Abstract
The Notch signal transduction pathway regulates the decision to proliferate versus differentiate. Although there are a myriad of mouse models for the Notch pathway, surprisingly little is known about how these genes regulate early eye development, particularly in the anterior lens. We employed both gain-of-function and loss-of-function approaches to determine the role of Notch signaling in lens development. Here we analyzed mice containing conditional deletion of the Notch effector Rbpj or overexpression of the activated Notch1 intracellular domain during lens formation. We demonstrate distinct functions for Notch signaling in progenitor cell growth, fiber cell differentiation and maintenance of the transition zone. In particular, Notch signaling controls the timing of primary fiber cell differentiation and is essential for secondary fiber cell differentiation. Either gain or loss of Notch signaling leads to formation of a dysgenic lens, which in loss-of-function mice undergoes a profound postnatal degeneration. Our data suggest both Cyclin D1 and Cyclin D2, and the p27Kip1 cyclin-dependent kinase inhibitor act downstream of Notch signaling, and define multiple critical functions for this pathway during lens development.
Keywords: lens development, Notch signaling, Rbpj, activated Notch, growth, cyclins, CKI
INTRODUCTION
Cellular organization into patterned structures is fundamental during animal development, with growth, patterning, morphogenesis and differentiation essential components of this process. Each event is spatiotemporally integrated, ensuring tissues and organs achieve proper size, shape and composition. Like other tissues and organs, vertebrate lens development requires careful coordination of these four components. Epithelial cells in the ocular lens undergo two temporally distinct modes of differentiation into fiber cells. First, primary fiber cells differentiate shortly after the lens invaginates from a placode to a lens pit and then into a lens vesicle. At this time, posterior lens progenitors closest to the central retina exit the cell cycle and initiate fiber cell differentiation, which is marked by the expression of alpha, beta, and gamma crystallin genes, and other fiber cell components. Secondary fiber cell differentiation directly follows, and is distinguished by a concerted migration of lens anterior epithelial layer (AEL) cells around the periphery to the equatorial region, wherein cells exit mitosis and migrate into the central lens. This equatorial region of the lens, where multiple signaling molecules converge on lens precursors, constitutes the transition zone, which remains the organizing center of lens fiber differentiation throughout the life of a vertebrate organism.
For both primary and secondary fiber cell differentiation, a highly conserved hierarchy of transcription factors orchestrates terminal differentiation into enucleated and organelle-deficient lens fiber cells. These final steps are critical for normal vision, as light must pass through an optically transparent lens to activate phototransduction within the retina. The transcription factors Pax6, Prox1, Maf, and Sox1 are essential regulators in the lens, since they directly regulate crystallin expression and fiber cell differentiation is blocked in their absence (Ashery-Padan et al., 2000; Cvekl et al., 1995; Glaser et al., 1994; Grindley et al., 1995; Kim et al., 1999; Nishiguchi et al., 1998; Ring et al., 2000; Wigle et al., 1999). These same factors, most notably Prox1, each promote the expression of cell cycle inhibitory molecules, including the Cyclin-dependent kinase inhibitors (CKIs) p27Kip1 (Cdkn1b) and p57Kip2 (Cdkn1c) (Wigle et al., 1999). CKIs have complex functions in the cell cycle, not only to inhibit Cyclin-CDK function, but also to promote S-phase in a context-dependent fashion (Besson et al., 2007). Lens cells lacking both p27Kip1 and p57Kip2 are unable to exit the cell cycle at the transition zone and fail to terminally differentiate and elongate, resulting in a propensity for apoptotic cell death via a p53-dependent pathway (Zhang et al., 1998). However, lens cell mitogens have remained elusive, either because they act redundantly or are broadly required throughout the body, thereby causing early embryonic lethality when mutated. Nonetheless, in vivo misexpression studies have pointed to Cyclin D1 (Ccnd1), Cyclin D2 (Ccnd2), and Cdk4 as likely targets of such a pro-mitogenic pathway (Gómez Lahoz et al., 1999).
FGF and BMP signaling at the transition zone are critical for lens fiber cell differentiation and survival (Beebe et al., 2004; Belecky-Adams et al., 2002; Faber et al., 2002; Robinson, 2006). Also, signaling through a Ras-MAPK pathway regulates some aspects of lens proliferation (Lovicu and McAvoy, 2001). Activated Ras signaling, by transgenic misexpression of dominant-active H-Ras, or the upstream ligand Pdgfa, causes over-proliferation of the lens epithelium (Reneker and Overbeek, 1996; Reneker et al., 2004). Conversely, transgenic expression of dominant-negative H-Ras impairs lens growth, thereby causing a small (microphthalmic) lens (Xie et al., 2006). Surprisingly, these perturbations in lens proliferation do not result in fiber cell defects, suggesting that other molecular pathway(s) coordinate the decision to proliferate versus differentiate.
The Notch signal transduction pathway is one of the major metazoan signaling networks. Canonical activation of this pathway occurs when a Notch receptor is engaged from a neighboring cell via the Delta-like (Dll) or Jagged (Jag) ligands. The Notch receptor undergoes proteolytic cleavage that liberates an intracellular domain (NotchIC), which translocates to the nucleus and acts in a transcriptional complex with Mastermind (Maml) and the Rbpj DNA-binding transcription factor (also known as RBP-Jκ1, CSL, or CBF-1) to activate Hairy-related transcriptional repressors (Fischer and Gessler, 2007; Ilagan and Kopan, 2007). Notch activation generally prevents differentiation and maintains progenitor or stem cell proliferation and is a classical mediator of lateral inhibition during cell fate determination (Bolós et al., 2007; Yoon and Gaiano, 2005). But Notch signaling has diverse, almost unlimited cellular outcomes, since it can regulate cell cycle progression, survival, fate determination, and morphogenesis in different organs and cellular contexts (Artavanis-Tsakonas et al., 1999; Thomas, 2005).
In the CNS and pancreas, disruption of Notch signaling causes premature progenitor cell differentiation, often leading to altered timing of differentiation of early-born cell types and a rapid depletion of the progenitor pool. Such phenotypes in the CNS, retina, and pancreas occur in mice lacking the Notch effector gene, Hes1 (Hatakeyama et al., 2004; Ishibashi et al., 1995; Jensen et al., 2000; Kageyama et al., 2000; Lee et al., 2005; Tomita et al., 1996). In a recent study of frog lens induction, Ogino and colleagues demonstrated that a Delta1-Notch signal from the optic vesicle to the lens placode helps regulates the progression of lens induction via Otx2 and Rbpj-mediated activation of Foxe3 transcription (Ogino et al., 2008). Intriguingly, Hes1 mutant mice also display defects in early lens development that range from complete loss to a microphthalmic lens, with reduced proliferation as early as the lens pit stage (Lee et al., 2005; Tomita et al., 1996). Recently, a conditional deletion of Rbpj in the developing lens was reported, resulting in a smaller lens and possible upregulation of p57Kip2 (Jia et al., 2007). The minor alterations in fiber cell differentiation reported in this study are inconsistent with the Hes1 mutant phenotype, and thus do not fully resolve the question of what processes Notch signaling regulates in the lens.
Here, we evaluate the consequences of both loss and gain of Notch signaling during mammalian lens development. Mice lacking Notch signaling, through tissue-specific removal of Rbpj, exhibit accelerated primary fiber cell differentiation and hypoproliferation accompanied by reduced levels of Pax6, Cyclin D1, and Cyclin D2. These defects result in the essentially complete loss of the lens (aphakia) in postnatal Rbpj conditionally mutant mice. Moreover, mice with constitutive Notch signaling through tissue-specific expression of the Notch1IC, show abnormal lens morphogenesis, hyperproliferation of the AEL, and inappropriate maintenance of Pax6 and other AEL-expressed genes. This causes severely delayed primary fiber cell differentiation. In both genetic manipulations of Notch signaling, the transition zone is malformed and secondary fiber cell differentiation is lost. Together, our data demonstrate that Notch signaling is essential for lens growth and differentiation.
MATERIALS AND METHODS
Animals
Rosa26Notch1IC mice were described previously (Murtaugh et al., 2003) and maintained as homozygotes. The P0-3.9-GFPCre construct was generated by replacing the NotI fragment containing the lacZ reporter from P0-3.9-lacZ (Zhang et al., 2002) with an XhoI-XbaI fragment, containing GFPCre from pBS-592 (Le et al., 1999). These regulatory elements are largely overlapping those of Le-Cre (including the EE). The linearized insert was injected into the male pronuclei of fertilized FVB eggs using standard techniques. The P0-3.9-GFPCre line is maintained on an FVB background and genotyped using a standard PCR protocol. Rbpjtm1Hon mice (termed RbpjCKO), were generated by Han et al., and maintained on a 129/SvJ background and genotyped as described (Han et al., 2002). Le-Cre mice, generated by Ashery-Padan et al., were maintained on a CD-1 background and PCR genotyped as described (Ashery-Padan et al., 2000). Images of adult heads or eyeballs were captured with a Leica dissecting microscope and Optronics digital camera.
Tissue Analyses
Embryonic and postnatal tissue was fixed in 4% paraformaldehyde/PBS for 15 minutes – 1 hour at 4°C and processed by stepwise sucrose/PBS incubation for 10 µm frozen sections in OCT by standard techniques. Primary antibodies used include anti-BrdU (BD Laboratories clone B44 1:100 or Serotec clone BU1/75 1:500), anti-cleaved PARP (Cell Signaling 1:500), anti-Cre (Novagen 1:5000), anti-Cyclin D1 (Neomarkers SP4 1:100; Sigma DCS-6 1:100 or Santa Cruz 72-13G 1:500), anti-Cyclin D2 (Santa Cruz 34B1-3 1:200), anti-E cadherin (Zymed ECCD-2 1:500), anti-Foxe3 (gift from Peter Carlsson 1:1000), anti-beta crystallin (gift from Richard Lang 1:8000), anti-gamma crystallin (Santa Cruz 1:1000), anti-GFP (Molecular Probes 1:1000 or Abcam 1:1000), anti-Hes1 (1:1000), anti-Jagged1 (Santa Cruz 1:1000), anti-p27Kip1 (BD Laboratories Clone 57 1:100 or Assay Designs 1:500), anti-p57Kip2 (Abcam 1:500 or Santa Cruz 1:50), anti-Pax6 (Covance 1:1000 or DSHB 1:20), anti-Prox1 (Covance 1:1000 or Chemicon 1:2000), anti-Pitx3 (gift from Marten Smidt 1:1000), anti-Six3 (gift from Guillermo Oliver 1:1000), anti-Sox1 (Affinity BioReagents 1:500), and anti-Sox2 (Chemicon 1:500). Detailed staining protocols are available upon request and generally followed those in (Lee et al., 2005) and (Zhang et al., 2003). Secondary antibodies were generated in donkey or goat versus the appropriate species and directly conjugated with Cy3 (Jackson Immunologicals), Alexa Fluor 488, Alexa Fluor 594 (Molecular Probes) or biotinylated (Jackson Immunologicals) and sequentially labeled with streptavidin Alexa 488 or 594 (Molecular Probes). Labeled sections were visualized with a Zeiss fluorescent microscope equipped with either a Leica or Zeiss camera and Apotome deconvolution device. Whole-mount or cryosection in situ hybridization was performed as described (Brown et al., 1998) using Rbpj, Hes1, Notch1, Notch2 and Jagged1 digoxygenin-labeled antisense riboprobes. For S-phase analyses, BrdU (Sigma) was injected intraperitoneally as described (Mastick and Andrews, 2001) and animals were sacrificed 1.5–4 hours later for tissue processing. Tissue sections were treated with 2N hydrochloric acid prior to standard antibody staining. TUNEL staining was performed using the in situ cell death detection kit, fluorescein according to the manufacturer’s instructions (Roche). Standard histologic staining of frozen or paraffin embedded sections was also performed. All images were processed using Axiovision (v5.0) and/or Adobe Photoshop software (v7.0) and manipulated electronically to adjust brightness and contrast as well as pseudocoloring.
Cell Counting
Tissue sections, separated by at least 60 µm, were antibody-stained and counted using NIH ImageJ or Axiovision software. Between 3–5 animals were analyzed per genotype and age and at least two independent sections through the center of the lens per animal quantified. Labeling indices were generated by dividing the number of antibody-positive cells by total DAPI-labeled nuclei, and compared against one other additional genotype using the Student T-test, or among multiple genotypes using Instat (v3.0) software to perform ANOVA and a Bonferroni posthoc test to determine p values.
Cell Culture
17EM15 mouse lens epithelial cells were cultured and passaged as described previously (Donner et al., 2007). 17ppuro cells were generated by transfecting 17EM15 cells with pPur DNA (Clontech) using Fugene according to manufacturer’s protocols. Cells were selected with 3.6 µg/mL puromycin and surviving cell colonies were pooled and passaged in the presence of 2 µg/mL puromycin. 17NotchΔE cells were generated similarly using pCS2+NotchΔE-pPur DNA, which was generated by cloning the PvuII-BamHI insert of pPur into pCS2+NotchΔE (Kopan et al., 1996).
RESULTS
Loss of Notch function via inactivation of Rbpj in the lens causes postnatal lens degeneration
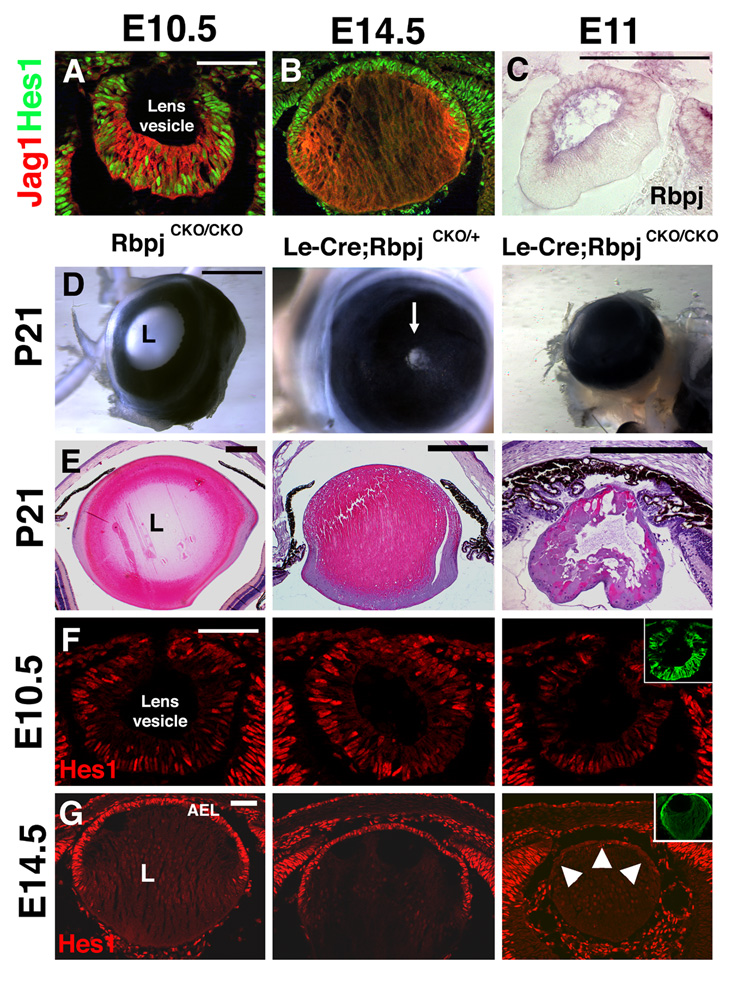
Multiple papers have reported Notch pathway gene expression in the embryonic eye, which indicated expression of Notch1, Notch2, and Jag1 in the developing lens. (Bao and Cepko, 1997; Bettenhausen et al., 1995; Ishibashi et al., 1995; Weinmaster et al., 1991; Weinmaster et al., 1992). To extend our previous observations that Hes1 is expressed in the developing eye (Lee et al., 2005), we performed double antibody staining for Hes1 and Jag1. Both factors colocalize during primary fiber cell formation (Fig. 1A), but show complementary expression domains after AEL formation (Fig. 1B). Rbpj, a downstream Notch effector that activates Hes1 (Jarriault et al., 1995), has a similar broad lens expression domain within the lens vesicle that becomes restricted to the AEL (Fig. 1C and data not shown).
Figure 1. Loss of Notch function via lens-specific deletion of Rbpj causes lens degeneration.
(A,B) Jag1 and Hes1 proteins are co-expressed in the lens pit at E10.5 (A), but exhibit complementary expression at E14.5 (B). Jag1 protein appears graded across the lens fibers, with highest expression at the equator and where fiber cells abut the AEL. (C) In situ hybridization for Rbpj RNA at E11 shows expression becoming restricted to the forming AEL. (D) P21 Le-Cre;RbpjCKO/+ eyes have a reduced pupillary opening (arrow) that is completely lacking in Le-Cre;RbpjCKO/CKO mutants. (E) Histological analysis of P21 eyes shows that Le-Cre;RbpjCKO/CKO eyes are aphakic and lack an anterior chamber, with rare remnants of lens tissue found in a subset of sections. (F,G) Hes1 protein is present mosaically in the Le-Cre;RbpjCKO/CKO lens at E10.5 (F), but is completely absent in the AEL at E14.5 (arrowheads, G). Cre expression reported by anti-GFP labeling is in the insets. Anterior is up in A-G; L = lens. Bar in A,C,F,G = 20 microns, in D = 500 microns, and in E = 5 microns (note different sized bars among genotypes).
Because Notch1 and Notch2 may function redundantly during lens development, we conditionally deleted the common Notch effector gene Rbpj, using the Le-Cre transgenic mouse line, where Cre recombinase and Green Fluorescent Protein (GFP) are expressed in the lens epithelium via the Pax6 ectoderm enhancer (EE) (Ashery-Padan et al., 2000). Le-Cre mice were crossed to RbpjCKO mice, in which exons 6 and 7 encoding the DNA binding domain are flanked by loxP sites (Han et al., 2002). Cre-mediated excision via the Le-Cre mouse in this study occurred mainly after lens induction (Fig. 1F), allowing us to test the role of canonical Notch signaling specifically during lens growth and differentiation.
Adult Le-Cre;RbpjCKO/+ and Le-Cre;RbpjCKO/CKO mice were generated in the expected Mendelian ratios. Le-Cre;RbpjCKO/+ and Le-Cre;RbpjCKO/CKO 3-week old mice have profound eye defects (Fig. 1D,E; n= 11/12 CKO hets and 17/17 CKO mutants). Le-Cre;RbpjCKO/+ animals are microphthalmic, with variable, but reduced pupillary eye openings, while Le-Cre;RbpjCKO/CKO mice have no visible external eye and completely lack pupillary eye openings (Fig. 1D). Histological sections from E18.5 to P21 littermates indicate a progressive loss of the lens, termed aphakia (Fig. S1), and by P21, Le-Cre;RbpjCKO/CKO lenses are largely disintegrated, with only rare remnants of disorganized lens tissue (Fig. 1E).
Hes1 expression, as a readout of Notch signaling, is mosaic in E10.5 Le-Cre;RbpjCKO/CKO lenses (Fig. 1F). This indicates either incomplete excision of RbpjCKO or an alternate Rbpj-independent mode of expression. However, by E14.5, Hes1 expression almost completely lost from the Le-Cre;RbpjCKO/CKO lenses, but not in the retina, suggesting conditional deletion of Rbpj had now occurred, and that Rbpj is required for Hes1 expression in the AEL (Fig. 1G).
Fiber cell differentiation occurs precociously in Le-Cre;RbpjCKO/CKO embryos
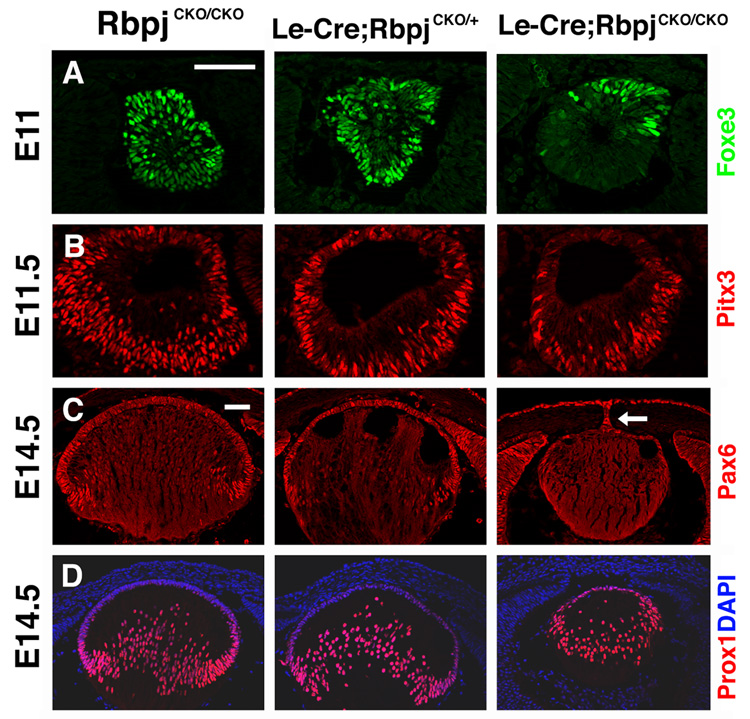
To understand the developmental events that caused aphakia in Le-Cre;RbpjCKO/CKO mice, we analyzed the expression of key lens developmental regulatory proteins. Foxe3 and Pitx3 encode transcription factors required for lens formation in humans and mice respectively (Blixt et al., 2000; Medina-Martinez et al., 2005; Rieger et al., 2001; Semina et al., 2000; Shi et al., 2005; Valleix et al., 2006). In Le-Cre;RbpjCKO/CKO lens vesicles, Foxe3 and Pitx3 expression domains are reduced in complementary patterns, with Foxe3 specifically lost in the posterior and Pitx3 lost in the anterior lens (Fig. 2A,B). Reduction of Foxe3 is particularly striking, since this factor is expressed by lens progenitors from E9.5 to adulthood and required for their proliferation (Blixt et al., 2000). Therefore, Foxe3 loss in Le-Cre;RbpjCKO/CKO lens vesicles strongly suggests premature differentiation of primary fiber cells, as occurs in Foxe3dyl/dyl and Foxe3−/− mutants (Blixt et al., 2000; Medina-Martinez et al., 2005). E14.5 Le-Cre;RbpjCKO/CKO lenses are markedly smaller than their littermates and exhibit dramatic reduction, but not total absence of Pax6, Sox2 and Six3 from the AEL (Fig. 2C and data not shown). In some Le-Cre;RbpjCKO/CKO eyes, a remnant lens stalk is observed between the AEL and cornea (arrow in Fig. 2C), a phenotype common to Pax6Sey/+, Foxe3dyl/dyl, and Pitx3ak/ak lenses (Blixt et al., 2000; Grimm et al., 1998; Makhani et al., 2007).
Figure 2. Lens-specific deletion of Rbpj affects expression of lens regulatory proteins.
(A) Antibody staining for Foxe3 shows dramatic downregulation in posterior Le-Cre;RbpjCKO/CKO E11 lenses while (B) Pitx3 is downregulated in anterior Le-Cre;RbpjCKO/CKO E11.5 lenses. (C) Late-phase Pax6 AEL expression is almost completely reduced at E14.5. The arrow points to a remnant lens stalk that is Pax6-positive. (D) Early differentiation marker Prox1 is inappropriately retained by anterior epithelial cells and highlights posterior fiber cell disorganization, in Le-Cre;RbpjCKO/CKO mutant lenses. Bar in A,C = 20 microns, anterior is up in A–D.
We analyzed the state of fiber cell differentiation at E14.5 by examining the transcription factor Prox1, which directly activates βB1-crystallin in the chick lens (Cui et al., 2004) and is required for lens fiber cell elongation in mice (Wigle et al., 1999). Prox1 activates normally in Le-Cre;RbpjCKO/+ and Le-Cre;RbpjCKO/CKO fiber cells indicating that early steps of fiber cell differentiation are unperturbed by loss of Rbpj function (Fig 2D). Consistent with this finding, β-crystallin is expressed normally in Rbpj mutant lenses (Fig S3). However, the progression of primary fiber cell development is abnormal in Le-Cre;RbpjCKO/CKO lenses, since AEL cells inappropriately express Prox1 as compared to RbpjCKO/CKO control and Le-Cre;RbpjCKO/+ littermates (Fig 2D). This occurs well ahead of the eventual degeneration of Le-Cre;RbpjCKO/CKO lenses.
Gain of function Notch signaling in the lens via constitutive expression of Notch1IC
Although our Rbpj conditional loss-of-function experiments show that Notch signaling is required for development of the mammalian lens, we wished to identify which specific steps of lens development Notch function is sufficient to regulate. Thus, we took advantage of a constitutively activated Notch1 allele (Rosa26Notch1IC), whose expression is initiated by Cre recombinase to drive high levels of Notch1IC (Murtaugh et al., 2003). For these experiments, we employed a novel transgenic mouse line that expresses a GFP-Cre fusion protein in the AEL under control of Pax6 regulatory elements present in the Pax6 P0 promoter and 3.9 Kb upstream region (P0-3.9-GFPCre). E14.5 P0- 3.9-GFPCre;Rosa26Notch1IC eyes are roughly normal in size, but at E18.5, are microphthalmic and the lens fails to opacify following fixation, suggesting defective fiber cell differentiation (Fig S2).
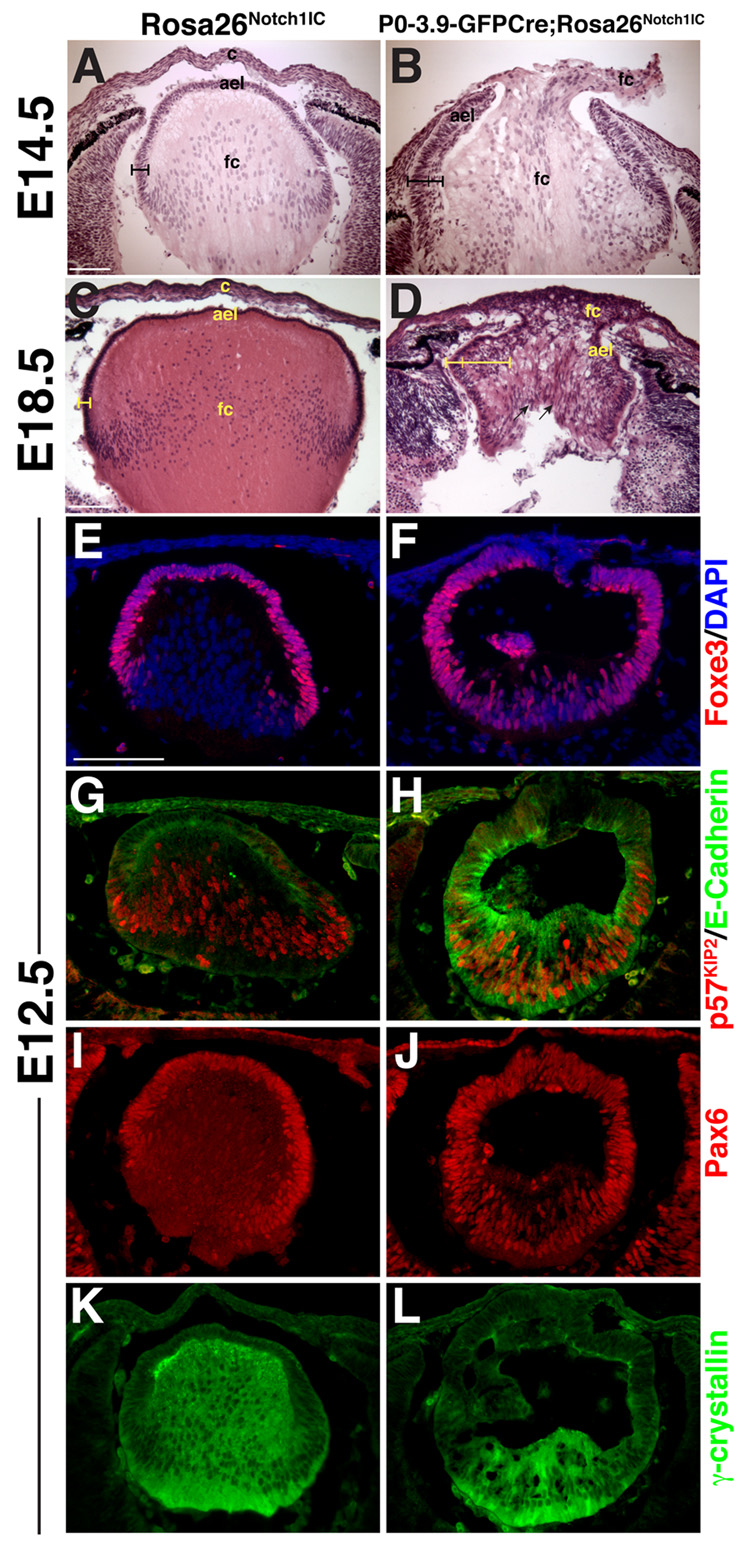
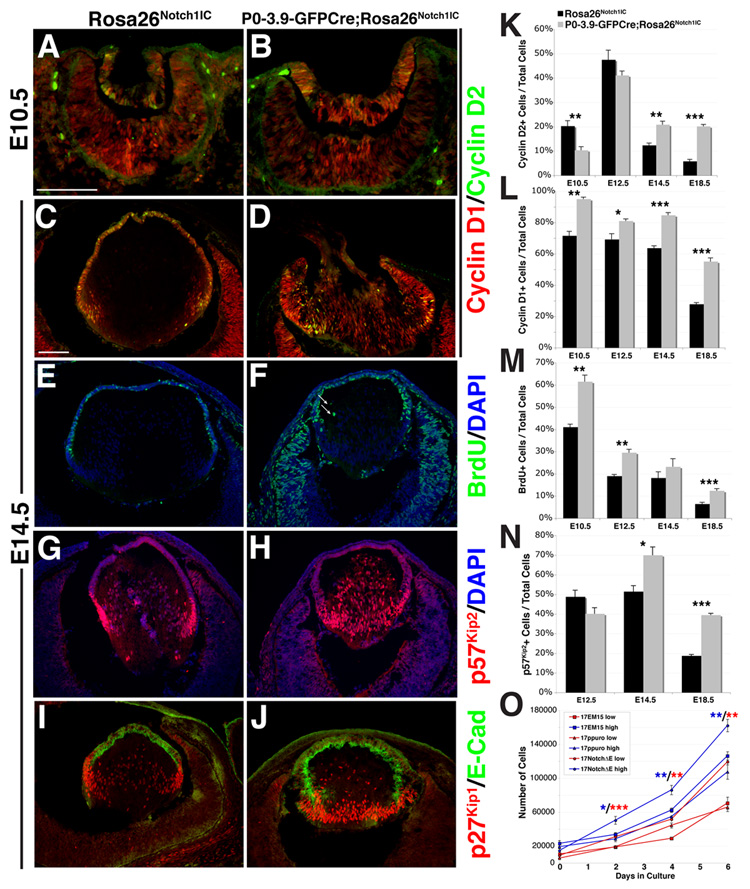
In E14.5-E18.5 histologic sections, P0-3.9-GFPCre;Rosa26Notch1IC lens AEL tissue is dramatically thickened into two distinct layers (brackets in Fig. 3A–D). In severely affected lenses, the cornea and AEL remain contiguous, leading to a large central opening where cells overflow from the lens cavity (Fig. 3B,D). From E14.5 onwards, the P0-3.9-GFPCre;Rosa26Notch1IC AEL remains inappropriately attached to the cornea.
Figure 3. Constitutive expression of Notch signaling alters lens growth and differentiation.
(A–D) Histological sections from the P0-3.9-GFPCre;Rosa26Notch1IC eyes show a thickening and multilayering of the AEL (indicated by brackets), as well as loss of a definitive cornea. At E18.5 (C,D), radially-aligned nuclei are observed in the P0-3.9-GFPCre;Rosa26Notch1IC lens (arrows in D). ael–anterior epithelial layer, c–cornea, fc–presumptive fiber cells. (E,F) Foxe3, (G,H) E-Cadherin, and (I,J) Pax6 expression inappropriately persist in E12.5 posterior P0-3.9-GFPCre;Rosa26Notch1IC lenses, indicating delayed primary fiber cell differentiation, accompanied by a (K,L) highly reduced γ-crystallin domain. (G,H) p57Kip2 initiates normally in the P0-3.9-GFPCre;Rosa26Notch1IC posterior lens in E-Cadherin-positive cells. Bar in A,C,E = 100 microns, anterior is up in A–L.
Delayed primary fiber cell differentiation and loss of secondary fiber cells in P0-3.9-GFPCre;Rosa26Notch1IC lenses
To understand if Notch1 constitutive activation affects primary fiber genesis oppositely to Rbpj loss-of-function, we looked at E12.5 when only primary fiber cells are differentiating and can be distinguished by their loss of the AEL markers Foxe3 and E-Cadherin. In E12.5 P0-3.9-GFPCre;Rosa26Notch1IC lenses, both Foxe3 (Fig. 3E,F) and E-Cadherin (Fig. 3G,H) expression are retained in presumptive primary fiber cells. Similarly, Pax6 is incompletely repressed in the posterior lens (Fig. 3I,J). E12.5, P0-3.9-GFPCre;Rosa26Notch1IC lenses also have dramatically reduced γ-crystallin domains (Fig. 3K,L). Expression of the CKI p57Kip2 is associated with cell cycle exit of primary fiber cells. In P0-3.9-GFPCre;Rosa26Notch1IC lenses, p57Kip2 expression initiates normally (Fig. 3G,H), but fiber cells that do form have defective elongation, similar to the phenotypes of Sox1, Prox1 and Maf mouse mutants (Kim et al., 1999; Nishiguchi et al., 1998; Ring et al., 2000; Wigle et al., 1999).
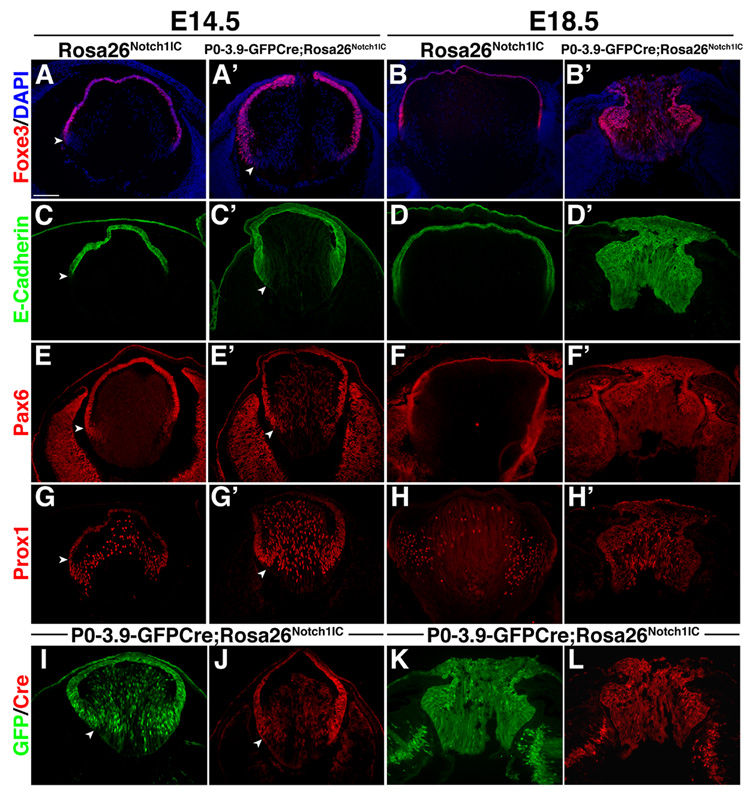
To address secondary fiber cell differentiation, we analyzed regulatory proteins later in development. Foxe3 (Fig. 4A,B) and E-Cadherin (Fig. 4C,D), are increased and posteriorly shifted in P0-3.9-GFPCre;Rosa26Notch1IC eyes. However, the sharp AEL/fiber cell boundary that these proteins highlight is maintained (arrowheads in Fig. 4), indicating that primary fiber cell specification does finally occur. By E18.5, P0-3.9-GFPCre;Rosa26Notch1IC lenses display an even greater expansion of the Foxe3 and E-Cadherin domains, including within the posterior lens, at the expense of presumptive secondary fiber cells (Fig. 4B,D). Similarly, other AEL and transition zone markers, Pax6, Hes1, and Sox2 (Fig. 4E,F and data not shown), are expanded and inappropriately expressed in the posterior lens in E14.5 and E18.5 P0-3.9-GFPCre;Rosa26Notch1IC lenses. In particular, Pax6 is strongly derepressed in E14.5 P0-3.9-GFPCre;Rosa26Notch1IC fiber cells (Fig. 4E’), as is the Pax6 EE (evidenced by Cre or GFP expression in (Fig. 4I–L). This suggests that Pax6 perdurance may occur at the transcriptional level in P0-3.9-GFPCre;Rosa26Notch1IC. Prox1 displays normal nuclear expression in E14.5 P0-3.9-GFPCre;Rosa26Notch1IC fiber cells (Fig. 4G), along with activation of γ-crystallin expression (Fig. S3), but at E18.5 Prox1 is co-expressed with E-Cadherin in P0-3.9-GFPCre;Rosa26Notch1IC lenses (Fig. 4D’,H’), a pattern normally only seen in the transition zone (Fig. 4H). Overall, alterations in AEL and fiber cells marker expression, combined with histologic analyses, show disrupted fiber cell differentiation in P0-3.9-GFPCre;Rosa26Notch1IC lenses.
Figure 4. Constitutive expression of Notch signaling alters expression of lens regulatory proteins.
(A,B) Lens epithelial cells expressing Foxe3 or (C,D) E-Cadherin are inappropriately expanded posteriorly in E14.5 and E18.5 P0-3.9-GFPCre;Rosa26Notch1IC lenses. Arrowheads denote the boundary between the AEL and fiber cells at E14.5. (E,F) Pax6 expression persists in E14.5 or E18.5 P0-3.9-GFPCre;Rosa26Notch1IC posterior lenses. (G,H) Prox1 is highly expressed in E14.5 and E18.5 P0-3.9-GFPCre;Rosa26Notch1IC posterior lenses. (I,K) GFP fluorescence or (J,L) Cre protein directed from P0-3.9-GFPCre is expressed throughout the AEL and posterior lens of E14.5 and E18.5 P0-3.9-GFPCre;Rosa26Notch1IC lenses. Some GFP expression may be directed from the Notch1IC allele, while Cre is only directed from P0-3.9-GFPCre. Note that sections in I and J are from different lenses, while K and L are colabelings of the same section. Bar in A = 100 microns; anterior is up in A–L.
Both primary and secondary fiber cell differentiation require Notch signaling
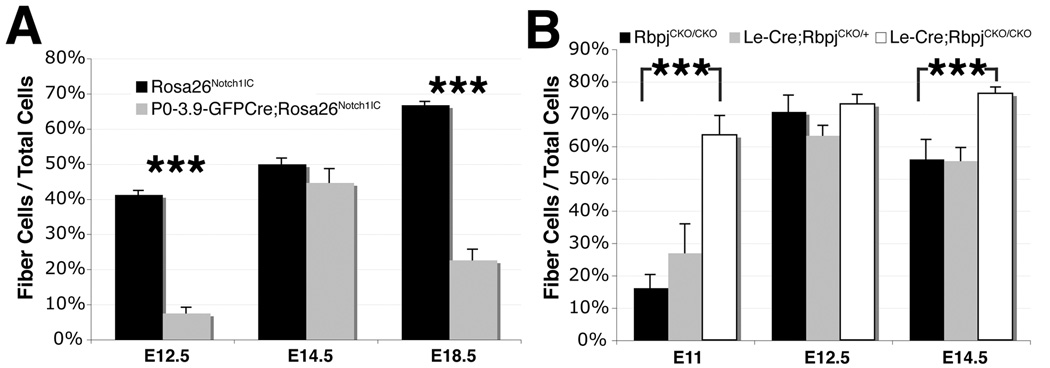
To assess changes in fiber cell differentiation directly, we quantified the Foxe3-negative cell population as a proxy for fiber cells, throughout the P0-3.9-GFPCre;Rosa26Notch1IC lens, at several ages between E12.5 and E18.5 (Fig. 5A). In the presence of excess activated Notch, the number of fiber cells is reduced at E12.5, but rebounds to wildtype numbers by E14.5 (Fig. 5A). At E18.5, however, fiber cell numbers are again significantly reduced in P0-3.9-GFPCre;Rosa26Notch1IC lenses, indicating distinct defects during primary and secondary fiber cell differentiation. Likewise, we also quantified fiber cells in Le-Cre;RbpjCKO/+ and Le-Cre;RbpjCKO/CKO lenses across a comparable time period (Fig. 5B). In contrast to the above delay in fiber cell differentiation, we observe a striking increase in fiber cells in Le-Cre;RbpjCKO/CKO lenses at E11. Then at E14.5, Le-Cre;RbpjCKO/CKO lenses undergo a second increase in fiber cells (Fig. 5B). Although alterations in fiber cell numbers onset at slightly different ages, conditional deletion of Rbpj and overexpression of activated Notch1IC affect fiber cell differentiation oppositely. Together, these data indicate that Notch signaling acts during both primary and secondary fiber cell differentiation.
Figure 5. Both gain and loss of Notch function affect primary and secondary fiber cell formation.
(A,B) Quantification of the percentage of fiber cells (Foxe3-negative/total cells) at the indicated times in P0-3.9-GFPCre;Rosa26Notch1IC (A) and Le-Cre;RbpjCKO/CKO lenses (B). Bar graphs show mean + s.e.m. *** = P<0.001.
Lens cell cycle progression requires Notch signaling
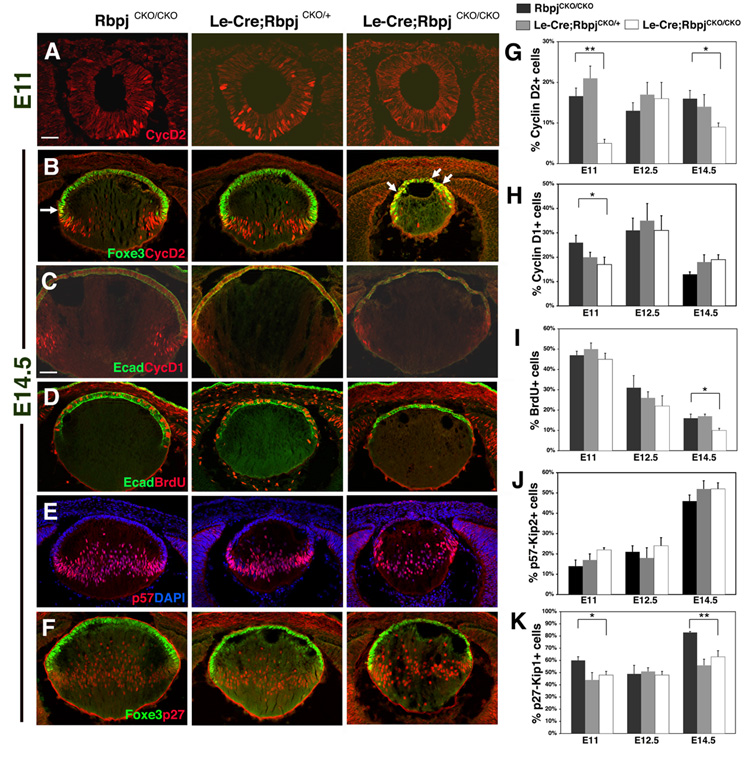
In both the Rbpj loss-of-function and Notch1IC gain-of-function animals, lenses are abnormally small in size, which could be due to increased cell death or reduced progenitor proliferation. For Rbpj conditional mutants, no increase in apoptosis from E11 to E18.5 was found using the marker cleaved PARP (data not shown). Therefore, we searched for changes in cell cycle progression during lens development. First, Le-Cre;RbpjCKO/CKO mutants display a dramatic loss of Cyclin D2-expressing cells at E11 (Fig. 6A,G) but, at E14.5, Cyclin D2 expression is more profoundly affected since it is both reduced and abnormally distributed (Fig. 6B,G). At this age, Foxe3 and Cyclin D2 are normally only coexpressed in a subset of transition zone cells (arrow in left panel, Fig. 6B). However, in Le-Cre;RbpjCKO/CKO embryos, we observe a marked increase in Foxe3+Cyclin D2+ AEL cells (arrows in right panel, Fig. 6B), coincident with a profound loss of Cyclin D2+ cells in the fiber cell compartment. This alteration in Cyclin D2 localization suggests that cell cycle progression is compromised in Le-Cre;RbpjCKO/CKO lenses.
Figure 6. Lens progenitors are reduced in conditional Rbpj mutant lenses.
(A,B) Cyclin D2+ cells are decreased in E11 and E14.5 Le-Cre;RbpjCKO/CKO mutant lenses. At E14.5, Cyclin D2 is improperly expressed throughout the AEL, determined by Foxe3 co-expression (arrows). (C) Cyclin D1+ cells are decreased in E14.5 Le-Cre;RbpjCKO/CKO lenses, especially around the exit point of the transition zone. (D) BrdU+ S-phase cells are decreased in E14.5 Le-Cre;RbpjCKO/CKO eyes. (E) p57Kip2 expression is unaltered in Le-Cre;RbpjCKO/CKO lenses at E11, E12.5 and E14.5. (F) p27Kip1 cells are decreased in Le-Cre;RbpjCKO/CKO lenses, especially at the transition zone in Foxe3+ cells (not shown). (G–K) Quantification of proliferation markers as indicated. Bar in A = 40 microns, in C = 20 microns; anterior is up in A–F. Bar graphs show mean + s.e.m. * = P<0.05. ** = P<0.01.
Next, we examined other markers of cell cycle progression or exit in Le-Cre;RbpjCKO/CKO embryos, and quantified Cyclin D1-expressing and BrdU pulse-labeled cells (Fig. 6C,D,H,I). Here we found that Cyclin D1+ cells are significantly reduced in E11 mutants (Fig. 6H). The combined reductions of Cyclin D1 and Cyclin D2 led to an eventual significant decrease in numbers of BrdU-abeled S-phase in Le-Cre;RbpjCKO/CKO lenses during secondary fiber cell genesis (Fig. 6D,I). Subsequently, we examined two markers of cell cycle exit, the CKIs p57Kip2 and p27Kip1. In contrast to the results of Jia et al. (Jia et al., 2007), we observed normal numbers and expression pattern of p57Kip2+ cells at E11, E12.5 and E14.5 in both Le-Cre;RbpjCKO/+ and Le-Cre;RbpjCKO/CKO embryos (Fig. 6E,J). However, both Le-Cre;RbpjCKO/+ and Le-Cre;RbpjCKO/CKO lenses have reduced numbers of p27Kip1+ cells at E11 and E14.5 (Fig. 6F,K). These data suggest that Cyclin D1, Cyclin D2, p27Kip1, but not p57Kip2, are regulated, directly or indirectly, by Notch signaling during lens growth.
We then evaluated the same markers of proliferation in P0-3.9-GFPCre;Rosa26Notch1IC lenses. Cyclin D2 is initially reduced in P0-3.9-GFPCre;Rosa26Notch1IC lenses at E10.5 (Fig. 7A,B,K), but subsequently rebounds to excess expression from E14.5 to E18.5, prominently in the transition zone (Fig. 7C,D,K), a phenotype opposite that of the loss-of-function mutants (Fig. 6B,G). Cyclin D1 is dramatically upregulated from at least E10.5 (Fig. 7A,B,L) to E18.5 (Fig. 7C,D,L) when these animals die. Like Pax6, strong Cyclin D1 expression persists in the posterior P0-3.9-GFPCre;Rosa26Notch1IC lens. We next assessed proliferation by examining S-phase cells via BrdU pulse labeling. At E10.5, P0-3.9-GFPCre;Rosa26Notch1IC lenses have a significant increase in proliferation that continues throughout development (Fig. 7E,F,M). Proliferating cells with excess activated Notch1IC are almost always localized to the AEL, although ectopically-dividing cells are infrequently present in the posterior lens (arrows in Fig. 7F). The increased proliferation of P0-3.9-GFPCre;Rosa26Notch1IC lenses is due to an increased number of dividing progenitor cells at E10.5 that expand the pool of lens progenitors (Fig. S4).
Figure 7. Lens proliferation is increased in activated Notch lenses.
(A–D) Cyclin D1 (red) is highly expressed throughout E10.5 P0-3.9-GFPCre;Rosa26Notch1IC lenses and inappropriately maintained in E14.5 P0-3.9-GFPCre;Rosa26Notch1IC posterior lenses. Cyclin D2 (green) is decreased in E10.5 P0-3.9-GFPCre;Rosa26Notch1IC lenses, but increased in E14.5 P0-3.9-GFPCre;Rosa26Notch1IC posterior cells, where it is normally absent. (E–F) BrdU+ S phase cells are increased in P0-3.9-GFPCre;Rosa26Notch1IC lenses, with rare dividing cells in E14.5 P0-3.9-GFPCre;Rosa26Notch1IC posterior lenses (arrows, approximately 0–2 cells per section). (G,H) Highest levels of p57Kip2 or (I,J) p27Kip1 expression are normally observed at the transition zone, but are expanded through the entire posterior lens in E14.5 P0-3.9-GFPCre;Rosa26Notch1IC eyes. (K–N) Quantification of proliferation markers in E10.5 – E18.5 lenses as indicated. (O) Quantification of proliferation over 6-days in culture shows increased proliferation of 17NotchΔE cells relative to controls. Cells were plated at low density (10,000 cells, red) or high density (20,000 cells, blue). Data points show mean +/− s.e.m. * = P<0.05. ** = P<0.01. *** = P<0.001. Bar in A,C = 100 microns; anterior is up in panels A–J.
No changes were observed for p57Kip2 expression (Figs. 5G,H, 7N) or p27Kip1 expression (data not shown) prior to E14.5, after which both CKIs become increased in expression continuing through E18.5 (Fig. 7G–J,N). Normally p57Kip2 and p27Kip1 expression are most prominent at the transition zone (Fig. 7G,I), however this expression domain is expanded throughout the posterior lens of P0-3.9- GFPCre;Rosa26Notch1IC eyes (Fig. 7H,J). This suggests that cells are unable to properly exit the transition zone at the lens equator. To determine if Notch activation directly regulates lens cell proliferation, we engineered a lens epithelial cell line, 17EM15, to overexpress the activated Notch1 receptor (17NotchΔE). We found that 17NotchΔE cells proliferate more rapidly than control cultures, independent of initial cell density (Fig. 7O).Together with our in vivo experiments, these data demonstrate that Notch signaling regulates lens epithelial cell proliferation.
Functional loss of the lens transition zone in Notch pathway mutants
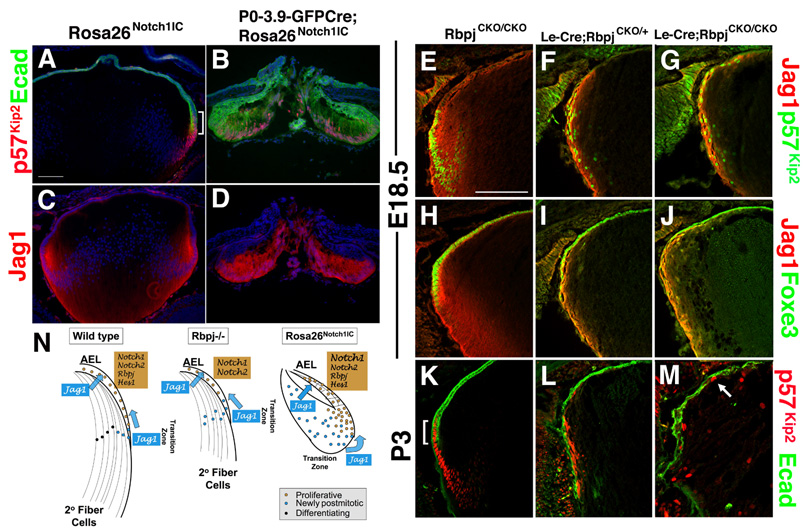
Altered expression of p27Kip1 suggests abnormalities are occurring to the transition zone in both types of Notch mutants, and these changes mirror defects in fiber cell differentiation. So, the transition zone was evaluated in P0-3.9-GFPCre;Rosa26Notch1IC lenses, through comparison of p57Kip2 and E- Cadherin expression patterns. At E18.5 and beyond, these markers are co-expressed only within the transition zone, thereby demarcating the boundary between post-mitotic epithelial (E- Cadherin+/p57Kip2)+ and non-epithelial (E-Cadherin-/p57Kip2)+ lens cells (bracket in Fig. 8A,K). In E18.5 P0-3.9-GFPCre;Rosa26Notch1IC lenses, the E-Cadherin+/ p57Kip2+ cells are expanded throughout the posterior lens, illustrating an enlarged transition zone consisting of epithelial cells (Fig. 8B). Another transition zone marker, Jag1 (Fig. 1B), is similarly expanded (Fig. 8D). In stark contrast, the transition zone of E18.5 Le-Cre;RbpjCKO/CKO lenses is reduced, with thinner Jag1 (Fig. 8E–G) and p57Kip2 (Fig. 8H–J) expression domains that are elongated around the lens periphery. By P3, the Le-Cre;RbpjCKO/+ transition zone is greatly degenerated (Fig. 8L) and completely missing from Le-Cre;RbpjCKO/CKO lenses (Fig. 8M). In these mutants, the few remaining p57Kip2+ cells are randomly arranged, with some mispositioned within the AEL, along with noncontiguous E-Cadherin+ lens progenitors (Fig. 8M).
Figure 8. Notch signaling controls the vertebrate lens transition zone.
(A,B) p57Kip2 is normally activated in a narrow domain that partially overlaps E-cadherin (Ecad) expression in the AEL, at the lens equator (bracket in panel A). In E18.5 P0-3.9-GFPCre;Rosa26Notch1IC lenses, the Ecad+p57Kip2+ domain is dramatically expanded. (C,D) Jag1 expression in the transition zone is abnormally enlarged in E18.5 P0-3.9-GFPCre;Rosa26Notch1IC lens. (E–G) Conversely, loss of Notch signaling causes reduced p57Kip2 and Jag1 expression domains that are elongated outside of the lens equator. p57Kip2+ nuclei are mislocalized. (H–M) By P3, Le-Cre;RbpjCKO/CKO lenses have almost completely lost p57Kip2 expression. The remaining p57Kip2+ cells are randomly arranged, including within the AEL (arrow). (N) Summary diagram of Notch signaling loss- and gain-of function phenotypes in the E18.5 lens. See Discussion section for details. Bar in A = 100 microns, in E = 50 microns; Anterior is up in all panels.
Together, Rbpj loss-of-function and Notch1IC gain-of-function experiments demonstrate an essential role for Notch signaling in the mammalian lens for proper cell cycle progression and fiber cell differentiation. We also reveal a late embryonic requirement for Notch signaling for the migration of lens progenitors from the AEL through the transition zone and into the fiber cell compartment.
DISCUSSION
Here we demonstrate that Notch signaling acts during lens development and is critically required for its growth, and differentiation. Furthermore, the phenotypes observed in Le-Cre;RbpjCKO/CKO mutants (termed loss-of-function (LOF) mutants) and P0-3.9-GFPCre;Rosa26Notch1IC mutants (termed gain-of-function (GOF) mutants) provide important insights for several human ophthalmologic diseases.
Notch signaling in lens development
Our expression data indicate that a functional unit of canonical Notch signaling is present during embryonic lens development. While the Jag1 ligand and Notch1/2 receptor domains overlap significantly in the lens pit, they soon separate spatially, with Jag1 restricted to the transition zone and Notch1/2 to the AEL. The implications of this separation are two-fold, as summarized in Fig. 7N. First, the ligand-receptor domains correlate well with Notch signaling regulation of fiber cell differentiation, initially in the lens vesicle for primary fiber cells, but later at the interface of the AEL and transition zone, where it controls secondary fiber cell migration and differentiation. Second, Jag1 protein in early differentiating fiber cells is expressed in a peripheral-to-central graded pattern. As fiber cells migrate out of the peripheral transition zone and differentiate centrally, Jag1 protein levels decrease. Thus, this second mode of Notch signaling is ideally configured to act as a feedback loop where differentiated fiber cells signal back to proliferative progenitors. Indeed, the pattern of proliferation in the AEL, as assessed in a sector-based analysis, correlates well with the amount of Jag1 ligand at the anterior surface of fiber cells (Shirke et al., 2001). This feedback model can be tested in the future through transgenic misexpression and/or conditional deletion of Jag1.
Evidence presented here support the idea that Notch regulates aspects of Pax6 lens expression. The persistence of Pax6 and Pax6 EE activity upon Notch1IC expression suggest Notch as an upstream regulator of Pax6. Analyses in Drosophila and Xenopus eye development also point to important roles for Notch signaling in eye specification, upstream of Pax6 orthologues (Kumar and Moses, 2001; Kurata et al., 2000; Onuma et al., 2002). In Xenopus embryos, ectopic expression of activated Notch causes complete lens duplication (Onuma et al., 2002). Intriguingly, Ogino and colleagues now show that Notch signaling regulates aspects of lens induction in Xenopus embryos and appears to directly regulate Foxe3 expression in the presumptive lens epithelium (Ogino et al., 2008). In that study Pax6 function is implicated upstream of Notch signaling, but here our data suggest that Notch signaling may subsequently act upon Pax6 late expression in the AEL. Thus the two studies complement one another by addressing Notch signaling during early and late embryonic lens development. Because of the temporal limitations of Cre-mediated excision using the Pax6 EE (Liu et al., 2006 and this paper), future studies will test the mechanism of Notch regulation of Pax6 in the mouse lens using earlier acting Cre drivers, and biochemical assays of direct binding to the Pax6 EE enhancer.
Notch signaling regulation of lens cell proliferation versus differentiation
The lens is a powerful tissue for cell cycle studies during development because this process is under tight spatial control (Griep, 2006). Our experiments provide several insights into Notch regulation of self-renewing proliferation in the AEL and cell cycle exit at the transition zone. First, the GOF mutants indicate that Notch is a potent mitogenic signal in the early lens epithelium. Hyperproliferation of the AEL was evident as early as E10.5 in the GOF mutants and caused a thickened, multilayered AEL. The rapidly dividing lens epithelium at E10.5 also exhibited abnormally uniform and high expression of Cyclin D1. The GOF mutant AEL maintains a high rate of proliferation, but one proportional to the number of AEL cells, suggesting that early Notch signaling determines the number of mitotic progenitors in the lens. Our in vitro studies support these conclusions, as lens cell lines overexpressing constitutively-activated Notch proliferate more than control cells. But cell cycle exit still occurs in vivo with excess activated Notch1IC, since CKI expression properly initiates in the posterior lens. Such strong compensation may explain the lack of lens tumors in humans (Seigel and Kummer, 2001).
The GOF and LOF analyses demonstrate that Cyclin D1/Cyclin D2 and p27Kip1 are downstream of Notch in the lens, as in other tissues (Kiaris et al., 2004; Ronchini and Capobianco, 2001; Sarmento et al., 2005; Stahl et al., 2006). Whether Notch promotes proliferation or cell cycle exit is strongly context-dependent and this switch can be tightly controlled by regulating the stoichiometry of both cyclins and CKIs. Our combined LOF and GOF analyses reveal different regulatory mechanisms utilized between Cyclin D1 and Cyclin D2. While Notch signaling appears necessary for proper expression of both Cyclins, Notch signaling alone seems insufficient to activate Cyclin D2, particularly during primary fiber cell development. The identification of other factors regulating Cyclin D2 may reveal a novel proliferative pathway. This more complex level of regulation may also be imposed at the transition zone, which is a rich source of other signaling molecules. In particular BMP and/or FGF signaling may act in concert with Notch to control lens cell cycle exit/differentiation or migration through this equatorial region.
The third putative Notch downstream gene revealed in our experiments is the CKI p27Kip1. Although logically proliferation defects in LOF and GOF mutants might indirectly cause p27Kip1 expression changes, this should occur oppositely (p27Kip1+ cells should increase when Cyclin D2+ cells decrease). Because the p27Kip1+ and Cyclin D2+ populations shifted identically in LOF experiments, we conclude that Notch signaling is simultaneously required for both Cyclin and CKI expression during lens development. This particularly evident in E14.5 and E18.5 GOF lenses, where expansion of the Cyclin D1, Cyclin D2, p27Kip1, and p57Kip2 expression domains are all observed.
However, none of these alterations in cell cycle regulation are sufficient to explain the fiber cell defects observed. Instead, a more complex model involving Notch regulation of the lens factors Foxe3, Pitx3, Pax6 and/or Prox1, which in turn also influences Cyclin and/or CKI expression is plausible, since all were affected in the LOF mutants along with Cyclin D1, Cyclin D2, and p27Kip1. For example, Rbpj-mediated Notch signaling is required for Foxe3 expression in the posterior lens during primary cell genesis, but once the AEL forms, other factors or pathways must maintain it in proliferating progenitors. But does Notch signaling directly control fiber cell differentiation? In the GOF mutants, the proportion of secondary fiber cells decreased dramatically between E14.5 and E18.5, while posterior lens cells maintained (or reacquired) AEL characteristics. While this suggests Notch signaling controls some aspect of fiber cell differentiation it can also be interpreted that excess Notch1IC drives an expansion of the AEL and transition zone compartments at the expense of fiber cells. Likewise, in LOF mutants, simultaneous defects in progenitor cell growth and secondary fiber cell differentiation occur. While it is clearer here that cell cycle exit, migration through the transition zone and fiber cell differentiation become uncoupled, our analyses are insufficient to demonstrate direct regulation of fiber cell differentiation by the Notch pathway. Definitive proof will require the identification of pro-fiber cell factors and examination of their expression and regulation in Notch pathway mutants.
Novel insights into human ocular disorders
Both GOF and LOF mutants resemble distinct human lens diseases. GOF mutant mice have a very strong Peter’s Anomaly, a condition postulated to occur when apoptotic cell death is blocked at the peripheral lens pit. In extreme cases, the cornea and AEL of GOF mutants fuse into a contiguous epithelium, preventing separation of lens cavity from the anterior chamber of the eye. Thus, hyperproliferation of the AEL may also be a culprit in human patients with Peter’s Anomaly. Alternatively, defects in the corneal endothelium can cause Peter’s Anomaly (Reneker et al., 2000), and in our GOF mutants, a profound loss of the corneal substratum occurs. As the Pax6 EE directs expression in both the early cornea and lens AEL, we cannot rule out that part of this phenotype may be manifested by Notch signaling in the cornea. Recently, Notch1 was shown to maintain corneal epithelial fate following injury (Vauclair et al., 2007). Thus, dysregulation of Notch signaling could be manifested as Peter’s Anomaly via multiple mechanisms.
LOF mutants strongly phenocopied congenital secondary aphakia, particularly progressive perinatal resorption. It is quite likely that earlier removal of Rbpj will confer severe primary aphakia in the mammalian eye. Only mutations in the human FOXE3 gene are reported to cause a nearly complete congenital lens dysgenesis (Valleix et al., 2006). But, mutations in human PITX3 result in cataracts and anterior segment dysgenesis (Semina et al., 1998), and deletion of an upstream region of Pitx3 in mice causes aphakia (Rieger et al., 2001; Semina et al., 2000). The aphakia of Rbpj LOF mutant mice may therefore be a combinatorial effect of reduced expression of Foxe3, Pitx3, Pax6 and possibly other genes. Because there is such a pronounced degeneration in postnatal Rbpj LOF lenses, these animals are particularly well suited to address mechanisms of lens degeneration and loss. Therefore, we propose that mutations in Notch pathway genes should be investigated in lens degeneration syndromes.
The state of Notch signaling in the lens
Recently Jia et al., characterized a lens defect utilizing a similar Rbpj conditional mutant strategy (Jia et al., 2007). However, their lens phenotypes are significantly milder than ours, lacking adult-onset aphakia. This is likely due to the different Cre drivers used; ours is Le-Cre in which the Pax6 EE drives Cre expression, while Jia et al., used a Cre line driven by the αA-crystallin promoter that additionally contains a Pax6 consensus binding site (Zhao et al., 2004). Thus, differences in the onset of Cre expression and/or the degree of Rbpj excision likely explain the phenotypic differences between the studies, and may explain the discrepancies seen for p27Kip1 (this paper) versus p57Kip2 (Jia et al.) Additionally, Jia et al. propose Hey1 as a major Notch effector gene, while our study shows dramatic effects on Hes1. Although multiple Hes/Hey genes may function as Notch effectors in the lens, we believe the strong expression of Hes1 throughout lens development, (Figs 1A,B,K,L 2B,E, and (Lee et al., 2005)) which is Rbpj-dependent at E14.5 (Fig 2E), coupled with dramatic loss-of-function ocular phenotype of Hes1 mutant mice (Lee et al., 2005), but not Hey1 or Hey1/Hey2 mutant mice (Fischer et al., 2004; Kokubo et al., 2005) point to Hes1 as the major Notch effector gene in the developing lens. Nonetheless, fundamental conclusions are similar between the two studies. Identification of Notch signaling as one long-sought-after integrator of the cell cycle with differentiation in the lens predicts many rapid and exciting new discoveries in the foreseeable future.
Supplementary Material
(A–C) Both Le-Cre;RbpjCKO/+ and Le-Cre;RbpjCKO/CKO animals have abnormally small eyes. Le-Cre;RbpjCKO/CKO mutants have missing fur and whiskers bilaterally, malformed eyelids and no externally visible eye. Hematoxylin staining of (D–F) E18.5, (G–I) P3, and (J–L) P21 sections shows progressive loss of Le-Cre;RbpjCKO/CKO mutant lenses and total lack of an anterior chamber (asterisk in J,K), while Le-Cre;RbpjCKO/+ heterozygotes have an intermediate phenotype. At E18.5, both Le-Cre;RbpjCKO/+ and Le-Cre;RbpjCKO/CKO eyes had small lenses with recognizable AEL and fiber cell compartments (E,F), although Le-Cre;RbpjCKO/CKO lenses were heavily vacuolated and the fiber cells exhibited abnormal posterior-anterior fiber cell distribution (F). By P21, Le-Cre;RbpjCKO/CKO eyes were aphakic, with remnants of highly disorganized and rosetted lens tissue (L). Bar in A = 500 microns, in D,J = 20 microns; Anterior is up in panels D-L, with C = cornea; L = lens and R = retina.
(A–C) GFP fluorescence in P0-3.9-GFPCre lenses is normally peripheral (A), while P0-3.9-GFPCre;Rosa26Notch1IC eyes show either uniform lens expression (B), or a prominent central gap of lens expression (C). (D–F) Microscopic examination of E18.5 eyes indicates microphthalmia and lack of opacity in P0-3.9-GFPCre;Rosa26Notch1IC lenses, as well as extruded lens material in panel F (arrows).
(A–C) No differences in the spatiotemporal expression of β-crystallin within fiber cells were found among the three Rbpj genotypes at E11, E12 (not shown) or E14.5. GFP expression indicates the Cre expression domain in panels B and C. (D–G) γ-crystallin is expressed strongly in E14.5 and E18.5 posterior lenses in all genotypes. Bar in A= 20 microns, D=100 microns.
The increased proliferation of P0-3.9-GFPCre;Rosa26Notch1IC lenses is due to an increased number of dividing progenitor cells at E10.5 that gives rise to an increased pool of progenitor cells. At E12.5 and beyond, the percentage of dividing Foxe3+ cells is unaltered.
ACKNOWLEDGEMENTS
The authors thank Tasuku Honjo and Tom Gridley for RbpjCKO mutant mice; Ruth Ashery-Padan and Richard Lang for Le-Cre transgenic mice; Charles Murtaugh, Jose Rivera-Feliciano and Cliff Tabin for Rosa26Notch1IC mice; Yingzi Yue for technical assistance; Peter Carlsson, Marten Smidt, Richard Lang, and Guillermo Oliver for antibody reagents; Tom Gridley, Tiffany Cook, Charles Murtaugh, Richard Lang and Daniel O’Connell for valuable discussion and/or critical reading of this manuscript. This work was supported by a fellowship from the Canadian Institutes of Health Research to SR; NIH grant EY10123 to RLM; and NIH grants EY13612, EY18097 and CHRF research funds to NLB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Ashery-Padan R, Marquardt T, Zhou X, Gruss P. Pax6 activity in the lens primordium is required for lens formation and for correct placement of a single retina in the eye. Genes Dev. 2000;14:2701–2711. doi: 10.1101/gad.184000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao ZZ, Cepko CL. The expression and function of Notch pathway genes in the developing rat eye. Journal of Neuroscience. 1997;17:1425–1434. doi: 10.1523/JNEUROSCI.17-04-01425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beebe D, Garcia C, Wang X, Rajagopal R, Feldmeier M, Kim JY, Chytil A, Moses H, Ashery-Padan R, Rauchman M. Contributions by members of the TGFbeta superfamily to lens development. Int J Dev Biol. 2004;48:845–856. doi: 10.1387/ijdb.041869db. [DOI] [PubMed] [Google Scholar]
- Belecky-Adams TL, Adler R, Beebe DC. Bone morphogenetic protein signaling and the initiation of lens fiber cell differentiation. Development. 2002;129:3795–3802. doi: 10.1242/dev.129.16.3795. [DOI] [PubMed] [Google Scholar]
- Besson A, Hwang HC, Cicero S, Donovan SL, Gurian-West M, Johnson D, Clurman BE, Dyer MA, Roberts JM. Discovery of an oncogenic activity in p27Kip1 that causes stem cell expansion and a multiple tumor phenotype. Genes Dev. 2007;21:1731–1746. doi: 10.1101/gad.1556607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettenhausen B, de Angelis MH, Simon D, Guenet J, Gossler A. Transient and restricted expression during mouse embryogenesis of Dll1, a murine gene closely related to Drosophila Delta. Development. 1995;121:2407–2418. doi: 10.1242/dev.121.8.2407. [DOI] [PubMed] [Google Scholar]
- Blixt A, Mahlapuu M, Aitola M, Pelto-Huikko M, Enerback S, Carlsson P. A forkhead gene, FoxE3, is essential for lens epithelial proliferation and closure of the lens vesicle. Genes Dev. 2000;14:245–254. [PMC free article] [PubMed] [Google Scholar]
- Bolós V, Grego-Bessa J, de la Pompa JL. Notch signaling in development and cancer. Endocr Rev. 2007;28:339–363. doi: 10.1210/er.2006-0046. [DOI] [PubMed] [Google Scholar]
- Brown NL, Kanekar S, Vetter ML, Tucker PK, Gemza DL, Glaser T. Math5 encodes a murine basic helix-loop-helix transcription factor expressed during early stages of retinal neurogenesis. Development. 1998;125:4821–4833. doi: 10.1242/dev.125.23.4821. [DOI] [PubMed] [Google Scholar]
- Cui W, Tomarev SI, Piatigorsky J, Chepelinsky AB, Duncan MK. Mafs, Prox1, and Pax6 can regulate chicken betaB1-crystallin gene expression. J Biol Chem. 2004;279:11088–11095. doi: 10.1074/jbc.M312414200. [DOI] [PubMed] [Google Scholar]
- Cvekl A, Kashanchi F, Sax CM, Brady JN, Piatigorsky J. Transcriptional regulation of the mouse alpha A-crystallin gene: activation dependent on a cyclic AMP-responsive element (DE1/CRE) and a Pax-6-binding site. Mol Cell Biol. 1995;15:653–660. doi: 10.1128/mcb.15.2.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner AL, Episkopou V, Maas RL. Sox2 and Pou2f1 interact to control lens and olfactory placode development. Dev Biol. 2007;303:784–799. doi: 10.1016/j.ydbio.2006.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber SC, Robinson ML, Makarenkova HP, Lang RA. Bmp signaling is required for development of primary lens fiber cells. Development. 2002;129:3727–3737. doi: 10.1242/dev.129.15.3727. [DOI] [PubMed] [Google Scholar]
- Fischer A, Gessler M. Delta-Notch--and then? Protein interactions and proposed modes of repression by Hes and Hey bHLH factors. Nucleic Acids Res. 2007;35:4583–4596. doi: 10.1093/nar/gkm477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Schumacher N, Maier M, Sendtner M, Gessler M. The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes Dev. 2004;18:901–911. doi: 10.1101/gad.291004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser T, Jepeal L, Edwards JG, Young SR, Favor J, Maas RL. PAX6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat Genet. 1994;7:463–471. doi: 10.1038/ng0894-463. [DOI] [PubMed] [Google Scholar]
- Gómez Lahoz E, Liégeois NJ, Zhang P, Engelman JA, Horner J, Silverman A, Burde R, Roussel MF, Sherr CJ, Elledge SJ, DePinho RA. Cyclin D- and E- dependent kinases and the p57(KIP2) inhibitor: cooperative interactions in vivo. Mol Cell Biol. 1999;19:353–363. doi: 10.1128/mcb.19.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griep AE. Cell cycle regulation in the developing lens. Semin Cell Dev Biol. 2006;17:686–697. doi: 10.1016/j.semcdb.2006.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C, Chatterjee B, Favor J, Immervoll T, Löster J, Klopp N, Sandulache R, Graw J. Aphakia (ak), a mouse mutation affecting early eye development: fine mapping, consideration of candidate genes and altered Pax6 and Six3 gene expression pattern. Dev Genet. 1998;23:299–316. doi: 10.1002/(SICI)1520-6408(1998)23:4<299::AID-DVG5>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Grindley JC, Davidson DR, Hill RE. The role of Pax-6 in eye and nasal development. Development. 1995;121:1433–1442. doi: 10.1242/dev.121.5.1433. [DOI] [PubMed] [Google Scholar]
- Han H, Tanigaki K, Yamamoto N, Kuroda K, Yoshimoto M, Nakahata T, Ikuta K, Honjo T. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int Immunol. 2002;14:637–645. doi: 10.1093/intimm/dxf030. [DOI] [PubMed] [Google Scholar]
- Hatakeyama J, Bessho Y, Katoh K, Ookawara S, Fujioka M, Guillemot F, Kageyama R. Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development. 2004;131:5539–5550. doi: 10.1242/dev.01436. [DOI] [PubMed] [Google Scholar]
- Ilagan MX, Kopan R. SnapShot: notch signaling pathway. Cell. 2007;128:1246. doi: 10.1016/j.cell.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Ishibashi M, Ang S-L, Shiota K, Nakanishi S, Kageyama R, Guillemot F. Targeted disruption of mammalian hairy and Enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix-lop-helix factors, premature neurogenesis, and severe neural tube defects. Genes and Development. 1995;9:9136–9148. doi: 10.1101/gad.9.24.3136. [DOI] [PubMed] [Google Scholar]
- Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- Jensen J, Pedersen EE, Galante P, Hald J, Heller RS, Ishibashi M, Kageyama R, Guillemot F, Serup P, Madsen OD. Control of endodermal endocrine development by Hes-1. Nat Genet. 2000;24:36–44. doi: 10.1038/71657. [DOI] [PubMed] [Google Scholar]
- Jia J, Lin M, Zhang L, York JP, Zhang P. The notch signaling pathway controls the size of the ocular lens by directly suppressing p57Kip2 expression. Mol Cell Biol. 2007;27:7236–7247. doi: 10.1128/MCB.00780-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T, Tomita K. The bHLH gene Hes1 regulates differentiation of multiple cell types. Molecules and Cells. 2000;10:1–7. doi: 10.1007/s10059-000-0001-0. [DOI] [PubMed] [Google Scholar]
- Kiaris H, Polit K, Grimm LM, Szabolcs M, Fisher P, Efstratiadis A, Artavanis-Tsakonas S. Modulation of notch signaling elicits signature tumors and inhibits hras1-induced oncogenesis in the mouse mammary epithelium. Am J Pathol. 2004;165:695–705. doi: 10.1016/S0002-9440(10)63333-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JI, Li T, Ho IC, Grusby MJ, Glimcher LH. Requirement for the c-Maf transcription factor in crystallin gene regulation and lens development. Proc Natl Acad Sci USA. 1999;96:3781–3785. doi: 10.1073/pnas.96.7.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokubo H, Miyagawa-Tomita S, Nakazawa M, Saga Y, Johnson RL. Mouse hesr1 and hesr2 genes are redundantly required to mediate Notch signaling in the developing cardiovascular system. Dev Biol. 2005;278:301–309. doi: 10.1016/j.ydbio.2004.10.025. [DOI] [PubMed] [Google Scholar]
- Kopan R, Schroeter EH, Weintraub H, Nye JS. Signal transduction by activated mNotch: importance of proteolytic processing and its regulation by the extracellular domain. Proc Natl Acad Sci U S A. 1996;93:1683–1688. doi: 10.1073/pnas.93.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar JP, Moses K. EGF receptor and Notch signaling act upstream of Eyeless/Pax6 to control eye specification. Cell. 2001;104:687–697. doi: 10.1016/s0092-8674(01)00265-3. [DOI] [PubMed] [Google Scholar]
- Kurata S, Go MJ, Artavanis-Tsakonas S, Gehring WJ. Notch signaling and the determination of appendage identity. Proc Natl Acad Sci USA. 2000;97:2117–2122. doi: 10.1073/pnas.040556497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Y, Miller JL, Sauer B. GFPcre fusion vectors with enhanced expression. Anal Biochem. 1999;270:334–336. doi: 10.1006/abio.1999.4110. [DOI] [PubMed] [Google Scholar]
- Lee HY, Wroblewski E, Philips GT, Stair CN, Conley K, Reedy M, Mastick GS, Brown NL. Multiple requirements for Hes 1 during early eye formation. Dev Biol. 2005;284:464–478. doi: 10.1016/j.ydbio.2005.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Lagutin OV, Mende M, Streit A, Oliver G. Six3 activation of Pax6 expression is essential for mammalian lens induction and specification. EMBO J. 2006;25:5383–5395. doi: 10.1038/sj.emboj.7601398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovicu FJ, McAvoy JW. FGF-induced lens cell proliferation and differentiation is dependent on MAPK (ERK1/2) signalling. Development. 2001;128:5075–5084. doi: 10.1242/dev.128.24.5075. [DOI] [PubMed] [Google Scholar]
- Makhani LF, Williams T, West-Mays JA. Genetic analysis indicates that transcription factors AP-2alpha and Pax6 cooperate in the normal patterning and morphogenesis of the lens. Mol Vis. 2007;13:1215–1225. [PubMed] [Google Scholar]
- Mastick GS, Andrews GL. Pax6 regulates the identity of embryonic diencephalic neurons. Mol Cell Neurosci. 2001;17:190–207. doi: 10.1006/mcne.2000.0924. [DOI] [PubMed] [Google Scholar]
- Medina-Martinez O, Brownell I, Amaya-Manzanares F, Hu Q, Behringer RR, Jamrich M. Severe defects in proliferation and differentiation of lens cells in Foxe3 null mice. Mol Cell Biol. 2005;25:8854–8863. doi: 10.1128/MCB.25.20.8854-8863.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proc Natl Acad Sci U S A. 2003;100:14290–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiguchi S, Wood H, Kondoh H, Lovell-Badge R, Episkopou V. Sox1 directly regulates the gamma-crystallin genes and is essential for lens development in mice. Genes Dev. 1998;12:776–781. doi: 10.1101/gad.12.6.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino H, Fisher M, Grainger RM. Convergence of a head-field selector Otx2 and Notch signaling: a mechanism for lens specification. Development. 2008;135:249–258. doi: 10.1242/dev.009548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onuma Y, Takahashi S, Asashima M, Kurata S, Gehring WJ. Conservation of Pax 6 function and upstream activation by Notch signaling in eye development of frogs and flies. Proc Natl Acad Sci USA. 2002;99:2020–2025. doi: 10.1073/pnas.022626999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reneker LW, Overbeek PA. Lens-specific expression of PDGF-A alters lens growth and development. Dev Biol. 1996;180:554–565. doi: 10.1006/dbio.1996.0328. [DOI] [PubMed] [Google Scholar]
- Reneker LW, Silversides DW, Xu L, Overbeek PA. Formation of corneal endothelium is essential for anterior segment development - a transgenic mouse model of anterior segment dysgenesis. Development. 2000;127:533–542. doi: 10.1242/dev.127.3.533. [DOI] [PubMed] [Google Scholar]
- Reneker LW, Xie L, Xu L, Govindarajan V, Overbeek PA. Activated Ras induces lens epithelial cell hyperplasia but not premature differentiation. Int J Dev Biol. 2004;48:879–888. doi: 10.1387/ijdb.041889lr. [DOI] [PubMed] [Google Scholar]
- Rieger DK, Reichenberger E, McLean W, Sidow A, Olsen BR. A double-deletion mutation in the Pitx3 gene causes arrested lens development in aphakia mice. Genomics. 2001;72:61–72. doi: 10.1006/geno.2000.6464. [DOI] [PubMed] [Google Scholar]
- Ring BZ, Cordes SP, Overbeek PA, Barsh GS. Regulation of mouse lens fiber cell development and differentiation by the Maf gene. Development. 2000;127:307–317. doi: 10.1242/dev.127.2.307. [DOI] [PubMed] [Google Scholar]
- Robinson ML. An essential role for FGF receptor signaling in lens development. Semin Cell Dev Biol. 2006;17:726–740. doi: 10.1016/j.semcdb.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronchini C, Capobianco AJ. Induction of cyclin D1 transcription and CDK2 activity by Notch(ic): implication for cell cycle disruption in transformation by Notch(ic) Mol Cell Biol. 2001;21:5925–5934. doi: 10.1128/MCB.21.17.5925-5934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmento LM, Huang H, Limon A, Gordon W, Fernandes J, Tavares MJ, Miele L, Cardoso AA, Classon M, Carlesso N. Notch1 modulates timing of G1-S progression by inducing SKP2 transcription and p27 Kip1 degradation. J Exp Med. 2005;202:157–168. doi: 10.1084/jem.20050559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seigel GM, Kummer A. The enigma of lenticular oncology. Digital Journal of Ophthalmology. 2001:7. [Google Scholar]
- Semina EV, Ferrell RE, Mintz-Hittner HA, Bitoun P, Alward WL, Reiter RS, Funkhauser C, Daack-Hirsch S, Murray JC. A novel homeobox gene PITX3 is mutated in families with autosomal-dominant cataracts and ASMD. Nat Genet. 1998;19:167–170. doi: 10.1038/527. [DOI] [PubMed] [Google Scholar]
- Semina EV, Murray JC, Reiter R, Hrstka RF, Graw J. Deletion in the promoter region and altered expression of Pitx3 homeobox gene in aphakia mice. Hum Mol Genet. 2000;9:1575–1585. doi: 10.1093/hmg/9.11.1575. [DOI] [PubMed] [Google Scholar]
- Shi X, Bosenko DV, Zinkevich VS, Foley S, Hyde DR, Semina EV, Vihtelic TS. Zebrafish pitx3 is necessary for normal lens and retinal development. Mech Dev. 2005;122:513–527. doi: 10.1016/j.mod.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Shirke S, Faber SC, Hallem E, Makarenkova HP, Robinson ML, Overbeek PA, Lang RA. Misexpression of IGF-I in the mouse lens expands the transitional zone and perturbs lens polarization. Mech Dev. 2001;101:167–174. doi: 10.1016/s0925-4773(00)00584-0. [DOI] [PubMed] [Google Scholar]
- Stahl M, Ge C, Shi C, Pestell RG, Stanley P. Notch1-induced transformation of RKE-1 cells requires up-regulation of cyclin D1. Cancer Res. 2006;66:7562–7570. doi: 10.1158/0008-5472.CAN-06-0974. [DOI] [PubMed] [Google Scholar]
- Thomas BJ. Cell-cycle control during development: taking it up a notch. Dev Cell. 2005;8:451–452. doi: 10.1016/j.devcel.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Tomita K, Ishibashi M, Nakahara K, Ang S-L, Nakanishi S, Guillemot F, Kageyama R. Mammalian hairy and Enhancer of split homolog 1 regulates differentiation of retinal neurons and is essential for eye morphogenesis. Neuron. 1996;16:723–734. doi: 10.1016/s0896-6273(00)80093-8. [DOI] [PubMed] [Google Scholar]
- Valleix S, Niel F, Nedelec B, Algros MP, Schwartz C, Delbosc B, Delpech M, Kantelip B. Homozygous nonsense mutation in the FOXE3 gene as a cause of congenital primary aphakia in humans. Am J Hum Genet. 2006;79:358–364. doi: 10.1086/505654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vauclair S, Majo F, Durham AD, Ghyselinck NB, Barrandon Y, Radtke F. Corneal epithelial cell fate is maintained during repair by Notch1 signaling via the regulation of vitamin A metabolism. Dev Cell. 2007;13:242–253. doi: 10.1016/j.devcel.2007.06.012. [DOI] [PubMed] [Google Scholar]
- Weinmaster G, Roberts VJ, Lemke G. A homolog of Drosophila Notch expressed during mammalian development. Development. 1991;113:199–205. doi: 10.1242/dev.113.1.199. [DOI] [PubMed] [Google Scholar]
- Weinmaster G, Roberts VJ, Lemke G. Notch2: a second mammalian Notch gene. Development. 1992;116:931–941. doi: 10.1242/dev.116.4.931. [DOI] [PubMed] [Google Scholar]
- Wigle JT, Chowdhury K, Gruss P, Oliver G. Prox1 function is crucial for mouse lens-fibre elongation. Nat Genet. 1999;21:318–322. doi: 10.1038/6844. [DOI] [PubMed] [Google Scholar]
- Xie L, Overbeek PA, Reneker LW. Ras signaling is essential for lens cell proliferation and lens growth during development. Dev Biol. 2006;298:403–414. doi: 10.1016/j.ydbio.2006.06.045. [DOI] [PubMed] [Google Scholar]
- Yoon K, Gaiano N. Notch signaling in the mammalian central nervous system: insights from mouse mutants. Nat Neurosci. 2005;8:709–715. doi: 10.1038/nn1475. [DOI] [PubMed] [Google Scholar]
- Zhang P, Wong C, DePinho RA, Harper JW, Elledge SJ. Cooperation between the Cdk inhibitors p27(KIP1) and p57(KIP2) in the control of tissue growth and development. Genes Dev. 1998;12:3162–3167. doi: 10.1101/gad.12.20.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Friedman A, Heaney S, Purcell P, Maas RL. Meis homeoproteins directly regulate Pax6 during vertebrate lens morphogenesis. Genes Dev. 2002;16:2097–2107. doi: 10.1101/gad.1007602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Heaney S, Maas RL. Cre-loxp fate-mapping of Pax6 enhancer active retinal and pancreatic progenitors. Genesis. 2003;35:22–30. doi: 10.1002/gene.10160. [DOI] [PubMed] [Google Scholar]
- Zhao H, Yang Y, Rizo CM, Overbeek PA, Robinson ML. Insertion of a Pax6 consensus binding site into the alphaA-crystallin promoter acts as a lens epithelial cell enhancer in transgenic mice. Invest Ophthalmol Vis Sci. 2004;45:1930–1939. doi: 10.1167/iovs.03-0856. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A–C) Both Le-Cre;RbpjCKO/+ and Le-Cre;RbpjCKO/CKO animals have abnormally small eyes. Le-Cre;RbpjCKO/CKO mutants have missing fur and whiskers bilaterally, malformed eyelids and no externally visible eye. Hematoxylin staining of (D–F) E18.5, (G–I) P3, and (J–L) P21 sections shows progressive loss of Le-Cre;RbpjCKO/CKO mutant lenses and total lack of an anterior chamber (asterisk in J,K), while Le-Cre;RbpjCKO/+ heterozygotes have an intermediate phenotype. At E18.5, both Le-Cre;RbpjCKO/+ and Le-Cre;RbpjCKO/CKO eyes had small lenses with recognizable AEL and fiber cell compartments (E,F), although Le-Cre;RbpjCKO/CKO lenses were heavily vacuolated and the fiber cells exhibited abnormal posterior-anterior fiber cell distribution (F). By P21, Le-Cre;RbpjCKO/CKO eyes were aphakic, with remnants of highly disorganized and rosetted lens tissue (L). Bar in A = 500 microns, in D,J = 20 microns; Anterior is up in panels D-L, with C = cornea; L = lens and R = retina.
(A–C) GFP fluorescence in P0-3.9-GFPCre lenses is normally peripheral (A), while P0-3.9-GFPCre;Rosa26Notch1IC eyes show either uniform lens expression (B), or a prominent central gap of lens expression (C). (D–F) Microscopic examination of E18.5 eyes indicates microphthalmia and lack of opacity in P0-3.9-GFPCre;Rosa26Notch1IC lenses, as well as extruded lens material in panel F (arrows).
(A–C) No differences in the spatiotemporal expression of β-crystallin within fiber cells were found among the three Rbpj genotypes at E11, E12 (not shown) or E14.5. GFP expression indicates the Cre expression domain in panels B and C. (D–G) γ-crystallin is expressed strongly in E14.5 and E18.5 posterior lenses in all genotypes. Bar in A= 20 microns, D=100 microns.
The increased proliferation of P0-3.9-GFPCre;Rosa26Notch1IC lenses is due to an increased number of dividing progenitor cells at E10.5 that gives rise to an increased pool of progenitor cells. At E12.5 and beyond, the percentage of dividing Foxe3+ cells is unaltered.