Abstract
Despite the fundamental importance and high level of compartmentation of mitochondrial nucleotide metabolism in plants, our knowledge concerning the transport of nucleotides across intracellular membranes remains far from complete. Study of a previously uncharacterized Arabidopsis (Arabidopsis thaliana) gene (At4g01100) revealed it to be a novel adenine nucleotide transporter, designated ADNT1, belonging to the mitochondrial carrier family. The ADNT1 gene shows broad expression at the organ level. Green fluorescent protein-based cell biological analysis demonstrated targeting of ADNT1 to mitochondria. While analysis of the expression of β-glucuronidase fusion proteins suggested that it was expressed across a broad range of tissue types, it was most highly expressed in root tips. Direct transport assays with recombinant and reconstituted ADNT1 were utilized to demonstrate that this protein displays a relatively narrow substrate specificity largely confined to adenylates and their closest analogs. ATP uptake was markedly inhibited by the presence of other adenylates and general inhibitors of mitochondrial transport but not by bongkrekate or carboxyatractyloside, inhibitors of the previously characterized ADP/ATP carrier. Furthermore, the kinetics are substantially different from those of this carrier, with ADNT1 preferring AMP to ADP. Finally, isolation and characterization of a T-DNA insertional knockout mutant of ADNT1, alongside complementation and antisense approaches, demonstrated that although deficiency of this transporter did not seem to greatly alter photosynthetic metabolism, it did result in reduced root growth and respiration. These findings are discussed in the context of a potential function for ADNT1 in the provision of the energy required to support growth in heterotrophic plant tissues.
Mitochondria are ubiquitously found in eukaryotes. In addition to their well-established role in respiration and cellular energy supply, mitochondria fulfill a variety of metabolic tasks and play a critical role in several processes that are essential for cell viability (Sweetlove et al., 2007). This is particularly true in autotrophic organisms such as plants, in which mitochondria provide precursors for a number of essential biosynthetic processes such as nitrogen fixation and the biosynthesis of amino acids and vitamin cofactors (Douce, 1985; Douce and Neuberger, 1989; MacKenzie and McIntosh, 1999; Giege et al., 2003; Fernie et al., 2004; Raschke et al., 2007; Sweetlove et al., 2007). The concerted function of intramitochondrial and extramitochondrial metabolism relies on the presence of a family of related proteins that catalyze the transport of various metabolites, nucleotides, and cofactors across the inner mitochondrial membrane: the mitochondrial carrier family (MCF). The MCF contains as many as 58 members (Picault et al., 2004). In recent years, several of these transporters have been cloned and investigated at the molecular and biochemical levels (Picault et al., 2002; Catoni et al., 2003; Considine et al., 2003; Hoyos et al., 2003; Smith et al., 2004; Bedhomme et al., 2005; Leroch et al., 2005; Bouvier et al., 2006; Palmieri et al., 2006b, 2006c, 2008). These studies revealed that some members of the MCF in plants can be present in the plastidic membranes (Bedhomme et al., 2005; Leroch et al., 2005; Bouvier et al., 2006; Thuswaldner et al., 2007) and, with few exceptions, confirmed previous gene annotations based on homologies to nonplant species. However, the transport functions of most members of this family remain unknown. A recent proteomic survey of the MCF led to the identification of six carrier proteins as belonging to a small subset of highly abundant carrier proteins in Arabidopsis (Arabidopsis thaliana) mitochondria (Millar and Heazlewood, 2003). These carrier proteins consist of two isoforms of the ADP/ATP carrier (At3g08580 and At5g13490), the dicarboxylate/tricarboxylate carrier (At5g19760), the phosphate carrier (At5g14040), the uncoupling protein (At3g54110), and a carrier protein of unknown function (At4g01100) for which no paralog/ortholog is known. In this study, we provide evidence that the gene product of At4g01100, named ADNT1, is a novel adenine nucleotide transporter in Arabidopsis mitochondria.
Localization experiments using GFP fused to ADNT1 suggested that this protein is targeted exclusively to mitochondria. Moreover, expression studies revealed a tissue-constitutive expression, although GUS staining of promoter-GUS fusions suggested that expression was much stronger in root tips and senescing tissues. When ADNT1 was overexpressed in Escherichia coli, purified, and characterized on the basis of its transport properties, it became apparent that this protein is a narrow-range carrier for ATP, AMP, and, to a lesser extent, ADP. The uptake of ATP by this carrier was markedly inhibited by the presence of other adenylates and general inhibitors of mitochondrial transport but not by inhibitors known to affect the ADP/ATP carrier. On the basis of its transport properties, we postulate that the carrier could play a role in oxidative phosphorylation, a hypothesis further supported by the phenotype of a T-DNA insertion in the At4g01100 gene mutant (and associated complementary transgenic lines), which exhibits a largely unaltered photosynthetic phenotype but reduced root respiration and growth compared with the wild type. These results are discussed in the context of current models of the operation of adenylate exchange between the cytosol and mitochondria of plant cells.
RESULTS
Subcellular Localization of ADNT1
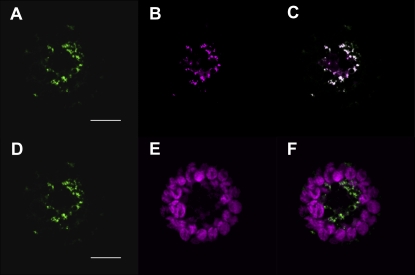
At4g01100, hereafter named ADNT1, encodes a previously uncharacterized member of the MCF in Arabidopsis (Picault et al., 2004). Recently, it was shown that ADNT1 resides in the mitochondrial inner membrane (Millar and Heazlewood, 2003). To confirm the localization of ADNT1, we prepared an ADNT1-GFP fusion construct and transiently transformed Arabidopsis protoplasts (Fig. 1). ADNT1-GFP-expressing cells displayed a punctate pattern of green fluorescence typical of mitochondrial networks (Fig. 1, A and D). The mitochondrial identity of these structures was demonstrated by colocalization with the mitochondria-specific dye MitoTracker Orange (Fig. 1, B and C) and was similar to that of the previously characterized SAMC1-GFP (Palmieri et al., 2006b). In contrast, the green fluorescence of ADNT1-GFP did not colocalize with the red autofluorescence of chlorophyll (Fig. 1, E and F). Taken together with earlier evidence, these data demonstrate that ADNT1 was localized to mitochondria.
Figure 1.
Subcellular localization of ADNT1. Transient expression in Arabidopsis protoplasts of GFP fused in frame with full-length ADNT1. Cells were labeled with Mitotracker Orange as a mitochondrial fluorescent marker, while chlorophyll red autofluorescence was used as a chloroplast marker. A and D, GFP fluorescence. B, Mitotracker Orange. C, Merged image of GFP fluorescence with MitoTracker Orange fluorescence. E, Chlorophyll autofluorescence. F, Merged image of GFP fluorescence and chlorophyll autofluorescence. Bars = 10 μm.
Expression Analysis of ADNT1
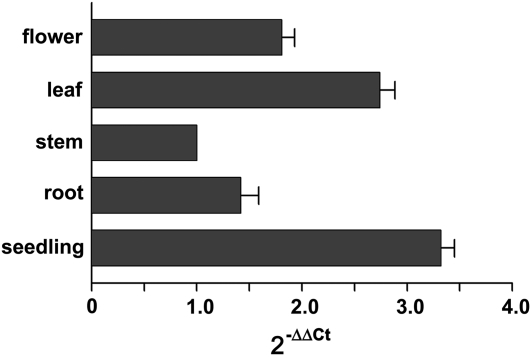
ADNT1 gene expression levels in different organs were determined by real-time reverse transcription (RT)-PCR. Relative expression levels were determined in various organs by RT-PCR using the housekeeping elongation factor EF1α gene as an internal control (Fig. 2). ADNT1 mRNA was expressed strongly in seedlings; it also exhibited considerable expression in leaves and flowers and to a lesser extent in roots and stems. These results are in line with the data housed in publicly available microarray data collections (Zimmermann et al., 2004).
Figure 2.
Expression of ADNT1 in various organs. Real-time RT-PCR experiments were conducted on cDNAs prepared by RT of total RNAs from various Arabidopsis organs, using gene-specific primers. Three independent preparations of total RNA (100 ng) from each organ were assayed in triplicate (se < 10%). The relative quantification of ADNT1 was performed according to the comparative method (2−ΔΔCt). EF1α was employed as a reference gene. For the calibrator (stem), ΔΔCt = 0 and 2−ΔΔCt = 1. For the remaining organs, the value of 2−ΔΔCt indicates the fold change in gene expression relative to the calibrator.
Expression Patterns of ADNT1 in Arabidopsis
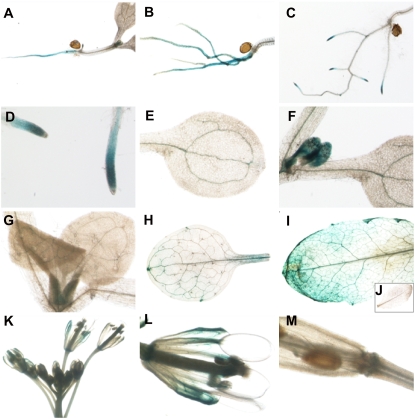
To complement the tissue-specific expression by semiquantitative RT-PCR reported above, we next investigated organ specificity by analyzing ADNT1 promoter-GUS fusion expression in transgenic Arabidopsis by fusing approximately 1 kb of the ADNT1 5′ upstream promoter region to the E. coli GUS reporter gene. During vegetative growth, ADNT1 expression was observed predominantly in seedling radicles and roots (Fig. 3, A and B); in a small portion of seedlings examined (approximately 20%), ADNT1 expression was found exclusively in root tips (Fig. 3, C and D). High expression, however, was additionally observed in the vascular tissue of cotyledons and in leaf primordia (Fig. 3, E and F), while reduced levels of staining were also apparent in the primary veins of the first two true leaves (Fig. 3G). No detectable expression was observed in seedling hypocotyls or root hairs (Fig. 3, A–D).
Figure 3.
GUS staining of Arabidopsis plants transformed with the ADNT1 promoter-GUS fusion. Histochemical analysis of promoter activity in shoot radicles (A), roots (B), root tips (C and D), vascular tissue of cotyledon (E), vascular tissue in leaf primordia (F), primary veins of the first two true leaves (G), adult leaf (H), senescent leaf (I) in contrast to wild-type control plant (J), immature flower (K), mature flower (L), and silique (M).
In adult plants, lower ADNT1 expression was detected in both young and adult leaves (a representative adult leaf is shown in Fig. 3H); while upon wounding or senescence, leaves displayed high GUS activity (a representative senescent leaf is shown in Fig. 3I) that was not apparent in wild-type control plants (Fig. 3J). During early flower development, no detectable expression was found in immature flowers up to stage 11 (following the terminology of Smyth et al. [1990]; Fig. 3K). At stage 12 of flower development and in mature flowers, high expression of ADNT1 was observed in sepals, predominantly in vascular tissues (Fig. 3L). Weak GUS staining was observed in the basal parts of mature flowers and siliques (Fig. 3M), but expression was apparently absent in other floral organs and in stems.
Bacterial Expression of ADNT1
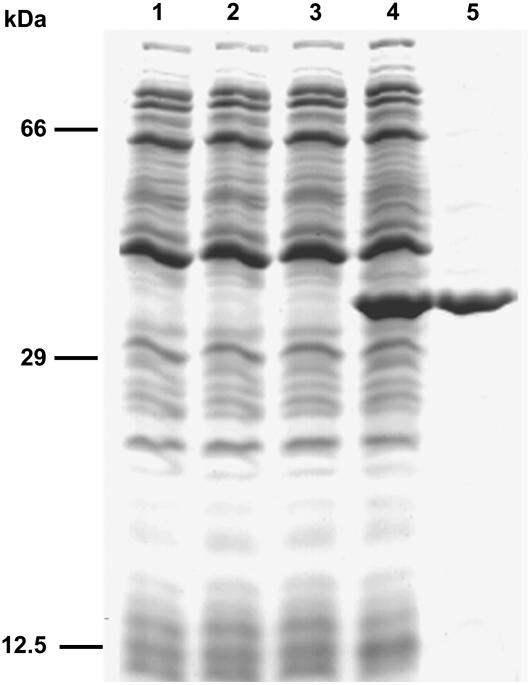
To identify its biochemical function, ADNT1 was expressed at high levels in E. coli C0214(DE3) (Fig. 4, lane 4). It accumulated as inclusion bodies and was purified by centrifugation and washing (Fig. 4, lane 5). The apparent molecular mass of the recombinant protein was about 38 kD (the calculated value with initiator Met was 38,322 D). The identity of the purified protein was confirmed by N-terminal sequencing. About 50 mg of purified protein were obtained per liter of culture. The protein was detected neither in bacteria harvested immediately before induction of expression (Fig. 4, lane 2) nor in cells harvested after induction but lacking the coding sequence in the expression vector (Fig. 4, lane 3).
Figure 4.
Overexpression in E. coli and purification of ADNT1. Proteins were separated by SDS-PAGE and stained with Coomassie Brilliant Blue dye. Lanes 1 to 4, E. coli C0214(DE3) containing the expression vector with (lanes 2 and 4) and without (lanes 1 and 3) the coding sequence for ADNT1. Samples were taken at the time of induction (lanes 1 and 2) and 5 h later (lanes 3 and 4). Equivalent samples were analyzed. Lane 5, 10 μg of ADNT1 purified from bacteria in lane 4. The positions of molecular mass markers are indicated at left.
Functional Characterization of Recombinant ADNT1
ADNT1 was reconstituted into liposomes, and its transport activities were tested for a variety of potential substrates in homoexchange experiments (i.e. with the same substrate inside and outside). Using external and internal substrate concentrations of 1 and 10 mm, respectively, the reconstituted protein catalyzed an active [14C]ATP/ATP exchange, inhibitable by a mixture of bathophenanthroline and pyridoxal 5′-phosphate. It did not catalyze homoexchanges for GTP, GDP, CTP, sulfate, phosphate, pyruvate, malate, oxoglutarate, citrate, Glu, Asp, Pro, His, Lys, Arg, Ser, Thr, Trp, glutathione, carnitine, and choline. No [14C]ATP/ATP exchange activity was observed with ADNT1 that had been boiled before incorporation into liposomes or by reconstitution of sarkosyl-solubilized material from bacterial cells either lacking the expression vector for ADNT1 or harvested immediately before the induction of expression.
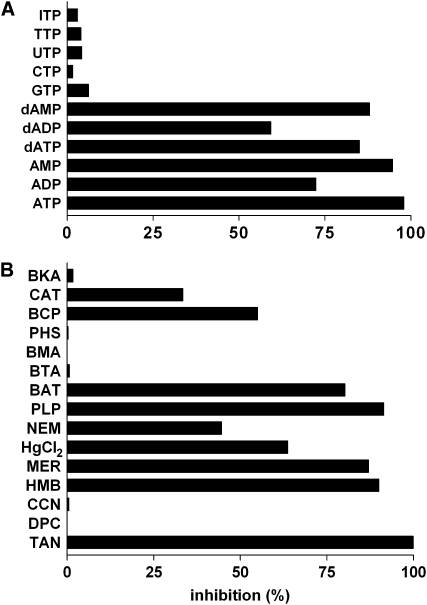
The substrate specificity of ADNT1 was examined in greater detail by measuring the uptake of [14C]ATP or [14C]AMP into proteoliposomes that had been preloaded with various potential substrates (Fig. 5). High rates of [14C]ATP and [14C]AMP uptake into proteoliposomes were observed with internal ATP, ADP, AMP, and the corresponding deoxynucleotides. Both ATP and AMP were also transported at a considerable rate in exchange for internal adenosine 5′-sulfophosphate. Negligible activity was found with internal nucleotides and deoxynucleotides of the bases I, C, U, T, and G as well as with cAMP, phosphate, pyrophosphate, adenosine, thiamine pyrophosphate, folate, NAD+, coenzyme A, and (data not shown) citrate, malate, oxoglutarate, thiamine monophosphate, nicotinamide mononucleotide, FMN, FAD, NADP, and S-adenosylmethionine (Fig. 5). Consistently, [14C]ATP uptake in the presence of 10 mm ATP inside the proteoliposomes was strongly inhibited by the external addition of ATP, AMP, dATP, and dAMP (Fig. 6A). Lower inhibition was found with ADP and dADP. Almost no effect was exerted by external GTP, CTP, UTP, TTP, and ITP and (data not shown) nucleoside monophosphates and diphosphates of the same bases, phosphate, malate, citrate, oxoglutarate, Arg, thiamine monophosphate and diphosphate, S-adenosylmethionine, nicotinamide mononucleotide, NAD, and FMN.
Figure 5.
Dependence of ADNT1 activity on internal substrate. Proteoliposomes were preloaded internally with various substrates (concentration, 10 mm). Transport was started by adding 30 μm [14C]ATP (A) or 30 μm [14C]AMP (B) and terminated after 1 min. Ads, Adenosine; APS, adenosine 5′-sulfophosphate; Pi, phosphate; PiPi, pyrophosphate; ThPP, thiamine pyrophosphate. Values are means ± sd of at least four experiments.
Figure 6.
Effects of inhibitors on the [14C]ATP/ATP exchange mediated by ADNT1. Proteoliposomes were preloaded internally with 10 mm ATP; transport was initiated by adding 30 μm [14C]ATP and terminated after 1 min. A, Effects of external substrates. The external substrates (concentration, 2 mm) were added together with [14C]ATP. B, Effects of mitochondrial carrier inhibitors. Thiol reagents and α-cyano-4-hydroxycinnamate (CCN) were added 2 min before the labeled substrate; the other inhibitors were added together with [14C]ATP. The final concentrations of the inhibitors were 0.1% tannic acid (TAN); 0.1 mm diethylpyrocarbonate (DPC), CCN, p-hydroxymercuribenzoate (HMB), and mersalyl (MER); 0.01 mm mercuric chloride (HgCl2); 1 mm N-ethylmaleimide (NEM), pyridoxal 5′-phosphate (PLP), bathophenanthroline (BAT), benzene-1,2,3-tricarboxylate (BTA), butylmalonate (BMA), and phenylsuccinate (PHS); 0.3 mm bromocresol purple (BCP); and 10 μm carboxyatractyloside (CAT) and bongkrekic acid (BKA). The extent of inhibition (%) from a representative experiment is reported. Similar results were obtained in at least three experiments.
The reaction catalyzed by reconstituted ADNT1 was inhibited strongly by tannic acid, pyridoxal 5′-phosphate, bathophenanthroline, and the sulfhydryl reagents mersalyl and p-hydroxymercuribenzoate and partially by N-ethylmalemide, mercuric chloride, and bromocresol purple (Fig. 6B). Carboxyatractyloside, a powerful inhibitor of the mitochondrial ADP/ATP carrier, only inhibited the ADNT1-mediated [14C]ATP/ATP exchange by 35% of its maximal level even at a concentration in excess of that which completely inhibits the ADP/ATP carrier (Klingenberg, 1989; Fiore et al., 1998). Other inhibitors of mitochondrial carriers, such as bongkrekate, 1,2,3-benzenetricarboxylate, butylmalonate, phenylsuccinate, diethylpyrocarbonate, and α-cyano-4 hydroxycinnamate, had no effect on ADNT1 activity (Fig. 6B).
Kinetic Characteristics of Recombinant ADNT1
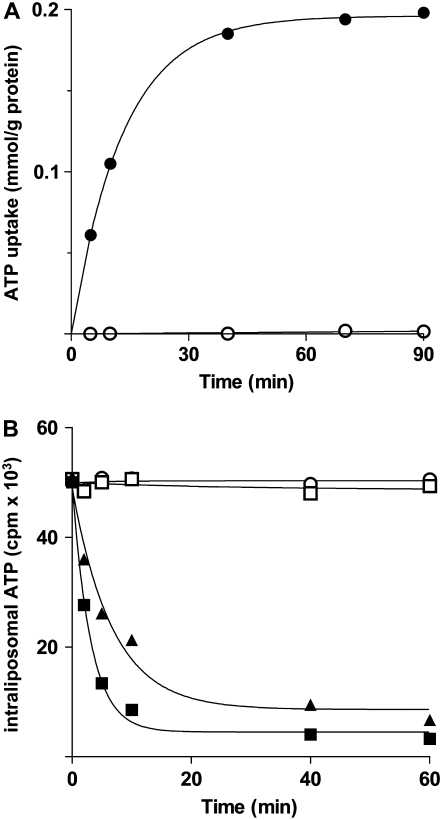
In Figure 7, the kinetics are compared for the uptake of 30 μm [14C]ATP into proteoliposomes either in the presence or in the absence of internal 10 mm ATP. The uptake of ATP by exchange followed a first-order kinetics (rate constant, 0.20 min−1; initial rate, 14.8 mol min−1 g−1 protein), with isotopic equilibrium being approached exponentially (Fig. 7A). In contrast, no [14C]ATP uptake was observed without internal substrate, indicating that ADNT1 does not catalyze a unidirectional transport (uniport) of ATP but only the exchange reaction. The uniport mode of transport was further investigated by measuring the efflux of [14C]ATP from prelabeled active proteoliposomes, as it provides a more convenient assay for unidirectional transport. In the absence of external substrate, no efflux was observed even after incubation for 60 min (Fig. 7B). However, upon addition of external AMP or ADP, an extensive efflux of radioactivity occurred, and this efflux was prevented completely by the presence of the inhibitors pyridoxal 5′-phosphate and bathophenanthroline (Fig. 7B). These results show clearly that reconstituted ADNT1 catalyzes an obligatory exchange reaction of substrates.
Figure 7.
Kinetics of [14C]ATP transport in proteoliposomes reconstituted with ADNT1. A, Uptake of ATP. A concentration of 30 μm [14C]ATP was added to proteoliposomes containing 10 mm ATP (exchange; black circles) or 10 mm NaCl and no substrate (uniport; white circles). Similar results were obtained in three independent experiments. B, Efflux of [14C]ATP from proteoliposomes containing 2 mm ATP. The internal substrate pool was labeled with [14C]ATP by carrier-mediated exchange equilibration. Then the proteoliposomes were passed through Sephadex G-75. The efflux of [14C]ATP was started by adding buffer A alone (white squares), 2 mm AMP in buffer A (black squares), 2 mm ADP in buffer A (black diamonds), or 2 mm AMP, 15 mm bathophenanthroline, and 20 mm pyridoxal 5′-phosphate in buffer A (white circles). Similar results were obtained in three independent experiments.
The kinetic constants of the recombinant purified ADNT1 were determined by measuring the initial transport rate at various external [14C]ATP, [14C]ADP, or [14C]AMP concentrations in the presence of a constant saturating internal concentration of the same substrate (homoexchanges). In double reciprocal plots for all three homoexchanges, linear functions were obtained that intersected the ordinate closer to a common point. For ATP, ADP, and AMP, the transport affinities (Km) were 26 ± 3 μm, 48 ± 4 μm, and 26 ± 3 μm (mean values of 20, six, and seven experiments), respectively. The average value of Vmax for ATP, ADP, and AMP homoexchanges was 30 ± 7 μmol min−1 g−1 protein. Adenosine 5′-sulfophosphate and dATP inhibited the [14C]ATP/ATP exchange competitively (data not shown), and their Ki values (for dissociation constant of an enzyme-inhibitor complex) were 60 ± 7 μm and 440 ± 64 μm, respectively (means of three experiments for each inhibitor).
Isolation and Genetic Characterization of an Arabidopsis Mutant Harboring a T-DNA Insertion within ADNT1
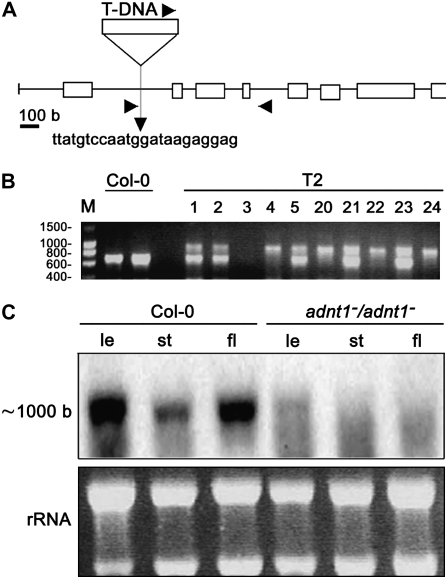
As a first analysis of the in vivo role of the Arabidopsis ADNT1 protein, a PCR-based strategy was used to screen the GABI-Kat mutant population of T-DNA-tagged Arabidopsis plants (Rosso et al., 2003) for disruption of the ADNT1 gene. The 451B06 GABI-Kat line, carrying a T-DNA insertion in the ADNT1 gene, has been characterized (Fig. 8). Segregating analysis of the selectable marker (data not shown) and sequencing of a PCR fragment encompassing the T-DNA/genomic DNA junction revealed that this line carries a single T-DNA insertion within the first intron of the ADNT1 gene (Fig. 8A). The zygosity of the T2 and T3 generations was analyzed by PCR screening. As shown in Figure 8B, lack of the wild-type band at 600 bp in T2 lines indicates a homozygous plant. Homozygous seeds of these knockout lines showed no apparent defect in germination rate compared with wild-type plants.
Figure 8.
Isolation and genetic characterization of an Arabidopsis mutant harboring a T-DNA insertion within the ADNT1 gene. A, Schematic representation of the ADNT1 gene. The mutant obtained by PCR screening of a T-DNA mutant collection (GABI-Kat) carries an insertion into the first intron of ADNT1. The scale of the T-DNA (approximately 4.5 kb) was not preserved for the sake of convenience. Boxes represent gene exons, and arrows on T-DNA, intron I, and intron IV denote primer positions for T2 and T3 population screening. B, PCR screening of the T2 population. M, DNA marker; T2, screened T2 population lines. Numbers at left indicate molecular masses in kilobases. C, Expression analysis of ADNT1. Northern blot of total RNA extracted from different tissues of the wild type (Col-0) and homozygous mutants (ADNT1−/ADNT1−); le, leaf; st, stem; fl, flower. The bottom section shows rRNA as a loading control.
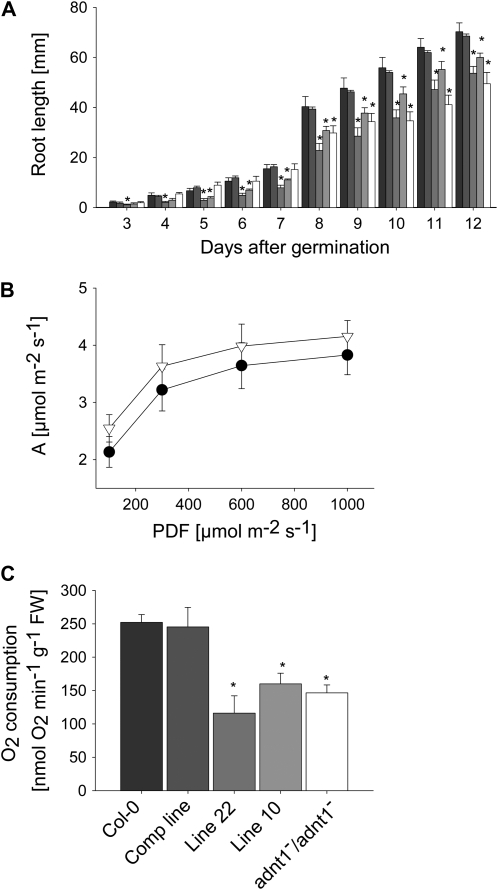
ADNT1 expression in T4 homozygous plants was verified in leaves, stems, and flowers by northern blotting. As shown in Figure 8C, lack of ADNT1 expression in the homozygous plants (ADNT1−/ADNT1−) confirms that they are null mutants. After the molecular identity of the T-DNA insertional mutant was established, homozygous plants were grown in soil under long-day conditions alongside the corresponding wild-type controls. Under these conditions, no difference between mutant and wild-type plants was observed in germination rate, rosette development, or bolting and flowering time. To analyze root growth, we germinated seeds on vertical agar plates and recorded root length of seedlings growing under a long-day regime every 1 or 2 d. Roots of homozygous plants were slightly shorter than those of wild-type plants during the first days after germination, with the difference in length becoming statistically significant (P < 0.05) at 8 d after germination (Fig. 9A). In order to gain insight into why the root growth was stunted, we grew the plants on soil and evaluated the rate of photosynthesis in aerial parts of the plants and the rate of respiration in the roots of the plants. Despite the fact that many perturbations in mitochondrial metabolism have been reported to result in alterations in photosynthetic metabolism (for review, see Sweetlove et al., 2007), the ADNT1 mutant displayed unaltered rates of assimilation (Fig. 9B) as well as unaltered rates of chloroplastic electron transport and transpiration (data not shown). In contrast, however, the rate of respiration in roots of the mutant was significantly lower than that observed in the wild-type control (Fig. 9C).
Figure 9.
Phenotypic characterization of Arabidopsis ADNT1 homozygous mutants. A, Root length of the wild type (Col-0), complemented line, antisense lines (lines 10 and 22), and homozygous ADNT1−/ADNT1− plants at 13 d after germination. Six to eight seeds per genotype were germinated on vertical plates for 3 d in the dark and then transferred to a 16-h-light/8-h-dark regime. Root lengths were recorded at the indicated day after germination. Black bars, Col-0; dark gray bars, complemented line; gray bars, antisense line 22; light gray bars, antisense line 10; white bars, ADNT1−/ADNT1− homozygous plants. Values are means ± se of three measurements from six different plates. B and C, Leaf assimilation rate as a function of light intensity (B) and root oxygen consumption in darkness (C). Values are presented as means ± se of six individual determinations per genotype. All measurements were performed in 5-week-old plants. The genotypes used for assimilation rate measurements were as follows: Col-0, black circles; ADNT1−/ADNT1− homozygous plants, white triangles. Asterisks indicate values determined by Student's t test to be significantly different (P < 0.05) from the control values. FW, Fresh weight.
In order to provide further genetic evidence for the role of ADNT1 in root morphology and respiration, we both complemented the mutant by expressing the full-length ADNT1 coding region between the constitutive 35S promoter and the t-nos terminator and created antisense plants by expressing the complete ADNT1 coding sequence in the opposite orientation in the Gateway pK2WG7 vector. In both cases, transgenic lines were selected on kanamycin and screened at the expression level. The complemented lines displayed wild-type level expression, while the antisense lines displayed decreases in expression to 42% (line 10) and 35% (line 22) of wild-type levels. In keeping with the results already reported, the complemented line exhibited wild-type-like growth, whereas the antisense line exhibited a decrease in root growth (Fig. 9A). Similarly, respiration in the complemented line was restored but was decreased with respect to the wild type in the antisense lines.
DISCUSSION
ADNT1 Transports ATP, AMP, and, to a Lesser Extent, ADP
In this work, ADNT1 was shown, by direct transport assays, to be capable of transporting ATP, AMP, and, to a lesser extent, ADP upon expression in E. coli and reconstitution into liposomes. This approach, which has previously been used for the identification of mitochondrial carriers from yeast (Palmieri et al., 2006a), mammals (Palmieri, 2004), and plants (Palmieri et al., 2006b and references therein, 2008), revealed that ADNT1 is different from any previously described mitochondrial carrier protein. Intriguingly, ADNT1 does not show strong sequence homology with any other mitochondrial carrier functionally identified until now in yeast, mammals, and plants. However, sequence-based phylogeny of the Arabidopsis members of the MCF (Picault et al., 2004) revealed that ADNT1 clusters together with At1g14560 (34% of identical amino acids) and At4g26180 (36%). However, the function of these genes has also not yet been identified. Furthermore, many protein sequences encoded by the genomes of plants and lower eukaryotes are likely orthologs of ADNT1. These sequences include, but are not limited to, TC53774 from Populus deltoides (86% identity at the amino acid level), TC171432 from Solanum lycopersicum (82%), Q6I583 from Oryza sativa (81%), TC102713 from Sorghum bicolor (81%), TC139822 from Hordeum vulgare (81%), Q2PYY0 from Solanum tuberosum (80%), TC7358 from Nicotiana tabacum (80%), TC265717 from Triticum aestivum (80%), and YPR011c from Saccharomyces cerevisiae (37%). It is doubtful, however, that there is an orthologous carrier in other higher eukaryotes, as the closest sequences to ADNT1 in Caenorhabditis elegans (Q20799; only 18% identical at the amino acid level), Drosophila melanogaster (Q26365; 24%), and Homo sapiens (the Graves's disease carrier protein; 31%; known to transport coenzyme A; Prohl et al., 2001) exhibit a relatively low degree of homology with ADNT1. Indeed, the level of homology is very similar to the basic homology existing between all of the different members of the MCF.
ADNT1 transports ATP, AMP, and, less efficiently, ADP, the corresponding deoxynucleotides, and adenosine 5′-sulfophosphate but virtually none of the other compounds tested. The substrate specificity of ADNT1 is distinct from that of any other previously characterized member of the MCF. In particular, ADNT1 differs from the well-known yeast, human, and Arabidopsis ADP/ATP carriers (Klingenberg, 1989; Fiore et al., 1998). These well-characterized carriers do not transport AMP, are strongly inhibited by carboxyatractyloside and bongkrekic acid, and share only 21% to 24% amino acid identity to ADNT1. The newly identified transporter also differs markedly from the yeast and human adenine nucleotide transporters ANT1 (Palmieri et al., 2001; Visser et al., 2002), which are exclusively localized in peroxisomes and exhibit only 15% to 23% amino acid identity with ADNT1. Furthermore, in contrast to ADNT1, ANT1 catalyzes both the uniport and the exchange modes of transport. ADNT1 is also quite different from the other characterized mitochondrial nucleotide transporters, such as the ATP-Mg/Pi carrier (Fiermonte et al., 2004), the GTP/GDP carrier (Vozza et al., 2004), the pyrimidine nucleotide carrier (Marobbio et al., 2006), the NAD+ carrier (Todisco et al., 2006), and the thiamine pyrophosphate carrier (Marobbio et al., 2002; Lindhurst et al., 2006).
ADNT1 Is a Mitochondrially Localized Transporter
The green fluorescence of the GFP-tagged ADNT1 completely overlapped with the fluorescent mitochondrion-selective dye MitoTracker Orange, demonstrating that the Arabidopsis ADNT1 transporter has a mitochondrial location. This finding is consistent with bioinformatic studies identifying it as a member of the MCF (Picault et al., 2004) and with direct evidence obtained via mitochondrial proteomics approaches (Millar and Heazlewood, 2003). Consistent with the fact that ADNT1 displays considerably different biochemical properties from the ADP/ATP carrier, its expression patterns seem to be somewhat variable to the isoforms of this protein and also with GUS-promoter fusion proteins, revealing ADNT1 to be particularly highly expressed in both bulky tissues and in regions of rapid heterotrophic growth in relation to its expression in other tissues. Detailed analysis of public microarray data sets (Zimmermann et al., 2004) supported these findings, since they suggest that ADNT1 displays a highly similar pattern of expression to ADP/ATP carrier 2. The only exceptions are those in floral organs such as the sepal, stamen, and pollen. ADNT1 is expressed at considerably lower levels than ADP/ATP carrier 2, whereas in senescent leaves, hypocotyl, xylem, and cork tissues, the expression of ADNT1 was considerable higher than that of ADP/ATP carrier 2. In contrast, ADNT1's transcripts are always present at lower levels than those of ADP/ATP carrier 1.
A Functional Role for ADNT1 in Facilitating Exchange of Cytosolic AMP and Intramitochondrial ATP
Given the transport characteristics of the recombinant ADNT1, its primary function is probably to catalyze the exchange between cytosolic AMP and intramitochondrial ATP. An ADNT1-mediated AMP/ATP exchange is likely to occur across the inner mitochondrial membrane when AMP is the predominant adenine nucleotide present in the cytosol. Under such conditions, AMP and ATP, exported from the mitochondria via ADNT1, would be expected to be converted by the intermembrane space isoform of adenylate kinase into two ADPs. It is conceivable that these ADP molecules would reenter the mitochondrial matrix via the ADP/ATP carrier, where they are likely converted to ATP in the process of oxidative phosphorylation. It is known that cytosolic AMP increases markedly in plant tissues during emergence from dormancy and during stresses such as anoxia and is primarily converted to ATP during recovery from these stresses (Saglio et al., 1980; Standard et al., 1983; Raymond et al., 1985). Furthermore, a conversion of AMP to ATP was shown by Roberts et al. (1997), in isolated and carefully washed S. tuberosum mitochondria, in the absence of ADP and ATP. These authors demonstrated that following the addition of AMP, there was a lag phase before respiration and ATP synthesis reached maximal rates. It was postulated that this lag period was the result of a limitation of adenylate kinase activity due to a shortage of ATP and that during this period, considerable ATP exits from the mitochondrial matrix. Our identification of ADNT1 provides a molecular entity capable of exporting ATP (in exchange for cytosolic AMP) from the mitochondria to the cytosol in amounts sufficient to enable the adenylate kinase-mediated phosphorylation of AMP and necessary to initiate the conversion of this nucleotide to ATP. The fact that knocking this protein out resulted in a restricted rate of root respiration, as determined by measuring the rate of oxygen consumption, and a reduced root growth provides further support for this model. It should be noted that, because of the presumed absence of adenylate kinase in the mitochondrial matrix, this model would require a transporter capable of catalyzing the export of AMP. However, such transporters are not without precedence, since mitochondrial AMP exporters were recently reported in both yeast and mammals (Fiermonte et al., 2004; Todisco et al., 2006). Moreover, a protein capable of performing this function was recently reported in Arabidopsis (Kirchberger et al., 2008). That there was little observable effect on photosynthesis or growth of the aerial parts of the mutants suggests that ADNT1 plays a less important role in photosynthetic tissues, a fact supported by the expression pattern of this gene. In addition to its potential role in oxidative phosphorylation, the availability of ADP in the intermembrane space is of high importance for the homeostatic maintenance of respiration, and a good supply of this substrate should serve to prevent side effects due to the accumulation of NADH and other reduced products of metabolism. These side effects could include the formation of potentially harmful superoxide and other reactive oxygen species known to cause wide-ranging damage to the cellular machinery and even to promote cell death (Mittler, 2002; Halliwell, 2006).
Physiological Function of ADNT1
The unique demands placed on the plant cell are reflected by the presence of a number of plant-specific respiratory chain components (Moller and Lin, 1986; Vanlerberghe and McIntosh, 1997; Rasmusson et al., 2008) and plant-specific nucleotide transporters between the mitochondria and the cytosol (Douce and Neuberger, 1989; Millar and Heazlewood, 2003; Picault et al., 2004). Our results here allow us to add ADNT1 to this list and to identify it as an AMP/ATP transporter catalyzing the exchange between cytosolic AMP and mitochondrial ATP. That said, the transport properties of the protein suggest that it can, in theory, additionally function as an ADP/ATP carrier. However, considerations such as the fact that the Km (ADP) of ADNT1 is 1 order of magnitude higher than that of the ADP/ATP carrier (Haferkamp et al., 2002), the fact that ADP/ATP1 is always expressed at higher levels than ADNT1, and the fact that all three carriers exhibit ubiquitous expression render the physiological significance of such a function minor at best. In keeping with this statement is the fact that recent preliminary evidence has suggested that mutants in the two mitochondrial isoforms of the ADP/ATP carrier have distinctive phenotypes from that described here for ADNT1 (with the ADP/ATP carrier mutants displaying stunted photosynthetic growth; L.J. Sweetlove, unpublished data). The results presented here, however, do allow us to assign a role for ADNT1 within the oxidative phosphorylation process. Indirect evidence in support of this comes from the fact that the characterization of the ADNT1 knockout mutant revealed a significantly reduced rate of root growth similar to that previously observed in S. lycopersicum plants deficient in the expression of enzymes of the tricarboxylic acid cycle (Carrari et al., 2003; Nunes-Nesi et al., 2005, 2007). However, further more direct evidence was supplied by the measurement of the rate of root respiration itself, which was revealed to be impaired in the mutant. The fact that both phenotypes were recovered by complementation and phenocopied by antisense lines revealed that they were directly attributable to the loss of function of ADNT1 in the mutant. Finally, our results clearly demonstrate that this protein is not functionally redundant to the ADP/ATP carriers and as such reveal new complexities in the subcellular regulation of adenylate metabolism and even oxidative phosphorylation itself. Since our bioinformatic analysis revealed that homologs of ADNT1 appear to be present in a wide range of plant and nonplant species, it may ultimately be highly informative to determine whether their function is conserved across species or has evolved to adapt to the specific demands imposed by the plant cell.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana ecotype Columbia [Col-0]) seeds were germinated on Murashige and Skoog plates (Murashige and Skoog, 1962) containing 1% Suc in a growth chamber (250 μmol photons m−2 s−1; 22°C) under a long-day regime (16 h of light/8 h of dark) before transfer to soil in a climate-controlled chamber under the same photoperiod. To analyze the accumulation of mRNA encoding ADNT1, plant tissues were collected 2 to 5 weeks after germination and immediately frozen in liquid nitrogen until use.
Promoter-GUS Fusion Experiments
For promoter-GUS fusion experiments, the promoter region of ADNT1 (from −1,056 to +36 bp) was amplified by PCR from Arabidopsis Col-0 genomic DNA using the following primers: forward, 5′-CACCTTTATCTTTTTGTCGTTCCTTCCAT-3′; reverse, 5′-TGCTGATTCTGTTCTTTTCACATCC-3′. The PCR products were first cloned into the shuttle vector pENTR/D-TOPO (Invitrogen) and then transferred into the binary Gateway vector pKGWFS7 in frame with the GUS gene (Karimi et al., 2002). The resulting constructs were introduced into Arabidopsis Col-0 plants by Agrobacterium-mediated transformation according to the floral dip method (Clough and Bent, 1998). To select transgenic plants, seeds were germinated on Murashige and Skoog plates (Murashige and Skoog, 1962) containing 1% Suc and supplemented with kanamycin in a growth chamber (250 μmol photons m−2 s−1; 22°C) under a 16-h-light/8-h-dark regime before transfer to soil in a climate-controlled chamber under the same photoperiod. Three lines showing strong expression were analyzed at various stages of development, namely after germination, at the two- to four-seedling stage, and in adult plants. Samples for histochemical GUS assay were harvested and stained overnight according to standard protocols (Jefferson et al., 1987) using 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid as substrate. Parallel assays were performed with corresponding nontransgenic material to rule out the possibility of localization artifacts caused by in toto incubation. Staining patterns were observed and documented using a Nikon digital microscope (SMZ1500) equipped with a Nikon digital camera (DXM1200).
Mutant Population Screening
The 451B06 GABI-Kat line, carrying a T-DNA insertion in the ADNT1 gene, was selected from the GABI-Kat T-DNA insertional mutant population (Rosso et al., 2003). Screening and selection within the mutant population were performed following the GABI-Kat instructions (http://www.mpiz-koeln.mpg.de/GABI-Kat/General_Information/GABI-Kat-sul-selection.html). The mutant was received from the GABI-Kat stock center as a hemizygous transgenic line (T2 generation), as judged from a 3:1 segregation of seed resistance to sulfadiazin (4-amino-N-[2-pyrimidinyl] benzene-sulfonamide-Na), the encoded resistance marker of the T-DNA insertion element. The number of T-DNA loci was assayed in the derived progeny of self-pollinated heterozygous lines (T3 generation) by analyzing the segregation pattern of resistance to sulfadiazin. Approximately 50 T3 generation seeds of the 451B06 GABI-Kat line were sterilized and sown on petri dishes containing Murashige and Skoog (Murashige and Skoog, 1962) medium supplemented with 11.25 mg L−1 sulfadiazin. After 24 h in the dark at 4°C, plates were transferred to a long-day regime, and selection was carried out according to the criterion of survival until 7 d after germination.
T2 and T3 resistant lines were screened for zygosity status by PCR analysis using a pair of primers annealing on the first and fourth introns of the ADNT1 gene (forward, 5′-GCAGTCAAATATAGATACGGG-3′; reverse, 5′-TTTAAAGGCTGAAGCGATAGGTAA-3′) and a third primer annealing of the left border of the T-DNA (5′-CCCATTTGGACGTGAATGTAGACAC-3′). All physiological measurements were carried out with homozygous plants obtained after two self-crosses of the initial T2 hemizygous line and therefore showed 100% resistance to sulfadiazin (T4 generation). The frequency of segregation, however, was in agreement with the 16:1 (resistant:susceptible) ratio, suggesting that insertion did not occur at a single Mendelian locus, leading us to create transgenic lines in order to confirm that the transporter was directly responsible for the phenotype.
Generation of ADNT1 Transgenics
ADNT1 was cloned taking the full-length coding region and subcloned into the Gateway vector pK2GW7 or pK2WG7 (Karimi et al., 2002) between the cauliflower mosaic virus 35S promoter and the t-nos terminator in the sense or antisense orientation, respectively. The coding sequence of ADNT1 was amplified by PCR using the following primers: forward, 5′-CACCATGGCATCAGAGGATGTGAAAAGAA-3′; reverse, 5′-TCAGTCTGATATCCTAAACTCCATCCTAAAA-3′. The resulting product was cloned into the entry vector pENTR/D-TOPO (Invitrogen) using the Gateway recombination system and subsequently recombined into the destination sense vector pK2GW7 or the antisense vector pK2WG7. The resulting constructs were introduced into Arabidopsis Col-0 plants by Agrobacterium-mediated transformation according to the floral dip method (Clough and Bent, 1998). To select transgenic plants, seeds were germinated on Murashige and Skoog plates (Murashige and Skoog, 1962) containing 1% Suc and supplemented with kanamycin in a growth chamber (250 μmol photons m−2 s−1; 22°C).
For complementation of the ADNT1−/ADNT1− mutant, homozygous plants were transformed with a pK2GW7 vector containing a full-length ADNT1 coding region between the constitutive 35S promoter and the t-nos terminator, and kanamycin-resistant transformants were screened on plates for further experiments. Antisense lines were screened at the expression level by real-time PCR using primers based on the cDNA sequence of ADNT1 and the 3′ untranslated region sequence in order to discriminate the endogenous ADNT1 mRNA from the introduced antisense ADNT1 mRNA. The forward and reverse primers were designed with Primer Express (Applied Biosystems) and corresponded to nucleotides 5′-ATTGCGATTGCATTTGTGACA-3′ and 5′-TTCTTGACGGCATTGAGATATCA-3′ for ADNT1 and to nucleotides 618 to 639 and 698 to 718 for the reference gene EF1α. The expression of the antisense transcript was also confirmed via real-time PCR using specific primers for the Gateway vector pK2WG7 and the ADNT1 cDNA sequence (forward, 5′-TCGTTGACACAGCTGCTGATT-3′; reverse, 5′-CCGCCCCCTTCACCAT-3′). Real-time PCR experiments were carried out as indicated in “Expression Analysis by Real-Time PCR.”
RNA Isolation and Northern-Blot Analysis
Total RNA was isolated from frozen organs using the commercially available Trizol kit (Gibco BRL) according to the manufacturer's instructions. RNA concentration was measured and its integrity was checked on a 1.5% agarose gel (w/v). Membrane hybridization was performed as described by Sambrook et al. (1989) using an ADNT1-specific probe of 328 bp (from +675 to +1,003 nucleotides of ADNT1 cDNA). The probe was labeled with [32P]dCTP by random priming using the Random Prime Labeling System (Amersham Bioscience), and RNA loading was checked with ethidium bromide. The membrane was autoradiographed at −70°C for several days with an intensifying screen.
Expression Analysis by Real-Time PCR
Total RNAs from different organs were reverse transcribed using the GeneAmp RNA PCR core kit (Applied Biosystems) with random hexamers as primers. For real-time PCR, primers based on the cDNA sequence of ADNT1 were designed with Primer Express (Applied Biosystems). The forward and reverse primers corresponded to nucleotides 512 to 532 and 398 to 418 for ADNT1 and to nucleotides 618 to 639 and 698 to 718 for EF1α. Real time-PCR was performed on an optical 96-well plate using the automated ABI Prism 7000 sequence detection system (Applied Biosystems). Fifty microliters of reaction volume contained 2 μL of template (reverse transcribed, first-strand cDNA), 25 μL of SYBR Green PCR Master Mix (Applied Biosystems), and 300 nm of each primer. The specificity of PCR amplification was checked with the heat dissociation protocol following the final cycle of PCR. Separation of real-time PCR products on 4% (w/v) agarose gels revealed single bands of the expected size whose identities were confirmed by direct sequencing. To correct for differences in the amount of starting first-strand cDNAs, the Arabidopsis EF1α gene was amplified in parallel as a reference gene. The relative quantification of ADNT1 in various organs was performed according to the comparative method (2−ΔΔCt; Bustin, 2000; Fiermonte et al., 2002), with the ΔCt stem as internal calibrator. 2−ΔΔCt = 2ΔCt sample−ΔCt calibrator, where ΔCt sample is the Ct sample – the Ct reference gene and Ct is the threshold cycle (i.e. the PCR cycle number at which emitted fluorescence exceeds 10 times the sd of baseline emissions).
cDNA Cloning and Construction of Expression Plasmids
The coding sequence for ADNT1 was amplified by PCR from an Arabidopsis cDNA library (Minet et al., 1992). Oligonucleotide primers (forward, 5′-GGATCCATGGCATCAGAGGATGTGAAAAGAA-3′; reverse, 5′-GAATTCTCAGTCTGATATCCTAAACTCCACT-3′) were synthesized corresponding to the extremities of the coding sequence (GenBank accession no. AF412085) with additional BamHI and EcoRI restriction sites. Product amplified using these primers was cloned into the pMWT7 expression vector (derived from pKN172; Fiermonte et al., 1993), which was subsequently transformed into Escherichia coli TOP 10 cells (Invitrogen). Transformants were selected on 2xTY plates containing ampicillin (100 μg mL−1) and screened by direct colony PCR and restriction digestion followed by sequence verification. For subcellular localization of ADNT1, the GFP fusion construct ADNT1-GFP was prepared. The coding sequence of ADNT1 without the terminal codon was amplified by PCR using the following primers: forward, 5′-CACCATGGCATCAGAGGATGTGAAAAGAA-3′; reverse, 5′-GTCTGATATCCTAAACTCCACTCCTAAAACG-3′. The resulting product was cloned into the entry vector pENTR/D-TOPO (Invitrogen) using the Gateway recombination system. Subsequently, the ADNT1 coding sequence was recombined into the destination vector pK7FWG2 encoding a C-terminally enhanced GFP under the control of the cauliflower mosaic virus 35S promoter (Karimi et al., 2002). The insert was sequenced to confirm identity prior to protoplast transformation.
Transient Expression of GFP Fusion Constructs
Protoplasts were prepared from 3-week-old Arabidopsis Col-0 plants (Yoo et al., 2007) grown as described above in the absence of kanamycin. Polyethylene glycol-calcium-mediated transfection was used to deliver the pK7FWG2 vector containing the coding sequence of ADNT1-GFP into protoplasts, followed by 18 to 36 h of incubation in the dark at 26°C to allow gene expression. After incubation, protoplasts were imaged using a laser-scanning confocal microscope (Leica DM IRBE microscope, TCS SPII confocal scanner). In some experiments, the transformed protoplasts were incubated in the presence of 500 nm MitoTracker Orange CM-H2TMRos (Molecular Probes) at 37°C for 10 min and subsequently at 26°C for 20 min before confocal analysis. After incubation, protoplasts were washed once. For the detection of GFP, excitation was at 488 nm and detection was between 498 and 534 nm. MitoTracker staining was detected between 553 and 600 nm, with excitation at 543 nm; chloroplast autofluorescence was detected between 664 and 700 nm, with excitation at 488 nm.
Bacterial Expression of ADNT1
The overproduction of ADNT1 as inclusion bodies in the cytosol of E. coli C0214(DE3) was accomplished as described previously (Fiermonte et al., 1993; Picault et al., 2002). Control cultures with empty vector were processed in parallel. Inclusion bodies were purified on a Suc density gradient, washed at 4°C with TE buffer (10 mm Tris and 1 mm EDTA, pH 7.0), then twice with a buffer containing Triton X-114 (3%, w/v), 1 mm EDTA, and 10 mm PIPES, pH 7.0, and once again with TE buffer. Protein was analyzed by SDS-PAGE on 17.5% gels; the identity of ADNT1 was confirmed by N-terminal sequencing and matrix-assisted laser desorption ionization-time of flight mass spectrometry of a trypsin digest of the purified protein excised from a Coomassie Brilliant Blue-stained gel. The yield of recombinant ADNT1 was estimated by laser densitometry as described previously (Fiermonte et al., 1998).
Reconstitution of the Recombinant ADNT1 into Liposomes
Purified ADNT1 was solubilized in the presence of 1.45% sarkosyl (w/v), and a small residue was removed by centrifugation (258,000g for 30 min). Solubilized protein was diluted 6-fold with a buffer containing 20 mm Na2SO4 and 10 mm PIPES, pH 7.0, and then reconstituted by cyclic removal of detergent (Palmieri et al., 1995). The reconstitution mixture consisted of protein solution (75 μL, 0.09 mg), 10% Triton X-114 (75 μL), 10% phospholipids (egg lecithin from Fluka) as sonicated liposomes (100 μL), 10 mm ATP, AMP, or ADP (except where indicated otherwise), cardiolipin (0.6 mg; Sigma), 20 mm PIPES, pH 7.0, and water (final volume, 700 μL). The mixture was recycled 13 times through an Amberlite column (3.2 cm × 0.5 cm) preequilibrated with buffer containing 20 mm PIPES, pH 7.0, and substrate at the same concentration as in the reconstitution mixture. All operations were performed at 4°C, except for the passages through Amberlite, which were carried out at room temperature.
Transport Measurements
External substrate was removed from the proteoliposomes on Sephadex G-75 columns preequilibrated with buffer A (50 mm NaCl and 10 mm PIPES, pH 7.0). Transport at 25°C was initiated by the addition of [14C]ATP, [14C]ADP, or [14C]AMP (NEN Life Science Products) to the eluted proteoliposomes and terminated by the “inhibitor-stop” method (Palmieri et al., 1995). In controls, the inhibitors were added simultaneously to the labeled substrate. Finally, the external radioactivity was removed on Sephadex G-75 and radioactivity in the liposomes was measured (Palmieri et al., 1995). Transport activity was calculated by subtracting the control values from the experimental values. The initial rate of transport was calculated in millimoles per minute per gram of protein from the time course of isotope equilibration (Palmieri et al., 1995). Various other transport activities were assayed by the inhibitor-stop method. For efflux measurements, the internal substrate pool of the proteoliposomes was made radioactive by carrier-mediated exchange equilibration (Palmieri et al., 1995) with 0.1 mm [14C]ATP added at high specific radioactivity. After 60 min, residual external radioactivity was removed by passing the proteoliposomes again through a column of Sephadex G-75 eluted with buffer A. Efflux was started by adding unlabeled external substrate or buffer A alone and terminated by adding the inhibitors indicated above.
Physiological Characterization of the Plants
Root growth was studied exactly as described by Carrari et al. (2005), photosynthesis was measured as detailed by Sweetlove et al. (2006), and root respiration was measured following the protocol described by Geigenberger et al. (2000).
Statistical Analysis
The t tests were performed using the algorithm embedded into Microsoft Excel. The term “significant” is used in the text only when the difference between the data sets in analysis was confirmed statistically (P < 0.05) by the application of Student's t tests.
Sequence data reported in this article as ADNT1 have been deposited in the EMBL Data Bank with accession number AM931440.
This work was supported by grants from the Ministero dell'Istruzione dell'Università e della Ricerca-PRIN, the Consiglio Nazionale delle Ricerche, the Centro di Eccellenza Geni in Campo Biosanitario ed Agroalimentare, and the Consorzio Italiano per le Biotecnologie as well as from the Max-Planck-Gesellschaft.
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Ferdinando Palmieri (fpalm@farmbiol.uniba.it).
Open access articles can be viewed online without a subscription.
References
- Bedhomme M, Hoffmann M, McCarthy EA, Gambonnet B, Moran RG, Rebeille F, Ravanel S (2005) Folate metabolism in plants: an Arabidopsis homolog of the mammalian folate transporter mediates folate import into chloroplasts. J Biol Chem 280 34823–34831 [DOI] [PubMed] [Google Scholar]
- Bouvier F, Linka N, Isner JC, Mutterer J, Weber APM, Camara B (2006) Arabidopsis SAMT1 defines a plastid transporter regulating plastid biogenesis and plant development. Plant Cell 18 3088–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA (2000) Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol 25 169–193 [DOI] [PubMed] [Google Scholar]
- Carrari F, Coll-Garcia D, Schauer N, Lytovchenko A, Palacios-Rojas N, Balbo I, Rosso M, Fernie AR (2005) Deficiency of a plastidial adenylate kinase in Arabidopsis results in elevated photosynthetic amino acid biosynthesis and enhanced growth. Plant Physiol 137 70–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrari F, Nunes-Nesi A, Gibon Y, Lytovchenko A, Ehlers Loureiro M, Fernie AR (2003) Reduced expression of aconitase results in an enhanced rate of photosynthesis and marked shifts in carbon partitioning in illuminated leaves of wild species tomato. Plant Physiol 133 1322–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catoni E, Schwab R, Hilpert M, Desimone M, Schwacke R, Flugge UI, Schumacher K, Frommer WB (2003) Identification of an Arabidopsis mitochondrial succinate-fumarate translocator. FEBS Lett 53 87–92 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Considine MJ, Goodman M, Echtay KS, Laloi M, Whelan J, Brand MD, Sweetlove LJ (2003) Superoxide stimulates a proton leak in potato mitochondria that is related to the activity of uncoupling protein. J Biol Chem 278 22298–22302 [DOI] [PubMed] [Google Scholar]
- Douce R (1985) Mitochondria in Higher Plants: Structure, Function and Biogenesis. Academic Press, New York
- Douce R, Neuberger M (1989) The uniqueness of plant mitochondria. Annu Rev Plant Physiol Plant Mol Biol 40 371–414 [Google Scholar]
- Fernie AR, Carrari F, Sweetlove LJ (2004) Respiratory metabolism: glycolysis, the TCA cycle and mitochondrial electron transport. Curr Opin Plant Biol 7 254–261 [DOI] [PubMed] [Google Scholar]
- Fiermonte G, De Leonardis F, Todisco S, Palmieri L, Lasorsa FM, Palmieri F (2004) Identification of the mitochondrial ATP-Mg/Pi transporter: bacterial expression, reconstitution, functional characterization, and tissue distribution. J Biol Chem 279 30722–30730 [DOI] [PubMed] [Google Scholar]
- Fiermonte G, Dolce V, Palmieri F (1998) Expression in Escherichia coli, functional characterization, and tissue distribution of isoforms A and B of the phosphate carrier from bovine mitochondria. J Biol Chem 273 22782–22787 [DOI] [PubMed] [Google Scholar]
- Fiermonte G, Palmieri L, Todisco S, Agrimi G, Palmieri F, Walker JE (2002) Identification of the mitochondrial glutamate transporter: bacterial expression, reconstitution, functional characterization, and tissue distribution of two human isoforms. J Biol Chem 277 19289–19294 [DOI] [PubMed] [Google Scholar]
- Fiermonte G, Walker JE, Palmieri F (1993) Abundant bacterial expression and reconstitution of an intrinsic membrane transport protein from bovine mitochondria. Biochem J 294 293–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore C, Trezeguet V, Le Saux A, Roux P, Schwimmer C, Dianoux AC, Noel F, Lauquin GJ, Brandolin G, Vignais PV (1998) The mitochondrial ADP/ATP carrier: structural, physiological and pathological aspects. Biochimie 80 137–150 [DOI] [PubMed] [Google Scholar]
- Geigenberger P, Fernie AR, Gibon Y, Christ M, Stitt M (2000) Metabolic activity decreases as an adaptive response to low internal oxygen in growing potato tubers. Biol Chem 381 723–740 [DOI] [PubMed] [Google Scholar]
- Giege P, Heazlewood JL, Roessner-Tunali U, Millar AH, Fernie AR, Leaver CJ, Sweetlove LJ (2003) Enzymes of glycolysis are functionally associated with the mitochondrion in Arabidopsis cells. Plant Cell 15 2140–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haferkamp I, Hackstein JH, Voncken FG, Schmit G, Tjaden J (2002) Functional integration of mitochondrial and hydrogenosomal ADP/ATP carriers in the Escherichia coli membrane reveals different biochemical characteristics for plants, mammals and anaerobic chytrids. Eur J Biochem 269 3172–3181 [DOI] [PubMed] [Google Scholar]
- Halliwell B (2006) Reactive species and antioxidants: redox biology is a fundamental theme of aerobic life. Plant Physiol 141 312–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyos ME, Palmieri L, Wertin T, Arrigoni R, Polacco JC, Palmieri F (2003) Identification of a mitochondrial transporter for basic amino acids in Arabidopsis thaliana by functional reconstitution into liposomes and complementation in yeast. Plant J 33 1027–1035 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A (2002) Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 5 193–195 [DOI] [PubMed] [Google Scholar]
- Kirchberger S, Tjaden J, Neuhaus HE (2008) Characterization of the Arabidopsis Brittle1 transport protein and impact of reduced activity on plant metabolism. Plant J 56 51–63 [DOI] [PubMed] [Google Scholar]
- Klingenberg M (1989) Molecular aspects of the adenine nucleotide carrier from mitochondria. Arch Biochem Biophys 270 1–14 [DOI] [PubMed] [Google Scholar]
- Leroch M, Kirchberger S, Haferkamp I, Wahl M, Neuhaus HE, Tjaden J (2005) Identification and characterization of a novel plastidic adenine nucleotide uniporter from Solanum tuberosum. J Biol Chem 280 17992–18000 [DOI] [PubMed] [Google Scholar]
- Lindhurst MJ, Fiermonte G, Song S, Struys F, De Leonardis F, Schwartzberg PL, Chen A, Castegna A, Verhoeven N, Mathews CK, et al (2006) Knockout of Slc25a19 causes mitochondrial thiamine pyrophosphate depletion, embryonic lethality, CNS malformations, and anemia. Proc Natl Acad Sci USA 103 15927–15932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie S, McIntosh L (1999) Higher plant mitochondria. Plant Cell 11 571–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marobbio CM, Di Noia MA, Palmieri F (2006) Identification of a mitochondrial transporter for pyrimidine nucleotides in Saccharomyces cerevisiae: bacterial expression, reconstitution and functional characterization. Biochem J 393 441–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marobbio CM, Vozza A, Harding M, Bisaccia F, Palmieri F, Walker JE (2002) Identification and reconstitution of the yeast mitochondrial transporter for thiamine pyrophosphate. EMBO J 21 5653–5661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Heazlewood JL (2003) Genomic and proteomic analysis of mitochondrial carrier proteins in Arabidopsis. Plant Physiol 131 443–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minet M, Dufour ME, Lacroute F (1992) Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Plant J 2 417–22 [DOI] [PubMed] [Google Scholar]
- Mittler R (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7 405–410 [DOI] [PubMed] [Google Scholar]
- Moller IM, Lin W (1986) Membrane-bound NAD(P)H dehydrogenases in higher plant cells. Annu Rev Plant Physiol 37 309–334 [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15 473–497 [Google Scholar]
- Nunes-Nesi A, Carrari F, Gibon Y, Sulpice R, Lytovchenko A, Fisahn J, Graham J, Ratcliffe RG, Sweetlove LJ, Fernie AR (2007) Deficiency of mitochondrial fumarase activity in tomato plants impairs photosynthesis via an effect on stomatal function. Plant J 50 1093–1106 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A, Carrari F, Lytovchenko A, Smith AMO, Ehlers-Loureiro M, Ratcliffe RG, Sweetlove LJ, Fernie AR (2005) Enhanced photosynthetic performance and growth as a consequence of decreasing mitochondrial malate dehydrogenase activity in transgenic tomato plants. Plant Physiol 137 611–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri F (2004) The mitochondrial transporter family (SLC25): physiological and pathological implications. Pflugers Arch 447 689–709 [DOI] [PubMed] [Google Scholar]
- Palmieri F, Agrimi G, Blanco E, Castegna A, Di Noia MA, Iacobazzi V, Lasorsa FM, Marobbio CMT, Palmierei L, Scarcia P, et al (2006. a) Identification of mitochondrial carriers in Saccharomyces cerevisiae by transport assay of reconstituted recombinant proteins. Biochim Biophys Acta 1757 1249–1262 [DOI] [PubMed] [Google Scholar]
- Palmieri F, Indiveri C, Bisaccia F, Iacobazzi V (1995) Mitochondrial metabolite carrier proteins: purification, reconstitution, and transport studies. Mitochondrial biogenesis and genetics part A. Methods Enzymol 260 349–369 [DOI] [PubMed] [Google Scholar]
- Palmieri L, Arrigoni R, Blanco E, Carrari F, Zanor MI, Studart-Guimaraes C, Fernie AR, Palmieri F (2006. b) Molecular identification of an Arabidopsis S-adenosylmethionine transporter: analysis of organ distribution, bacterial expression, reconstitution into liposomes, and functional characterization. Plant Physiol 142 855–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri L, Picault N, Arrigoni R, Besin E, Palmieri F, Hodges M (2008) Molecular identification of three Arabidopsis thaliana mitochondrial dicarboxylate carrier isoforms: organ distribution, bacterial expression, reconstitution into liposomes and functional characterization. Biochem J 410 621–629 [DOI] [PubMed] [Google Scholar]
- Palmieri L, Rottensteiner H, Girzalsky W, Scarcia P, Palmieri F, Erdmann R (2001) Identification and functional reconstitution of the yeast peroxisomal adenine nucleotide transporter. EMBO J 20 5049–5059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri L, Todd CD, Arrigoni R, Hoyos ME, Santoro A, Polacco JC, Palmieri F (2006. c) Arabidopsis mitochondria have two basic amino acid transporters with partially overlapping specificities and differential expression in seedling development. Biochim Biophys Acta 1757 1277–1283 [DOI] [PubMed] [Google Scholar]
- Picault N, Hodges M, Palmieri L, Palmieri F (2004) The growing family of mitochondrial carriers in Arabidopsis. Trends Plant Sci 9 138–146 [DOI] [PubMed] [Google Scholar]
- Picault N, Palmieri L, Pisano I, Hodges M, Palmieri F (2002) Identification of a novel transporter for dicarboxylates and tricarboxylates in plant mitochondria: bacterial expression, reconstitution, functional characterization, and tissue distribution. J Biol Chem 277 24204–24211 [DOI] [PubMed] [Google Scholar]
- Prohl C, Pelzer W, Diekert K, Kmita H, Bedekovics T, Kispal G, Lill R (2001) The yeast mitochondrial carrier Leu5p and its human homologue Graves' disease protein are required for accumulation of coenzyme A in the matrix. Mol Cell Biol 21 1089–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschke M, Bürkle L, Müller N, Nunes-Nesi A, Fernie AR, Arigoni D, Amrhein N, Fitzpatrick TB (2007) Vitamin B1 biosynthesis in plants requires the essential iron-sulfur cluster protein, THIC. Proc Natl Acad Sci USA 104 19637–19642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmusson AG, Geisler DA, Møller IM (2008) The multiplicity of dehydrogenases in the electron transport chain of plant mitochondria. Mitochondrion 8 47–60 [DOI] [PubMed] [Google Scholar]
- Raymond P, Alani A, Pradet A (1985) ATP production by respiration and fermentation, and energy charge during aerobiosis and anaerobiosis in 12 fatty and starchy germinating seeds. Plant Physiol 79 879–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts JKM, Aubert S, Gout E, Bligny R, Douce R (1997) Cooperation and competition between adenylate kinase, nucleoside diphosphokinase, electron transport and ATP synthase in plant mitochondria studied by 31P-nuclear magnetic resonance. Plant Physiol 113 191–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53 247–259 [DOI] [PubMed] [Google Scholar]
- Saglio PH, Raymond P, Pradet A (1980) Metabolic activity and energy charge of excised maize root tips under anoxia control by soluble sugars. Plant Physiol 66 1053–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Smith AMO, Ratcliffe RG, Sweetlove LJ (2004) Activation and function of mitochondrial uncoupling protein in plants. J Biol Chem 279 51944–51952 [DOI] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standard SA, Perret D, Bray CM (1983) Nucleotide levels and loss of vigor and viability in germinating wheat embryos. J Exp Bot 34 1047–1054 [Google Scholar]
- Sweetlove LJ, Fait A, Nunes-Nesi A, Williams T, Fernie AR (2007) The mitochondrion: an integration point of cellular metabolism and signalling. Crit Rev Plant Sci 26 17–43 [Google Scholar]
- Sweetlove LJ, Lytovchenko A, Morgan M, Nunes-Nesi A, Taylor NL, Baxter CJ, Eickmeier I, Fernie AR (2006) Mitochondrial uncoupling protein is required for efficient photosynthesis. Proc Natl Acad Sci USA 103 19587–19592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuswaldner S, Lagerstedt JO, Rojas-Stütz M, Bouhidel K, Der C, Leborgne-Castel N, Mishra A, Marty F, Schoefs B, Adamska I, et al (2007) Identification, expression, and functional analyses of a thylakoid ATP/ADP carrier from Arabidopsis. J Biol Chem 282 8848–8859 [DOI] [PubMed] [Google Scholar]
- Todisco S, Agrimi G, Castegna A, Palmieri F (2006) Identification of the mitochondrial NAD+ transporter in Saccharomyces cerevisiae. J Biol Chem 281 1524–1531 [DOI] [PubMed] [Google Scholar]
- Vanlerberghe GC, McIntosh L (1997) ALTERNATIVE OXIDASE: from gene to function. Annu Rev Plant Physiol Plant Mol Biol 48 703–734 [DOI] [PubMed] [Google Scholar]
- Visser WF, van Roermund CW, Waterham HR, Wanders RJ (2002) Identification of human PMP34 as a peroxisomal ATP transporter. Biochem Biophys Res Commun 299 494–497 [DOI] [PubMed] [Google Scholar]
- Vozza A, Blanco E, Palmieri L, Palmieri F (2004) Identification of the mitochondrial GTP/GDP transporter in Saccharomyces cerevisiae. J Biol Chem 279 20850–20857 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protocols 2 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR, Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]