Abstract
Rice stripe virus (RSV) is the type member of the genus Tenuivirus. RSV has four single-stranded RNAs and causes severe disease in rice fields in different parts of China. To date, no reports have described how RSV spreads within host plants or the viral and/or host factor(s) required for tenuivirus movement. We investigated functions of six RSV-encoded proteins using trans-complementation experiments and biolistic bombardment. We demonstrate that NSvc4, encoded by RSV RNA4, supports the intercellular trafficking of a movement-deficient Potato virus X in Nicotiana benthamiana leaves. We also determined that upon biolistic bombardment or agroinfiltration, NSvc4:enhanced green fluorescent protein (eGFP) fusion proteins localize predominantly near or within the walls of onion and tobacco epidermal cells. In addition, the NSvc4:eGFP fusion protein can move from initially bombarded cells to neighboring cells in Nicotiana benthamiana leaves. Immunocytochemistry using tissue sections from RSV-infected rice leaves and an RSV NSvc4-specific antibody showed that the NSvc4 protein accumulated in walls of RSV-infected leaf cells. Gel retardation assays revealed that the NSvc4 protein interacts with single-stranded RNA in vitro, a common feature of many reported plant viral movement proteins (MPs). RSV NSvc4 failed to interact with the RSV nucleocapsid protein using yeast two-hybrid assays. Taken together, our data indicate that RSV NSvc4 is likely an MP of the virus. This is the first report describing a tenuivirus MP.
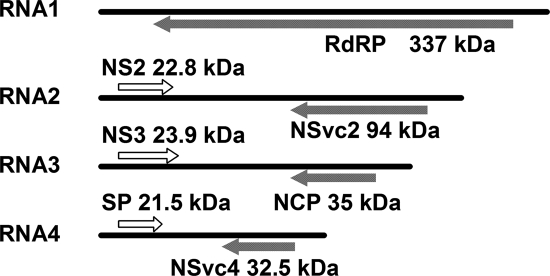
Rice stripe virus (RSV) is the type member of the genus Tenuivirus (Association of Applied Biologists Descriptions of Plant Viruses website [http://www.dpvweb.net/dpv/showdpv.php?dpvno=375]). RSV causes severe diseases in rice fields, especially in China, and is known to be transmitted by the small brown plant hopper (Laodelphax striatellus) in a persistent, circulative-propagative manner (8, 46). RSV is an RNA virus with four segmented single-stranded genomes, contains seven open reading frames (ORFs), and uses a negative and ambisense coding strategy for replication and infection in plants (Fig. 1) (32). RNA1 is negative sense and encodes a putative protein with a molecular mass of approximately 337 kDa; this protein is reported to be part of the RNA-dependent RNA polymerase and is associated with the RSV filamentous ribonucleoprotein (42). RNAs 2 to 4 are ambisense, and each RNA contains two ORFs, one in the 5′ half of the viral RNA and the other in the 5′ half of the viral cRNA. RNA2 encodes NS2 (about 22.8 kDa, with an unknown function) from the viral RNA and NSvc2 (94 kDa, a putative membrane glycoprotein) from the viral cRNA. RNA3 encodes a 23-kDa protein that functions as a suppressor of gene silencing (R. Xiong et al., unpublished data) and the nucleocapsid protein (NCP) (35 kDa). The nonstructural disease-specific protein (SP) (21.5 kDa) and NSvc4 (32.5 kDa, with unknown function) are encoded by RNA4 (14, 15, 31, 38, 47, 48).
FIG. 1.
Genome organization of RSV. ORFs are displayed as white boxes on viral-sense RNAs and shaded boxes on viral complementary-sense RNAs. Arrowheads indicate the direction of translation. RdRp, RNA-dependent RNA polymerase; SP, disease-specific protein.
Numerous studies have indicated that plant viruses encode specific proteins known as movement proteins (MPs) to control their spread through plasmodesmata (PD) in walls between cells as well as from leaf to leaf via vascular-dependent transport (24, 28, 35, 43). During this movement process, the virally encoded MPs interact with viral genomes for transport from the viral replication sites to the PDs in the walls of infected cells along the cytoskeleton and/or endoplasmic reticulum (ER) network. The virus is then thought to move through the PDs in the form of MP-associated ribonucleoprotein complexes or as virions (2, 21, 22, 25). The first model has been studied most with the MP of Tobacco mosaic virus (TMV), which interacts with the viral RNA and traverses the PD as a ribonucleoprotein complex. Viral coat protein (CP) is not required for localized cell-to-cell movement in TMV and also is not required for the systemic or localized movement of some other viruses such as Tobacco rattle virus, Tomato bushy stunt virus, or Barley stripe mosaic virus (9, 30, 36). However, CP is required for the movement of other viruses that move as nucleoprotein complexes through the PD, including the potexviruses (43), potyviruses (10), and several other viruses (4, 35). The second model includes viruses that move through the PD in the forms of encapsidated particles. This cell-to-cell movement model normally requires interactions between the viral MP and CP (5, 16, 17). Recently, it was shown that the actin/ER network is involved in viral intracellular trafficking to PD (21, 45).
Due to the lack of a reverse genetics system, there is no direct genetic information about the spread of tenuiviruses in their host plants, and no tenuivirus-encoded proteins have been identified as being viral MPs. We recently analyzed six RSV-encoded proteins for their abilities to traffic the viral genome between cells. Our results presented here demonstrate for the first time that the RSV NSvc4 protein, but not the other five RSV proteins, can move between cells and complement the cell-to-cell movement of a movement-defective mutant of potato virus X (PVX) in N. benthamiana leaves. The NSvc4:enhanced green fluorescent protein (GFP) (eGFP) fusion protein was also found to accumulate predominantly near or within the walls of bombarded or agroinfiltrated onion and tobacco epidermal cells. We propose that the RSV NSvc4 protein is an MP of RSV, and the role of the protein in RSV intercellular movement is discussed.
MATERIALS AND METHODS
Constructs and procedures for complementation experiments.
The movement-defective PVX tagged with the β-glucuronidase (GUS) gene (pPVX.GUS-Bsp) (27) was used for complementation experiments. Construct pBI-P25, expressing the PVX p25 protein, was provided by R. X. Fang (Institute of Microbiology, Chinese Academy of Sciences) (11). Six ORFs of RSV (i.e., NS2, NSvc2, NS3, NCP, NSvc4, and SP) were amplified individually from total RNA extracted from RSV-infected rice tissues collected from a rice field in the Jiangsu province using Pfu DNA polymerase and specific primer pairs (NS2-F/NS2-R for NS2, NSvc2-F/NSvc2-R for NSvc2, NS3-F/NS3-R for NS3, NCP-F/NCP-R for NCP, NSvc4-F/NSvc4-R for NSvc4, and SP-F/SP-R for SP) (Table 1). The resulting reverse transcription (RT)-PCR products were cloned individually into the BamHI/SalI site in a transient expression vector, pBin438, under the control of a Cauliflower mosaic virus 35S promoter (6). The recombinant plasmids containing various RSV ORFs were designated 35S-NS2, 35S-NSvc2, 35S-NS3, 35S-NCP, 35S-SP, and 35S-NSvc4, respectively. To prepare a mutant NSvc4 construct, the start codon for the NSvc4 ORF was altered to ATC by PCR with primers NSvc4M-F and NSvc4-R. The amplified PCR product was ligated into the pBin438 vector that had been previously digested with the BamHI and SalI restriction enzymes, and the resulting plasmid was designated 35S-NSvc4M. All constructs were sequenced to confirm their authenticity using an automated dye terminator sequencing system (model 377; PE Applied Biosystems, Foster City, CA) according to the manufacturer's protocol.
TABLE 1.
Sequences and restriction sites of PCR primers
| Primer and purpose | Sequence (5′→3′)a | Modification(s) |
|---|---|---|
| Construction of expression vectors | ||
| NS2-F | TCGGATCCATGGCATTACTCCTTTTCAATG | BamHI |
| NS2-R | GCGTCGACTCACATTAGAATAGGACACTCA | SalI |
| NSvc2-F | GAGAATTCATGCATTTTAAATCATATTTC | EcoRI |
| NSvc2-R | TGGTCGACTTAATCAACCTGTCTGATGTC | SalI |
| NS3-F | TCGGATCCATGAACGTGTTCACATCGTC | BamHI |
| NS3-R | CAGTCGACCTACAGCACAGCTGGAGAG | SalI |
| NCP-F | GAGGATCCATGGGTACCAACAAGCCAG | BamHI |
| NCP-R | TCGTCGACCTAGTCATCTGCACCTTCTG | SalI |
| SP-F | TGGGATCCATGCAAGACGTACAAAGGAC | BamHI |
| SP-R | CTGTCGACCTATGTTTTATGAAGAAGAGGT | SalI |
| NSvc4-F | GCGGATCCATGGCTTTGTCTCGACTTTT | BamHI |
| NSvc4-R | ACGTCGACCTACATGATGACAGAAACTTC | SalI |
| NSvc4m-F | GCGGATCCATCGCTTTGTCTCGACTTTT | BamHI |
| Construction of NSvc4-GFP fusion clones | ||
| NSvc4-Kpn-F | GCGGTACCATGGCTTTGTCTCGACTTTT | KpnI |
| NSvc4-Bam-R | ACGGATCCCATGATGACAGAAACTTC | BamHI |
| GFP-Bam-F | GCGGATCCATGAGTAAAGGAGAAGA | BamHI |
| GFP-Pst-R | CCCTGCAGTTATTTGTATAGTTCAT | PstI |
| Production of in vitro transcription templates | ||
| RNA1-T7-F | TAATACGACTCACTATAGGGGGATCCGTCACTTGAGACAGATTTTGATC | T7 promoter, BamHI |
| RNA1-T7-R | TAATACGACTCACTATAGGGGTTTTTCAGAGGGTCTGGC | T7 promoter |
| RNA2-T7-F | TAATACGACTCACTATAGGGGGATCCGTATGCATGAGTGAACCTATTG | T7 promoter, BamHI |
| RNA2-T7-R | TAATACGACTCACTATAGGGAATTCATATACTCTGGTGATAC | T7 promoter |
| RNA3-T7-F | TAATACGACTCACTATAGGGGGATCCGTGATCTATATAGAGTCTTCC | T7 promoter, BamHI |
| RNA3-T7-R | TAATACGACTCACTATAGGGGATCTGGATTCTGTCCCATGA | T7 promoter |
| RNA4-T7-F | TAATACGACTCACTATAGGGGGATCCACACAAAGTCCAGGGCATTTG | T7 promoter, BamHI |
| RNA4-T7-R | TAATACGACTCACTATAGGGATGAAAAGGGTGTCAGTCTCC | T7 promoter |
| PVX-T7-F | TAATACGACTCACTATAGGGGGATCCGTTCGACAGAGAGATCCACTC | T7 promoter, BamHI |
| PVX-T7-R | TAATACGACTCACTATAGGGAAGCTTCCCAGACCTCTTGTG | T7 promoter |
| GFP-T7-F | TAATACGACTCACTATAGGGATGAGTAAAGGAGAAGAAC | T7 promoter |
| GFP-F | ATGAGTAAAGGAGAAGA | |
| GFP-T7-R | TAATACGACTCACTATAGGGCGGGCATGGCACTCTTG | T7 promoter |
| Construction of YTHS plasmids | ||
| NSvc4-EcoR-F | GC GAATTC ATG GCT TTG TCT CGA CTT TT | EcoRI |
| NCP-Nde-F | GA CATATG ATG GGT ACC AAC AAG CCA G | NdeI |
| NCP-Bam-R | TC GGATCCCTA GTC ATC TGC ACC TTC TG | BamHI |
Boldface type indicates a T7 promoter, and underlining indicates a restriction enzyme site.
The constructs were introduced into cells of N. benthamiana leaves through particle bombardment as described previously (18). The bombarded leaves were harvested at 40 h postbombardment (hpb) and examined for GUS expression using a histochemical GUS detection method with specific modifications as indicated below. Leaf samples were infiltrated with 600 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (X-Gluc) diluted in a solution containing 0.115 M phosphate buffer (pH 7.0), 3 mM potassium ferricyanide, and 10 mM EDTA. The phosphate-ferricyanide-EDTA mixture was used to limit the diffusion of the intermediate products of the reaction. After overnight incubation at 37°C, the leaf samples were fixed in 70% ethanol and examined under a light microscope to assess GUS staining.
Localization of the NSvc4 protein in tobacco and onion cells.
To determine the localization of RSV NSvc4 in cells, we PCR amplified the eGFP gene (18) using primers GFP-Bam-F and GFP-Pst-R (Table 1). The PCR product was digested with the BamHI and PstI enzymes and inserted into the BamHI/PstI site within the pCHF3 vector as described previously (3) to produce pCHF3-eGFP. We then amplified the NSvc4 gene without the stop codon using primers NSvc4-Kpn-F and NSvc4-Bam-R. After digestion with the KpnI and BamHI restriction enzymes, the PCR product was cloned into the KpnI/BamHI site within the pCHF3-eGFP construct to produce pCHF3-NSvc4:eGFP. These constructs (pCHF3-eGFP and pCHF3-NSvc4:eGFP) were bombarded individually into N. benthamiana epidermal cells as described above or into onion epidermal cells as described previously (6). The bombarded tobacco or onion cells were harvested at 24 hpb and examined for free eGFP or NSVc4:eGFP fusion expression using a Leica TCS SP5 inverted confocal imaging system (Leica Microsystems, Mannheim, Germany).
To further confirm the localization of NSvc4:eGFP in cells, pCHF3-eGFP and pCHF3-NSvc4:eGFP were also introduced individually into Agrobacterium tumefaciens strain EHA105 using an electroporation method (6). Leaves of 4-week-old N. benthamiana plants were infiltrated with A. tumefaciens cells harboring the constructs as previously described (39). At 2 days postinfiltration, the infiltrated leaves were harvested and examined for eGFP fluorescence under the Leica confocal microscope set at 488 nm.
Expression and purification of recombinant NSvc4.
The NSvc4 gene was PCR amplified, cloned into vector pET-32a according to the manufacturer's instructions (Novagen, Madison, WI), and expressed as a recombinant protein with an N-terminal His6 fusion in Escherichia coli BL21(DE3) cells after induction with isopropyl-β-d-thiogalactopyranoside. The E. coli cells were lysed in a solution containing 50 mM NaH2PO4, 300 mM NaCl, and 10 mM imidazole (pH 8.0), and the fusion protein was purified through affinity chromatography on Ni-nitrilotriacetic acid agarose according to the manufacturer's instructions (Qiagen, Hilden, Germany). The His6-tagged NSvc4 (His-NSvc4) fusion protein was determined by immunoblotting with an anti-His monoclonal antibody followed by an alkaline phosphatase-conjugated anti-mouse immunoglobulin G according to instructions provided by the manufacturer (Sigma, St. Louis, MO). The detection signal was developed with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate according to instructions provided by the manufacturer (Promega, Madison, WI).
Gel retardation assay.
For gel retardation assays, we prepared several DNA fragments through RT-PCR. Four fragments (275 bp per fragment) representing partial RSV RNAs 1, 2, 3, and 4 were chosen arbitrarily and amplified individually using primer pairs RNA1-T7-F/RNA1-T7-R (for the RNA1 fragment), RNA2-T7-F/RNA2-T7-R (for RNA2), RNA3-T7-F/RNA3-T7-R (for RNA3), and RNA4-T7-F/RNA4-T7-R (for RNA4). Each resulting PCR fragment contains a T7 promoter and a BamHI site (Table 1) at its 5′ end and a second T7 promoter at its 3′ end. Radioactive double-stranded RNAs (dsRNAs) were made from the PCR fragments through in vitro transcription using [α-32P]UTP and T7 RNA polymerase according to instructions provided by the manufacturer (Takara, Dalian, China). The contaminating cDNA templates were removed through DNase digestion according to instructions provided by the manufacturer (NEB Co., Beverly, MA). RNA transcripts were then further purified using G-50 Nick columns according to the manufacturer's instructions (GE Healthcare, Bucks, United Kingdom). The quality of each RNA transcript was determined using sequencing gels. RNA transcripts prepared from a construct carrying a partial PVX sequence (nucleotides 3183 to 3457) were used as a control for the assay. To prepare radioactive single-stranded RNA, the PCR products were digested with BamHI to remove one T7 promoter prior to in vitro transcription. Long double-stranded or single-stranded GFP RNA (268 bp) was transcribed from the PCR fragments amplified with primers GFP-T7-F and GFP-T7-R or GFP-F and GFP-T7-R (Table 1). The resulting radioactive RNA transcripts were mixed with purified His6-tagged NSvc4 in 13.5 μl binding buffer containing 83 mM Tris-HCl (pH 7.5), 0.8 mM MgCl2, 66 mM KCl, 100 mM NaCl, 10 mM dithiothreitol, and 5 U RNasin. The mixtures were incubated at room temperature for 60 min and then separated by electrophoresis on a native 8% polyacrylamide gel in 0.5× TBE (90 mM Tris base-90 mM boric acid-EDTA [pH 8.0]). The gels were dried and analyzed using a Typhoon 9200 imager according to instructions provided by the manufacturer (GE Healthcare). The radioactive RNA transcripts were also mixed with bovine serum albumin (BSA) and used as negative controls during the assay.
Immunocytochemistry and electron microscopy.
Small tissue samples (approximately 1 mm by 3 mm) were excised from RSV-infected and mock-inoculated Oryza sativa cv. Wuyujing 3 leaves and fixed in 1% (vol/vol) glutaraldehyde and 2% (vol/vol) formaldehyde in 50 mM phosphate-buffered saline (PBS) (pH 6.8) for 3 h at 4°C. After dehydration in a graded ethanol series (30%, 50%, 70%, 90%, and 100%), the fixed samples were embedded in Lowicryl K4M agar (Electron Microscopy Sciences, Fort Washington, PA) as described previously (6). For immunogold labeling, ultrathin tissue sections (about 70 nm thick) were cut from the embedded tissues and placed onto 200-mesh nickel grids. After blocking for 30 min at room temperature with 50 mM PBS (pH 6.8) containing 1% (wt/vol) BSA and 0.02% polyethylene glycol 2000 (blocking solution), the grids were incubated for 1 h at room temperature with a polyclonal antibody raised against the RSV NSvc4 protein for 20 to 60 min at room temperature. The grids were washed with several changes of PBS followed by 1 h of incubation in protein A-gold (Sigma) diluted in blocking solution. After incubation, the grids were washed several times with PBS followed by double-distilled H2O. The grids were stained with uranyl acetate and then lead citrate prior to examination under an electron microscope (JEM-1200EX; Jeol, Japan). RSV-infected tissue sections probed with a preimmune serum and then protein A-gold conjugate were used as negative controls during the experiment.
YTHS.
To prepare plasmids for a yeast two-hybrid screen (YTHS), the full-length coding sequence of RSV NSvc4, which had been fused to the GAL4 DNA-binding domain or the GAL4 activation domain, was amplified by RT-PCR with primers NSvc4-EcoR-F and NSvc4-Bam-R. The product was then cloned into pGBKT7 or pGADT7 at the EcoRI/BamHI digestion site to produce pGAD-NSvc4 and pGBK-NSvc4, respectively. The full-length coding sequences of RSV NCP were amplified by RT-PCR with primers NCP-Nde-F and NCP-Bam-R and fused to the GAL4 DNA-binding domain or the GAL4 activation domain. The products were then cloned into pGBKT7 or pGADT7 at the NdeI/BamHI site to produce pGAD-NCP and pGBK-NCP, respectively. Two plasmids, pGAD-NSvc4 and pGBK-NCP, or pGBK-NSvc4 and pGAD-NCP, were cotransformed into Saccharomyces cerevisiae AH109 cells using the BD Matchmaker library construction and screening kits according to the manufacturer's instructions (Clontech, Palo Alto, CA). At the same time, the two plasmids pGBKT7-53 and pGADT7-RecT (Clontech) were cotransformed into S. cerevisiae AH109 cells and used as positive controls. Plasmids pGBKT7-Lam and pGADT7-RecT were also cotransformed into the AH109 cells and used as negative controls.
RESULTS
Complementation of movement-defective PVX by RSV NSvc4.
To identify the viral gene(s) involved in RSV cell-to-cell movement, we inserted RSV ORFs individually into plant expression vector pBin438. We then introduced each of the six constructs together with the movement-defective PVX construct (pPVX.GUS-Bsp) into mature leaves of N. benthamiana via particle bombardment. In addition, construct pBI-P25, expressing the PVX p25 triple-gene-block MP, was cobombarded with pPVX.GUS-Bsp into the N. benthamiana leaves and used as a positive control. When only pPVX.GUS-Bsp, which has a deletion in p25, was bombarded into the leaves of N. benthamiana, GUS activity was observed in single cells at 40 hpb under a stereomicroscope (Fig. 2A). This result supports the previously reported data showing that mutant PVX is able to express GUS in initially infected cells but is defective in cell-to-cell movement (27). The cobombardment of pPVX.GUS-Bsp with one of the six constructs (e.g., 35S-NS2, 35S-NSvc2, 35S-NS3, 35S-NCP, 35S-SP, and 35S-NSvc4) showed that only the NSvc4 from 35S-NSvc4 was able to complement the cell-to-cell movement of mutant PVX in bombarded leaves of N. benthamiana, as determined by the observation of blue spots at 40 hpb. Microscopic examination of the pPVX.GUS-Bsp/35S-NSvc4-cobombarded N. benthamiana leaves confirmed the presence of GUS expression in clusters of multiple epidermal cells (Fig. 2B). This observation suggests that NSvc4 facilitated mutant PVX spreading from initially infected cells into the surrounding cells. Further analysis of pPVX.GUS-Bsp/35S-NSvc4-cobombarded mature and young developing N. benthamiana leaves showed that the ability of RSV NSvc4 to complement the movement of mutant PVX in these leaves was similar (data not shown). These results provide the first evidence that RSV NSvc4 has a function similar to that reported previously for other viral MPs. RSV NSvc4 also appeared to be less efficient in facilitating mutant PVX movement in N. benthamiana leaves than the PVX p25 gene based on the size of GUS-containing foci in cobombarded N. benthamiana leaves (Fig. 2C). To further confirm that the complementation of the movement-defective PVX was controlled by the NSvc4 protein, we altered the translational start codon of the NSvc4 ORF from ATG to ATC. After the cobombardment of N. benthamiana leaves with pPVX.GUS-Bsp and 35S-NSvc4M, multicell GUS foci were not seen in any bombardment experiment (data not shown). These results strongly indicate that the NSvc4 protein can function to complement PVX GUS-Bsp movement.
FIG. 2.
Histochemical analysis of GUS activity in Nicotiana benthamiana leaves bombarded with pPVX-GUS-BSP (A), 35S-NSvc4 plus pPVX-GUS-BSP (B), or pBI-P25 plus pPVX.GUS-BSP (C). The assay was conducted at 40 hpb. Images were photographed using an Olympus stereomicroscope with enlargement.
Subcellular localizations of RSV NSvc4-GFP in leaves of onion and tobacco and of NSvc4 in RSV-infected O. sativa.
To determine the localization of RSV NSvc4 in plant cells, the NSvc4 ORF was fused to the GFP ORF, and the plasmid (pCHF3-NSvc4:eGFP) was bombarded into onion epidermal cells. By 24 hpb, green fluorescence from the NSvc4:eGFP fusion protein, visualized using confocal microscopy, was present in the cytoplasm and nucleus and also as punctate spots at the cell periphery in over 90% of the cells examined (Fig. 3A). The amount of green fluorescence from NSvc4:eGFP appeared to be present at levels in the cytoplasm and nuclei of bombarded cells similar to those in cells bombarded with the control vector pCHF3-eGFP (Fig. 3B). The transient expression of the NSvc4:eGFP fusion in N. benthamiana epidermal cells through agroinfiltration also resulted in the formation of intense fluorescing punctate foci near the walls of infiltrated cells (Fig. 3C), indicating a possible association of NSvc4 with the PDs. In contrast, fluorescence was generally cytoplasmic, and cell wall-associated punctate spots were not observed in control N. benthamiana epidermal cells infiltrated with pCHF3-eGFP (Fig. 3D).
FIG. 3.
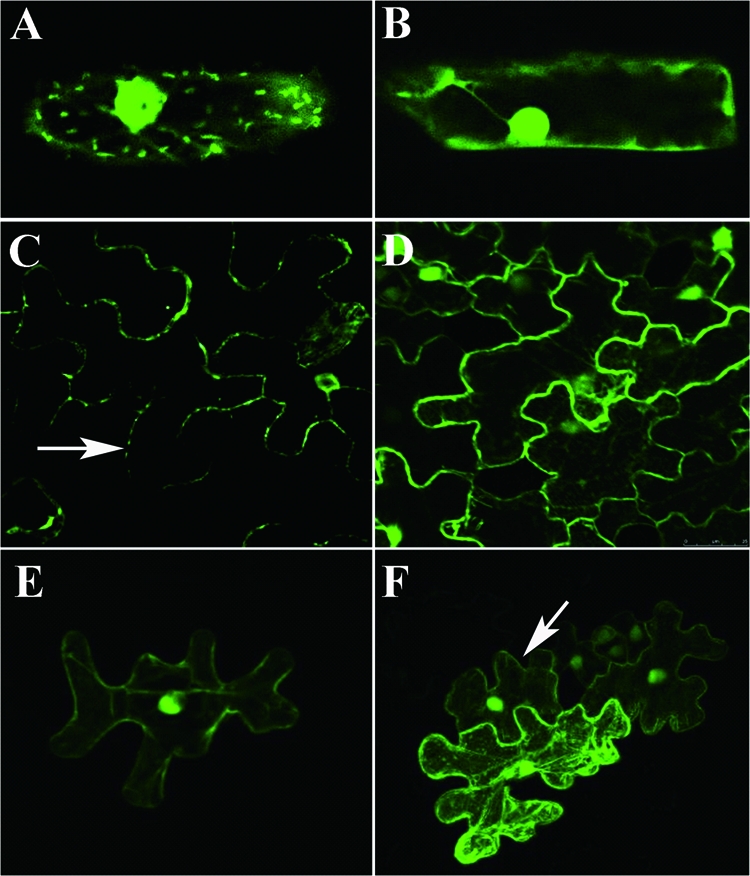
Green fluorescence in bombardment or agroinfiltration-treated onion and Nicotiana benthamiana cells with pCHF3-NSvc4:eGFP or pCHF3-eGFP. Cells were imaged under a confocal microscope at 24 h after bombardment or agroinfiltration. The NSvc4:eGFP fusion protein accumulated in the cytoplasm and nucleus and as punctate spots adjacent to cell walls of bombarded onion (A) and agroinfiltrated tobacco cells (C). GFP alone accumulated evenly in the cytoplasm and nucleus in bombarded onion (B) and agroinfiltrated tobacco cells (D). N. benthamiana leaves bombarded with pCHF3-eGFP showed green fluorescence only in single epidermal cells (E), while N. benthamiana leaves bombarded with pCHF3-NSvc4:eGFP showed green fluorescence in multiple epidermal cells (F).
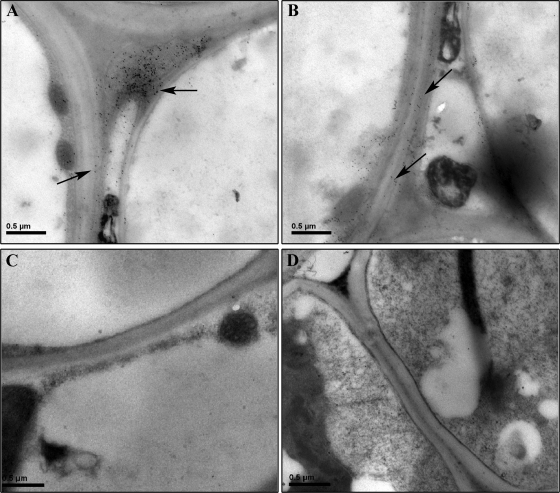
To further confirm that RSV NSvc4 associates with the cell wall of RSV-infected O. sativa cells, we prepared thin sections from fixed and embedded O. sativa leaves. The sections were then analyzed for NSvc4 localization using a gold-conjugated antibody against the protein. Under an electron microscope, the conjugated gold particles were seen primarily in cell walls of infected young leaves (Fig. 4A and B). This gold labeling pattern is similar to that reported for other viral MPs. Wall-specific gold labeling was not observed in RSV-infected sections probed with buffer or with a preimmune serum followed by the gold-conjugated antibody (Fig. 4C and data not shown) or in cells from mock-inoculated rice leaf sections (Fig. 4D). These results support the findings from the bombardment and agroinfiltration experiments and show that RSV NSvc4 is capable of targeting walls of different types of cells.
FIG. 4.
Detection of NSvc4 in RSV-infected rice cells using immunogold labeling and electron microscopy. The NSvc4 protein was detected near or in cell walls of infected cells. Arrows in A and B indicate immunogold labeling in cells. No immunogold labeling was seen in RSV-infected rice sections treated with buffer followed by the gold-conjugated secondary antiserum (C). No gold labeling was detected in healthy rice leaf sections probed with the NSvc4 antiserum (D). Bars represent 0.5 μm.
Trafficking GFP from cell to cell by NSvc4.
To test if the NSvc4:eGFP fusion protein can traffic from one cell to another, plasmids pCHF3-NSvc4:eGFP and pCHF3-eGFP were delivered individually into epidermal cells of N. benthamiana leaves through biolistic bombardment. The bombarded leaves were harvested 1 day after bombardment and examined for GFP expression in cells using a confocal microscope. Green fluorescence was seen in single N. benthamiana epidermal cells bombarded with plasmid pCHF3-eGFP (Fig. 3E). In N. benthamiana leaves bombarded with plasmid pCHF3-NSvc4:eGFP, however, the green fluorescence from the fusion protein was seen in clusters of two to five cells in approximately 30% of bombarded sites (Fig. 3F). This indicates that the fusion protein has the ability to spread from the originally bombarded cells into the adjacent cells through PDs.
RNA-binding capability of the recombinant NSvc4 protein.
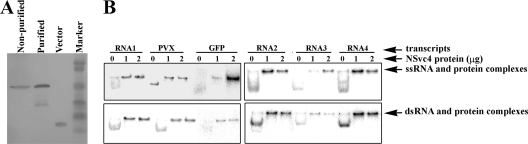
The His6-tagged NSvc4 protein was expressed in E. coli cells, purified under native conditions (Fig. 5A), and tested for RNA-binding properties. In vitro transcripts corresponding to a 275-bp segment of RSV RNA1, RNA2, RNA3, or RNA4 were used for the assay, and transcripts representing a 275-bp segment of PVX and a 268-bp segment of GFP were used as controls. Results of the experiments show that all the partial RSV transcripts, as well as the partial PVX and GFP transcripts, interacted with the recombinant RSV NSvc4 protein as determined by the reduced mobilities of the RNA/NSvc4 complexes during electrophoresis (Fig. 5B, top). Similar binding activities were also observed when NSvc4 was incubated with dsRNA (Fig. 5B, bottom). When BSA was incubated with RNA transcripts under comparable concentrations, no retarded electrophoretic mobility was found with any of the treatments (data not shown). Thus, we conclude that the observed RNA/NSvc4 binding resulted from a unique RNA-binding property of the NSvc4 protein.
FIG. 5.
Electrophoretic mobility shift assay of RNA-NSvc4 complexes. (A) His6-tagged NSvc4 protein before and after purification. (B) Different concentrations of His6-tagged NSvc4 were incubated with [α-33P]UTP-labeled RNA transcripts, and RNA-NSvc4 complexes were resolved individually on native gels. Gels were analyzed by phosphorimaging. [α-33P]UTP-labeled RNA transcripts were a 275-bp fragment representing partial sequences of RSV RNA1, RNA2, RNA3, RNA4, or PVX or a 268-bp fragment representing a partial sequence of the GFP gene. ssRNA, single-stranded RNA.
Interaction between NSvc4 and the NCP.
To determine whether NSvc4 interacts with the viral nucleocapsid protein during RSV infection, the NSvc4 ORF and the NCP ORF were separately inserted in frame into the GAL4 DNA-binding domain in vector pGBKT7 and also GAL4 activation domain vector pGADT7. Plasmids pGAD-NSvc4 and pGAD-NCP, both encoding the fusions from the GAL4 activation domain, were cotransformed with pGBK-NCP (e.g., pGAD-NSvc4 and pGBK-NCP) and pGBK-NSvc4 (e.g., pGBK-NSvc4 and pGAD-NCP) into yeast cells. Yeast cells cotransformed with plasmids pGBKT7-53 and pGADT7-RecT, and pGBKT7-Lam and pGADT7-RecT, served as positive and negative controls. Our results show that only yeast cells cotransformed with pGBKT7-53/pGADT7-RecT were able to grow on selective media (Fig. 6), indicating that no specific interactions occurred between NSvc4 and the NCP in the yeast two-hybrid system.
FIG. 6.
Analysis of the interaction between NSvc4 and NCP using a yeast two-hybrid assay. Yeast cells cotransformed with pGADT7-RecT and pGBKT7-53 (row 1) and pGADT7-RecT and pGBKT7-Lam (row 2) were included as positive and negative controls. Row 3 shows yeast cells cotransformed with pGADT7-NSvc4 and pGBKT7-NCP. Row 4 shows yeast cells cotransformed with pGADT7-NCP and pGBKT7-NSvc4. Only yeast cells that cotransformed with pGADT7-RecT and pGBKT7-53 were able to grow on selective medium.
DISCUSSION
To establish systemic infections in plants, plant viruses need to traverse PDs in walls between adjacent cells, followed by long-distance transport through vascular bundles. To move between cells, plant viruses evolved different strategies utilizing a specific MP(s) encoded by the viral genome (5, 16). Although several tenuiviruses have been reported to cause severe diseases in cereal crops in different countries including China (8, 31, 33, 46; Association of Applied Biologists Descriptions of Plant Viruses website [http://www.dpvweb.net/dpv/showdpv.php?dpvno=375]), little is known about the viral and/or host factors involved in tenuivirus movement between plant cells. In this study, we investigated six proteins encoded by RSV and their ability to traffic virus or infectious viral material between cells. Our results provide the first evidence that a fusion protein between NSvc4 and GFP is able to accumulate in multicell clusters following transient expression in the epidermal cells of N. benthamiana leaves. This multicell accumulation indicated that the NSvc4:eGFP fusion had the ability to move through PDs into neighboring cells. Our complementation experiments demonstrated that RSV NSvc4 can support a movement-defective PVX for cell-to-cell movement in N. benthamiana leaves, possibly through directing the mutant PVX to the site adjacent to PDs and/or through regulating the size exclusion limit of the PDs. We suggest that the same action may be responsible for RSV RNA movement in infected rice cells.
Complementation of mutant virus movement has been carried out in various host plants using MPs encoded by several different viruses belonging to the same or different genera (27). For example, MPs of Rice dwarf virus and Rice yellow stunt virus were used to complement PVX mutant movement in N. benthamiana plants (11, 18). Even more interesting, Flock House virus, an insect virus, can spread in N. benthamiana plants expressing MP of TMV or Red clover necrotic mosaic virus (7). Thus, it is not surprising that an MP encoded by a tenuivirus could facilitate the movement of a movement-defective PVX in N. benthamiana plants. The complementation of mutant PVX using the RSV NSvc4 protein appeared to be less effective than that supported by the PVX 25-kDa MP, as determined by the size of the blue GUS foci. This difference might be due to RSV primarily infecting rice (a monocotyledonous plant) in nature, and the RSV NSvc4 protein may not interact efficiently with a specific host factor(s) in a dicotyledonous plant like N. benthamiana.
The NSvc4:eGFP fusion protein was able to move from the initially bombarded cells to the neighboring cells in Nicotiana benthamiana leaves (Fig. 3F) but was unable to traffic in onion cells (Fig. 3A). Mechanical inoculation assays indicate that Nicotiana benthamiana is a host of RSV, while RSV cannot infect onion mechanically or by insect transmission (our unpublished data). The lack of trafficking in onion cells suggests that in this nonhost, either the mechanism to modify the MP to allow functional moving is lacking, a key host protein is absent, or a defense mechanism prevents MP function or results in its elimination (34).
It has been reported that MPs encoded by tobamoviruses and dianthoviruses can bind single-stranded RNAs and transport them through PD in the form of ribonucleoprotein complexes (23, 37, 44). Liang et al. (19) previously showed single- and double-stranded RNA-binding activities of RSV NSvc4 and suggested that the RNA-binding activities of NSvc4 may relate to a movement function. Our gel mobility shift assays, using purified NSvc4 protein and radioactively labeled RNA transcripts, also showed a sequence-nonspecific single-stranded RNA- and dsRNA-binding property for NSvc4. The nucleocapsid is the minimal infectious unit for negative-strand viruses like RSV, so it is likely that the movement of negative-strand viruses like RSV occurs as a viral nucleocapsid complex. Hence, we speculate that during RSV infection in rice, NSvc4 interacts with the viral nucleocapsid for transport from cell to cell rather than interacting directly with the viral RNA in the form of viral ribonucleoprotein complexes. However, yeast two-hybrid assays did not support an interaction between NSvc4 and NCP. It is still not clear which model best fits RSV movement. Future studies are necessary to investigate the possibility that NSvc4 participates in the transit of infectious nucleocapsids by associating with the RNA within core complexes.
Using electron microscopy, we have further demonstrated that RSV NSvc4 accumulated in areas adjacent to and within cell walls. The pattern of NSvc4 accumulation is similar to that observed previously for TMV and Cucumber mosaic virus MPs and for the Soilborne wheat mosaic virus 37K protein (1, 12, 13, 29, 40). Our observation of NSvc4 in cell walls though immunocytochemistry suggests that RSV NSvc4 may interact with the cytoskeleton and/or ER network in RSV-infected rice cells and travel along the network from viral replication sites to PDs in walls.
Liang et al. (20) previously described the immunolocalization of several RSV gene products including NSvc4 in infected rice plants and in virulent leafhoppers. NSvc4 was identified in a ring-like structure in infected plant cells, and cell wall localization was not mentioned. The differences between the patterns shown by previous work and those shown in this study are not clear, but our results clearly show the presence of NSvc4 at the cell wall.
It was previously suggested that the protein encoded by the complementary strand of tenuivirus RNA4 had a predicted secondary structure that corresponded with viral 30K movement proteins (26). Analysis of RSV NSvc4 potential structures also revealed that the protein has core regions common to the 30K MP superfamily (data not shown). Taken together, our study provides the first experimental evidence demonstrating that RSV NSvc4 functions as an MP during RSV infection in its host plant.
Acknowledgments
This research was supported by the National Natural Science Foundation of China (grant no. 30870110), the National Key Basic Research and Development Program (grant no. 2006CB101903), the Natural Science Foundation of Zhejiang Province (grant no. Y305013), and the Foundation from Jiangsu Academy of Agricultural Sciences (grant no. 6110731).
Footnotes
Published ahead of print on 25 September 2008.
REFERENCES
- 1.An, H. B., U. Melcher, P. Doss, M. Payton, A. C. Guenzi, and J. Verchot-lubicz. 2003. Evidence that the 37 kDa protein of soil-borne wheat mosaic virus is a virus movement protein. J. Gen. Virol. 843153-3163. [DOI] [PubMed] [Google Scholar]
- 2.Ashby, J., E. Boutant, M. Seemanpillai, A. Sambade, C. Ritzenthaler, and M. Heinlein. 2006. Tobacco mosaic virus movement protein functions as a structural microtubule-associated protein. J. Virol. 808329-8344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai, X. Z., X. Zhou, Y. P. Xu, M. H. A. J. Joosten, and P. J. G. M. de Wit. 2007. Cladosporium fulvum CfHNNI1 induces hypersensitive necrosis, defence gene expression and disease resistance in both host and nonhost plants. Plant Mol. Biol. 6489-101. [DOI] [PubMed] [Google Scholar]
- 4.Callaway, A., D. Giesman-Cookmeyer, E. T. Gillock, T. L. Sit, and S. A. Lommel. 2001. The multifunctional capsid proteins of plant RNA viruses. Annu. Rev. Phytopathol. 39419-460. [DOI] [PubMed] [Google Scholar]
- 5.Carrington, J. C., K. D. Kasschau, S. K. Mahajan, and M. C. Schaad. 1996. Cell-to-cell and long-distance transport of viruses in plants. Plant Cell 81669-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui, X. F., G. X. Li, D. W. Wang, D. W. Hu, and X. P. Zhou. 2005. A begomovirus DNAβ-encoded protein binds DNA, functions as a suppressor of RNA silencing, and targets the cell nucleus. J. Virol. 7910764-10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dasgupta, R., B. H. Garcia II, and R. M. Goodman. 2001. Systemic spread of an RNA insect virus in plants expressing plant viral movement protein genes. Proc. Natl. Acad. Sci. USA 984910-4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falk, B. W., and J. H. Tsai. 1998. Biology and molecular biology of viruses in the genus Tenuivirus. Annu. Rev. Phytopathol. 36139-163. [DOI] [PubMed] [Google Scholar]
- 9.Hamilton, W. D. O., and D. C. Baulcombe. 1989. Infectious RNA produced by in vitro transcription of a full-length tobacco rattle virus RNA-1 cDNA. J. Gen. Virol. 70963-968. [Google Scholar]
- 10.Hofius, D., A. T. Maier, C. Dietrich, I. Jungkunz, F. Börnke, E. Maiss, and U. Sonnewald. 2007. Capsid protein-mediated recruitment of host DnaJ-like proteins is required for Potato virus Y infection in tobacco plants. J. Virol. 8111870-11880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang, Y.-W., Y.-F. Geng, X.-B. Ying, X.-Y. Chen, and R.-X. Fang. 2005. Identification of a movement protein of rice yellow stunt rhabdovirus. J. Virol. 792108-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Itaya, A., H. Hickman, Y. Bao, R. Nelson, and B. Ding. 1997. Cell-to-cell trafficking of cucumber mosaic virus movement protein: green fluorescent protein fusion produced by biolistic gene bombardment in tobacco. Plant J. 121223-1230. [Google Scholar]
- 13.Itaya, A., Y. M. Woo, C. Masuta, Y. Bao, R. S. Nelson, and B. Ding. 1998. Developmental regulation of intercellular protein trafficking through plasmodesmata in tobacco leaf epidermis. Plant Physiol. 118373-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kakutani, T., Y. Hayano, T. Hayashi, and Y. Minobe. 1990. Ambisense segment 4 of rice stripe virus: possible evolutionary relationship with phleboviruses and uukuviruses (Bunyaviridae). J. Gen. Virol. 711427-1432. [DOI] [PubMed] [Google Scholar]
- 15.Kakutani, T., Y. Hayano, T. Hayashi, and Y. Minobe. 1991. Ambisense segment 3 of rice stripe virus: the first instance of a virus containing two ambisense segments. J. Gen. Virol. 72465-468. [DOI] [PubMed] [Google Scholar]
- 16.Lazarowitz, S. G. 1999. Probing plant cell structure and function with viral movement proteins. Curr. Opin. Plant Biol. 2332-338. [DOI] [PubMed] [Google Scholar]
- 17.Lazarowitz, S. G., and R. N. Beachy. 1999. Viral movement proteins as probes for intracellular and intercellular trafficking in plants. Plant Cell 11535-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, Y., Y. M. Bao, C. H. Wei, Z. S. Kang, Y. W. Zhong, P. Mao, G. Wu, Z. L. Chen, J. Schiemann, and R. S. Nelson. 2004. Rice dwarf phytoreovirus segment S6-encoded nonstructural protein has a cell-to-cell movement protein. J. Virol. 785382-5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang, D. L., X. Q. Ma, Z. C. Qu, and R. Hull. 2005. Nucleic acid binding property of the gene products of rice stripe virus. Virus Genes 31203-209. [DOI] [PubMed] [Google Scholar]
- 20.Liang, D. L., Z. C. Qu, X. Q. Ma, and R. Hull. 2005. Detection and localization of rice stripe virus gene products in vivo. Virus Genes 31211-221. [DOI] [PubMed] [Google Scholar]
- 21.Liu, J. Z., E. B. Blancaflor, and R. S. Nelson. 2005. The tobacco mosaic virus 126-kilodalton protein, a constituent of the virus replication complex, alone or within the complex aligns with and traffics along microfilaments. Plant Physiol. 1381853-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lough, T. J., N. E. Netzler, S. J. Emerson, P. Sutherland, F. Carr, D. L. Beck, W. J. Lucas, and R. L. Forster. 2000. Cell-to-cell movement of potexviruses: evidence for a ribonucleoprotein complex involving the coat protein and first triple gene block protein. Mol. Plant-Microbe Interact. 13962-974. [DOI] [PubMed] [Google Scholar]
- 23.Lucas, W. J. 1995. Plasmodesmata: intercellular channels for macromolecular transport in plants. Curr. Opin. Cell Biol. 7673-680. [DOI] [PubMed] [Google Scholar]
- 24.Lucas, W. J. 2006. Plant viral movement proteins: agents for cell-to-cell trafficking of viral genomes. Virology 344169-184. [DOI] [PubMed] [Google Scholar]
- 25.Lucas, W. J., and S. Wolf. 1999. Connections between virus movement, macromolecular signaling and assimilate allocation. Curr. Opin. Plant Biol. 2192-197. [DOI] [PubMed] [Google Scholar]
- 26.Melcher, U. 2000. The ‘30 K’ superfamily of viral movement protein. J. Gen. Virol. 81257-266. [DOI] [PubMed] [Google Scholar]
- 27.Morozov, S. Y., O. N. Fedorkin, G. Juttner, J. Schiemann, D. C. Baulcombe, and J. G. Atabekov. 1997. Complementation of a potato virus X mutant mediated by bombardment of plant tissues with cloned viral movement protein genes. J. Gen. Virol. 782077-2083. [DOI] [PubMed] [Google Scholar]
- 28.Nelson, R. S., and V. Citovsky. 2005. Plant viruses: invaders of cells and pirates of cellular pathways. Plant Physiol. 1381809-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oparka, K. J., D. A. M. Prior, S. Santa Cruz, H. S. Padgett, and R. N. Beachy. 1997. Gating of epidermal plasmodesmata is restricted to the leading edge of expanding infection sites of Tobacco mosaic virus (TMV). Plant J. 12781-789. [DOI] [PubMed] [Google Scholar]
- 30.Petty, I. T. D., and A. O. Jackson. 1989. Mutational analysis of barley stripe mosaic virus RNAβ. Virology 179712-718. [DOI] [PubMed] [Google Scholar]
- 31.Qu, Z. C., D. Liang, G. Harper, and R. Hull. 1997. Comparison of sequences of RNAs 3 and 4 of rice stripe virus from China with those of Japanese isolates. Virus Genes 1599-103. [DOI] [PubMed] [Google Scholar]
- 32.Ramirez, B. C., and A. L. Haenni. 1994. Molecular biology of tenuiviruses: a remarkable group of plant viruses. J. Gen. Virol. 75467-475. [DOI] [PubMed] [Google Scholar]
- 33.Ramirez, B. C., G. Macaya, L. A. Calvert, and A. L. Haenni. 1992. Rice hoja blanca virus genome characterization and expression in vitro. J. Gen. Virol. 731457-1464. [DOI] [PubMed] [Google Scholar]
- 34.Reichel, C., and R. N. Beachy. 2000. Degradation of tobacco mosaic virus movement protein by the 26S proteosome. J. Virol. 743330-3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scholthof, H. B. 2005. Plant virus transport: motions of functional equivalence. Trends Plant Sci. 10376-382. [DOI] [PubMed] [Google Scholar]
- 36.Scholthof, H. B., T. J. Morris, and A. O. Jackson. 1993. The capsid protein gene of tomato bushy stunt virus is dispensable for systemic movement and can be replaced for localized expression of foreign genes. Mol. Plant-Microbe Interact. 6309-322. [Google Scholar]
- 37.Soellick, T. R., J. F. Uhrig, G. L. Bucher, J. W. Kellmann, and P. H. Schreier. 2000. The movement protein NSm of tomato spotted wilt tospovirus (TSWV): RNA binding, interaction with the TSWV N protein, and identification of interacting plant proteins. Proc. Natl. Acad. Sci. USA 972373-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takahashi, M., S. Toriyama, C. Hamamatsu, and A. Ishihama. 1993. Nucleotide sequence and possible ambisense coding strategy of rice stripe virus RNA segment 2. J. Gen. Virol. 74769-773. [DOI] [PubMed] [Google Scholar]
- 39.Thomas, C. L., E. M. Bayer, C. Ritzenthaler, L. Fernandez-Calvino, and A. J. Maule. 2008. Specific targeting of a plasmodesmal protein affecting cell-to-cell communication. PLoS Biol. 6e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomenius, K., D. Claphan, and T. Meshi. 1987. Localization by immunogold cytochemistry of the virus-coded 30 K protein in plasmodesmata of leaves infected with Tobacco mosaic virus. Virology 160363-371. [DOI] [PubMed] [Google Scholar]
- 41.Reference deleted.
- 42.Toriyama, S., M. Takahashi, Y. Sano, T. Shimizu, and A. Ishihama. 1994. Nucleotide sequence of RNA1, the largest genomic segment of rice stripe virus, the prototype of the tenuiviruses. J. Gen. Virol. 753569-3579. [DOI] [PubMed] [Google Scholar]
- 43.Verchot-Lubicz, J. 2005. A new cell-to-cell transport model for potexviruses. Mol. Plant-Microbe Interact. 18283-290. [DOI] [PubMed] [Google Scholar]
- 44.Wolf, S., W. J. Lucas, C. M. Deom, and R. N. Beachy. 1989. Movement protein of tobacco mosaic virus modifies plasmodesmatal size exclusion limit. Science 246377-379. [DOI] [PubMed] [Google Scholar]
- 45.Wright, K. M., N. T. Wood, A. G. Roberts, S. Chapman, P. Boevink, K. M. Mackenzie, and K. J. Oparka. 2007. Targeting of TMV movement protein to plasmodesmata requires the actin/ER network: evidence from FRAP. Traffic 821-31. [DOI] [PubMed] [Google Scholar]
- 46.Xiong, R. Y., Z. B. Cheng, J. X. Wu, Y. J. Zhou, T. Zhou, and X. P. Zhou. 2008. First report of an outbreak of Rice stripe virus on wheat in China. Plant Pathol. 57397. [Google Scholar]
- 47.Zhu, Y., T. Hayakawa, and S. Toriyama. 1992. Complete nucleotide sequence of RNA4 of rice stripe virus isolate T and comparison with another isolate and with maize stripe virus. J. Gen. Virol. 731309-1312. [DOI] [PubMed] [Google Scholar]
- 48.Zhu, Y., T. Hayakawa, S. Toriyama, and M. Takahashi. 1991. Complete nucleotide sequence of RNA3 of rice stripe virus: an ambisense coding strategy. J. Gen. Virol. 72763-767. [DOI] [PubMed] [Google Scholar]