Abstract
XPC is responsible for DNA damage sensing in nucleotide excision repair (NER). Mutations in XPC lead to a defect in NER and to xeroderma pigmentosum (XP-C). Here, we analyzed the biochemical properties behind mutations found within three patients: one amino acid substitution (P334H, XP1MI, and GM02096), one amino acid incorporation in a conserved domain (697insVal, XP8BE, and GM02249), and a stop mutation (R579St, XP67TMA, and GM14867). Using these mutants, we demonstrated that HR23B stabilizes XPC on DNA and protects it from degradation. XPC recruits the transcription/repair factor TFIIH and stimulates its XPB ATPase activity to initiate damaged DNA opening. In an effort to understand the severity of XP-C phenotypes, we also demonstrated that single mutations in XPC perturb other repair processes, such as base excision repair (e.g., the P334H mutation prevents the stimulation of Ogg1 glycosylase because it thwarts the interaction between XPC and Ogg1), thereby leading to a deeper understanding of the molecular repair defect of the XP-C patients.
Genome stability and integrity are continually challenged by internal and external genotoxic attacks such as UV light or environmental and therapeutic chemicals. These can lead to high-distorting DNA injuries that can block either replication and/or transcription (11). To remove such lesions, eukaryotic cells possess nucleotide excision repair (NER), a dynamic and versatile repair mechanism that is divided in two discerned subpathways: one performing the repair of lesions in the overall genome (GGR), the other being responsible for the fast DNA repair observed in the template strand of actively transcribed genes (TCR) (21, 26). The main difference between these pathways resides in the initial detection of the DNA damage: in TCR, the elongating RNA polymerase blocked by the lesion induces the assembly of repair complexes (17, 26). On the contrary in GGR, the damage is detected by XPC. The latter was shown to be associated with HR23B (the mammalian homolog of Saccharomyces cerevisiae Rad23) and centrin2 (37, 49, 54). TFIIH is then recruited to the damage, allowing its XPB ATPase and XPD helicase to open the DNA to create a DNA bubble (16). The single-stranded DNA structure is protected and expanded by the arrival of the RPA and XPA NER factors (51). The two repair endonucleases, XPF-ERCC1 and XPG cut the 30-nucleotide damaged oligonucleotide before the gap-filing process occurs (15, 31, 38, 48).
Molecular defects in NER cause the autosomal-recessive disorders xeroderma pigmentosum (XP), trichothiodystrophy (TTD), and Cockayne syndrome (CS). Cell complementation analysis has revealed eight XP complementation groups: seven NER defective (XP-A to G) and a variant (XP-V) proficient in NER but presenting defects in translesion DNA synthesis (4). The complementation group C is the most common form of XP in the United States, and numerous causative mutations over the entire XPC gene have been identified (42). XPC patients display skin sensitivity to sunlight and a 1,000-fold increased susceptibility to developing skin cancer. However, they rarely exhibit neurological disorders or developmental defects.
Recent studies point to a role for XPC in other DNA repair pathways such as the double-strand break (13) or base excision repair (BER) (12). In line with these observations, XPC-null mice displayed pathologies also related to oxidative DNA damage persistence (32).
Thus far, investigations on the mechanistic defects leading to XP or TTD have been beneficial in understanding the function of the corresponding proteins in NER (7, 8, 14, 19). Here we have investigated the biochemical defect of three XP-C patients presenting homozygous mutations. XP67TMA presents a point mutation leading to the truncated XPC/R579St (20). XP1MI exhibits a single amino acid change at position 334 (P334H) located in a nonconserved domain. Cells from this patient presented low levels of UV-induced unscheduled DNA synthesis. This patient is also one of the rare XP-C patients who exhibits neurological problems (42). XP8BE presents an insertion of valine at position 697; among five cell lines examined, XP8BE was the least sensitive to UV irradiation and exhibited a near-normal level of XPC mRNA (6, 28). Using biochemical and in vivo approaches we delineated and precised the role of XPC in NER and BER.
MATERIALS AND METHODS
Plasmids and construction of XPC mutants for protein expression.
His6-XPC and HR23B (49) were constructed in pVL1393 vector. XPC point mutations and deletions were obtained by site-directed mutagenesis (QuikChange; Stratagene), using pVL1393-His6-XPC as the template. The resulting vectors were recombined with baculovirus DNA (BaculoGold DNA; Pharmingen) and used to infect Sf9 cells.
The pEGFP-C1 plasmid expressing the green fluorescent protein (GFP) protein, and its derivative pEGFP-XPC expressing the EGFP-XPC fusion proteins have been previously described (44). The cDNAs coding the wild-type and mutated XPC were cloned between SalI and KpnI restriction sites of pEGFP-C1. Cells were plated onto 24-well plates with coverslips and transfected by using JetPEI transfection reagent (Polyplus Transfection, Illkirch, France) with 1 μg of pEGFP-XPC DNA, according to manufacturer's protocol. At 24 h after transfection, cells were UV treated.
Cell line culture.
Human primary fibroblasts GM02096 (XP-C), GM14867 (XP-C), and FB789 (wild type, NER proficient) were cultured in Ham F-10 medium supplemented with 15% fetal calf serum and antibiotics. XPC-deficient GM02249 lymphoblasts and GM02184 (lymphoblasts, NER proficient) were maintained in RPMI 1640 medium supplemented with 15% heat-inactivated fetal calf serum. Cell survival studies were performed as described previously (22).
Fluorescence and confocal microscopy.
After local UV irradiation (8), the different cell lines were fixed in 3% paraformaldehyde for 15 min at room temperature and permeabilized with phosphate-buffered saline (PBS)-0.5% Triton X-100 for 5 min. After being washed with PBS-Tween (0.05%), the slides were incubated for 1 h with the indicated primary antibodies. After extensive washing with PBS-Tween, the cells were incubated for 1 h with the corresponding secondary antibodies in PBS-Tween (0.05%). The slides were counterstained for DNA with DAPI prepared in Vectashield mounting medium (Vector Laboratories). All images were collected by using a Leica Confocal TCS 4D microscope equipped with both UV laser and an argon/krypton laser and standard filters to allow collection of the data at 488 and 568 nm.
Protein purification.
Recombinant wild-type or mutated XPC/HR23B complexes were obtained by two-step purification. XPC/HR23B insect cell extracts (1 liter) were loaded on 2 ml of a phosphocellulose column (Whatman P11) (30), being the XPC/HR23B eluted at 1 M KCl. After having adjusted to 0.3 M KCl with buffer A (25 mM Tris-HCl [pH 7.8], 10% glycerol, 1 mM dithiothreitol [DTT]), the protein fraction was further incubated for 2 h at 4°C with 350 μl of Ni2+-NTA agarose beads (Qiagen). The packed resin was subsequently washed with buffer B (25 mM Tris-HCl [pH 7.8], 10% glycerol, 1 mM DTT, 0.25 M NaCl) containing 20 mM imidazole. The XPC/HR23B complex was eluted with the same buffer containing 200 mM imidazole. The fractions were pooled and dialyzed against buffer C (25 mM Tris-HCl [pH 7.8], 20% glycerol, 0.5 mM DTT, 0.1 mM EDTA, 50 mM KCl). XPB and p52 were expressed in insect cells and purified as described previously (9). Ogg1 was purified as described in reference 3.
Protein binding studies on immobilized DNA.
The covalently closed circular DNA-Pt containing a single 1,3-intrastrand d(GpTpG) cisplatin-DNA cross-link was prepared as described previously (47), based on the 105.TS plasmid (18). The immobilized damaged DNA template is generated by digesting the DNA-Pt plasmid (27). For each sample, 50 ng of purified biotinylated DNA-Pt was bound to 10 μg of magnetic streptavidin beads (Dynabeads M-280 Streptavidin; Dynal) and equilibrated first in buffer D (20 mM Tris-HCl [pH 7.8], 10% glycerol, 0.1 mM EDTA, 0.5 mM DTT, 400 mM KCl) and then in buffer E (same as buffer D but with 50 mM KCl). The immobilized Cax-Pt was then incubated with the various purified XPC/HR23B proteins at 30°C as indicated. Bound proteins were further analyzed by Western blotting. Gels were scanned with the Genetool software (Sygene), and the amount of proteins retained on the lesion DNA was calculated in relation to the loading input.
In vitro incision of 8-OH-Gua containing oligonucleotides.
To test the stimulatory effect of XPC, we used a gel-purified DNA fragment (36 pmol) containing a single 8-OH-Gua lesion, dephosphorylated and 5′ end labeled with γ-32P. This DNA fragment was generated by enzymatic restriction (Bsu36I/ApaI) of a single 8-OH-Gua-containing plasmid (18). The incision reaction was performed, adding 60 fmol of recombinant Ogg1 and an amount of recombinant XPC-HR23B complex as indicated, in a final volume of 10 μl. The incision products were loaded into 8% denaturing PAGE gels. The quantification of the autoradiograms was performed by using Genetool software (Sygene).
In vitro assays.
The following procedures were previously used and described in our laboratory: NER (dual incision) (41), KMnO4 footprinting (51), far-Western analysis (12), ATPase assay (14), and chromatin immunoprecipitation (ChIP) (10).
Antibodies.
A polyclonal rabbit antibody against XPC N-terminal was raised against the polypeptide TPEQAKTRERSEKIKLEF corresponding to residues 169 to 186. Anti-Ogg1 was described in reference 12. Anti-polyclonal full-length XPC (41) recognizes the XPC protein both in vitro and in vivo. Mouse anti-HR23B was from BD Biosciences. Fluorescent labeling was performed with rabbit immunoglobulin G (IgG) polyclonal anti-XPB (S-19; 1:200; Santa Cruz Biotechnology), rabbit IgG polyclonal anti-XPC (1:200), rabbit IgG polyclonal anti-XPA (FL-273; 1:200; Santa Cruz Biotechnology), mouse IgG monoclonal anti-CPD (TDM2; 1:2,000; MBL International Corp.), and rabbit polyclonal anti-GFP (TP401; Torrey Pines Biolabs). The secondary antibodies were Cy3-conjugated anti-mouse IgG and Cy3-conjugated anti-rabbit IgG.
RESULTS
XPC mutations disturb both NER and BER.
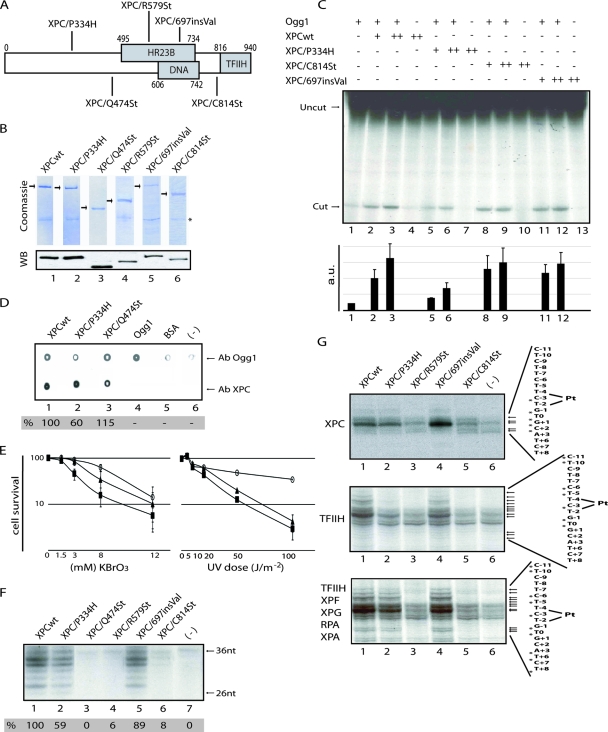
P334H, R579stop, and 697insVal mutations (Fig. 1A) found within XP-C patients were introduced into the XPC cDNA. We also designed two C-terminal truncations: XPC/Q474St and XPC/C814St, knowing that the 814-940 XPC domain interacts with TFIIH, while the 495-734 and 606-742 domains interact with HR23B and DNA, respectively (29, 49, 52). Wild-type and mutated His-tagged XPC protein were expressed and purified from insect cells (Fig. 1B; see also below).
FIG. 1.
DNA repair activities of the recombinant XPC-HR23B. (A) Diagram of XPC mutations and domains. (B) Purified XPC-HR23B complexes were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and stained with Coomassie blue (upper panel) or detected by immunoblotting with a XPC antibody that recognizes the N-terminal region (lower panel). The asterisk corresponds to HR23B. (C) A DNA fragment containing a single 8-OH-Gua lesion was incubated with 60 fmol of Ogg1 and 100 (+) or 200 (++) fmol of purified XPCwt and XPC mutants, as indicated, for 30 min at 37°C. The Ogg1 activity was measured by densitometric scanning and is expressed in arbitrary units from at least three independent experiments. (D) Far Western analysis of Ogg1 binding capacities of XPC proteins was performed as described previously (12). Ogg1 and XPC levels were tested by probing either with anti-XPC or anti-Ogg1 polyclonal antibodies (lanes 1 to 4). Ogg1, bovine serum albumin (BSA), and buffer alone were used as negative controls (lanes 4 to 6). (E) Cell survival of GM02096 (XPC/P334H, closed rectangles), GM14867 (XPC/R579St, closed triangles), and wild-type fibroblasts (FB789, open circles) following either KBrO3 (left graph) or UV-C (right graph) treatments. (F) Portions (10 ng) of each of the purified XPC-HR23B complexes (lanes 1 to 6) were tested in an in vitro NER assay (lane 7, no XPC added). (G) KMnO4 footprint using 10 ng of XPC-HR23B (upper), plus 300 ng of TFIIH (middle), as well as all of the other NER factors, except XPF-ERCC1 (lower panel). Arrows indicate KMnO4 sensitive sites (3′ sense is denoted as +N, and 5′ sense is denoted as -N).
We first investigated whether the mutations in XPC would impair the stimulation of the glycosylase activity of the human 8-oxoguanine DNA glycosylase, Ogg1 (1, 3, 12, 40, 43, 46). XPC/C814St and XPC/697insVal stimulated the hydrolysis of a linearized plasmid containing a single 8-OH-Gua lesion to a level comparable to that of the XPCwt, (Fig. 1C, compare lanes 8 to 13 and lanes 1 to 4). In contrast, addition of XPC/P334H showed a lower hydrolysis of the 8-OH-Gua containing DNA template compared to XPCwt (lanes 5 to 7). XPC/Q474St, which lacks the C-terminal region including the DNA-binding domain, does not stimulate the glycosylase activity of Ogg1 (data not shown and see below). To further document the interaction of XPC with Ogg1, we blotted XPCwt, XPC/P334H, and the C-terminal truncated XPC/Q474St onto a membrane and incubated it with Ogg1 in a far-Western assay. After extensive washes, the membrane was blotted with an antibody directed toward Ogg1. XPC/Q474St and XPCwt interacted similarly with Ogg1 (Fig. 1D, lanes 3 and 1). In contrast, Ogg1 showed a significantly weaker interaction with XPC/P334H (lane 2) than with either XPCwt or XPC/Q474St. We then investigated the sensitivity of XP-C cells to the killing effects of the oxidizing agent KBrO3. GM02096 cells (XPC/P334H) displayed increased sensitive to KBrO3 compared to either GM14867 (XPC/R579St) or wild-type fibroblasts (Fig. 1E, left graph). In contrast, both XP-C cell lines presented a similar and strong sensitivity to UV-C irradiation compared to wild-type cells (Fig. 1E, right graph). These results indicate that the P334H mutation, located in the N-terminal domain of XPC, thwarts XPC-Ogg1 interaction, thereby leading to a reduced BER activity.
Next, we examined the functionality of the XPC proteins in NER. XPC mutants, together with HR23B, were added to a reconstituted NER system containing, in addition to a single 1,3-intrastrand d(GpTpG) cisplatin-DNA, the highly purified DNA repair factors TFIIH, XPA, RPA, XPG, and ERCC1-XPF (41). The amount of excised damaged oligonucleotides was reduced by ca. 40% in the presence of XPC/P334H, while no excision activity was detected with the truncated XPC/Q474St and XPC/R579St compared to the XPCwt (Fig. 1F). In contrast, the valine insertion at position 697 did not significantly modify the level of excised damaged oligonucleotide (lane 5). Moreover, and contrary to what was observed in BER (Fig. 1C), XPC/C814St was inactive in NER (Fig. 1F, lane 6).
XPC has an essential role in DNA opening, a crucial step for the subsequent recruitment of the following NER factors (56). To investigate whether the XPC mutations disturb the opened complex formation, we used a permanganate (KMnO4) footprint assay. KMnO4 preferentially oxidizes the single-strand residues over those in the double-stranded conformation. The formation of strand breaks after piperidine treatment indicates changes in the cisplatinated DNA structure (bending) after protein binding. In the absence of any NER factors (Fig. 1G, upper panel, lane 6), permanganate-sensitive sites at positions C−3, G−1, T0, G+1, and C+2 indicated a distortion and the presence of a bubble generated by the cisplatin lesion itself (51). In the presence of either XPCwt, XPC/P334H, or XPC/697insVal, we observed an increase in DNA bending (lanes 1, 2, and 4) that can be visualized by the increase in the intensity of the hypersensitive sites at positions G−1 and T0, compared to the XPCwt protein (lane 4). Note also that XPC/R579St and XPC/C814St do not significantly modify the bending of the DNA at the site of lesion as deduced from the decreased sensitivity to permanganate at position C−3, G−1, and T0, compared to XPCwt (lanes 1, 3, and 5).
TFIIH contains two ATP-dependent helicases that, once associated to the XPC/damaged DNA, cause a structural change in the complex (51), allowing it to be targeted by other NER factors (41). TFIIH, when added to the damaged DNA in the presence of either XPCwt or XPC/697insVal, promoted an ATP-dependent increase of the DNA bubble opening at positions C−11, T−10, C−6, T−5, C−3, G−1, and T0 (Fig. 1G, middle panel, lanes 1 and 4). The opening of damaged DNA was reduced in the presence of XPC/P334H (lane 2). Addition of the remaining NER factors enhanced the opening generated by XPCwt, XPC/P334H and XPC/697insVal (lower panel, lanes 1, 2 and 4) and further expanded it, as we noticed the appearance of specific sensitive sites at positions A+3, T+6, and T+8. Addition of the NER factors to XPC/R579St or XPC/C814St and TFIIH did not lead to the appearance of any specific footprint sensitive site, showing the inability of both proteins to stimulate the opening by TFIIH. Altogether, the data presented above point out the crucial and specific role of the C-terminal domain of XPC in the interaction with and in the bending of the UV-damaged DNA. It also shows that the C-terminal domain, together with the N-terminal domain, mediates the stimulation of the glycosylase activity of Ogg1 by XPC.
In vivo NER activity of mutated XPC.
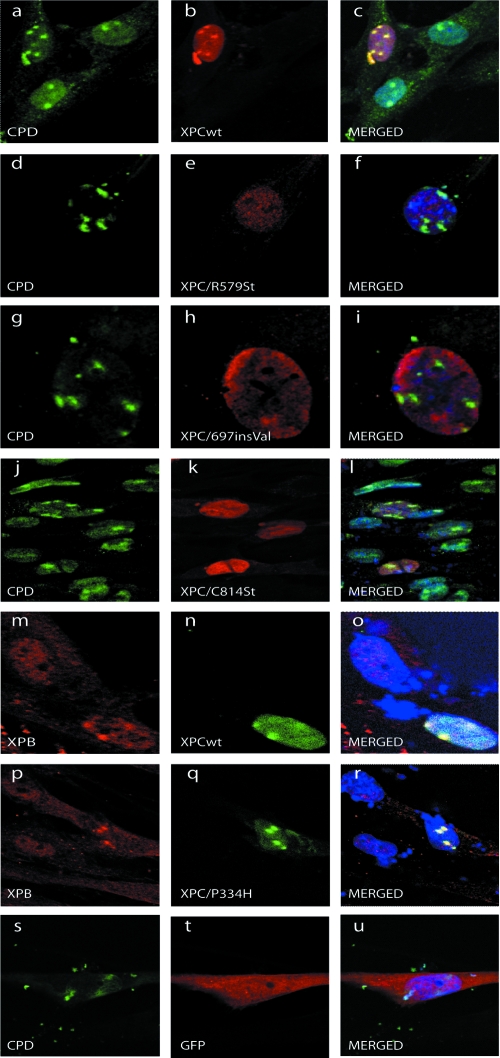
Using local UV irradiation combined with either immunofluorescent staining or direct GFP fluorescence (53), we explored the ability of the XPCwt or mutants, as well as TFIIH and XPA, to bind to the damaged DNA in vivo. We locally UV irradiated XPC-deficient fibroblasts (GM14867) that were transiently expressing either GFP-XPCwt, GFP-XPC/P334H, GFP-XPC/R579St, GFP-XPC/697insVal, or GFP-XPC/C814St fusion proteins (see Fig. S1A in the supplemental material). Confocal microscopy showed that 15 min after UV irradiation, XPCwt and XPC/P334H accumulated at UV-induced spots (Fig. 2a to c; see also Fig. S1B in the supplemental material). Moreover, XPB colocalized with XPCwt or XPC/P334H after UV irradiation, indicating the ability to recruit TFIIH to the damaged site (Fig. 2m to r). In contrast, neither XPC/R579St lacking the C-terminal region nor XPC/697insVal was recruited to damaged sites (Fig. 2d to i) and thus both were unable to recruit TFIIH (data not shown). Interestingly, XPC/C814St that still possesses an intact DNA-binding domain hardly bound the damaged sites (Fig. 2j to l).
FIG. 2.
Recruitment of XPC and TFIIH at sites of UV damage. GM14867 fibroblasts were transiently transfected with pEGFP-XPC constructs and locally UV irradiated (100 J/m2). Cells were allowed to repair for 15 min at 37°C and immunoblotted with monoclonal anti-CPD (a, d, g, j, and s), polyclonal anti-full-length XPC (b and k), polyclonal anti-GFP (e, h, and t), or monoclonal anti-XPB (m and p). Panels n and q correspond to the direct GFP signal.
These data demonstrate that the C-terminal domain of XPC is responsible for the binding of the protein to the UV-damaged DNA and the recruitment of TFIIH in vivo.
The mutated XPC disturbs the recruitment of the other NER factors.
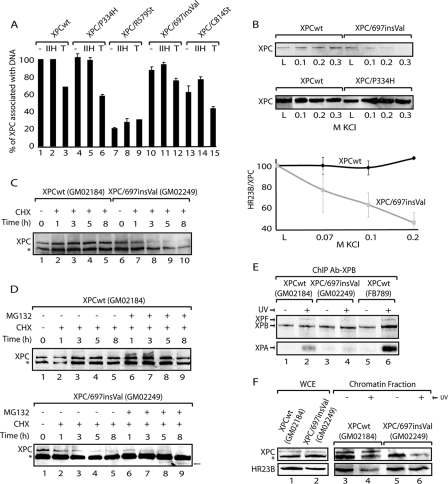
To further examine whether the XPC mutations prevent the interaction with either HR23B or the damaged DNA, we carried out two sets of experiments. First, XPC proteins were overexpressed together with HR23B in insect cells and purified by a two-step procedure including a phosphocellulose and nickel-chelating agarose adsorption chromatography (see Materials and Methods). All of the XPC proteins presented similar levels of expression according to Coomassie blue staining and immunoblotting with an antibody raised against the N-terminal domain of XPC (Fig. 1B). With the exception of XPC/Q474St and XPC/R579St (Fig. 1A), all of the tagged XPC proteins adsorbed on the Ni2+ chelate column retained HR23B in the presence of 0.25 M KCl, demonstrating how critical the XPC C terminus is for HR23B binding. Next, we set up an assay in which a biotinylated DNA fragment containing a single cisplatin lesion was immobilized on magnetic beads (26, 41) and incubated for 30 min with the XPC-HR23B complex either alone, with TFIIH, or with all of the dual incision factors (Fig. 3A, -, IIH, and T columns, respectively). The immobilized DNA was washed with a buffer containing 0.07 M KCl, and the remaining XPC proteins were analyzed by immunoblotting. We observed that the three C-terminal truncated proteins XPC/Q474St (data not shown), XPC/R579St, and XPC/C814St bound to the damaged DNA with a lower affinity than did XPCwt, XPC/P334H, or XPC/697insVal (Fig. 3A, columns 7 and 13). Under our experimental conditions, the addition of TFIIH maintained XPCwt, XPC/P334H, and XPC/697insVal on the damaged DNA (columns 2, 5, and 11). However, incubation of all of the dual incision factors in the presence of ATP partially removed the XPC proteins from the damaged DNA (Fig. 3A, columns 3, 6, and 12). These data suggest a role for the other NER factors in the accurate positioning of XPC on the damaged DNA (35, 41).
FIG. 3.
DNA-binding capacities of XPC proteins. (A) Purified XPC-HR23B complexes were incubated with a DNA fragment containing a cisplatin lesion immobilized on streptavidin beads (- columns), in the presence of TFIIH (IIH columns) or all of the dual incision factors (T columns). The values correspond to the quantification of the immunoblot bands of the XPC retained in the DNA, as a percentage of the protein input. (B) Amounts of either XPCwt and XPC/697insVal (upper panel) or XPCwt and XPC/P334H (lower panel), remaining on the immobilized damaged DNA after washing at 0.1, 0.2, and 0.3 M KCl. The graph represents the association between HR23B and XPCwt (⧫) or XPC/697insVal ( ), when washed with 0.07 to 0.2 M KCl as indicated (lower graph). Error bars correspondent to the standard error of the mean of two independent experiments. (C) GM02184 (XPCwt) or GM02249 (XPC/697insVal) human lymphoblasts were incubated overtime with 0.1 mM CHX. Cell extracts were then analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting with an XPC antibody. The asterisk corresponds to a nonspecific band. (D) GM02184 (XPCwt) or GM02249 (XPC/697insVal) human lymphoblasts were incubated during the indicated times with CHX either alone or in combination with MG132. Cell extracts were analyzed as described in panel C. The arrow indicates a probable degradation product (55). (E) ChIP followed by Western blot analysis of Ab-XPB immunoprecipitated samples from GM02184 (XPCwt), GM02249 (XPC/697insVal), and FB789 (XPCwt) cell lines fixed at t = 0 (no UV) or t = 15 min after UV irradiation (20 J/m2). A total of 400 μg of formaldehyde cross-linked protein extract was used per immunoprecipitation. (F) Portions (100 μg) of whole-cell extracts (WCE) or 40 μg of chromatin fraction extract from GM02184 or GM02249 cells were immunoblotted for the presence of XPC and HR23B. The asterisk corresponds to a nonspecific band.
), when washed with 0.07 to 0.2 M KCl as indicated (lower graph). Error bars correspondent to the standard error of the mean of two independent experiments. (C) GM02184 (XPCwt) or GM02249 (XPC/697insVal) human lymphoblasts were incubated overtime with 0.1 mM CHX. Cell extracts were then analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting with an XPC antibody. The asterisk corresponds to a nonspecific band. (D) GM02184 (XPCwt) or GM02249 (XPC/697insVal) human lymphoblasts were incubated during the indicated times with CHX either alone or in combination with MG132. Cell extracts were analyzed as described in panel C. The arrow indicates a probable degradation product (55). (E) ChIP followed by Western blot analysis of Ab-XPB immunoprecipitated samples from GM02184 (XPCwt), GM02249 (XPC/697insVal), and FB789 (XPCwt) cell lines fixed at t = 0 (no UV) or t = 15 min after UV irradiation (20 J/m2). A total of 400 μg of formaldehyde cross-linked protein extract was used per immunoprecipitation. (F) Portions (100 μg) of whole-cell extracts (WCE) or 40 μg of chromatin fraction extract from GM02184 or GM02249 cells were immunoblotted for the presence of XPC and HR23B. The asterisk corresponds to a nonspecific band.
When added to an in vitro assay, XPC/697insVal induced the same specific repair activity as did XPCwt (Fig. 1F), despite the inability of the mutant XPC protein to target the UV-damaged DNA in vivo (Fig. 2Ag to i). To resolve this discrepancy, we measured the stability of both XPCwt and XPC/697insVal on the damaged DNA in the presence of increasing salt concentrations. Both proteins, together with HR23B, were incubated with the immobilized damaged DNA for 10 min, and the beads were subsequently washed with increasing salt concentrations. The proteins remaining on the damaged template were analyzed by immunoblotting. Increasing salt concentrations up to 0.3 M KCl released XPC/697insVal from the damaged DNA, while XPCwt was still bound (Fig. 3B, upper panel). XPC/C814St, but not XPC/P334H, showed the same weakened capacity to be stabilized at the DNA damage template (data not shown and Fig. 3B, lower panel). Similar DNA-binding experiments showed that the XPC/697insVal mutation reduced either directly or indirectly the binding of HR23B around the damaged DNA (graph in Fig. 3B and see Fig. S1C in the supplemental material). We observed a rapid release of HR23B from damaged DNA when incubated with XPC/697insVal at 0.07 to 0.2 M KCl, whereas XPCwt and XPC/C814St (data not shown) still remained associated with HR23B and DNA. Altogether, our data suggest that the XPC/697insVal mutation might disturb the formation and/or the stabilization of an intermediate NER complex underlining the multifunction of the C-terminal part of XPC.
The XPC/697insVal is unstable in vivo.
Next, we examined the stability of XPC/697insVal in the GM02249 primary lymphoblastoid cell line, which expresses endogenous XPC/697insVal (42). Whereas the level of XPC protein in normal human lymphoblasts (GM02184) was barely changed when the cells were treated with cycloheximide, an inhibitor of de novo protein synthesis, similar treatment of the XPC/697insVal deficient cells resulted in a rapid reduction of endogenous XPC/697insVal level (Fig. 3C). To investigate whether such reduction was dependent on the proteasome pathway, these cells were treated both with MG132, an inhibitor of the proteasome degradation pathway, and with cycloheximide. We observed that MG132 failed to stabilize XPC/697insVal (Fig. 3D, lower and upper panels). A similar result was observed before with the XPC/W690S mutation (55), underlining the importance of this XPC conserved region for protein stability in vivo.
We next investigated the initial assembly of the NER proteins by ChIP (17). Using an antibody directed against XPB (Ab-XPB), we isolated the complex from the chromatin of wild-type lymphoblasts (GM02184) and fibroblasts (FB789) 20 min after UV treatment. At this time, we observed that the TFIIH complex, visualized by XPB, was associated with XPA and XPF, two NER factors required for dual incision (Fig. 3E, lanes 1 and 2 and lanes 5 and 6). On the contrary, this association was greatly reduced in XPC/697insVal cells (lanes 3 to 4). Moreover, the cellular XPC/697insVal protein was not detected in the chromatin fraction (Fig. 3F, lanes 2, 5, and 6). Interestingly, we noticed that in the XP-C mutant cell line the concentration of HR23B in the chromatin fraction was maintained after UV irradiation, which was not the case in wild-type cells (Fig. 3F, compare lanes 5 and 6 to lanes 3 and 4). Conversely, ChIP with an antibody toward HR23B revealed a decreased capacity of HR23B to coimmunoprecipitate repair proteins after UV (data not shown). These data point out the significance of the role that HR23B plays in the transient stabilization of XPC on damaged DNA.
The XPC/P334H retards the recruitment of XPA.
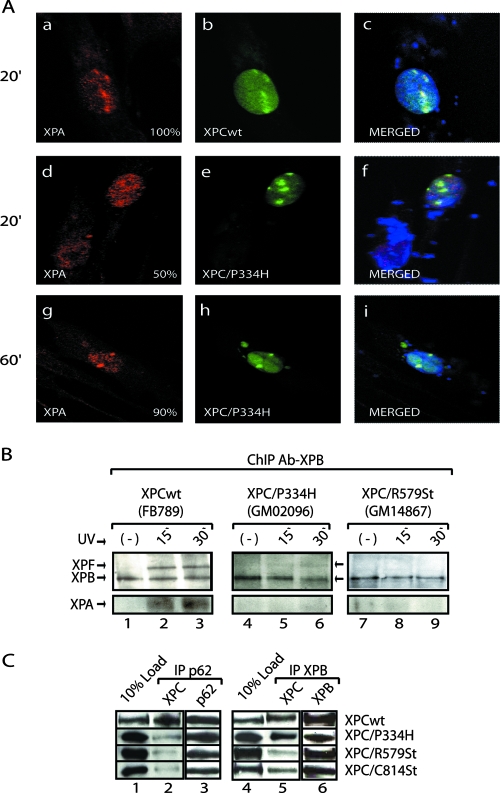
We next focused on the behavior of XPC/P334H protein in vivo. This protein is active in NER in vitro (Fig. 1F), although its ability to attract or position TFIIH is altered (Fig. 1G, middle panel). Using confocal microscopy, we found that while XPC/P334H colocalized with damaged DNA and recruited TFIIH (Fig. 2p to r and see Fig. S1B in the supplemental material), it delayed the recruitment of XPA. Indeed, 20 min after UV irradiation, we observed a deficient recruitment of XPA to UV damage sites in cells expressing XPC/P334H compared to cells expressing XPCwt (Fig. 4Aa to f). Optimal colocalization of XPA with XPB (TFIIH) only occurred at 60 min postirradiation (Fig. 4g to i). We conclude from these experiments that the XPC/P334H mutation, which allows both the binding to damaged DNA and the recruitment of TFIIH, does not permit a proper recruitment of XPA.
FIG. 4.
Recruitment of XPC and XPA at sites of UV damage. (A) XPC deficient human primary fibroblasts (GM14867) were transiently transfected with either pEGFP-XPCwt or pEGFP-XPC/P334H and UV irradiated 24 h posttransfection. Cells were allowed to repair for the indicated times and immunoblotted with a monoclonal anti-XPA antibody (a, d, and g). The XPC signal corresponds to direct fluorescence from GFP (b, e, and h). Values correspond to increased fluorescent signal at UV spots in relation to the background. (B) ChIP followed by Western blot analysis (17) of Ab-XPB immunoprecipitated samples from FB789, GM02096 (XPC/P334H), and GM14867 (XPC/R579St) cell lines and fixed at t = 0, 15, and 30 min after UV irradiation. Portions (400 μg) of formaldehyde cross-linked extract were used per immunoprecipitation. (C) Portions (100 μg) of whole-cell extracts from XPC and XPB or p62 transfected Sf9 cells were immunoprecipitated with specific anti-p62 or anti-XPB antibodies (lanes 1 and 4, 10% of load; lane 2 and 5, XPC coimmunoprecipitated blotted with an antibody against XPC; lane 3, XPB; and lane 6, p62 immunoprecipitated).
We then investigated the composition of the NER complexes by ChIP in XPC/P334H (GM02096) and XPC/R579St (GM14867), as well as in XPCwt (FB789) primary fibroblasts, 15 and 30 min after UV treatment. In the GM02096 cell line, XPC/P334H is expressed (data not shown). In NER-proficient cells (FB789), ChIP performed with Ab-XPB and subsequent Western blotting showed that XPF and XPA coprecipitated with TFIIH 15 min after UV treatment (Fig. 4B, lanes 1 to 3). These factors could not be visualized in ChIP with XPC/R579St cell extracts (lanes 7 to 9). However, in ChIP with XPC/P334H-deficient cell extracts, a weak signal of XPF was visualized 30 min after treatment (Fig. 4B, lane 6) that was not increased 60 min after treatment (data not shown), confirming the delay in the formation of the initial NER complex as previously observed by immunofluorescence (Fig. 4A).
To further examine the interactions between XPC and TFIIH, wild-type or mutated XPC proteins were overexpressed in Sf9 cells, together with either the XPB or the p62 subunits of TFIIH (2). Antibodies directed toward XPB or p62 were able to coimmunoprecipitate significant amounts of XPCwt but neither XPC/R579St or XPC/C814St (Fig. 4C, lanes 2 and 5). Interestingly, we observed that XPC/P334H mutation did not impair the interaction with XPB (lane 5) but affected the interaction with p62 (lane 2). Altogether, these assays demonstrate that p62 interacts with both the C- and the N-terminal regions of XPC, while XPB only targets the C-terminal region. Any XPC mutations that weaken such interactions will potentially modify the accurate positioning of TFIIH and the recruitment of the subsequent factors.
XPC mutants impair TFIIH enzymatic activities.
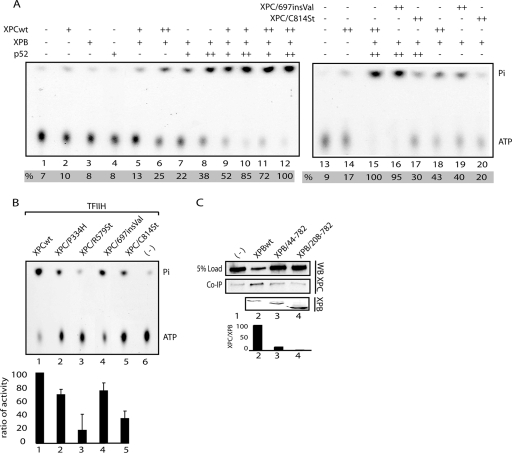
We then investigated how XPC mutations modulate TFIIH ATPase activity. Indeed, XPC together with the p52 subunit of TFIIH stimulates the ATPase activity of XPB (9). Addition of increasing amounts of either XPC or p52 to XPB led to an increase of its DNA-dependent ATPase activity (Fig. 5A, lanes 2 to 8) in an additive way (lanes 9 to 12). XPC/697insVal and XPC/P334H (data not shown) were also able to stimulate the ATPase activity of XPB in contrast to the truncated XPC/C814St that cannot optimally target XPB (Fig. 5A, lanes 15 to 20). However, the stimulation of the ATPase activity of the entire TFIIH complex by either XPC/697insVal or XPC/P334H was significantly reduced compared to XPCwt (Fig. 5B). These data highlight a possible destabilization of the complex formed between the mutated XPC and TFIIH on the damaged DNA.
FIG. 5.
Mutations in XPC impair the stimulation of TFIIH ATPase. (A) A portion (10 ng) of purified XPB was tested in an ATPase assay in the presence of 20 (+) or 50 (++) ng of p52 and of 20 (+) or 40 (++) ng of XPC proteins as indicated (lanes 1 to 12). Quantification of the release of the inorganic phosphate (Pi) and ATP was done by using a Bio-Imaging analyzer. (B) Portions (50 ng) of XPC-HR23B complexes were tested in an ATPase assay in the presence of 100 ng of TFIIH and 120 ng of DNA. Lane 6, ATP only. Graph corresponds to the quantification of two independent experiments. (C) Coimmunoprecipitation experiments between XPC and different truncated XPB proteins, using a specific monoclonal anti-XPB antibody.
Since we know that p52 interacts with the N-terminal part of XPB (24), we sought to define the XPC/XPB interacting domain. Whole-cell extracts from baculovirus-infected cells containing the N-terminal truncated XPB/44-782, XPB/208-782, or XPBwt together with XPCwt were immunoprecipitated with an antibody recognizing the C-terminal region of XPB. Western blot analysis showed that deletion of the N-terminal part of XPB drastically weakened its interaction with XPC (Fig. 5C, WB and histogram), explaining also the inability of XPC to stimulate the ATPase activity of the truncated XPB/44-782 and XPB/208-782 polypeptides (data not shown).
Altogether, our results show that XPC regulates the ATPase activity of XPB through an interaction with the first 40 amino acids of the N-terminal domain of XPB, in a region adjacent to the p52-binding site.
DISCUSSION
Using mutated XPC proteins, three of them found in XP-C patients, we have dissected the role of XPC in two major DNA repair pathways, BER and NER.
Distinct role for XPC in BER.
Here we demonstrated that the Ogg1 DNA glycosylase activity (1, 3) is stimulated by XPC through a direct interaction with its N-terminal part that encompasses the P334 surrounding region. The XPC/P334H mutation found in patient XP1MI weakens the interaction with Ogg1, resulting in a decreased capacity to regulate the glycosylase activity. This was demonstrated by the Ogg1 enzymatic and the far-Western assays (Fig. 1C and D), as well as by the high sensitivity of XPC/P334H cells to oxidative DNA-damaging agents such as KBrO3 (Fig. 1E). However, the 474 first amino acids of the N-terminal part of XPC are not sufficient for an optimal Ogg1 stimulation. It seems that an XPC domain located between amino acid Q474 and amino acid C814 (Fig. 1C and D) is required either to directly interact with the oxidative damaged DNA and/or to simply allow an optimal positioning of Ogg1 near the lesion site. Under our experimental conditions, XPC was not able to recognize or bend the DNA structure induced by the oxidative damage (data not shown). However, this bend was observed in the case of UV- or cisplatin-induced DNA damage (Fig. 1G).
The severe clinical features developed by XP-C patients probably result from the involvement of XPC not only in NER but also in BER. We cannot exclude that these mutations or the absence of both the DNA and the HR23B binding domains in the XPC protein could perturb the regulation of other DNA glycosylases such as TDG (thymine-DNA glycosylase) or MPG (3-methyladenine DNA glycosylase) (33, 45).
XPC and the recognition of DNA damage.
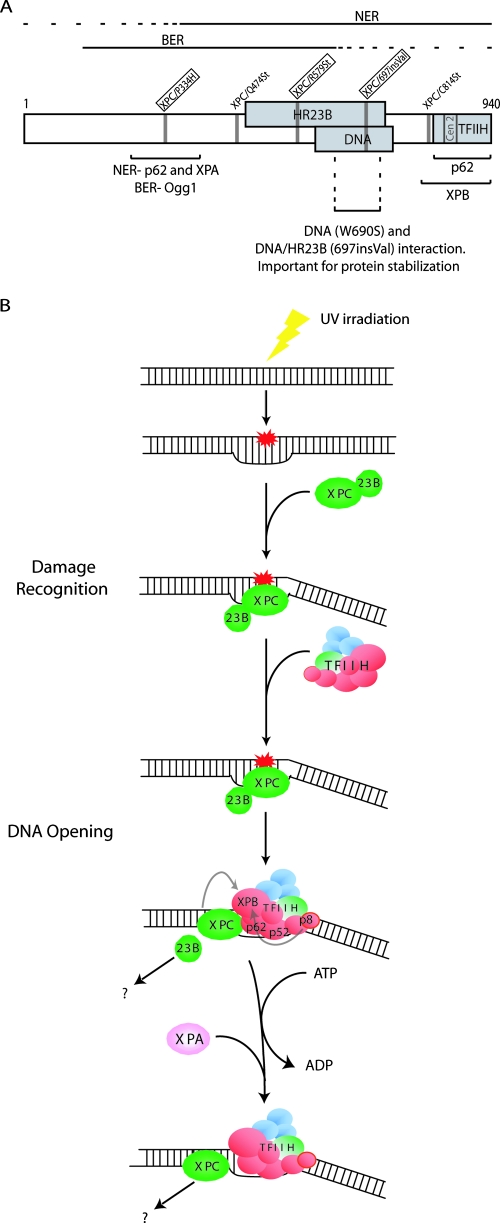
The analysis of the biochemical defects resulting from mutations found in XP-C patients (6, 42) helped us to define the exact function of XPC in the first steps of the NER reaction (Fig. 6). After recognition and bending of the damaged DNA (23, 34), XPC needs to be both stabilized and targeted by other partners, such as HR23B (36). We found that the XPC/697insVal mutation weakens the interaction with HR23B (Fig. 3B), making the complex unstable and thus explaining the defect in chromatin-dependent DNA-damaged binding in vivo (Fig. 3F). Previous studies revealed that Rad23 (the yeast HR23B homolog) could stabilize Rad4 (the yeast XPC homolog) independently of DNA damage (54). Interestingly, we also found that the single 697insVal mutation significantly destabilizes the human XPC in vivo, leading to its proteasome-independent degradation (as observed in the XPC/W690S patient) (55). Therefore, HR23B could be the link that enables XPC to associate with the damaged DNA within the chromatin context to further recruit additional factors on the damaged DNA.
FIG. 6.
(A) Scheme showing the XPC protein, indicating the mutations studied in the present study (boxes correspond to patient mutations), the interacting domains/regions identified thus far, and the role of some regions identified in the present study (the XPA binding region was described in reference 5). (B) First steps of the NER reaction. XPC, after UV irradiation, is stabilized by HR23B and recognizes and bends locally the damaged DNA; this bending precedes the recruitment of TFIIH through a double contact between XPC and the subunits p62 and XPB. This is followed by the release of HR23B concomitantly after. The ATPase of XPB is then regulated by XPC and p52. This results in a reorganization of the XPC/TFIIH/damaged DNA, which is then ready to recruit XPA and the other NER factors.
We next observed a partial release of HR23B from the chromatin fraction after UV irradiation in wild-type cells but not in XP-C cells. This suggests that HR23B may be important for the recognition step and the immediate subsequent steps in which XPC is involved postrecognition. However, we cannot exclude that XPC could be rapidly degraded in the absence of stabilization by HR23B. For instance, the truncated XPC mutants, such as XPC/R579St, are not detected in vivo (see Fig. S1A in the supplemental material) and the XPC/697insVal mutant proteins are extremely unstable (Fig. 3C and D). Moreover, the lack of in vivo XPC truncated transcripts was already demonstrated and seemingly involves nonsense-mediated mRNA decay (25).
XPC initiates TFIIH DNA opening activity.
TFIIH is recruited after XPC recognition and stabilization on the damaged DNA (41, 50). We showed that the last 130 amino acids of XPC are crucial for both binding to the damaged DNA (even if this C-terminal sequence does not encompass the DNA-binding domain previously identified) (Fig. 3A) (53) and the subsequent recruitment of TFIIH. KMnO4 footprinting experiments clearly demonstrated that XPC/C814St was unable to bind and bend the damaged DNA in vitro, therefore causing the inhibition of the recruitment of TFIIH and the subsequent NER factors in vivo. Moreover, our study details how TFIIH further links the damaged DNA: we found that the recruitment of TFIIH to XPC/damaged DNA involves the XPB and p62 subunits of TFIIH. The p62 protein binds the domain surrounding the P334 residue, as well as the C-terminal region of XPC that is also targeted by XPB.
In the absence of this multidomain binding, as observed within the XPC/P334H and XPC/R579St (Fig. 4C), the accurate positioning of TFIIH is impaired. Despite being recruited by XPC/P334H (Fig. 1G), TFIIH does not exhibit optimal XPB ATPase activity (Fig. 5B), leading to the delay in the arrival of XPA (Fig. 4A). This suggests that the stimulation of the ATPase activity of XPB relies on the accurate positioning of TFIIH on the damaged DNA, since the ATPase activity of XPB is stimulated by its interaction with both XPC itself and the p52 subunit of TFIIH (9). Next, upon arrival of TFIIH and ATP hydrolysis (51), there is a reorganization of the XPC/TFIIH/damaged DNA complex that liberates the space for the recognition of the damage complex by XPA and the arrival of the other NER factors (Fig. 4B and 6). Interestingly, the delay in the arrival of XPA, as observed in XPC/P334H cells, may partially mimic the biochemical properties observed in XPA-mutated cell lines (39).
In conclusion, we have illustrated the importance of XPC not only in NER but also in BER (Fig. 6A). In addition, we have provided explanations for the biochemical defects found within the three XP-C patients, helping us to illustrate the key role that XPC plays in NER (Fig. 6B). Indeed, we have shown how single mutations perturb protein stability, thus affecting repair processes at the level of recognition and/or stabilization at damage sites and the subsequent recruitment of repair proteins.
Supplementary Material
Acknowledgments
We thank Sascha Feuerhahn, Vincent Mocquet, Colm Ryan, and Renier Velez-Cruz for fruitful discussions and critical reading of the manuscript.
This study was supported by funds from the French League against Cancer (CDP 589111) and the French National Research Agency (NR-05-MRAR-005-01). B.B.D.J. was supported by a Marie Curie Fellowship (EEC grant MRTN-CT-2003-503618), as well as by the Association de la Recherche Contre le Cancer.
Footnotes
Published ahead of print on 22 September 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aburatani, H., Y. Hippo, T. Ishida, R. Takashima, C. Matsuba, T. Kodama, M. Takao, A. Yasui, K. Yamamoto, and M. Asano. 1997. Cloning and characterization of mammalian 8-hydroxyguanine-specific DNA glycosylase/apurinic, apyrimidinic lyase, a functional mutM homologue. Cancer Res. 572151-2156. [PubMed] [Google Scholar]
- 2.Araujo, S. J., E. A. Nigg, and R. D. Wood. 2001. Strong functional interactions of TFIIH with XPC and XPG in human DNA nucleotide excision repair, without a preassembled repairosome. Mol. Cell. Biol. 212281-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bjoras, M., L. Luna, B. Johnsen, E. Hoff, T. Haug, T. Rognes, and E. Seeberg. 1997. Opposite base-dependent reactions of a human base excision repair enzyme on DNA containing 7,8-dihydro-8-oxoguanine and abasic sites. EMBO J. 166314-6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bootsma, D., K. H. Kraemer, J. E. Cleaver, and J. H. J. Hoeijmakers. 2002. Nucleotide excision repair syndromes: xeroderma pigmentosum, Cockayne syndrome, and trichothiodystrophy, p. 211-237. In B. Vogelstein and K. W. Kinzler (ed.), The genetic basis of human cancer, 2nd ed. McGraw-Hill, New York, NY.
- 5.Bunick, C. G., M. R. Miller, B. E. Fuller, E. Fanning, and W. J. Chazin. 2006. Biochemical and structural domain analysis of xeroderma pigmentosum complementation group C protein. Biochemistry 4514965-14979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chavanne, F., B. C. Broughton, D. Pietra, T. Nardo, A. Browitt, A. R. Lehmann, and M. Stefanini. 2000. Mutations in the XPC gene in families with xeroderma pigmentosum and consequences at the cell, protein, and transcript levels. Cancer Res. 601974-1982. [PubMed] [Google Scholar]
- 7.Coin, F., E. Bergmann, A. Tremeau-Bravard, and J. M. Egly. 1999. Mutations in XPB and XPD helicases found in xeroderma pigmentosum patients impair the transcription function of TFIIH. EMBO J. 181357-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coin, F., L. P. De Santis, T. Nardo, O. Zlobinskaya, M. Stefanini, and J. M. Egly. 2006. p8/TTD-A as a repair-specific TFIIH subunit. Mol. Cell 21215-226. [DOI] [PubMed] [Google Scholar]
- 9.Coin, F., V. Oksenych, and J. M. Egly. 2007. Distinct roles for the XPB/p52 and XPD/p44 subcomplexes of TFIIH in damaged DNA opening during nucleotide excision repair. Mol. Cell 26245-256. [DOI] [PubMed] [Google Scholar]
- 10.Coin, F., V. Oksenych, V. Mocquet, S. Groh, C. Blattner, and J. M. Egly. 2008. Nucleotide excision repair driven by the dissociation of CAK from TFIIH. Mol. Cell 319-20. [DOI] [PubMed] [Google Scholar]
- 11.De Laat, W. L., N. G. Jasper, and J. H. Hoejmakers. 1999. Molecular mechanism of nucleotide excision repair. Genes Dev. 13768-785. [DOI] [PubMed] [Google Scholar]
- 12.D'Errico, M., E. Parlanti, M. Teson, B. M. de Jesus, P. Degan, A. Calcagnile, P. Jaruga, M. Bjoras, M. Crescenzi, A. M. Pedrini, J. M. Egly, G. Zambruno, M. Stefanini, M. Dizdaroglu, and E. Dogliotti. 2006. New functions of XPC in the protection of human skin cells from oxidative damage. EMBO J. 254305-4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Despras, E., P. Pfeiffer, B. Salles, P. Calsou, S. Kuhfittig-Kulle, J. F. Angulo, and D. S. Biard. 2007. Long-term XPC silencing reduces DNA double-strand break repair. Cancer Res. 672526-2534. [DOI] [PubMed] [Google Scholar]
- 14.Dubaele, S., L. Proietti De Santis, R. J. Bienstock, A. Keriel, M. Stefanini, B. Van Houten, and J. M. Egly. 2003. Basal transcription defect discriminates between xeroderma pigmentosum and trichothiodystrophy in XPD patients. Mol. Cell 111635-1646. [DOI] [PubMed] [Google Scholar]
- 15.Evans, E., J. Fellows, A. Coffer, and R. D. Wood. 1997. Open complex formation around lesion during nucleotide excision repair provides a structure for cleavage by human XPG protein. EMBO J. 16625-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans, E., J. G. Moggs, J. R. Hwang, J. M. Egly, and R. D. Wood. 1997. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J. 166559-6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fousteri, M., W. Vermeulen, A. A. van Zeeland, and L. H. Mullenders. 2006. Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol. Cell 23471-482. [DOI] [PubMed] [Google Scholar]
- 18.Frit, P., K. Kwon, F. Coin, J. Auriol, S. Dubaele, B. Salles, and J. Egly. 2002. Transcriptional activators stimulate DNA repair. Mol. Cell 101391-1401. [DOI] [PubMed] [Google Scholar]
- 19.Giglia-Mari, G., F. Coin, J. A. Ranish, D. Hoogstraten, A. Theil, N. Wijgers, N. G. Jaspers, A. Raams, M. Argentini, P. J. van der Spek, E. Botta, M. Stefanini, J. M. Egly, R. Aebersold, J. H. Hoeijmakers, and W. Vermeulen. 2004. A new, tenth subunit of TFIIH is responsible for the DNA repair syndrome trichothiodystrophy group A. Nat. Genet. 36714-719. [DOI] [PubMed] [Google Scholar]
- 20.Gozukara, E. M., S. G. Khan, A. Metin, S. Emmert, D. B. Busch, T. Shahlavi, D. M. Coleman, M. Miller, N. Chinsomboon, M. Stefanini, and K. H. Kraemer. 2001. A stop codon in xeroderma pigmentosum group C families in Turkey and Italy: molecular genetic evidence for a common ancestor. J. Investig. Dermatol. 117197-204. [DOI] [PubMed] [Google Scholar]
- 21.Hanawalt, P. 2002. Subpathways of nucleotide excision repair and their regulation. Oncogene 218949-8956. [DOI] [PubMed] [Google Scholar]
- 22.Harhaji, L., D. Popadic, D. Miljkovic, I. Cvetkovic, A. Isakovic, and V. Trajkovic. 2006. Acidosis affects tumor cell survival through modulation of nitric oxide release. Free Radic. Biol. Med. 40226-235. [DOI] [PubMed] [Google Scholar]
- 23.Janicijevic, A., K. Sugasawa, Y. Shimizu, F. Hanaoka, N. Wijgers, M. Djurica, J. H. Hoeijmakers, and C. Wyman. 2003. DNA bending by the human damage recognition complex XPC-HR23B. DNA Repair 2325-336. [DOI] [PubMed] [Google Scholar]
- 24.Jawhari, A., J. P. Laine, S. Dubaele, V. Lamour, A. Poterszman, F. Coin, D. Moras, and J. M. Egly. 2002. p52 mediates XPB function within the transcription/repair factor TFIIH. J. Biol. Chem. 27731761-31767. [DOI] [PubMed] [Google Scholar]
- 25.Khan, S. G., K. S. Oh, T. Shahlavi, T. Ueda, D. B. Busch, H. Inui, S. Emmert, K. Imoto, V. Muniz-Medina, C. C. Baker, J. J. DiGiovanna, D. Schmidt, A. Khadavi, A. Metin, E. Gozukara, H. Slor, A. Sarasin, and K. H. Kraemer. 2006. Reduced XPC DNA repair gene mRNA levels in clinically normal parents of xeroderma pigmentosum patients. Carcinogenesis 2784-94. [DOI] [PubMed] [Google Scholar]
- 26.Laine, J. P., and J. M. Egly. 2006. Initiation of DNA repair mediated by a stalled RNA polymerase IIO. EMBO J. 25387-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laine, J. P., V. Mocquet, and J. M. Egly. 2006. TFIIH enzymatic activities in transcription and nucleotide excision repair. Methods Enzymol. 408246-263. [DOI] [PubMed] [Google Scholar]
- 28.Li, L., E. S. Bales, C. A. Peterson, and R. J. Legerski. 1993. Characterization of molecular defects in xeroderma pigmentosum group C. Nat. Genet. 5413-417. [DOI] [PubMed] [Google Scholar]
- 29.Masutani, C., M. Araki, K. Sugasawa, P. J. van der Spek, A. Yamada, A. Uchida, T. Maekawa, D. Bootsma, J. H. Hoeijmakers, and F. Hanaoka. 1997. Identification and characterization of XPC-binding domain of hHR23B. Mol. Cell. Biol. 176915-6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masutani, C., K. Sugasawa, J. Yanagisawa, T. Sonoyama, M. Ui, T. Enomoto, K. Takio, K. Tanaka, P. J. van der Spek, D. Bootsma, et al. 1994. Purification and cloning of a nucleotide excision repair complex involving the xeroderma pigmentosum group C protein and a human homologue of yeast RAD23. EMBO J. 131831-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsunaga, T., D. Mu, C. H. Park, J. T. Reardon, and A. Sancar. 1995. Human DNA repair excision nuclease: analysis of the roles of the subunits involved in dual incisions by using anti-XPG and anti-ERCC1 antibodies. J. Biol. Chem. 27020862-20869. [DOI] [PubMed] [Google Scholar]
- 32.Melis, J. P., S. W. Wijnhoven, R. B. Beems, M. Roodbergen, J. van den Berg, H. Moon, E. Friedberg, G. T. van der Horst, J. H. Hoeijmakers, J. Vijg, and H. van Steeg. 2008. Mouse models for xeroderma pigmentosum group A and group C show divergent cancer phenotypes. Cancer Res. 681347-1353. [DOI] [PubMed] [Google Scholar]
- 33.Miao, F., M. Bouziane, R. Dammann, C. Masutani, F. Hanaoka, G. Pfeifer, and T. R. O'Connor. 2000. 3-Methyladenine-DNA glycosylase (MPG protein) interacts with human RAD23 proteins. J. Biol. Chem. 27528433-28438. [DOI] [PubMed] [Google Scholar]
- 34.Min, J. H., and N. P. Pavletich. 2007. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature 449570-575. [DOI] [PubMed] [Google Scholar]
- 35.Mocquet, V., J. P. Laine, T. Riedl, Z. Yajin, M. Y. Lee, and J. M. Egly. 2008. Sequential recruitment of the repair factors during NER: the role of XPG in initiating the resynthesis step. EMBO J. 27155-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ng, J. M. Y., W. Vermeulen, G. T. J. van der Horst, S. Bergink, K. Sugasawa, H. Vrieling, and J. H. J. Hoeijmakers. 2003. A novel regulation mechanism of DNA repair by damage-induced and RAD23-dependent stabilization of xeroderma pigmentosum group C protein. Genes Dev. 131630-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishi, R., Y. Okuda, E. Watanabe, T. Mori, S. Iwai, C. Masutani, K. Sugasawa, and F. Hanaoka. 2005. Centrin 2 stimulates nucleotide excision repair by interacting with xeroderma pigmentosum group C protein. Mol. Cell. Biol. 255664-5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Donnovan, A., A. A. Davies, J. G. Moggs, S. C. West, and R. D. Wood. 1994. XPG endonuclease makes the 3′ incision in human DNA nucleotide excision repair. Nature 371432-435. [DOI] [PubMed] [Google Scholar]
- 39.Oh, K. S., K. Imoto, J. Boyle, S. G. Khan, and K. H. Kraemer. 2007. Influence of XPB helicase on recruitment and redistribution of nucleotide excision repair proteins at sites of UV-induced DNA damage. DNA Repair 61359-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Radicella, J. P., C. Dherin, C. Desmaze, M. S. Fox, and S. Boiteux. 1997. Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 948010-8015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riedl, T., F. Hanaoka, and J. M. Egly. 2003. The comings and goings of nucleotide excision repair factors on damaged DNA. EMBO J. 225293-5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rivera-Begeman, A., L. D. McDaniel, R. A. Schultz, and E. C. Friedberg. 2007. A novel XPC pathogenic variant detected in archival material from a patient diagnosed with xeroderma pigmentosum: a case report and review of the genetic variants reported in XPC. DNA Repair 6100-114. [DOI] [PubMed] [Google Scholar]
- 43.Roldan-Arjona, T., Y. F. Wei, K. C. Carter, A. Klungland, C. Anselmino, R. P. Wang, M. Augustus, and T. Lindahl. 1997. Molecular cloning and functional expression of a human cDNA encoding the antimutator enzyme 8-hydroxyguanine-DNA glycosylase. Proc. Natl. Acad. Sci. USA 948016-8020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Santagati, F., E. Botta, M. Stefanini, and A. M. Pedrini. 2001. Different dynamics in nuclear entry of subunits of the repair/transcription factor TFIIH. Nucleic Acids Res. 291574-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shimizu, Y., S. Iwai, F. Hanaoka, and K. Sugasawa. 2003. Xeroderma pigmentosum group C protein interacts physically and functionally with thymine DNA glycosylase. EMBO J. 22164-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shinmura, K., H. Kasai, A. Sasaki, H. Sugimura, and J. Yokota. 1997. 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) DNA glycosylase and AP lyase activities of hOGG1 protein and their substrate specificity. Mutat. Res. 38575-82. [DOI] [PubMed] [Google Scholar]
- 47.Shivji, M. K. K., V. N. Podust, U. Hubsher, and R. D. Wood. 1995. Nucleotide excision repair DNA synthesis by DNA polymerase epsilon in the presence of PCNA, RFC, and RPA. Biochemistry 345011-5017. [DOI] [PubMed] [Google Scholar]
- 48.Sijbers, A. M., W. L. de Laat, R. R. Ariza, M. Biggerstaff, Y. F. Wei, J. G. Moggs, K. C. Carter, B. K. Shell, E. Evans, M. C. de Jong, S. Rademakers, J. de Rooij, N. G. Jaspers, J. H. Hoeijmakers, and R. D. Wood. 1996. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell 86811-822. [DOI] [PubMed] [Google Scholar]
- 49.Sugasawa, K., C. Masutani, A. Uchida, T. Maekawa, P. J. van der Spek, D. Bootsma, J. H. Hoeijmakers, and F. Hanaoka. 1996. HHR23B, a human Rad23 homolog, stimulates XPC protein in nucleotide excision repair in vitro. Mol. Cell. Biol. 164852-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sugasawa, K., T. Okamoto, Y. Shimizu, C. Masutani, S. Iwai, and F. Hanaoka. 2001. A multistep damage recognition mechanism for global genomic nucleotide excision repair. Genes Dev. 15507-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tapias, A., J. Auriol, D. Forget, J. H. Enzlin, O. D. Scharer, F. Coin, B. Coulombe, and J. M. Egly. 2004. Ordered conformational changes in damaged DNA induced by nucleotide excision repair factors. J. Biol. Chem. 27919074-19083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uchida, A., K. Sugasawa, C. Masutani, N. Dohmae, M. Araki, M. Yokoi, Y. Ohkuma, and F. Hanaoka. 2002. The carboxy-terminal domain of the XPC protein plays a crucial role in nucleotide excision repair through interactions with transcription factor IIH. DNA Repair 1449-461. [DOI] [PubMed] [Google Scholar]
- 53.Volker, M., M. J. Mone, P. Karmakar, A. van Hoffen, W. Schul, W. Vermeulen, J. H. Hoeijmakers, R. van Driel, A. A. van Zeeland, and L. H. Mullenders. 2001. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell 8213-224. [DOI] [PubMed] [Google Scholar]
- 54.Xie, Z., S. Liu, Y. Zhang, and Z. Wang. 2004. Roles of Rad23 protein in yeast nucleotide excision repair. Nucleic Acids Res. 325981-5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yasuda, G., R. Nishi, E. Watanabe, T. Mori, S. Iwai, D. Orioli, M. Stefanini, F. Hanaoka, and K. Sugasawa. 2007. In vivo destabilization and functional defects of the xeroderma pigmentosum C protein caused by a pathogenic missense mutation. Mol. Cell. Biol. 276606-6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yokoi, M., C. Masutani, T. Maekawa, K. Sugasawa, Y. Ohkuma, and F. Hanaoka. 2000. The xeroderma pigmentosum group C protein complex XPC-HR23B plays an important role in the recruitment of transcription factor IIH to damaged DNA. J. Biol. Chem. 2759870-9875. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.