Abstract
Summary: The RecBCD enzyme of Escherichia coli is a helicase-nuclease that initiates the repair of double-stranded DNA breaks by homologous recombination. It also degrades linear double-stranded DNA, protecting the bacteria from phages and extraneous chromosomal DNA. The RecBCD enzyme is, however, regulated by a cis-acting DNA sequence known as Chi (crossover hotspot instigator) that activates its recombination-promoting functions. Interaction with Chi causes an attenuation of the RecBCD enzyme's vigorous nuclease activity, switches the polarity of the attenuated nuclease activity to the 5′ strand, changes the operation of its motor subunits, and instructs the enzyme to begin loading the RecA protein onto the resultant Chi-containing single-stranded DNA. This enzyme is a prototypical example of a molecular machine: the protein architecture incorporates several autonomous functional domains that interact with each other to produce a complex, sequence-regulated, DNA-processing machine. In this review, we discuss the biochemical mechanism of the RecBCD enzyme with particular emphasis on new developments relating to the enzyme's structure and DNA translocation mechanism.
INTRODUCTION
One key function of the RecBCD enzyme is to salvage broken replication forks via homologous, or template-directed, recombinational DNA repair. This crucial repair process allows the completion of DNA replication and bacterial cell division. A second function stems from its seemingly contradictory capacity to degrade linear duplex DNA. This activity is responsible for the degradation of unwanted linear pieces of chromosomal DNA that otherwise could block proper replication restart and lead to spurious replication or misplaced recombination. The enzyme will be introduced and discussed mainly in these contexts, a perspective that reflects the growing recognition that recombination and degradation are integral parts of accurate chromosome duplication and maintenance and are required for a cell's viability (for reviews, see references 76, 147, 148, 159, and 165). The participation of RecBCD in various aspects of Escherichia coli DNA metabolism relates to its ability to process duplex DNA ends. This characteristic has played an important role in the discovery and biochemical analysis of the RecBCD enzyme.
The RecBCD enzyme has been reviewed previously (6, 24, 98, 159, 161, 212, 262, 269, 285). This review will differ in its focus on biochemical mechanisms. In particular, we will discuss three important new developments in our understanding of the RecBCD mechanism: firstly, the identification of RecBCD as a bipolar DNA helicase that employs a unique mechanism for tracking along a DNA duplex; secondly, the use of new biophysical methods, including single-molecule and rapid-reaction techniques, to study the DNA translocation activity; and finally, the determination of a crystal structure of RecBCD bound to a DNA break. Earlier biochemical results will be discussed in the context of these new data. We will see that RecBCD is a prototypical example of a molecular machine, which nature has constructed by combining several autonomous protein domains. The temporal and spatial coupling of functional domains in the RecBCD complex produces a regulated helicase-nuclease activity that is capable of more than the sum of its parts. Finally, we discuss the distinctive AddAB-like helicase-nuclease enzymes that are found mainly in the gram-positive bacteria. Although relatively little is currently known about these proteins, they may offer new insight into how the DNA break-processing reaction is coupled to defects in DNA replication and the interplay between homologous recombination and alternative mechanisms for the repair of double-stranded DNA (dsDNA) breaks (DSBs).
CELLULAR EVENTS INVOLVING DUPLEX DNA ENDS
Production of DNA Ends
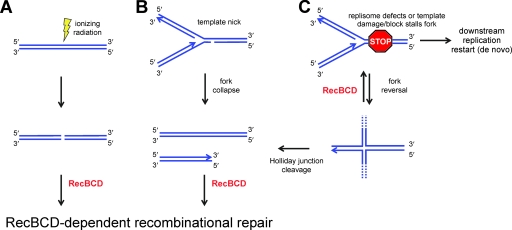
The physiological substrate for the RecBCD enzyme is a free blunt or nearly blunt duplex DNA end (292). These occur as intermediates in a variety of DNA transactions but are also formed as a result of damage. Indeed, DSBs are a potentially lethal form of damage and, accordingly, mechanisms have evolved for their repair. DSBs are caused, either directly or indirectly, by a wide variety of endogenous and exogenous agents, including ionizing radiation, UV light, oxygen radicals, and DNA-damaging agents (e.g., alkylating agents). Inappropriate chromosomal DNA cleavage may also occur via the activity of host restriction-modification systems, particularly if the restriction alleviation mechanisms that act as the first line of defense for the host DNA are inactivated (194). Importantly, in normally growing cells in the absence of any exogenous source of DNA damage, DSBs are formed in almost every cell cycle as a consequence of replication through imperfect DNA templates. The variety of potential lesions is huge, but some that result in broken dsDNA that could serve as substrates for RecBCD-dependent recombinational repair are shown in Fig. 1. For example, ionizing radiation can directly produce breaks through dsDNA (Fig. 1A). Alternatively, the dsDNA break can form indirectly due to replication on a nicked DNA template (Fig. 1B). The progression of the replisome through the nick will disrupt the continuity of the replication fork, leaving behind one intact chromosome and one broken daughter chromosome with a single DNA end; this has been referred to as “replication fork collapse” (119, 164, 165, 260). In addition, defective replisome components, DNA damage, or physical blocks (e.g., bound proteins) in the template strand may halt the replication fork (for reviews, details, and opinions, see references 127, 202, 205, and 206). In vitro studies have established that, in some cases, the replication machinery can itself reinitiate de novo leading- and lagging-strand synthesis downstream of the lesion (126) but that, in others, it may dissociate. Such stalled forks may undergo either spontaneous reversal (205, 249) (the supercoiling-promoted reversal of the fork to anneal the nascent DNA strands [225]) or active (e.g., RecG-catalyzed) regression (200, 201) to form a four-way junction resembling a Holliday junction (74, 166, 208, 251). The fork reversal itself does not correct the lesion, but it potentially provides additional time for the template damage to be repaired. The fate of this Holliday junction depends on the nature of the replication stall and on the many competing cellular processes; the ultimate pathway, or combination of pathways, utilized in vivo remains unclear in most cases. In some cases (i.e., when the replication machinery is compromised), the replication fork can be reestablished by the RecBCD-dependent nucleolytic degradation of the newly formed DNA end in a process termed “replication fork resetting” (205, 249). In other cases (e.g., in some fraction of UV-induced DNA damage), the Holliday junction is proposed to migrate back to reconstitute a replication fork (127, 200-202). When not degraded by RecBCD, due to either mutation (249) or UV irradiation (201), the Holliday junction can be cleaved by RuvABC, resulting in the production of a double-stranded DNA break. Last but not least, another major class of DNA lesions is single-stranded DNA (ssDNA) gaps that result from incomplete replication (e.g., DNA replication stalled by UV damage), or other DNA repair processes, and that do not produce a DSB. These ssDNA gaps are repaired (postreplication gap repair) by enzymes of the RecF pathway, which promote recombination with the daughter strand (69, 71-73, 110, 130, 131, 138, 144, 176, 242-245, 310, 314, 318). RecBCD is not involved in ssDNA gap repair, and hence the RecF pathway of recombination is not discussed further (see references 168, 205, and 269). The frequency of interrupted replication is high, occurring in most replication cycles, even under conditions where the cells are not challenged with DNA-damaging agents (164, 168, 195). The restart of DNA replication is essential for cell viability, and hence DSB repair coupled to replication restart mechanisms should be regarded as an evolutionarily essential chromosome maintenance function (127).
FIG. 1.
Direct and indirect sources of DSBs. (A) A DSB can be formed directly by, for example, ionizing radiation. This results in the production of two free DNA ends. (B) If a replication fork encounters a nick in the template strand, an arm will detach to generate a single DSB. This is commonly referred to as replication fork collapse. (C) Following replication fork stalling due to replisome defects, or because there is damage or a block in the template strands, several fates are possible (see references 127, 202, and 205). Replication may restart de novo downstream of the damage without repair. Alternatively, the replication fork may undergo reversal to form a Holliday junction. In this case, the free end of the Holliday junction can be degraded by RecBCD to reform a fork and allow replisome reassembly. Replication can also continue after DNA repair by resorbing the Holliday junction to reform a fork. Otherwise, the Holliday junction may be cleaved to produce a single-ended DSB as in B. All of these DSBs are substrates for RecBCD-dependent recombinational repair.
In addition to linear dsDNA being generated by DNA-damaging events, dsDNA ends are also present as transient cellular intermediates in many biological processes. For example, during conjugative recombination, donor DNA enters the cell as a single strand that is converted to linear duplex DNA by replication. This conversion does not necessarily produce the nearly blunt dsDNA ends required by RecBCD, and in vivo, many of E. coli's ssDNA-specific nucleases (particularly the 3′-specific nucleases) are used to degrade the extraneous ssDNA tails that limit RecBCD binding (80, 297). This process of genetic exchange, critical for the long-term evolution of bacteria, uses the same recombination machinery that probably evolved to deal with the acute problem of repairing DNA. Likewise, the free DNA ends that are present in the life cycle of many phages attract the attention of RecBCD. In this case, however, the function of the enzyme is to degrade the phage DNA, serving an antiviral function for the bacterium. Thus, the challenge for the RecBCD enzyme is to distinguish between friend (broken chromosomal DNA) and foe (linear phage DNA). As will be discussed below, the recombination hotspot sequence Chi provides this information. Thus, the cellular functions of the RecBCD enzyme are diverse, but all of these cellular processes are united by the presence of a dsDNA end created as a biological intermediate that is processed by RecBCD enzyme.
Recombinational Repair of Double-Stranded DNA Breaks
Nature has evolved two general strategies for repairing DSBs: homologous recombination and nonhomologous end joining. The nonhomologous end joining process is designed to directly reattach two broken DNA ends, and it involves DNA end-bridging proteins and a specific DNA ligase (79, 253, 317). Unlike homologous recombination, nonhomologous end joining does not require the presence of a homologous donor molecule and is error prone; there is potential to lose information near the sites of the DNA breaks and, even worse, to rejoin the wrong two DNA ends to produce a DNA translocation. Interestingly, the interplay and relative importance of the homologous recombination and nonhomologous end joining pathways probably reflect the availability of homologous donor DNA for the repair process. For instance, nonhomologous end joining is the dominant mechanism for DSB repair in the G1 phase of the eukaryotic cell cycle; on the other hand, homologous recombination plays a more prominent role at late S/G2 phase (141, 219). The nonhomologous end joining pathway does not appear to exist in E. coli (234). However, it is present in a range of other bacteria, where it may play an important role during prolonged stationary phase, for example, during sporulation and biofilm formation (317).
Unlike nonhomologous end joining, homologous recombination is a relatively error-free process, which can repair DNA lesions by using an intact homologous donor molecule (e.g., the sister chromatid) as a template (Fig. 2). Whereas nonhomologous end joining is limited to the reattachment of any two broken DNA ends, homologous recombination can either rejoin the two opposed ends of a broken DNA molecule (“classical” dsDNA break repair) (281) (Fig. 2A) or restart DNA replication from a detached replication fork (recombination-dependent replication) (148, 159) (Fig. 2B), a process that involves only a single DNA end (164, 260). In either case, the central step in the process is a “synapsis” between homologous DNA molecules, resulting in the exchange of DNA strands. DNA strand exchange is catalyzed by the RecA protein, which forms a filament on ssDNA (77, 233, 252). This nucleoprotein filament is the active species in the homology search and the subsequent invasion of a homologous duplex DNA (see references 32, 109, 159, and 161). Consequently, DNA lesions requiring recombinational repair must first be processed into ssDNA by the action of helicases and nucleases.
FIG. 2.
RecBCD-dependent homologous recombination pathways. (A) In recombination-dependent DSB repair, a broken DNA molecule containing two (previously contiguous) DNA ends is rejoined in several steps involving recombination with an intact homologous donor DNA (“ends-in” recombination) (261). The pathway shown is the classical dsDNA gap repair model invoking the resolution of two Holliday junctions in the recombination intermediates formed following DNA pairing (281). In the first step, catalyzed by the RecBCD enzyme, the DNA ends are resected to form 3′ ssDNA overhang structures terminated at the Chi sequences; the DNA upstream of Chi is degraded by RecBCD, whereas the downstream Chi-containing ssDNA is preserved due to the attenuation of RecBCD nuclease activity by Chi (see reference 269). The RecA protein is loaded onto these Chi-containing ssDNA overhangs by RecBCD and promotes the homology search and DNA strand invasion with a donor duplex. The donor DNA acts as a template for DNA synthesis, resulting in the formation of two Holliday junctions. These are resolved to yield intact duplex products. (B) In recombination-dependent replication, a single DNA end is formed upon the collapse of a replication fork. The end is processed by RecBCD as in A to create a single 3′ ssDNA overhang with Chi at its terminus. RecA catalyzes the reattachment of the broken processed DNA arm to the sister DNA duplex, reforming a fork structure. Replication restart then proceeds in several steps and eventually results in the loading of the replicative helicase in a PriA-dependent process and resumption of replication in an origin-independent manner (127). (C) Integration of linear dsDNA (e.g., in conjugation and transduction) occurs by an “ends-out” mechanism (261). Both ends of the linear duplex DNA are resected by RecBCD to form 3′ ssDNA overhangs that end at Chi sequences as in A. RecA-promoted DNA strand invasion of each processed end into homologous donor DNA primes PriA-dependent DNA replication in a bidirectional manner. The replication of the entire chromosome results in the integration of the linear dsDNA piece at a homologous locus, as shown; alternatively, the resolution of the Holliday junctions can also result in integration (not shown). Note that in all three panels, for clarity, the location of the Chi sequence is shown only during the initial stages of each pathway.
In E. coli, the two partially overlapping pathways for recombinational DNA repair are the RecBCD and RecF pathways (68, 130, 167, 168), which act on DSBs and ssDNA gaps, respectively (see reference 269). The RecBCD enzyme initiates the repair of DSBs by converting a blunt dsDNA end into a duplex DNA molecule possessing a 3′-terminated ssDNA tail. In addition, as part of this processing step, the RecBCD enzyme directs the RecA protein onto this ssDNA (17) (Fig. 3). An important component in this process is the octameric regulatory sequence called Chi, which was originally identified as a recombination hotspot (128, 172). Chi (crossover hotspot instigator), or χ, is the sequence on one strand of DNA, 5′-GCTGGTGG-3′ (33, 271). The binding of the RecA protein to the Chi-terminated 3′ ssDNA tail is not a passive process but is facilitated by the action of the RecBCD protein (17, 91-93). In the absence of such a loading mechanism, the resulting ssDNA is rapidly and tightly bound by the ssDNA binding (SSB) protein, which binds ssDNA nonspecifically. Once formed, the RecA nucleoprotein filament searches for a homologous donor and catalyzes DNA strand invasion (28, 35, 192). Following the invasion and exchange of DNA strands, replication can ensue from the 3′ end of the invading ssDNA (322), eventually resulting in the loading of the replicative DNA helicase (127). This process requires many components, including the PriA, PriB, and PriC proteins (127, 195, 247).
FIG. 3.
The RecBCD enzyme is a sequence-regulated DNA helicase-nuclease. Shown are details of the RecBCD-catalyzed DNA end-processing reaction. (1) RecBCD (light blue) binds tightly to a blunt (or nearly blunt) DNA end of a linear DNA duplex. (2) RecBCD couples the hydrolysis of ATP to DNA translocation and unwinding (i.e., helicase activity). The ssDNA products are cleaved asymmetrically, with the degradation of the 3′-terminated ssDNA tail being much more vigorous than the degradation of the complementary tail. (3) The enzyme continues to translocate until it pauses at a correctly oriented Chi sequence. The recognition of the Chi sequence dramatically alters the biochemical properties of the enzyme (indicated by color change to pink). (4) The enzyme continues to translocate, but the nuclease polarity is switched; the degradation of the 3′ ssDNA tail is attenuated, whereas the hydrolysis of the 5′ ssDNA tail is upregulated. After Chi recognition, RecBCD facilitates the loading of the RecA protein onto the 3′ ssDNA tail. (5) RecBCD repeatedly deposits RecA protomers, which act as nucleation points for filament growth primarily in the 5′→3′ direction. (6) The RecBCD enzyme dissociates from the DNA. The product of the enzyme is a recombinogenic nucleoprotein complex of the RecA protein bound to the 3′ ssDNA tail with Chi at its terminus. This product is able to invade homologous duplex DNA to promote the recombinational repair of a DSB or to restart DNA replication as appropriate. Note that this cartoon is not to scale; several thousand base pairs of DNA may be processed before and after the Chi sequence, and the RecA filament consists of many more protomers than are shown.
Although RecBCD is responsible for the initiation of DSB repair in wild-type E. coli, the recombinational DSB repair pathways are partially redundant, demonstrating the remarkable plasticity of bacterial DSB repair and the evolutionary drive to repair these breaks. For example, in the absence of RecBCD activity (and with the suppressor mutations sbcB and sbcC or sbcD) (112, 130, 163, 182) or, alternatively, in the absence of exonuclease I (xonA) and exonuclease VII (xseA) functions (306), the RecF pathway can promote recombinational DNA repair of DSBs. In this situation, the DSB is resected by the combined activities of the RecQ helicase and RecJ exonuclease (125, 186, 214, 269), and the overall biochemical process bears mechanistic similarity to eukaryotic DSB repair (see references 269 and 279). Furthermore, in otherwise wild-type cells, when the RecBCD enzyme has lost the RecA-loading function but not its helicase activity, the proteins of the RecF pathway can provide the essential RecA-loading capability, and breaks are repaired by a “hybrid” pathway (6, 136, 137). In wild-type E. coli cells, however, the RecBCD enzyme is responsible for 95 to 99% of all recombination events occurring at dsDNA breaks (102, 132, 315) but not at ssDNA gaps (see reference 269). Interestingly, the capacity to efficiently repair DSBs plays an important role in the pathogenicity of Salmonella (44, 50). Systemic infection with Salmonella requires growth inside phagocytic cells, an environment in which extensive DNA damage is caused by reactive oxygen species and nitric oxide; the loss of RecBCD function decreases both viability and virulence for E. coli and Salmonella (50, 265).
The view of the RecBCD enzyme as an essential housekeeping repair factor that supports DNA replication ties in well with the observation that cells lacking RecBCD function are largely inviable (∼70%) (51, 53, 182, 207). In this context, the cells that do not survive represent mostly those cells in which DSBs were formed from a disrupted replication fork, were not repaired, and, hence, did not complete replication (52).
Bacterial Conjugation and Transduction
Conjugal recombination is the process by which genes can be transferred from donor bacteria to recipients in a process mediated by fertility factor F (261). This process allows the horizontal transfer of genes and recombination between partially homologous sequences. DNA enters the host cell in a single-stranded form but is converted to linear duplex DNA by replication. Linear duplex DNA acts as a substrate for the RecBCD enzyme to promote “ends-out” recombination with the host genomic DNA (Fig. 2C). Transductional recombination describes the horizontal transfer of DNA via a phage vector (196). Some lytic phages frequently misincorporate bacterial genomic DNA into their phage heads during packaging. When these phages subsequently “infect” another host, they inject the packaged bacterial DNA. This transduced DNA is injected as a linear duplex, and it can therefore recombine with the chromosome of the new host via the RecBCD pathway (261). Because conjugation and transduction are particularly amenable to genetic analysis, they have proven to be important for the dissection of RecBCD enzyme function, and differences in these two processes have also helped to reveal the roles that other nucleases play in the RecBCD pathway of recombination (297).
Host DNA Degradation
The RecBCD functions described above all involve RecA-dependent homologous recombination. However, the simple observation that either ΔrecBCD cells (∼30% viability) or ΔrecA ΔrecBCD cells (∼20% viability) are significantly less viable than ΔrecA cells (∼50%) argues for a role of RecBCD in DNA metabolism that does not involve recombination (51, 53, 207). This additional drop in viability is related to the loss of the DNA-degradative capacity of RecBCD and reflects an inability either to degrade the detached arms of collapsed replication forks (avoiding the “sigma replication trap”) (170) or to reset reversed replication forks by removing the newly formed linear dsDNA from the end of a reversed replication fork (Fig. 1C) (166, 249). Current results more strongly support the latter possibility as an important cellular function for the degradative activity of RecBCD (207).
The cellular degradative capacity of RecBCD is, however, saturable since it is present at only about 10 molecules per cell (82, 285). Extensive chromosomal breakage induced by DNA-damaging agents, for example, leads to the inhibition of RecBCD in vivo (42, 150, 170, 232, 298). Further studies with plasmids led to the conclusion that the interaction with Chi sequences produced inactivation and that a RecA mutation affected the phenomenon (169, 170). Although not fully understood in vivo, the interaction with Chi in vitro causes a disassembly of RecBCD into subunits when it dissociates from the DNA (290). As a result, these subunits are either unstable in vivo or sequestered by binding to chromosomal DNA (81), resulting in either a complete loss of RecBCD activity or a partial loss (loss of RecD function) (170). Regardless of the mechanism, RecBCD has a finite degradative capacity in vivo, a trait that can limit the wanton destruction of DNA in cells.
Foreign DNA Degradation
Bacteria expend a considerable amount of effort in keeping themselves free of invading DNA. The well-known type II restriction-modification systems constitute the primary defensive mechanism against foreign DNA and can, remarkably, account for greater than 4% of the genome for some bacteria (Helicobacter pylori) (179). The type II restriction enzymes cleave foreign DNA into linear fragments that are substrates for more extensive nucleolytic degradation by the RecBCD enzyme (85, 121, 171, 255). Moreover, RecBCD can act directly upon the genomes of phages, such as lambda or T4, that contain free DNA ends (27, 30). Indeed, any phage that exposes free DNA ends as part of its life cycle must find a means to evade destruction by RecBCD. The ensuing evolutionary battle between phage and bacteria has created some interesting “weapons,” including phage-encoded inhibitors of RecBCD that block its activity either by binding and protecting the ends of the linear phage genomes (18, 254, 316) or by binding directly to RecBCD (209, 210). The crystal structure of the Gam protein from lambda phage was recently solved and suggests that Gam acts as a competitive inhibitor of DNA binding by mimicking the structure of a duplex DNA end (75). Thus, RecBCD also contributes to the important cellular function of degrading foreign DNA that has free dsDNA ends (27, 30, 255).
The DNA Repair versus DNA Degradation Paradox
The juxtaposition of this degradative capacity against its recombinational DNA repair function has been a historical paradox. The key to understanding the enzyme's split personality is the regulatory sequence Chi (see below). Before Chi recognition, the enzyme is a voracious destructive nuclease-helicase, whereas following recognition, it is a repair helicase-nuclease that recruits the RecA protein onto nascent Chi-containing ssDNA. In this context, the regulation of RecBCD by the Chi sequence can be viewed as a self-recognition mechanism analogous to, but mechanistically distinct from, the protection of host DNA from type II restriction enzymes afforded by methylation at their target sequences (121). The Chi sequence is the most overrepresented octamer in the E. coli genome that also displays the property of being asymmetrically oriented with regard to the replication origin (38). Consequently, DSBs that arise during the replication of the genome are recognized as self-DNA and are directed for repair by the homologous recombination pathway (78, 169), whereas foreign DNA up to 170 kb in size (e.g., T4 gene 2− phage) is degraded and rendered nonfunctional by the RecBCD enzyme.
RecBCD ENZYME
Discovery
Our current understanding of RecBCD and its role in E. coli DNA metabolism results from the confluence of several lines of research. The enzyme itself was discovered as a potent exonuclease activity in E. coli (26, 113, 114, 143, 177, 215, 216, 282, 300, 320, 321). It is maintained at a low copy number of about 10 RecBCD molecules per cell (82, 285), because overproduction actually impairs recombinational DNA repair and increases chromosomal degradation (82).
This uniquely ATP-dependent nuclease activity was named exonuclease V and was subsequently shown to result from coupled helicase and ssDNA-endonuclease activities (99, 142, 143, 181, 193, 241, 283, 284). The link between RecBCD and recombination was first demonstrated by the absence of exonuclease V activity from the recB and recC strains of E. coli that were isolated from screens for defects in conjugational recombination (26, 321). Subsequently, it was shown that mutations that inactivate the recB or recC gene lead to defects in conjugational, transductional, and phage recombination; a loss of SOS induction; sensitivity to DNA-damaging agents that cause DSBs; and low cell viability (15, 26, 51-53, 162, 177, 207, 274, 307, 313, 320). Note that all of these phenotypes result from defects in processes involving free DNA ends, which act as the entry points for the RecBCD enzyme. The importance of the RecD subunit was only fully appreciated when it was identified as being a physical component of the “RecBC” holoenzyme required for the exonuclease activity (7, 37, 105, 178). This late discovery was, in part, due to the complexity of interpreting the recD phenotype. In contrast to the recB and recC phenotypes, recD cells are fully viable, are resistant to DNA-damaging agents, and display hyperrecombinogenic behavior (7, 37). However, recombination in recD cells becomes heavily dependent on the activities of other exonucleases, in particular, RecJ and exonuclease VII, both capable of ssDNA degradation in the 5′→3′ direction (80, 83, 183, 184, 187). RecD function also appears to be important when restriction alleviation mechanisms are impaired (194), for the degradation of restricted phage (255), and for survival in high-pressure or low-temperature environments (36, 229). In species that lack RecBCD homologues, the function of an apparently solo RecD-like protein is needed for resistance to oxidants (250, 327), gamma rays, and UV light (250) or for pilin variation (56).
Crossover Hotspot Instigator
The Chi sequence is a critical cis-acting DNA element for the processing of DNA by the RecBCD enzyme (Fig. 3). The Chi sequence was identified independently of research on the RecBCD enzyme during genetic analysis of coliphage lambda (128, 172, 272). Wild-type lambda does not contain a Chi sequence within its 48.5-kbp genome. The Chi sequence was isolated as a spontaneously occurring mutation in the phage genome that allowed the formation of large plaques on a lawn of E. coli when the phage had been forced to use the host's recombination machinery. (This involved shutting down the phage recombination system red, which comprises the Redα exonuclease and Redβ protein, and inactivating the phage protein Gam, which is an inhibitor of the RecBCD protein [128, 172, 272].) Chi stimulates recombination in a directional manner, with a stimulatory gradient that decreases with a half-distance of approximately 3.2 kbp when the DNA sequences are fully homologous (64, 172, 273, 275), but it can also act for more than 10 kbp when a heterology is present on one of the recombining molecules (213). Stimulation requires both a DNA end and the RecBCD enzyme (212). Sequencing of lambda-pBR322 hybrids revealed the identity of Chi as the sequence 5′-GCTGGTGG-3′, its complement, or both; recognition of Chi occurred when the enzymes approached from the 3′ side of the sequence as written (263). In vitro analysis would later reveal that Chi was the single-stranded DNA sequence 5′-GCTGGTGG-3′ (33). Analysis of the E. coli genome reveals that Chi is the third most overrepresented octamer: E. coli contains 1,008 Chi sequences (the original sequence of E. coli MG1655 reported 1,009 sequences [38], but the recent database shows only 1,008) (19); Chi sequences are four- to eightfold more frequent than expected by chance and appear on average once every 4.5 kb (38, 100, 302). Even more significant is their skew: 75% are oriented toward the origin of replication, making Chi the most directionally biased 8-nucleotide sequence in E. coli (100, 246). Furthermore, there is a statistically significant association between Chi sequences and GT-rich “islands”; GT-rich DNA is the preferred substrate for RecA homology-dependent pairing (302). These observations tie in nicely with the role of the RecBCD enzyme as a repair factor functioning during DNA replication (19, 100). As noted previously (173), codon usage does not directly explain the frequency and distribution of Chi sequences (70). Instead, the overrepresentation and skewing of Chi sequences reflect characteristics of GT-rich sequences that coincide with an underlying bias in codon usage and a bias in transcription polarity (29, 70, 299, 303); however, these characteristics statistically correlate most significantly with replication direction (19, 100). This implies that the RecBCD enzyme selected Chi largely from the overrepresented recombinogenic GT-rich sequences, which arise from both codon usage and genome base composition, for use as a regulatory switch and recombination hotspot in recombination-dependent replication (19, 100, 302, 304), rather than there being strong prior selective pressure for the Chi sequence to become overrepresented due to its role in recombinational repair. In other words, the RecBCD enzyme adapted largely to the genome and not vice versa.
In vitro analysis of the purified RecBCD enzyme (discussed below) would go on to reveal the intimate relationship between RecBCD, Chi, and RecA by demonstrating that the Chi sequence directly modulates both the DNA translocation and nuclease activities of the holoenzyme, and it regulates the loading of RecA onto the Chi-containing ssDNA.
BIOCHEMICAL ACTIVITIES OF RecBCD
Isolated Subunits
The RecBCD enzyme is a heterotrimer consisting of three different polypeptides (Fig. 4). RecB is a 134-kDa protein containing motifs characteristic of superfamily 1 (SF1) DNA helicases toward the N terminus (115) and those characteristic of diverse families of nucleases at the C terminus (20). The RecC protein is also large (129 kDa), but analysis of its primary sequence does not reveal any significant similarities to protein domains with known activities. The smaller RecD subunit (67 kDa), which was identified as a component of the holoenzyme later than the RecB and RecC subunits (7, 37, 105), also contains motifs characteristic of SF1 DNA helicases (115). Consideration of primary structure alone suggests the presence of two helicase domains, a single nuclease domain, and a substantial amount of protein with unassigned function in the RecBCD holoenzyme. The work of several groups has employed a reductionist approach to correlate structure and function in the RecBCD enzyme. Indeed, all of the component polypeptides as well as identifiable domains therein have been expressed, purified, and characterized.
FIG. 4.
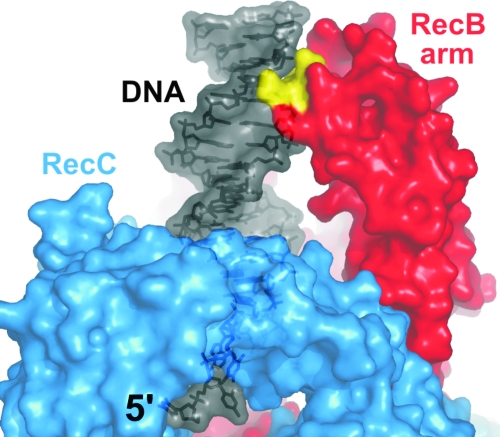
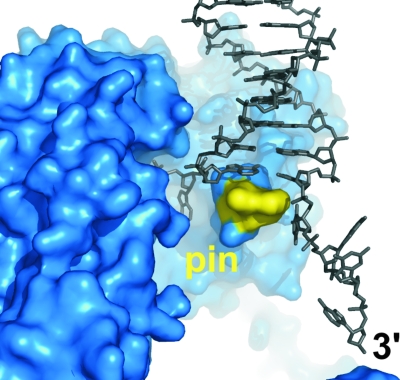
Primary structure of the RecBCD enzyme. The RecB protein is modular. The N-terminal domain (red) contains seven motifs (numbered) characteristic of SF1 helicases. The C-terminal domain (magenta) contains motifs characteristic of a diverse family of nucleases. “Nuc” marks the position of nuclease motif 3, which contains key catalytic aspartate and lysine residues (D1067, D1080, and K1082) (308, 325). The RecC protein (blue) does not show significant homology to other known proteins. A region of RecC implicated in Chi recognition is indicated (see the text). The RecD protein (green) contains the seven conserved SF1 helicase motifs (numbered). The total number of amino acids (aa) in each polypeptide is indicated in parentheses. This color scheme is used in figures of the RecBCD-DNA crystal structure.
The RecB protein is a DNA-dependent ATPase (129) and a weak DNA helicase operating with a 3′→5′ translocation polarity (40). DNA helicases commonly, but not exclusively, unwind duplex DNA using a flanking ssDNA as a loading site. (The RecBCD holoenzyme is one exception to this rule, whereas the isolated RecB and RecD proteins are not; the reason for the apparent complexity will become clear in the section below on bipolar DNA translocation.) A helicase is described as displaying 3′→5′ polarity if it requires that the flanking ssDNA tail is 3′ terminated and vice versa (198). The simple explanation for this polarity is that the helicase binds to the ssDNA loading site and translocates unidirectionally along it (89), displacing the complementary strand when it reaches the duplex. In accord with the primary structure, the 3′→5′ helicase activity of the RecB protein resides in a distinct N-terminal domain (324). The N-terminal domain interacts with the RecC protein to form a rapid and processive DNA helicase (197). A C-terminal domain in RecB can function independently as a DNA endo- and exonuclease and is responsible for all nuclease activities associated with the RecBCD complex (278, 308, 324, 325). The C-terminal nuclease domain of RecB was shown to interact directly with the RecA protein (67, 270). This interaction plays an essential role in the mechanism of loading the RecA protein onto the Chi-containing ssDNA produced by the RecBCD enzyme (13, 17, 23). Modeling of the interaction between RecA and the nuclease domain of RecB suggests that it is similar to the RecA-RecA interface in the nucleoprotein filament (270). Consequently, it was suggested that RecA loading may be achieved by nucleating a RecA filament through molecular mimicry, as was also proposed for analogous systems (160, 221).
There is rather little published work on the biochemical properties of the RecC protein. The isolated subunit does not possess ATPase, helicase, or nuclease activity (197), but it does bind ssDNA, and it can stimulate the ATPase and helicase activities of the RecB protein, with which it forms a complex of 1:1 stoichiometry (197) (our unpublished observations). Limited proteolysis of the RecC protein suggests that a 35-kDa C-terminal domain is required for interaction with the RecD protein (9). Certain mutations in the RecC protein are responsible for RecBCD holoenzymes with altered Chi recognition specificity, arguing that the RecC protein is involved in Chi recognition (22, 124); we will return to this point in the discussion of the RecBCD-DNA crystal structure.
Study of the RecD subunit has proven rather more challenging because it is largely insoluble upon overexpression, and this problem continues throughout purification. Although native soluble RecD has been purified, it was found to be inactive for all assays tested (197). The solubility and activity issues were resolved by refolding either in solution (62) or on a column (87). Experiments with this material revealed that the enzyme possesses ssDNA-dependent ATPase activity (62, 87) and 5′→3′ DNA helicase activity (87).
In general, the biochemical properties of each subunit marry well with those predicted from a careful examination of the primary structure. However, simply understanding which activities are associated with which polypeptides yields little information on the complex reaction mechanism of the holoenzyme. As described below, the key to understanding the reaction mechanism lies in understanding how these activities are coupled within the whole RecBCD complex.
RecBCD Complex
The purified RecBCD enzyme possesses an impressive arsenal of biochemical activities, including DNA binding, RecA binding, DNA-dependent ATPase, DNA helicase, ssDNA endonuclease, ssDNA exonuclease, dsDNA exonuclease, and Chi-regulated nuclease and helicase activities. These properties, which can all be measured independently in vitro, are coupled to generate a complex DNA-processing reaction (see the legend of Fig. 3 for details).
Analytical ultracentrifugation and gel filtration demonstrated that RecBCD exists as a heterotrimer in solution (197, 216, 319), and biochemical approaches show that a single heterotrimer bound to a DNA end is the active species (287). Binding to a DNA end is at least 2 orders of magnitude tighter than that to an internal site (238). The dissociation constant is in the subnanomolar or low-nanomolar range depending on the salt concentration and on whether the DNA ends contain short ssDNA overhangs on either the 3′- or 5′-terminated strands (107, 237, 238, 291). This affinity is much greater than that of other common SF1 helicases, such as Rep helicase, whose affinity for ssDNA is in the micromolar range, depending on conditions (see reference 185 and references therein). Thus, given that the concentration of a single DNA end in a cell the size of E. coli can be estimated to be ∼1 nM, and the concentration of 10 RecBCD molecules would be ∼10 nM, the interaction between RecBCD and a DSB is clearly physiologically appropriate, whereas helicases with affinities comparable to those of Rep would not be effective competitors unless present at high (micromolar) concentrations. DNase I-footprinting and permanganate sensitivity experiments demonstrate that the RecBCD complex protects about 20 bp of the DNA duplex (111) and that 4 to 6 bases of ssDNA at a blunt duplex end are separated due to binding alone (103) (i.e., in the absence of ATP binding/hydrolysis). In agreement with these observations, a recent thermodynamic analysis of the RecBCD interaction with DNA ends demonstrated that the optimal binding substrate for the enzyme contains unpaired ssDNA overhangs of 6 bases and 10 bases in the 3′- and 5′-terminated strands, respectively (319). The 5′-terminated strand can be cross-linked to the RecC and RecD polypeptides, and the 3′-terminated strand can be cross-linked to RecB (111). The results of these studies of the RecBCD-DNA complex provide a consistent picture of the initiation complex that is, in general, in excellent agreement with the high-resolution structural information that is discussed below.
The ATPase and helicase activities are coupled. ATP hydrolysis is strongly dependent on linear ssDNA or dsDNA, is extremely fast (∼1,000 bp s−1 at 25°C) (237), and supports a rapid and processive DNA-unwinding activity (238). To our knowledge, the RecBCD enzyme remains the fastest (1,000 to 2,000 bp s−1) (31, 88, 97, 120, 237, 238, 266, 267) and most processive (∼30,000 bp) (31, 88, 120, 236) bona fide helicase reported in the literature. Importantly, DNA unwinding by the RecBCD holoenzyme is substantially faster and more processive than DNA unwinding by either the purified RecB or RecD subunit alone (87) or the isolated helicase subunits of closely related proteins such as PcrA, Rep, and UvrD (see references 88 and 257 for discussion). The macroscopic efficiency of ATP hydrolysis is between 1.3 and 3 molecules of ATP per base pair unwound, depending on reaction conditions (152, 237).
Although named exonuclease V because the vigorous dsDNA degradation requires and initiates from a DNA end, from a mechanistic standpoint, the RecBCD enzyme is a nonspecific ssDNA endonuclease, and both the purified holoenzyme and the isolated RecB nuclease domain function as such in vitro (193, 278, 308, 312). However, on linear duplex DNA substrates, the nuclease activity is manifest as an ATP-dependent dsDNA exonuclease due to the coupling of the endonucleolytic cleavage of ssDNA produced by DNA unwinding to the helicase activity (i.e., the helicase activity processively converts linear duplex DNA into an ssDNA substrate, which is concomitantly fed to the nuclease domain) (92, 240, 284, 294). The nuclease activity requires Mg2+ ions as an essential cofactor and is inhibited by Ca2+ ions (241, 278, 321). The two DNA strands are degraded asymmetrically (92, 93, 284). The ssDNA products generated by this combined helicase-nuclease reaction are heterogeneous in size but can range from just 2 nucleotides in length (111a) to many kilobase pairs (240, 241, 284, 294, 321), depending on the Mg2+ and ATP concentrations (Fig. 5). At physiological concentrations of free (not total) Mg2+ (1 to 2 mM) and ATP (1 to 3 mM) (2, 39, 168), the 3′-terminated strand is hydrolyzed into fragments tens to hundreds of nucleotides in length, and the 5′-terminated strand is cut into much longer segments (approximately a few kilobases) (93), especially if SSB protein is present (16). Upon recognition of the Chi sequence, cleavage of the 3′-terminated strand at the entry site is attenuated at least 500-fold, and endonucleolytic activity is switched to the opposite strand (14, 92) (Fig. 3): the degradation of the 3′-terminated strand is reduced substantially, whereas the hydrolysis of the 5′-terminated strand is slightly upregulated. This “nuclease attenuation and polarity switch” is part of the underlying molecular mechanism that transforms RecBCD from a destructive nuclease-helicase into a productive helicase-nuclease, which initiates the DNA repair process by promoting the formation of a recombinogenic 3′-terminated ssDNA overhang.
FIG. 5.
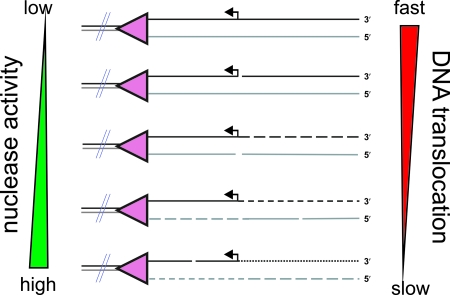
Effect of ATP and Mg2+ ions on the cleavage pattern observed upon DNA processing by RecBCD. During DNA translocation and unwinding, RecBCD cleaves both nascent single strands using a single nuclease active site. The frequency of cleavage on each strand (i.e., the average distance between cleavage points) is dependent on the voracity of the nuclease domain (which increases with increasing free Mg2+ ion concentrations) and the translocation rate (which increases with increasing ATP:Mg2+ concentrations) as indicated. For example, a fast-moving enzyme with low nuclease activity will produce mostly full-length ssDNA (top DNA molecule shown), whereas a slow-moving enzyme with high nuclease activity will produce very closely spaced cleavage points in the unwound DNA (last DNA molecule shown). The relative accessibility of each strand to the nuclease domain is also an important factor, and this is controlled by Chi recognition. Consequently, RecBCD can produce a spectrum of cleavage products dependent upon the precise in vitro solution conditions. Under physiological conditions (1 to 3 mM ATP [39] and 1 to 2 mM free magnesium ion [2]), the processing of Chi-containing DNA by RecBCD results largely in products similar to those depicted in the third and fourth DNA molecules illustrated.
During DNA translocation and unwinding, RecBCD cleaves the DNA using a single nuclease active site (278, 308, 325, 326). Extensive in vitro studies have established that the nuclease and translocation activities are independent (92, 284, 294). Furthermore, the endonucleolytic cleavage of ssDNA that accompanies unwinding is stochastic, and the frequency increases with an increasing free Mg2+ concentration (92, 93). Consequently, RecBCD can produce a spectrum of cleavage products that depend on solution conditions. The density of cleavage positions along each of the nascent ssDNA strands (and, hence, the size of the resultant ssDNA fragments) is a function of (i) the translocation rate, (ii) the ability of the nuclease active site to access each of the ssDNA strands, and (iii) the voracity of the nuclease active site. Note that the free Mg2+ and effective ATP concentrations, which govern the cleavage and translocation kinetics, respectively, are themselves interdependent because ATP chelates Mg2+ ions to form the Mg2+-ATP complex that is the cofactor required for helicase activity. These considerations are fully consistent with the spectrum of different cleavage patterns that can be produced by RecBCD under different solution conditions in vitro (Fig. 5). It was originally observed in vitro that the RecBCD enzyme could uniquely cleave at or near Chi (224), which was an anticipated intermediate step in some early models for recombination initiation (204, 262). This observation led to the “nick-at-Chi” model, which set the stage for mechanistic studies of the Chi-RecBCD interaction. However, it subsequently became clear that the “nick-at-Chi” products are observed only with a narrow set of nonphysiological reaction conditions, and their formation can now be appreciated in the broader context of RecBCD enzymatic behavior and its regulation by Chi. Instead, the various products of DNA processing by RecBCD can be understood as the expected (and predictable) consequences of quantitative differences in translocation and cleavage kinetics. As described below, the recognition of Chi results in a pause in enzyme translocation (92, 93, 120, 267). It is important to recognize that the “desirable” effect of the RecBCD enzyme pausing at Chi is a high probability of (randomly) cleaving DNA in the vicinity of Chi, even under conditions of limited nuclease activity. In agreement, the precise locations of the cleavage events that occur at Chi move closer to Chi when endonucleolytic activity increases (224, 286, 291). The first publication on this phenomenon reported that a nick is introduced on the 3′ strand about 4 to 6 bp upstream of Chi (224, 286). Subsequently, when the magnesium ion concentration was increased to more physiological concentrations, the position of the final, high-frequency cleavage event was seen to occur within the sequence itself (291). These characteristics, and others, were seen to reflect both the attenuation of nuclease activity and the switch in degradation polarity at Chi, which are manifest in the product profiles seen when reaction conditions are systematically varied (93). Because there is a continuum in nucleolytic degradative behavior, both before and after interactions with Chi, these properties do not simply represent two alternative forms or activities of the enzyme; hence, the nick-at-Chi products represent a subset of this general behavior of RecBCD. Thus, the products of processing Chi-containing dsDNA arise from the nonspecific degradative capacity of RecBCD superimposed on a pause at the Chi sequence and the resulting attenuation and polarity switch of endonucleolytic action.
The recognition of Chi is a stochastic event, with the probability of a successful response to a single Chi sequence being around 30 to 40% under optimal conditions (91, 92, 289). For this reason, many substrates used to analyze RecBCD in vitro use three closely spaced Chi sequences in direct repeats, in which case the overall probability of recognition at the “triple-Chi” locus is measured at 80 to 90% (267). Experiments with DNA substrates that had noncomplementary DNA strands at the Chi locus demonstrated that Chi is recognized as ssDNA and specifically as the sequence 5′-GCTGGTGG-3′ by the enzyme approaching from the 3′ side (33). The recognition of Chi is not absolutely sequence specific, and sequences that are similar to Chi (e.g., seven of the eight canonical nucleotides) are also recognized albeit with reduced efficiencies (21, 63, 65). Under conditions of limited free Mg2+, RecBCD displays the property of being reversibly inactivated after an encounter with the Chi sequence (90, 289). The enzyme completes the unwinding of the DNA molecule to which it is bound, but it is incapable of unwinding a new DNA molecule. Consequently, under these unique conditions, the enzyme does not act catalytically on Chi-containing DNA substrates, and a stoichiometric amount of enzyme relative to DNA is required for complete unwinding. However, the Chi-inactivated enzyme can be reversed by the addition of excess Mg2+ to restore normal DNA-processing activities. This inactivation is related to the Chi-dependent disassembly of the holoenzyme into individual subunits (290). However, because the processivity of the individual motor subunits is exceedingly small (tens of base pairs), subunit disassembly clearly cannot occur at Chi and cannot be the molecular basis for regulation by Chi (290), since the enzyme continues to function as a processive helicase-nuclease downstream of the recombination hotspot (91, 92).
The RecBCD enzyme has yet another cryptic activity that is revealed upon Chi recognition and that assists in the next, DNA strand invasion, step of recombinational DNA repair, which requires the RecA protein. Early studies showed that RecA could polymerize on the ssDNA produced by the RecBCD enzyme and could use that ssDNA to produce homologously paired joint molecules (235, 239). Subsequently, it was demonstrated that Chi stimulated this joint molecule formation in a reaction that optimally required the RecA, RecBCD, and SSB proteins (91-93). The stimulation was a consequence of two different effects. As detailed above, the first effect was that the nuclease activity was attenuated and switched, leading to the preservation of the Chi-containing ssDNA (14, 91-93). However, the second stimulatory effect resulted from the loading of the RecA protein by the RecBCD enzyme onto the 3′-terminated Chi-containing ssDNA tail which it produces, thereby handing off to the next step in the homologous recombination pathway (13, 14, 17, 23, 167). This facilitated loading of RecA is essential for RecBCD-mediated recombination in vitro and in vivo due to the competition for ssDNA from the SSB protein (16, 23). Several studies show that the nuclease domain of RecB is required for this process (10, 13, 67, 270). The RecA protein interacts directly with the nuclease domain of RecB, and the modeling of the interaction interface suggests that it mimics the contacts formed between two neighboring RecA monomers within a RecA nucleoprotein filament (270). Since nucleation is known to be rate limiting for RecA nucleoprotein filament formation (54, 108, 157, 158), a simple model for RecA loading envisions the RecB nuclease domain depositing RecA monomers on the 3′ ssDNA tail following Chi recognition. These RecA protomers would then act as nucleation points for net polymerization in the 5′→3′ direction (Fig. 3).
The properties of the RecBC enzyme (the holoenzyme lacking the RecD subunit) have also been extensively studied. Interestingly, the RecBC enzyme is largely devoid of nuclease activity (218), which originally led to the mistaken (but reasonable) conclusion that the RecD subunit harbored the nuclease active site. The RecBC enzyme loads RecA constitutively (i.e., in a Chi-independent manner) onto the 3′-terminated DNA strand at its entry end, and it supports Chi-independent recombination in vivo (55, 66, 187, 295, 296). These observations led to the suggestion that the RecBC enzyme was a phenocopy of the Chi-modified RecBCD enzyme. Clearly, this was not strictly true because the Chi-modified RecBCD enzyme retains 5′→3′ exonuclease activity (12). However, the idea led to a long-standing model for Chi recognition in which a conformational change resulted in the ejection of RecD from the holoenzyme complex (90, 150, 211, 276). This view was supported by the observation that the overproduction of the RecD polypeptide in trans in some cases antagonizes or reverses the effect of Chi on RecBCD (43, 150, 211). The RecD ejection model was eventually disproved by in vitro single-molecule analysis, which is described below (120), but there remains evidence for some form of conformational change involving the RecD subunit following Chi recognition (232, 298). Recent in vivo work suggests that damage to the E. coli chromosome results in a transient loss of all RecBCD-related activities due to the simple titration of RecBCD by the DNA breaks (81). The presence of the free RecD protein in trans somehow prevents the dissociation of the RecBCD enzyme from the damaged chromosomes so that this titration effect becomes permanent. This finding indicates that the recycling of the RecBCD enzyme onto new substrates following Chi recognition requires the disassembly of the complex (290) and that this disassembly is blocked by excess free RecD. Finally, even though its genetic deletion still allows productive recombination, the RecD subunit serves three important functions. First, RecD is needed to “activate” the nucleolytic functions contained within the RecB subunit, since the RecBC enzyme lacking the RecD subunit has comparably little nuclease activity (12, 152, 154). Second, RecD contributes to the processivity of the holoenzyme since its processivity is higher than that of the RecBC enzyme (88, 154). Third, as mentioned above, it also negatively regulates the RecA-loading ability of the RecBCD enzyme (10). This is evident not only for the wild-type RecBCD enzyme but also for the RecB(D1080A)CD mutant (see below), which can recognize Chi but fails to respond appropriately (13); by removing the RecD subunit, the resulting RecB(D1080A)C enzyme shows wild-type RecBC-like behavior, with constitutive Chi-independent RecA-loading ability (10).
By the end of the last decade, a clear picture of the DNA end-processing reaction catalyzed by the RecBCD enzyme was emerging. Importantly, the in vitro biochemical behavior of RecBCD correlated well with its known in vivo functions, both qualitatively and quantitatively, making RecBCD an excellent target for mechanistic interrogation.
DNA Translocation and Unwinding Mechanism
Bipolar DNA translocation mechanism.
The RecBCD enzyme is a rare example of a protein complex containing two DNA motors of opposite polarity. This “bipolar” organization allows the enzyme to translocate along both strands of the DNA duplex in a rapid and highly processive fashion (Fig. 6). The first hints of the dual-motor design came from electron microscopic observation of the unwinding intermediates of RecBCD (284, 293, 294). Under conditions in which nuclease activity is minimized (e.g., with added Ca2+ [241, 321] and/or SSB protein [16]), the enzyme does not produce the Y-shaped unwinding intermediates expected of classical DNA helicase activity. Rather, the enzyme unwinds the duplex to produce one long 5′-terminated ssDNA overhang (41) and a shorter ssDNA loop with an associated ssDNA tail on the 3′-terminated strand (“loop-2-tails” intermediate) (Fig. 6). This observation led to proposals that the RecBCD enzyme could unwind and “rewind” DNA by translocation along one DNA strand (41, 284) or that it possessed two DNA translocation activities, RecB and RecD, which operated at different speeds on opposite strands to unwind the DNA and produce the loop-tail structures (238). However, it was decades later that the underlying mechanism behind these unusual unwinding intermediates was elucidated (87, 288).
FIG. 6.
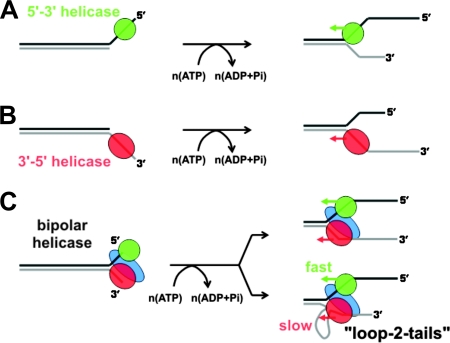
Unwinding polarity of SF1 DNA helicases. (A) Unwinding of DNA by a 5′→3′ DNA helicase. The enzyme moves in a 5′→3′ direction on ssDNA. To unwind duplex DNA, the helicase first binds to a 5′ ssDNA tail neighboring the duplex and then translocates along that strand and into the duplex DNA. (B) Likewise, a 3′→5′ DNA helicase can bind to a 3′ ssDNA overhang and translocate unidirectionally toward the 5′ end and into the duplex portion of the substrate. (C) RecBCD is a bipolar DNA helicase. It contains two DNA helicase subunits of opposite polarity and can initiate unwinding from a blunt duplex end. In the initiation complex, the 3′→5′ subunit (red) binds to the 3′ side of the DNA end, and the 5′→3′ helicase (green) binds to the 5′ side. Although the translocation polarities of the two DNA motors are opposite, they move in the same overall direction on the antiparallel DNA duplex. If the two DNA motors move at unequal speeds, then the faster motor will be associated with a longer ssDNA tail, and the slower motor will be associated with an ssDNA loop and a shorter ssDNA tail. This unwinding intermediate (referred to as a “loop-2-tails” structure) has been observed by electron microscopy (see the text for details).
Using RecBCD mutants in which ATP hydrolysis in either the RecB or RecD subunit was inactivated, electron microscopy was again employed to visualize unwinding intermediates (288). If either subunit is inactivated, a single ssDNA tail is associated with a loop of similar length, broadly consistent with the idea that both RecB and RecD act as independent DNA motors, and each one translocates on one strand of the antiparallel DNA duplex. The inactive motor does not translocate along the DNA, and so an ssDNA loop forms ahead of that subunit. Labeling of the termini of the unwound ssDNA strands revealed that the RecB subunit is responsible for translocation along the 3′-terminated ssDNA strand and the RecD subunit for translocation along the 5′-terminated strand. In parallel, experiments on the isolated RecB and RecD subunits showed directly that they were DNA helicases displaying opposite polarities of unwinding (87). Moreover, either RecB or RecD alone was shown to support DNA unwinding in the RecBCD complex. Together, these results strongly supported a model for DNA translocation in which both the RecB and RecD subunits functioned as ssDNA motors to propel the holoenzyme along the 3′-terminated and 5′-terminated strands, respectively. If the motors traveled at unequal speeds, this would result in the production of “loop-2-tails” unwinding intermediates as observed in the original electron microscopic experiments; if the two motors were to move at the same speeds, more conventional “Y-tailed” intermediates would be observed (Fig. 6). In the first scenario, the leading motor is acting as a true DNA helicase, while the second slower motor is simply an ssDNA translocase. However, it is important to note that this distinction is superficial because experiments with mutants containing only one active motor subunit clearly demonstrate that either RecB or RecD can act alone as an efficient DNA helicase in the context of the holoenzyme (88). This experiment also demonstrates that the two motors are (at least substantially) autonomous. The bipolar DNA translocation model fits well with models for SF1 DNA helicase activity in which the role of the protein architecture specified by the helicase “signature motifs” is to couple ATP hydrolysis to unidirectional ssDNA translocation (89, 257, 305).
The existence of dual motors in RecBCD raises the obvious question of what benefits this apparently energetically costly arrangement may confer. This problem was addressed empirically by studying the properties of mutant RecBCD enzymes in which either the RecB or RecD motor was inactivated by mutagenesis. (Doubly mutated protein, with both motors disabled, is inactive as an ATPase and DNA helicase [288].) Both motors are required for the maximal rate and, in particular, for the high processivity observed in the wild-type enzyme (88). The inactivation of either RecD or RecB results in DNA-unwinding rate decreases of 2- or 3-fold and processivity decreases of 6- or 25-fold, respectively. The fact that the “single-motor” variants of the RecBCD enzyme are slower and less processive than the dual-motor wild-type enzyme implies some cooperation between the two translocating subunits. This cooperation cannot be of a fully concerted type, where the movement of each motor alternates successively, because such a mechanism would be inconsistent with their autonomous behavior and the loop formation. However, their cooperation might be more akin to the “cooperating-monomers” model proposed for the T4-encoded Dda helicase, where the action of a lead helicase monomer is enhanced by a second Dda monomer that is translocating independently behind the first one (48, 49): in the case of RecBCD, the two translocation monomers are within the same complex. Nonetheless, either motor is capable of powering effective DNA translocation alone. Even with just a single active RecB or RecD motor subunit, the holoenzyme remains much faster and more processive than many other DNA helicases studied in isolation. The theoretical basis for the improved DNA translocation and unwinding in bipolar helicases has been discussed (277). It was also suggested that the dual-motor system may allow the RecBCD holoenzyme to bypass gaps or damage on both strands of the DNA duplex (87), a prediction that has been verified experimentally (L. Yang and S. C. Kowalczykowski, unpublished observations).
Additional experiments with the single-motor mutant enzymes demonstrated that the RecB motor activity is absolutely required for the recognition and response to Chi, whereas RecD motor activity is dispensable for this recognition (268). This finding implies that the single strand of DNA containing the Chi sequence must be translocated into the RecBCD enzyme by RecB to permit interactions with Chi; this view is in full accord with the crystal structure and will be discussed below.
Relative speeds of the two motors.
The finding that the RecB and RecD subunits are largely autonomous motor subunits raised the question as to which is the faster motor in the dual-motor holoenzyme and whether the faster motor is the same subunit at all times and conditions. Biochemical analysis of the single-motor mutants showed that each motor subunit displayed a characteristic dependence on the Mg2+ concentration (268). When driven only by the RecD motor, RecBCD moved fastest at limiting Mg2+ concentrations. In contrast, high free Mg2+ concentrations favored a holoenzyme driven only by the RecB motor. For the wild-type holoenzyme, electron microscopy showed conclusively that for the conditions examined (which were limiting for Mg2+), the RecD subunit is the lead motor and, hence, the helicase; this meant that the RecB subunit is the slower motor and, hence, the translocase (41, 288). This conclusion is supported by recent single-molecule analyses, which demonstrated that a small loop of ssDNA must exist ahead of the RecB motor before Chi recognition (266). Quite unexpectedly, however, following Chi recognition, the holoenzyme moves more slowly because the roles of these two motor subunits reverse, and RecBCD is now driven by RecB as the lead motor. The finding that RecD is the lead subunit and is ahead of the RecB subunit leads to the conclusion that RecD is actually past the Chi sequence when RecC, which is behind RecB, recognizes Chi. Furthermore, because RecB assumes the lead after Chi, the recognition event must slow or stop the RecD subunit, since it remains part of the holoenzyme beyond Chi (120). Thus, Chi regulates holoenzyme translocation speed by inactivating the faster motor subunit and switching to the slower motor (266). If the frequency of RecA loading events is independent of RecBCD enzyme speed, then this switch to a slower motor may help to increase the coverage of the RecA protein on the Chi-containing ssDNA by decreasing the spacing of RecA nucleation events.
It is possible that the subunit used as the lead motor by the RecBCD enzyme can be changed, at least in vitro, by a simple manipulation of the solution conditions. It is also expected that the speed of each motor can be independently affected by the ATP concentration, because the affinities of each subunit for ATP, as measured by 8-azido-ATP cross-linking, are 30 μM and 120 μM for RecD and RecB, respectively (140). Moreover, based on precedents in the literature that were established for other SF1 DNA helicases (106, 139), it is also possible that the trailing motor subunit will be moving faster on ssDNA than it does while unwinding duplex DNA.
Recently, it was shown that the speed of the RecB motor can be reduced by novel mutations in one of the conserved helicase motifs (motif 6, Y803H and V804E) (8). In vivo, the mutant RecBCD enzyme failed to respond to Chi and was deficient in recombination, but it retained helicase-nuclease activity. The resulting mutant RecBCD comprised a normal fast RecD motor and the slow RecB motor. As a consequence, on linear dsDNA, the RecD subunit traveled to the end of its strand (the 5′ strand) well before the RecB subunit finished translocating to the end of its respective strand (the 3′ strand). Interestingly, the 3′ strand was degraded only to the position where the RecB subunit would have been located when the faster RecD subunit reached the far end of the DNA. This finding meant that the RecB subunit, with its associated nuclease domain, must have stopped when RecD stopped translocation, and, perhaps as a result of a pause, the RecB nuclease cleaved the ssDNA for a last time at that same location. These findings were interpreted as evidence for intersubunit communication wherein RecD communicated its translocation status to RecB. However, these findings are also consistent with the nonspecific degradation of the ssDNA until the RecD subunit reaches the end of the DNA (or is otherwise stopped), after which the holoenzyme dissociates, during which time the potentially paused RecB motor and nuclease subunit cleave one last time.
The above-described data highlight just some of the complexities and challenges associated with studies of a DNA helicase comprising two (mostly) autonomous motor subunits that have translocation velocities which vary with reaction conditions, and either subunit could be unwinding dsDNA or simply translocating on ssDNA.
Step size.
Helicases are directional motors that use either a stepping mechanism (e.g., inchworm) (305) or Brownian ratchet (175) to move along DNA. In the simplest case, the step size is nothing more than the physical distance through which the enzyme advances, due to conformational changes, when it moves on the ssDNA; the steps may or may not be of uniform length. However, the precise value and meaning of the measured step size depend on the experimental approach used, and there has been little consistency in the measured values, even for work on the same enzyme. Nowhere is this better illustrated than for the RecBCD enzyme. Furthermore, in thinking about the stepping activity of RecBCD, it should be remembered that the interpretation of any data pertaining to the step size is potentially complicated by the fact that the complex employs two motor subunits.
The simplest approach to measuring a step size is to investigate the macroscopic relationship between ATP hydrolysis and DNA unwinding. Two studies of this type suggested that 1.4 to 3.0 ATP molecules are consumed for every base pair translocated (152, 237). This value is reduced if the RecD motor is inactivated (1.1 to 1.2 ATP molecules consumed per base pair) or removed (1.3 to 1.4 ATP molecules consumed per base pair) (152). A simple interpretation of these results is that each motor makes a 1-base step for each ATP hydrolyzed, consistent with models for the related PcrA and UvrD helicases (89, 174, 301, 305). However, it should be noted that the experiments ignore possible contributions to the step size value from ATP hydrolysis that is uncoupled from DNA translocation and unwinding.
An alternative method for step size determination employs rapid reaction techniques to monitor the unwinding kinetics of a series of duplexes of different lengths (188). The distribution of a translocating motor on a one-dimensional lattice as a function of time is dependent on the distance moved per rate-limiting step (190, 280). Consequently, global fitting of DNA-unwinding time courses to an appropriate stepping model can yield a “kinetic” step size. The kinetic step size measures the average distance between rate-limiting enzymatic events; the kinetic step size could be a simple parameter, being the same size as the physical step size, or it can be a complex term, comprising multiple translocation steps. Moreover, in deriving this value, it is assumed that each step is associated with a single, strongly rate-limiting kinetic event and that the enzyme-DNA complexes investigated are kinetically homogenous (190). Thus, the kinetic step size does not necessarily relate to the net distance traveled per ATP molecule. This value was measured at 3.4 (±0.6) bp for the RecBCD enzyme and was shown to be independent of temperature and ATP concentration (189).
An altogether different approach was again employed with the RecBC enzyme to measure another type of physical step size (34). By challenging the translocating enzyme with ssDNA gaps of various lengths, it was shown that RecBC (lacking the 5′→3′ RecD helicase subunit), which translocates along DNA on the 3′-terminated strand relative to the entry point, could traverse ssDNA gaps up to ∼23 nucleotides in size. This and other findings implied that the enzyme translocates in discrete physical steps of 23 (±2) bp, leading to the development of a “quantized” inchworm model for DNA translocation wherein the RecBC enzyme takes a large “spring-loaded” physical translocation step on the duplex DNA ahead of itself while recognizing that DNA unwinding was occurring by translocation along ssDNA in smaller steps powered by the RecB subunit at the rear of the complex.
Single-molecule approaches.
The discussion above highlights the difficulties associated with measurements of the stepping motion of a molecular motor. Progress in this area is likely to depend heavily on the application of new single-molecule techniques that have already been applied successfully to the “classical” motor proteins such as myosin and kinesin (135, 220). Indeed, the last 10 years have witnessed remarkable progress in single-molecule analysis as a new and powerful tool for the study of DNA and its interactions with proteins (47). Such methods have the potential to remove the complications of heterogeneity and asynchrony in ensemble kinetic measurements. The processive stepping activity of DNA motors (including helicases, translocases, and polymerases) is particularly amenable to single-molecule analysis, and researchers have quickly capitalized on the robust activity of RecBCD to exploit the new technology (Fig. 7).
FIG. 7.
Single-molecule analysis of the RecBCD enzyme. (A) Direct visualization of DNA translocation using fluorescently labeled DNA. A single lambda phage DNA molecule is attached to a polystyrene bead, and YOYO-1 (a fluorescent DNA binding dye) is then bound. The bead is held in an optical trap, with the DNA molecule stretched out behind it by solution flow. A single molecule of the RecBCD enzyme is able to bind to the free DNA end. Upon the addition of ATP, the RecBCD enzyme unwinds and degrades the duplex. This is observed as a progressive shortening of the fluorescent DNA in an epifluorescence microscope. (A movie of this experiment can be viewed at http://microbiology.ucdavis.edu/sklab/kowalczykowskilab.htm.) The cartoon is not to scale; the stretched lambda DNA is 48.5 kbp (∼15 to 16 μM) long and binds several thousand molecules of YOYO-1 dye. (B) Direct monitoring of RecBCD translocation by tracking of a fluorescent nanoparticle (40-nm diameter). The instrument setup is as described above for A except that the DNA is not labeled. Instead of monitoring DNA degradation, the fluorescent nanoparticle is attached to the translocating enzyme via a biotin moiety on the RecD subunit. The position of the nanoparticle relative to the position of the optical laser trap measures DNA translocation. (C) RecBCD enzyme translocation on DNA monitored by tethered-particle light microscopy. The biotinylated RecBCD enzyme is bound to a streptavidin-coated bead. The RecBCD enzyme then binds to the free end of a surface-attached DNA molecule (∼1.4 kbp), effectively tethering the bead to the surface with the DNA as a linker. The translocation of RecBCD along the DNA pulls the bead toward the surface, which places an increasing constraint on the bead's Brownian motion that can be monitored by light microscopy. (D) RecBCD enzyme translocation on DNA monitored by high-resolution optical trapping. A biotinylated RecBCD enzyme is attached to a streptavidin-coated surface (alternatively, but not shown, a second optical trap can be used). RecBCD, attached to a bead, can capture the free end of a 7-kbp DNA molecule. The bead is held in an optical trap with a force clamp. Upon DNA translocation, the RecBCD enzyme pulls on the DNA and generates force against the trapped bead. A feedback mechanism moves the surface stage toward the trapped bead to maintain a constant force. The instrument can assess the effects of applied force against translocation and measure the movement of the enzyme with nanometer resolution.
The first measurements of the activity of single RecBCD enzymes were reported in 2001 using optical tweezer and tethered-particle light microscopy approaches. In the optical tweezer experiments (31), DNA molecules are attached to polystyrene beads and labeled uniformly with YOYO-1, a fluorescent DNA binding dye (Fig. 7A). The beads are injected into a flow cell and caught in a laser trap. The trailing duplex DNA, bound by a single RecBCD enzyme, is observed by fluorescence microscopy. Upon the addition of ATP, the DNA strand shortens as the enzyme tracks along, displacing the florescent dye and degrading the duplex. Unexpectedly, although the mean translocation rate is similar to that determined by bulk measurements, those of individual RecBCD enzymes vary widely, indicating static disorder within the population. The source of this heterogeneity is currently unclear. Importantly, static disorder in the translocation rate complicates the meaning of the step size derived from the analysis of transient kinetics. Variation of the translocation rate of individual RecBCD enzymes as a function of time (dynamic disorder) is not detected at the resolution of these experiments. On Chi-free DNA, the enzyme moves at a constant rate, without detectable pauses, regardless of the local DNA sequence. However, when the DNA contains a Chi sequence, the translocation is unexpectedly altered (267). The RecBCD enzyme pauses at the Chi sequence (for between 1 and 15 s) and then resumes translocation at about one-half of the rate observed before the encounter with Chi. The velocity after Chi is not proportionally related to the velocity prior to Chi but, rather, is randomly distributed around the new average rate. The lower rate of translocation post-Chi is associated with a switch in the lead motor subunit, and the pause prior to the switch is the kinetic lifetime associated with the associated conformational change (266). Although conventional bulk measurements had hinted at a role for Chi in controlling DNA translocation (92), this result highlights the type of information that becomes easily accessible by single-molecule analysis.
In the tethered-particle light microscopy experiment (94), DNA is attached to a glass surface at one end. The biotinylated RecBCD enzyme, itself attached to a streptavidin-coated bead, is able to bind the free DNA end (Fig. 7C). Upon the addition of ATP, the bead is pulled toward the glass surface as the enzyme translocates along the DNA. Movement is observed by light microscopy as an increasing constraint on the bead's Brownian motion. Because the RecBCD enzyme was attached to the bead via the RecD subunit, and a negligible dissociation of the bead was observed at the position of a Chi sequence, it was argued that RecD is not ejected at Chi. Unfortunately, because neither the pause nor velocity change was detected, it remains unclear whether Chi was actually being recognized in these experiments. Indeed, consideration of the relative translocation speeds of RecB and RecD may provide an explanation for the apparent lack of Chi recognition: the experimental protocol employed low concentrations of ATP (5 to 50 μM) and a short DNA substrate (∼1.4 kbp) (94). These conditions favor rather slow translocation by the RecB motor, which would result in RecD reaching the end of the short DNA substrate before RecB has reached the Chi sequence.
Neither of the above-described experiments had the spatial resolution to measure the step size of the single translocating RecBCD enzyme. An alternative experimental system is to attach RecBCD, biotinylated on RecD, to a glass surface and monitor translocation along a DNA duplex that is attached at its distal end to a bead (Fig. 7D) (222). The bead is held in a laser trap, and a feedback mechanism is used to move the stage toward the trapped bead under constant force to compensate for the enzyme translocation. Because discrete steps were not detected, and the translocation is measured with a 2-nm spatial resolution, these experiments excluded step sizes for translocation (for the RecD subunit) of greater than 5 bp. Under the applied force, which acts against the direction of translocation, spontaneous pauses and backsliding of the complex were observed. These probably relate to the dissociation/pausing of the leading motor, followed by a slippage of the enzyme along the DNA (the RecBCD enzyme encircles the DNA strands within channels) (see below) to the position bound by the slower DNA motor.
Finally, the movement of the RecBCD enzyme was imaged directly on bead-immobilized DNA stretched by flow (Fig. 7B) (120). A fluorescent nanoparticle is attached to the RecD subunit, allowing the direct observation of RecBCD translocation. The bead was seen to move along the DNA, pause at Chi, and then resume slower translocation exactly as observed previously (267). Because the nanoparticle is still associated with the translocating enzyme after Chi recognition, these experiments provide the clearest possible evidence that the recognition and response to Chi do not involve RecD ejection.
STRUCTURE AND MECHANISM OF RecBCD
The crystal structure of the entire RecBCD enzyme bound to a DNA substrate was solved to 3.1 Å in the absence of nucleotide cofactor (256) (Fig. 8). The DNA substrate consisted of a single self-complementary 43-base oligonucleotide that forms a 19-bp duplex with a 5-base hairpin at one end. This DNA substrate can bind RecBCD in only one orientation because a hairpin structure is known to prevent RecBCD binding (287). As discussed above, the SF1 helicase motors found in the RecB and RecD subunits are expected to function as ssDNA translocases. Despite this fact, the enzyme uses blunt-ended dsDNA as an initiation substrate, which raises the question of how ssDNA is generated for the helicase domains. The observation that the DNA in an initiation complex is sensitive to permanganate modification (103) argues that the binding event alone generates the ssDNA that is required to drive translocation using the SF1 motor design. The crystal structure of the initiation complex confirms that this is indeed the case. Despite the fact that the DNA substrate forms a dsDNA end that can be fully duplex, the DNA substrate in the crystals is unwound by 4 bp, with each ssDNA tail inserted into a different entry tunnel in the complex (Fig. 8B). The two tunnels propagate right through the RecBCD complex. As the enzyme translocates, DNA enters as duplex, is split into two nascent ssDNA strands that pass through these tunnels, and is presented to the nuclease domain at the rear of the enzyme. As DNA passes from the front to the back of the enzyme, it encounters several important structural features in the complex (Fig. 8B). These include an “arm,” which stretches ahead of the enzyme to contact incoming duplex DNA; a “pin” upon which the duplex is split into two nascent ssDNA strands that pass into tunnels; two helicase motors, which drive the translocation; a “Chi-scanning site,” which searches for a correctly orientated recombination hotspot; and, finally, the nuclease active site, which cleaves ssDNA as it emerges from the rear of the complex. It was reported that a stimulatory RNA is present in the RecBCD complex (11). No such RNA was observed in the structure despite the preparation used for crystallography being 100% active for DNA unwinding and capable of efficient Chi recognition (M. S. Dillingham, unpublished observations).
FIG. 8.
Crystal structure of a RecBCD-DNA initiation complex. (A) The RecBCD-DNA complex. Alpha-helices are shown as cylinders, and beta-sheets are shown as arrows. The color scheme is the same as that in the primary structure diagram (Fig. 4). The helicase domain of RecB (RecBHel) is shown in red, and the nuclease domain (RecBNuc) is in magenta. RecC is in blue, RecD is in green, and the hairpin DNA substrate is in orange. The hairpin end of the DNA is not visible, as it is disordered in the structure, but would occupy the space to the left of the DNA end as shown here. (B) Cutaway view of the RecBCD-DNA complex showing the tunnels running through the complex. The color scheme is described above. The protein is shown as a space-filling model, which has been cut back (dark surfaces) to reveal the ssDNA tails in the tunnel entrances. The DNA substrate has not been cut away and is shown as a partially transparent space-filling model. Architectural features on the path of the ssDNA strands as they are pulled through the complex are highlighted (see the text for details).
RecB Subunit
Biochemical analysis had suggested that the RecB protein was modular in structure, and this is borne out in the crystal structure. The N terminus of RecB forms the four-subdomain structure expected for a UvrD-like SF1 helicase (257) (Fig. 9). Two of these subdomains display a RecA-like fold (subdomains 1A and 2A). These tandem RecA domains are the core helicase domains and contain all seven helicase “signature” motifs (257). The helicase motifs are found at the interface between subdomains 1A and 2A and form the nucleotide binding pocket and part of the ssDNA binding site. Motifs 3 and 6 are known to be important for coupling ATP hydrolysis to ssDNA translocation (116). Contacts between the RecB protein and the 3′ ssDNA tail are essentially the same as those found in complexes of related helicases with DNA (156, 174, 305), although the 3′ tail does not extend completely across the expected ssDNA binding site. The protein binds the ssDNA via aromatic stacking interactions with the nucleobases and electrostatic contacts with the phosphate backbone. Subdomains 1B and 2B are commonly found in related helicases and were suggested to play general roles as “auxiliary domains” that are responsible for protein-protein interactions or for modifying the basic helicase activity by, for example, targeting to specific DNA structures (257, 259). In agreement with this idea, the RecB auxiliary domains play roles in dsDNA binding and protein-protein interactions. Subdomain 1B forms an “arm” that contacts intact duplex DNA about 12 bp ahead of the ssDNA/dsDNA junction (Fig. 10). The role of the arm is currently unclear, but it may direct the helicase to DNA ends, act as a guide for the duplex DNA during translocation, mediate the large translocation step across ssDNA gaps, or play a direct role in destabilizing the duplex ahead of the translocating enzyme (analogous to the role suggested for auxiliary domain 2B of the PcrA helicase) (305). The RecB 2B domain inserts into a large hole in the RecC protein to form an unusual, intimate, and stable protein-protein interaction.
FIG. 9.
Crystal structure of the RecB helicase domain. (A) Cartoon representation of the RecB helicase domain showing the four subdomain structures characteristic of SF1 DNA helicases. The nuclease domain, which is attached via a long linker, is partially shown in magenta. The DNA substrate is shown in deep purple. (B) RecB helicase with the location of the seven conserved helicase “signature” motifs shown. (C) Surface representation colored as described above for A. Note the cleft at the interface of subdomains 1A and 2A, which forms the ATP-binding pocket. Also particularly evident is the 70-amino-acid linker that connects the helical domain to the nuclease (Nuc) domain (the linker is colored rose and runs upward from left to right). (D) Surface representation colored as described above for B. Note that the helicase motifs line the ATP binding cleft and extend to the ssDNA binding site that is formed across the top surface of subdomains 1A and 2A.
FIG. 10.
The RecB arm. Shown is a close-up view of the “arm” structure (red) formed by an auxiliary subdomain (subdomain 1B) of RecB. The arm contacts duplex DNA ahead of the translocating enzyme. A conserved patch of residues in close proximity to the minor groove of the DNA substrate (black semitransparent model) is shown in yellow. The RecC protein is in the foreground (blue). The 5′ ssDNA tail is pointing toward the viewer through a hole in the RecC protein.
The C terminus of RecB is found at the rear of the enzyme complex and is connected to the helicase domain by a ∼70-amino-acid linker packed neatly against the RecC protein. As predicted previously (20), the C terminus forms a single discrete domain with structural homology to lambda exonuclease. The nuclease active site is revealed by the coordination of a Ca2+ ion by the highly conserved residues found in the nuclease motifs (20), including aspartate residues from motifs 2 and 3 (Fig. 11). A mutation of either aspartate residue (D1067 and D1080) to alanine or lysine (K1082) to glutamine results in the complete elimination of the nuclease activity (278, 308, 325), whereas a mutation of the conserved tyrosines (T1081 and T1114) to alanine has no effect on nuclease activity. Ca2+ ions inhibit the nuclease activity of RecBCD (241, 278, 283), presumably by competing with Mg2+ ions for the active site.
FIG. 11.
The active site of the RecB nuclease domain. The active site of the RecB nuclease domain is marked out by the coordination of a Ca2+ ion (yellow) with conserved nuclease motifs. The three motifs characteristic of a diverse collection of nuclease enzymes are numbered (20). Motif 1 (orange) contains a glutamate residue (E1020) that may be involved in the coordination of a second divalent cation. Motifs 2 (blue) and 3 (red) contain aspartate residues (D1067 and D1080) that coordinate the bound Ca2+ ion. A motif specific to the “RecB-like nuclease” family is shown in green and contains highly conserved tyrosine and glutamine residues (Q1110 and Y1114). A histidine residue (magenta) that is completely conserved, but does not form part of a defined nuclease motif, also coordinates the bound Ca2+ ion (H956).
RecC Subunit
Because so little was known about RecC from biochemical analysis, the structure of the RecC protein has proven to be particularly informative. The overall architecture of RecC is striking; surface representations reveal a large hole in the center of the protein flanked by two smaller tunnels on either side (Fig. 12). As described above, the large hole accommodates domain 2B of the RecB protein. One of the small tunnels contains the 5′-terminated ssDNA strand, and the other is positioned immediately “behind” the RecB motor domains so as to accept the 3′-terminated strand following the initiation of DNA translocation and unwinding. Consequently, during DNA unwinding, the RecBCD complex threads itself onto both nascent ssDNA strands via the tunnels in the RecC protein. Dissociation from the nucleic acid lattice is minimized because it would require the backsliding of the whole complex to the end of the remaining DNA. Therefore, the tunnels in RecC provide a simple structural basis for the observation that the RecC protein dramatically stimulates the processivity of RecB helicase activity. The tertiary structure of RecC shows unexpected structural homology to DNA helicase domains in the N terminus (256) and to nuclease domains in the C terminus (146, 231) (Fig. 12). However, within the RecC primary structure, there has been a complete loss of the active-site amino acid motifs that are normally associated with helicase and nuclease activity. The implication of this is that the RecC protein is a catalytically dead helicase-nuclease and that the recB and recC genes probably arose from a gene duplication event (146).
FIG. 12.
Locations of tunnels in the RecC protein for the nascent ssDNA strands. (A) Cartoon representation of the RecC protein revealing its subdomain structure. Four subdomains (green, red, yellow, and blue) are characteristic of SF1 DNA helicases, although the RecC protein is not active as a DNA helicase and does not possess the conserved motifs. The C-terminal domain (magenta) (“Nuc”) has a nuclease-like fold, but note that the RecC protein is not a nuclease and does not possess the conserved motifs expected for a nuclease. The 5′ ssDNA tail of the DNA substrate (black) is pointing toward the viewer and is in a tunnel in the RecC protein. The 3′ ssDNA tail runs underneath RecC (where it is in contact with the RecB helicase domain) (not shown) and toward a second small tunnel in RecC. The duplex DNA points away from the viewer below the 5′ ssDNA strand and the RecC protein. (B) Space-filling model of the RecC protein, colored as described above for A. This illustration clearly shows the two small tunnels for the nascent ssDNA strands and the large central hole that accommodates RecB auxiliary subdomain 2B. In particular, note that the 3′ tunnel is formed at the top surface of the core helicase-like domains (green and red) and that mutations (recC1004 allele) associated with altered Chi recognition specificity (black) map to this region. The 5′ tunnel is formed by the nuclease-like domain. (C) View equivalent to those of A and B with the RecC protein shown as a space-filling representation completely in blue. Cartoon representations of the RecB and RecD proteins have been added. The color scheme is described in the legend of Fig. 4. Note that the RecD protein and the RecB nuclease are positioned above the 5′ ssDNA and 3′ ssDNA tunnels, respectively. RecB subdomain 2B occupies the large hole in the middle of the RecC protein.
The inactive helicase and nuclease domains of RecC form parts of the 3′ and 5′ tunnels, respectively. Several lines of evidence support the view that the 3′ tunnel in RecC is the Chi recognition site. Firstly, Chi is known to be recognized as ssDNA on the 3′ strand (33), and the tunnel in RecC is on the path of this strand. Secondly, the tunnel in RecC is located at the correct distance from the nuclease active site to explain the location of the final cleavage sites at the Chi sequence (see the RecBCD enzyme mechanism section below). Thirdly, the 3′ tunnel in RecC is, in structural terms, a nonfunctional equivalent of a helicase motor (it is formed by the core helicase domains 1A and 2A), which might provide an ideal protein architecture to function as a scanning site for a specific ssDNA sequence. Finally, members of the RecC* class of mutations, which alter the efficiency and specificity of Chi recognition, map to the 3′ tunnel in the RecC subunit (22, 124, 248) (Fig. 12 and see below). Indeed, recent site-directed mutagenesis experiments confirm the identity of this region as the Chi recognition locus (N. Handa, L. Yang, M. S. Dillingham, D. B. Wigley, and S. C. Kowalczykowski, unpublished data).
The RecC protein also appears to assist in the DNA-unwinding activity by contributing a dual-methionine “pin” that acts as a wedge to split the duplex at the junction between ssDNA and dsDNA (Fig. 13). Related architectural features were observed previously at the DNA junctions of other DNA translocases (228, 257, 258). Given that the methionine pin is such a striking feature in the structure, and one with an apparently central function in the enzyme's unwinding mechanism, it is surprising that the methionine dyad is poorly conserved. However, it is possible that the steric role of the methionine pin can be accomplished by a wide range of amino acid substitutions. Site-directed mutagenesis on the RecC pin should help resolve this issue. The RecC protein acts as a core scaffolding protein in the holoenzyme because, in addition to both directing the paths of nascent single strands during unwinding and contacting the RecB protein, it also forms protein-protein contacts with RecD. Note that there are no direct contacts between RecB and RecD in this initiation complex.
FIG. 13.
The methionine pin of the RecC protein. A methionine dyad (yellow) in RecC (blue) forms a pin structure at the junction of ssDNA and dsDNA (black). The translocation of the two single strands of DNA by the RecB and RecD helicase motors will split the duplex over the methionine pin. The free 3′ end of the DNA substrate is indicated. The 5′ ssDNA tail disappears into a tunnel in the RecC protein.
RecD Subunit
The RecD component of the RecBCD complex represents the first crystal structure of a 5′→3′ SF1 DNA helicase. The structure reveals three ordered domains, two of which form the tandem RecA-like fold responsible for DNA motor activity (equivalent to core domains 1A and 2A of RecB) and the third of which is responsible for protein-protein interactions with RecC (Fig. 14). RecD is located immediately at the exit point of the 5′ tunnel in RecC. In this position, the 5′→3′ ssDNA motor is ideally located to propel the 5′-terminated ssDNA strand through the tunnel and on toward the nuclease domain at the rear of the enzyme. However, the 5′-terminated ssDNA strand in the crystal structure does not reach the RecD motor and therefore probably cannot be engaged by the RecA-like helicase domains until translocation is initiated by the RecB motor. This suggestion is supported by the observation that a mutant RecBCD enzyme containing an inactive RecB motor cannot initiate the unwinding of DNA substrates unless they possess short 5′-terminated ssDNA overhangs (88). The optimum binding substrate for RecBCD possesses 10 nucleotides on the 5′-terminated DNA strand (319). This suggests that the RecD helicase binds several nucleotides, as is expected for an SF1 DNA helicase (257). The RecD subunit may be required to position the RecB nuclease domain in an active state, thereby explaining the very low nuclease activity observed in the RecBC complex. There is no direct contact between RecD and RecB evident in the structure. However, the binding of ATP to RecD or its engagement with ssDNA could cause the small movement that could result in a direct interaction between the two, which would explain the regulation of nuclease activity by RecD; alternatively, the regulation could be indirect via structural changes mediated through RecC.
FIG. 14.
Locations of subdomains and motifs in the RecD protein. (A) Cartoon representation of the three-subdomain structure of RecD. The small alpha-helical N-terminal domain 1 (wheat) is responsible for contacts with the RecC protein. Subdomains 2 and 3 (green and red) form a tandem RecA fold and are equivalent to subdomains 1A and 2A of SF1 DNA helicases. The 5′ ssDNA tail (deep purple) is shown approaching these core helicase domains. A portion of the protein comprising residues 466 to 523 is not resolved in the structure. (B) Cartoon representation with the seven conserved helicase motifs indicated. (C) Surface representation colored as described above for A. (D) Surface representation colored as described above for B. Notice the location of the helicase motifs at the interface and top surface of the core helicase domains. Their general location is similar to that seen in the RecB protein. Two regions are shown for motif 1a. The lower patch (*) represents the motif as defined previously (223). However, these residues are poorly conserved in RecD, do not resemble a canonical motif 1a, and do not occupy a similar position to the motif 1a of other SF1 helicases. The upper patch of residues (L201 to A208) represents a strongly conserved region that occupies a position similar to that of motif 1a of other SF1 helicases.
RecBCD Holoenzyme
By revealing the relative positions of its functional domains, the crystal structure of RecBCD contributes a key piece to the puzzle of the RecBCD enzyme mechanism. In particular, the location of a putative Chi-scanning site between the RecB motor and the RecB nuclease domain suggests a simple minimal model for the nuclease polarity switch at Chi (Fig. 15A). Before Chi is recognized, the enzyme translocates along the DNA duplex with both nascent single strands of DNA being fed through the tunnels in RecC and exiting at the rear of the enzyme close to the RecB nuclease domain (Fig. 15B). Rapid and processive DNA translocation is powered by the bipolar ssDNA motors located in the RecB and RecD subunits. The RecD motor is faster, resulting in the formation of a small ssDNA loop ahead of RecB (288). The nascent 3′-terminated single strand is positioned favorably for nucleolytic degradation and is cut frequently. The 5′-terminated strand only occasionally accesses the nuclease active site, and accordingly, it is cut rather less frequently. Note that even though the single strands of DNA would be presented to the nuclease active site from opposite sides, their geometries in the active site may be identical because they are in an antiparallel orientation as they exit the enzyme.
FIG. 15.
Model for RecBCD enzyme mechanism (see the text for details). (A) Cartoon representation of the initiation complex as seen in Fig. 8. The three subunits are color coded as described above, with important functional regions in the structure also labeled. (B) Pre-Chi recognition. RecBCD travels along duplex DNA powered by ATP hydrolysis in the RecB and RecD helicase motors. RecD can be the faster motor, and consequently, a loop of ssDNA may form ahead of the slower RecB motor. The RecB nuclease domain is most favorably positioned to cleave the 3′ ssDNA tail, and this strand is therefore hydrolyzed much more vigorously than the 5′ ssDNA tail. Between the RecB motor and the nuclease domain, the 3′ ssDNA tail is scanned for the Chi sequence as it passes through a tunnel in the RecC protein. (C) The Chi sequence is recognized and remains tightly bound in a tunnel in the RecC protein. The complex pauses before translocation resumes at a reduced rate. Exit of the 3′ ssDNA tail is prevented, and so a final cleavage event takes place on this strand just upstream of the Chi sequence. The 5′ ssDNA strand continues to exit from the rear of the enzyme, where, in the absence of competition from the complementary strand, it is now cleaved more readily by the nuclease domain. A loop of ssDNA accumulates downstream of the Chi sequence. The RecB nuclease domain undocks, recruits RecA proteins from solution, and loads them onto the growing ssDNA loop to promote RecA nucleoprotein filament formation. This process is proposed to be triggered by the release of the RecB nuclease domain from RecC following Chi recognition.
As the enzyme translocates, the 3′-terminated strand is continually passed through the Chi-scanning site formed by the helicase-like domains of RecC (22, 124). When a Chi sequence enters this recognition domain, it has a finite probability of being successfully recognized. It was suggested that the recognition of the Chi sequence is manifest as the tight binding of the Chi sequence before it exits the RecC tunnel and the prevention of its escape toward the nuclease domain (Fig. 15C) (58, 256). The tight binding of the Chi sequence would prevent further translocation of the 3′-terminated tail into the nuclease domain and, thereby, ensure a final cleavage event near Chi (291). This final cleavage at Chi is reinforced by the pause that accompanies Chi recognition (120, 267). The upregulation of degradation of the 5′-terminated strand may simply reflect the lack of competition from the bound 3′-terminated strand, or the slower translocation of the RecBCD enzyme after Chi recognition may also contribute to this effect (267). Alternatively, Chi recognition may produce a conformational change in the enzyme that assists in the nuclease polarity switch by repositioning the nuclease domain toward the 5′ ssDNA exit site. The location of a Chi-scanning site immediately behind the RecB motor neatly explains the observation that the RecD motor is dispensable for Chi recognition, whereas the RecB motor is essential (268). If the RecB motor is inactivated, the Chi sequence cannot easily pass through to the scanning site for recognition, likely because in the absence of the ATP hydrolysis-induced changes in RecB structure and DNA affinity, the ssDNA remains relatively stably bound to one conformation. Indeed, electron microscopic analysis shows that the mutation of the RecB motor impedes the movement of ssDNA through the subunit (288).
Following Chi recognition, the enzyme continues to translocate (Fig. 15C). Because the Chi sequence remains bound to RecC for some time, an ssDNA loop is produced downstream of Chi on the 3′-terminated strand. Evidence for the presence of such a loop has been provided by single-molecule analysis (267). The formation of the ssDNA loop is apparently not an essential aspect of RecBCD function because the RecBC enzyme does not form an ssDNA loop (288), and it is both recombination proficient (7) and a constitutive RecA loader (66); rather, the formation of the ssDNA loop reflects the underlying longer-lived Chi-RecC subunit interaction that mediates the Chi-induced enzymatic changes in RecBCD. RecA loading involves the nuclease domain of RecB, which would need to be released from the surface of RecC following Chi recognition to expose the RecA interaction surface (270). This structural change would permit interaction with RecA and the subsequent deposition of RecA prenuclei on the growing ssDNA loop (270). The nucleation of a RecA filament requires up to four to five monomers (108) that could be delivered by RecB in one step or that could be transiently stabilized by RecB. Successful nucleation would promote the growth of the nucleoprotein filament required for DNA strand exchange. Finally, because the unwound ssDNA is produced in the 3′→5′ direction but net RecA polymerization is 5′→3′ (230), nucleoprotein filament assembly must necessarily be discontinuous (66). The frequency of nucleation by RecB is unknown, but the stimulatory distance over which Chi affects recombination is at least 10 kb (213).
Mutants of the RecBCD Enzyme
The crystal structure and proposed mechanism of RecBCD provide a framework for an understanding of the behavior of mutant RecBCD proteins (see reference 4). In the case of the RecB subunit, many of the mutations occur in the helicase motifs (Fig. 9B and 16) and would be expected to reduce the translocation activity of the RecB motor to various extents. Indeed, site-specific mutagenesis of the conserved Walker A motif lysine (helicase motif 1) in RecB (K29) and RecD (K177) to a glutamine demonstrated its need for ATP hydrolysis, DNA unwinding, and translocation in each subunit (61, 87, 133, 151, 153, 155, 266, 268, 288). Many mutations in recB that were identified in genetic screens, recB2109 (T807I), recB2152 (T807I), recB2154 (R800C), and recB2155 (R794C), contain mutations in or near the highly conserved residues that form helicase motif 6 (S. K. Amundsen and G. R. Smith, personal communication) (4, 23). Studies of related SF1 helicases demonstrate that this motif is involved in ATP hydrolysis and in coupling ATP hydrolysis to DNA translocation (116, 117, 264). The mutants are recombination deficient and lack detectable Chi-dependent cleavage activities. Likewise, the purified RecB2109CD enzyme has helicase and nuclease activities, but the nuclease activity is aberrant in that the 3′-terminated ssDNA is not degraded. The mutant also does not respond to Chi and, hence, does not load the RecA protein onto ssDNA, resulting in very poor joint-molecule formation in vitro (23, 95, 96). These observations are all consistent with a specific loss or reduction in the RecB motor activity that is required for both translocation of the 3′-terminated ssDNA and delivery of the Chi sequence to its recognition domain within the RecC subunit of the complex. The residual activities of the RecB2109CD enzyme likely result from RecD motor activity. recB344 (A68V) contains a mutation in helicase motif 1a of RecB (S. K. Amundsen and G. R. Smith, personal communication), which is known to be involved in the binding and translocation of ssDNA by SF1 helicases (86). This “Tex-class” mutant retains helicase-nuclease activity but is only partially defective for Chi-dependent activities (191), probably because the RecB motor retains partial function.
FIG. 16.
Mutants of the RecBCD enzyme. The crystal structure of RecBCD is shown, with the main chains of RecB, RecC, and RecD shown in red/magenta, blue/cyan/grayish blue, and green, respectively. The DNA is shown in orange. This color coding is essentially the same as that described in the legend of Fig. 8, as is the orientation of the structure. Note that the RecC backbone is shown in three different shades of blue. The light blue/gray section represents residues 981 to 1084; deletion from the C terminus to within this region yields the “double-dagger” phenotype. The cyan section represents residues 790 to 922; further deletion from the C terminus to within this region yields the “dagger” phenotype. The remainder of RecC is dark blue. The locations of other individual amino acids that are mutated in several well-studied recBCD alleles are indicated by labeled surface representations of the wild-type residues shown in black. The “RecC1004” region (shown in orange) defines residues in the partial frameshift mutation that resulted in an altered specificity of Chi recognition. The “RecB motif 6” region includes R794, R800, Y803, V804, and T807. See the text for details of the phenotypes and/or in vitro enzyme properties associated with each mutation.
In the RecC subunit, several mutations map at or near the Chi recognition tunnel, as defined by the recC1004 mutant (22, 124) (Fig. 16). The recC2145 allele (R186H/G304S) contains two amino acid substitutions, which lie in subdomains 1B and 1A of RecC, respectively (S. K. Amundsen and G. R. Smith, personal communication). This complex retains helicase-nuclease activity but is defective for Chi-dependent activities and recombination, consistent with an inability to recognize Chi (4). RecBC1004D, a member of the recC* class of mutations (248), is a well-studied partial frameshift mutant that causes the substitution of amino acid residues between positions 647 and 655 (Fig. 12B, black surface, and 16, orange residues) (21, 22, 122-124). These are surface residues at the interface of domains 2A and 2B of RecC that form part of the proposed Chi-scanning tunnel (256). This mutant is partially recombination proficient but displays reduced Chi recognition efficiency and altered Chi specificity (21, 22, 124); consequently, its ability to participate in DNA pairing is reduced but can be rescued by a hyperactive mutant RecA protein (122). Other alleles of the recC* class, recC1001 and recC1003, have frameshifts that result in similar, though not identical, mutant proteins, which likely recognize different variants of the canonical Chi sequence (22). Similarly, recC343 (P666L), a member of the “Tex class” of mutants (191), contains a defect at the surface of the Chi-scanning site (Fig. 16) (Amundsen and Smith, personal communication). Accordingly, it displays a reduced stimulation of recombination by the Chi sequence.
In some cases, mutations affect the formation of the RecBCD complex. For example, the recC1010 (G905E) mutation maps to the surface of the RecC “nuclease-like” domain that packs against RecC domain 2B (Fig. 12A and 16) (9). Thus, this point mutation indirectly prevents RecD from associating with RecBC (9) and results in the “double-dagger” (‡) phenotype: recombination proficiency that is independent of Chi and the absence of nuclease activities (55). These properties are shared by RecBC (ΔrecD) and by C-terminal RecC deletions that prevent the binding of RecD to RecBC (9). Deletion analysis revealed that at least 4 amino acids at the C terminus are required to maintain the interaction with RecD and that the removal of the C terminus from residue 1084 to as far as residue 981 eliminates RecD binding; deletions that were more N terminal than residue 678 resulted in a recBCD null phenotype (9). The definition of a region comprising residues 981 to 1084 as being essential to the RecC-RecD interaction is consistent with the crystal structure (256) (Fig. 16, light grayish blue region).
Further genetic dissection of the C-terminal region of the RecC subunit led to the discovery of a new class of RecBCD mutants, designated the “dagger” (†) class (5). Phenotypically, the prototypic member of this class, the recC1041 allele, is recombination proficient, defective for nuclease activity, and partially responsive to Chi. Reexamination of the RecC deletion set revealed that the † class comprised a subset of the original ‡ class: further removal of the C terminus to residue 922 (200 residues), or to as far as residue 790 (332 residues), results in the † phenotype (residues 790 to 922) (Fig. 16, cyan region). A distinguishing feature of the † class is their responsiveness to Chi, a property that normally requires RecD function (5, 9). Thus, the † class differs from the ‡ class, which fails to respond to Chi and fails to interact with RecD (9). A likely interpretation of this difference is that the † class of mutants is only partially defective in their interaction with RecD, perhaps due to indirect effects since there are no known contacts between RecD and the remaining region of RecC. The low hotspot activity of Chi is likely a consequence of a mixture of Chi-independent (i.e., RecBC1041) and Chi-dependent (i.e., RecBC1041D) recombination (5). Preliminary biochemical analyses failed to detect RecD (<3%) in the mutant enzyme, but processing of dsDNA with a Chi site in vitro produced a trace amount of Chi-containing ssDNA (5). However, the yield of Chi-containing ssDNA produced by the mutant enzyme preparation was only about 0.01% of the amount produced by the wild-type enzyme, leaving open the possibility that Chi-specific fragments are the result of a trace amount of the RecBC1041D enzyme (5). The contribution of contaminating holoenzyme to DNA processing would be exacerbated by the 250-fold-greater affinity of RecBCD for DNA than that of RecBC (5). Collectively, these RecD interaction-defective mutants, which are almost devoid of nuclease activity, reveal that the elimination of nuclease activity by mutation (or attenuation of the wild-type enzyme by interactions with Chi) is necessary but insufficient for recombination function. Importantly, because the resultant RecBC enzymes are constitutive for RecA loading, they show that RecA loading is also essential to recombination (10, 13, 17, 23, 66, 67).
Another interesting nuclease-deficient mutant is the RecB(D1080A)CD enzyme, which is mutated not in the RecB motor domain but rather in its nuclease domain (Fig. 11 and 16). The holoenzyme is defective in recombination, whereas the RecB(D1080A)C enzyme is proficient, showing the same characteristics as the RecBC enzyme. As expected from the mutation of a conserved residue in the single nucleolytic site, the purified enzyme lacks nuclease activity; however, it is a processive helicase (13). The processing of dsDNA by RecB(D1080A)CD produces only intact ssDNA and no Chi-specific ssDNA products. Nonetheless, the enzyme can recognize the Chi sequence because it is reversibly inactivated by Chi, but RecB(D1080A)CD cannot coordinate the loading of RecA. Thus, even though the mutant enzyme can recognize Chi, the recognition event cannot be transmitted into the conformation change that is required to load RecA (13). Thus, RecB(D1080A)CD also reveals RecA loading as an essential recombination function of RecBCD. Furthermore, since RecB(D1080A)C is recombinationally proficient, the conformational change required for loading must be possible when RecD is absent (10). Although the basis for the failure of RecB(D1080A)CD to load RecA is not known, the structure of RecBCD offers an interesting hypothesis (M. R. Singleton, M. S. Dillingham, S. C. Kowalczykowski, and D. B. Wigley, unpublished results). To load RecA, the nuclease domain of RecB is postulated to swing free of its docking site on RecC (270). However, if the 3′ strand were to enter inside the ring-like RecB nuclease domain prior to exiting the enzyme, then the nuclease/RecA-loading domain could be topologically trapped by the ssDNA since it is not being degraded in the nuclease-deficient mutant. This domain would not be able to swing free, and the loading of RecA would be prevented. It remains to be determined whether this proposition can explain the defect of RecB(D1080A)CD.
Just as for the RecB motor, a mutation of motif 1 in RecD(K177Q) eliminates ATPase and helicase activity (87, 151). However, in contrast to the equivalent RecB subunit mutant holoenzyme, the RecBCD(K177Q) enzyme is fully functional in vivo. The RecBCD(K177Q) enzyme has both helicase and nuclease activities, it recognizes Chi, and it loads the RecA protein onto the Chi-containing ssDNA which it produces (268). The nuclease- and Chi-dependent behavior of the RecBCD(K177Q) enzyme is in distinct contrast to the enzyme lacking RecD, RecBC, that was discussed above. However, the two RecD motor-defective enzymes, RecBCD(K177Q) and RecBC, display comparably reduced processivities and rates of ATP hydrolysis and dsDNA unwinding (88, 151, 152, 154). Thus, the RecD subunit has a largely dispensable motor activity, but it is required for both nuclease activity and the manifestation of Chi-dependent regulation.
The RecBCD structure and mechanism illustrate how nature can develop complex reaction mechanisms via the mixing and matching of functional protein domains into multifunctional molecular machines. The integration of genetic, biochemical, biophysical, and structural analyses is finally revealing the interrelated functions of this machine. We can now understand why mutations in the RecB motor affect Chi recognition: it is not because RecB is involved in the recognition process directly but, rather, because it translocates the Chi-containing ssDNA to the recognition site in RecC. The RecC subunit is revealed structurally as a defunct helicase that has been adapted to function uniquely as a “scanning head” to read a specific sequence in the form of ssDNA. Finally, the RecD subunit is not the nuclease subunit but, rather, is a second dispensable motor subunit that is needed to regulate the nuclease activity, which resides in, of all places, a separate domain tethered to the RecB motor. Individually, each subunit is a poor ATPase, helicase, or nuclease, but together, they comprise a remarkably complex piece of regulatable biological engineering. In fact, there are other examples of helicase-translocase domains acting in concert with nuclease domains (25, 46, 146, 199, 217). Moreover, helicase domains are often found in complexes with a range of other DNA modification activities including primase, topoisomerase, and methylase. It will be of interest to unveil the relevant protein architectures to see how these activities are coupled to produce new functionality.
THE AddAB FAMILY OF HELICASE-NUCLEASE ENZYMES
Primary Structure and Phylogeny
The canonical RecBCD-like helicase-nucleases are found in gram-negative bacteria. An alternative class of enzymes, the AddAB family (also referred to as RexAB), was originally thought to fulfill the same break-processing function in a restricted niche of gram-positive bacteria (57, 59, 60, 101, 118). However, AddAB-type complexes are now appreciated to be more prevalent, having also been found widely in the proteobacteria (3, 203, 234, 328). Like RecBCD, the AddAB enzyme is implicated in recombinational repair, and the inactivation of either subunit renders cells sensitive to DNA damage and reduces viability. The suggestion that AddAB-like enzymes are functional analogues of RecBCD is strongly supported by the observation that the DNA repair defects of a ΔrecBCD E. coli strain can be partially rescued by the heterologous expression of Bacillus subtilis AddAB or Lactococcus lactis RexAB (101, 149, 207).
As the name suggests, the AddAB (ATP-dependent DNase) enzyme is thought to function as a heterodimer. The AddA subunit is clearly homologous to RecB, containing an N-terminal SF1 helicase domain and a C-terminal nuclease domain (Fig. 17). However, the AddB subunit is not a clear homologue of either the RecC or the RecD protein. Interestingly, primary sequence analysis suggests weak homology at the extreme N terminus to the SF1 DNA helicases, of which both RecB and RecD are members, and an apparently intact Walker A motif (equivalent to helicase motif 1) is also found in this region. Weak homology to RecC is also detected over a separate and more central segment of the N-terminal region, and well-characterized nuclease motifs suggest the presence of another nuclease domain at the C terminus of AddB. Thus, AddB has a domain architecture that is unique but somewhat resembles a hybrid of the RecB and RecC proteins.
FIG. 17.
Primary structure of the AddAB enzyme. The AddA protein (yellow) is structurally homologous to RecB; the N-terminal domain contains seven motifs (numbered) characteristic of SF1 helicases, and the C-terminal domain contains nuclease motifs. The position of a putative catalytic aspartate residue is marked “Nuc.” The AddB protein (magenta) shows weak homology to UvrD-like SF1 DNA helicases (which include RecB and AddA) and to the RecC protein in the regions indicated. A Walker A motif involved in the binding and hydrolysis of NTP, and equivalent to helicase motif 1, is found at the extreme N terminus. Nuclease motifs equivalent to those found in RecB and AddA are present toward the C terminus. The total number of amino acids (aa) in each polypeptide is indicated in parentheses.
Biochemical Analysis of AddAB
Remarkably, although the RecBCD and AddAB enzymes possess an altogether different primary structure arrangement (Fig. 4 and 17), they catalyze a similar (but not identical) processing reaction involving Chi-regulated helicase and nuclease activities (57) (Fig. 3). The specific sequence that acts to attenuate the 3′-strand-specific exonuclease activity of Bacillus subtilis AddAB (ChiBs) differs from that of E. coli and is only 5 bp in length (5′-AGCGG-3′). The presence of alternative Chi sequences in different organisms has led to the suggestion that the helicase-nuclease activity and unique Chi sequences have coevolved in different bacterial species (59, 227). A further difference in the reaction mechanism is that DNA cleavage before Chi is symmetric: the 3′- and 5′-terminated nascent ssDNA strands are cleaved with similar frequencies. Modification of the nuclease activity by Chi simply shuts off the 3′ strand cleavage activity rather than “switching” polarity, as observed for RecBCD-like enzymes (57). The observation of symmetric DNA cleavage and the presence of two nuclease domains in AddAB have led to the attractive hypothesis that each nuclease domain is exclusively responsible for the cleavage of one ssDNA strand (57, 59, 226, 227). Experiments with the RexAB enzyme (a Lactococcus lactis AddAB homologue) confirmed that both nuclease domains are active in vivo (226). Recent work on Bacillus subtilis AddAB demonstrated that each nuclease domain is dedicated to the cleavage of just one DNA strand (AddA cleaves the 3′ strand, and AddB cleaves the 5′ strand) and that the generation of Chi-specific fragments requires the AddA, but not the AddB, nuclease domain (323). It is unclear whether or not the AddAB enzyme employs a bipolar DNA translocation mechanism. Only one clear set of SF1 helicase motifs is present in the primary structure, which suggests that the AddAB enzyme translocates along DNA with a “conventional” single-motor mechanism. Perhaps the shorter and more frequent Chi sequences that are recognized by AddAB are less demanding in terms of the enzyme's processivity. However, the presence of an intact Walker A motif (equivalent to helicase motif 1) near the N terminus of the AddB subunit implies that AddB may also bind and/or hydrolyze ATP. This putative ATPase activity and its role in the enzyme mechanism remain to be rigorously investigated. An interesting feature of AddAB-dependent DNA processing is that the enzyme forms a highly stable (half-life of ∼15 min) nucleoprotein complex at Chi (58, 323). This observation is consistent with a mechanism for Chi recognition in which the translocating AddAB enzyme binds tightly to Chi on the 3′-terminated strand and prevents its access to the nuclease domain. Importantly, this mechanism is in accord with the Chi-scanning and binding hypothesis proposed on the basis of the RecBCD crystal structure.
AddAB-dependent recombination in Bacillus subtilis is a less prominent pathway than is that of its E. coli (RecBCD) counterpart, and several degenerate pathways for the initiation of homologous recombination from DSBs exist (104, 145). A previously reported interaction between AddB and the ScpA subunit of the Bacillus structural maintenance of chromosomes (SMC) complex hints that AddAB may play a specialized role in the repair of dsDNA ends generated by disrupted replication forks (84). The SMC complex is involved in the segregation and compaction of the bacterial nucleoid at cell division and is localized to the same region of dividing cells as is the replisome (180). The cell has a limited spatial and temporal window in which to repair collapsed replication forks by homologous recombination because the nascent daughter chromosomes are rapidly separated following replication. Therefore, an interaction between AddAB and SMC may serve to recruit AddAB at the right place and time to fulfill its break-processing role. This would tie in nicely with the observation that Chi sequences are oriented toward the origin of replication and with the current view of RecBCD/AddAB performing an evolutionarily conserved and essential function in support of chromosome duplication. In this respect, it is of particular interest that bacilli were recently shown to possess an active nonhomologous end joining pathway (311), which could act to repair DSBs in the absence of a homologous donor, when the homologous recombination pathway is unable to operate.
UNRESOLVED QUESTIONS
The bacterial RecBCD enzyme illustrates principles of modularity in the design of complex enzyme mechanism. However, some aspects of this mechanism remain poorly understood. Although recent work on the Chi recognition mechanism of AddAB family enzymes supports the allosteric Chi binding hypothesis presented above, further support from mutational analysis, guided by the crystal structure, is desirable. If the Chi sequence is held bound in the RecC scanning tunnel and the RecB motor remains active after Chi recognition, then it is not clear where the resulting ssDNA loop would emerge from the enzyme complex, although the RecB/RecC interface is a probable location. Furthermore, the conformational change that accompanies Chi recognition is not known. Crystal structures with specific and nonspecific ssDNA bound in the relevant site would be particularly helpful in resolving these issues. Because Chi is recognized during translocation, producing a structurally homogenous preparation of the Chi-modified RecBCD enzyme is technically challenging. However, given the paucity of structures involving sequence-specific ssDNA recognition, it may prove to be particularly rewarding. The fact that RecBCD mutants with altered Chi recognition specificities, as well as RecBCD analogues which bind alternative Chi sequences, exist suggests that this system may provide valuable insights into these fundamental sequence-specific ssDNA-protein interactions.
There is currently no consensus on the translocation step size, and the crystal structure provides no obvious basis for the 3- to 4-base kinetic step size and only a wishful suggestion that the arm is involved in the large 23-base physical step size. However, the step size issue seems likely to yield to ever-improving single-molecule techniques (1) or structural analysis of complexes with nonhydrolyzable nucleotide analogues bound in the helicase motor active sites.
More work is required to understand the sequence of events that occur following Chi recognition. The exact mechanism of RecA loading, the translocation status of the RecD motor subunit, the timing of the initiation of DNA strand invasion, and the manner in which the RecBCD enzyme is released from the recombinogenic DNA are all unclear. It will be of interest to determine if the AddAB enzyme is able to recruit the RecA protein to see whether the RecBCD paradigm for RecA loading extends to all bacterial helicase-nucleases.
The links between the broken DNA and the initiation of repair are not fully understood. Different types of DNA breaks, or those produced in different cellular contexts, may be optimally repaired by specialized mechanisms. If this is indeed the case, then there are likely to be regulatory mechanisms to ensure that DSBs are directed to the appropriate repair machinery. The interaction of the RecBCD enzyme with other protein partners, such as components of the replication machinery, remains a largely unexplored area of research, and this line of questioning could reveal how RecBCD-dependent recombinational repair is coordinated with the cell cycle, replication, and alternative pathways for the repair of DSBs and why a helicase as fast as the replisome is involved in the repair of DSBs that accompany normal DNA replication.
Many bacteria possess either a RecBCD- or an AddAB-like enzyme. However, there are clear examples (e.g., Deinococcus radiodurans) of those that possess neither, but a RecD homologue is often found (234, 309). It will be of considerable interest to determine the mechanisms of DSB repair in these organisms. Eukaryotic organisms are not considered to possess a RecBCD-like function (although the possibility that they do, but it is currently undiscovered, cannot be excluded). They certainly do, however, possess helicases with associated nuclease domains (25, 45, 134). These might perform different biological functions but through a similar mechanism, demonstrating the biological utility of a combined helicase-nuclease architecture. Eukaryotic organisms do not employ restriction-modification systems and do not deal with viral invasions in the same manner as do the Bacteria, so perhaps there was no need to maintain the remarkable but dangerous ability to control a potentially destructive nuclease for use in the template-directed repair of DNA.
Acknowledgments
We thank Maria Spies, Wolf Heyer, Bénédicte Michel, Andrei Kuzminov, Piero Bianco, Joe Yeeles, Emma Longman, Naofumi Handa, Ichiro Amitani, Clarke Conant, Petr Cejka, Aura Carreira, Jason Wong, Katsumi Morimatsu, Bian Liu, Jason Bell, Taeho Kim, Liang Yang, Ryan Jensen, Edgar Valencia-Morales, Tony Forget, Christopher Dombrowski, Behzad Rad, and Jody Plank for their comments on the manuscript. We are particularly grateful to Gerry Smith and Sue Amundsen for generously sharing unpublished data and for thought-provoking discussions regarding the RecBCD enzyme.
REFERENCES
- 1.Abbondanzieri, E. A., W. J. Greenleaf, J. W. Shaevitz, R. Landick, and S. M. Block. 2005. Direct observation of base-pair stepping by RNA polymerase. Nature 438460-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alatossava, T., H. Jutte, A. Kuhn, and E. Kellenberger. 1985. Manipulation of intracellular magnesium content in polymyxin B nonapeptide-sensitized Escherichia coli by ionophore A23187. J. Bacteriol. 162413-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amundsen, S. K., J. Fero, L. M. Hansen, G. A. Cromie, J. V. Solnick, G. R. Smith, and N. R. Salama. 2008. Helicobacter pylori AddAB helicase-nuclease and RecA promote recombination-related DNA repair and survival during stomach colonization. Mol. Microbiol. 69994-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amundsen, S. K., A. M. Neiman, S. M. Thibodeaux, and G. R. Smith. 1990. Genetic dissection of the biochemical activities of RecBCD enzyme. Genetics 12625-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amundsen, S. K., and G. R. Smith. 2007. Chi hotspot activity in Escherichia coli without RecBCD exonuclease activity: implications for the mechanism of recombination. Genetics 17541-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amundsen, S. K., and G. R. Smith. 2003. Interchangeable parts of the Escherichia coli recombination machinery. Cell 112741-744. [DOI] [PubMed] [Google Scholar]
- 7.Amundsen, S. K., A. F. Taylor, A. M. Chaudhury, and G. R. Smith. 1986. recD: the gene for an essential third subunit of exonuclease V. Proc. Natl. Acad. Sci. USA 835558-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amundsen, S. K., A. F. Taylor, M. Reddy, and G. R. Smith. 2007. Intersubunit signaling in RecBCD enzyme, a complex protein machine regulated by Chi hot spots. Genes Dev. 213296-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amundsen, S. K., A. F. Taylor, and G. R. Smith. 2002. A domain of RecC required for assembly of the regulatory RecD subunit into the Escherichia coli RecBCD holoenzyme. Genetics 161483-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amundsen, S. K., A. F. Taylor, and G. R. Smith. 2000. The RecD subunit of the Escherichia coli RecBCD enzyme inhibits RecA loading, homologous recombination, and DNA repair. Proc. Natl. Acad. Sci. USA 977399-7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amundsen, S. K., A. F. Taylor, and G. R. Smith. 1998. A stimulatory RNA associated with RecBCD enzyme. Nucleic Acids Res. 262125-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson, D. G., J. J. Churchill, and S. C. Kowalczykowski. 1997. Chi-activated RecBCD enzyme possesses 5′→3′ nucleolytic activity, but RecBC enzyme does not: evidence suggesting that the alteration induced by Chi is not simply ejection of the RecD subunit. Genes Cells 2117-128. [DOI] [PubMed] [Google Scholar]
- 13.Anderson, D. G., J. J. Churchill, and S. C. Kowalczykowski. 1999. A single mutation, RecB(D1080A), eliminates RecA protein loading but not Chi recognition by RecBCD enzyme. J. Biol. Chem. 27427139-27144. [DOI] [PubMed] [Google Scholar]
- 14.Anderson, D. G., and S. C. Kowalczykowski. 1997. The recombination hot spot χ is a regulatory element that switches the polarity of DNA degradation by the RecBCD enzyme. Genes Dev. 11571-581. [DOI] [PubMed] [Google Scholar]
- 15.Anderson, D. G., and S. C. Kowalczykowski. 1998. Reconstitution of an SOS response pathway: derepression of transcription in response to DNA breaks. Cell 95975-979. [DOI] [PubMed] [Google Scholar]
- 16.Anderson, D. G., and S. C. Kowalczykowski. 1998. SSB protein controls RecBCD enzyme nuclease activity during unwinding: a new role for looped intermediates. J. Mol. Biol. 282275-285. [DOI] [PubMed] [Google Scholar]
- 17.Anderson, D. G., and S. C. Kowalczykowski. 1997. The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a χ-regulated manner. Cell 9077-86. [DOI] [PubMed] [Google Scholar]
- 18.Appasani, K., D. S. Thaler, and E. B. Goldberg. 1999. Bacteriophage T4 gp2 interferes with cell viability and with bacteriophage lambda Red recombination. J. Bacteriol. 1811352-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arakawa, K., R. Uno, Y. Nakayama, and M. Tomita. 2007. Validating the significance of genomic properties of Chi sites from the distribution of all octamers in Escherichia coli. Gene 392239-246. [DOI] [PubMed] [Google Scholar]
- 20.Aravind, L., K. S. Makarova, and E. V. Koonin. 2000. Holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 283417-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arnold, D. A., P. R. Bianco, and S. C. Kowalczykowski. 1998. The reduced levels of χ recognition exhibited by the RecBC1004D enzyme reflect its recombination defect in vivo. J. Biol. Chem. 27316476-16486. [DOI] [PubMed] [Google Scholar]
- 22.Arnold, D. A., N. Handa, I. Kobayashi, and S. C. Kowalczykowski. 2000. A novel, 11 nucleotide variant of χ, χ*: one of a class of sequences defining the Escherichia coli recombination hotspot χ. J. Mol. Biol. 300469-479. [DOI] [PubMed] [Google Scholar]
- 23.Arnold, D. A., and S. C. Kowalczykowski. 2000. Facilitated loading of RecA protein is essential to recombination by RecBCD enzyme. J. Biol. Chem. 27512261-12265. [DOI] [PubMed] [Google Scholar]
- 24.Arnold, D. A., and S. C. Kowalczykowski. 19 April 2001, posting date. RecBCD helicase/nuclease. In Encyclopedia of life sciences. John Wiley & Sons, Chichester, United Kingdom.http://www.els.net.
- 25.Bae, S. H., and Y. S. Seo. 2000. Characterization of the enzymatic properties of the yeast dna2 helicase/endonuclease suggests a new model for Okazaki fragment processing. J. Biol. Chem. 27538022-38031. [DOI] [PubMed] [Google Scholar]
- 26.Barbour, S. D., and A. J. Clark. 1970. Biochemical and genetic studies of recombination proficiency in Escherichia coli. I. Enzymatic activity associated with recB+ and recC+ genes. Proc. Natl. Acad. Sci. USA 65955-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Behme, M. T., G. D. Lilley, and K. Ebisuzaki. 1976. Postinfection control by bacteriophage T4 of Escherichia coli recBC nuclease activity. J. Virol. 1820-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bell, C. E. 2005. Structure and mechanism of Escherichia coli RecA ATPase. Mol. Microbiol. 58358-366. [DOI] [PubMed] [Google Scholar]
- 29.Bell, S. J., Y. C. Chow, J. Y. K. Ho, and D. R. Forsdyke. 1998. Correlation of chi orientation with transcription indicates a fundamental relationship between recombination and transcription. Gene 216285-292. [DOI] [PubMed] [Google Scholar]
- 30.Benzinger, R., L. W. Enquist, and A. Skalka. 1975. Transfection of Escherichia coli spheroplasts. V. Activity of recBC nuclease in rec+ and rec− spheroplasts measured with different forms of bacteriophage DNA. J. Virol. 15861-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bianco, P. R., L. R. Brewer, M. Corzett, R. Balhorn, Y. Yeh, S. C. Kowalczykowski, and R. J. Baskin. 2001. Processive translocation and DNA unwinding by individual RecBCD enzyme molecules. Nature 409374-378. [DOI] [PubMed] [Google Scholar]
- 32.Bianco, P. R., and S. C. Kowalczykowski. 23 September 2005, posting date. RecA protein. In Encyclopedia of life sciences. John Wiley & Sons, Chichester, United Kingdom. http://www.els.net.
- 33.Bianco, P. R., and S. C. Kowalczykowski. 1997. The recombination hotspot Chi is recognized by the translocating RecBCD enzyme as the single strand of DNA containing the sequence 5′-GCTGGTGG-3′. Proc. Natl. Acad. Sci. USA 946706-6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bianco, P. R., and S. C. Kowalczykowski. 2000. Translocation step size and mechanism of the RecBC DNA helicase. Nature 405368-372. [DOI] [PubMed] [Google Scholar]
- 35.Bianco, P. R., R. B. Tracy, and S. C. Kowalczykowski. 1998. DNA strand exchange proteins: a biochemical and physical comparison. Front. Biosci. 3D570-D603. [DOI] [PubMed] [Google Scholar]
- 36.Bidle, K. A., and D. H. Bartlett. 1999. RecD function is required for high-pressure growth of a deep-sea bacterium. J. Bacteriol. 1812330-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biek, D. P., and S. N. Cohen. 1986. Identification and characterization of recD, a gene affecting plasmid maintenance and recombination in Escherichia coli. J. Bacteriol. 167594-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 2771453-1474. [DOI] [PubMed] [Google Scholar]
- 39.Bochner, B. R., and B. N. Ames. 1982. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J. Biol. Chem. 2579759-9769. [PubMed] [Google Scholar]
- 40.Boehmer, P. E., and P. T. Emmerson. 1992. The RecB subunit of the Escherichia coli RecBCD enzyme couples ATP hydrolysis to DNA unwinding. J. Biol. Chem. 2674981-4987. [PubMed] [Google Scholar]
- 41.Braedt, G., and G. R. Smith. 1989. Strand specificity of DNA unwinding by RecBCD enzyme. Proc. Natl. Acad. Sci. USA 86871-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brcic-Kostic, K., E. Salaj-Smic, N. Marsic, S. Kajic, I. Stojiljkovic, and Z. Trgovcevic. 1991. Interaction of RecBCD enzyme with DNA damaged by gamma radiation. Mol. Gen. Genet. 228136-142. [DOI] [PubMed] [Google Scholar]
- 43.Brcic-Kostic, K., I. Stojiljkovic, E. Salaj-Smic, and Z. Trgovcevic. 1992. Overproduction of the RecD polypeptide sensitizes Escherichia coli cells to gamma-radiation. Mutat. Res. 281123-127. [DOI] [PubMed] [Google Scholar]
- 44.Buchmeier, N. A., C. J. Lipps, M. Y. So, and F. Heffron. 1993. Recombination-deficient mutants of Salmonella typhimurium are avirulent and sensitive to the oxidative burst of macrophages. Mol. Microbiol. 7933-936. [DOI] [PubMed] [Google Scholar]
- 45.Budd, M. E., and J. L. Campbell. 1995. A yeast gene required for DNA replication encodes a protein with homology to DNA helicases. Proc. Natl. Acad. Sci. USA 927642-7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Budd, M. E., W. Choe, and J. L. Campbell. 2000. The nuclease activity of the yeast DNA2 protein, which is related to the RecB-like nucleases, is essential in vivo. J. Biol. Chem. 27516518-16529. [DOI] [PubMed] [Google Scholar]
- 47.Bustamante, C., Z. Bryant, and S. B. Smith. 2003. Ten years of tension: single-molecule DNA mechanics. Nature 421423-427. [DOI] [PubMed] [Google Scholar]
- 48.Byrd, A. K., and K. D. Raney. 2005. Increasing the length of the single-stranded overhang enhances unwinding of duplex DNA by bacteriophage T4 Dda helicase. Biochemistry 4412990-12997. [DOI] [PubMed] [Google Scholar]
- 49.Byrd, A. K., and K. D. Raney. 2004. Protein displacement by an assembly of helicase molecules aligned along single-stranded DNA. Nat. Struct. Mol. Biol. 11531-538. [DOI] [PubMed] [Google Scholar]
- 50.Cano, D. A., M. G. Pucciarelli, F. Garcia-del Portillo, and J. Casadesus. 2002. Role of the RecBCD recombination pathway in Salmonella virulence. J. Bacteriol. 184592-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Capaldo-Kimball, F., and S. D. Barbour. 1971. Involvement of recombination genes in growth and viability of Escherichia coli K-12. J. Bacteriol. 106204-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Capaldo, F. N., and S. D. Barbour. 1975. The role of the rec genes in the viability of Escherichia coli K12. Basic Life Sci. 5A405-418. [DOI] [PubMed] [Google Scholar]
- 53.Capaldo, F. N., G. Ramsey, and S. D. Barbour. 1974. Analysis of the growth of recombination-deficient strains of Escherichia coli K-12. J. Bacteriol. 118242-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chabbert, M., C. Cazenave, and C. Helene. 1987. Kinetic studies of recA protein binding to a fluorescent single-stranded polynucleotide. Biochemistry 262218-2225. [DOI] [PubMed] [Google Scholar]
- 55.Chaudhury, A. M., and G. R. Smith. 1984. A new class of Escherichia coli recBC mutants: implications for the role of RecBC enzyme in homologous recombination. Proc. Natl. Acad. Sci. USA 817850-7854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chaussee, M. S., J. Wilson, and S. A. Hill. 1999. Characterization of the recD gene of Neisseria gonorrhoeae MS11 and the effect of recD inactivation on pilin variation and DNA transformation. Microbiology 145389-400. [DOI] [PubMed] [Google Scholar]
- 57.Chédin, F., S. D. Ehrlich, and S. C. Kowalczykowski. 2000. The Bacillus subtilis AddAB helicase/nuclease is regulated by its cognate Chi sequence in vitro. J. Mol. Biol. 2987-20. [DOI] [PubMed] [Google Scholar]
- 58.Chédin, F., N. Handa, M. S. Dillingham, and S. C. Kowalczykowski. 2006. The AddAB helicase/nuclease forms a stable complex with its cognate χ sequence during translocation. J. Biol. Chem. 28118610-18617. [DOI] [PubMed] [Google Scholar]
- 59.Chédin, F., and S. C. Kowalczykowski. 2002. A novel family of regulated helicases/nucleases from gram-positive bacteria: insights into the initiation of DNA recombination. Mol. Microbiol. 43823-834. [DOI] [PubMed] [Google Scholar]
- 60.Chédin, F., P. Noirot, V. Biaudet, and S. D. Ehrlich. 1998. A five-nucleotide sequence protects DNA from exonucleolytic degradation by AddAB, the RecBCD analogue of Bacillus subtilis. Mol. Microbiol. 291369-1377. [DOI] [PubMed] [Google Scholar]
- 61.Chen, H. W., D. E. Randle, M. Gabbidon, and D. A. Julin. 1998. Functions of the ATP hydrolysis subunits (RecB and RecD) in the nuclease reactions catalyzed by the RecBCD enzyme from Escherichia coli. J. Mol. Biol. 27889-104. [DOI] [PubMed] [Google Scholar]
- 62.Chen, H. W., B. Ruan, M. Yu, J. Wang, and D. A. Julin. 1997. The RecD subunit of the RecBCD enzyme from Escherichia coli is a single-stranded DNA-dependent ATPase. J. Biol. Chem. 27210072-10079. [DOI] [PubMed] [Google Scholar]
- 63.Cheng, K. C., and G. R. Smith. 1987. Cutting of Chi-like sequences by the RecBCD enzyme of Escherichia coli. J. Mol. Biol. 194747-750. [DOI] [PubMed] [Google Scholar]
- 64.Cheng, K. C., and G. R. Smith. 1989. Distribution of Chi-stimulated recombinational exchanges and heteroduplex endpoints in phage lambda. Genetics 1235-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cheng, K. C., and G. R. Smith. 1984. Recombinational hotspot activity of Chi-like sequences. J. Mol. Biol. 180371-377. [DOI] [PubMed] [Google Scholar]
- 66.Churchill, J. J., D. G. Anderson, and S. C. Kowalczykowski. 1999. The RecBC enzyme loads RecA protein onto ssDNA asymmetrically and independently of Chi, resulting in constitutive recombination activation. Genes Dev. 13901-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Churchill, J. J., and S. C. Kowalczykowski. 2000. Identification of the RecA protein-loading domain of RecBCD enzyme. J. Mol. Biol. 297537-542. [DOI] [PubMed] [Google Scholar]
- 68.Clark, A. J., and S. J. Sandler. 1994. Homologous genetic recombination: the pieces begin to fall into place. Crit. Rev. Microbiol. 20125-142. [DOI] [PubMed] [Google Scholar]
- 69.Clark, A. J., M. R. Volkert, and L. J. Margossian. 1979. A role for recF in repair of UV damage to DNA. Cold Spring Harb. Symp. Quant. Biol. 43887-892. [DOI] [PubMed] [Google Scholar]
- 70.Colbert, T., A. F. Taylor, and G. R. Smith. 1998. Genomics, Chi sites and codons: ‘islands of preferred DNA pairing’ are oceans of ORFs. Trends Genet. 14485-488. [DOI] [PubMed] [Google Scholar]
- 71.Cole, R. S. 1971. Properties of F′ factor deoxyribonucleic acid transferred from ultraviolet-irradiated donors: photoreactivation in the recipient and the influence of recA, recB, recC, and uvr genes. J. Bacteriol. 106143-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Courcelle, J. 2005. Recs preventing wrecks. Mutat. Res. 577217-227. [DOI] [PubMed] [Google Scholar]
- 73.Courcelle, J., C. Carswell-Crumpton, and P. C. Hanawalt. 1997. recF and recR are required for the resumption of replication at DNA replication forks in Escherichia coli. Proc. Natl. Acad. Sci. USA 943714-3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Courcelle, J., J. R. Donaldson, K. H. Chow, and C. T. Courcelle. 2003. DNA damage-induced replication fork regression and processing in Escherichia coli. Science 2991064-1067. [DOI] [PubMed] [Google Scholar]
- 75.Court, R., N. Cook, K. Saikrishnan, and D. Wigley. 2007. The crystal structure of lambda-Gam protein suggests a model for RecBCD inhibition. J. Mol. Biol. 37125-33. [DOI] [PubMed] [Google Scholar]
- 76.Cox, M. M., M. F. Goodman, K. N. Kreuzer, D. J. Sherratt, S. J. Sandler, and K. J. Marians. 2000. The importance of repairing stalled replication forks. Nature 40437-41. [DOI] [PubMed] [Google Scholar]
- 77.Cox, M. M., and I. R. Lehman. 1981. RecA protein of Escherichia coli promotes branch migration, a kinetically distinct phase of DNA strand exchange. Proc. Natl. Acad. Sci. USA 783433-3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dabert, P., S. D. Ehrlich, and A. Gruss. 1992. Chi sequence protects against RecBCD degradation of DNA in vivo. Proc. Natl. Acad. Sci. USA 8912073-12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Della, M., P. L. Palmbos, H. M. Tseng, L. M. Tonkin, J. M. Daley, L. M. Topper, R. S. Pitcher, A. E. Tomkinson, T. E. Wilson, and A. J. Doherty. 2004. Mycobacterial Ku and ligase proteins constitute a two-component NHEJ repair machine. Science 306683-685. [DOI] [PubMed] [Google Scholar]
- 80.Dermić, D. 2006. Functions of multiple exonucleases are essential for cell viability, DNA repair and homologous recombination in recD mutants of Escherichia coli. Genetics 1722057-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dermić, D., E. Dermić, D. Zahradka, M. Petranović, and N. Lerš. 2006. Gamma-irradiated RecD overproducers become permanent recB−/C− phenocopies for extrachromosomal DNA processing due to prolonged titration of RecBCD enzyme on damaged Escherichia coli chromosome. Biochimie 88379-386. [DOI] [PubMed] [Google Scholar]
- 82.Dermić, D., E. Halupecki, D. Zahradka, and M. Petranović. 2005. RecBCD enzyme overproduction impairs DNA repair and homologous recombination in Escherichia coli. Res. Microbiol. 156304-311. [DOI] [PubMed] [Google Scholar]
- 83.Dermić, D., D. Zahradka, and M. Petranović. 2006. Exonuclease requirements for recombination of λ-phage in recD mutants of Escherichia coli. Genetics 1732399-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dervyn, E., M. F. Noirot-Gros, P. Mervelet, S. McGovern, S. D. Ehrlich, P. Polard, and P. Noirot. 2004. The bacterial condensin/cohesin-like protein complex acts in DNA repair and regulation of gene expression. Mol. Microbiol. 511629-1640. [DOI] [PubMed] [Google Scholar]
- 85.Dharmalingam, K., and E. B. Goldberg. 1976. Mechanism localisation and control of restriction cleavage of phage T4 and lambda chromosomes in vivo. Nature 260406-410. [DOI] [PubMed] [Google Scholar]
- 86.Dillingham, M. S., P. Soultanas, P. Wiley, M. R. Webb, and D. B. Wigley. 2001. Defining the roles of individual residues in the single-stranded DNA binding site of PcrA helicase. Proc. Natl. Acad. Sci. USA 988381-8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dillingham, M. S., M. Spies, and S. C. Kowalczykowski. 2003. RecBCD enzyme is a bipolar DNA helicase. Nature 423893-897. [DOI] [PubMed] [Google Scholar]
- 88.Dillingham, M. S., M. R. Webb, and S. C. Kowalczykowski. 2005. Bipolar DNA translocation contributes to highly processive DNA unwinding by RecBCD enzyme. J. Biol. Chem. 28037069-37077. [DOI] [PubMed] [Google Scholar]
- 89.Dillingham, M. S., D. B. Wigley, and M. R. Webb. 2000. Demonstration of unidirectional single-stranded DNA translocation by PcrA helicase: measurement of step size and translocation speed. Biochemistry 39205-212. [DOI] [PubMed] [Google Scholar]
- 90.Dixon, D. A., J. J. Churchill, and S. C. Kowalczykowski. 1994. Reversible inactivation of the Escherichia coli RecBCD enzyme by the recombination hotspot χ in vitro: evidence for functional inactivation or loss of the RecD subunit. Proc. Natl. Acad. Sci. USA 912980-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dixon, D. A., and S. C. Kowalczykowski. 1991. Homologous pairing in vitro stimulated by the recombination hotspot, Chi. Cell 66361-371. [DOI] [PubMed] [Google Scholar]
- 92.Dixon, D. A., and S. C. Kowalczykowski. 1993. The recombination hotspot χ is a regulatory sequence that acts by attenuating the nuclease activity of the E. coli RecBCD enzyme. Cell 7387-96. [DOI] [PubMed] [Google Scholar]
- 93.Dixon, D. A., and S. C. Kowalczykowski. 1995. Role of the Escherichia coli recombination hotspot, χ, in RecABCD-dependent homologous pairing. J. Biol. Chem. 27016360-16370. [DOI] [PubMed] [Google Scholar]
- 94.Dohoney, K. M., and J. Gelles. 2001. Chi-sequence recognition and DNA translocation by single RecBCD helicase/nuclease molecules. Nature 409370-374. [DOI] [PubMed] [Google Scholar]
- 95.Eggleston, A. K., and S. C. Kowalczykowski. 1993. Biochemical characterization of a mutant recBCD enzyme, the recB2109CD enzyme, which lacks χ-specific, but not non-specific, nuclease activity. J. Mol. Biol. 231605-620. [DOI] [PubMed] [Google Scholar]
- 96.Eggleston, A. K., and S. C. Kowalczykowski. 1993. The mutant recBCD enzyme, recB2109CD enzyme, has helicase activity but does not promote efficient joint molecule formation in vitro. J. Mol. Biol. 231621-633. [DOI] [PubMed] [Google Scholar]
- 97.Eggleston, A. K., N. A. Rahim, and S. C. Kowalczykowski. 1996. A helicase assay based on the displacement of fluorescent, nucleic acid-binding ligands. Nucleic Acids Res. 241179-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Eggleston, A. K., and S. C. West. 1997. Recombination initiation: easy as A, B, C, D, χ? Curr. Biol. 7R745-R749. [DOI] [PubMed] [Google Scholar]
- 99.Eichler, D. C., and I. R. Lehman. 1977. On the role of ATP in phosphodiester bond hydrolysis catalyzed by the recBC deoxyribonuclease of Escherichia coli. J. Biol. Chem. 252499-503. [PubMed] [Google Scholar]
- 100.El Karoui, M., V. Biaudet, S. Schbath, and A. Gruss. 1999. Characteristics of Chi distribution on different bacterial genomes. Res. Microbiol. 150579-587. [DOI] [PubMed] [Google Scholar]
- 101.El Karoui, M., D. Ehrlich, and A. Gruss. 1998. Identification of the lactococcal exonuclease/recombinase and its modulation by the putative Chi sequence. Proc. Natl. Acad. Sci. USA 95626-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Emmerson, P. T. 1968. Recombination deficient mutants of Escherichia coli K12 that map between thyA and argA. Genetics 6019-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Farah, J. A., and G. R. Smith. 1997. The RecBCD enzyme initiation complex for DNA unwinding: enzyme positioning and DNA opening. J. Mol. Biol. 272699-715. [DOI] [PubMed] [Google Scholar]
- 104.Fernandez, S., A. Sorokin, and J. C. Alonso. 1998. Genetic recombination in Bacillus subtilis 168: effects of recU and recS mutations on DNA repair and homologous recombination. J. Bacteriol. 1803405-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Finch, P. W., A. Storey, K. Brown, I. D. Hickson, and P. T. Emmerson. 1986. Complete nucleotide sequence of recD, the structural gene for the alpha subunit of exonuclease V of Escherichia coli. Nucleic Acids Res. 148583-8594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fischer, C. J., N. K. Maluf, and T. M. Lohman. 2004. Mechanism of ATP-dependent translocation of E. coli UvrD monomers along single-stranded DNA. J. Mol. Biol. 3441287-1309. [DOI] [PubMed] [Google Scholar]
- 107.Gabbidon, M. R., V. E. Rampersaud, and D. A. Julin. 1998. Salt-stable complexes of the Escherichia coli RecBCD enzyme bound to double-stranded DNA. Arch. Biochem. Biophys. 350266-272. [DOI] [PubMed] [Google Scholar]
- 108.Galletto, R., I. Amitani, R. J. Baskin, and S. C. Kowalczykowski. 2006. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature 443875-878. [DOI] [PubMed] [Google Scholar]
- 109.Galletto, R., and S. C. Kowalczykowski. 2007. RecA. Curr. Biol. 17R395-R397. [DOI] [PubMed] [Google Scholar]
- 110.Ganesan, A. K. 1974. Persistence of pyrimidine dimers during post-replication repair in ultraviolet light-irradiated Escherichia coli K12. J. Mol. Biol. 87103-119. [DOI] [PubMed] [Google Scholar]
- 111.Ganesan, S., and G. R. Smith. 1993. Strand-specific binding to duplex DNA ends by the subunits of the Escherichia coli RecBCD enzyme. J. Mol. Biol. 22967-78. [DOI] [PubMed] [Google Scholar]
- 111a.Ghatak, A., and D. A. Julin. 2006. Kinetics of ATP-stimulated nuclease activity of the Escherichia coli RecBCD enzyme. J. Mol. Biol. 361954-968. [DOI] [PubMed] [Google Scholar]
- 112.Gibson, F. P., D. R. Leach, and R. G. Lloyd. 1992. Identification of sbcD mutations as cosuppressors of recBC that allow propagation of DNA palindromes in Escherichia coli K-12. J. Bacteriol. 1741222-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Goldmark, P. J., and S. Linn. 1970. An endonuclease activity from Escherichia coli absent from certain rec− strains. Proc. Natl. Acad. Sci. USA 67434-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Goldmark, P. J., and S. Linn. 1972. Purification and properties of the recBC DNase of Escherichia coli K-12. J. Biol. Chem. 2471849-1860. [PubMed] [Google Scholar]
- 115.Gorbalenya, A. E., and E. V. Koonin. 1993. Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 3419-429. [Google Scholar]
- 116.Hall, M. C., and S. W. Matson. 1999. Helicase motifs: the engine that powers DNA unwinding. Mol. Microbiol. 34867-877. [DOI] [PubMed] [Google Scholar]
- 117.Hall, M. C., A. Z. Ozsoy, and S. W. Matson. 1998. Site-directed mutations in motif VI of Escherichia coli DNA helicase II result in multiple biochemical defects: evidence for the involvement of motif VI in the coupling of ATPase and DNA binding activities via conformational changes. J. Mol. Biol. 277257-271. [DOI] [PubMed] [Google Scholar]
- 118.Halpern, D., A. Gruss, J. P. Claverys, and M. El-Karoui. 2004. rexAB mutants in Streptococcus pneumoniae. Microbiology 1502409-2414. [DOI] [PubMed] [Google Scholar]
- 119.Hanawalt, P. C. 1966. The U.V. sensitivity of bacteria: its relation to the DNA replication cycle. Photochem. Photobiol. 51-12. [PubMed] [Google Scholar]
- 120.Handa, N., P. R. Bianco, R. J. Baskin, and S. C. Kowalczykowski. 2005. Direct visualization of RecBCD movement reveals cotranslocation of the RecD motor after χ recognition. Mol. Cell 17745-750. [DOI] [PubMed] [Google Scholar]
- 121.Handa, N., A. Ichige, K. Kusano, and I. Kobayashi. 2000. Cellular responses to postsegregational killing by restriction-modification genes. J. Bacteriol. 1822218-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Handa, N., and S. C. Kowalczykowski. 2007. A RecA mutant, RecA(730), suppresses the recombination deficiency of the RecBC(1004)D-chi* interaction in vitro and in vivo. J. Mol. Biol. 3651314-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Handa, N., S. Ohashi, and I. Kobayashi. 1997. Clustering of chi sequence in Escherichia coli genome. Microb. Comp. Genomics 2287-298. [DOI] [PubMed] [Google Scholar]
- 124.Handa, N., S. Ohashi, K. Kusano, and I. Kobayashi. 1997. Chi-star, a chi-related 11-mer sequence partially active in an E. coli recC1004 strain. Genes Cells 2525-536. [DOI] [PubMed] [Google Scholar]
- 125.Harmon, F. G., and S. C. Kowalczykowski. 1998. RecQ helicase, in concert with RecA and SSB proteins, initiates and disrupts DNA recombination. Genes Dev. 121134-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Heller, R. C., and K. J. Marians. 2006. Replication fork reactivation downstream of a blocked nascent leading strand. Nature 439557-562. [DOI] [PubMed] [Google Scholar]
- 127.Heller, R. C., and K. J. Marians. 2006. Replisome assembly and the direct restart of stalled replication forks. Nat. Rev. Mol. Cell Biol. 7932-943. [DOI] [PubMed] [Google Scholar]
- 128.Henderson, D., and J. Weil. 1975. The nature and origin of a class of essential gene substitutions in bacteriophage lambda. Virology 67124-135. [DOI] [PubMed] [Google Scholar]
- 129.Hickson, I. D., C. N. Robson, K. E. Atkinson, L. Hutton, and P. T. Emmerson. 1985. Reconstitution of RecBC DNase activity from purified Escherichia coli RecB and RecC proteins. J. Biol. Chem. 2601224-1229. [PubMed] [Google Scholar]
- 130.Horii, Z.-I., and A. J. Clark. 1973. Genetic analysis of the recF pathway to genetic recombination in Escherichia coli K12: isolation and characterization of mutants. J. Mol. Biol. 80327-344. [DOI] [PubMed] [Google Scholar]
- 131.Howard-Flanders, P. 1973. DNA repair and recombination. Br. Med. Bull. 29226-235. [DOI] [PubMed] [Google Scholar]
- 132.Howard-Flanders, P., and L. Theriot. 1966. Mutants of Escherichia coli K-12 defective in DNA repair and in genetic recombination. Genetics 531137-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hsieh, S., and D. A. Julin. 1992. Alteration by site-directed mutagenesis of the conserved lysine residue in the consensus ATP-binding sequence of the RecB protein of Escherichia coli. Nucleic Acids Res. 205647-5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Huang, S., B. Li, M. D. Gray, J. Oshima, I. S. Mian, and J. Campisi. 1998. The premature ageing syndrome protein, WRN, is a 3′→5′ exonuclease. Nat. Genet. 20114-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ishii, Y., A. Ishijima, and T. Yanagida. 2001. Single molecule nanomanipulation of biomolecules. Trends Biotechnol. 19211-216. [DOI] [PubMed] [Google Scholar]
- 136.Ivancic-Bace, I., P. Peharec, S. Moslavac, N. Skrobot, E. Salaj-Smic, and K. Brcic-Kostic. 2003. RecFOR function is required for DNA repair and recombination in a RecA loading-deficient recB mutant of Escherichia coli. Genetics 163485-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ivancic-Bace, I., E. Salaj-Smic, and K. Brcic-Kostic. 2005. Effects of recJ, recQ, and recFOR mutations on recombination in nuclease-deficient recB recD double mutants of Escherichia coli. J. Bacteriol. 1871350-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Iyer, V. N., and W. D. Rupp. 1971. Usefulness of benzoylated naphthoylated DEAE-cellulose to distinguish and fractionate double-stranded DNA bearing different extents of single-stranded regions. Biochim. Biophys. Acta 228117-126. [DOI] [PubMed] [Google Scholar]
- 139.Jeong, Y. J., M. K. Levin, and S. S. Patel. 2004. The DNA-unwinding mechanism of the ring helicase of bacteriophage T7. Proc. Natl. Acad. Sci. USA 1017264-7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Julin, D. A., and I. R. Lehman. 1987. Photoaffinity labeling of the recBCD enzyme of Escherichia coli with 8-azidoadenosine 5′-triphosphate. J. Biol. Chem. 2629044-9051. [PubMed] [Google Scholar]
- 141.Karathanasis, E., and T. E. Wilson. 2002. Enhancement of Saccharomyces cerevisiae end-joining efficiency by cell growth stage but not by impairment of recombination. Genetics 1611015-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Karu, A. E., and S. Linn. 1972. Uncoupling of the recBC ATPase from DNase by DNA crosslinked with psoralen. Proc. Natl. Acad. Sci. USA 692855-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Karu, A. E., V. Mackay, P. J. Goldmark, and S. Linn. 1973. The recBC deoxyribonuclease of Escherichia coli K-12. Substrate specificity and reaction intermediates. J. Biol. Chem. 2484874-4884. [PubMed] [Google Scholar]
- 144.Khidhir, M. A., S. Casaregola, and I. B. Holland. 1985. Mechanism of transient inhibition of DNA synthesis in ultraviolet-irradiated E. coli: inhibition is independent of recA whilst recovery requires recA protein itself and an additional, inducible SOS function. Mol. Gen. Genet. 199133-140. [DOI] [PubMed] [Google Scholar]
- 145.Kidane, D., and P. L. Graumann. 2005. Dynamic formation of RecA filaments at DNA double strand break repair centers in live cells. J. Cell Biol. 170357-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kinch, L. N., K. Ginalski, L. Rychlewski, and N. V. Grishin. 2005. Identification of novel restriction endonuclease-like fold families among hypothetical proteins. Nucleic Acids Res. 333598-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kogoma, T. 1996. Recombination by replication. Cell 85625-627. [DOI] [PubMed] [Google Scholar]
- 148.Kogoma, T. 1997. Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol. Mol. Biol. Rev. 61212-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kooistra, J., B. J. Haijema, and G. Venema. 1993. The Bacillus subtilis addAB genes are fully functional in Escherichia coli. Mol. Microbiol. 7915-923. [DOI] [PubMed] [Google Scholar]
- 150.Koppen, A., S. Krobitsch, B. Thoms, and W. Wackernagel. 1995. Interaction with the recombination hot spot χ in vivo converts the RecBCD enzyme of Escherichia coli into a χ-independent recombinase by inactivation of the RecD subunit. Proc. Natl. Acad. Sci. USA 926249-6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Korangy, F., and D. A. Julin. 1992. Alteration by site-directed mutagenesis of the conserved lysine residue in the ATP-binding consensus sequence of the RecD subunit of the Escherichia coli RecBCD enzyme. J. Biol. Chem. 2671727-1732. [PubMed] [Google Scholar]
- 152.Korangy, F., and D. A. Julin. 1994. Efficiency of ATP hydrolysis and DNA unwinding by the RecBC enzyme from Escherichia coli. Biochemistry 339552-9560. [DOI] [PubMed] [Google Scholar]
- 153.Korangy, F., and D. A. Julin. 1992. Enzymatic effects of a lysine-to-glutamine mutation in the ATP-binding consensus sequence in the RecD subunit of the RecBCD enzyme from Escherichia coli. J. Biol. Chem. 2671733-1740. [PubMed] [Google Scholar]
- 154.Korangy, F., and D. A. Julin. 1993. Kinetics and processivity of ATP hydrolysis and DNA unwinding by the RecBC enzyme from Escherichia coli. Biochemistry 324873-4880. [DOI] [PubMed] [Google Scholar]
- 155.Korangy, F., and D. A. Julin. 1992. A mutation in the consensus ATP-binding sequence of the RecD subunit reduces the processivity of the RecBCD enzyme from Escherichia coli. J. Biol. Chem. 2673088-3095. [PubMed] [Google Scholar]
- 156.Korolev, S., J. Hsieh, G. H. Gauss, T. M. Lohman, and G. Waksman. 1997. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell 90635-647. [DOI] [PubMed] [Google Scholar]
- 157.Kowalczykowski, S. C. 1991. Biochemical and biological function of Escherichia coli RecA protein: behavior of mutant RecA proteins. Biochimie 73289-304. [DOI] [PubMed] [Google Scholar]
- 158.Kowalczykowski, S. C. 1991. Biochemistry of genetic recombination: energetics and mechanism of DNA strand exchange. Annu. Rev. Biophys. Biophys. Chem. 20539-575. [DOI] [PubMed] [Google Scholar]
- 159.Kowalczykowski, S. C. 2000. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 25156-165. [DOI] [PubMed] [Google Scholar]
- 160.Kowalczykowski, S. C. 2002. Molecular mimicry connects BRCA2 to Rad51 and recombinational DNA repair. Nat. Struct. Biol. 9897-899. [DOI] [PubMed] [Google Scholar]
- 161.Kowalczykowski, S. C., D. A. Dixon, A. K. Eggleston, S. D. Lauder, and W. M. Rehrauer. 1994. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 58401-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kushner, S. R. 1974. In vivo studies of temperature-sensitive recB and recC mutants. J. Bacteriol. 1201213-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kushner, S. R., H. Nagaishi, A. Templin, and A. J. Clark. 1971. Genetic recombination in Escherichia coli: the role of exonuclease I. Proc. Natl. Acad. Sci. USA 68824-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kuzminov, A. 1995. Collapse and repair of replication forks in Escherichia coli. Mol. Microbiol. 16373-384. [DOI] [PubMed] [Google Scholar]
- 165.Kuzminov, A. 2001. DNA replication meets genetic exchange: chromosomal damage and its repair by homologous recombination. Proc. Natl. Acad. Sci. USA 988461-8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kuzminov, A. 1995. Instability of inhibited replication forks in E. coli. Bioessays 17733-741. [DOI] [PubMed] [Google Scholar]
- 167.Kuzminov, A. 1996. Recombinational repair of DNA damage. R. G. Landes Company, Austin, TX.
- 168.Kuzminov, A. 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage λ. Microbiol. Mol. Biol. Rev. 63751-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kuzminov, A., E. Schabtach, and F. W. Stahl. 1994. Chi sites in combination with RecA protein increase the survival of linear DNA in Escherichia coli by inactivating exoV activity of RecBCD nuclease. EMBO J. 132764-2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kuzminov, A., and F. W. Stahl. 1997. Stability of linear DNA in recA mutant Escherichia coli cells reflects ongoing chromosomal DNA degradation. J. Bacteriol. 179880-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lackey, D., and S. Linn. 1980. Assay for type II restriction endonucleases using the Escherichia coli recBC DNase and duplex circular DNA. Methods Enzymol. 6526-28. [DOI] [PubMed] [Google Scholar]
- 172.Lam, S. T., M. M. Stahl, K. D. McMilin, and F. W. Stahl. 1974. Rec-mediated recombinational hot spot activity in bacteriophage lambda. II. A mutation which causes hot spot activity. Genetics 77425-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lao, P. J., and D. R. Forsdyke. 2000. Crossover hot-spot instigator (Chi) sequences in Escherichia coli occupy distinct recombination/transcription islands. Gene 24347-57. [DOI] [PubMed] [Google Scholar]
- 174.Lee, J. Y., and W. Yang. 2006. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell 1271349-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Levin, M. K., M. Gurjar, and S. S. Patel. 2005. A Brownian motor mechanism of translocation and strand separation by hepatitis C virus helicase. Nat. Struct. Mol. Biol. 12429-435. [DOI] [PubMed] [Google Scholar]
- 176.Ley, R. D. 1973. Postreplication repair in an excision-defective mutant of Escherichia coli: ultraviolet light-induced incorporation of bromodeoxyuridine into parental DNA. Photochem. Photobiol. 1887-95. [DOI] [PubMed] [Google Scholar]
- 177.Lieberman, R. P., and M. Oishi. 1973. Formation of the recB-recC DNase by in vitro complementation and evidence concerning its subunit nature. Nat. New Biol. 24375-77. [PubMed] [Google Scholar]
- 178.Lieberman, R. P., and M. Oishi. 1974. The recBC deoxyribonuclease of Escherichia coli: isolation and characterization of the subunit proteins and reconstitution of the enzyme. Proc. Natl. Acad. Sci. USA 714816-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lin, L. F., J. Posfai, R. J. Roberts, and H. Kong. 2001. Comparative genomics of the restriction-modification systems in Helicobacter pylori. Proc. Natl. Acad. Sci. USA 982740-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lindow, J. C., M. Kuwano, S. Moriya, and A. D. Grossman. 2002. Subcellular localization of the Bacillus subtilis structural maintenance of chromosomes (SMC) protein. Mol. Microbiol. 46997-1009. [DOI] [PubMed] [Google Scholar]
- 181.Linn, S., and V. MacKay. 1975. The degradation of duplex DNA by the recBC DNase of Escherichia coli. Basic Life Sci. 5A293-299. [DOI] [PubMed] [Google Scholar]
- 182.Lloyd, R. G., and C. Buckman. 1985. Identification and genetic analysis of sbcC mutations in commonly used recBC sbcB strains of Escherichia coli K-12. J. Bacteriol. 164836-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Lloyd, R. G., and C. Buckman. 1991. Overlapping functions of recD, recJ and recN provide evidence of three epistatic groups of genes in Escherichia coli recombination and DNA repair. Biochimie 73313-320. [DOI] [PubMed] [Google Scholar]
- 184.Lloyd, R. G., M. C. Porton, and C. Buckman. 1988. Effect of recF, recJ, recN, recO and ruv mutations on ultraviolet survival and genetic recombination in a recD strain of Escherichia coli K12. Mol. Gen. Genet. 212317-324. [DOI] [PubMed] [Google Scholar]
- 185.Lohman, T. M. 1993. Helicase-catalyzed DNA unwinding. J. Biol. Chem. 2682269-2272. [PubMed] [Google Scholar]
- 186.Lovett, S. T., and A. J. Clark. 1984. Genetic analysis of the recJ gene of Escherichia coli K-12. J. Bacteriol. 157190-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lovett, S. T., C. Luisi-DeLuca, and R. D. Kolodner. 1988. The genetic dependence of recombination in recD mutants of Escherichia coli. Genetics 12037-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lucius, A. L., C. J. Wong, and T. M. Lohman. 2004. Fluorescence stopped-flow studies of single turnover kinetics of E. coli RecBCD helicase-catalyzed DNA unwinding. J. Mol. Biol. 339731-750. [DOI] [PubMed] [Google Scholar]
- 189.Lucius, A. L., and T. M. Lohman. 2004. Effects of temperature and ATP on the kinetic mechanism and kinetic step-size for E.coli RecBCD helicase-catalyzed DNA unwinding. J. Mol. Biol. 339751-771. [DOI] [PubMed] [Google Scholar]
- 190.Lucius, A. L., N. K. Maluf, C. J. Fischer, and T. M. Lohman. 2003. General methods for analysis of sequential “n-step” kinetic mechanisms: application to single turnover kinetics of helicase-catalyzed DNA unwinding. Biophys. J. 852224-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Lundblad, V., A. F. Taylor, G. R. Smith, and N. Kleckner. 1984. Unusual alleles of recB and recC stimulate excision of inverted repeat transposons Tn10 and Tn5. Proc. Natl. Acad. Sci. USA 81824-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Lusetti, S. L., and M. M. Cox. 2002. The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu. Rev. Biochem. 7171-100. [DOI] [PubMed] [Google Scholar]
- 193.MacKay, V., and S. Linn. 1974. The mechanism of degradation of duplex deoxyribonucleic acid by the recBC enzyme of Escherichia coli K-12. J. Biol. Chem. 2494286-4294. [PubMed] [Google Scholar]
- 194.Makovets, S., L. M. Powell, A. J. Titheradge, G. W. Blakely, and N. E. Murray. 2004. Is modification sufficient to protect a bacterial chromosome from a resident restriction endonuclease? Mol. Microbiol. 51135-147. [DOI] [PubMed] [Google Scholar]
- 195.Marians, K. J. 2004. Mechanisms of replication fork restart in Escherichia coli. Philos. Trans. R. Soc. Lond. B Biol. Sci. 35971-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Masters, M. 1985. Generalized transduction, p. 197-215. In F. Scaife, D. Leach, and A. Galizzi (ed.), Genetics of bacteria. Academic Press, London, United Kingdom.
- 197.Masterson, C., P. E. Boehmer, F. McDonald, S. Chaudhuri, I. D. Hickson, and P. T. Emmerson. 1992. Reconstitution of the activities of the RecBCD holoenzyme of Escherichia coli from the purified subunits. J. Biol. Chem. 26713564-13572. [PubMed] [Google Scholar]
- 198.Matson, S. W., S. Tabor, and C. C. Richardson. 1983. The gene 4 protein of bacteriophage T7. Characterization of helicase activity. J. Biol. Chem. 25814017-14024. [PubMed] [Google Scholar]
- 199.McClelland, S. E., and M. D. Szczelkun. 2004. The type I and III restriction endonucleases: structural elements in molecular motors that process DNA, p. 111-132. In A. Pingoud (ed.), Restriction endonucleases. Springer, Berlin, Germany.
- 200.McGlynn, P., and R. G. Lloyd. 2002. Genome stability and the processing of damaged replication forks by RecG. Trends Genet. 18413-419. [DOI] [PubMed] [Google Scholar]
- 201.McGlynn, P., and R. G. Lloyd. 2000. Modulation of RNA polymerase by (p)ppGpp reveals a RecG-dependent mechanism for replication fork progression. Cell 10135-45. [DOI] [PubMed] [Google Scholar]
- 202.McGlynn, P., and R. G. Lloyd. 2002. Recombinational repair and restart of damaged replication forks. Nat. Rev. Mol. Cell Biol. 3859-870. [DOI] [PubMed] [Google Scholar]
- 203.Mertens, K., L. Lantsheer, D. G. Ennis, and J. E. Samuel. 2008. Constitutive SOS expression and damage-inducible AddAB-mediated recombinational repair systems for Coxiella burnetii as potential adaptations for survival within macrophages. Mol. Microbiol. 691411-1426. [DOI] [PubMed] [Google Scholar]
- 204.Meselson, M. S., and C. M. Radding. 1975. A general model for genetic recombination. Proc. Natl. Acad. Sci. USA 72358-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Michel, B., H. Boubakri, Z. Baharoglu, M. LeMasson, and R. Lestini. 2007. Recombination proteins and rescue of arrested replication forks. DNA Repair (Amsterdam) 6967-980. [DOI] [PubMed] [Google Scholar]
- 206.Michel, B., G. Grompone, M. J. Flores, and V. Bidnenko. 2004. Multiple pathways process stalled replication forks. Proc. Natl. Acad. Sci. USA 10112783-12788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Miranda, A., and A. Kuzminov. 2003. Chromosomal lesion suppression and removal in Escherichia coli via linear DNA degradation. Genetics 1631255-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Morgan, A. R., and A. Severini. 1990. Interconversion of replication and recombination structures: implications for terminal repeats and concatemers. J. Theor. Biol. 144195-202. [DOI] [PubMed] [Google Scholar]
- 209.Murphy, K. C. 2000. Bacteriophage P22 Abc2 protein binds to RecC increases the 5′ strand nicking activity of RecBCD and together with lambda bet, promotes Chi-independent recombination. J. Mol. Biol. 296385-401. [DOI] [PubMed] [Google Scholar]
- 210.Murphy, K. C. 1991. Lambda Gam protein inhibits the helicase and Chi-stimulated recombination activities of Escherichia coli RecBCD enzyme. J. Bacteriol. 1735808-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Myers, R. S., A. Kuzminov, and F. W. Stahl. 1995. The recombination hot spot χ activates RecBCD recombination by converting Escherichia coli to a recD mutant phenocopy. Proc. Natl. Acad. Sci. USA 926244-6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Myers, R. S., and F. W. Stahl. 1994. Chi and the RecBC D enzyme of Escherichia coli. Annu. Rev. Genet. 2849-70. [DOI] [PubMed] [Google Scholar]
- 213.Myers, R. S., M. M. Stahl, and F. W. Stahl. 1995. Chi recombination activity in phage lambda decays as a function of genetic distance. Genetics 141805-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Nakayama, H., K. Nakayama, R. Nakayama, N. Irino, Y. Nakayama, and P. C. Hanawalt. 1984. Isolation and genetic characterization of a thymineless death-resistant mutant of Escherichia coli K-12: identification of a new mutation (recQ1) that blocks the recF recombination pathway. Mol. Gen. Genet. 195474-480. [DOI] [PubMed] [Google Scholar]
- 215.Nobrega, F. G., F. H. Rola, M. Pasetto-Nobrega, and M. Oishi. 1972. Adenosine triphosphatase associated with adenosine triphosphate-dependent deoxyribonuclease (recB-recC enzyme-E. coli-ATP to phosphodiester hydrolysis ratio-DNA-dependent ATPase activity). Proc. Natl. Acad. Sci. USA 6915-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Oishi, M. 1969. An ATP-dependent deoxyribonuclease from E. coli with a possible role in genetic recombination. Proc. Natl. Acad. Sci. USA 641292-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Opresko, P. L., M. Otterlei, J. Graakjaer, P. Bruheim, L. Dawut, S. Kolvraa, A. May, M. M. Seidman, and V. A. Bohr. 2004. The Werner syndrome helicase and exonuclease cooperate to resolve telomeric D-loops in a manner regulated by TRF1 and TRF2. Mol. Cell 14763-774. [DOI] [PubMed] [Google Scholar]
- 218.Palas, K. M., and S. R. Kushner. 1990. Biochemical and physical characterization of exonuclease V from Escherichia coli. Comparison of the catalytic activities of the RecBC and RecBCD enzymes. J. Biol. Chem. 2653447-3454. [PubMed] [Google Scholar]
- 219.Pâques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Park, H., E. Toprak, and P. R. Selvin. 2007. Single-molecule fluorescence to study molecular motors. Q. Rev. Biophys. 4087-111. [DOI] [PubMed] [Google Scholar]
- 221.Pellegrini, L., D. S. Yu, T. Lo, S. Anand, M. Lee, T. L. Blundell, and A. R. Venkitaraman. 2002. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature 420287-293. [DOI] [PubMed] [Google Scholar]
- 222.Perkins, T. T., H. W. Li, R. V. Dalal, J. Gelles, and S. M. Block. 2004. Forward and reverse motion of single RecBCD molecules on DNA. Biophys. J. 861640-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Phillips, R. J., D. C. Hickleton, P. E. Boehmer, and P. T. Emmerson. 1997. The RecB protein of Escherichia coli translocates along single-stranded DNA in the 3′ to 5′ direction: a proposed ratchet mechanism. Mol. Gen. Genet. 254319-329. [DOI] [PubMed] [Google Scholar]
- 224.Ponticelli, A. S., D. W. Schultz, A. F. Taylor, and G. R. Smith. 1985. Chi-dependent DNA strand cleavage by RecBC enzyme. Cell 41145-151. [DOI] [PubMed] [Google Scholar]
- 225.Postow, L., C. Ullsperger, R. W. Keller, C. Bustamante, A. V. Vologodskii, and N. R. Cozzarelli. 2000. Positive torsional strain causes the formation of a four-way junction at replication forks. J. Biol. Chem. 2762790-2796. [DOI] [PubMed] [Google Scholar]
- 226.Quiberoni, A., I. Biswas, M. El Karoui, L. Rezaiki, P. Tailliez, and A. Gruss. 2001. In vivo evidence for two active nuclease motifs in the double-strand break repair enzyme RexAB of Lactococcus lactis. J. Bacteriol. 1834071-4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Quiberoni, A., L. Rezaiki, M. El Karoui, I. Biswas, P. Tailliez, and A. Gruss. 2001. Distinctive features of homologous recombination in an ‘old’ microorganism, Lactococcus lactis. Res. Microbiol. 152131-139. [DOI] [PubMed] [Google Scholar]
- 228.Rafferty, J. B., S. E. Sedelnikova, D. Hargreaves, P. J. Artymiuk, P. J. Baker, G. J. Sharples, A. A. Mahdi, R. G. Lloyd, and D. W. Rice. 1996. Crystal structure of DNA recombination protein RuvA and a model for its binding to the Holliday junction. Science 274415-421. [DOI] [PubMed] [Google Scholar]
- 229.Regha, K., A. K. Satapathy, and M. K. Ray. 2005. RecD plays an essential function during growth at low temperature in the Antarctic bacterium Pseudomonas syringae Lz4W. Genetics 1701473-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Register, J. C., III, and J. Griffith. 1985. The direction of RecA protein assembly onto single strand DNA is the same as the direction of strand assimilation during strand exchange. J. Biol. Chem. 26012308-12312. [PubMed] [Google Scholar]
- 231.Rigden, D. J. 2005. An inactivated nuclease-like domain in RecC with novel function: implications for evolution. BMC Struct. Biol. 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Rinken, R., and W. Wackernagel. 1992. Inhibition of the recBCD-dependent activation of Chi recombinational hot spots in SOS-induced cells of Escherichia coli. J. Bacteriol. 1741172-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Roca, A. I., and M. M. Cox. 1997. RecA protein: structure, function, and role in recombinational DNA repair. Prog. Nucleic Acid Res. Mol. Biol. 56129-223. [DOI] [PubMed] [Google Scholar]
- 234.Rocha, E. P., E. Cornet, and B. Michel. 2005. Comparative and evolutionary analysis of the bacterial homologous recombination systems. PLoS Genet. 1e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Roman, L. J., D. A. Dixon, and S. C. Kowalczykowski. 1991. RecBCD-dependent joint molecule formation promoted by the Escherichia coli RecA and SSB proteins. Proc. Natl. Acad. Sci. USA 883367-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Roman, L. J., A. K. Eggleston, and S. C. Kowalczykowski. 1992. Processivity of the DNA helicase activity of Escherichia coli recBCD enzyme. J. Biol. Chem. 2674207-4214. [PubMed] [Google Scholar]
- 237.Roman, L. J., and S. C. Kowalczykowski. 1989. Characterization of the adenosinetriphosphatase activity of the Escherichia coli RecBCD enzyme: relationship of ATP hydrolysis to the unwinding of duplex DNA. Biochemistry 282873-2881. [DOI] [PubMed] [Google Scholar]
- 238.Roman, L. J., and S. C. Kowalczykowski. 1989. Characterization of the helicase activity of the Escherichia coli RecBCD enzyme using a novel helicase assay. Biochemistry 282863-2873. [DOI] [PubMed] [Google Scholar]
- 239.Roman, L. J., and S. C. Kowalczykowski. 1989. Formation of heteroduplex DNA promoted by the combined activities of Escherichia coli RecA and RecBCD proteins. J. Biol. Chem. 26418340-18348. [PubMed] [Google Scholar]
- 240.Rosamond, J., B. Endlich, K. M. Telander, and S. Linn. 1979. Mechanisms of action of the type-I restriction endonuclease EcoB and the RecBC DNase from E. coli. Cold Spring Harb. Symp. Quant. Biol. 431049-1057. [DOI] [PubMed] [Google Scholar]
- 241.Rosamond, J., K. M. Telander, and S. Linn. 1979. Modulation of the action of the recBC enzyme of Escherichia coli K-12 by Ca2+. J. Biol. Chem. 2548646-8652. [PubMed] [Google Scholar]
- 242.Rothman, R. H., and A. J. Clark. 1977. Defective excision and postreplication repair of UV-damaged DNA in a recL mutant strain of E. coli K-12. Mol. Gen. Genet. 155267-277. [DOI] [PubMed] [Google Scholar]
- 243.Rothman, R. H., and A. J. Clark. 1977. The dependence of postreplication repair on uvrB in a recF mutant of Escherichia coli K-12. Mol. Gen. Genet. 155279-286. [DOI] [PubMed] [Google Scholar]
- 244.Rothman, R. H., T. Kato, and A. J. Clark. 1975. The beginning of an investigation of the role of recF in the pathways of metabolism of ultraviolet-irradiated DNA in Escherichia coli. Basic Life Sci. 5A283-291. [DOI] [PubMed] [Google Scholar]
- 245.Rupp, W. D., and P. Howard-Flanders. 1968. Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J. Mol. Biol. 31291-304. [DOI] [PubMed] [Google Scholar]
- 246.Salzberg, S. L., A. J. Salzberg, A. R. Kerlavage, and J. F. Tomb. 1998. Skewed oligomers and origins of replication. Gene 21757-67. [DOI] [PubMed] [Google Scholar]
- 247.Sandler, S. J. 2000. Multiple genetic pathways for restarting DNA replication forks in Escherichia coli K-12. Genetics 155487-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Schultz, D. W., A. F. Taylor, and G. R. Smith. 1983. Escherichia coli RecBC pseudorevertants lacking Chi recombinational hotspot activity. J. Bacteriol. 155664-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Seigneur, M., V. Bidnenko, S. D. Ehrlich, and B. Michel. 1998. RuvAB acts at arrested replication forks. Cell 95419-430. [DOI] [PubMed] [Google Scholar]
- 250.Servinsky, M. D., and D. A. Julin. 2007. Effect of a recD mutation on DNA damage resistance and transformation in Deinococcus radiodurans. J. Bacteriol. 1895101-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Severini, A., A. R. Morgan, D. R. Tovell, and D. L. Tyrrell. 1994. Study of the structure of replicative intermediates of HSV-1 DNA by pulsed-field gel electrophoresis. Virology 200428-435. [DOI] [PubMed] [Google Scholar]
- 252.Shibata, T., C. DasGupta, R. P. Cunningham, and C. M. Radding. 1979. Purified Escherichia coli recA protein catalyzes homologous pairing of superhelical DNA and single-stranded fragments. Proc. Natl. Acad. Sci. USA 761638-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Shuman, S., and M. S. Glickman. 2007. Bacterial DNA repair by non-homologous end joining. Nat. Rev. Microbiol. 5852-861. [DOI] [PubMed] [Google Scholar]
- 254.Silverstein, J. L., and E. B. Goldberg. 1976. T4 DNA injection. II. Protection of entering DNA from host exonuclease V. Virology 72212-223. [DOI] [PubMed] [Google Scholar]
- 255.Simmon, V. F., and S. Lederberg. 1972. Degradation of bacteriophage lambda deoxyribonucleic acid after restriction by Escherichia coli K-12. J. Bacteriol. 112161-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Singleton, M. R., M. S. Dillingham, M. Gaudier, S. C. Kowalczykowski, and D. B. Wigley. 2004. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature 432187-193. [DOI] [PubMed] [Google Scholar]
- 257.Singleton, M. R., M. S. Dillingham, and D. B. Wigley. 2007. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 7623-50. [DOI] [PubMed] [Google Scholar]
- 258.Singleton, M. R., S. Scaife, and D. B. Wigley. 2001. Structural analysis of DNA replication fork reversal by RecG. Cell 10779-89. [DOI] [PubMed] [Google Scholar]
- 259.Singleton, M. R., and D. B. Wigley. 2002. Modularity and specialization in superfamily 1 and 2 helicases. J. Bacteriol. 1841819-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Skalka, A. 1974. A replicator's view of recombination (and repair), p. 421-432. In R. F. Grell (ed.), Mechanisms in recombination. Plenum Press, New York, NY.
- 261.Smith, G. R. 1991. Conjugational recombination in Escherichia coli: myths and mechanisms. Cell 6419-27. [DOI] [PubMed] [Google Scholar]
- 262.Smith, G. R. 2001. Homologous recombination near and far from DNA breaks: alternative roles and contrasting views. Annu. Rev. Genet. 35243-274. [DOI] [PubMed] [Google Scholar]
- 263.Smith, G. R., S. M. Kunes, D. W. Schultz, A. Taylor, and K. L. Triman. 1981. Structure of Chi hotspots of generalized recombination. Cell 24429-436. [DOI] [PubMed] [Google Scholar]
- 264.Soultanas, P., M. S. Dillingham, S. S. Velankar, and D. B. Wigley. 1999. DNA binding mediates conformational changes and metal ion coordination in the active site of PcrA helicase. J. Mol. Biol. 290137-148. [DOI] [PubMed] [Google Scholar]
- 265.Spek, E. J., T. L. Wright, M. S. Stitt, N. R. Taghizadeh, S. R. Tannenbaum, M. G. Marinus, and B. P. Engelward. 2001. Recombinational repair is critical for survival of Escherichia coli exposed to nitric oxide. J. Bacteriol. 183131-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Spies, M., I. Amitani, R. J. Baskin, and S. C. Kowalczykowski. 2007. RecBCD enzyme switches lead motor subunits in response to χ recognition. Cell 131694-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Spies, M., P. R. Bianco, M. S. Dillingham, N. Handa, R. J. Baskin, and S. C. Kowalczykowski. 2003. A molecular throttle: the recombination hotspot χ controls DNA translocation by the RecBCD helicase. Cell 114647-654. [DOI] [PubMed] [Google Scholar]
- 268.Spies, M., M. S. Dillingham, and S. C. Kowalczykowski. 2005. Translocation by the RecB motor is an absolute requirement for χ-recognition and RecA protein loading by RecBCD enzyme. J. Biol. Chem. 28037078-37087. [DOI] [PubMed] [Google Scholar]
- 269.Spies, M., and S. C. Kowalczykowski. 2005. Homologous recombination by RecBCD and RecF pathways, p. 389-403. In N. P. Higgins (ed.), The bacterial chromosome. ASM Press, Washington, DC.
- 270.Spies, M., and S. C. Kowalczykowski. 2006. The RecA binding locus of RecBCD is a general domain for recruitment of DNA strand exchange proteins. Mol. Cell 21573-580. [DOI] [PubMed] [Google Scholar]
- 271.Sprague, K. U., D. H. Faulds, and G. R. Smith. 1978. A single base-pair change creates a Chi recombinational hotspot in bacteriophage lambda. Proc. Natl. Acad. Sci. USA 756182-6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Stahl, F. W. 2005. Chi: a little sequence controls a big enzyme. Genetics 170487-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Stahl, F. W., and M. M. Stahl. 1975. Rec-mediated recombinational hot spot activity in bacteriophage lambda. IV. Effect of heterology on Chi-stimulated crossing over. Mol. Gen. Genet. 14029-37. [DOI] [PubMed] [Google Scholar]
- 274.Stahl, F. W., and M. M. Stahl. 1974. A role for recBC nuclease in the distribution of crossovers along unreplicated chromosomes of phage lambda. Mol. Gen. Genet. 13127-30. [DOI] [PubMed] [Google Scholar]
- 275.Stahl, F. W., M. M. Stahl, R. E. Malone, and J. M. Crasemann. 1980. Directionality and nonreciprocality of Chi-stimulated recombination in phage lambda. Genetics 94235-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Stahl, F. W., L. C. Thomason, I. Siddiqi, and M. M. Stahl. 1990. Further tests of a recombination model in which Chi removes the RecD subunit from the RecBCD enzyme of Escherichia coli. Genetics 126519-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Stukalin, E. B., H. Phillips III, and A. B. Kolomeisky. 2005. Coupling of two motor proteins: a new motor can move faster. Phys. Rev. Lett. 94238101. [DOI] [PubMed] [Google Scholar]
- 278.Sun, J. Z., D. A. Julin, and J. S. Hu. 2006. The nuclease domain of the Escherichia coli RecBCD enzyme catalyzes degradation of linear and circular single-stranded and double-stranded DNA. Biochemistry 45131-140. [DOI] [PubMed] [Google Scholar]
- 279.Symington, L. S. 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66630-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Szczelkun, M. D. 2002. Kinetic models of translocation, head-on collision, and DNA cleavage by type I restriction endonucleases. Biochemistry 412067-2074. [DOI] [PubMed] [Google Scholar]
- 281.Szostak, J. W., T. L. Orr-Weaver, R. J. Rothstein, and F. W. Stahl. 1983. The double-strand-break repair model for recombination. Cell 3325-35. [DOI] [PubMed] [Google Scholar]
- 282.Tanner, D., F. G. Nobrega, and M. Oishi. 1972. 5′-oligonucleotides as the acid-soluble products of the ATP-dependent DNase form Escherichia coli. J. Mol. Biol. 67513-516. [DOI] [PubMed] [Google Scholar]
- 283.Taylor, A., and G. R. Smith. 1980. Unwinding and rewinding of DNA by exonuclease V, p. 909-919. In B. Alberts and C. F. Fox (ed.), Mechanistic studies of DNA replication and genetic recombination, vol. 19. Academic Press, New York, NY. [Google Scholar]
- 284.Taylor, A., and G. R. Smith. 1980. Unwinding and rewinding of DNA by the RecBC enzyme. Cell 22447-457. [DOI] [PubMed] [Google Scholar]
- 285.Taylor, A. F. 1988. The RecBCD enzyme of Escherichia coli, p. 231-263. In R. Kucherlapati and G. R. Smith (ed.), Genetic recombination. American Society for Microbiology, Washington, DC.
- 286.Taylor, A. F., D. W. Schultz, A. S. Ponticelli, and G. R. Smith. 1985. RecBC enzyme nicking at Chi sites during DNA unwinding: location and orientation-dependence of the cutting. Cell 41153-163. [DOI] [PubMed] [Google Scholar]
- 287.Taylor, A. F., and G. R. Smith. 1995. Monomeric RecBCD enzyme binds and unwinds DNA. J. Biol. Chem. 27024451-24458. [DOI] [PubMed] [Google Scholar]
- 288.Taylor, A. F., and G. R. Smith. 2003. RecBCD enzyme is a DNA helicase with fast and slow motors of opposite polarity. Nature 423889-893. [DOI] [PubMed] [Google Scholar]
- 289.Taylor, A. F., and G. R. Smith. 1992. RecBCD enzyme is altered upon cutting DNA at a Chi recombination hotspot. Proc. Natl. Acad. Sci. USA 895226-5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Taylor, A. F., and G. R. Smith. 1999. Regulation of homologous recombination: Chi inactivates RecBCD enzyme by disassembly of the three subunits. Genes Dev. 13890-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Taylor, A. F., and G. R. Smith. 1995. Strand specificity of nicking of DNA at Chi sites by RecBCD enzyme: modulation by ATP and magnesium levels. J. Biol. Chem. 27024459-24467. [DOI] [PubMed] [Google Scholar]
- 292.Taylor, A. F., and G. R. Smith. 1985. Substrate specificity of the DNA unwinding activity of the RecBC enzyme of Escherichia coli. J. Mol. Biol. 185431-443. [DOI] [PubMed] [Google Scholar]
- 293.Telander-Muskavitch, K. M., and S. Linn. 1980. Electron microscopy of E. coli recBC enzyme reaction intermediates, p. 901-918. In B. Alberts (ed.), Mechanistic studies of DNA replication and genetic recombination, vol. 19. Academic Press, New York, NY. [Google Scholar]
- 294.Telander-Muskavitch, K. M., and S. Linn. 1982. A unified mechanism for the nuclease and unwinding activities of the recBC enzyme of Escherichia coli. J. Biol. Chem. 2572641-2648. [PubMed] [Google Scholar]
- 295.Thaler, D. S., E. Sampson, I. Siddiqi, S. M. Rosenberg, F. W. Stahl, and M. Stahl. 1988. A hypothesis: Chi-activation of recBCD enzyme involves removal of the recD subunit, p. 413-422. In E. C. Friedberg and P. C. Hanawalt (ed.), Mechanisms and consequences of DNA damage processing. Alan R. Liss, Inc., New York, NY.
- 296.Thaler, D. S., E. Sampson, I. Siddiqi, S. M. Rosenberg, L. C. Thomason, F. W. Stahl, and M. M. Stahl. 1989. Recombination of bacteriophage lambda in recD mutants of Escherichia coli. Genome 3153-67. [DOI] [PubMed] [Google Scholar]
- 297.Thoms, B., I. Borchers, and W. Wackernagel. 2008. Effects of single-strand DNases ExoI, RecJ, ExoVII, and SbcCD on homologous recombination of recBCD+ strains of Escherichia coli and roles of SbcB15 and XonA2 ExoI mutant enzymes. J. Bacteriol. 190179-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Thoms, B., and W. Wackernagel. 1998. Interaction of RecBCD enzyme with DNA at double-strand breaks produced in UV-irradiated Escherichia coli: requirement for DNA end processing. J. Bacteriol. 1805639-5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Tillier, E. R., and R. A. Collins. 2000. The contributions of replication orientation, gene direction, and signal sequences to base-composition asymmetries in bacterial genomes. J. Mol. Evol. 50249-257. [DOI] [PubMed] [Google Scholar]
- 300.Tomizawa, J., and H. Ogawa. 1972. Structural genes of ATP-dependent deoxyribonuclease of Escherichia coli. Nat. New Biol. 23914-16. [DOI] [PubMed] [Google Scholar]
- 301.Tomko, E. J., C. J. Fischer, A. Niedziela-Majka, and T. M. Lohman. 2007. A nonuniform stepping mechanism for E. coli UvrD monomer translocation along single-stranded DNA. Mol. Cell 26335-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Tracy, R. B., F. Chédin, and S. C. Kowalczykowski. 1997. The recombination hot spot Chi is embedded within islands of preferred DNA pairing sequences in the E. coli genome. Cell 90205-206. [DOI] [PubMed] [Google Scholar]
- 303.Uno, R., Y. Nakayama, K. Arakawa, and M. Tomita. 2000. The orientation bias of Chi sequences is a general tendency of G-rich oligomers. Gene 259207-215. [DOI] [PubMed] [Google Scholar]
- 304.Uno, R., Y. Nakayama, and M. Tomita. 2006. Over-representation of Chi sequences caused by di-codon increase in Escherichia coli K-12. Gene 38030-37. [DOI] [PubMed] [Google Scholar]
- 305.Velankar, S. S., P. Soultanas, M. S. Dillingham, H. S. Subramanya, and D. B. Wigley. 1999. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell 9775-84. [DOI] [PubMed] [Google Scholar]
- 306.Viswanathan, M., J. J. Lacirignola, R. L. Hurley, and S. T. Lovett. 2000. A novel mutational hotspot in a natural quasipalindrome in Escherichia coli. J. Mol. Biol. 302553-564. [DOI] [PubMed] [Google Scholar]
- 307.Wackernagel, W. 1973. Genetic transformation in E. coli: the inhibitory role of the recBC DNase. Biochem. Biophys. Res. Commun. 51306-311. [DOI] [PubMed] [Google Scholar]
- 308.Wang, J., R. Chen, and D. A. Julin. 2000. A single nuclease active site of the Escherichia coli RecBCD enzyme catalyzes single-stranded DNA degradation in both directions. J. Biol. Chem. 275507-513. [DOI] [PubMed] [Google Scholar]
- 309.Wang, J., and D. A. Julin. 2004. DNA helicase activity of the RecD protein from Deinococcus radiodurans. J. Biol. Chem. 27952024-52032. [DOI] [PubMed] [Google Scholar]
- 310.Wang, T.-C. V., and K. C. Smith. 1984. recF-dependent and recF recB-independent DNA gap-filling repair processes transfer dimer-containing parental strands to daughter strands in Escherichia coli K-12 uvrB. J. Bacteriol. 158727-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Weller, G. R., B. Kysela, R. Roy, L. M. Tonkin, E. Scanlan, M. Della, S. K. Devine, J. P. Day, A. Wilkinson, F. di Fagagna, K. M. Devine, R. P. Bowater, P. A. Jeggo, S. P. Jackson, and A. J. Doherty. 2002. Identification of a DNA nonhomologous end-joining complex in bacteria. Science 2971686-1689. [DOI] [PubMed] [Google Scholar]
- 312.Wilcox, K. W., and H. O. Smith. 1976. Mechanism of DNA degradation by the ATP-dependent DNase from Hemophilus influenzae Rd. J. Biol. Chem. 2516127-6134. [PubMed] [Google Scholar]
- 313.Wilkins, B. M. 1969. Chromosome transfer from F-lac+ strains of Escherichia coli K-12 mutant at recA, recB, or recC. J. Bacteriol. 98599-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Wilkins, B. M., and P. Howard-Flanders. 1968. The genetic properties of DNA transferred from ultraviolet-irradiated Hfr cells of Escherichia coli K-12 during mating. Genetics 60243-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Willetts, N. S., and D. W. Mount. 1969. Genetic analysis of recombination-deficient mutants of Escherichia coli K-12 carrying rec mutations cotransducible with thyA. J. Bacteriol. 100923-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Williams, J. G., and C. M. Radding. 1981. Partial purification and properties of an exonuclease inhibitor induced by bacteriophage Mu-1. J. Virol. 39548-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Wilson, T. E., L. M. Topper, and P. L. Palmbos. 2003. Non-homologous end-joining: bacteria join the chromosome breakdance. Trends Biochem. Sci. 2862-66. [DOI] [PubMed] [Google Scholar]
- 318.Witkin, E. M., V. Roegner-Maniscalco, J. B. Sweasy, and J. O. McCall. 1987. Recovery from ultraviolet light-induced inhibition of DNA synthesis requires umuDC gene products in recA718 mutant strains but not in recA+ strains of Escherichia coli. Proc. Natl. Acad. Sci. USA 846805-6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Wong, C. J., A. L. Lucius, and T. M. Lohman. 2005. Energetics of DNA end binding by E. coli RecBC and RecBCD helicases indicate loop formation in the 3′-single-stranded DNA tail. J. Mol. Biol. 352765-782. [DOI] [PubMed] [Google Scholar]
- 320.Wright, M., and G. Buttin. 1969. Mechanisms of enzyme degradation of bacterial chromosomes and their regulation. Bull. Soc. Chim. Biol. (Paris) 511373-1383. (In French.) [PubMed] [Google Scholar]
- 321.Wright, M., G. Buttin, and J. Hurwitz. 1971. The isolation and characterization from Escherichia coli of an adenosine triphosphate-dependent deoxyribonuclease directed by recB,C genes. J. Biol. Chem. 2466543-6555. [PubMed] [Google Scholar]
- 322.Xu, L., and K. J. Marians. 2002. A dynamic RecA filament permits DNA polymerase-catalyzed extension of the invading strand in recombination intermediates. J. Biol. Chem. 27714321-14328. [DOI] [PubMed] [Google Scholar]
- 323.Yeeles, J. T., and M. S. Dillingham. 2007. A dual-nuclease mechanism for DNA break processing by AddAB-type helicase-nucleases. J. Mol. Biol. 37166-78. [DOI] [PubMed] [Google Scholar]
- 324.Yu, M., J. Souaya, and D. A. Julin. 1998. The 30-kDa C-terminal domain of the RecB protein is critical for the nuclease activity, but not the helicase activity, of the RecBCD enzyme from Escherichia coli. Proc. Natl. Acad. Sci. USA 95981-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Yu, M., J. Souaya, and D. A. Julin. 1998. Identification of the nuclease active site in the multifunctional RecBCD enzyme by creation of a chimeric enzyme. J. Mol. Biol. 283797-808. [DOI] [PubMed] [Google Scholar]
- 326.Zhang, X. J., and D. A. Julin. 1999. Isolation and characterization of the C-terminal nuclease domain from the RecB protein of Escherichia coli. Nucleic Acids Res. 274200-4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Zhou, Q., X. Zhang, H. Xu, B. Xu, and Y. Hua. 2007. A new role of Deinococcus radiodurans RecD in antioxidant pathway. FEMS Microbiol. Lett. 271118-125. [DOI] [PubMed] [Google Scholar]
- 328.Zuniga-Castillo, J., D. Romero, and J. M. Martinez-Salazar. 2004. The recombination genes addAB are not restricted to gram-positive bacteria: genetic analysis of the recombination initiation enzymes RecF and AddAB in Rhizobium etli. J. Bacteriol. 1867905-7913. [DOI] [PMC free article] [PubMed] [Google Scholar]