Abstract
During the early stages of seed development, Arabidopsis (Arabidopsis thaliana) endosperm is syncytial and proliferates rapidly through repeated rounds of mitosis without cytokinesis. This stage of endosperm development is important in determining final seed size and is a model for studying aspects of cellular and molecular biology, such as the cell cycle and genomic imprinting. However, the small size of the Arabidopsis seed makes high-throughput molecular analysis of the early endosperm technically difficult. Laser capture microdissection enabled high-resolution transcript analysis of the syncytial stage of Arabidopsis endosperm development at 4 d after pollination. Analysis of Gene Ontology representation revealed a developmental program dominated by the expression of genes associated with cell cycle, DNA processing, chromatin assembly, protein synthesis, cytoskeleton- and microtubule-related processes, and cell/organelle biogenesis and organization. Analysis of core cell cycle genes implicates particular gene family members as playing important roles in controlling syncytial cell division. Hormone marker analysis indicates predominance for cytokinin signaling during early endosperm development. Comparisons with publicly available microarray data revealed that approximately 800 putative early seed-specific genes were preferentially expressed in the endosperm. Early seed expression was confirmed for 71 genes using quantitative reverse transcription-polymerase chain reaction, with 27 transcription factors being confirmed as early seed specific. Promoter-reporter lines confirmed endosperm-preferred expression at 4 d after pollination for five transcription factors, which validates the approach and suggests important roles for these genes during early endosperm development. In summary, the data generated provide a useful resource providing novel insight into early seed development and identify new target genes for further characterization.
Most of the world's food calories come from seeds, and extensive research has been directed at improving nutritional value and traits such as seed size and number. However, seeds are complex organs, and improvement by rational design requires an understanding of the contribution of specific tissues during important stages of seed development.
Seed development in most angiosperms begins with double fertilization, where the haploid egg cell and the double haploid central cell are both fertilized by identical haploid sperm cells contributed by a single pollen grain. This generates the diploid embryo and the triploid endosperm, respectively. The embryo and the endosperm grow rapidly in a coordinated manner that is heavily influenced by the surrounding maternal integument tissues that later form the seed coat (Olsen, 2004; Garcia et al., 2005). During early stages of seed development, maternal resources are mainly used for rapid cell division and growth of new tissues. Once cell division slows, the seed enters a maturation phase during which resources are reallocated to the synthesis of storage compounds, such as starch followed by the accumulation of oils and proteins. Biosynthetic activity then slows as the seed moves through a late maturation phase and prepares to desiccate prior to dormancy.
The developing endosperm plays several important roles during seed development (Berger, 2003; Olsen, 2004). In many plant species, including Arabidopsis (Arabidopsis thaliana), the triploid primary endosperm nucleus undergoes several rounds of free nuclear division, growing rapidly as a syncytium (Olsen, 2004). During the first phase of endosperm development, nuclei divisions are synchronous, but by the fourth division, three mitotic domains are apparent that differ with regard to the rate of nuclei divisions (Boisnard-Lorig et al., 2001; Brown et al., 2003). These are termed the micropylar endosperm, which surrounds the embryo, the peripheral endosperm, which lines the wall of the developing embryo sac, and the chalazal endosperm, which develops adjacent to the vascular connection with the seed parent (Boisnard-Lorig et al., 2001). Nuclear division continues through a third phase of endosperm development, which is marked by a migration of nuclei to the chalazal region, leading to the formation of chalazal nodules and cysts. Toward the end of this phase, the endosperm consists of approximately 200 nucleocytoplasmic domains and the embryo reaches the globular stage of development. Phase 4 sees the initiation of endosperm cellularization and reduced rates of mitosis (Scott et al., 1998; Boisnard-Lorig et al., 2001; Ingouff et al., 2005).
A role for endosperm in supporting the formation and growth of the embryo during early stages of development is suggested by the positioning of the chalazal endosperm and by the fact that seeds with severely defective endosperm cannot complete development (Scott et al., 1998). During the maturation stages, endosperm cellularizes and storage reserves are produced that accumulate in the endosperm cells (Sorensen et al., 2002). In plants that have ephemeral endosperms, such as Arabidopsis and oilseed rape (Brassica napus), the embryo develops at the expense of the endosperm and absorbs these reserves, storing them in the cotyledons (Scott et al., 1998; Olsen, 2004). Seeds that generate large endosperms during the early stages of development produce large embryos at maturity. The early proliferation of the endosperm, therefore, is associated with the growth of seeds and final seed size (Scott et al., 1998; Bushell et al., 2003). The alteration of the rate and duration of cell division in the endosperm has been proposed as a biotechnological strategy for altering seed size (Tiwari et al., 2006).
Arabidopsis provides an important model system for studying the underlying mechanisms of early seed development. Extensive and rapid analysis of many aspects of seed biology can be conducted in Arabidopsis due to the established protocols for producing and analyzing mutant and transgenic lines and the availability of a genome sequence facilitating the generation of tools for high-throughput molecular analysis. However, a major drawback to studying seed biology in Arabidopsis is its very small seeds. Laser microdissection is an important method for obtaining individual tissues or cell types for biochemical analysis. Originally developed for isolating cancerous cells from normal tissue (Emmert-Buck et al., 1996), laser microdissection has been used successfully to obtain DNA, RNA, proteins, and metabolites from a range of plant species and tissue types (for review, see Day et al., 2005, 2007a; Nelson et al., 2006). Therefore, it provides an ideal tool for analyzing gene expression changes in specific cell types during the early stages of Arabidopsis seed development (Spencer et al., 2007).
In a previous study, we compared different methods of transcriptome amplification from small amounts of RNA for use with printed long-oligonucleotide microarrays (Day et al., 2007b). A two-round IVT-based amplification was selected and used to obtain array data for proliferating syncytial endosperm at 4 d after pollination (DAP). This corresponded to the end of the third phase of endosperm development, when the syncytial endosperm contains many nucleocytoplasmic domains, but is prior to cellularization at 5 to 6 DAP (Scott et al., 1998; Boisnard-Lorig et al., 2001; Ingouff et al., 2005). The microarray data that formed the basis of this study were generated by hybridizing laser capture microdissected (LCM) endosperm-derived target alongside target from similarly treated silique tissues using a two-color microarray approach (Day et al., 2007b). A total of 18,220 unique probes gave signal higher than 2-fold background, and t testing identified 12,710 probes as being significantly differentially expressed between the whole silique and endosperm samples using a P value cutoff of <0.05. Analysis of embryo, seed coat, and endosperm markers within the data indicated that a 2-fold differential expression in the endosperm direction provided a stringent cutoff for identification of endosperm-preferred expression (Day et al., 2007b). This procedure identified 2,568 individual loci as being preferentially expressed in the endosperm. Here, we present extensive validation of the LCM endosperm array data by quantitative reverse transcription (qRT)-PCR and GUS reporter lines and provide a comprehensive analysis of the endosperm transcriptome in the context of existing online resources. This analysis has enabled novel insight into early endosperm development and has identified 793 genes as having early seed-specific and endosperm-preferred expression.
RESULTS
Microarray Analysis from Laser-Microdissected Endosperm Reliably Identifies Differential Expression in the Endosperm
To ensure that the microarray platform was correctly measuring differential expression between the endosperm and silique samples, 16 differentially expressed genes from the array data were selected for concurrent qRT-PCR analysis. Excess amplified RNA produced during the microarray target preparation was used to provide template for the qRT-PCR. The expression ratios produced by qRT-PCR and the microarray experiments were very similar (Table I), and all genes were confirmed as preferentially expressed in the endosperm sample by both the microarray and qRT-PCR.
Table I.
Validation of array results by qPCR and marker distribution
| Typea | Locus | Description | Log Ratio Arrayb | Log Ratio qPCRbc |
|---|---|---|---|---|
| MV1 | At1g19320 | Thaumatin family protein | 3.11 | 3.78 |
| MV2 | At1g65300 | MADS box transcription factor (PHE2/AGL38) | 2.97 | 3.3 |
| MV3 | At3g23060 | Zinc finger (C3HC4-type RING finger) family | 2.91 | 1.7 |
| MV4 | At5g27000 | Kinesin-like protein (ATK4) | 2.78 | 3.91 |
| M1 | At1g65330 | MADS box transcription factor (PHE1/AGL37) | 2.45 | ND |
| MV5 | At2g32460 | R2R3 MYB transcription factor (MYB101) | 2.45 | 3.98 |
| M2 | At4g25530 | Homeodomain-containing transcription factor (FWA) | 2.43 | 3.16 |
| MV6 | At2g32370 | Homeodomain-containing transcription factor | 2.37 | 2.59 |
| M3 | At3g24220 | 9-cis-Epoxycarotenoid dioxygenase (NCED6) | 2.24 | ND |
| M4 | At1g71890 | Suc-proton symporter (SUC5) | 2.17 | ND |
| MV7 | At1g09500 | Alcohol dehydrogenase | 1.89 | 2.69 |
| MV8 | At3g05310 | GTP and calcium ion-binding protein | 1.88 | 2.35 |
| M5 | At2g35670 | C2H2 transcription factor (FIS2) | 1.83 | ND |
| MV9 | At3g54560 | Histone H2a protein (HTA11) | 1.76 | 2.73 |
| M6 | At1g02580 | Polycomb family protein (MEA) | 1.74 | 1.68 |
| MV10 | At1g69770 | Chromomethylase family protein (CMT3) | 1.55 | 2.86 |
| MV11 | At3g11400 | G-subunit of eukaryotic initiation factor (EIF3) | 1.16 | 1.48 |
| MV12 | At1g17130 | Cell cycle control protein-related | 1.13 | 2.95 |
| MV13 | At3g55010 | Cycloligase (PUR5) | 1.12 | 1.2 |
| M7 | At3g19160 | Cytokinin synthase (IPT8) | 1.12 | ND |
| MV14 | At4g28840 | Unknown protein | 1.06 | 2.11 |
| MV15 | At3g61300 | C2 domain-containing protein | 1.05 | 0.74 |
| MV16 | At5g65710 | ATP-binding kinase (HSL2) | 0.86 | 0.33 |
MV1 to MV16 are genes that gave a range of differential expression in the array data, and M1 to M7 are endosperm marker genes identified from the literature.
Log ratio array and log ratio qPCR correspond to log2 values for the ratio of expression between whole silique and LCM endosperm samples.
Relative expression by qRT-PCR was calculated by comparison with the ACTIN2 (At3g18780) reference gene. ND, Not determined.
Identification of Endosperm-Preferred Genes Specifically Expressed during Early Seed Development Using Online Data Sets
To help identify genes with early endosperm-specific roles, we searched three online data sets that included a wide range of different tissues and at least one early seed or silique sample. This identified many genes with apparent early silique/seed-specific expression.
Massively parallel signature sequencing (MPSS) data (available from http://mpss.udel.edu/at/GeneQuery.php; Meyers et al., 2004) include two independent silique libraries (1–2 DAP) that are consistent with the early proliferative stage of endosperm development as well as data for inflorescences, leaves, roots, and germinating seedlings. We identified 200 genes that showed enrichment in the silique libraries. Of these, 68 were preferentially expressed in the endosperm and were MPSS silique library specific. A summary of the 35 endosperm-preferred genes with the highest MPSS tag frequencies and silique-specific MPSS profiles is given in Supplemental Figure S1.
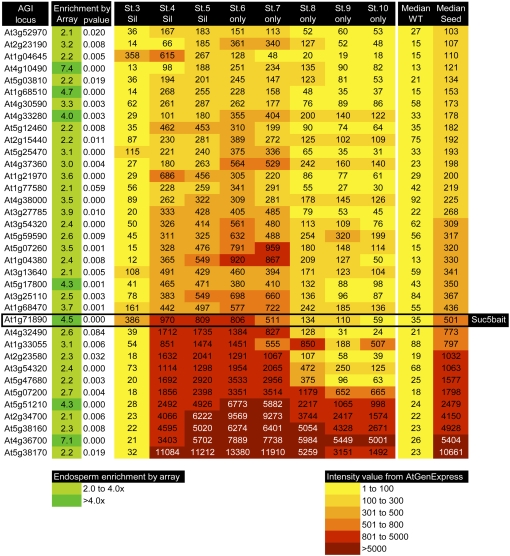
The AtGenExpress developmental series (Schmid et al., 2005) uses Affymetrix GeneChip technology to profile Arabidopsis transcripts from different organs and at different stages of development. This library contains data for seeds dissected from the silique 6 DAP onward, but the earlier stages (2–3, 3–4, and 4 DAP) use whole silique material. SUC5 has strong endosperm-preferred expression during the proliferative stages of seed development (Baud et al., 2005) and was used as an expression template to pull out 196 genes with similar expression (based on an r value cutoff of 0.75; Supplemental Table S1). This list was filtered based on endosperm-preferred expression in our data, a median expression level in AtGenExpress of <100 units in non-seed-containing tissues, and a median expression of >100 across the seed series to generate entries for Figure 1.
Figure 1.
Heat map showing endosperm-preferred genes in the AtGenExpress seed data. This heat map includes genes with endosperm-preferred expression from our arrays that correlate with SUC5 expression (based on an r value cutoff of 0.75) across the AtGenExpress expression library using online tools provided at the University of Toronto BAR Web site. The DataMetaFormatter tool created a heat map of probe intensities for genes in AtGenExpress seed development series and also calculated the median expression intensities in all other wild-type (WT) tissues. This heat map only includes genes that had a median expression level in AtGenExpress of <100 units in non-seed-containing tissues and a median expression of >100 across the seed series. Stages 3, 4, 5, 6, 7, 8, 9, and 10 correspond to siliques containing seeds at 48 to 66 h after flowering (HAF), siliques containing seeds at 66 to 84 HAF, siliques containing seeds at 84 to 90 HAF, isolated seeds at 90 to 96 HAF, isolated seeds at 96 to 108 HAF, isolated seeds at 108 to 120 HAF, isolated seeds at 120 to 144 HAF, and isolated seeds at 144 to 192 HAF, respectively. [See online article for color version of this figure.]
A more recent Affymetrix GeneChip data set profiles a range of tissues, including ovules and seeds dissected from gynoecia and siliques, respectively (available at http://estdb.biology.ucla.edu/genechip/). This data set includes immature seeds at 1, 3 to 4, and 7 to 8 DAP and was generated in the Goldberg (University of California, Los Angeles [UCLA]) and Harada (University of California, Davis) laboratories by Brandon Le (UCLA), Anhthu Bui (UCLA), and Julie Pelletier (University of California, Davis). We refer to it here as GHL data. We identified genes with similar expression patterns to early endosperm markers (see “Materials and Methods”) and cross-referenced these to genes with differential expression from our arrays. This created a subgroup of 2,608 putative early seed-specific genes (Supplemental Table S2).
Partitioning the Data for Further Analysis
To gain insight into the differential processes in operation during early silique and endosperm development at 4 DAP, we analyzed endosperm-preferred (EP; >2-fold differentially expressed in the endosperm sample compared with the silique sample) and other silique tissue-preferred (OST; >2-fold differentially expressed in the silique sample compared with the endosperm sample) gene groups. We also looked at subgroups containing genes that were thought to be early seed specific from our analysis of the GHL data that we termed ESS-EP and ESS-OST. The GHL data were found to be the most reliable source for identifying early seed-specific expression, since they displayed high sensitivity toward known endosperm markers compared with the AtGenExpress data (see “Materials and Methods”; Supplemental Fig. S2). The list of Arabidopsis Genome Initiative (AGI) numbers used for each partition is available in Supplemental Table S3.
Analysis of the Representation of Gene Ontologies and Functional Categories
The Arabidopsis genome has been extensively annotated. The Arabidopsis Information Resource (TAIR), as part of the Gene Ontology (GO) Consortium (Rhee et al., 2003), and the Munich Information Center for Protein Sequences (MIPS; Mewes et al., 2008) both provide annotation schemes that use controlled vocabularies aimed at providing descriptions of the roles of genes that are applicable to all organisms. We identified statistically significant enrichment of annotation terms using both the TAIR GO and FunCat schemes. Since both gave similar insight into our microarray data, we present the analysis based only on the TAIR GO terms. However, complete analysis using both vocabularies is provided in Supplemental Tables S4 to S7. Analysis of the EP partition indicated enrichment for GO terms associated with the rapidly proliferating nature of the endosperm at 4 DAP (i.e. molecular biosynthesis, protein formation, the cell cycle, DNA metabolism, replication, and microtubule-based movement; Supplemental Tables S4).
The OST partition represents genes that are predominantly expressed in nonendosperm tissues of the silique. The less proliferative nature of the growth and development of the majority of nonendosperm tissues manifests as an enrichment of GO terms for growth, development, cell communication, signal transduction, and hormone-mediated signaling (Supplemental Table S4). We also saw an enrichment of a large number of GO terms associated with endogenous and environmental stimuli that presumably reflect the need for the silique to provide a buffered environment for immature seeds to develop. Unlike the syncytial endosperm, the nonendosperm tissues of the silique are mostly composed of cells encased in a cell wall matrix, a difference that is corroborated in our data by enrichment for cell wall organization/biogenesis and cell wall loosening. Also enriched in this partition are terms for carbohydrate metabolism and biomolecular transport (Supplemental Table S4).
The GO analysis was refined to only include genes expressed specifically during the early stages of seed development. The ESS-EP partition was heavily enriched for genes associated with aspects of the cell cycle, DNA and chromatin biochemistry, microtubule-associated processes, and protein synthesis. The ESS-OST partition was enriched for relatively few GO terms (development, ovule development, carpel development, gynoecium development, and organ development), consistent with a less proliferative type of tissue development in nonendosperm seed tissues (ESS-EP and ESS-OST GO analysis shown in Table II).
Table II.
GO analysis of early seed-specific endosperm-preferred genes
TAIR GO terms showing significant enrichment in the ESS-EP and ESS-OST partitions using a P value cutoff of <0.01 without correction for multiple testing.
| Term | Observeda | Expectedb | P |
|---|---|---|---|
| TAIR ESS-EP partition | |||
| Organelle organization and biogenesis | 8.10% | 2.20% | 0.0000 |
| DNA metabolism | 6.60% | 1.50% | 0.0000 |
| DNA replication | 2.90% | 0.40% | 0.0000 |
| Cell cycle | 3.20% | 0.60% | 0.0000 |
| Cell organization and biogenesis | 10.70% | 4.80% | 0.0000 |
| Chromosome organization and biogenesis | 3.20% | 0.60% | 0.0000 |
| Microtubule-based process | 2.40% | 0.40% | 0.0000 |
| Microtubule-based movement | 1.90% | 0.20% | 0.0000 |
| Cytoskeleton-dependent intracellular transport | 2.00% | 0.30% | 0.0000 |
| Protein biosynthesis | 6.10% | 2.50% | 0.0000 |
| Cytoskeleton organization and biogenesis | 2.90% | 0.70% | 0.0000 |
| Chromatin assembly or disassembly | 2.00% | 0.40% | 0.0000 |
| DNA-dependent DNA replication | 1.50% | 0.20% | 0.0000 |
| Chromosome organization and biogenesis (sensu Eukaryota) | 2.40% | 0.60% | 0.0000 |
| DNA packaging | 2.30% | 0.50% | 0.0000 |
| Establishment and/or maintenance of chromatin architecture | 2.30% | 0.50% | 0.0000 |
| Regulation of progression through the cell cycle | 1.80% | 0.40% | 0.0001 |
| Regulation of the cell cycle | 1.80% | 0.40% | 0.0001 |
| Nucleobase, nucleoside, nucleotide, and nucleic acid metabolism | 16.40% | 10.80% | 0.0001 |
| Chromatin assembly | 1.60% | 0.30% | 0.0002 |
| Macromolecule biosynthesis | 6.60% | 3.20% | 0.0003 |
| Intracellular transport | 4.30% | 1.80% | 0.0003 |
| Establishment of cellular localization | 4.30% | 1.80% | 0.0004 |
| Cellular localization | 4.30% | 1.80% | 0.0004 |
| Nucleosome assembly | 1.40% | 0.30% | 0.0019 |
| Macromolecule metabolism | 24.20% | 18.30% | 0.0028 |
| M phase | 1.00% | 0.20% | 0.0080 |
| TAIR ESS-OST partition | |||
| Ovule development | 1.10% | 0.00% | 0.0001 |
| Carpel development | 1.10% | 0.10% | 0.0015 |
| Gynoecium development | 1.10% | 0.10% | 0.0021 |
| Development | 7.50% | 3.40% | 0.0031 |
| Organ development | 2.70% | 0.60% | 0.0063 |
The frequency of occurrence of a GO term in the tested gene list.
The frequency of occurrence of a GO term in the list of genes for which probes were present on the array.
Representation Analysis of Selected Gene Families
Several gene families of interest have been characterized in the recent literature or collected in online resources. Representation analyses of selected gene families are summarized in Table III, and details are given below. Significance during this stage of the analysis was based on a P value cutoff of <0.005, unless otherwise stated.
Table III.
Representation analysis of selected gene families
Differential representation of selected gene families in the EP and OST gene groups. The underrepresentation (↓), overrepresentation (↑), or not significantly different representation (↔) was calculated by hypergeometric testing using a P value of <0.005.
| Hormone | EP | OST |
|---|---|---|
| Hormone markers | ↓ | ↑ |
| ACC responsive | ↔ | ↑ |
| ABA responsive | ↔ | ↑ |
| BL responsive | ↓ | ↑ |
| CK responsive | ↑ | ↑ |
| GA responsive | ↔ | ↔ |
| IAA responsive | ↓ | ↑ |
| MJ responsive | ↓ | ↑ |
| Transcription factors | ↔ | ↑ |
| DNA methylation sensitive | ↔ | ↑ |
| Chromatin related | ↑ | ↓ |
| Core cell cycle | ↑ | ↔ |
| E2F regulated | ↑ | ↓ |
Analysis of Cell Cycle Genes
Plant syncytial development requires a rapid progression through the cell cycle, suppression of phragmoplast formation, and an uncoupling of cytokinesis from mitosis (Otegui and Staehelin, 2000). To gain further insights into this process, we analyzed our data to identify endosperm-preferred genes that have been implicated in controlling cell cycle progression (Table IV). The core cell cycle genes of Arabidopsis (Vandepoele et al., 2002) and genes shown to be regulated by the E2F members of this family were overrepresented in the EP data (Table IV; Supplemental Table S8). Motifs associated with E2F binding were also highly enriched in this partition, such that the “E2FAT motif” (TYTCCCGCC) was enriched at the P < 1 × 10−5 level in both the EP and ESS-EP partitions and the “E2F-binding site motif” (TTTCCCGC) was enriched in the EP and ESS-EP partitions at the P < 1 × 10−9 and P < 1 × 10−10 levels, respectively.
Table IV.
The distribution of core cell cycle genes in the endosperm-preferred data
| Locus | Name | Description | Pa | Fold Changeb |
|---|---|---|---|---|
| At5g14960 | DEL2 | DP-E2F-related 2 | 0.002 | 6.65 |
| At1g16330 | CYCB3;1 | Cyclin family | 0.002 | 4.40 |
| At5g06150 | CYCB1;2 | Cyclin (cyc1b) | 0.000 | 3.96 |
| At5g25380 | CYCA2;1 | Cyclin 3a (cyc3a) | 0.001 | 3.56 |
| At2g17620 | CYCB2;1 | Cyclin, putative | 0.004 | 3.40 |
| At1g44110 | CYCA1;1 | Cyclin, putative | 0.001 | 2.98 |
| At1g20610 | CYCB2;3 | Cyclin, putative | 0.000 | 2.92 |
| At3g11520 | CYCB1;3 | Cyclin, putative | 0.002 | 2.87 |
| At1g20930 | CDKB2;2 | Cell division control protein, putative | 0.001 | 2.78 |
| At1g73690 | CDKD;1 | Cell division protein kinase, putative | 0.003 | 2.74 |
| At4g37490 | CYCB1;1 | G2/mitosis-specific cyclin | 0.005 | 2.63 |
| At3g54180 | CDKB1;1 | Cell division control protein (CDC2B) | 0.008 | 2.55 |
| At1g77390 | CYCA1;2 | Cyclin, putative | 0.000 | 2.48 |
| At5g11300 | CYCA2;2 | Cyclin, putative | 0.007 | 2.31 |
| At5g02470 | DPa | DPA transcription factor | 0.002 | 2.29 |
| At2g32710 | KRP4 | Kip-related protein 4 | 0.000 | 2.15 |
| At2g38620 | CDKB1;2 | Cell division control protein, putative | 0.000 | 2.12 |
| At5g43080 | CYCA3;1 | Cyclin, putative | 0.012 | 2.12 |
| At2g27970 | CKS2 | CDK-subunit 2 | 0.018 | 2.11 |
| At3g50070 | CYCD3;3 | Cyclin family | 0.004 | 2.06 |
| At1g18040 | CDKD;3 | Cell division protein kinase, putative | 0.022 | 1.99 |
| At3g01330 | DEL3 | DP-E2F-related protein 3 | 0.026 | 1.87 |
| At5g10270 | CDKC;1 | Cyclin-dependent kinase C;1 | 0.000 | 1.76 |
| At5g64960 | CDKC;2 | Cyclin-dependent kinase C;2 | 0.001 | 1.69 |
| At2g36010 | E2Fa | E2FA transcription factor | 0.022 | 1.69 |
| At4g03270 | CYCD6;1 | Cyclin family | 0.024 | 1.62 |
| At1g47870 | E2Fc | E2FC transcription factor | 0.020 | 1.45 |
Core cell cycle genes that showed consistent signal on the arrays were identified using a P value of <0.05.
All fold changes represent differential expression in the endosperm direction. The solid line represents a fold change cutoff of 2.0.
Progression through the cell cycle occurs via coordinated sequential activation of distinct phases. M phase-specific expression is associated with an M-specific activator sequence (MSA) in the promoter region of a gene. A total of 161 differentially expressed genes from our array analysis contained the MSA sequence in the 500 bp upstream of the ATG. Of these putative M phase-specific genes, 27 were in the EP partition and 16 were in the ESS-EP partition (Supplemental Table S8). Hypergeometric testing of these putative M phase-specific transcripts indicated significant enrichment in the EP and ESS-EP partitions at the P < 0.01 and P < 0.0005 levels, respectively.
Analysis of Hormone Response Pathways
The varied distributions of phytohormones and their well-documented ability to regulate growth and development of the seed make them obvious candidates for identifying important components in the control of early endosperm development (Lur and Setter, 1993; Yang et al., 2002, 2006). A recent study by Nemhauser et al. (2006) used the AtGenExpress hormone series to identify genes that were only expressed in response to particular hormones, suggesting that these genes can be used as markers for hormone action. To gain insight into the influence of plant hormones in the developing silique at 4 DAP, we looked for the presence of these markers in our gene groups. The OST partition (representing many tissues) was significantly enriched for hormone markers, whereas the EP partition (single tissue) had significant underrepresentation (Table III).
Analysis of the full lists of genes responsive to the hormones ethylene, abscisic acid, brassinosteroid, cytokinin, gibberellin, auxin, and methyl jasmonate (1-aminocyclopropane-1-carboxylic acid [ACC], ABA, brassinolide [BL], CK, GA, indole-3-acetic acid [IAA], and MJ, respectively) showed that all but one of the hormone-responsive gene groups (GA) were significantly enriched in the OST partition (Table III; Supplemental Table S9), reinforcing the observations made using the marker list. The ACC-, ABA-, and GA-responsive genes were well represented in the endosperm-preferred partition, but only the CK-responsive genes were significantly enriched (Table III). Endosperm-preferred genes involved in CK signaling are given in Table V.
Table V.
Distribution of CK signaling genes in the endosperm-preferred data
| Locus | Name | Type | Pa | Fold Changeb |
|---|---|---|---|---|
| At5g07210 | ARR21 | B type | 0.001 | 5.19 |
| At4g11140 | CRF1/ERF | Response factor | 0.000 | 3.93 |
| At1g49190 | ARR19 | B type | 0.035 | 3.62 |
| At4g23750 | CRF2/ERF | Response factor | 0.000 | 3.60 |
| At5g53290 | CRF3/ERF | Response factor | 0.002 | 2.91 |
| At4g00760 | APRR8 | B type | 0.001 | 2.38 |
| At5g58080 | ARR18 | B type | 0.002 | 2.28 |
| At1g03430 | AHP5 | AHP | 0.003 | 1.99 |
| At3g29350 | AHP2 | AHP | 0.006 | 1.89 |
| At3g62670 | ARR20 | B type | 0.004 | 1.61 |
CK signaling genes that showed consistent signal on the arrays were identified using a P value of <0.05.
All fold changes represent differential expression in the endosperm direction. The solid line represents a fold change cutoff of 2.0.
The hormone-responsive gene lists include genes that are up-regulated, down-regulated, or have a complex regulatory pattern in response to exogenous hormone application. The distribution of up-regulated, down-regulated, and complex CK-regulated genes in the data partitions were compared using a χ2 test. Significant differences from the expected distribution for CK-regulated genes were seen for both the EP and OST partitions. The EP partition included a much larger than expected number of CK up-regulated genes (observed, 94%; expected, 66%) and fewer than expected CK down-regulated genes (observed, 4%; expected, 32%). Conversely, in the OST partition, we saw a much larger than expected number of CK down-regulated genes (observed, 52%; expected, 32%) and fewer than expected CK up-regulated genes (observed, 47%; expected, 66%).
Interestingly, none of the 48 ARF and AUX-IAA transcription factors represented in the differentially expressed gene list gave evidence for endosperm-preferred expression (data not shown). Conversely, 19 of these transcription factors were present in the OST partition. Interactions between these two groups of proteins mediate auxin-dependent transcriptional regulation, and when taken together as an “auxin signaling group” (ARFs plus AUX-IAAs), hypergeometric testing showed that the underrepresentation in the EP partition was significant (P = 0.0032).
Analysis of Chromatin-Related and DNA Methylation-Sensitive Genes
Transcriptional regulation is closely related to chromatin structure, and during syncytial development, endosperm has a high proportion of euchromatin, with small chromocenters and distinct heterochromatic foci (Baroux et al., 2007). Euchromatin is associated with active transcription, and alterations in chromatin structure have been associated with the onset of cell division, morphogenesis, and differentiation (Zhao et al., 2001; Berger and Gaudin, 2003; Williams et al., 2003; Baroux et al., 2007; De Veylder et al., 2007). Enrichment analysis revealed a significant overrepresentation of chromatin-related genes (obtained from ChromDB [http://www.chromdb.org/]) in the endosperm (Table III; Supplemental Table S10). Dynamic changes in chromatin structure are associated with epigenetic alterations, such as DNA methylation and histone modifications. DNA methylation tends to be associated with transcriptional repression, and a recent study has identified a number of genes that appear to have methylation-sensitive transcription (Zhang et al., 2006). Enrichment analysis of the methylation-sensitive genes in our partitioned data generated a similar distribution pattern to that observed for transcription factors (Table III; Supplemental Table S11), which implies that DNA methylation is a widespread form of transcriptional control throughout developing siliques.
Analysis of Transcription Factors
The Database of Arabidopsis Transcription Factors (DATF [http://datf.cbi.pku.edu.cn/]) includes information about 1,922 transcription factors classified into 64 families (Guo et al., 2005). Our analysis identified differential expression for 943 of these, 187 of which were endosperm preferred (Supplemental Table S12). Furthermore, 71 transcription factors were found to be endosperm preferred and early seed specific (Supplemental Table S12). Table VI highlights the transcription factors that our analysis validates (see below) to be early seed specific, with strong evidence for endosperm-preferred expression from the microarrays. To see if any transcription factor families were overrepresented in the EP and ESS-EP lists, we calculated the frequency of each family in our data (our analysis was limited to the 28 families that had 10 or more members showing differential expression in our array data). However, little evidence for enrichment of particular types of transcription factors was observed (data not shown).
Table VI.
Validated early seed-specific, endosperm-preferred transcription factors
Transcription factors (27) that were endosperm preferred in the LCM endosperm array data and called early seed specific by searching online data resources and then confirmed by qRT-PCR and/or reporter lines.
| Locus | Description | P | Fold Changea |
|---|---|---|---|
| At5g40430 | Myb family transcription factor (MYB22) | 0.000 | 8.77 |
| At1g65300 | MADS box protein (PHE2/AGL38)b | 0.000 | 7.84 |
| At5g26630 | MADS box protein (AGL35) | 0.000 | 7.77 |
| At5g14960 | DP-E2F-related 2 (DEL2) | 0.002 | 6.65 |
| At3g56520 | No apical meristem protein family | 0.004 | 6.30 |
| At4g18870 | Heat shock transcription factor familyb | 0.001 | 5.69 |
| At1g65330 | MADS box protein (PHE1/AGL37) | 0.002 | 5.48 |
| At4g25530 | Homeodomain protein (FWA) | 0.000 | 5.40 |
| At5g07210 | Two-component response regulator protein (ARR21) | 0.001 | 5.19 |
| At2g32370 | Homeodomain proteinb | 0.000 | 5.19 |
| At4g21080 | Dof zinc finger proteinb | 0.001 | 5.10 |
| At2g26320 | MADS box protein (AGL33) | 0.006 | 4.97 |
| At5g56200 | C2H2-type zinc finger protein family | 0.000 | 4.59 |
| At2g15740 | C2H2-type zinc finger protein family | 0.024 | 4.51 |
| At5g17800 | Myb family transcription factor (MYB56) | 0.001 | 4.33 |
| At4g11400 | ARID/BRIGHT DNA-binding domain protein family | 0.000 | 4.29 |
| At2g01810 | PHD finger protein family | 0.000 | 4.23 |
| At5g11510 | Myb family transcription factor (MYB3R4) | 0.001 | 3.87 |
| At3g27785 | Myb family transcription factor (MYB118) | 0.010 | 3.86 |
| At4g36590 | MADS box protein (AGL40) | 0.000 | 3.85 |
| At1g49190 | Response regulator protein family (ARR19)b | 0.035 | 3.62 |
| At4g23750 | Cytokinin response factor (CRF2) | 0.000 | 3.60 |
| At5g60440 | MADS box protein (AGL62) | 0.003 | 3.57 |
| At2g35670 | C2H2-type zinc finger protein (FIS2) | 0.016 | 3.55 |
| At4g38000 | Dof zinc finger protein | 0.000 | 3.50 |
| At3g03260 | Homeodomain protein | 0.021 | 3.46 |
| At2g34880 | Maternal effect embryo arrest 27 (MEE27) | 0.004 | 3.32 |
All fold changes represent differential expression in the endosperm direction.
Seed-specific genes for which promoter-GUS reporter gene analysis is shown in Figure 4.
Evidence for Biological Significance of Protein Interactions for MADS Box Transcription Factors Expressed during Proliferative Endosperm Development
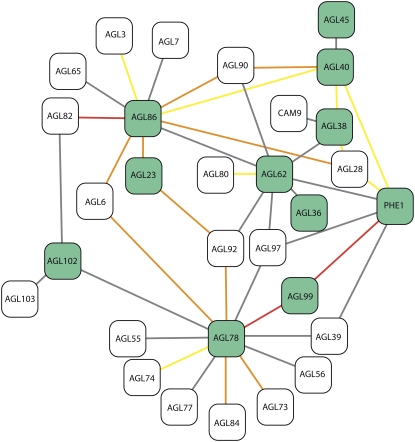
Twelve MADS box genes were found in the EP partition and, interestingly, all but one were type I MADS box genes (Table VII). MADS box proteins often form homodimers and heterodimers, and a comprehensive analysis using yeast two-hybrid technology has identified interactions within the members of the Arabidopsis family (de Folter et al., 2005). By combining these data with our cell type-specific experiments, we can help infer in planta biological significance during endosperm development for the following MADS box transcription factor interactions: AGL45-AGL40, AGL40-PHE2, AGL40-PHE1, PHE2-AGL62, AGL62-PHE1, AGL62-AGL36, PHE1-AGL99, AGL99-AGL78, AGL78-AGL102, AGL86-AGL23, and AGL86-AGL40 (Table VII; Fig. 2). We also identified seven MADS box genes that gave strong evidence for endosperm-specific expression at 4 DAP (AGL35, PHE1, PHE2, AGL33, AGL40, AGL62, and AGL91; Table VII; Fig. 2).
Table VII.
Distribution of MADS box transcription factors in the endosperm-preferred data
| Locus | Name | Type | Pa | Fold Changeb |
|---|---|---|---|---|
| At1g65300 | AGL38/PHE2c | I | 0.000 | 7.84 |
| At5g26630 | AGL35c | I | 0.000 | 7.77 |
| At1g65360 | AGL23 | I | 0.001 | 6.45 |
| At1g47760 | AGL102 | I | 0.001 | 5.80 |
| At1g65330 | AGL37/PHE1c | I | 0.002 | 5.48 |
| At2g26320 | AGL33c | II | 0.006 | 4.97 |
| At4g36590 | AGL40c | I | 0.000 | 3.85 |
| At5g60440 | AGL62c | I | 0.003 | 3.57 |
| At3g66656 | AGL91c | I | 0.039 | 2.41 |
| At1g31630 | AGL86 | I | 0.000 | 2.23 |
| At5g65330 | AGL78 | I | 0.018 | 2.00 |
| At5g26650 | AGL36 | I | 0.004 | 2.00 |
| At3g05860 | AGL45 | I | 0.014 | 1.78 |
| At5g65070 | AGL69/MAF4 | II | 0.009 | 1.68 |
| At5g04640 | AGL99 | I | 0.038 | 1.50 |
MADS box genes showing consistent signal on the arrays were identified using a P value of <0.05.
All fold changes represent differential expression in the endosperm direction. The solid line represents a fold change cutoff of 2.0.
MADS box genes with seed-specific expression.
Figure 2.
Interaction summary of endosperm-preferred MADS box transcription factors. Interaction plot of endosperm-preferred MADS box transcription factors in the array data adapted from the output of the Arabidopsis Interactions Viewer at http://bar.utoronto.ca/. Black outline corresponds to genes differentially expressed in our array data. Dark and pale green fill corresponds to >2.0-fold endosperm-enriched and 1.5- to 2-fold endosperm-enriched transcripts, respectively. Gray outline represents genes not differentially expressed in the array data. Connecting lines indicate that a protein-protein interaction is evident in the literature. The more red the line, the better the correlation for transcript expression across the AtGenExpress expression series, as assessed by Pearson correlation. [See online article for color version of this figure.]
Validation of Early Seed and Early Seed-Specific Expression
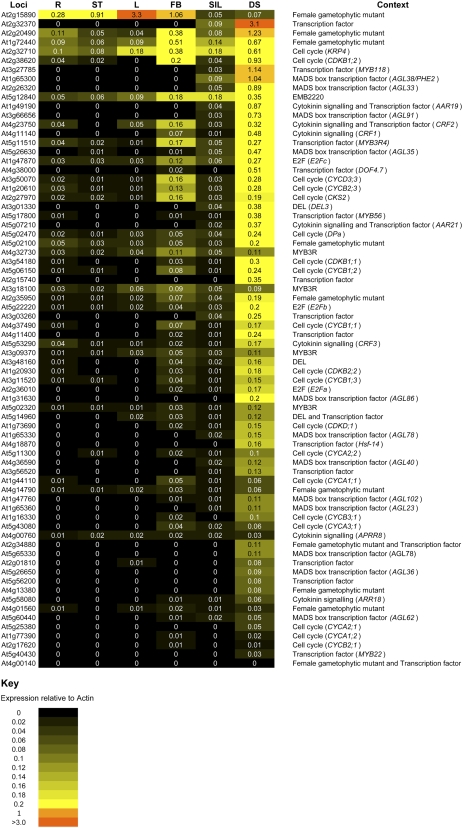
All of the endosperm-preferred genes discussed in detail as part of our analysis had their expression levels assessed in different plant tissues by qRT-PCR (Fig. 3). Samples were taken from leaves, stems, roots, flower buds, whole siliques, and seeds dissected from 4-DAP siliques. All genes detected showed higher expression in the seed sample than in whole siliques, consistent with the original LCM endosperm array data. Data analysis also predicted early seed-specific expression for a number of transcription factors (Table VI). Of the 25 novel candidates in Table VI, only two (At4g23750 and At5g11510) showed significant transcript expression in a nonseed tissue sample (Fig. 3). Both were only additionally expressed in the flower buds, perhaps suggesting prior expression in the male and/or female gametophytes prior to fertilization. At4g00140 was not detected in any conventional samples by qRT-PCR.
Figure 3.
Heat map showing qRT-PCR validation of early seed and/or early seed-specific expression. The expression level of each transcript was calculated relative to ACTIN2. R, ST, L, FB, SIL, and DS represents roots, stems, leaves, flower buds, siliques, and dissected seeds, respectively. Context relates to the section of the article in which a particular gene is discussed. [See online article for color version of this figure.]
Identification of Promoters Driving Expression in the Early Endosperm
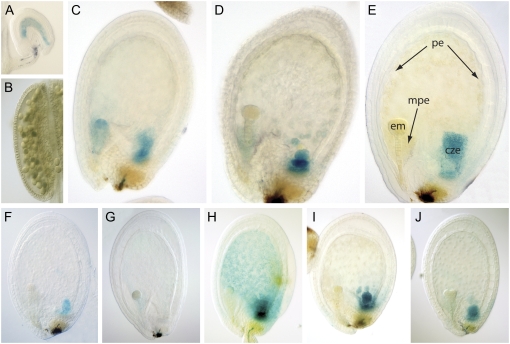
GUS reporter constructs were made for a selection of transcription factors to assess their use as markers for early endosperm development (Fig. 4). The promoter for At1g65300 (AGL38/PHE2) drove expression during very early embryo and endosperm development but became restricted to the chalazal endosperm region around the late globular stage of embryo development. Expression was also seen in pollen. The At1g49190 (ARR19) promoter was expressed specifically in the chalazal endosperm during globular and early heart stages of seed development, with some evidence of expression in stomatal guard cells of the silique. The At4g21080 (DOF4.5; Yanagisawa, 2002) promoter showed very strong chalaza-specific expression at the globular stage but then became more widespread throughout the endosperm, with the intensity of staining decreasing around the heart stage. The At4g18870 (HSF14; Guo et al., 2008) promoter also had very strong expression in the chalazal endosperm plus strong expression in the peripheral endosperm, but staining was not apparent in the micropylar domain. At around the heart stage, expression became restricted to the chalazal endosperm. Expression was also apparent in the cotyledons of the embryo during the mature green stages of seed development. At5g60440 (AGL62) was expressed specifically throughout the early developing endosperm at low levels, but staining was not apparent from the heart stage (Fig. 4).
Figure 4.
Promoter-GUS expression during early seed development. GUS reporter constructs were made for a selection of transcription factors to assess their use as markers for early endosperm development. A to E, Expression of the At1g65300 (AGL38/PHE2) construct was seen in mature pollen (B) and subsequently in the central cell after fertilization (A). Expression then persisted in the very early embryo and endosperm (C) but became restricted to the endosperm around the late globular stage (D). Expression was then specific to the chalazal region during transition (E). F, The At1g49190 (ARR19) promoter was expressed in the chalazal endosperm during globular and early heart stages of seed development. G, The At5g60440 (AGL62) promoter was expressed throughout the syncytial endosperm, albeit at low levels, and is shown here during the globular stage. H, The At4g18870 (HSF15) promoter had very strong expression in the chalazal endosperm plus strong expression in the peripheral endosperm at the globular stage of development. Staining was not apparent in the micropylar domain. I and J, The At4g21080 (DOF4.5) promoter showed very strong chalaza-specific expression at the globular stage (I), with expression becoming more widespread in the endosperm at the heart stage (J). The labels on E are as follows: cze, chalazal endosperm; em, embryo; mpe, micropylar endosperm; pe, peripheral endosperm.
DISCUSSION
Using laser microdissection and microarray analysis, we have obtained the transcriptome of the syncytial endosperm (Day et al., 2007b). Here, we present a detailed analysis of the transcriptome, focusing on the genes differentially expressed in the endosperm compared with other silique tissues. We also identified a subset of approximately 800 of these genes that are specifically expressed in the seed and therefore probably play key roles in seed development. Analysis of our data was consistent with the idea that the syncytial endosperm at 4 DAP is locked into a proliferative state dominated by transcripts associated with the regulation of the cell cycle, DNA processes, chromatin assembly, protein synthesis, cytoskeleton/microtubule-related processes, and cell/organelle biogenesis and organization. In the discussion below, we focus on the biological significance of the endosperm-preferred expression of particular genes involved in the cell cycle, hormone biology, transcriptional regulation, and early endosperm development.
Analysis of Cell Cycle Genes Suggests Roles for Gene Family Members in the Regulation of Syncytial Division
In Arabidopsis, eight mitotic divisions occur during the syncytial phase of development, until there are approximately 200 nuclei (Boisnard-Lorig et al., 2001). Plant syncytial development requires a rapid progression through the cell cycle, suppression of phragmoplast formation, and an uncoupling of cytokinesis from mitosis (Otegui and Staehelin, 2000). To gain further insights into this process, we analyzed our data to identify endosperm-expressed genes that have been implicated in controlling cell cycle progression.
Cyclin-Dependent Kinase/Cyclin Regulation of the Cell Cycle
The components of the cell cycle are largely conserved across the eukaryotes (Mironov et al., 1999). Fluctuations of distinct combinations of cyclin-dependent kinases (CDKs) and cyclins are necessary for progression through the different phases of the cell cycle (Gutierrez, 2005; De Veylder et al., 2007). In Arabidopsis, there are 12 CDKs and at least 49 cyclins divided into nine classes (Vandepoele et al., 2002; Wang et al., 2004). Different CDK/cyclin heterodimer combinations phosphorylate different protein targets, which then coordinate entry into the next phase of the cell cycle. A- and B-type CDKs are the main drivers of the plant cell cycle, and CDKB1;1, CDKB1;2, CDKB2;2, and CDKD1;1 (CDKD3 is just below the 2-fold cutoff) are present in the endosperm-preferred partition and probably play a key role in controlling endosperm proliferation (Table IV).
A most striking aspect of our analysis of core cell cycle genes (Vandepoele et al., 2002) is the predominance of cyclin A and B genes in the endosperm-preferred partition. Five of the seven A-type cyclins (CYCA1;1, CYCA1;2, CYCA2;1, CYCA2;2, and CYCA3;1) and all six of the B-type cyclins (CYCB1;1, CYCB1;2, CYCB1;3, CYCB2;1, CYCB2;3, and CYCB3;1) with differential expression on our arrays were endosperm preferred (Table IV). In contrast to the A and B types, only one of seven D-type cyclins present on the array (CYCD3;3; Vandepoele et al., 2002) was detected in the endosperm-preferred partition. Both A- and B-type cyclins are associated with mitotic cycles, and it has been predicted that cyclin accumulation is part of the overall program responsible for syncytial development (Boisnard-Lorig et al., 2001). The maize (Zea mays) CYCA1 is concentrated in the phragmoplast during cytokinesis (John et al., 2001). The Arabidopsis CYCA1 ortholog is endosperm preferred in our analysis, yet the precise nuclear localization of CYCA1 is not known and formation of the phragmoplast is suppressed during syncytial endosperm development. CYCB1;1 has been studied extensively during early endosperm development (Boisnard-Lorig et al., 2001), and CYCB1;1 to -3 are endosperm preferred in our analysis. It has been shown that a nondegradable form of CYCB1 suppresses phragmoplast formation at the end of each nuclear division (Weingartner et al., 2004). Therefore, studies on the regulation of CYCB1 genes should provide further insights into their possible roles in syncytial endosperm development.
Regulation of CDK/cyclin complexes through the cell cycle is also mediated through inhibitors of cyclin-dependent kinases. In plants, inhibitors of cyclin-dependent kinases are more similar to Kip protein (KRP, for Kip-related protein; De Veylder et al., 2001). There are seven KRPs present in the Arabidopsis genome. The putative CDK inhibitory protein KRP4 is endosperm preferred (Table IV), which is consistent with its reported expression in mitotically active cells (Ormenese et al., 2004). This indicates that genes capable of negatively regulating the cell cycle are also present in the EP partition.
Regulation of G2-M Phase Expression by Myb3R Genes
Proliferating plant cells show periodic transcription of a large portion of their genes (Breyne et al., 2002). In tobacco (Nicotiana tabacum), M phase-specific expression is regulated by Myb3R genes, with Myb3RA1 and Myb3RA2 showing phase-dependent transcription and their proteins binding MSA sequences in the promoter region of their target genes (Ito et al., 1998, 2001). The third tobacco gene, Myb3RB, which is thought to antagonize Myb3RA to control expression of the G2-M phase-specific genes, is expressed at a constant level during the cell cycle and is incapable of activating MSA-containing promoters (Araki et al., 2004). All five Arabidopsis Myb3R genes show evidence of endosperm expression in our data, and AtMyb3R2 and AtMyb3R4 have endosperm-preferred expression, suggesting that they play an important role in the regulation of G2-M phase-specific genes in the syncytial endosperm. Furthermore, transcripts of their putative M phase-specific target genes were enriched in the endosperm. Analysis of reporter constructs during early endosperm development should help elucidate the role of individual AtMyb3R genes, such that phase-dependent expression would indicate an MSA-activating role and phase-independent expression would indicate an inhibitory role.
E2F Transcription Factors
The orderly fluctuation of particular CDK/cyclins is required for progression through the cell cycle. These fluctuations are mediated in part by E2F transcription factors (Otegui and Staehelin, 2000; Kosugi and Ohashi, 2002; Mariconti et al., 2002). Consistent with a pronounced role during early endosperm development, E2F target genes (and upstream E2F-binding motifs) were significantly enriched in the EP and ESS-EP groups, indicating that some E2F-responsive genes have endosperm-specific roles (Table III). All six Arabidopsis E2F transcription factors (E2Fa to E2Fc and DEL1 to DEL3) gave signals consistent with endosperm expression, although data for E2Fb and DEL1 were relatively variable. E2Fa to E2Fc have important roles regulating transcription during the G1-S transition, with E2Fa and E2Fb acting as positive regulators of the cell cycle (De Veylder et al., 2002; Kosugi and Ohashi, 2003; Sozzani et al., 2006), activating targets that are necessary for DNA replication during S phase (Ramirez-Parra et al., 2003; del Pozo et al., 2006), and E2Fc suppressing cell division by acting as a transcriptional repressor (del Pozo et al., 2002; Vandepoele et al., 2005). E2F-DP-retinoblastoma protein complexes bind but do not activate the expression of S phase factor targets until the retinoblastoma protein becomes phosphorylated by a G1-specific cyclin/CDK (Nakagami et al., 2002).
Unlike E2Fa to E2Fc, DEL1 to DEL3 do not interact with DPa and DPb and bind E2F-binding sites in a monomeric form (Kosugi and Ohashi, 2002; De Veylder et al., 2007). The DEL proteins have been shown to be abundant in meristematic cells and were thought to balance the activities of E2Fa/bDPa/b transcription factors by restraining cell proliferation (Ohashi-Ito et al., 2002). DEL3 and DEL2 were present in the endosperm (Table IV), indicating a role for DEL proteins during proliferative syncytial endosperm development, in which repeated rounds of mitosis occur in the absence of cytokinesis and cell wall formation. Indeed, recent data suggest that DEL genes function as important negative regulators of cell differentiation (Ramirez-Parra et al., 2004; Vlieghe et al., 2005), consistent with a role during the syncytial phase of endosperm development. Intriguingly, DEL1 inhibits the endocycle and preserves the mitotic state of proliferating cells, and DEL3 represses transcripts associated with cell wall biosynthesis and expansion (Ramirez-Parra et al., 2004; Vlieghe et al., 2005).
In summary, analysis of the core cell cycle genes and their putative downstream targets reveals genes that appear to play important roles in the proliferation of the endosperm. Further insight into the roles of particular cell cycle genes, such as DEL2 and KRP4, in the endosperm could be obtained using reporter constructs, in situ hybridization, or region-specific endosperm LCM to associate gene expression with either the proliferating or endoreduplicating domains of the endosperm (Boisnard-Lorig et al., 2001).
Analysis of Hormone Markers Indicates an Important Role for CKs in the Proliferating Endosperm
Analysis of hormone-responsive genes showed that only the CK-responsive genes were significantly enriched in the EP partition (Table III). This is consistent with studies in rice (Oryza sativa) and maize that show significant correlations between CK levels and the rate of cell division in the early endosperm (Lur and Setter, 1993; Yang et al., 2002). Moreover, the early endosperm appears to be a site of CK biosynthesis, with major components of the CK biosynthetic pathway (isopententyltransferase genes 4 and 8) being specifically expressed in the chalazal endosperm during early seed development (Miyawaki et al., 2006; Table I).
CK Signaling Genes Are Expressed in the Early Endosperm
The enrichment of CK-responsive genes and the presence of CK biosynthesis genes in the EP partition indicate that CK signaling is important during the early stages of Arabidopsis endosperm development (Table V). CK perception and signaling are similar to two-component phosphorelays in bacteria (Müller and Sheen, 2007). AHK4 shows evidence for endosperm expression in our data and is an example of a sensor His kinase (AHK) that is able to initiate a phosphorelay when bound to CKs. Arabidopsis Histidine-containing Phosphotransfer proteins (AHPs) are subsequently phosphorylated and translocated to the nucleus, where they transfer their phosphate to Arabidopsis type-B Response Regulators (ARRs; Müller and Sheen, 2007). These mediate the transcriptional response to CKs, which includes inducing the primary response genes, the type-A ARRs, which act as negative regulators of the primary signal transduction pathway, likely via interaction with the AHPs (Dortay et al., 2006). AHP genes have functional overlap, with only multiple AHP mutants displaying a reduced sensitivity to CKs. The AHP1-5 quintuple knockout plants show multiple abnormalities in growth and development, including seeds with uneven size and some seed abortion (Hutchison et al., 2006). The average size of nonabortive seeds was about 20% larger than in the wild type, although this might be a consequence of increased maternal resources (de Jong and Scott, 2007). Similar variation in seed development was seen in the AHP2,3,5 triple mutant, which indicated that these AHPs have roles during seed development (Hutchison et al., 2006). Interestingly, of the AHPs in the list of differentially expressed genes, AHP2 and AHP5 are very close to the arbitrary 2-fold cutoff (1.99 and 1.89, respectively; Table V), indicating relatively high expression in the endosperm compared with other silique tissues. This suggests that the reduced seed set and abortion in the triple mutant might be results of aberrant endosperm development.
CK-responsive genes were significantly overrepresented in both the EP partition and the OST partition (Fig. 4), but further analysis suggested a very different response to CK in the endosperm than in the other silique tissues, with CK predominantly activating genes in the EP group and down-regulating genes in the OST group. A differential response to CK in the endosperm compared with other silique tissues was also evident in our analysis of the ARR genes in the EP and OST partitions (Table V). The EP partition included three ARRs (ARR18, ARR19, and ARR21), all of which are B-type ARRs that act as transcriptional activators during CK signaling. In contrast, the majority of A-type ARRs (negative regulators) in our data were distributed in the OST partition and perhaps indicate a more inhibitory regulation of CK signaling in other silique tissues compared with endosperm at 4 DAP.
Recently, a second group of transcription factors, termed the cytokinin response factors (CRFs), were shown to act as part of the CK two-component pathway and to require the action of both the AHK and AHP genes to mediate a CK response (Rashotte et al., 2006). CRFs mediate a large fraction of the CK transcriptional response that functionally overlaps with the B-type ARR response. Five of the six CRFs are present in our data, confirming an important role for this gene family in developing siliques at 4 DAP. CRF5/6 double mutant embryos fail to develop past the early heart stage (Rashotte et al., 2006). This suggests that CRF5 and CRF6 are embryo expressed during early seed development, which is consistent with the presence CRF5 in our OST partition. The four other CRFs (CRF1 to CRF4) are included in our EP partition, indicating a role in early endosperm development (Table V). The roles of CRF1 and CRF3 are unclear, as these genes show little or no response to CK (Rashotte et al., 2006). However, like CRF5, CRF2 is unregulated by CK in a B-type ARR-dependent manner and likely plays an important role in CK-dependent transcriptional activation in the endosperm. CRF2 and the B-type response regulators ARR18, ARR19, and ARR21 are present in our ESS-EP partition, indicating a seed-specific role for these components of CK signaling.
In a study that characterized the expression of B-type ARRs, RT-PCR analysis showed that ARR19 and ARR21 are expressed specifically in silique tissues, although this expression was not detected using promoter-GUS reporter constructs (Mason et al., 2004). However, alternative promoter-GUS constructs (ARR21 in Tiwari et al. [2006] and ARR19 in Fig. 4 of this study) indicate expression in the chalazal endosperm during early endosperm development. Interestingly, overexpression experiments show that ARR21 induces expression of the key CK biosynthesis gene IPT4 (Kiba et al., 2005). As IPT4 is also expressed in the chalazal endosperm (Miyawaki et al., 2004), it seems likely that ARR21 and perhaps ARR19 act to ensure IPT gene expression and therefore CK biosynthesis during early wild-type seed development.
Although our analysis indicates that the chalazal endosperm plays an important role in directing proliferation of the endosperm via CK signaling, the chalazal endosperm does not appear to undergo mitosis and shows evidence of endoreduplication (Boisnard-Lorig et al., 2001). This is contrary to current thinking that high levels of CKs exist in mitotically dividing cells, the suggested sites of de novo CK biosynthesis (Moncaleán et al., 2001; Friml, 2003; Nordstrom et al., 2004; He et al., 2005). Another important function for CKs may be determining cell fate, as a Sinapis alba MADS box gene (Bonhomme et al., 2000) and a maize homolog of Arabidopsis KNAT1, important in the development and maintenance of the shoot apical meristem (Hewelt et al., 2000), are induced by CK treatment.
Auxin Signaling and Auxin-Responsive Genes Are Underrepresented in the Proliferating Endosperm
While our data suggest a primary role for CK signaling during syncytial endosperm development, there is likely to be significant interplay with other phytohormones. It is well documented that the ratio of auxins and CKs plays an important role in controlling tissue proliferation and differentiation. A study of maize endosperm showed CK levels at 9 DAP that corresponded to the maximal cell division rate in the endosperm and that were reduced sharply as auxin levels increased toward the mid to late stages of endosperm development. This is consistent with other studies on grain development, in which CK levels were maximal during early stages and auxin levels reached maximal levels later in seed development (Mengel et al., 1985; Lur and Setter, 1993). Furthermore, auxin has been shown to have a rapid negative control on CK levels by suppressing its biosynthesis (Nordstrom et al., 2004). Our data indicate strong CK-associated and weak auxin-associated transcriptional responses in the syncytial endosperm at 4 DAP (Table III), along with significant underrepresentation of auxin signaling genes in the EP partition. A hypothesis that is consistent with these observations is that at 4 DAP of endosperm development, auxin levels are low, enabling increased CK biosynthesis and high rates of cell division; subsequent increases in auxin might then promote endosperm cellularization.
The Roles of Other Hormones during Endosperm Development Are Indicated through the Representation of Ethylene-, ABA-, and GA-Related Genes and the Underrepresentation of Brassinosteroid-Responsive Genes
The distribution of both ACC- and ABA-responsive genes indicates that both ethylene and ABA signaling systems are active during early silique development and have roles during endosperm development (Table III; Supplemental Table S9). In a recent study, Yang et al. (2006) made direct comparisons of both ethylene and ABA levels with endosperm cell division in spikelets of rice. They concluded that cell division and grain-filling rates were positively associated with ABA and negatively associated with ethylene. However, during the earliest stages of endosperm development (0–12 DPA), their data show that ethylene evolution rates and ACC contents of the rice spikelets were at their highest. Conversely, ABA levels were low at anthesis and slowly increased to peak values at 12 DAP or later, depending on the cultivar. These observations are consistent with the idea that during the early syncytial stage of development (0–4 d in rice) ethylene levels are high, which may inhibit ABA biosynthesis (Ghassemian et al., 2000; del Pozo et al., 2005). It remains to be elucidated whether ethylene and ABA have an antagonistic effect during early endosperm development. The finding that the NCED6 gene, which cleaves cis-epoxycarotenoids in the first step of ABA biosynthesis, has early endosperm-specific expression suggests an important role for ABA synthesis during the proliferative stage of endosperm development in Arabidopsis (Table I; Lefebvre et al., 2006).
The other hormones, BL, MJ, and GA, play important roles in various aspects of plant development (del Pozo et al., 2005), and our silique/endosperm data would suggest a stronger influence of GA on transcription in the endosperm than of either BL and MJ (Table III; Supplemental Table S9). This is consistent with observations that jasmonic acid has a negative effect on G1/S and G2/M transitions (Świątek et al., 2002) and that GAs stimulate cell division (Fabian et al., 2000). However, the underrepresentation of BL-responsive genes was unexpected, since BLs have been shown to promote cell division and are able to up-regulate the expression of CYCD3;1 and CDKB1;1 (Yoshizumi et al., 1999; Hu et al., 2000). It is possible that the underrepresentation of BL-responsive genes in the endosperm is in some way related to the underrepresentation of auxin genes, since auxin and BLs can cooperate to promote organ growth via cell cycle activation (Bao et al., 2004).
Identification of Putative Early Seed-Specific Transcription Factors with Endosperm-Preferred Expression
Seventy-one transcription factors were found to be endosperm preferred and early seed specific. These genes likely play important roles within the endosperm, and representative genes were further characterized using promoter-GUS reporter lines (Fig. 4). This both implicates the genes (At1g65300, At1g49190, At4g21080, At4g18870, and At5g60440) as being important during proliferative endosperm development and also provides tools to misexpress genes that may promote endosperm proliferation as a rational approach to generate larger seeds.
Analysis Suggests Important Roles for Type-1 MADS Box Genes during Endosperm Development
A family of transcription factors that includes key regulators of proliferation of the syncytial endosperm in Arabidopsis are the MADS box genes. The best-known MADS box transcription factors have roles in flower development and flowering time and are classified as type II MADS box genes. However, although more than half of the MADS box transcription factors are type I, little is known about their roles in plant development (Parenicová et al., 2003). Of the 12 MADS box genes in the endosperm-preferred list, 11 are type I (Table VII). Recently, AGL23 was shown to play a role in female gametophyte and embryo development (Colombo, 2008). AGL23-GUS lines show that AGL23 is expressed throughout female gametophyte development and in the developing peripheral and chalazal endosperm, consistent with our expression data. Our ability to see biological significance in the interactions predicted by de Folter et al. (2005) using yeast two-hybrid analysis will guide future studies of type I MADS box genes and help elucidate their roles in endosperm development. For example, of the seven MADS box genes that are strongly endosperm preferred at 4 DAP, only four interact with other endosperm-preferred MADS box genes. Analysis of the MADS box interactome indicates that type I heterodimer formation requires a member of the Mα subclass (de Folter et al., 2005). Of the four endosperm-specific MADS box genes that can form dimers, two are Mα subclass genes (AGL40 and AGL62) and the other two are Mγ subclass genes (PHE1 and PHE2). AGL40 and AGL62 do not form heterodimers together in yeast, but both PHE1 and PHE2 can form heterodimers with both AGL40 and AGL62 (Fig. 2).
Since a prolonged syncytial growth pattern is limited to early endosperm development in Arabidopsis, it follows that key developmental switches that define syncytial competence are also early seed specific. This is the case for AGL62, which is required for normal syncytial endosperm development, since disruption of the AGL62 gene results in very early cellularization of the endosperm, approximately 24 h after fertilization (Kang et al., 2008). Both the Drews laboratory AGL62-GFP reporter line (Kang et al., 2008) and the AGL62-GUS reporter presented in this study confirm that AGL62 expression is specific to the syncytial endosperm. AGL62 interacts in vitro with many seed-expressed MADS box genes that may help mediate its ability to enable syncytial proliferation (which requires inhibition of cytokinesis and cell wall formation). As mentioned, AGL62 can form a heterodimer with PHE1, which has also been implicated as a positive regulator of endosperm proliferation and has endosperm-specific expression at 4 DAP (Köhler et al., 2003; this study). PHE2 has 72% homology to PHE1 at the amino acid level and has been reported to have a very similar expression pattern, although no data were provided to substantiate this claim (Köhler et al., 2003). Our PHE2 promoter-GUS analysis confirms that PHE2 expression is largely equivalent to PHE1 expression during wild-type seed development. Transcription of both AGL62 and PHE1 appears to be regulated by members of the fertilization-independent seed-Polycomb (FIS-PcG) complex, since levels of both AGL62 and PHE1 are increased and persist longer in FIS than during wild-type seed development (Köhler et al., 2003). Developing seeds of the FIS class mutant mea abort during heart stage, which coincides with an overproliferated endosperm. Abortion of mea seeds is partly due to abnormally high levels of PHE1, since reducing PHE1 transcript in mea using an antisense construct can rescue the abortion phenotype. Like PHE1, PHE2 expression was also reported to be increased in mea seeds, which is suggestive of similar regulation and perhaps a redundant function. Alternatively, PHE2 may be an antagonist of PHE1 that competes to form heterodimers with AGL62, thus acting to reduce the rate of syncytial proliferation. Our data also predict that the other endosperm-specific Mα MADS box gene AGL40 plays an important role in the molecular control circuit that enables syncytial proliferation of the early endosperm, perhaps by mediating the levels of PHE1 and/or PHE2 proteins available to form heterodimers with AGL62.
Genes Involved in Important Early Developmental Processes Are Expressed at 4 DAP
Pagnussat et al. (2005) identified 130 mutants in an extensive screen for female gametophytic mutants, some of which had aberrant endosperm. Sixty-one of the genes identified as having female gametophytic phenotypes are present in our differentially expressed gene list at 4 DAP. Eleven are in the EP partition, suggesting an important role during the proliferative stage of endosperm development at 4 DAP, as well as their prior roles in enabling fertilization (At5g02100), embryo sac development (At4g14790 and At2g35950), fusion of the polar nuclei (At2g20490, At1g72440, and At4g00140), and very early embryo and endosperm formation (At2g15890, At4g13380, At2g34880, and At4g01560).
Seedgenes.org (http://www.seedgenes.org/index.html), a database of Arabidopsis seed mutants, contains six genes described as having female gametophytic inheritance patterns (in which siliques produce approximately 50% mutant seeds following pollination of heterozygotes, regardless of pollen genotype; Tzafrir et al., 2003). These include the components of the FIS-PcG complex MEA, FIS2, FIE, and MSI1 and the DNA glycosylase DME. The sixth gene, EMB2220 (At5g12840), is not early seed specific but is approximately 2-fold enriched in our endosperm sample, similar to FIE (which is also not seed specific). EMB2220 mutants have not been fully characterized, but it is tempting to speculate that this gene is also involved in endosperm development.
The FIS-PcG complex is involved in repressing seed development prior to fertilization, the formation of distinct mitotic domains during syncytial endosperm development, and the timing of endosperm cellularization (Köhler and Makarevich, 2006). Histone modifications via the FIS-PcG complex and DNA methylation via the DNA methylase MET1 have been shown to be important components of allele-specific repression of imprinted genes in Arabidopsis (Feil and Berger, 2007). The DNA glycosylase DME is involved in activation of the maternal allele of the imprinted genes MEA, FWA, and FIS2 (Feil and Berger, 2007). This parent of origin-dependent differential repression and activation of alleles appears to be limited to the endosperm in plants (Feil and Berger, 2007). Concordantly, all confirmed imprinted genes in Arabidopsis (i.e. PHE1, MEA, FIS2, and FWA [Luo et al., 2000; Kinoshita et al., 2004; Köhler et al., 2005]) are present in our EP partition. Therefore, it is likely that our EP partition is enriched for imprinted genes and provides a short list for identifying new members of this gene family.
CONCLUSION
The use of laser-assisted microdissection technology has enabled the isolation and high-resolution transcript analysis of early endosperm. This, and work by Casson et al. (2005), Day et al. (2007b), Le et al. (2007), and Spencer et al. (2007), demonstrates the power of laser-assisted microdissection for studying tissue-specific expression during early seed development, even in Arabidopsis, which has relatively small seeds. The proliferative stage of endosperm development is unusual in that rapid nuclear division occurs in the absence of cell division. Since the proliferative stage of endosperm development is important for determining final seed size, understanding the processes that control endosperm proliferation should lead to novel ways of improving seed yield. The data generated provide a useful resource enabling novel insight into this process, as illustrated by the examples discussed here, and provide a starting point for further study.
MATERIALS AND METHODS
LCM Endosperm Microarray Data
The LCM endosperm microarray data analyzed in this study have been described (Day et al., 2007b) and deposited at the National Center for Biotechnology Information gene expression omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE6703.
Preparation of qRT-PCR cDNA Template and PCR Cycling Conditions for Analysis of LCM-Derived Samples
Total RNA was obtained from LCM-dissected endosperm as described (Day et al., 2007b), and total RNA was obtained using the Picopure RNA isolation kit (Arcturus) with an optional on-column DNase step (Qiagen). The purified total RNA was quantified using the Ribogreen RNA quantification kit (Invitrogen) according to the manufacturer's instructions. IVT-based amplifications were carried out using the Message Amp II kit (Ambion) following the manufacturer's instructions. Comparisons between the ability of qRT-PCR and the microarrays to measure differential expression of 17 genes in the same samples used cDNA made from amplified RNA (aRNA) generated using one round of IVT. The remaining first-round aRNA was then used as the basis for a second round of IVT, which generated target for the microarray study. For the qRT-PCR, the aRNA was primed with random hexamers and first-strand cDNA was synthesized using SuperScript III (Invitrogen) according to the manufacturer's instructions. Real-time qRT-PCR was carried out using reagents from the LightCycler FastStart DNA MasterPlus SYBR Green I kit (Roche) in 20-μL volumes using a LightCycler 1.0 (Roche). The amplification conditions for qPCR were as follows: denaturation at 95°C for 10 min; cycling at 94°C for 5 s, 58°C for 17 s, and 72°C for 10 s (single acquire); melting at 95°C for 0 s, 55°C for 20 s, and 95°C for 0 s, with ramp at 0.2°C s−1 (continuous acquire); and cooling at 40°C for 20 s. Reaction products were confirmed by melting curve analysis and by running out the product on a 1.2% agarose gel. The primers used for qRT-PCR are listed in Supplemental Table S13.
Preparation of qRT-PCR cDNA Template, and PCR Cycling Conditions for Analysis of Conventional Fresh Tissue Samples
RNA was extracted from fresh Arabidopsis (Arabidopsis thaliana) tissues using the Qiagen Plant RNeasy kit as per the manufacturer's instructions with some alterations for the seed samples. Disruption of the silique, stem, leaf, root, and flower bud tissues was carried out by harvest into 1.5-mL Eppendorf tubes and flash freezing in liquid nitrogen. Tissues were then quickly ground to a powder in Eppendorf tubes using a precooled plastic pestle on dry ice. RNA extraction reagent was added before the samples thawed. For the dissected seed samples, developing siliques were removed from plants using tweezers and cut open using a dissecting microscope with a hypodermic needle, being careful not to damage the seed within. The majority of seeds were scraped onto the back of the needle and deposited into a precooled Eppendorf tube on dry ice. Frozen seeds were transferred to precooled plastic bags embedded in dry ice, and RNA extraction reagent was pipetted into the frozen bag and allowed to thaw. Individual developing seeds (visualized through the plastic using a dissecting microscope) were completely disrupted using pressure from the tip of blunt tweezers and used as input for the Plant RNeasy kit.
RNA was quantified using a Nanodrop spectrophotometer, and cDNA was generated using the VILO cDNA synthesis kit (Invitrogen) according to the manufacturer's instructions. Real-time qRT-PCR was carried out using reagents from the Express SYBR Green ER MasterMix kit (Invitrogen) in 7-μL volumes using a LightCycler 480 (Roche). The amplification conditions for qPCR were as follows: denaturation at 95°C for 10 min; cycling at 94°C for 5 s, 61°C or 58°C for 17 s, and 72°C for 10 s (single acquire); melting at 95°C for 0 s, 55°C for 20 s, and 95°C for 0 s, with ramp at 0.2°C s−1 (continuous acquire); and cooling at 40°C for 20 s. Reaction products were confirmed by melting curve analysis and by 1.2% agarose gel electrophoresis. The primers used for qRT-PCR are listed in Supplemental Table S13 and include primers designed by Han and Kim (2006).
Identification of Genes with Evidence of Early Silique/Seed-Specific Expression
We selected the top approximately 3,200 genes that reported preferred endosperm expression in our array data to manually search the Arabidopsis MPSS database. This database contains libraries of signatures generated from several tissue types, each containing approximately 2 million signature tags (Meyers et al., 2004). These libraries are normalized so that transcript abundance can be compared between libraries. At the time of query, the database could be searched using several AGI locus identifiers per search. The resulting output was used to create a summary file that was imported into The Institute for Genomic Research Multiexperiment Viewer (MeV) 3.0 software and searched using pattern matching (Pavlidis and Noble, 2001).
To identify genes with early endosperm-specific expression, we used the known marker genes FIS2, FWA, PHE1, and SUC5 (Luo et al., 2000; Kinoshita et al., 2004; Baud et al., 2005; Köhler et al., 2005) as bait to search the AtGenExpress developmental series (Arabidopsis transcript profiles from different organs and at different stages of development were determined using Affymetrix GeneChip technology) using the Expression Angler (Bio-Array Resource for Arabidopsis Functional Genomics [BAR]; http://bar.utoronto.ca/). The software calculates the similarity of expression patterns to the marker genes for the other genes in the database using a Pearson correlation coefficient (r value). FIS2, FWA, and PHE1 had very few genes correlated with their expression patterns, probably caused by ill-defined expression patterns. In contrast, SUC5, which has strong endosperm-preferred expression during the proliferative stages of development (Baud et al., 2005), gave a well-defined pattern in the AtGenExpress seed data. The SUC5 expression template was used to identify genes similarly expressed during early seed development in the AtGenExpress data. This list was then entered into the BAR DataMetaFormatter tool (http://bar.utoronto.ca/) to create a heat map of probe intensities for genes in the AtGenExpress seed development series.
The GHL Affymetrix GeneChip data sets were downloaded from the National Center for Biotechnology Information gene expression omnibus (http://www.ncbi.nlm.nih.gov/geo/), imported into The Institute for Genomic Research MeV software, and screened using the pattern-matching function to identify those genes with similar expression patterns to the early endosperm markers SUC5, PHE1, FWA, and FIS2 (Supplemental Table S2). Pattern-matched genes for all markers were combined and cross-referenced to genes with differential expression on our arrays. This ESS subgroup was used in subsequent analysis due to its enhanced sensitivity toward known endosperm markers. For example, Supplemental Figure S2 shows a heat map of normalized intensity data from both the GHL GeneChip data and the AtGenExpress seed development series. Negligible signal was apparent for the known endosperm markers PHE1, IPT8, FWA, FIS2, and MEA in the AtGenExpress data, whereas expression of all of these markers was easily apparent in the GHL data. This likely reflects the fact that the GHL data were derived from seeds removed from the surrounding silique tissues, whereas the samples representing early seed development in the AtGenExpress data included the whole silique.
Other Data Processing and Analysis
The BAR (http://bar.utoronto.ca/) duplicate remover, DataMetaFormatter, Expression Angler, and Arabidopsis Interactions Viewer (Toufighi et al., 2005) were used for list processing, heat map generation, pattern matching, and interaction plot generation, respectively. TAIR (http://www.Arabidopsis.org/) was used to gather gene annotation information and to assemble some gene lists. Hormone-responsive genes were obtained from Nemhauser et al. (2006). Arabidopsis transcription factors were obtained from DATF (http://datf.cbi.pku.edu.cn/). Core cell cycle genes were obtained from TAIR (Vandepoele et al., 2002), and E2F-regulated genes were obtained from Vandepoele et al. (2005). We assessed the representation of E2F target genes in our data using a list of genes shown (1) to be induced by ectopic expression of E2Fa and DPa transcription factors, (2) to contain a distinctive cis-acting element in their promoters (E2F box), and (3) to have rice orthologs that also have promoters containing an E2F box (Vandepoele et al., 2005). Arabidopsis genes that contain the MSA sequence in the 500 bp upstream of the ATG were identified using the Patmatch tool from the TAIR Web site (Garcia-Hernandez et al., 2002). The Athena promoter analysis database (http://www.bioinformatics2.wsu.edu/cgi-bin/Athena/cgi/home.pl; O'Connor et al., 2005) was used to assess for enrichment of E2F-binding motifs in the 500-bp upstream regions of our EP and ESS-EP partitions.
Genes with methylation-sensitive transcription were obtained from Zhang et al. (2006). Chromatin-related genes were obtained from ChromDB (http://www.chromdb.org/). In some instances, a particular locus was represented by more than one probe on our array. In a few cases, different probes for the same locus were observed in different partitions, since cutoff values were based on a specific signal ratio. For subsequent analysis, therefore, we included the locus identifier in both partitions.
TAIR Gene Ontology (TAIR-GO) and MIPS Functional Category (MIPS-FunCat) enrichment analyses were performed using the Virtual Plant 0.9 Web site (http://virtualplant-prod.bio.nyu.edu/cgi-bin/virtualplant.cgi). This enabled us to calculate the frequency of genes with a particular TAIR-GO or MIPS-FunCat as a percentage of the whole genome. If a subgroup of genes contains significantly different proportions of a GO or FunCat, biological importance can be inferred. To assess the presence of inherent bias in the GO or FunCat groups represented by the probes on our array platform, we queried the whole genome with the list of loci on the array. This analysis revealed that several GO and FunCat groups were already overrepresented on our arrays (data not shown). To get an accurate assessment of the representation of biological processes in our partitioned data, therefore, we used our array list as the background population. t tests were carried out in MeV with no correction for multiple testing.
Construction and Analysis of GUS Reporter Lines
Five promoters were selected to validate the endosperm-preferred expression of the downstream genes predicted by the LCM endosperm microarray data. The genomic regions between the 2,356, 2,089, 1,110, and 582 nucleotide positions upstream of the translational start codons were amplified by PCR for At1g65300/PHE2, At1g49190/ARR19, At4g21080, and At4g18870, respectively. These fragments were cloned upstream of the uidA gene in the binary vector pCAMBIA1391-Z (CAMBIA) and transformed into Agrobacterium tumefaciens strain GV3101. The genomic region between −1,453 and +787 of At5g60440/AGL62 (relative to the translational start codon) was amplified and cloned in frame upstream of the uidA gene in the binary vector pCAMBIA1381xc (CAMBIA) and transformed into A. tumefaciens strain LBA4404. A list of the primers used is provided in Supplemental Table S13. Arabidopsis was transformed using the standard floral dipping protocol (Clough and Bent, 1998). Developing seeds from hygromycin-resistant primary transformants were assessed for GUS activity essentially following Stangeland and Salehian (2002). Briefly, fruits at various stages of development were dissected and placed in GUS staining buffer (50 mm phosphate buffer [pH 7.2], 0.5 mm potassium ferricyanide/ferrocyanide, and 1 mg mL−1 5-bromo-4-chloro-3-indolyl-β-glucuronic acid) overnight at 37°C. Younger fruits were cleared in Hoyer's medium (100 g chloral hydrate in 30 mL of water) for 2 to 4 h. Older fruits were placed in 3:1 ethanol:acetic acid for 4 to 8 h and cleared overnight in Hoyer's medium. Developing seeds were fully dissected from the fruits and mounted in Hoyer's mounting medium with a coverslip. Prepared slides were viewed using differential interference contrast optics on an Olympus BX51 microscope equipped with an Optronics Magnafire 2.1A digital imaging system.
The microarray data discussed in this article can be found in the National Center for Biotechnology Information gene expression omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE6703.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Heat map showing endosperm-preferred genes in MPSS data.
Supplemental Figure S2. Heat maps comparing endosperm marker detection in online data sets.
Supplemental Table S1. Genes with AtGenExpress expression profiles correlating with the early endosperm-specific gene SUC5.
Supplemental Table S2. List of putative ESS genes from the GHL data.
Supplemental Table S3. AGI list for gene partitions.
Supplemental Tables S4 to S7. Frequency of genes within a particular TAIR-GO or MIPS-FunCat from the different partitions.
Supplemental Table S8. Representations of putative M phase-specific and E2F target genes within the different partitions.
Supplemental Table S9. List of hormone-responsive genes in each partition.
Supplemental Table S10. List of chromatin-related genes (obtained from ChromDB; http://www.chromdb.org/) in each partition.
Supplemental Table S11. List of genes affected by reduced methylation in each partition.
Supplemental Table S12. List of transcription factor genes in each partition.
Supplemental Table S13. Sequences of the primers used for qRT-PCR and to generate the genomic fragments for the GUS constructs.
Supplementary Material
Acknowledgments
We thank Peter Stockwell for formatting MPSS data for pattern matching and R. Kaji for making the AGL62-GUS plants.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Richard C. Macknight (richard.macknight@otago.ac.nz).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Araki S, Ito M, Soyano T, Nishihama R, Machida Y (2004) Mitotic cyclins stimulate the activity of c-Myb-like factors for transactivation of G2/M phase-specific genes in tobacco. J Biol Chem 279 32979–32988 [DOI] [PubMed] [Google Scholar]
- Bao F, Shen J, Brady SM, Muday GK, Asami T, Yang Z (2004) Brassinosteroids interact with auxin to promote lateral root development in Arabidopsis. Plant Physiol 134 1624–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroux C, Pien S, Grossniklaus U (2007) Chromatin modification and remodeling during early seed development. Curr Opin Genet Dev 17 473–479 [DOI] [PubMed] [Google Scholar]
- Baud S, Wuilleme S, Lemoine R, Kronenberger J, Caboche M, Lepiniec L, Rochat C (2005) The AtSUC5 sucrose transporter specifically expressed in the endosperm is involved in early seed development in Arabidopsis. Plant J 43 824–836 [DOI] [PubMed] [Google Scholar]
- Berger F (2003) Endosperm: the crossroad of seed development. Curr Opin Plant Biol 6 42–50 [DOI] [PubMed] [Google Scholar]
- Berger F, Gaudin V (2003) Chromatin dynamics and Arabidopsis development. Chromosome Res 11 277–304 [DOI] [PubMed] [Google Scholar]
- Boisnard-Lorig C, Colon-Carmona A, Bauch M, Hodge S, Doerner P, Bancharel E, Dumas C, Haseloff J, Berger F (2001) Dynamic analyses of the expression of the HISTONE:YFP fusion protein in Arabidopsis show that syncytial endosperm is divided in mitotic domains. Plant Cell 13 495–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonhomme F, Kurz B, Melzer S, Bernier G, Jacqmard A (2000) Cytokinin and gibberellin activate SaMADS A, a gene apparently involved in regulation of the floral transition in Sinapis alba. Plant J 24 103–111 [DOI] [PubMed] [Google Scholar]
- Breyne P, Dreesen R, Vandepoele K, De Veylder L, Van Breusegem F, Callewaert L, Rombauts S, Raes J, Cannoot B, Engler G, et al (2002) Transcriptome analysis during cell division in plants. Proc Natl Acad Sci USA 99 14825–14830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RC, Lemmon BE, Nguyen H (2003) Events during the first four rounds of mitosis establish three developmental domains in the syncytial endosperm of Arabidopsis thaliana. Protoplasma 222 167–174 [DOI] [PubMed] [Google Scholar]
- Bushell C, Spielman M, Scott RJ (2003) The basis of natural and artificial postzygotic hybridization barriers in Arabidopsis species. Plant Cell 15 1430–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson S, Spencer M, Walker K, Lindsey K (2005) Laser capture microdissection for the analysis of gene expression during embryogenesis of Arabidopsis. Plant J 42 111–123 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Colombo M, Masiero S, Vanzulli S, Lardelli P, Kater MM, Colombo L (2008) AGL23, a type I MADS-box gene that controls female gametophyte and embryo development in Arabidopsis. Plant J 23 1037–1048 [DOI] [PubMed] [Google Scholar]
- Day RC, Grossniklaus U, Macknight RC (2005) Be more specific! Laser-assisted microdissection of plant cells. Trends Plant Sci 10 397–406 [DOI] [PubMed] [Google Scholar]
- Day RC, McNoe L, Macknight R (2007. a) Transcript analysis of laser microdissected plant cells. Physiol Plant 129 267–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day RC, McNoe L, Macknight R (2007. b) Evaluation of global RNA amplification and its use for high-throughput transcript analysis of laser-microdissected endosperm. Int J Plant Genomics 2007 61028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Folter S, Immink RG, Kieffer M, Parenicová L, Henz SR, Weigel D, Busscher M, Kooiker M, Colombo L, Kater MM, et al (2005) Comprehensive interaction map of the Arabidopsis MADS box transcription factors. Plant Cell 17 1424–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong TJ, Scott RJ (2007) Parental conflict does not necessarily lead to the evolution of imprinting. Trends Plant Sci 12 439–443 [DOI] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GT, de Almeida Engler J, Ormenese S, Maes S, Naudts M, Van Der Schueren E, Jacqmard A, Engler G, et al (2002) Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J 21 1360–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GT, Krols L, Terras F, Landrieu I, van der Schueren E, Maes S, Naudts M, Inze D (2001) Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13 1653–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Inze D (2007) The ins and outs of the plant cell cycle. Nat Rev Mol Cell Biol 8 655–665 [DOI] [PubMed] [Google Scholar]
- del Pozo JC, Boniotti MB, Gutierrez C (2002) Arabidopsis E2Fc functions in cell division and is degraded by the ubiquitin-SCF(AtSKP2) pathway in response to light. Plant Cell 14 3057–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Diaz-Trivino S, Cisneros N, Gutierrez C (2006) The balance between cell division and endoreplication depends on E2FC-DPB, transcription factors regulated by the ubiquitin-SCFSKP2A pathway in Arabidopsis. Plant Cell 18 2224–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Lopez-Matas MA, Ramirez-Parra E, Gutierrez C (2005) Hormonal control of the plant cell cycle. Physiol Plant 123 173–183 [Google Scholar]
- Dortay H, Mehnert N, Bürkle L, Schmülling T, Heyl A (2006) Analysis of protein interactions within the cytokinin-signaling pathway of Arabidopsis thaliana. FEBS J 273 4631–4644 [DOI] [PubMed] [Google Scholar]
- Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, Weiss RA, Liotta LA (1996) Laser capture microdissection. Science 274 998–1001 [DOI] [PubMed] [Google Scholar]
- Fabian T, Lorbiecke R, Umeda M, Sauter M (2000) The cell cycle genes cycA1;1 and cdc2Os-3 are coordinately regulated by gibberellin in planta. Planta 211 376–383 [DOI] [PubMed] [Google Scholar]
- Feil R, Berger F (2007) Convergent evolution of genomic imprinting in plants and mammals. Trends Genet 23 192–199 [DOI] [PubMed] [Google Scholar]
- Friml J (2003) Auxin transport: shaping the plant. Curr Opin Plant Biol 6 7–12 [DOI] [PubMed] [Google Scholar]
- Garcia D, Fitz Gerald JN, Berger F (2005) Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell 17 52–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Hernandez M, Berardini TZ, Chen G, Crist D, Doyle A, Huala E, Knee E, Lambrecht M, Miller N, Mueller LA, et al (2002) TAIR: a resource for integrated Arabidopsis data. Funct Integr Genomics 2 239–253 [DOI] [PubMed] [Google Scholar]
- Ghassemian M, Nambara E, Cutler S, Kawaide H, Kamiya Y, McCourt P (2000) Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell 12 1117–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo A, He K, Liu D, Bai S, Gu X, Wei L, Luo J (2005) DATF: a database of Arabidopsis transcription factors. Bioinformatics 21 2568–2569 [DOI] [PubMed] [Google Scholar]
- Guo J, Wu J, Ji Q, Wang C, Luo L, Yuan Y, Wang Y, Wang J (2008) Genome-wide analysis of heat shock transcription factor families in rice and Arabidopsis. J Genet Genomics 35 105–118 [DOI] [PubMed] [Google Scholar]
- Gutierrez C (2005) Coupling cell proliferation and development in plants. Nat Cell Biol 7 535–541 [DOI] [PubMed] [Google Scholar]
- Han S, Kim D (2006) AtRTPrimer: database for Arabidopsis genome-wide homogeneous and specific RT-PCR primer-pairs. BMC Bioinformatics 7 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He SS, Hoelscher A, Liu J, O'Neill D, Layton J, McCarroll R, Dotson S (2005) Cell cycle specific isopentenyl transferase expression led to coordinated enhancement of cell division, cell growth and plant development in transgenic Arabidopsis. Plant Biotechnol 22 261–270 [Google Scholar]
- Hewelt A, Prinsen E, Thomas M, Van Onckelen H, Meins F (2000) Ectopic expression of maize knotted1 results in the cytokinin-autotrophic growth of cultured tobacco tissues. Planta 210 884–889 [DOI] [PubMed] [Google Scholar]
- Hu Y, Bao F, Li J (2000) Promotive effect of brassinosteroids on cell division involves a distinct CycD3-induction pathway in Arabidopsis. Plant J 24 693–701 [DOI] [PubMed] [Google Scholar]
- Hutchison CE, Li J, Argueso C, Gonzalez M, Lee E, Lewis MW, Maxwell BB, Perdue TD, Schaller GE, Alonso JM, et al (2006) The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 18 3073–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingouff M, Haseloff J, Berger F (2005) Polycomb group genes control developmental timing of endosperm. Plant J 42 663–674 [DOI] [PubMed] [Google Scholar]
- Ito M, Araki S, Matsunaga S, Itoh T, Nishihama R, Machida Y, Doonan JH, Watanabe A (2001) G2/M-phase-specific transcription during the plant cell cycle is mediated by c-Myb-like transcription factors. Plant Cell 13 1891–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Iwase M, Kodama H, Lavisse P, Komamine A, Nishihama R, Machida Y, Watanabe A (1998) A novel cis-acting element in promoters of plant B-type cyclin genes activates M phase-specific transcription. Plant Cell 10 331–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John PC, Mews M, Moore R (2001) Cyclin/Cdk complexes: their involvement in cell cycle progression and mitotic division. Protoplasma 216 119–142 [DOI] [PubMed] [Google Scholar]
- Kang IH, Steffen JG, Portereiko MF, Lloyd A, Drews GN (2008) The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell 20 635–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T, Naitou T, Koizumi N, Yamashino T, Sakakibara H, Mizuno T (2005) Combinatorial microarray analysis revealing Arabidopsis genes implicated in cytokinin responses through the His→Asp phosphorelay circuitry. Plant Cell Physiol 46 339–355 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Miura A, Choi Y, Kinoshita Y, Cao X, Jacobsen SE, Fischer RL, Kakutani T (2004) One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science 303 521–523 [DOI] [PubMed] [Google Scholar]
- Köhler C, Hennig L, Spillane C, Pien S, Gruissem W, Grossniklaus U (2003) The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev 17 1540–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C, Makarevich G (2006) Epigenetic mechanisms governing seed development in plants. EMBO Rep 7 1223–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C, Page DR, Gagliardini V, Grossniklaus U (2005) The Arabidopsis thaliana MEDEA Polycomb group protein controls expression of PHERES1 by parental imprinting. Nat Genet 37 28–30 [DOI] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y (2002) E2Ls, E2F-like repressors of Arabidopsis that bind to E2F sites in a monomeric form. J Biol Chem 277 16553–16558 [DOI] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y (2003) Constitutive E2F expression in tobacco plants exhibits altered cell cycle control and morphological change in a cell type-specific manner. Plant Physiol 132 2012–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le BH, Wagmaister JA, Kawashima T, Bui AQ, Harada JJ, Goldberg RB (2007) Using genomics to study legume seed development. Plant Physiol 144 562–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V, North H, Frey A, Sotta B, Seo M, Okamoto M, Nambara E, Marion-Poll A (2006) Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J 45 309–319 [DOI] [PubMed] [Google Scholar]
- Luo M, Bilodeau P, Dennis ES, Peacock WJ, Chaudhury A (2000) Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc Natl Acad Sci USA 97 10637–10642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lur HS, Setter TL (1993) Role of auxin in maize endosperm development (timing of nuclear DNA endoreduplication, zein expression, and cytokinin). Plant Physiol 103 273–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariconti L, Pellegrini B, Cantoni R, Stevens R, Bergounioux C, Cella R, Albani D (2002) The E2F family of transcription factors from Arabidopsis thaliana: novel and conserved components of the retinoblastoma/E2F pathway in plants. J Biol Chem 277 9911–9919 [DOI] [PubMed] [Google Scholar]
- Mason MG, Li J, Mathews DE, Kieber JJ, Schaller GE (2004) Type-B response regulators display overlapping expression patterns in Arabidopsis. Plant Physiol 135 927–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengel K, Friedrich B, Judel GK (1985) Effect of light intensity on the concentrations of phytohormones in developing wheat grains. J Plant Physiol 120 255–266 [Google Scholar]
- Mewes HW, Dietmann S, Frishman D, Gregory R, Mannhaupt G, Mayer KF, Münsterkötter M, Ruepp A, Spannagl M, Stümpflen V, et al (2008) MIPS: analysis and annotation of genome information in 2007. Nucleic Acids Res 36 D196–D201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Vu TH, Tej SS, Ghazal H, Matvienko M, Agrawal V, Ning J, Haudenschild CD (2004) Analysis of the transcriptional complexity of Arabidopsis thaliana by massively parallel signature sequencing. Nat Biotechnol 22 1006–1011 [DOI] [PubMed] [Google Scholar]
- Mironov V, De Veylder L, Van Montagu M, Inze D (1999) Cyclin-dependent kinases and cell division in plants: the nexus. Plant Cell 11 509–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki K, Matsumoto-Kitano M, Kakimoto T (2004) Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J 37 128–138 [DOI] [PubMed] [Google Scholar]
- Miyawaki K, Tarkowski P, Matsumoto-Kitano M, Kato T, Sato S, Tarkowska D, Tabata S, Sandberg G, Kakimoto T (2006) Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc Natl Acad Sci USA 103 16598–16603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncaleán P, López-Iglesias C, Fernández B, Rodriguez A (2001) Immunocytochemical location of endogenous cytokinins in buds of kiwifruit (Actinidia deliciosa) during the first hours of in vitro culture. Histochem J 33 403–411 [DOI] [PubMed] [Google Scholar]
- Müller B, Sheen J (2007) Arabidopsis cytokinin signaling pathway. Sci STKE 2007 cm5. [DOI] [PubMed] [Google Scholar]
- Nakagami H, Kawamura K, Sugisaka K, Sekine M, Shinmyo A (2002) Phosphorylation of retinoblastoma-related protein by the cyclin D/cyclin-dependent kinase complex is activated at the G1/S-phase transition in tobacco. Plant Cell 14 1847–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson T, Tausta SL, Gandotra N, Liu T (2006) Laser microdissection of plant tissue: what you see is what you get. Annu Rev Plant Biol 57 181–201 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126 467–475 [DOI] [PubMed] [Google Scholar]
- Nordstrom A, Tarkowski P, Tarkowska D, Norbaek R, Astot C, Dolezal K, Sandberg G (2004) Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: a factor of potential importance for auxin-cytokinin-regulated development. Proc Natl Acad Sci USA 101 8039–8044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor TR, Dyreson C, Wyrick JJ (2005) Athena: a resource for rapid visualization and systematic analysis of Arabidopsis promoter sequences. Bioinformatics 21 4411–4413 [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K, Demura T, Fukuda H (2002) Promotion of transcript accumulation of novel zinnia immature xylem-specific HD-Zip III homeobox genes by brassinosteroid. Plant Cell Physiol 43 1146–1153 [DOI] [PubMed] [Google Scholar]
- Olsen OA (2004) Nuclear endosperm development in cereals and Arabidopsis thaliana. Plant Cell 16 S214–S227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormenese S, de Almeida Engler J, De Groodt R, De Veylder L, Inze D, Jacqmard A (2004) Analysis of the spatial expression pattern of seven Kip related proteins (KRPs) in the shoot apex of Arabidopsis thaliana. Ann Bot (Lond) 93 575–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui M, Staehelin LA (2000) Syncytial-type cell plates: a novel kind of cell plate involved in endosperm cellularization of Arabidopsis. Plant Cell 12 933–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat GC, Yu HJ, Ngo QA, Rajani S, Mayalagu S, Johnson CS, Capron A, Xie LF, Ye D, Sundaresan V (2005) Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132 603–614 [DOI] [PubMed] [Google Scholar]
- Parenicová L, de Folter S, Kieffer M, Horner DS, Favalli C, Busscher J, Cook HE, Ingram RM, Kater MM, Davies B, et al (2003) Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15 1538–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlidis P, Noble WS (2001) Analysis of strain and regional variation in gene expression in mouse brain. Genome Biol 2 RESEARCH0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Parra E, Frundt C, Gutierrez C (2003) A genome-wide identification of E2F-regulated genes in Arabidopsis. Plant J 33 801–811 [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra E, Lopez-Matas MA, Frundt C, Gutierrez C (2004) Role of an atypical E2F transcription factor in the control of Arabidopsis cell growth and differentiation. Plant Cell 16 2350–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte AM, Mason MG, Hutchison CE, Ferreira FJ, Schaller GE, Kieber JJ (2006) A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc Natl Acad Sci USA 103 11081–11085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SY, Beavis W, Berardini TZ, Chen G, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, Montoya M, et al (2003) The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res 31 224–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37 501–506 [DOI] [PubMed] [Google Scholar]
- Scott RJ, Spielman M, Bailey J, Dickinson HG (1998) Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 125 3329–3341 [DOI] [PubMed] [Google Scholar]
- Sorensen MB, Mayer U, Lukowitz W, Robert H, Chambrier P, Jurgens G, Somerville C, Lepiniec L, Berger F (2002) Cellularisation in the endosperm of Arabidopsis thaliana is coupled to mitosis and shares multiple components with cytokinesis. Development 129 5567–5576 [DOI] [PubMed] [Google Scholar]
- Sozzani R, Maggio C, Varotto S, Canova S, Bergounioux C, Albani D, Cella R (2006) Interplay between Arabidopsis activating factors E2Fb and E2Fa in cell cycle progression and development. Plant Physiol 140 1355–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer MW, Casson SA, Lindsey K (2007) Transcriptional profiling of the Arabidopsis embryo. Plant Physiol 143 924–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stangeland B, Salehian Z (2002) An improved clearing method for GUS assay in Arabidopsis endosperm and seeds. Plant Mol Biol Rep 20 107–114 [Google Scholar]
- Świątek A, Lenjou M, Van Bockstaele D, Inzé D, Van Onckelen H (2002) Differential effect of jasmonic acid and abscisic acid on cell cycle progression in tobacco BY-2 cells. Plant Physiol 128 201–211 [PMC free article] [PubMed] [Google Scholar]
- Tiwari S, Spielman M, Day RC, Scott RJ (2006) Proliferative phase endosperm promoters from Arabidopsis thaliana. Plant Biotechnol J 4 393–407 [DOI] [PubMed] [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ (2005) The botany array resource: e-northerns, expression angling, and promoter analyses. Plant J 43 153–163 [DOI] [PubMed] [Google Scholar]
- Tzafrir I, Dickerman A, Brazhnik O, Nguyen Q, McElver J, Frye C, Patton D, Meinke D (2003) The Arabidopsis SeedGenes Project. Nucleic Acids Res 31 90–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, Raes J, De Veylder L, Rouzé P, Rombauts S, Inzé D (2002) Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14 903–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, Vlieghe K, Florquin K, Hennig L, Beemster GT, Gruissem W, Van de Peer Y, Inze D, De Veylder L (2005) Genome-wide identification of potential plant E2F target genes. Plant Physiol 139 316–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlieghe K, Boudolf V, Beemster GT, Maes S, Magyar Z, Atanassova A, de Almeida Engler J, De Groodt R, Inze D, De Veylder L (2005) The DP-E2F-like gene DEL1 controls the endocycle in Arabidopsis thaliana. Curr Biol 15 59–63 [DOI] [PubMed] [Google Scholar]
- Wang G, Kong H, Sun Y, Zhang X, Zhang W, Altman N, DePamphilis CW, Ma H (2004) Genome-wide analysis of the cyclin family in Arabidopsis and comparative phylogenetic analysis of plant cyclin-like proteins. Plant Physiol 135 1084–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingartner M, Criqui MC, Mészáros T, Binarova P, Schmit AC, Helfer A, Derevier A, Erhardt M, Bögre L, Genschik P (2004) Expression of a nondegradable cyclin B1 affects plant development and leads to endomitosis by inhibiting the formation of a phragmoplast. Plant Cell 16 643–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L, Zhao J, Morozova N, Li Y, Avivi Y, Grafi G (2003) Chromatin reorganization accompanying cellular dedifferentiation is associated with modifications of histone H3, redistribution of HP1, and activation of E2F-target genes. Dev Dyn 228 113–120 [DOI] [PubMed] [Google Scholar]
- Yanagisawa S (2002) The Dof family of plant transcription factors. Trends Plant Sci 7 555–560 [DOI] [PubMed] [Google Scholar]
- Yang J, Zhang J, Huang Z, Wang Z, Zhu Q, Liu L (2002) Correlation of cytokinin levels in the endosperms and roots with cell number and cell division activity during endosperm development in rice. Ann Bot (Lond) 90 369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhang J, Wang Z, Liu K, Wang P (2006) Post-anthesis development of inferior and superior spikelets in rice in relation to abscisic acid and ethylene. J Exp Bot 57 149–160 [DOI] [PubMed] [Google Scholar]
- Yoshizumi T, Nagata N, Shimada H, Matsui M (1999) An Arabidopsis cell cycle-dependent kinase-related gene, CDC2b, plays a role in regulating seedling growth in darkness. Plant Cell 11 1883–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, et al (2006) Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126 1189–1201 [DOI] [PubMed] [Google Scholar]
- Zhao J, Morozova N, Williams L, Libs L, Avivi Y, Grafi G (2001) Two phases of chromatin decondensation during dedifferentiation of plant cells: distinction between competence for cell fate switch and a commitment for S phase. J Biol Chem 276 22772–22778 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.