Abstract
Summary: The modulation of the immune response is a common practice of many highly pathogenic viruses. The emergence of the highly pathogenic coronavirus severe acute respiratory virus (SARS-CoV) serves as a robust model system to elucidate the virus-host interactions that mediate severe end-stage lung disease in humans and animals. Coronaviruses encode the largest positive-sense RNA genome of ∼30 kb, encode a variety of replicase and accessory open reading frames that are structurally unique, and encode novel enzymatic functions among RNA viruses. These viruses have broad or specific host ranges, suggesting the possibility of novel strategies for targeting and regulating host innate immune responses following virus infection. Using SARS-CoV as a model, we review the current literature on the ability of coronaviruses to interact with and modify the host intracellular environment during infection. These studies are revealing a rich set of novel viral proteins that engage, modify, and/or disrupt host cell signaling and nuclear import machinery for the benefit of virus replication.
INTRODUCTION
The integration of modern genetics, biochemistry, and molecular biology techniques into the field of virology over the last 30 years has led to quantum leaps in our understanding of the molecular mechanisms by which different virus infections cause novel pathogenic disease outcomes. The ability to analyze the effects of each viral protein on the host transcriptome and proteome coupled with the ability to identify the interaction partners that impact host cell signaling pathways is enabling the current generation of scientists to interrogate host-pathogen interactions at the molecular level, developing a system-wide view of disease processes. The use of viral reverse genetic systems for model viruses and gene knockout mice allows virologists to investigate the detailed interactions that occur during infection to regulate complex disease outcomes. Our understanding of coronavirus pathogenesis, especially that of mouse hepatitis virus and severe acute respiratory syndrome coronavirus (SARS-CoV), has increased exponentially since the creation of reverse genetic clones for each virus in recent years (2, 159, 160).
Using this growing suite of new molecular tools, our laboratory and others have been investigating the interactions between the virus and host during the initial phases of disease. The initial host response to infection is controlled by the innate immune system, a complex, highly regulated network of cytokines, chemokines, complement proteins, macrophages, and dendritic cells (DCs), which serves as a robust barrier to the establishment and maintenance of a productive virus infection. However, viruses have coevolved an armament of components that evade and/or block host innate immune responses, or they express components that shield infection from detection (3, 5, 22, 35, 81, 92, 93, 127, 151, 156). Given the diverse array of novel genetic functions, it is not surprising that coronaviruses also encode several proteins that modulate the host innate immune response during infection.
HUMAN CORONAVIRUSES AND DISEASE
Classically, human coronaviruses 229E and OC43 were viewed as causing mild upper respiratory tract infections. Moreover, animal models of disease were lacking, sparking minimal interest in identifying the determinants that regulate disease outcomes. OC43 has been found throughout the world to be a cause of both the common cold and other more severe respiratory diseases, especially in children and the elderly (8, 30, 137). More recently, Patrick et al. found that in an elder care facility, almost 50% of staff and patients were OC43 positive during an outbreak of severe respiratory disease. Infected elderly patients showed severe respiratory distress and pulmonary symptoms, resulting in a 10% mortality rate (94). Human coronavirus 229E has also been found to cause lower respiratory tract disease, mostly in the elderly, infants, and individuals with chronic underlying immune conditions (6). These new revelations, coupled with the emergence of the highly pathogenic SARS-CoV, demonstrated that coronavirus genomes likely encode unusual determinants that promote severe disease outcomes, especially in the elderly. The advent of SARS-CoV resulted in the identification of two new human coronaviruses, NL63 and HKU1, which produce severe lower respiratory tract infections in infants and young children and in aged adults, respectively (100, 150). In particular, the first report of HKU1 infection was initially documented as being a SARS-CoV case in an elderly Hong Kong patient (150). The growing recognition that human coronaviruses are important and potentially highly pathogenic has revitalized interest in understanding pathogenic pathways to disease as well as the development of vaccines and therapeutics for controlling and preventing severe disease outcomes. Among the human coronaviruses, SARS-CoV is the best characterized both biochemically and molecularly (146), displays robust growth in primary and continuous cell lines, has well-characterized young and/or aged animal models of human disease (106-108, 147), and has well-documented reverse genetic approaches (2, 160), and vaccine efficacy has been studied in young and aged animal models (26, 155). Importantly, the first coronavirus antagonists of innate immunity were documented in the SARS-CoV genome (59), pioneering similar studies in other important human and animal CoV genomes (156).
CORONAVIRUS VIRION AND GENOME ORGANIZATION
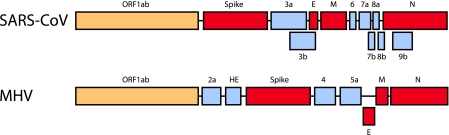
The coronavirus genome is about 30 kb in size and generally encodes three broad protein classes (76, 112) (Fig. 1). Virions are roughly 90 to 120 nm in diameter and contain a lipid bilayer surrounding a helical nucleocapsid structure that protects the genome. Several structural proteins are encoded within the intact virion, and these include the 180/90-kDa spike (S) protein, a ∼50- to 60-kDa nucleocapsid (N) protein, an 8-kDa envelope (E) protein, and the ∼23-kDa membrane (M) protein. A second class of genes encodes the accessory or group-specific proteins. These proteins are typically unique to each particular coronavirus strain and differ among MHV, SARS-CoV, and the other human coronaviruses. In the case of SARS-CoV, ORF3a, ORF6, and ORF7a/b are reported virion proteins (49, 117, 121), although the literature lacks consensus (86). The exact functions for most of these proteins are unclear; however, these proteins may influence viral pathogenesis and disease outcomes, regulate specific virus-host interactions, and/or promote the development of an intracellular environment that is conducive for efficient virus growth. In many but not all cases, the deletion of the group-specific open reading frames (ORFs) has a minimal impact on in vitro replication but may or may not attenuate pathogenesis in vivo, depending on the virus strain (24).
FIG. 1.
SARS-CoV and MHV genome structure. The genome structure of coronaviruses is very conserved among all known coronaviruses. In each coronavirus, the N-terminal two-thirds of the genome encodes the nonstructural proteins, also called the replicase proteins (orange box). The C-terminal one-third of the genome encodes the structural (red boxes) and accessory (gray boxes) ORFs. The structural ORFs encode the spike, envelope (E), membrane (M), and nucleocapsid (N) proteins. Each coronavirus has similar structural ORFs in their genomes. The accessory ORFs, in gray, are unique to each coronavirus. There is no sequence or structural similarities between the MHV and SARS-CoV accessory proteins.
The final broad category of coronavirus genes encodes the replicase proteins, also called nonstructural proteins. These viral proteins are encoded in the 5′-most two-thirds of the coronavirus genome, which is essential for polyprotein processing, replicase complex formation, and efficient virus replication (9). More recently, it is becoming clear that the replicase proteins may also encode critical virulence determinants that not only regulate virus growth efficiency but also directly engage the host proteome to directly potentiate pathogenic mechanisms and regulate disease severity (27, 170).
INNATE IMMUNE INDUCTION
The innate immune response is a coordinated series of signaling pathways in all nucleated cells that function to thwart an invading pathogen's replication and disease potential. From interferon (IFN) induction and secretion to the recruitment of macrophages and DCs to sites of infection, the system functions to restrict tissue tropism and spread, dampen virus replication efficiency, and eliminate virally infected cells (reviewed in references 36, 119, and 133). In addition to IFN regulatory factor 3 (IRF3) in the IFN pathway, another critical signaling protein for the innate immune response is nuclear factor of kappa light polypeptide gene enhancer in B cells (NF-κB). NF-κB is activated during viral infection from the sensing of viral replication products and via cytokine secretion from macrophages and DCs (37). This leads to a broad induction of the innate immune response while also fine-tuning the response to remove virus while not harming the cells.
The modulation of these pathways is critical for virus survival, as evidenced by the many viruses that express proteins that block various key effector proteins in these pathways and from the increased disease severity noted in many gene knockout animals. Protein products from many viruses including the NSP1, ORF6, and N proteins from SARS-CoV (34, 56, 59, 84, 144, 156), the NS1 protein from influenza virus (114, 164), the VP35 and VP24 proteins from Ebola virus (5, 104), the leader protein from picornaviruses (25, 42, 138), and the V proteins from Nipah and Hendra viruses (3, 111) have each been identified as being immunomodulating proteins. Each protein blocks one or more key signaling proteins in the IFN and NF-κB pathways to enhance viral replication and pathogenesis. The influenza virus NS1 protein affects the IRF3 signaling pathway as well as mRNA stability and trafficking (60, 164). In contrast, VP35 from Ebola virus and ORF6 from SARS-CoV block nuclear import (34, 59), while the V proteins from Nipah and Hendra viruses induce signal transducer and activator of transcription (STAT) protein degradation (111). Picornavirus leader blocks by binding to promoter regions of IFN genes via a zinc finger domain and inhibits transcription (25, 42, 138). Each protein antagonizes the innate immune response but uses different tools and targets to achieve these goals. An understanding of how each antagonist affects the innate immune response illuminates key interactions between the host signaling pathway components and the virus. In addition, these studies pinpoint key host cell components that function to regulate virus replication and pathogenesis, providing novel targets for the development of antiviral compounds.
Interferon
Initially identified as a protein that interfered with virus replication by Isaacs and Lindenmann in 1957, IFNs are key regulators of viral replication in mammals and birds (52, 125, 128). Recently, several new types of IFN have been identified, adding to the previously known type I IFNs (alpha and beta), type II IFN (gamma), and now type III IFN (lambda) (4, 58, 136) (Table 1). The lesser known omega IFN (IFN-ω), IFN-δ, and IFN-τ are part of the type I IFN family (7, 67, 77, 122, 148), although the role of these new and lesser-studied IFNs in coronavirus replication and pathogenesis is unknown.
TABLE 1.
Types of IFN identified
| IFN type | IFN class(es) | Receptor | Pathway |
|---|---|---|---|
| I | α, β, δ, ω | IFN-α/β (IFNAR1, IFNAR2) | ISGF3 (STAT1/STAT2/IRF9) |
| I | τ (also known as ε) | IFN-α/β (IFNAR1, IFNAR2) | Pregnancy specific |
| II | γ | IFN-γ (IFNGR1, IFNGR2) | STAT1/STAT1 homodimers |
| III | λ | 1L-28α/IL-1ORβ | ISGF3 |
The induction of IFN by various cell types is an initial signal to the host that a foreign invader has infected its cells. The production of IFN by cells induces neighboring cells to remodel the intracellular environment by producing a range of antiviral proteins, aiding in a block of viral replication (115). Several hundred proteins are induced following IFN production and signaling (125). The mechanism of how these IFN-induced proteins block virus infection is mostly unknown, but some well-studied players include the RNA-dependent protein kinase (PKR), RNase L, the Mx gene product, and the IFN-stimulated gene 15 (ISG15) protein (45). Different IFNs induce different genes both quantitatively and qualitatively in cell culture. For example, IFN-α and IFN-λ (also called interleukin-28 [IL-28]) use the same signaling pathway to induce ISGs, although they induce distinct but overlapping sets of genes at different time points after stimulation (58, 136). How the architecture of the signaling pathways regulates differential gene expression is unknown, especially in cell types derived from different tissues, and represents a major unresolved question in the regulation of the antiviral response (75). Some ISGs have been shown to affect coronavirus replication and will be discussed below in more detail. IFN induction is known to occur from any nucleated cell, although the type and amount of IFN produced vary with the cell type (19).
Interferon Induction
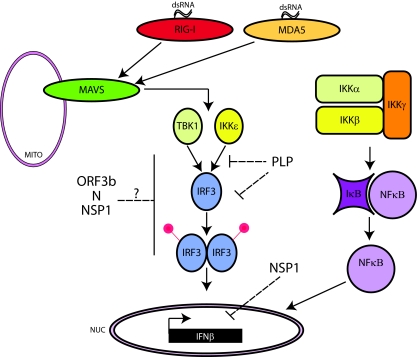
There are several pathways by which virus infection and IFN alter host gene expression patterns in the cell (Fig. 2). The most direct pathway is via the presence of viral double-stranded RNA (dsRNA) in the cell. Two dsRNA-sensing molecules, RIG-I and MDA5, reside in the cytoplasm of nucleated cells and are able to discern viral dsRNA from host dsRNA (57, 169). The mechanism of discrimination is thought to be regulated by the recognition of the 5′ cap on the mRNA by RIG-I, while MDA5 may recognize uncapped mRNAs that are often produced during the replication of some viruses. Host mRNA is always capped and thus invisible to the dsRNA-sensing machinery (97). There is new evidence that different viruses activate each sensor uniquely, although the details of the recognition and activation processes are still under study (46, 70, 73, 97, 129). Once one or both of these sensors are activated, they interact with a mitochondrial membrane protein called MAVS (mitochondrial antiviral) (also called IPS1, Cardif, and VISA) (73, 97, 129). MAVS function is dependent on mitochondrial localization; however, the reason for that dependence is unknown (46, 70). Once MAVS becomes activated, the signaling cascade continues through the kinases TBK1 and IKKɛ (reviewed in reference 118). IKKɛ signaling has also been shown to induce many IFN-inducible proteins via the STAT1 pathway as well (132). These kinases integrate the upstream signaling events and then directly phosphorylate the transcription factor IRF3, which normally resides in the cytoplasm. IRF3 phosphorylation promotes dimerization, and the IRF3 dimer is imported into the nucleus by a karyopherin (importin) complex where, with the help of other transcription factors like NF-κB, the complex initiates the transcription of IFN-β. The IFN-β protein is then secreted from the cell and can act in either an autocrine or a paracrine fashion to amplify the IFN response (125).
FIG. 2.
The innate immune induction pathway and SARS-CoV. The major proteins in the innate induction pathway are shown as they signal from sensing a pathogen to induction of IFN-β. Initially, RIG-I and MDA5 sense dsRNA in the cytoplasm, produced as a by-product of RNA virus replication. They signal to the mitochondrial membrane protein MAVS, which in turn activates the kinases TBK1 and IKKɛ. These kinases then phosphorylate IRF3, causing it to dimerize and traffic to the nucleus, where it, along with NF-κB, induces the transcription of IFN-β. SARS-CoV proteins actively modulate this pathway. ORF3b, N, and NSP1 affect the signal transduction pathway that activates IRF3 by an unknown mechanism. NSP1 also affects the mRNA stability of IFN-β transcript. The PLP of SARS-CoV also affects IRF3 and NF-κB. PLP blocks the phosphorylation of IRF3 and its activation while also blocking the activation of NF-κB. This results in a block in IFN-β induction.
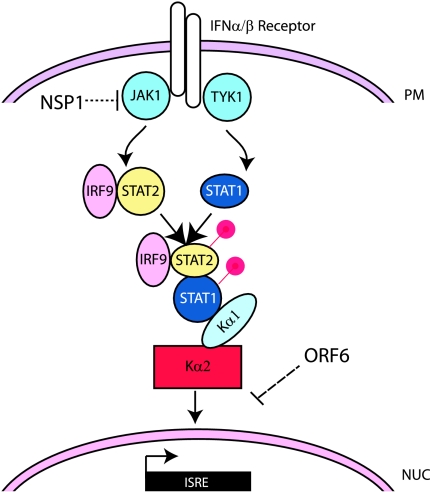
On the surface of most nucleated cells, the type I IFN receptor functions to detect and bind IFN-α and -β and initiate the IFN signal transduction pathway (Fig. 3). This pathway begins with the recruitment of kinases (JAK1 and TYK2) to the cytoplasmic tail of the activated receptor. Once activated, these kinases phosphorylate the cytoplasmic STAT proteins. When IFN-β is the inducer, STAT1 and STAT2 are phosphorylated, leading to the formation of the ISGF3 complex, consisting of STAT1, STAT2, and IRF9. This complex, now active, is imported into the nucleus via the karyopherin alpha 1 (KPNA1)/karyopherin beta 1 (KPNB1) proteins (80). Once in the nucleus, RAN-GTP is hydrolyzed in the karyopherin complex, and the cargo is released. In the nucleus, ISGF3 induces the expression of several hundred genes, the promoters of which all contain a specific element, the IFN-stimulated response element (ISRE). The many genes induced in response to IFN-β prime the cell to block the replication of the virus. This may affect either the cells already infected or neighboring cells, which bind and induce antiviral proteins in response to secreted IFN (68).
FIG. 3.
The JAK/STAT signaling pathway and SARS-CoV. The JAK/STAT pathway responds to type I IFN secreted from neighboring cells. The IFN-α/β receptor binds to either IFN-α or -β and signals to the Jak1 or TYK1 kinase. These kinases phosphorylate both STAT1 and STAT2. This phosphorylation induces the complex formation of STAT1/STAT2/IRF9 (the ISGF3 complex) and targets the complex to the nucleus with the help of the import factors KPNA1 (Kα1) and KPNB1 (Kβ1). Once in the nucleus, the complex turns on genes containing an ISRE in their promoter. SARS-CoV proteins have been shown to affect this pathway. NSP1 reduces the levels of Jak1 in the cytoplasm and affects it kinase activity. ORF6 blocks the nuclear import of ISGF3 by reducing the free Kβ1 in the cytoplasm and retaining it on the ER/Golgi membrane.
TLR Signaling
The Toll-like receptor (TLR) sensors also function as a major pathway for the induction of IFN. The TLR family is composed of 13 related paralogs in the human genome, each with overlapping but distinct binding and signaling potentials. From single-stranded RNA, dsRNA, CpG DNA, bacterial, and viral protein components, the TLRs signal the cell to activate various antipathogen defenses (133). Viral sensing is mediated primarily via recognition patterns encrypted in TLRs 3, 7, 8, and 9 (1). TLRs are membrane-spanning proteins and recognize pathogen-associated molecular patterns (1). Pathogen-associated molecular patterns are small structures and motifs common to viruses or bacteria, such as lipopolysaccharides, dsRNA, or unmethylated CpG DNA. The key to their recognition is that TLRs reside in the endosomal membrane, with their receptor ends facing the lumen of the vesicles, which allows the proteins to sense the incoming virus after it is endocytosed into the cell. TLRs, which use different adapter proteins, in comparison to RIGI and MDA5, use the scaffolding proteins TRIF and MyD88 to signal through MAVS and IRF3 before inducing the expression of IFN. Cervantes-Barrangan et al. demonstrated that TLR7 is critical for IFN induction in plasmacytoid DCs (pDCs) when infected with MHV (12). These pathways may work in concert or independently to induce IFN expression and establish the antiviral response. Continued investigations into the similarities and differences of TLR interactions with SARS-CoV and other coronaviruses will be beneficial for understanding their role in coronavirus pathogenesis.
Innate Immune Response and Coronaviruses
SARS-CoV and MHV, both group II coronaviruses, have been shown to interact intimately with the innate immune response. As reported for other viruses, the coronavirus-host interplay and response outcome are highly dependent on cell type, virus concentration, and whether the results are obtained from in vitro or in vivo experimentations. MHV and SARS-CoV induce various degrees of IFN as well, depending on all of these variables (139, 168); type I IFN expression was found to be induced following SARS-CoV infection of macrophages (157), DCs (66), and epithelial cell lines (Caco2, MA104, and 293) (90) as well as in vivo in mice, macaques, and humans (38, 106, 141). Various degrees of induction have been found in these cell lines, from very high early levels that diminish during infection, to very high levels only late in infection, to no induction of IFN at all. The reasons behind these differences are currently not well understood.
The interaction between SARS-CoV and the innate immune system appears to be tightly balanced during infection. In macrophages and DCs, SARS-CoV has been shown to induce type I IFN mRNA production (11, 123); however, the extent of induction and the number of cells inducing IFN were not analyzed. From these experiments, it is unclear whether the virus is actually entering the cells or just binding signaling molecules on the surface. Additionally, several groups have performed microarrays on infected cells, and despite finding little, if any, type I IFN mRNA expression, they noted a significant induction of many downstream cytokines and chemokine genes that were normally induced after the induction of type I IFN (18). Although it is currently unclear which signaling pathways are most responsible for protection from SARS-CoV pathogenesis, these data illustrate the complexity and wealth of undiscovered signaling networks in this area.
SARS-CoV AND HUMAN INFECTIONS
During and after the SARS epidemic in 2003, patient samples were analyzed to determine the molecular basis for viral pathogenesis in humans. Using these samples, the age of the patient was shown to dramatically influence disease outcome and host cytokine responses following infection. For example, levels of IFN-γ, IL-4, and IL-10 were increased only in convalescent SARS-CoV patients, while IL-6 was upregulated in most SARS-CoV patients (167). There was a correlation between the expression of the cytokines IL-6, IL-8, and monocyte chemoattractant protein 1 and IP-10 with higher mortality and severity of disease (103).
In those same studies, microarray-based quantification of cytokine transcripts was performed using peripheral blood mononuclear cells from infected patients. Increased levels of proinflammatory cytokines were seen across all infected patients, but interestingly, little IFN-α or -β was found. This may be due to the stability of the transcripts in the infected cells, or it may be due to the IFN-antagonistic differences in SARS-CoV-infected cells. It is unknown how cytokine induction during infection is modified by SARS-CoV and how this relates to the pathogenesis of the virus. Analyses of human samples have identified markers of severity such as human leukocyte antigen class I (B*0703) and class II (DRB1*0301), mannose binding protein, OAS1, and MxA (43, 87, 163, 165). Reproduction of these findings has been difficult due to small sample sizes and limitations of access of the samples (153, 163). More recently, IL-12 RB1, Rantes, and IFN-γ polymorphisms were linked to SARS disease severity (16, 88). Undoubtedly, more genomic links to disease will be found in the future.
IFN Evasion Mechanisms
Targeted gene knockout studies have demonstrated the critical importance of the innate immune system in regulating virus-induced disease outcomes. Consequently, it is very likely that this network of host genes has exerted considerable pressure to select for coevolutionary changes in viral gene function. One way that viruses have evolved to combat the innate immune system is to encode protein functions that block various aspects of the host response to viral infection. SARS-CoV and MHV have been shown to produce large amounts of IFN-inducing dsRNA while not inducing IFN in those cells (139, 145). Although mechanistically unclear, several possibilities are under investigation. The viruses may be encoding proteins (IFN antagonists) that directly inhibit the signaling pathways that are responsible for IFN induction, as described above. Significant amounts of data support the hypothesis that coronaviruses encode one or more IFN antagonist genes. The virus may also be able to evade detection by replicating in privileged sites that are compartmentalized from the sensing and signaling machinery. For example, coronaviruses have been shown to replicate on double-membrane vesicles which may protect the viral RNA from sensing (98, 126). One hypothesis is that double-membrane vesicles protect the dsRNA by internalizing the RNA into the vesicle, while others hypothesized that the viral replication machinery somehow binds and protects this RNA. In support of this hypothesis, when cells are infected with either MHV or SARS-CoV, there is no induction of IFN-β. However, when superinfected with Sendai virus or treated with poly(I:C), after the initial coronavirus infection, there is substantial IFN-β induction (33, 168). When MHV was used to infect conventional DCs in vitro (12), low levels of type I IFN mRNA were reported. Similarly low levels of type I IFN were found in the brains of infected mice in vivo (105). Spiegel and Weber showed that in SARS-CoV-infected Caco2 and HEK 293 cells, little, if any, IFN-α, -β, or -λ was induced (124). Interestingly, IP-10 and IL-8 were also strongly induced in Caco2 cells but not HEK 293 cells. The difference in cytokine induction between these two cell lines is not understood but may be important for the underlying differences seen in humans and mice as well. These data suggest that one or more pathways inhibited by coronavirus infection can still be activated by other inducers or that when tested in cell culture models, the blocking of IFN signaling pathways is incomplete. Continued studies of the location and dynamics of replicating and translated viral RNA in the cell will aid in understanding this phenomenon.
Structural Proteins
Coronavirus virions are made up of several virally encoded proteins including the spike, envelope, membrane, and nucleocapsid proteins. The spike (S) protein is a heavily glycosylated type I membrane protein that decorates the outside of the virion and is essential for attachment to the host receptor. For SARS-CoV, angiotensin-converting enzyme 2 (ACE2) (69) is the receptor. For MHV, the receptor is Ceacam1 (carcinoembryonic antigen-related cell adhesion molecule 1) (149). Importantly, the S glycoprotein has been shown to affect several cell signaling pathways in vitro. Using overexpression plasmids in HeLa cells, the S glycoprotein was proposed to inhibit host cell translation by interaction with eIF3f (152). In addition, S glycoprotein induces apoptosis in VeroE6 cells (17), stimulates macrophages to produce proinflammatory cytokines (IL-6 and tumor necrosis factor alpha) in Raw 264.7 cells (143), and activates COX-2 expression in VeroE6, HEK 293, and Cos7 cells (72). Spike has also been shown to downregulate ACE2 on the surface of cells. After binding on the surface, both ACE2 and the virion are internalized (63), potentially reducing the amount of ACE2 on the surface and preventing the cleavage of ACE. Importantly, ACE is a proinflammatory molecule that normally functions in wound repair and healing of acute lung injury (62). After SARS-CoV infection, the amount of ACE2 on the surface of cells is reduced via internalization with the SARS-CoV virion (51). This reduces the level of signaling that ACE would normally induce when bound to cells and slows the repair process, potentially exacerbating the lung pathogenesis of SARS-CoV (62).
A correlation with ACE2 binding/internalization and pathogenesis has recently been demonstrated for SARS-CoV. Rockx et al. and Sheahan et al. were able generate viruses that contain the Urbani genetic backbone but that have different zoonotic or human strain spike glycoproteins. Their spike sequences were based on the evolutionary changes that occurred during animal-to-human SARS-CoV cross-species transmission and adaptation to the human host (110, 120). Those authors showed that while different spike proteins can utilize the same ACE2 receptor for efficient entry, different spike proteins cause various levels of pathology in the lung, especially in aged rodent models. They hypothesized that not only are the binding and internalization of ACE2 an important aspect of SARS-CoV pathogenesis, but the modulation of the ACE2 signaling network is critical for the acute lung injury seen in survivors of SARS-CoV infection.
The envelope (E) protein is a small transmembrane protein that is expressed abundantly but incorporated in small quantities into the virion. In cells, it was proposed to coordinate with the M glycoprotein to regulate efficient budding via direct or indirect interactions with the nucleocapsid (9). The E protein has also been shown to form ion channel-like pores on membranes in infected cells, but the role of the pores in pathogenesis is unknown (135). While essential for group I coronavirus virion formation, the E protein can be deleted from group II coronaviruses like SARS-CoV and MHV, curtailing efficient assembly, release, and pathogenesis in vivo, presumably by limiting virus titers and the rate of spread (32, 65). Consonant with these findings, recombinant SARS-CoVs lacking the E protein are attenuated in vivo (23).
The membrane (M) protein is an abundant small protein that is essential for virion assembly (170). Recently, a SARS-CoV M mutation that confers efficient dissemination and increased replication in primary human kidney cells, presumably by interactions with key cellular targets that efficiently enhance the assembly and release of infectious virions, has been identified (91). Unfortunately, the mechanism by which these mutations enhance virion yields and/or infectivity is unknown. The nucleocapsid protein is an internal virion protein that binds to and packages the viral RNA into the virion (39). The N protein is also tightly associated with replication complexes and may function as a chaperone protein to regulate efficient subgenomic mRNA synthesis (171).
The viral and cellular components of the virion proteome are the subject of some controversy. A comprehensive proteomic approach was undertaken by Neuman et al. using a mass spectrophoretic analysis of purified virions (86). This comprehensive approach demonstrated that ORF6 and ORF7a were not localized in purified SARS-CoV virions, challenging previous reports in the literature (48, 49). Surprisingly, the replicase proteins NSP2, NSP3, and NSP5 and group-specific proteins ORF3b and ORF9b were found abundantly in the purified virions (86). Among these, NSP2, ORF3b, and ORF9b are nonessential and can be deleted from the genome without seriously affecting replication efficiency in vitro (40, 162). Importantly, NSP3 and NSP5 encode proteolytic activities that process the ORF1a/b polyprotein into individual replicase components and potentially process a variety of cellular proteins as well (170). At this time, the precise location and function of these proteins in the virion are unclear and under investigation. Additionally, many host cellular proteins were identified in the virion, including many ribosomal, nuclear, endoplasmic reticulum (ER), Golgi apparatus, and plasma membrane proteins. The reason for the disparity among these three studies is unknown but may represent differences in the stringency of virion preparation or may be due to differences in the culturing conditions. One complication is that SARS-CoV produces noninfectious particles as well as fully functional virions, potentially confounding interpretations of data from mass spectrophoretic versus immunoelectron microscopy studies. Continued work using antibodies to each identified protein in virions will be necessary to determine viral and cellular proteome participation in virion assembly, maturation, and infectivity. The role of this plethora of host proteins in coronavirus virion formation, function, infection, and pathogenesis remains unknown but might help virions escape immune recognition.
Nucleocapsid Protein
The SARS-CoV and MHV nucleocapsid proteins have been shown to affect different aspects of the innate immune response and appear to modulate several signaling pathways in the cell. Many of these studies were performed using overexpression constructs in isolation and have not been confirmed in the context of virus infection. He et al. showed that N is able to induce AP-1 signaling in vitro (44). Kopecky-Bromberg et al. showed that SARS-CoV N was able to block the induction of reporter gene expression from an IFN-β promoter and also block NF-κB signaling (59). They indicated that the N protein is also able to inhibit an ISRE promoter in response to Sendai virus infection but not IFN-β treatment. The mechanism by which this inhibition is occurring is unknown and under investigation. When expressed in a chimeric vaccinia virus, the MHV N protein was also shown to inhibit the activation of PKR, a strongly antiviral protein, in the cytoplasm (156). Ye et al. further demonstrated that type I IFN was not induced and that PKR was not activated in MHV-infected cells (156). PKR activation normally leads to a block in protein synthesis by phosphorylating the alpha subunit of the translation factor eIF2. While N does not itself prevent PKR activation, it alters PKR's function such that it no longer signals properly. The N proteins between these two group 2 coronaviruses are quite conserved, so it will be interesting to determine if they both encode overlapping functions during infection or whether there really are distinct inhibitory mechanisms.
Of note, the various domains of coronavirus nucleocapsid may affect many pathways similar to that of influenza virus NS1. Both proteins have RNA binding and protein-protein interaction domains that may be important for their host cell functions. NS1 has been shown to affect mRNA nuclear export, mRNA stability, IRF3 activation, and STAT1 and NF-κB signaling (14, 60, 61, 82, 101, 102, 164). Various domains of NS1 affect each pathway differently but together produce a multilayered modulation of various host signaling pathways. Thus, each domain of nucleocapsid may be performing multiple functions on multiple pathways during infection as well. The MHV nucleocapsid protein has also been shown to have a role in the induction of viral hepatitis (89). Ning et al. showed that the nucleocapsid from MHV type 3 induces fgl2 prothrombinase to induce fibrin deposition and fulminant liver failure (89). This leads to the hypothesis that nucleocapsids from other coronaviruses may encode IFN antagonism and signal transduction-modulating activities that are yet to be uncovered.
Many questions remain concerning the role of the N protein in replication and pathogenesis. Will each coronavirus nucleocapsid gene encode activities similar to those described here or encode distinct activities that target other pathways in the cell? Each coronavirus also targets and infects different cell types and organs. Consequently, N protein functions may play different roles in pathogenesis in different cell types and species. SARS-CoV infects primarily ciliated airway cells and type II pneumocytes, while MHV infects neurons, glia, and liver cells, depending on the strain. Is this difference correlated with different evolving functions of N in disease? Since N can be differentially phosphorylated on many sites in the protein, it is possible that different N isomers/posttranslational modifications function differently to potentiate disease outcomes? We believe that the current molecular virology and reverse genetic platforms provide an approach to elucidate the complex virus-host interactions that lead to disease.
Accessory Proteins
Each coronavirus genome encodes a distinct set of accessory proteins that are somehow important in the virus life cycle. SARS-CoV encodes ORFs 3a, 3b, 6, 7a, 7b, 8, and 9b and an ORF internal to N (31). MHV encodes ORFs 2a, 4, and 5a. There is no overlap or homology among these proteins across these two strains. To date, the only coronavirus accessory proteins shown to antagonize IFN pathways are derived from SARS-CoV (34, 59). In work described previously by Kopecky-Bromberg et al., ORF3b and ORF6 were shown to block IFN induction and IFN signaling. While the mechanism for ORF3b antagonism has not been identified, the mechanism by which ORF6 antagonizes host innate immune signaling has been clearly elucidated. Studies from our group and others (34, 59, 131) have shown how ORF6 blocks the IFN signaling arm of the innate immune response.
ORF6 is a 63-amino-acid ER/Golgi membrane protein that has its C-terminal tail facing the cytoplasm and its N terminus either in the ER lumen or associated with the ER membrane. We found that ORF6 was able to block the nuclear import of the STAT1/STAT2/IRF9 and STAT1/STAT1 complexes in the presence of IFN-β or IFN-γ treatment, respectively (34). ORF6 expression resulted in the reduction of STAT1-dependent gene induction. The C-terminal 10 amino acids were critical for this import block, mediated by a recruitment of nuclear import factors to the ER/Golgi membrane (Fig. 3). Using a variety of approaches, we found that KPNA2 was specifically bound to the C-terminal tail of ORF6. This retained KPNA2 at the ER/Golgi membrane and, subsequently, recruited KPNB1 to the ER/Golgi membrane as well. The recruitment of KPNB1 onto membrane complexes limited the bioavailability of KPNB1, an essential component that was needed for the nuclear import of STAT1 complexes. Consequently, ORF6 blocked the nuclear import of the STAT1 complexes. ORF6 may affect other signaling pathways since KPNB1 is a common component of the classical nuclear import pathways. ORF6 has also been shown to increase the pathogenesis of the normally nonlethal MHV-A59 virus (96). In vitro studies with the MHV/ORF6 virus showed that ORF6 expression increased the level of virus production from cells compared to the level of expression of wild-type (WT) virus. Recently, Hussain et al. showed that ORF6 expression is able to block proteins containing classical import signals but not proteins that use nonimportin nuclear import mechanisms, in agreement with our work showing that ORF6 blocks KPNB1-mediated nuclear import (50). MHV's accessory proteins have not been implicated in impacting the innate immune response; however, they may play a role in MHV's capacity to evade the innate immunosensing proteins, as the deletion of some but not all of these proteins impacts in vivo pathogenesis (24).
The clear association of SARS-CoV ORF6 and Ebola virus VP24 with the host nuclear import pathway identifies it as a key site of virus-host interactions that modulate and regulate the intracellular environment. By modulating the kinetics of nuclear import during infection, the virus controls innate immune, adaptive immune, apoptotic, and cell stress signaling networks. While the mechanism may vary, it would be surprising if other pathogenic viruses do not modulate the same pathways as those seen in SARS-CoV during their infection process. The levels of karyopherin proteins vary by cell type and age of the cells as well, so the effect of ORF6 in cells may depend on the amount of import factors available for binding and inhibition (99, 154).
Replicase Proteins
The 5′-end ∼20 kb of each coronavirus genome consists of two large, overlapping ORFs, which encode 15 to 16 proteins that are localized in replication complexes and are thought to function primarily in regulating viral replication and transcription in infected cells. About ∼13 kb from the 5′ end, a ribosomal frameshifting site is encoded, which allows the efficient translation of the downstream ORF1b, encoding replicase, helicase, and a variety of novel RNA-processing enzymes whose exact function in coronavirus transcription is unclear. Once translated, the polyprotein produced from ORF1a or ORF1a/b is processed by virally encoded proteases, the papain like-protease (PLP) encoded in NSP3 and the main protease (Mpro) encoded by NSP5. Each protease cleaves the polyprotein at specific sites to liberate the 16 polyproteins used in SARS-CoV replication (reviewed in references 9 and 79). It has been postulated that the intermediate versions of the cleaved polyprotein may also have some special functions, although direct support for this hypothesis has recently been challenged in the literature (28, 29, 116).
Although there is significant homology between the replicase proteins (or nonstructural proteins [NSPs]) of coronaviruses, in general, the replicase proteins nsp1 to nsp3 at the 5′ end of the genome are most heterogeneous (74). In contrast, nsp7 to nsp10 are highly conserved among coronaviruses. Different coronavirus family members also encode different numbers of functional PLPs. For example, SARS-CoV encodes a single PLP, while MHV encodes two PLPs in NSP3. In the latter, both PLPs are active and preferentially target different cleavage sites to liberate nsp1 to nsp3 from the ORF1a polyprotein. Most NSPs are essential for replication, although in SARS-CoV and MHV, NSP2 is nonessential, portions of NSP1 have also been deleted, and various mutations have been made in other replicase proteins (for example, NSP15) (41).
NSP1
Historically, the replicase proteins were thought to function exclusively in RNA synthesis, influencing viral disease outcomes as a function of virus load. The finding that several coronavirus replicase proteins function in innate immune evasion was surprising. This observation changes the old paradigm by directly implicating replicase protein function in pathogenesis and virulence. Structurally, SARS-CoV NSP1 is a 20-kDa protein that is localized in the cytoplasm of infected cells (20). While all coronaviruses encode an NSP1-like protein, SARS-CoV NSP1 may encode a variety of unique functions. Using overexpression plasmids, both NSP1 and NSP3 have been shown to encode innate immune antagonism activities. Kamitani et al. and Narayanan et al. demonstrated that NSP1 was able to block IFN-β mRNA induction but did not antagonize the IRF3 signaling pathway (56, 84). NSP1 expression degraded not only IFN-β mRNA but also several endogenous cellular mRNAs as well. As SARS-CoV-infected cells also degraded cellular mRNA, those authors proposed that the nsp1 degradation of host mRNA is an important mechanism of blocking host antiviral defenses. In a recent paper by Narayanan et al., they described an NSP1 mutant virus that loses its ability to degrade host mRNA and, interestingly, that induces a large amount of IFN-β during infection (84).
Other work suggested that SARS-CoV NSP1 can directly antagonize IFN induction. Wathelet et al. demonstrated that NSP1 inhibits the signal transduction pathways involving IRF3, STAT1, and NF-κB (144). They showed that NSP1 is able to block STAT1 phosphorylation and IRF3 dimerization and affects cell cycle progression without inducing apoptosis. Using an NSP1 mutant, those authors showed that the recombinant virus was more sensitive to IFN treatment, agreeing with the previously published work showing that NSP1 is an IFN antagonist. Interestingly, those authors did not observe the mRNA degradation phenotype seen in the previous studies, suggesting that additional work is needed to delineate the exact role of NSP1 as an innate immune antagonist and its potential role in selective host mRNA degradation.
Recent work by Zust et al. also showed that NSP1 in MHV affects virus survival in vivo (172). They found that the deletion of the first 99 amino acids of NSP1 in MHV produced a virus that grew efficiently in tissue culture but did not grow well in mice. In fact, 100% of the WT virus-infected mice died, while the mutant virus-infected mice all survived. IFN induction in the NSP1 mutant-infected macrophages was increased compared to that in WT virus-infected macrophages. Those authors showed that in type I IFN receptor knockout mice, the nsp1 mutant virus replicates to WT levels. These data suggest a role for NSP1 in MHV pathogenesis in mice; however, its exact mechanism of action is unknown.
NSP3
SARS-CoV NSP3 is one of the larger cleaved products of ORF1a in the SARS-CoV genome, encoding a multifunctional >200-kDa protein that includes several predicted glycosylation sites. Several different functional domains of NSP3 have been identified, including a poly(ADP-ribose) binding domain, protease, deubiquitinase, and de-ISGylase. Wathelet et al. also demonstrated that the overexpression of NSP3 antagonized host IFN responses as well (144). Using a luciferase-based screen, Wathelet et al. showed that SARS-CoV NSP3 expression blocked IFN-β induction after cells were infected with Sendai virus. This investigation did not, however, identify which domain(s) of NSP3 was responsible for that block. In support of these findings, Devaraj et al. showed that the SARS-CoV PLP domain of NSP3 was responsible for the antagonism of the IFN-β response (27). Using transfection experiments with only the PLP domain, those authors demonstrated that PLP blocked IFN-β induction via Sendai virus infection and poly(I:C) treatment by inhibiting phosphorylation, dimerization, and nuclear import of IRF3. Furthermore, those authors showed that PLP and IRF3 coprecipitated and that the interaction was independent from the enzymatic function of PLP. Mutations in the active site of the protease that blocked PLP activity did not affect IRF3 binding.
Recent unpublished work from our laboratory suggests that while PLP is an IFN antagonist, the mechanism of action is different than what was described previously by Devaraj et al. We find that PLP is able to inhibit IRF3-mediated IFN induction and block IRF3 phosphorylation; however, we also find that PLP is able to inhibit NF-κB gene induction (M. Frieman et al., unpublished data). Devaraj et al. showed that PLP had no effect on the NF-κB pathway (27). While PLP blocks IRF3 phosphorylation, it increases the phosphorylation of IκBα, the inhibitor of NF-κB activation. We also find that identical cleavage site mutants used in the study reported Devaraj et al. produce various degrees of IFN and NF-κB inhibition in our assays, with some mutants maintaining the IFN antagonist activity and others losing their antagonist activity. Interestingly, we can show that the ubiquitin-like domain at the N terminus of PLP is a determinant of IFN antagonist function. The deletion of the ubiquitin-like domain from PLP leads to a loss of IFN-antagonistic activity for both IRF3 and NF-κB pathways; however, the protease remains active. We are currently identifying the mechanism of action of PLP in vitro and in vivo.
Cell Types Involved in the SARS-CoV Interferon Response
During the initial infection with SARS-CoV, two large groups of cell types figure prominently in the innate immune response. SARS-CoV utilizes the ACE2 protein on the surface of epithelial cells as its cellular receptor, and its expression is essential for virus entry (69). Cell types that are prominent targets for SARS-CoV infection include bronchiolar epithelial cells, especially ciliated epithelial cells and alveolar type II pneumocytes. A second broad class of important contributors to pathogenesis includes the infiltrating immune cells that are composed primarily of macrophages, neutrophils, and DCs. DCs can be further divided into myeloid-type DCs (mDCs) and pDCs, both of which have been shown to be important in clearing viral infections (1, 53, 83). When activated, pDCs produce large amounts of type I IFN to signal neighboring cells of the incoming infection. mDCs do not produce type I IFN to the extent of pDCs but are important in the acquired immune response by secreting chemokines necessary for the activation of B cells and T cells.
SARS-CoV has also been shown to use DC-SIGN and L-SIGN as coreceptors for entry (54, 78). Each one is abundantly expressed in DC populations, and coronavirus infection of DCs in culture shows an interesting interaction between virus and cell. In pDCs, SARS-CoV and MHV did not progress to a productive infection and did not produce progeny viruses; however, incubating pDCs with virus induced the production of large amounts of type I IFN (12). Although SARS-CoV infection was not analyzed, MHV was shown to signal through TLR7 and MyD88, which were essential for IFN-β expression (12). TLR7, a membrane protein localized primarily in endosomes, may function as a crucial sentinel for the detection of coronavirus infection. TLR7 signals through IRF7 to induce IFN-β expression. In the absence of intracellular infection and gene expression, it is unlikely that the IFN antagonist proteins will be produced in quantities that would block IFN signaling or expression, although nsp3, the N protein, and perhaps ORF6 may be present in sufficient quantities with the virion to provide low-level antagonism. Importantly, pDC depletion with antibodies prior to MHV infection diminished the quantity of IFN produced during infection and resulted in increased viral replication and disease. These data demonstrated that the initial sensing of coronaviruses by the innate immune machinery might be the critical step in protecting the host from infection.
AGING AND PATHOGENESIS
The impact of host senescence on viral pathogenesis is beginning to be understood at the molecular level. In the case of influenza virus, the rates of hospitalizations of people under 5 and over 65 years of age are essentially the same, although importantly, those over 65 years of age are 35 times more likely to die from infection (21). During the SARS-CoV epidemic, the elderly were more likely to die from SARS-CoV infection than any other age group (13, 71). The elderly exhibit a decrease in their adaptive immune response with increasing age, which parallels their increase in morbidity and mortality after infection (64). Studies of the responsiveness of the innate immune response in the elderly are limited and have shown conflicting results. In an aged mouse infection with coxsackievirus B3 and Newcastle disease virus, the levels of IFN-α and -β were decreased in aged mice compared to those in young mice (113). The results with poly(I:C) treatment, a potent type I IFN inducer, in old mice have demonstrated either a decrease or an increase in IFN production (55, 166). A better understanding of the relationship between aging and both the innate and adaptive immune responses is necessary to understand the increased pathogenesis of SARS-CoV in the elderly population.
While investigating the aging phenomenon in SARS-CoV susceptibility, we have begun to identify how aged mice respond to SARS-CoV vaccines. We have shown that SARS-CoV vaccine efficacy wanes in aged mice when mice were both vaccinated when young and challenged when old. More importantly, vaccinated aged animals mounted extremely poor immune responses and were not protected from challenge with a homologous virus (26). Additionally, aged mice are more susceptible to SARS-CoV infection and produced more lung damage and pathology than young mice (106). The reasons for the increased pathogenesis of SARS-CoV in aged animals remains to be elucidated but reflects a more global issue of understanding the molecular mechanisms of disease and reversing disproportionate disease burdens that are noted in the elderly after many viral infections. Current work on identifying susceptibility alleles specific to aged individuals compared to young individuals may aid in our understanding of the increase in infectivity and pathogenesis in the elderly.
CONCLUSION
Coronaviruses have emerged as an important model system to study virus-host interactions. Their large genomic RNA encodes a variety of novel proteins, many of which have solved structures containing novel protein folds but only predicted functions in virus replication and pathogenesis. The tools of molecular virology, robust sequence databases of human and animal strains, well-characterized mouse models of severe end-stage lung disease, age-related susceptibility phenotypes, and the ability to develop systems biology and genetic approaches will allow a detailed understanding of coronavirus pathogenesis and emergence. This will make it a choice model for illuminating novel virus-host interactions that regulate severe disease outcomes in young and immunosenescent animals and humans.
The molecular biology tools of reverse genetics allow the unprecedented genetic interrogation of the large RNA virus genome. Infectious clones for SARS-CoV, bat SARS-CoV, MHV, transmissible gastroenteritis virus, infectious bronchitis virus, and human coronaviruses OC43, NL63, and 229E will enable the research community to manipulate these genomes and identify the role of each protein in virus-host interactions, pathogenesis, and disease (10, 26, 95, 110, 134, 158-161). Despite the identification of several innate immune antagonist genes in the Coronaviridae genome, the role of these genes in regulating disease outcomes and pathogenesis remains an important focus for future investigations. Although it is well recognized that changes within the S glycoprotein regulate the initial steps in cross-species transmission via receptor interactions, adaptation and virulence determinants are generally poorly understood.
SARS-CoV emergence was associated with a large number of adaptive changes within nsp3, nsp4, nsp9, NSP14, NSP15, S, M, ORF3a, E, N, and ORF8, which may influence disease outcomes (15). It is not clear whether innate coronavirus antagonism genes play a key role in adaptation and transmission to new host species as has been described for poxviruses (142). Surprisingly, SARS-CoV and the less virulent human coronavirus NL63 both utilize the same receptor, ACE2, for docking and entry into cells, yet NL63 is significantly less pathogenic than the SARS-CoV (47). These data suggest that novel functions are encoded within the SARS-CoV genome to enhance virulence. Do the novel accessory ORF functions encoded within the highly pathogenic SARS-CoV represent the key ingredients to high virulence? Equally important is the notion that the replicase proteins encoded within ORF1a and ORF1b are multifunctional, regulating replication efficiency while engaging select host cell signaling machinery to promote an intracellular environment conducive to virus growth and pathogenesis. Given the availability of novel tools, multiple novel proteins, robust genetic systems, and reagents, coronaviruses provide a novel opportunity to investigate interesting interactions between host cells and viral pathogens.
Future Directions
Comparison of these different systems necessitates a holistic view of disease processes. While we may start at the level of what happens in a tissue culture flask, it is the intimate interactions between the infected cell and its neighbors that determine the course of disease. The secretion of cytokines and chemokines from infected tissue and the subsequent recruitment of macrophages, DCs, neutrophils, and eosinophils will be critically important for the survival of the host. How each virus modulates this response may very well represent the distinguishing features between a lethal or mild, acute or chronic, highly pathogenic virus infection.
Systems Biology
Use of a systems biology approach may enable us to finally understand the intricate dynamics between the cell and virus proteome that eventually constitute a particular disease phenotype. By comparing the transcriptional profiles of a lung epithelial cell during the course of infection with SARS-CoV, influenza virus, Ebola virus, human coronavirus NL63, or respiratory syncytial virus may allow an understanding of the pathways and proteins that are involved in the host response. The integration of transcriptome data, proteomic profiles, and detailed interaction networks between viral and cellular proteins should provide a systems view of the intricate communication networks regulating virus infection at the cellular level. Platform approaches for each of these key system components are available, providing high-throughput identification of the interaction networks and the impact on host expression (reviewed in reference 130). Additionally, lipidomic and metabolomic profiles may be useful in adding more data to our understanding of the infection. Comparing the responses to infection in lungs from SARS-CoV-, H5N1 influenza virus-, Ebola virus-, and respiratory syncytial virus-infected hosts can further our understanding of how the each virus modulates the innate immune response.
Animal Models and Systems Genetics
The obvious limitation to all of these studies is the animal model system. For SARS-CoV, a mouse-adapted virus reproduces the acute disease progression found in the more severe cases of SARS-CoV infection in humans (106). Moreover, animal models that reproduce the complex age-related phenotype and that progress to acute respiratory distress syndrome provide novel resources for illuminating pathogenic mechanisms in vivo (110). In the various SARS-CoV models, the genetic background of the mouse heavily influences pathogenic outcomes. The role of host susceptibility alleles in coronavirus pathogenesis, as well as most viral pathogens, is heavily understudied and represents a major target for future research. Mouse resources that allow an understanding of the roles of polygenic traits in viral pathogenesis are now becoming available, like the Collaborative Cross mice (109). Using these mouse resources, detailed systems genetic approaches can be developed to understand the mechanisms by which host susceptibility alleles interact with the virus proteome to potentiate different disease outcomes.
The use of a systems genetic approach may be our best chance at identifying the host genes responsible for coronavirus disease and age-related susceptibility phenotypes. The ability to manipulate both the viral and host genetic systems will allow a further understanding of the parameters of pathogenesis and virus control for both mouse and human disease. With the power of systems biology and genetics, an intricate understanding of the elegant interplay between a pathogen and its host should result in novel targets for antiviral drugs and therapies in the future.
REFERENCES
- 1.Akira, S., and H. Hemmi. 2003. Recognition of pathogen-associated molecular patterns by TLR family. Immunol. Lett. 8585-95. [DOI] [PubMed] [Google Scholar]
- 2.Almazan, F., M. L. Dediego, C. Galan, D. Escors, E. Alvarez, J. Ortego, I. Sola, S. Zuniga, S. Alonso, J. L. Moreno, A. Nogales, C. Capiscol, and L. Enjuanes. 2006. Construction of a severe acute respiratory syndrome coronavirus infectious cDNA clone and a replicon to study coronavirus RNA synthesis. J. Virol. 8010900-10906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrejeva, J., K. S. Childs, D. F. Young, T. S. Carlos, N. Stock, S. Goodbourn, and R. E. Randall. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. USA 10117264-17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ank, N., H. West, and S. R. Paludan. 2006. IFN-lambda: novel antiviral cytokines. J. Interf. Cytok. Res. 26373-379. [DOI] [PubMed] [Google Scholar]
- 5.Basler, C. F., X. Wang, E. Muhlberger, V. Volchkov, J. Paragas, H. D. Klenk, A. Garcia-Sastre, and P. Palese. 2000. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc. Natl. Acad. Sci. USA 9712289-12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beckham, J. D., A. Cadena, J. Lin, P. A. Piedra, W. P. Glezen, S. B. Greenberg, and R. L. Atmar. 2005. Respiratory viral infections in patients with chronic, obstructive pulmonary disease. J. Infect. 50322-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bekisz, J., H. Schmeisser, J. Hernandez, N. D. Goldman, and K. C. Zoon. 2004. Human interferons alpha, beta and omega. Growth Factors 22243-251. [DOI] [PubMed] [Google Scholar]
- 8.Birch, C. J., H. J. Clothier, A. Seccull, T. Tran, M. C. Catton, S. B. Lambert, and J. D. Druce. 2005. Human coronavirus OC43 causes influenza-like illness in residents and staff of aged-care facilities in Melbourne, Australia. Epidemiol. Infect. 133273-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brian, D. A., and R. S. Baric. 2005. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 2871-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casais, R., V. Thiel, S. G. Siddell, D. Cavanagh, and P. Britton. 2001. Reverse genetics system for the avian coronavirus infectious bronchitis virus. J. Virol. 7512359-12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castilletti, C., L. Bordi, E. Lalle, G. Rozera, F. Poccia, C. Agrati, I. Abbate, and M. R. Capobianchi. 2005. Coordinate induction of IFN-alpha and -gamma by SARS-CoV also in the absence of virus replication. Virology 341163-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cervantes-Barragan, L., R. Züst, F. Weber, M. Spiegel, K. S. Lang, S. Akira, V. Thiel, and B. Ludewig. 2007. Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood 1091131-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan-Yeung, M., and R. H. Xu. 2003. SARS: epidemiology. Respirology 8S9-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chien, C. Y., Y. Xu, R. Xiao, J. M. Aramini, P. V. Sahasrabudhe, R. M. Krug, and G. T. Montelione. 2004. Biophysical characterization of the complex between double-stranded RNA and the N-terminal domain of the NS1 protein from influenza A virus: evidence for a novel RNA-binding mode. Biochemistry 431950-1962. [DOI] [PubMed] [Google Scholar]
- 15.Chinese SARS Molecular Epidemiology Consortium. 2004. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science 3031666-1669. [DOI] [PubMed] [Google Scholar]
- 16.Chong, W. P., W. K. Ip, G. H. Tso, M. W. Ng, W. H. Wong, H. K. Law, R. W. Yung, E. Y. Chow, K. L. Au, E. Y. Chan, W. Lim, J. S. Peiris, and Y. L. Lau. 2006. The interferon gamma gene polymorphism +874 A/T is associated with severe acute respiratory syndrome. BMC Infect. Dis. 682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chow, K. Y., Y. S. Yeung, C. C. Hon, F. Zeng, K. M. Law, and F. C. Leung. 2005. Adenovirus-mediated expression of the C-terminal domain of SARS-CoV spike protein is sufficient to induce apoptosis in Vero E6 cells. FEBS Lett. 5796699-6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cinatl, J., Jr., G. Hoever, B. Morgenstern, W. Preiser, J. U. Vogel, W. K. Hofmann, G. Bauer, M. Michaelis, H. F. Rabenau, and H. W. Doerr. 2004. Infection of cultured intestinal epithelial cells with severe acute respiratory syndrome coronavirus. Cell. Mol. Life Sci. 612100-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colonna, M., A. Krug, and M. Cella. 2002. Interferon-producing cells: on the front line in immune responses against pathogens. Curr. Opin. Immunol. 14373-379. [DOI] [PubMed] [Google Scholar]
- 20.Connor, R. F., and R. L. Roper. 2007. Unique SARS-CoV protein nsp1: bioinformatics, biochemistry and potential effects on virulence. Trends Microbiol. 1551-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Couch, R. B., J. A. Kasel, W. P. Glezen, T. R. Cate, H. R. Six, L. H. Taber, A. L. Frank, S. B. Greenberg, J. M. Zahradnik, and W. A. Keitel. 1986. Influenza: its control in persons and populations. J. Infect. Dis. 153431-440. [DOI] [PubMed] [Google Scholar]
- 22.Cruz, C. D., H. Palosaari, J. P. Parisien, P. Devaux, R. Cattaneo, T. Ouchi, and C. M. Horvath. 2006. Measles virus V protein inhibits p53 family member p73. J. Virol. 805644-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeDiego, M. L., E. Alvarez, F. Almazan, M. T. Rejas, E. Lamirande, A. Roberts, W. J. Shieh, S. R. Zaki, K. Subbarao, and L. Enjuanes. 2007. A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J. Virol. 811701-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Haan, C. A., P. S. Masters, X. Shen, S. Weiss, and P. J. Rottier. 2002. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology 296177-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Los Santos, T., S. de Avila Botton, R. Weiblen, and M. J. Grubman. 2006. The leader proteinase of foot-and-mouth disease virus inhibits the induction of beta interferon mRNA and blocks the host innate immune response. J. Virol. 801906-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deming, D., T. Sheahan, M. Heise, B. Yount, N. Davis, A. Sims, M. Suthar, J. Harkema, A. Whitmore, R. Pickles, A. West, E. Donaldson, K. Curtis, R. Johnston, and R. Baric. 2006. Vaccine efficacy in senescent mice challenged with recombinant SARS-CoV bearing epidemic and zoonotic spike variants. PLoS Med. 3e525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Devaraj, S. G., N. Wang, Z. Chen, Z. Chen, M. Tseng, N. Barretto, R. Lin, C. J. Peters, C. T. Tseng, S. C. Baker, and K. Li. 2007. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J. Biol. Chem. 28232208-32221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Donaldson, E. F., R. L. Graham, A. C. Sims, M. R. Denison, and R. S. Baric. 2007. Analysis of murine hepatitis virus strain A59 temperature-sensitive mutant TS-LA6 suggests that nsp10 plays a critical role in polyprotein processing. J. Virol. 817086-7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Donaldson, E. F., A. C. Sims, R. L. Graham, M. R. Denison, and R. S. Baric. 2007. Murine hepatitis virus replicase protein nsp10 is a critical regulator of viral RNA synthesis. J. Virol. 816356-6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.El-Sahly, H. M., R. L. Atmar, W. P. Glezen, and S. B. Greenberg. 2000. Spectrum of clinical illness in hospitalized patients with “common cold” virus infections. Clin. Infect. Dis. 3196-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fischer, F., D. Peng, S. T. Hingley, S. R. Weiss, and P. S. Masters. 1997. The internal open reading frame within the nucleocapsid gene of mouse hepatitis virus encodes a structural protein that is not essential for viral replication. J. Virol. 71996-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fischer, F., C. F. Stegen, P. S. Masters, and W. A. Samsonoff. 1998. Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J. Virol. 727885-7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frieman, M., M. Heise, and R. Baric. 2008. SARS coronavirus and innate immunity. Virus Res. 133101-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frieman, M., B. Yount, M. Heise, S. A. Kopecky-Bromberg, P. Palese, and R. S. Baric. 2007. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 819812-9824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gale, M. J., Jr., M. J. Korth, N. M. Tang, S. L. Tan, D. A. Hopkins, T. E. Dever, S. J. Polyak, D. R. Gretch, and M. G. Katze. 1997. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology 230217-227. [DOI] [PubMed] [Google Scholar]
- 36.Garcia-Sastre, A., and C. A. Biron. 2006. Type 1 interferons and the virus-host relationship: a lesson in detente. Science 312879-882. [DOI] [PubMed] [Google Scholar]
- 37.Ghosh, S., M. J. May, and E. B. Kopp. 1998. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 16225-260. [DOI] [PubMed] [Google Scholar]
- 38.Glass, W. G., K. Subbarao, B. Murphy, and P. M. Murphy. 2004. Mechanisms of host defense following severe acute respiratory syndrome-coronavirus (SARS-CoV) pulmonary infection of mice. J. Immunol. 1734030-4039. [DOI] [PubMed] [Google Scholar]
- 39.Goldsmith, C. S., K. M. Tatti, T. G. Ksiazek, P. E. Rollin, J. A. Comer, W. W. Lee, P. A. Rota, B. Bankamp, W. J. Bellini, and S. R. Zaki. 2004. Ultrastructural characterization of SARS coronavirus. Emerg. Infect. Dis. 10320-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Graham, R. L., A. C. Sims, S. M. Brockway, R. S. Baric, and M. R. Denison. 2005. The nsp2 replicase proteins of murine hepatitis virus and severe acute respiratory syndrome coronavirus are dispensable for viral replication. J. Virol. 7913399-13411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guarino, L. A., K. Bhardwaj, W. Dong, J. Sun, A. Holzenburg, and C. Kao. 2005. Mutational analysis of the SARS virus Nsp15 endoribonuclease: identification of residues affecting hexamer formation. J. Mol. Biol. 3531106-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hato, S. V., C. Ricour, B. M. Schulte, K. H. Lanke, M. de Bruijni, J. Zoll, W. J. Melchers, T. Michiels, and F. J. van Kuppeveld. 2007. The mengovirus leader protein blocks interferon-alpha/beta gene transcription and inhibits activation of interferon regulatory factor 3. Cell. Microbiol. 92921-2930. [DOI] [PubMed] [Google Scholar]
- 43.He, L., Y. Ding, Q. Zhang, X. Che, Y. He, H. Shen, H. Wang, Z. Li, L. Zhao, J. Geng, Y. Deng, L. Yang, J. Li, J. Cai, L. Qiu, K. Wen, X. Xu, and S. Jiang. 2006. Expression of elevated levels of pro-inflammatory cytokines in SARS-CoV-infected ACE2+ cells in SARS patients: relation to the acute lung injury and pathogenesis of SARS. J. Pathol. 210288-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He, R., A. Leeson, A. Andonov, Y. Li, N. Bastien, J. Cao, C. Osiowy, F. Dobie, T. Cutts, M. Ballantine, and X. Li. 2003. Activation of AP-1 signal transduction pathway by SARS coronavirus nucleocapsid protein. Biochem. Biophys. Res. Commun. 311870-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hiscott, J., N. Grandvaux, S. Sharma, B. R. Tenoever, M. J. Servant, and R. Lin. 2003. Convergence of the NF-kappaB and interferon signaling pathways in the regulation of antiviral defense and apoptosis. Ann. N. Y. Acad. Sci. 1010237-248. [DOI] [PubMed] [Google Scholar]
- 46.Hiscott, J., J. Lacoste, and R. Lin. 2006. Recruitment of an interferon molecular signaling complex to the mitochondrial membrane: disruption by hepatitis C virus NS3-4A protease. Biochem. Pharmacol. 721477-1484. [DOI] [PubMed] [Google Scholar]
- 47.Hofmann, H., K. Pyrc, L. van der Hoek, M. Geier, B. Berkhout, and S. Pohlmann. 2005. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. USA 1027988-7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang, C., N. Ito, C. T. Tseng, and S. Makino. 2006. Severe acute respiratory syndrome coronavirus 7a accessory protein is a viral structural protein. J. Virol. 807287-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang, C., C. J. Peters, and S. Makino. 2007. Severe acute respiratory syndrome coronavirus accessory protein 6 is a virion-associated protein and is released from 6 protein-expressing cells. J. Virol. 815423-5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hussain, S., S. Perlman, and T. M. Gallagher. 2008. Severe acute respiratory syndrome coronavirus protein 6 accelerates murine hepatitis virus infections by more than one mechanism. J. Virol. 827212-7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Imai, Y., K. Kuba, S. Rao, Y. Huan, F. Guo, B. Guan, P. Yang, R. Sarao, T. Wada, H. Leong-Poi, M. A. Crackower, A. Fukamizu, C. C. Hui, L. Hein, S. Uhlig, A. S. Slutsky, C. Jiang, and J. M. Penninger. 2005. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature 436112-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Isaacs, A., and J. Lindenmann. 1957. Virus interference. I. The interferon. Proc. R. Soc. Lond. B Biol. Sci. 147258-267. [PubMed] [Google Scholar]
- 53.Ito, T., Y. H. Wang, and Y. J. Liu. 2005. Plasmacytoid dendritic cell precursors/type I interferon-producing cells sense viral infection by Toll-like receptor (TLR) 7 and TLR9. Springer Semin. Immunopathol. 26221-229. [DOI] [PubMed] [Google Scholar]
- 54.Jeffers, S. A., S. M. Tusell, L. Gillim-Ross, E. M. Hemmila, J. E. Achenbach, G. J. Babcock, W. D. Thomas, Jr., L. B. Thackray, M. D. Young, R. J. Mason, D. M. Ambrosino, D. E. Wentworth, J. C. Demartini, and K. V. Holmes. 2004. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA 10115748-15753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang, J., D. Gross, S. Nogusa, P. Elbaum, and D. M. Murasko. 2005. Depletion of T cells by type I interferon: differences between young and aged mice. J. Immunol. 1751820-1826. [DOI] [PubMed] [Google Scholar]
- 56.Kamitani, W., K. Narayanan, C. Huang, K. Lokugamage, T. Ikegami, N. Ito, H. Kubo, and S. Makino. 2006. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc. Natl. Acad. Sci. USA 10312885-12890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kash, J. C., E. Muhlberger, V. Carter, M. Grosch, O. Perwitasari, S. C. Proll, M. J. Thomas, F. Weber, H.-D. Klenk, and M. G. Katze. 2006. Global suppression of the host antiviral response by Ebola- and Marburgviruses: increased antagonism of the type I interferon response is associated with enhanced virulence. J. Virol. 803009-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kontsek, P., G. Karayianni-Vasconcelos, and E. Kontsekova. 2003. The human interferon system: characterization and classification after discovery of novel members. Acta Virol. 47201-215. [PubMed] [Google Scholar]
- 59.Kopecky-Bromberg, S. A., L. Martinez-Sobrido, M. Frieman, R. A. Baric, and P. Palese. 2007. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 81548-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Krug, R. M. 1993. The regulation of export of mRNA from nucleus to cytoplasm. Curr. Opin. Cell Biol. 5944-949. [DOI] [PubMed] [Google Scholar]
- 61.Krug, R. M., W. Yuan, D. L. Noah, and A. G. Latham. 2003. Intracellular warfare between human influenza viruses and human cells: the roles of the viral NS1 protein. Virology 309181-189. [DOI] [PubMed] [Google Scholar]
- 62.Kuba, K., Y. Imai, and J. M. Penninger. 2006. Angiotensin-converting enzyme 2 in lung diseases. Curr. Opin. Pharmacol. 6271-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kuba, K., Y. Imai, S. Rao, H. Gao, F. Guo, B. Guan, Y. Huan, P. Yang, Y. Zhang, W. Deng, L. Bao, B. Zhang, G. Liu, Z. Wang, M. Chappell, Y. Liu, D. Zheng, A. Leibbrandt, T. Wada, A. S. Slutsky, D. Liu, C. Qin, C. Jiang, and J. M. Penninger. 2005. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 11875-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kumar, R., and E. A. Burns. 2008. Age-related decline in immunity: implications for vaccine responsiveness. Expert Rev. Vaccines 7467-479. [DOI] [PubMed] [Google Scholar]
- 65.Kuo, L., and P. S. Masters. 2003. The small envelope protein E is not essential for murine coronavirus replication. J. Virol. 774597-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Law, H. K., C. Y. Cheung, H. Y. Ng, S. F. Sia, Y. O. Chan, W. Luk, J. M. Nicholls, J. S. Peiris, and Y. L. Lau. 2005. Chemokine up-regulation in SARS-coronavirus-infected, monocyte-derived human dendritic cells. Blood 1062366-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lefevre, F., M. Guillomot, S. D'Andrea, S. Battegay, and C. La Bonnardiere. 1998. Interferon-delta: the first member of a novel type I interferon family. Biochimie 80779-788. [DOI] [PubMed] [Google Scholar]
- 68.Levy, D. E., D. S. Kessler, R. Pine, N. Reich, and J. E. Darnell, Jr. 1988. Interferon-induced nuclear factors that bind a shared promoter element correlate with positive and negative transcriptional control. Genes Dev. 2383-393. [DOI] [PubMed] [Google Scholar]
- 69.Li, W., M. J. Moore, N. Vasilieva, J. Sui, S. K. Wong, M. A. Berne, M. Somasundaran, J. L. Sullivan, K. Luzuriaga, T. C. Greenough, H. Choe, and M. Farzan. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426450-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li, X. D., L. Sun, R. B. Seth, G. Pineda, and Z. J. Chen. 2005. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc. Natl. Acad. Sci. USA 10217717-17722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu, M., W. N. Liang, Q. Chen, X. Q. Xie, J. Wu, X. He, and Z. J. Liu. 2006. Risk factors for SARS-related deaths in 2003, Beijing. Biomed. Environ. Sci. 19336-339. [PubMed] [Google Scholar]
- 72.Liu, M., Y. Yang, C. Gu, Y. Yue, K. K. Wu, J. Wu, and Y. Zhu. 2007. Spike protein of SARS-CoV stimulates cyclooxygenase-2 expression via both calcium-dependent and calcium-independent protein kinase C pathways. FASEB J. 211586-1596. [DOI] [PubMed] [Google Scholar]
- 73.Loo, Y. M., J. Fornek, N. Crochet, G. Bajwa, O. Perwitasari, L. Martinez-Sobrido, S. Akira, M. A. Gill, A. Garcia-Sastre, M. G. Katze, and M. Gale, Jr. 2008. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 82335-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Magiorkinis, G., E. Magiorkinis, D. Paraskevis, A. M. Vandamme, M. Van Ranst, V. Moulton, and A. Hatzakis. 2004. Phylogenetic analysis of the full-length SARS-CoV sequences: evidence for phylogenetic discordance in three genomic regions. J. Med. Virol. 74369-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Marcello, T., A. Grakoui, G. Barba-Spaeth, E. S. Machlin, S. V. Kotenko, M. R. MacDonald, and C. M. Rice. 2006. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology 1311887-1898. [DOI] [PubMed] [Google Scholar]
- 76.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 3001399-1404. [DOI] [PubMed] [Google Scholar]
- 77.Martal, J. L., N. M. Chene, L. P. Huynh, R. M. L'Haridon, P. B. Reinaud, M. W. Guillomot, M. A. Charlier, and S. Y. Charpigny. 1998. IFN-tau: a novel subtype I IFN1. Structural characteristics, non-ubiquitous expression, structure-function relationships, a pregnancy hormonal embryonic signal and cross-species therapeutic potentialities. Biochimie 80755-777. [DOI] [PubMed] [Google Scholar]
- 78.Marzi, A., T. Gramberg, G. Simmons, P. Moller, A. J. Rennekamp, M. Krumbiegel, M. Geier, J. Eisemann, N. Turza, B. Saunier, A. Steinkasserer, S. Becker, P. Bates, H. Hofmann, and S. Pohlmann. 2004. DC-SIGN and DC-SIGNR interact with the glycoprotein of Marburg virus and the S protein of severe acute respiratory syndrome coronavirus. J. Virol. 7812090-12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Masters, P. S. 2006. The molecular biology of coronaviruses. Adv. Virus Res. 66193-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McBride, K. M., and N. C. Reich. 2003. The ins and outs of STAT1 nuclear transport. Sci. STKE 2003RE13. [DOI] [PubMed] [Google Scholar]
- 81.Meylan, E., J. Curran, K. Hofmann, D. Moradpour, M. Binder, R. Bartenschlager, and J. Tschopp. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 4371167-1172. [DOI] [PubMed] [Google Scholar]
- 82.Min, J. Y., S. Li, G. C. Sen, and R. M. Krug. 2007. A site on the influenza A virus NS1 protein mediates both inhibition of PKR activation and temporal regulation of viral RNA synthesis. Virology 363236-243. [DOI] [PubMed] [Google Scholar]
- 83.Nakano, H., M. Yanagita, and M. D. Gunn. 2001. CD11c(+)B220(+)Gr-1(+) cells in mouse lymph nodes and spleen display characteristics of plasmacytoid dendritic cells. J. Exp. Med. 1941171-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Narayanan, K., C. Huang, K. Lokugamage, W. Kamitani, T. Ikegami, C. T. Tseng, and S. Makino. 2008. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J. Virol. 824471-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Reference deleted.
- 86.Neuman, B. W., J. S. Joseph, K. S. Saikatendu, P. Serrano, A. Chatterjee, M. A. Johnson, L. Liao, J. P. Klaus, J. R. Yates III, K. Wüthrich, R. C. Stevens, M. J. Buchmeier, and P. Kuhn. 2008. Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J. Virol. 825279-5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ng, M. H., K. M. Lau, L. Li, S. H. Cheng, W. Y. Chan, P. K. Hui, B. Zee, C. B. Leung, and J. J. Sung. 2004. Association of human-leukocyte-antigen class I (B*0703) and class II (DRB1*0301) genotypes with susceptibility and resistance to the development of severe acute respiratory syndrome. J. Infect. Dis. 190515-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ng, M. W., G. Zhou, W. P. Chong, L. W. Lee, H. K. Law, H. Zhang, W. H. Wong, S. F. Fok, Y. Zhai, R. W. Yung, E. Y. Chow, K. L. Au, E. Y. Chan, W. Lim, J. S. Peiris, F. He, and Y. L. Lau. 2007. The association of RANTES polymorphism with severe acute respiratory syndrome in Hong Kong and Beijing Chinese. BMC Infect. Dis. 750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ning, Q., M. Liu, P. Kongkham, M. M. Lai, P. A. Marsden, J. Tseng, B. Pereira, M. Belyavskyi, J. Leibowitz, M. J. Phillips, and G. Levy. 1999. The nucleocapsid protein of murine hepatitis virus type 3 induces transcription of the novel fgl2 prothrombinase gene. J. Biol. Chem. 2749930-9936. [DOI] [PubMed] [Google Scholar]
- 90.Okabayashi, T., H. Kariwa, S. Yokota, S. Iki, T. Indoh, N. Yokosawa, I. Takashima, H. Tsutsumi, and N. Fujii. 2006. Cytokine regulation in SARS coronavirus infection compared to other respiratory virus infections. J. Med. Virol. 78417-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pacciarini, F., S. Ghezzi, F. Canducci, A. Sims, M. Sampaolo, E. Ferioli, M. Clementi, G. Poli, P. G. Conaldi, R. Baric, and E. Vicenzi. 2008. Persistent replication of severe acute respiratory syndrome coronavirus in human tubular kidney cells selects for adaptive mutations in the membrane protein. J. Virol. 825137-5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Parisien, J. P., J. F. Lau, J. J. Rodriguez, B. M. Sullivan, A. Moscona, G. D. Parks, R. A. Lamb, and C. M. Horvath. 2001. The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2. Virology 283230-239. [DOI] [PubMed] [Google Scholar]
- 93.Park, M. S., M. L. Shaw, J. Munoz-Jordan, J. F. Cros, T. Nakaya, N. Bouvier, P. Palese, A. Garcia-Sastre, and C. F. Basler. 2003. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J. Virol. 771501-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Patrick, D. M., M. Petric, D. M. Skowronski, R. Guasparini, T. F. Booth, M. Krajden, P. McGeer, N. Bastien, L. Gustafson, J. Dubord, D. Macdonald, S. T. David, L. F. Srour, R. Parker, A. Andonov, J. Isaac-Renton, N. Loewen, G. McNabb, A. McNabb, S. H. Goh, S. Henwick, C. Astell, J. P. Guo, M. Drebot, R. Tellier, F. Plummer, and R. C. Brunham. 2006. An outbreak of human coronavirus OC43 infection and serological cross-reactivity with SARS coronavirus. Can. J. Infect. Dis. Med. Microbiol. 17330-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Penzes, Z., C. Wroe, T. D. Brown, P. Britton, and D. Cavanagh. 1996. Replication and packaging of coronavirus infectious bronchitis virus defective RNAs lacking a long open reading frame. J. Virol. 708660-8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pewe, L., H. Zhou, J. Netland, C. Tangadu, H. Olivares, L. Shi, D. Look, T. Gallagher, and S. Perlman. 2006. A SARS-CoV-specific protein enhances virulence of an attenuated strain of mouse hepatitis virus. Adv. Exp. Med. Biol. 581493-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pichlmair, A., O. Schulz, C. P. Tan, T. I. Naslund, P. Liljestrom, F. Weber, and C. Reis e Sousa. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314997-1001. [DOI] [PubMed] [Google Scholar]
- 98.Prentice, E., W. G. Jerome, T. Yoshimori, N. Mizushima, and M. R. Denison. 2004. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 27910136-10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pujol, G., H. Soderqvist, and A. Radu. 2002. Age-associated reduction of nuclear protein import in human fibroblasts. Biochem. Biophys. Res. Commun. 294354-358. [DOI] [PubMed] [Google Scholar]
- 100.Pyrc, K., M. F. Jebbink, B. Berkhout, and L. van der Hoek. 2004. Genome structure and transcriptional regulation of human coronavirus NL63. Virol. J. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Qian, X. Y., F. Alonso-Caplen, and R. M. Krug. 1994. Two functional domains of the influenza virus NS1 protein are required for regulation of nuclear export of mRNA. J. Virol. 682433-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Qiu, Y., and R. M. Krug. 1994. The influenza virus NS1 protein is a poly(A)-binding protein that inhibits nuclear export of mRNAs containing poly(A). J. Virol. 682425-2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Reghunathan, R., M. Jayapal, L.-Y. Hsu, H.-H. Chng, D. Tai, B. P. Leung, and A. J. Melendez. 2005. Expression profile of immune response genes in patients with severe acute respiratory syndrome. BMC Immunol. 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Reid, S. P., L. W. Leung, A. L. Hartman, O. Martinez, M. L. Shaw, C. Carbonnelle, V. E. Volchkov, S. T. Nichol, and C. F. Basler. 2006. Ebola virus VP24 binds karyopherin α1 and blocks STAT1 nuclear accumulation. J. Virol. 805156-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rempel, J. D., S. J. Murray, J. Meisner, and M. J. Buchmeier. 2004. Mouse hepatitis virus neurovirulence: evidence of a linkage between S glycoprotein expression and immunopathology. Virology 31845-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Roberts, A., D. Deming, C. D. Paddock, A. Cheng, B. Yount, L. Vogel, B. D. Herman, T. Sheahan, M. Heise, G. L. Genrich, S. R. Zaki, R. Baric, and K. Subbarao. 2007. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog. 3e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Roberts, A., E. W. Lamirande, L. Vogel, J. P. Jackson, C. D. Paddock, J. Guarner, S. R. Zaki, T. Sheahan, R. Baric, and K. Subbarao. 2008. Animal models and vaccines for SARS-CoV infection. Virus Res. 13320-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Roberts, A., C. Paddock, L. Vogel, E. Butler, S. Zaki, and K. Subbarao. 2005. Aged BALB/c mice as a model for increased severity of severe acute respiratory syndrome in elderly humans. J. Virol. 795833-5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Roberts, A., F. Pardo-Manuel de Villena, W. Wang, L. McMillan, and D. W. Threadgill. 2007. The polymorphism architecture of mouse genetic resources elucidated using genome-wide resequencing data: implications for QTL discovery and systems genetics. Mamm. Genome 18473-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rockx, B., T. Sheahan, E. Donaldson, J. Harkema, A. Sims, M. Heise, R. Pickles, M. Cameron, D. Kelvin, and R. Baric. 2007. Synthetic reconstruction of zoonotic and early human severe acute respiratory syndrome coronavirus isolates that produce fatal disease in aged mice. J. Virol. 817410-7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rodriguez, J. J., J. P. Parisien, and C. M. Horvath. 2002. Nipah virus V protein evades alpha and gamma interferons by preventing STAT1 and STAT2 activation and nuclear accumulation. J. Virol. 7611476-11483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 3001394-1399. [DOI] [PubMed] [Google Scholar]
- 113.Rytel, M. W. 1987. Effect of age on viral infections: possible role of interferon. J. Am. Geriatr. Soc. 351092-1099. [DOI] [PubMed] [Google Scholar]
- 114.Salvatore, M., C. F. Basler, J. P. Parisien, C. M. Horvath, S. Bourmakina, H. Zheng, T. Muster, P. Palese, and A. Garcia-Sastre. 2002. Effects of influenza A virus NS1 protein on protein expression: the NS1 protein enhances translation and is not required for shutoff of host protein synthesis. J. Virol. 761206-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sawicki, S. G., D. L. Sawicki, D. Younker, Y. Meyer, V. Thiel, H. Stokes, and S. G. Siddell. 2005. Functional and genetic analysis of coronavirus replicase-transcriptase proteins. PLoS Pathog. 1e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schaecher, S. R., J. M. Mackenzie, and A. Pekosz. 2007. The ORF7b protein of severe acute respiratory syndrome coronavirus (SARS-CoV) is expressed in virus-infected cells and incorporated into SARS-CoV particles. J. Virol. 81718-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sen, G. C. 2001. Viruses and interferons. Annu. Rev. Microbiol. 55255-281. [DOI] [PubMed] [Google Scholar]
- 119.Seth, R. B., L. Sun, and Z. J. Chen. 2006. Antiviral innate immunity pathways. Cell Res. 16141-147. [DOI] [PubMed] [Google Scholar]
- 120.Sheahan, T., B. Rockx, E. Donaldson, A. Sims, R. Pickles, D. Corti, and R. Baric. 2008. Mechanisms of zoonotic severe acute respiratory syndrome coronavirus host range expansion in human airway epithelium. J. Virol. 822274-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Shen, S., P. S. Lin, Y. C. Chao, A. Zhang, X. Yang, S. G. Lim, W. Hong, and Y. J. Tan. 2005. The severe acute respiratory syndrome coronavirus 3a is a novel structural protein. Biochem. Biophys. Res. Commun. 330286-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Spencer, T. E., T. L. Ott, and F. W. Bazer. 1996. Tau-interferon: pregnancy recognition signal in ruminants. Proc. Soc. Exp. Biol. Med. 213215-229. [DOI] [PubMed] [Google Scholar]
- 123.Spiegel, M., K. Schneider, F. Weber, M. Weidmann, and F. T. Hufert. 2006. Interaction of severe acute respiratory syndrome-associated coronavirus with dendritic cells. J. Gen. Virol. 871953-1960. [DOI] [PubMed] [Google Scholar]
- 124.Spiegel, M., and F. Weber. 2006. Inhibition of cytokine gene expression and induction of chemokine genes in non-lymphatic cells infected with SARS coronavirus. Virol. J. 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67227-264. [DOI] [PubMed] [Google Scholar]
- 126.Stertz, S., M. Reichelt, M. Spiegel, T. Kuri, L. Martinez-Sobrido, A. Garcia-Sastre, F. Weber, and G. Kochs. 2007. The intracellular sites of early replication and budding of SARS-coronavirus. Virology 361304-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Symons, J. A., A. Alcami, and G. L. Smith. 1995. Vaccinia virus encodes a soluble type I interferon receptor of novel structure and broad species specificity. Cell 81551-560. [DOI] [PubMed] [Google Scholar]
- 128.Takaoka, A., and H. Yanai. 2006. Interferon signalling network in innate defence. Cell. Microbiol. 8907-922. [DOI] [PubMed] [Google Scholar]
- 129.Takeuchi, O., and S. Akira. 2008. MDA5/RIG-I and virus recognition. Curr. Opin. Immunol. 2017-22. [DOI] [PubMed] [Google Scholar]
- 130.Tan, S. L., G. Ganji, B. Paeper, S. Proll, and M. G. Katze. 2007. Systems biology and the host response to viral infection. Nat. Biotechnol. 251383-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tangudu, C., H. Olivares, J. Netland, S. Perlman, and T. Gallagher. 2007. Severe acute respiratory syndrome coronavirus protein 6 accelerates murine coronavirus infections. J. Virol. 811220-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tenoever, B. R., S. L. Ng, M. A. Chua, S. M. McWhirter, A. Garcia-Sastre, and T. Maniatis. 2007. Multiple functions of the IKK-related kinase IKKepsilon in interferon-mediated antiviral immunity. Science 3151274-1278. [DOI] [PubMed] [Google Scholar]
- 133.Theofilopoulos, A. N., R. Baccala, B. Beutler, and D. H. Kono. 2005. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu. Rev. Immunol. 23307-336. [DOI] [PubMed] [Google Scholar]
- 134.Thiel, V., and S. G. Siddell. 2005. Reverse genetics of coronaviruses using vaccinia virus vectors. Curr. Top. Microbiol. Immunol. 287199-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Torres, J., K. Parthasarathy, X. Lin, R. Saravanan, A. Kukol, and D. X. Liu. 2006. Model of a putative pore: the pentameric alpha-helical bundle of SARS coronavirus E protein in lipid bilayers. Biophys. J. 91938-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Uze, G., and D. Monneron. 2007. IL-28 and IL-29: newcomers to the interferon family. Biochimie 89729-734. [DOI] [PubMed] [Google Scholar]
- 137.Vabret, A., T. Mourez, S. Gouarin, J. Petitjean, and F. Freymuth. 2003. An outbreak of coronavirus OC43 respiratory infection in Normandy, France. Clin. Infect. Dis. 36985-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.van Pesch, V., O. van Eyll, and T. Michiels. 2001. The leader protein of Theiler's virus inhibits immediate-early alpha/beta interferon production. J. Virol. 757811-7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Versteeg, G. A., P. J. Bredenbeek, S. H. van den Worm, and W. J. Spaan. 2007. Group 2 coronaviruses prevent immediate early interferon induction by protection of viral RNA from host cell recognition. Virology 36118-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Reference deleted.
- 141.Vogel, L. N., A. Roberts, C. D. Paddock, G. L. Genrich, E. W. Lamirande, S. U. Kapadia, J. K. Rose, S. R. Zaki, and K. Subbarao. 2007. Utility of the aged BALB/c mouse model to demonstrate prevention and control strategies for severe acute respiratory syndrome coronavirus (SARS-CoV). Vaccine 252173-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang, F., Y. Ma, J. W. Barrett, X. Gao, J. Loh, E. Barton, H. W. Virgin, and G. McFadden. 2004. Disruption of Erk-dependent type I interferon induction breaks the myxoma virus species barrier. Nat. Immunol. 51266-1274. [DOI] [PubMed] [Google Scholar]
- 143.Wang, W., L. Ye, B. Li, B. Gao, Y. Zeng, L. Kong, X. Fang, H. Zheng, Z. Wu, and Y. She. 2007. Up-regulation of IL-6 and TNF-alpha induced by SARS-coronavirus spike protein in murine macrophages via NF-kappaB pathway. Virus Res. 1281-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wathelet, M. G., M. Orr, M. B. Frieman, and R. S. Baric. 2007. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J. Virol. 8111620-11633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Weber, F., V. Wagner, S. B. Rasmussen, R. Hartmann, and S. R. Paludan. 2006. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 805059-5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Weiss, S. R., and S. Navas-Martin. 2005. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 69635-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wentworth, D. E., L. Gillim-Ross, N. Espina, and K. A. Bernard. 2004. Mice susceptible to SARS coronavirus. Emerg. Infect. Dis. 101293-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wilkinson, M., and A. Morris. 1983. Interferon with novel characteristics produced by human mononuclear leukocytes. Biochem. Biophys. Res. Commun. 111498-503. [DOI] [PubMed] [Google Scholar]
- 149.Williams, R. K., G. S. Jiang, S. W. Snyder, M. F. Frana, and K. V. Holmes. 1990. Purification of the 110-kilodalton glycoprotein receptor for mouse hepatitis virus (MHV)-A59 from mouse liver and identification of a nonfunctional, homologous protein in MHV-resistant SJL/J. mice. J. Virol. 643817-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Woo, P. C. Y., S. K. P. Lau, C.-M. Chu, K.-H. Chan, H.-W. Tsoi, Y. Huang, B. H. L. Wong, R. W. Poon, J. J. Cai, K.-W. Luk, L. L. M. Poon, S. S. Y. Wong, Y. Guan, J. S. M. Peiris, and K.-Y. Yuen. 2005. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 79884-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Xiang, Y., R. C. Condit, S. Vijaysri, B. Jacobs, B. R. Williams, and R. H. Silverman. 2002. Blockade of interferon induction and action by the E3L double-stranded RNA binding proteins of vaccinia virus. J. Virol. 765251-5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Xiao, H., L. H. Xu, Y. Yamada, and D. X. Liu. 2008. Coronavirus spike protein inhibits host cell translation by interaction with eIF3f. PLoS ONE 3e1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Xiong, P., X. Zeng, M. S. Song, S. W. Jia, M. H. Zhong, L. L. Xiao, W. Lan, C. Cai, X. W. Wu, F. L. Gong, and W. Wang. 2008. Lack of association between HLA-A, -B and -DRB1 alleles and the development of SARS: a cohort of 95 SARS-recovered individuals in a population of Guangdong, southern China. Int. J. Immunogenet. 3569-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yang, W., and S. M. Musser. 2006. Nuclear import time and transport efficiency depend on importin beta concentration. J. Cell Biol. 174951-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yang, Z. Y., W. P. Kong, Y. Huang, A. Roberts, B. R. Murphy, K. Subbarao, and G. J. Nabel. 2004. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature 428561-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ye, Y., K. Hauns, J. O. Langland, B. L. Jacobs, and B. G. Hogue. 2007. Mouse hepatitis coronavirus A59 nucleocapsid protein is a type I interferon antagonist. J. Virol. 812554-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yilla, M., B. H. Harcourt, C. J. Hickman, M. McGrew, A. Tamin, C. S. Goldsmith, W. J. Bellini, and L. J. Anderson. 2005. SARS-coronavirus replication in human peripheral monocytes/macrophages. Virus Res. 10793-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Youn, S., J. L. Leibowitz, and E. W. Collisson. 2005. In vitro assembled, recombinant infectious bronchitis viruses demonstrate that the 5a open reading frame is not essential for replication. Virology 332206-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Yount, B., K. M. Curtis, and R. S. Baric. 2000. Strategy for systematic assembly of large RNA and DNA genomes: transmissible gastroenteritis virus model. J. Virol. 7410600-10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yount, B., K. M. Curtis, E. A. Fritz, L. E. Hensley, P. B. Jahrling, E. Prentice, M. R. Denison, T. W. Geisbert, and R. S. Baric. 2003. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA 10012995-13000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yount, B., M. R. Denison, S. R. Weiss, and R. S. Baric. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 7611065-11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Yount, B., R. S. Roberts, A. C. Sims, D. Deming, M. B. Frieman, J. Sparks, M. R. Denison, N. Davis, and R. S. Baric. 2005. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J. Virol. 7914909-14922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Yuan, F. F., J. Tanner, P. K. Chan, S. Biffin, W. B. Dyer, A. F. Geczy, J. W. Tang, D. S. Hui, J. J. Sung, and J. S. Sullivan. 2005. Influence of FcgammaRIIA and MBL polymorphisms on severe acute respiratory syndrome. Tissue Antigens 66291-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Yuan, W., and R. M. Krug. 2001. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. EMBO J. 20362-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhang, H., G. Zhou, L. Zhi, H. Yang, Y. Zhai, X. Dong, X. Zhang, X. Gao, Y. Zhu, and F. He. 2005. Association between mannose-binding lectin gene polymorphisms and susceptibility to severe acute respiratory syndrome coronavirus infection. J. Infect. Dis. 1921355-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zhang, X., H. Fujii, H. Kishimoto, E. LeRoy, C. D. Surh, and J. Sprent. 2002. Aging leads to disturbed homeostasis of memory phenotype CD8(+) cells. J. Exp. Med. 195283-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zhang, Y., J. Li, Y. Zhan, L. Wu, X. Yu, W. Zhang, L. Ye, S. Xu, R. Sun, Y. Wang, and J. Lou. 2004. Analysis of serum cytokines in patients with severe acute respiratory syndrome. Infect. Immun. 724410-4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zhou, H., and S. Perlman. 2007. Mouse hepatitis virus does not induce beta interferon synthesis and does not inhibit its induction by double-stranded RNA. J. Virol. 81568-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhou, Z., K. Hoebe, X. Du, Z. Jiang, L. Shamel, and B. Beutler. 2005. Antagonism between MyD88- and TRIF-dependent signals in B7RP-1 up-regulation. Eur. J. Immunol. 351918-1927. [DOI] [PubMed] [Google Scholar]
- 170.Ziebuhr, J. 2005. The coronavirus replicase. Curr. Top. Microbiol. Immunol. 28757-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zuniga, S., I. Sola, J. L. Moreno, P. Sabella, J. Plana-Duran, and L. Enjuanes. 2007. Coronavirus nucleocapsid protein is an RNA chaperone. Virology 357215-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zust, R., L. Cervantes-Barragan, T. Kuri, G. Blakqori, F. Weber, B. Ludewig, and V. Thiel. 2007. Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. PLoS Pathog. 3e109. [DOI] [PMC free article] [PubMed] [Google Scholar]