for a long time (23). The evidence is derived from attempts to directly quantify the number of virus particles transmitted by aphids and from observations of changes in the genetic structure of virus populations after aphid transmission. Thus, using radiolabeled virus particles, Pirone and Thornbury (45) estimated that successful aphid transmission of the potyviruses Tobacco etch virus and Tobacco vein mottling virus could occur after acquisition of between 15 and 20 particles from virus suspensions. Also, random changes in the main genotype of Citrus tristeza virus observed after aphid transmission (1, 5) or random uptake of reduced numbers of Citrus tristeza virus genotypes by aphids (42) could be explained by, or result in, founder effects due to population bottlenecks. More recently, Ali et al. (3) reported experiments specifically designed to test if population bottlenecks occurred during transmission of CMV by the aphid species A. gossypii and Myzus persicae (Sulzer). Aphids acquired CMV from source plants infected by a mixture of 12 silent mutants, and the random changes in the genetic composition of the CMV population in test plants indicated severe population bottlenecks during transmission. However, no attempt was made to provide an estimate of the number of founders for aphid-inoculated plants. The only available estimate, to our knowledge, was published by Moury et al. (35), who analyzed the competition for transmission by M. persicae of two genotypes of PVY which the aphids acquired from suspensions of mixtures containing different proportions of the two genotypes. They found that the number of virus particles transmitted by the aphids was very low, between 0.5 and 3.2.
Here we report an estimate of the effective number of founders starting a new infection after aphid transmission, using a different virus, CMV, and based on data on the segregation of two genotypes or, more precisely, two alleles at two different loci in the CMV genome. Our data show a very small value for the effective number of founders, Nef, between 1 and 2. It should be pointed out that the estimates from three different experiments, using different analytical tools to monitor allele segregation at different loci, were highly coherent: estimates from experiments 2 and 3 were virtually identical and within the range of the estimate from experiment 1 (Table 4), which had a larger uncertainty due to the extremely large difference of accumulation of RNA1 from Fny-CMV and LS-CMV in coinfection with Fny-CMV RNAs 2 and 3, and to the much lower transmissibility of genotype L1F2F3 than F1F2F3 (Tables 1 and 2). While it cannot be discarded that the inoculum infectivities of L1F2F3 and F1F2F3, as estimated in C. quinoa, were different in tomato, the large difference in accumulation of L1 and F1 in systemically infected leaves is in agreement with previous results, which showed coadaptation of genes within the CMV genome (17), and can be explained by the heterologous gene combinations being outcompeted by the homologous ones as infection progresses. As the frequency of transmission of CMV depends on the level of virion accumulation (6, 15) (see also the data in Table 1), the large differences in accumulation result in very low values of transmission of L1F2F3 relative to F1F2F3. The same argument applies to F1L2F3 and F1F2F3, although differences in accumulation and transmission were lower. Because RNAs 1 and 2 of Fny-CMV outcompete RNAs 1 and 2 of LS-CMV when coinfected with Fny-CMV, there is a source of uncertainty in the above estimates of Nef, as the estimates of POB, i.e., of plants infected only with the F allele, is an overestimate, and the frequency of plants infected with both alleles F and L is an underestimate. The fact that segregation of F and L alleles was analyzed in the inoculated leaves of source plants should keep the bias associated with this asymmetry to a minimum, and also it should avoid other uncertainties associated with the occurrence of bottlenecks during systemic colonization (20, 26, 30, 50). Still, estimates derived from values of POA should be better estimates than those derived from POB values, which are overestimates.
The values of Nef estimated here are compatible with the results reported in reference 3, as an average of 2.8 mutants out of 12 present in source plants were recovered from their test plants. More significant is perhaps the similarity of our estimate for the Nef of CMV during aphid transmission and that of reported by Moury et al. in reference 35 for the number of PVY particles transmitted by M. persicae. Note that when the data in reference 35 are analyzed applying our model (equations 3 and 4), the similarity persists, with values of Nef for PVY of 0.5 to 2.2. It must be pointed out that the two studies followed different experimental and analytical approaches, with different limitations and uncertainties; for instance, our approach may better represent what occurs during aphid transmission in nature, but the estimation of the frequencies of the two genotypes in virus sources for aphid acquisition could be known without uncertainties in reference 35. That so similar estimates of Nef were obtained for two viruses when using different approaches is highly relevant. Although both PVY and CMV are plant RNA viruses transmitted nonpersistently by aphids, they use different strategies; transmission of PVY depends on two viral factors, the CP and the HC-Pro protein, while that of CMV depends only on CP (39). Moreover, PVY and CMV differ in genome organization, PVY being monopartite and CMV tripartite (32, 44). Our data show that values of Nef are not affected by the number of genomic segments, three or four, necessary to start an infection (compare estimates from experiments 2 and 3). The comparison of these values with those of PVY extend this conclusion to the range of one to four genomic segments. It has been hypothesized that multipartitism must have a biological cost for a virus, and different arguments and models have been proposed to explain the evolution of multipartitism in plant RNA viruses (8, 9, 19, 36, 37, 46). The way in which constancy of Nef values over a range of multipartite genome organizations would affect these arguments and models is an issue that should be explored. It can be hypothesized that aphid-transmitted viruses have converged to an optimal/minimal transmission bottleneck regardless of the number of particles required to start an infection, i.e., of mono- or multipartitism, so that they will have similar evolutionary dynamics. Still, to reach the values of Nef of monopartite viruses, more particles should be transmitted for multipartite viruses. Aphid transmission could provide the ways to compensate for this cost, which could provide an explanation for why multipartitism has evolved only in vector-transmitted plant viruses.
Severe bottlenecks during horizontal transmission of viruses could result in random genetic drift being as important as, or more important than, deterministic processes, such as selection, as has been documented for viruses infecting plants, animals, and bacteria (23, 51). With small founder numbers, the relative importance of selection and drift will depend on the number of transmission events, which is related to the aphid population density for aphid-transmitted viruses. The consequences for the evolution of plant virus pathogenicity and virulence, and hence for virus emergence, have been seldom analyzed formally, with a few exceptions (7, 16, 35). The results that we present here strongly suggest that the occurrence of severe bottlenecks during horizontal transmission is general for viruses nonpersistently transmitted by aphids, which are the largest group of plant viruses (39, 40), and indicate that the role of random genetic drift should be considered when modeling virus evolution.
Acknowledgments
This work was in part supported by grant AGL2005-01122, Comisión Interministerial de Ciencia y Tecnología, Spain, to F.G.A. We thank the University of Santa Rosa de Cabal—Colombia for the commission of studies of M.B.
We thank Soledad Sacristán for helpful discussions. Antolín López-Quiros, Miguel A. Mora, and María Plaza provided excellent technical support.
Footnotes
Published ahead of print on 8 October 2008.
REFERENCES
- 1.Albiach-Martí, M. R., J. Guerri, A. Hermoso de Mendoza, F. Laigret, J. F. Ballester-Olmos, and P. Moreno. 2000. Aphid transmission alters the genomic and defective RNA populations of Citrus tristeza virus isolates. Phytopathology 90134-138. [DOI] [PubMed] [Google Scholar]
- 2.Aldahoud, R., W. O. Dawson, and G. E. Jones. 1989. Rapid random evolution of the genetic structure of replicating Tobacco mosaic virus populations. Intervirology 30227-233. [DOI] [PubMed] [Google Scholar]
- 3.Ali, A., H. Li, W. L. Schneider, D. J. Sherman, S. Gray, D. Smith, and M. J. Roossinck. 2006. Analysis of genetic bottlenecks during horizontal transmission of Cucumber mosaic virus. J. Virol. 808345-8350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aranda, M. A., A. Fraile, J. Dopazo, J. M. Malpica, and F. García-Arenal. 1997. Contribution of mutation and RNA recombination to the evolution of a plant pathogenic RNA. J. Mol. Evol. 4481-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ayllón, M. A., L. Rubio, A. Moya, J. Guerri, and P. Moreno. 1999. The haplotype distribution of two genes of Citrus tristeza virus is altered after host change or aphid transmission. Virology 25532-39. [DOI] [PubMed] [Google Scholar]
- 6.Banik, M. T., and T. A. Zitter. 1990. Determination of cucumber mosaic virus titer in muskmelon by enzyme-linked immunosorbent assay and correlation with aphid transmission. Plant Dis. 74857-859. [Google Scholar]
- 7.Bergstrom, C. T., P. McElhany, and L. A. Real. 1999. Transmission bottlenecks as determinants of virulence in rapidly evolving pathogens. Proc. Natl. Acad. Sci. USA 965095-5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chao, L. 1988. Evolution of sex in RNA viruses. J. Theor. Biol. 13399-112. [DOI] [PubMed] [Google Scholar]
- 9.Chao, L. 1990. Fitness of RNA virus decreased by Muller's ratchet. Nature 348454-455. [DOI] [PubMed] [Google Scholar]
- 10.Chen, B., and R. I. B. Francki. 1990. Cucumovirus transmission by the aphid Myzus persicae is determined solely by the viral coat protein. J. Gen. Virol. 71939-944. [Google Scholar]
- 11.Crow, J. F., and M. Kimura. 1970. An introduction to population genetics theory. Harper & Row, Publishers, Inc., New York, NY.
- 12.Domingo, E., and J. J. Holland. 1997. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 51151-178. [DOI] [PubMed] [Google Scholar]
- 13.Domingo, E., E. Escarmís, E. Menéndez-Arias, and J. J. Holland. 1999. Viral quasispecies and fitness variations, p. 141-162. In E. Domingo, R. Webster, and J. J. Holland (ed.), Origin and evolution of viruses. Academic Press, Inc., San Diego, CA.
- 14.Escriu, F., A. Fraile, and F. García-Arenal. 2000. Evolution of virulence in natural populations of the satellite RNA of Cucumber mosaic virus. Phytopathology 90480-485. [DOI] [PubMed] [Google Scholar]
- 15.Escriu, F., K. Perry, and F. García-Arenal. 2000. Transmissibility of Cucumber mosaic virus by Aphis gossypii correlates with viral accumulation and is affected by the presence of its satellite RNA. Phytopathology 901068-1072. [DOI] [PubMed] [Google Scholar]
- 16.Escriu, F., A. Fraile, and F. García-Arenal. 2003. The evolution of virulence in a plant virus. Evolution 57755-765. [DOI] [PubMed] [Google Scholar]
- 17.Escriu, F., A. Fraile, and F. García-Arenal. 2007. Constraints to genetic exchange support gene coadaptation in a tripartite RNA virus. PLoS Pathog. 3e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fereres, A., P. Perez, C. Gemeno, and F. Ponz. 1993. Transmission of Spanish pepper-PVY isolates by aphid vectors: epidemiological implications. Environ. Entomol. 221260-1265. [Google Scholar]
- 19.Frank, S. A. 2001. Multiplicity of infection and the evolution of hybrid incompatibility in segmented viruses. Heredity 87522-529. [DOI] [PubMed] [Google Scholar]
- 20.French, R., and D. C. Stenger. 2003. Evolution of Wheat streak mosaic virus: dynamics of population growth within plants may explain limited variation. Annu. Rev. Phytopathol. 41199-214. [DOI] [PubMed] [Google Scholar]
- 21.French, R., and D. C. Stenger. 2005. Population structure within lineages of Wheat streak mosaic virus derived from a common founding event exhibits stochastic variation inconsistent with the deterministic quasi-species model. Virology 343179-189. [DOI] [PubMed] [Google Scholar]
- 22.Frost, S. D. W., M. J. Dumaurier, S. Wain-Hobson, and A. J. Leigh-Brown. 2001. Genetic drift and within host metapopulation dynamics of HIV-1 infection. Proc. Natl. Acad. Sci. USA 986975-6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.García-Arenal, F., A. Fraile, and J. M. Malpica. 2001. Variability and genetic structure of plant virus populations. Annu. Rev. Phytopathol. 39157-186. [DOI] [PubMed] [Google Scholar]
- 24.Harrison, B. D. 1956. The infectivity of extracts made from leaves at intervals after inoculation with viruses. J. Gen. Microbiol. 15210-220. [DOI] [PubMed] [Google Scholar]
- 25.Hull, R. 2004. Matthews' plant virology, 4th ed., p. 544. Elsevier-Academic Press, San Diego, CA.
- 26.Jridi, C., J. F. Martin, V. Marie-Jeanne, G. Labonne, and S. Blanc. 2006. Distinct viral populations differentiate and evolve independently in a single perennial host plant. J. Virol. 802349-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurath, G., and P. Palukaitis. 1990. Serial passage of infectious transcripts of a Cucumber mosaic virus satellite RNA clone results in sequence heterogeneity. Virology 1768-15. [DOI] [PubMed] [Google Scholar]
- 28.Kuss, S. K., C. A. Etheredge, and J. K. Pfeiffer. 2008. Multiple host barriers restrict poliovirus trafficking in mice. PLoS Pathog. 4e1000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leigh-Brown, A. J. 1997. Analysis of HIV-1 env gene sequences reveals evidence for a low effective number in the viral population Proc. Natl. Acad. Sci. USA 941862-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, H., and M. J. Roossinck. 2004. Genetic bottlenecks reduce population variation in an experimental RNA virus population. J. Virol. 7810582-10587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, S., X. He, G. Park, C. Josefsson, and K. L. Perry. 2002. A conserved capsid protein surface domain of Cucumber mosaic virus is essential for efficient aphid vector transmission. J. Virol. 769756-9762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.López-Moya, J. J., and J. A. García. 1999. Potyvirus (Potyviridae), p. 1369-1375. In A. Granoff and R. G. Webster (ed.), Encycopledia of virology, 2nd ed. Academic Press, San Diego, CA.
- 33.Lot, H., J. Marrou, J. B. Quiot, and C. Esvan. 1972. Contribution à l'étude du virus de la mosaïque du concombre (CMV). Méthode de purification rapide du virus. Ann. Phytopathol. 425-38. [Google Scholar]
- 34.Matthews, R. E. F. 1970. Plant virology, p. 201-204. Academic Press, Inc., San Diego, CA.
- 35.Moury, B., F. Fabre, and R. Senoussi. 2007. Estimation of the number of virus particles transmitted by an insect vector. Proc. Natl. Acad. Sci. USA 10417891-17896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nee, S. 1987. The evolution of multicompartmental genomes in viruses. J. Mol. Evol. 25277-281. [DOI] [PubMed] [Google Scholar]
- 37.Nee, S. 1989. On the evolution of sex in RNA viruses. J. Theor. Biol. 138407-412. [DOI] [PubMed] [Google Scholar]
- 38.Ng, J. C., S. Liu, and K. L. Perry. 2000. Cucumber mosaic virus mutants with altered physical properties and defective in aphid vector transmission. Virology 276395-403. [DOI] [PubMed] [Google Scholar]
- 39.Ng, J. C., and K. L. Perry. 2004. Transmission of plant viruses by aphid vectors. Mol. Plant Pathol. 5505-511. [DOI] [PubMed] [Google Scholar]
- 40.Ng, J. C., and B. W. Falk. 2006. Virus-vector interactions mediating nonpersistent and semipersistent transmission of plant viruses. Annu. Rev. Phytopathol. 44183-212. [DOI] [PubMed] [Google Scholar]
- 41.Nijhuis, M., C. A. B. Boucher, P. Scipper, T. Leitner, R. Schuurman, and J. Albert. 1998. Stochastic processes strongly influence HIV-1 evolution during suboptimal protease-inhibitor therapy. Proc. Natl. Acad. Sci. USA 9514441-14446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nolasco, G., F. Fonseca, and G. Silva. 2008. Occurrence of genetic bottlenecks during Citrus tristeza virus acquisition by Toxoptera citricida under field conditions. Arch. Virol. 153259-271. [DOI] [PubMed] [Google Scholar]
- 43.Palukaitis, P., M. J. Roossinck, R. G. Dietzgen, and R. I. B. Francki. 1992. Cucumber mosaic virus. Adv. Virus Res. 41281-348. [DOI] [PubMed] [Google Scholar]
- 44.Palukaitis, P., and F. García-Arenal. 2003. Cucumoviruses. Adv. Virus Res. 62241-323. [DOI] [PubMed] [Google Scholar]
- 45.Pirone, T. P., and D. W. Thornbury. 1988. Quantity of virus required for aphid transmission of a potyvirus. Phytopathology 78104-107. [Google Scholar]
- 46.Pressing, J., and D. C. Reanney. 1984. Divided genomes and intrinsic noise. J. Mol. Evol. 20135-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rizzo, T. M., and P. Palukaitis. 1990. Construction of full-length cDNA of Cucumber mosaic virus RNAs 1, 2 and 3: generation of infectious RNA transcripts. Mol. Gen. Genet. 222249-256. [DOI] [PubMed] [Google Scholar]
- 48.Roossinck, M. J., L. Zhang, and K. H. Hellwald. 1999. Rearrangements in the 5′-nontranslated region and phylogenetic analyses of Cucumber mosaic virus RNA 3 indicate radial evolution of three subgroups. J. Virol. 736752-6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rouzine, I. M., and J. M. Coffin. 1999. Linkage disequilibrium test implies a large effective population number for HIV in vivo. Proc. Natl. Acad. Sci. USA 9610758-10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sacristán, S., J. M. Malpica, A. Fraile, and F. García-Arenal. 2003. Estimation of population bottlenecks during systemic movement of Tobacco mosaic virus in tobacco plants. J. Virol. 779906-9911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sala, M., and S. Wain-Hobson. 2000. Are RNA viruses adapting or merely changing? J. Mol. Evol. 5112-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shukla, D. D., C. W. Ward, and A. A. Brunt. 1994. The Potyviridae. CAB International, Wallingford, United Kingdom.
- 53.Sokal, R. R., and F. J. Rohlf. 1995. Biometry, 3rd ed. W. H. Freeman and Company, New York, NY.
- 54.van Kammen, A. 1968. The relationship between the components of Cowpea mosaic virus. I. Two ribonucleoprotein particles necessary for the infectivity of CPMV. Virology 34312-318. [DOI] [PubMed] [Google Scholar]


