Abstract
Brefeldin A-inhibited guanine nucleotide-exchange protein (BIG) 1 activates human ADP-ribosylation factor (ARF) 1 and 3 by accelerating the replacement of ARF-bound GDP with GTP to initiate recruitment of coat proteins for membrane vesicle formation. Liquid chromatography MS/MS analysis of peptides from proteins that co-precipitated with BIG1 antibodies identified “kinesin family member 21A” (KIF21A), a plus-end-directed motor protein that moves cargo on microtubules away from the microtubule-organizing center. Reciprocal immunoprecipitation (IP) of endogenous proteins and microscopically apparent overlap of immunoreactive BIG1 with overexpressed GFP-KIF21A in the perinuclear region were consistent with an interaction of KIF21A–BIG1. Overexpression of full-length KIF21A and BIG1 and their fragments in HEK293 cells followed by reciprocal IP revealed that the C-terminal tail of KIF21A, with seven WD-40 repeats, may interact with structure in the C-terminal region of BIG1. Interfering with cyclic activation and inactivation of ARF1 by overexpressing constitutively active ARF1(Q71L) or dominant inactive ARF1(T31N) altered the distribution of BIG1 as well as its interaction with KIF21A. A requirement for ARF1 was confirmed by its selective depletion with siRNA. Unlike disruption of microtubules with nocodazole, selective inhibition of transport by depletion of KIF21A with specific siRNA altered BIG1 distribution without changing that of intrinsic Golgi membrane proteins. These newly recognized interactions of BIG1 and KIF21A should enable us to understand better the mechanisms through which, acting together, they may integrate local events in membrane trafficking with longer-range transport processes and to relate those processes to the diverse signaling and scaffold functions of BIG1.
Keywords: ADP-ribosylation factor 1, Golgi, microtubule
Brefeldin A-inhibited guanine nucleotide-exchange protein (BIG) 1, a member of the Sec7 family of ADP-ribosylation factor (ARF) guanine nucleotide-exchange proteins (GEP), activates class I ARFs (human ARF1 and 3) by catalyzing replacement of ARF-bound GDP with GTP, to initiate membrane vesicle formation (1–3). BIG1 had been localized at trans-Golgi structures (4, 5); by electron microscopy, Golgi membranes in BIG1-depleted cells were less sharply defined and had more vesicle-like structures at the trans face than those in control or BIG2 siRNA-treated cells (6). BIG1 also was found in nuclei of serum-deprived HepG2 cells or after their brief incubation with the immunosuppressive drug FK506 (7) or with cAMP (8).
In the ∼ 200-kDa BIG1 molecule, a central ARF-activating Sec7 domain was the first recognized functional structure (9). Elements identified later include a site in the N-terminal region that interacts with FK506-binding protein 13 (10) and an A kinase-anchoring protein sequence identical to one of three such sequences that differ in specificities for binding each of the four cAMP-dependent protein kinase regulatory (R) subunits and that were first identified in BIG2 (11), the very similar GEP initially co-purified with BIG1 (9). PKA-catalyzed phosphorylation of serine-883, a predicted substrate site in BIG1, was required for its cAMP-induced nuclear accumulation (8). Later experiments with recombinant proteins in vitro demonstrated that GEP activity of BIG1 was decreased after phosphorylation by PKA (12). Most recently, it was reported that BIG1, nucleolin, U3 small nucleolar RNA, the U3-binding protein fibrillarin, and the RNA-binding protein La may exist together in nuclear complexes, consistent with a potential role for BIG1 in nucleolar function (13). The C-terminal region of BIG1 includes a binding site for myosin IXb. This motor protein possesses GTPase-activating protein activity for Rho that is inhibited by competition with RhoA for binding to myosin IXb (14).
A role for myosin IXb in the translocation of BIG1 along actin fibers in cells remains to be demonstrated. We present here evidence that BIG1 interacts directly with the motor protein KIF21A, which is involved in its microtubule-dependent transport. KIF21A is a member of the family of kinesin proteins that control cell morphogenesis, survival, and other functions (15). The 45 KIF molecules are classified according to position of the motor domain as N-terminal, middle, and C-terminal types, respectively, N-, M-, and C-kinesins (16). KIF21A is an N-kinesin that acts as a plus-end-directed motor to move cargo away from the microtubule-organizing center. BIG1 activation of ARF1 also is implicated, presumably early, in the KIF21A-BIG1 interaction and initiation of transport vesicle formation at the Golgi.
Results
Identification of Proteins Co-Immunoprecipitated with BIG1.
Immunoblotting with anti-BIG1 antibodies confirmed IP of ∼ 85% of BIG1 from HepG2 cell extracts with anti-BIG1 but not with control normal rabbit IgG (data not shown). Coomassie R-250 staining, however, revealed very few unique bands [Fig. 1, supporting information (SI) Table S1]. To identify proteins not abundant enough to be detected in this way, gel lanes were divided into seven segments based on positions of molecular size marker proteins. After in-gel digestion with trypsin, peptides were analyzed using nanospray liquid chromatography (LC)-MS/MS, and compositions of control and BIG1 immunoprecipitation (IP) were compared. A protein of special interest to us, KIF21A, was found in the 150–250 kDa region, which also included BIG1 and BIG2 (Table S1). We chose to study this kinesin motor protein first, because it offered an opportunity to begin to understand mechanisms by which BIG1 and its associated multimolecular complexes are translocated along microtubule tracks in cells.
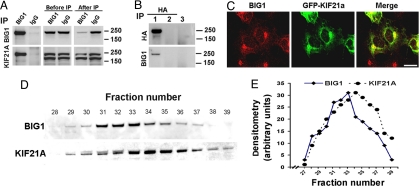
Fig. 1.
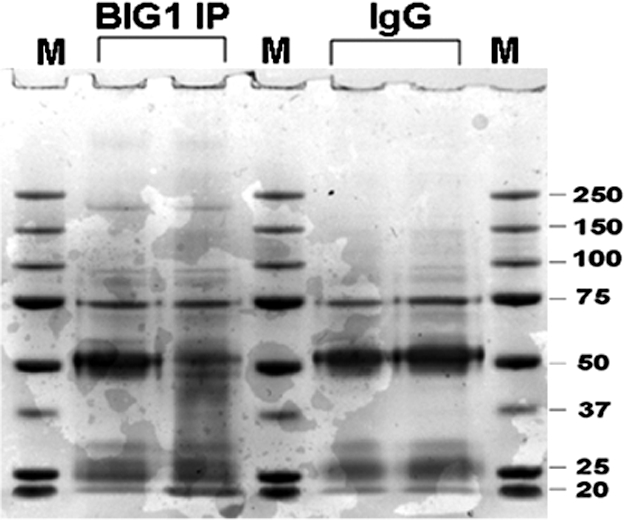
BIG1 Immunoprecipitation (IP) and Coomassie blue staining. Products of IP with control IgG or anti-BIG1 antibodies from 1 mg of HepG2 cell lysate protein were separated by 4–12% NuPAGE gel, and stained with Coomassie R250 blue. After destaining, the gel was divided into seven pieces according to the centers of molecular weight marker bands from top to bottom (>250, 250–150, 150–100, 100–75, 75–50, 50–37, <37 kDa). Proteins in gel blocks were subjected to in-gel trypsin digestion, and analyzed by LC-MS/MS.
Co-Immunoprecipitation and Intracellular Co-Localization of BIG1 and KIF21A.
Because antibodies for IP of endogenous KIF21A were not available, constructs for overexpression of full-length KIF21A with an N-terminal HA or GFP tag were used in some experiments. Also, these experiments used HEK293 rather than HepG2 cells, because they are more convenient for protein overexpression as well as for microscopy. IP of endogenous BIG1 also yielded endogenous KIF21A (Fig. 2A), just as endogenous BIG1 was precipitated by anti-HA antibodies from cells overexpressing HA-KIF21A but not by the empty vector (Fig. 2B). On confocal immunofluorescence microscopy, endogenous BIG1 in HEK293 cells appeared to be scattered in punctate collections throughout the cytoplasm with concentration in a perinuclear region reminiscent of Golgi structures (Fig. 2C). Overexpressed GFP-KIF21A also was concentrated at seemingly similar sites near the nucleus but perhaps was more linearly arranged throughout the cytoplasm. Apparently overlapping or coincident loci of BIG1 and KIF21A and their co-IP were consistent with their potential functional interaction.
Fig. 2.
Co-IP and cellular localization of BIG1 and KIF21A. (A) After IP of HEK293 cell extract with anti-BIG1 antibodies or control IgG, samples of precipitated proteins (total), of lysate (Before IP, 1/3), and supernatant (After IP, 1/3), were separated in 4–12% NuPAGE gels and immunoblotted with anti-BIG1 and anti-KIF21A antibodies. KIF21A antibodies react with KIF21A (187 kDa) and an unknown protein just below it that did not immunoprecipitate with BIG1. (B) IP with anti-HA antibodies of proteins from cells transfected with HA-KIF21A (lane 1) or with empty vector (lane 2), or with control IgG of proteins from cells transfected with HA-KIF21A (lane 3) 24 h before preparation of cell extracts. Products of IP with anti-HA antibodies (lanes 1 and 2) or IgG (lane 3) were separated in 4–12% NuPAGE gels and were immunoblotted with antibodies against HA and BIG1. (C) Cells transfected 24 h earlier with GFP-KIF21A were fixed and reacted with anti-BIG1 antibodies before preparation for confocal immunofluorescence microscopy. (Scale bar, 16 μm.) (D and E) Cell proteins (3 mg, 1 ml of cytosol) were separated on Superdex 200. Samples (20 μl) of fractions (250 μl) were analyzed by Western blotting with anti-BIG1 and anti-KIF21A antibodies (D) and densitometric quantification (E).
Earlier separation of cytosolic proteins on Sepharose CL-6B had shown peaks of both BIG1 and BIG2 in fractions containing molecules of >670 kDa (4). Similar gel filtration of HepG2 cell cytosol on Superdex 200 was followed by quantification of immunoreactive BIG1 and KIF21A in column fractions by Western blotting and densitometry (Fig. 2 D and E). The peak of KIF21A was two fractions later than that of BIG1, consistent with the presence, at least in part, of BIG1 and KIF21A together in multimolecular complexes.
Interaction of C-Terminal Regions of BIG1 and KIF21A Molecules in Cells.
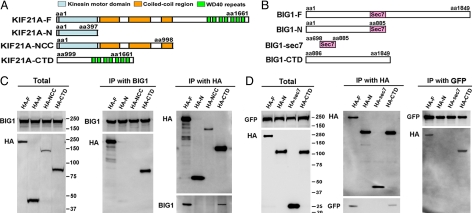
KIF21A molecules contain three recognized functional domains, the N-terminal kinesin family motor region, an ∼ 600-amino acid central region containing three predicted coiled-coil sequences, and the C-terminal tail with seven consensus WD-40 repeats (Fig. 3A). cDNA sequences encoding human KIF21A amino acids 1–397, 1–998, and 999-1661 (termed “N,” “NCC,” and “CTD,” respectively) were cloned into pCMV-HA vector for overexpression of recombinant proteins with N-terminal HA tags. HEK293 cells were transfected with the indicated constructs 24 h before cell extracts were prepared for IP with anti-BIG1 and anti-HA antibodies. For reasons that are not clear, the amounts of HA-NCC in cell extracts were always less than those of the other HA-KIF21A proteins (Fig. 3C, Total). Neither HA-NCC nor HA-N was ever seen after IP with BIG1, which did co-precipitate full-length HA-KIF21A and HA-KIF21A-CTD (Fig. 3C).
Fig. 3.
Identification of interacting regions of BIG1 and KIF21A. Constructs encoding full-length or partial sequences of KIF21A (A) or BIG1 (B) with N-terminal HA (KIF21A, BIG1) tags in pCMV-HA vectors were transfected alone (C) or co-transfected with GFP-KIF21A (D) into HEK293 cells 24 h before preparation of cell extracts. Samples (10 μg) of total extracts (Total) and proteins precipitated with anti-BIG1, anti-HA, or anti-GFP antibodies were separated and immunoblotted with indicated antibodies.
Constructs encoding BIG1 amino acids 1–885, 698–885 (Sec7), 886-1849, or 1–1849 with N-terminal HA tags were each co-transfected with GFP-KIF21A-F (full-length) into HEK293 cells 24 h before preparation of lysates for IP with anti-GFP or anti-HA antibodies. Only full-length BIG1 or its C-terminal portion (CTD) was found after IP of GFP-KIF21A (Fig. 3D). It appears that the C-terminal tail of KIF21A may interact directly with structural elements in the C-terminal region of BIG1.
Effect of ARF1 on Distribution and Interaction of BIG1 and KIF21A.
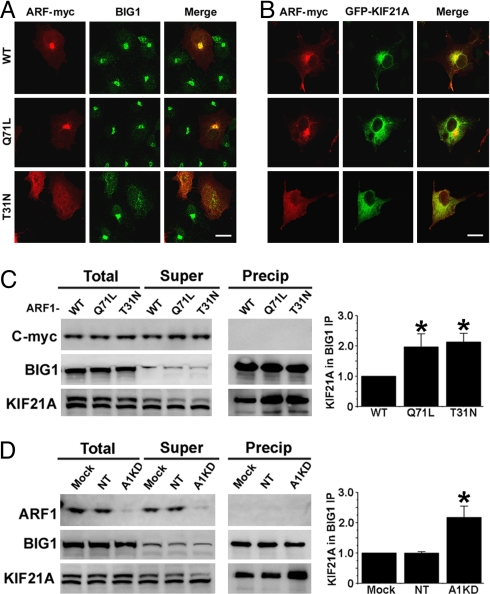
ARF1, as a target of BIG1 guanine nucleotide-exchange activity, has been implicated in its Golgi-associated functions. To assess the potential involvement of ARF1 in the BIG1-KIF21A interaction, constructs for overexpression of WT and two mutants with C-terminal myc tags were prepared. On confocal immunofluorescence microscopy, WT ARF1-myc appeared most concentrated at Golgi membranes near the nucleus, as were BIG1 and KIF21A (Fig. 4 A and B). In cells overexpressing ARF1(Q71L)-myc, which is assumed to be constitutively active with GTP-bound or WT ARF1-myc, distributions of all of these proteins were quite similar, although KIF21A may have been somewhat more diffuse. In contrast, in cells expressing inactive ARF1(T31N)-myc, there was no perinuclear concentration of BIG1 or GFP-KIF21A; each was scattered in punctate collections throughout the cytoplasm. BIG1 and KIF21A, although not clearly co-localized in any region, evidently were not released into cytosol when dominant negative ARF1 caused dispersion of Golgi fragments (Fig. 4, A and B). Interaction of BIG1 and KIF21A was altered, however. With overexpression of either ARF1 (Q71L) or (T31N), co-IP of endogenous KIF21A with BIG1 was approximately twice that seen with WT ARF1-myc, whereas total amounts of the three ARF1 proteins were not very different (Fig. 4C). The effect of ARF1 depletion with siRNA also was notable in that IP with anti-BIG1 antibodies yielded much more KIF21A from ARF1-depleted cells than from control cells (Fig. 4D), although total amounts of BIG1 and KIF21A in ARF1-depleted cells were similar to those in cells treated with siRNA or with vehicle alone (mock). Neither ARF1-myc nor endogenous ARF1 was detected in BIG1 IPs. All these findings are consistent with the notion that ARF1 function (i.e., cyclic activation/inactivation) is needed for the dynamic interaction of BIG1 and KIF21A.
Fig. 4.
Effect of ARF1 overexpression or depletion on interaction of BIG1 and KIF21A. (A) HEK293 cells were transfected with plasmids encoding ARF1-myc, WT or with Q71L or T31N replacements, 24 h before fixation and reaction with anti-myc (red) and anti-BIG1 (green) antibodies for confocal immunofluorescence microscopy. (Scale bar, 16 μm.) (B) Cells were co-transfected with GFP-KIF21A and ARF1-myc, WT or with Q71L or T31N mutations, 24 h before fixation and reaction with anti-myc (red) antibodies. (Scale bar, 16 μm.) (C) IP with anti-BIG1 antibodies of proteins from cells overexpressing ARF1-myc, WT or with Q71L or T31N mutations, samples of proteins (total, 1/3), supernatant (super, 1/3), and precipitate (precip) were separated for immunoblotting with anti-BIG1 and anti-KIF21A antibodies. Amounts of KIF21A in BIG1 IP in three experiments quantified by densitometry and expressed relative to that in WT cells (= 1) are reported as means ± SE. *, P ≤ 0.01 vs. WT. (D) IP with anti-BIG1 antibodies of proteins from cells treated with vehicle (Mock), or nontargeting (NT) or ARF1 siRNA (A1KD) for 48 h before preparation of cell extracts. Samples of proteins were prepared and treated as in Fig. 4C except for immunoblotting with anti-ARF1 to reveal endogenous ARF1 rather than overexpressed ARF1-myc. Amounts of KIF21A in BIG1 IP in three experiments quantified by densitometry and expressed relative to that of the protein in Mock cells (= 1) are reported as means ± SE. *, P ≤ 0.01 vs. mock.
Effects of Microtubule Disruption with Nocodazole and Specific Depletion of BIG1 or KIF21A with siRNA on Their Intracellular Localization.
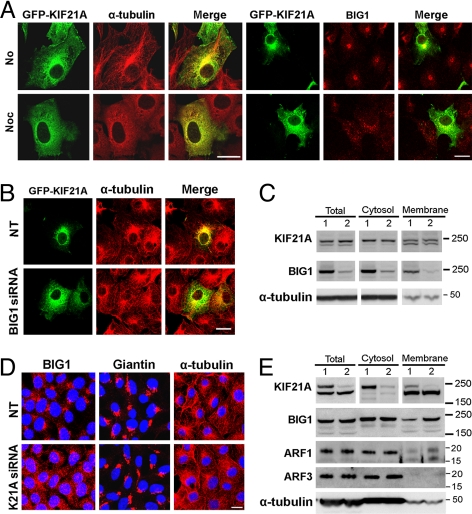
Networks of microtubules in HEK293 cells were seen as linear arrangements of anti-α-tubulin immunofluorescence that extensively overlapped GFP-KIF21A near the nuclei (Fig. 5A). In cells incubated for 1 h with microtubule-destabilizing nocodazole, microtubules were no longer discernible, and α-tubulin, as well as GFP-KIF21A, was scattered diffusely throughout the cytoplasm. Although both endogenous BIG1 and GFP-KIF21A were dispersed completely from their juxta-nuclear concentrations by nocodazole, the scattered collections of BIG1 appeared larger than those of GFP-KIF21A (Fig. 5A), perhaps reflecting their presence in different molecular complexes when no longer associated together with microtubules.
Fig. 5.
Effect nocodazole and siRNA on the intracellular localization of BIG1 and KIF21A. (A) HEK293 cells transfected 24 h earlier with GFP-KIF21A were incubated with DMSO vehicle (No) or nocodazole (Noc, 10 μg/ml) for 1 h before reaction with anti-BIG1 and anti α-tubulin antibodies. (Scale bar, 16 μm.) (B) Cells grown in two-well poly D-lysine-coated slides were transfected with GFP-KIF21A 24 h before incubation for 48 h with nontargeting (NT) or BIG1-specific siRNA, followed by fixation and immunostaining with α-tubulin antibodies (Scale bar, 16 μm). (C) Proteins in cell extracts Total (postnuclear), cytosol, and membrane fractions prepared 72 h after addition of nontargeting (lane 1) or BIG1-specific siRNA (lane 2) treatment, were separated in 4–12% NuPAGE gels and immunoblotted with indicated antibodies. (D) After incubation for 72 h with addition of nontargeting (NT) or KIF21A siRNA, cells were fixed and immunostained with anti-BIG1, giantin and α-tubulin antibodies. (Scale bar, 16 μm.) (E) Samples of cell extracts total (postnuclear), cytosol, and membrane fractions prepared 72 h after nontargeting (lane 1) or KIF21A siRNA (lane 2) treatment were separated in 4–12% NuPAGE gels, and immunoblotted with indicated antibodies.
Amounts of GFP-KIF21A in total, cytosol, and membrane fractions from cells depleted of BIG1 with siRNA were not obviously altered (Fig. 5C), but microscopically GFP-KIF21A appeared to be dispersed from a perinuclear concentration, whereas microtubules appeared intact (Fig. 5B). These observations are all consistent with the notion that an interaction of BIG1 with the KIF21A motor protein, presumably in multimolecular association at vesicle membranes, is necessary for their dynamic accumulation in the perinuclear region. Western blotting of endogenous proteins from cells to which specific KIF21A siRNA had been added 72 h earlier (Fig. 5E) revealed marked depletion of the less mobile of two immunoreactive bands, whereas the band with greater motility remained unchanged. This finding is assumed to offer additional evidence that the protein that was not co-precipitated with anti-BIG1 antibodies (Fig. 2A) is not KIF21A. Although the amounts of ARF3 and α-tubulin in cell fractions appeared unchanged by KIF21A depletion, the abundance of both BIG1 and ARF1 was greater in membrane fractions (Fig. 5E).
Effects of KIF21A depletion were also seen microscopically (Fig. 5D). Although distributions of giantin and TGN230 (data not shown) were unaffected, widespread dispersion of BIG1 in KIF21A-depleted cells resembled that seen after nocodazole treatment (Fig. 5A). Both effects could be a result of interference in different ways with the regulated translocation of BIG1 on vesicles propelled along microtubules by KIF21A. Microtubule disruption with nocodazole, which stops translocation of any molecules that are traveling on microtubules, has been found to cause dispersion of BIG1 throughout the cytoplasm and most strikingly to prevent completely its accumulation in nuclei (11), an effect we did not evaluate here. Overall distribution of BIG1 in KIF21A-depleted cells (Fig. 5D) reflects the presumably continued translocation of BIG1 (with different associated molecules) propelled by other motors, after prevention of its KIF21A-powered transit peripherally from the centrosomal region.
Discussion
Structure–function relationships in the BIG1 molecule were investigated first in the central Sec7 domain that activates class I ARFs (human ARF1 and 3) to initiate coat recruitment and vesicle formation by accelerating the replacement of ARF-bound GDP with GTP (1, 2). The C-terminal region of BIG1 contains a site at which RhoA competes for binding with myosin IXb, a unique motor protein that moves processively from the Golgi toward the peripheral minus-end of the actin filament on which it travels (14, 17). A role for myosin IXb in intracellular BIG1 translocation has not been described, however. We report here the apparently direct interaction of BIG1 with KIF21A, a plus-end-directed motor protein that moves on microtubules.
The dependence on intact microtubules for BIG1 accumulation in cell nuclei was already known (8), but the motor responsible had not been identified, and we have not shown a role for KIF21A in that process. It is known that protein transport in the Golgi complex requires both microtubule- and actin-associated motor proteins (18–21). In amphibian melanophores, long-range transport toward microtubule plus-ends is powered by kinesin-2, whereas myosin V serves in the actin-rich cell cortex to move melanosomes during dispersion (22). A model of cargo transport based on other studies (23) proposed that when a kinesin-propelled cargo moving toward the microtubule plus-end meets a myosin Va diffusing from a microtubule-actin intersection or from another site at which it arrived on the microtubule, the two molecules may associate directly via tail–tail interactions or by myosin Va binding to the cargo. Kinesin power then could continue toward the periphery, where myosin Va would become the motor on actin tracks. Myosin Va was shown to maneuver effectively through actin intersections and Arp2/3-branched filaments (23, 24). Delivery of cargo molecules to their destinations in BIG1-associated vesicles may well require both microtubule and actin transport via the appropriate motors, such as KIF21A and myosin IXb.
The KIF21A molecule comprises three functional domains: an N-terminal (head) motor domain of ∼ 400 aa, a central (predicted) coiled-coil stalk (∼ 600 aa), and C-terminal tail (amino acids 1,000–1,661) containing seven WD-40 repeats (25). The highly conserved motor domain is responsible for microtubule binding and ATP hydrolysis. The stalk domain is a flexible connector between motor and tail that also could participate in interactions with other molecules. Single amino acid mutations in the coiled-coil stalk of KIF21A have been identified in patients with congenital fibrosis of the extraocular muscles types 1 and 3. These conditions are autosomal dominant congenital disorders with strabismus that appears to result from abnormal development of ocular nuclei and nerves (26, 27).
Among KIFs, differences in tail domains typically involve sites of cargo loading, which often depends on an adaptor/scaffolding protein or complex (28). WD-40 repeats were first identified in β-transducin (29) and since then have been found in numerous, functionally unrelated proteins where they are believed to participate in protein–protein interactions (30, 31). Here, using co-IP of overexpressed fragments of the two proteins, we showed that BIG1, which has GEF activity for class I ARF GTPases, interacts with the tail domain of KIF21A. In this way, KIF21A might mediate interactions with BIG1-associated cargoes through its WD-40 domains,
Several workers showed BIG1 localization at trans-Golgi compartments, where it facilitated recruitment of AP and GGA proteins (4, 5, 32). Microscopically, we found significant overlap of BIG1 immunoreaction and GFP-KIF21A in the Golgi region, consistent with their molecular interaction. For a long time, transport processes were thought of and investigated separately from membrane-trafficking events, such as regulated budding and fusion. More recently, however, mechanistic links that integrate these processes in cells have been revealed by the demonstration of regulatory roles for specific Rab molecules in motor protein recruitment (18, 33). Because each of the Rab proteins functions in a specific cellular compartment, it can contribute to the precise spatial and temporal control of numerous molecular interactions required to regulate intracellular transport. Other small GTPases cooperate with specific molecular motors to recruit cargo for vesicular transport via scaffolding or adaptor proteins. For example, a mitochondrial Rho-like GTPase Miro is necessary for the recruitment of kinesin to mitochondria via the adaptor protein Milton, to which it binds (34, 35). Centaurin-α1, a GTPase-activating protein (GAP) for ARF6, associated directly through its GAP domain with the stalk region of KIF13B, an interaction that was essential to maintain its position at the leading edge of a migrating cell (36). These findings add to the evidence that cytoskeletal dynamics and motor protein actions are regulated through the Rab, ARF, and Rho families of GTPases.
Imaging of live cells had revealed that the Golgi complex is dynamic and related to several other organelles through bidirectional molecular transport (37). Disruption of microtubules or blockage of dynein function caused dramatic reorganization of Golgi architecture from a compact juxta-nuclear element at the centrosome to dispersed/tubulo-vesicular structures near endoplasmic reticulum exit sites (18, 20). The balance between molecular transport of molecules into and selective export out of the membranes determines transGolgi composition and morphology. Unlike disruption of microtubules with nocodazole treatment in our experiments, more limited, specific inhibition of microtubule-based transport by depletion of KIF21A content with specific siRNA altered BIG1 distribution without changing the microscopic appearance of intrinsic Golgi membrane proteins. However, similar depletion of BIG1 seemed not to affect KIF21A distribution.
Despite increasing recognition of the diverse molecular associations in which BIG1 participates, its ability to activate ARF1 by accelerating GTP replacement of bound GDP on the inactive cytosolic GTPase, which was the basis for its purification (9), is still a major function. Interfering with the cyclic activation and inactivation of ARF1 by overexpressing constitutively active ARF1 (Q71L) or dominant inactive ARF1 (T31N) altered the distribution of BIG1 as well as its dynamic interaction with KIF21A. The requirement for ARF1 action was confirmed by its selective depletion with siRNA. Dramatic effects of microtubule disruption with nocodazole on the distribution of BIG1 and KIF21A, along with effects of depletion of each protein on the other, emphasize the importance of learning when and how in the process of ARF1-initiated vesicle formation the kinesin motor is recruited (perhaps by BIG1) and incorporated along with other components of transport complexes. More information about the molecular interactions described here should enable us to understand better the mechanisms through which they together are related to the diverse signaling and scaffolding functions of proteins like BIG1, in addition to the mechanisms that integrate membrane trafficking with longer range transport processes.
Materials and Methods
Antibodies and Reagents.
Full-length human KIF21A cDNA (Origene Technologies) was subcloned into PCMV-HA and pEGFP-C3 vectors (Clontech). Plasmids encoding full-length human HA-BIG1 or ARF1-myc (WT and with Q71L, or T31L mutations) were kindly provided by Dr. Heather Jones and Toyoko Hiroi, who prepared them. Affinity-purified rabbit antibodies against human BIG1 have been described (1). Polyclonal rabbit antibodies against KIF21A and giantin were purchased from Aviva Systems Biology and Covance, respectively; mouse monoclonal antibody against c-myc was purchased from Zymed Laboratories Inc.; mouse monoclonal antibody against α-tubulin and rabbit anti-HA antibodies were purchased from Sigma; Alexa 488 and 594-conjugated secondary antibodies were purchased from Molecular Probes; EDTA-free protease-inhibitor mixture and GeneJuice reagent were purchased from Roche and Novagen, respectively.
Immunoprecipitation and Western Blotting.
To identify previously unrecognized proteins that interact with BIG1, we prepared extracts of HepG2 cells grown in DMEM containing 10% FBS, penicillin G (100 units/ml), and streptomycin (100 μg/ml) at 37 °C in an atmosphere of 5% CO2/95% air. Cells from two 10-cm plates (90% confluence) were harvested by scraping in cold MLB buffer (25 mM Hepes, pH 7.5, 150 mM NaCl, 1 mM EDTA, 10 mM MgCl2, 1% CA-630, 10% glycerol, and EDTA-free protease inhibitor mixture). After centrifugation of cell lysates (5 min, 14,000 × g, 4 °C), samples of supernatant termed “cell extract” were used for IP. Samples of total extract (500 μg of protein) plus 5 μg of BIG1 antibodies or normal rabbit IgG and 50 μl of protein A/G beads (50% slurry, Pierce) were incubated at 4 °C overnight with agitation. Beads were washed three times with MLB buffer before elution (95 °C, 5 min) of bound proteins in 100 μl of gel-loading buffer. Samples of eluted proteins were separated by electrophoresis in 4–12% NuPAGE gel, transferred to nitrocellulose membranes, and incubated with anti-BIG1 antibodies, followed by appropriate horseradish peroxidase-conjugated secondary antibodies and Super Signal Chemiluminescent substrate (Pierce).
Procedures used for IP and Western blotting were identical in all experiments with quantities adjusted for scale of each. All observations were replicated at least three times.
Identification of proteins immunoprecipitated with BIG1: in-gel trypsin digestion and nanospray LC-MS/MS analysis. Proteins immunoprecipitated with BIG1 antibodies or normal rabbit IgG (control) from 1 mg of total cell extract were separated in 4–12% NuPAGE gel, and stained with Coomassie blue R-250. After destaining, lanes were divided (by razor blade) into segments based on molecular size markers. Proteins in each gel block were reduced, alkylated with iodoacetamide, and trypsinized. Peptides were extracted, dried, and dissolved in 0.1% formic acid for LC-MS/MS analysis using a Thermo Finnigan ProteomeX workstation (LCQ Deca XP MS) and LCQ Deca XP mass spectrometer equipped with a nano-spray ion source. Three MS/MS scans using the dynamic exclusion option followed each MS survey scan. BioWorks 3.1 (Thermo Finnigan) was used for database search (38).
Depletion of Endogenous Proteins with siRNA and Immunofluorescence Microscopy.
All siRNAs and other reagents were purchased from Dharmacon Research and used as in previous experiments (39).
For immunofluorescence microscopy, cells grown on four-well slides were fixed with 3.7% paraformaldehyde in PBS (15 min, room temperature), blocked for 30 min with 10% normal goat serum and 0.1% saponin in PBS, and incubated 1 h at room temperature with primary antibodies diluted in blocking buffer (BIG1, 1:500, α-tubulin, 1:1000, Giantin, 1:1000). After washing with PBS, cells were incubated for 1 h with Alexa Fluor-labeled secondary antibodies (diluted 1:1,000 in blocking buffer) and washed with PBS. Coverslips were mounted in Prolong Gold anti-fade reagent with DAPI (Molecular Probes) and inspected with a confocal microscope (Leica model TCS4D/DMIRBE).
Superdex 200 Chromatography of HepG2 Cell Cytosolic Fractions.
To prepare cytosol, HepG2 cells (∼ 10 × 105) were washed, scraped, and homogenized (20 strokes, in a Dounce tissue grinder) in buffer A (50 mM Hepes, 50 mM sucrose, 1 mM EDTA, 10 mM PPi, 5 mM NaF, 100 mM NaCl, 1 μM okadaic acid, 1 mM Na3VO4, and Roche protease inhibitor mixture). Homogenates were centrifuged (800 × g, 10 min), and supernatants (postnuclear fractions) were centrifuged (105,000 × g, 1.5 h) to separate cytosol (supernatant) from membrane fractions (pellet). Samples of cytosol (1 ml), adjusted to a concentration of 3 mg/ml by concentration with Centriprep centrifugal filter device (Millipore), were applied to a Superdex 200 column (1 × 30 cm, Amersham) that was equilibrated and eluted with buffer A with 150 mM NaCl and no sucrose. Samples (20 μl) of fractions (250 μl) were used for immunoblotting with BIG1 and KIF21a antibodies.
Supplementary Material
Acknowledgments.
We thank Dr. Christian Combs and Dr. Daniela Malide (National Heart, Lung, and Blood Institute Confocal Microscopy Core Facility) for invaluable help. This research was supported by the Intramural Research Program of the National Institutes of Health, National Heart, Lung, and Blood Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810104105/DCSupplemental.
References
- 1.Moss J, Vaughan M. Molecules in the ARF orbit. J Biol Chem. 1998;273:21431–21434. doi: 10.1074/jbc.273.34.21431. [DOI] [PubMed] [Google Scholar]
- 2.Jackson CL, Casanova JE. Turning on ARF: The Sec7 family of guanine-nucleotide-exchange factors. Trends Cell Biol. 2000;10:60–67. doi: 10.1016/s0962-8924(99)01699-2. [DOI] [PubMed] [Google Scholar]
- 3.Mouratou B, et al. The domain architecture of large guanine nucleotide exchange factors for the small GTP-binding protein Arf. BMC Genomics. 2005;6:20. doi: 10.1186/1471-2164-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamaji R, et al. Identification and localization of two brefeldin A-inhibited guanine nucleotide-exchange proteins for ADP-ribosylation factors in a macromolecular complex. Proc Natl Acad Sci USA. 2000;97:2567–2572. doi: 10.1073/pnas.97.6.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao X, Lasell TK, Melancon P. Localization of large ADP-ribosylation factor-guanine nucleotide exchange factors to different Golgi compartments: Evidence for distinct functions in protein traffic. Mol Biol Cell. 2002;13:119–133. doi: 10.1091/mbc.01-08-0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen X, Hong MS, Moss J, Vaughan M. BIG1, a brefeldin A-inhibited guanine nucleotide-exchange protein, is required for correct glycosylation and function of integrin beta1. Proc Natl Acad Sci USA. 2007;104:1230–1235. doi: 10.1073/pnas.0610535104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Padilla PI, Pacheco-Rodriguez G, Moss J, Vaughan M. Nuclear localization and molecular partners of BIG1, a brefeldin A-inhibited guanine nucleotide-exchange protein for ADP-ribosylation factors. Proc Natl Acad Sci USA. 2004;101:2752–2757. doi: 10.1073/pnas.0307345101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Citterio C, et al. Effect of protein kinase A on accumulation of brefeldin A-inhibited guanine nucleotide-exchange protein 1 (BIG1) in HepG2 cell nuclei. Proc Natl Acad Sci USA. 2006;103:2683–2688. doi: 10.1073/pnas.0510571103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Togawa A, Morinaga N, Ogasawara M, Moss J, Vaughan M. Purification and cloning of a brefeldin A-inhibited guanine nucleotide-exchange protein for ADP-ribosylation factors. J Biol Chem. 1999;274:12308–12315. doi: 10.1074/jbc.274.18.12308. [DOI] [PubMed] [Google Scholar]
- 10.Padilla PI, et al. Interaction of FK506-binding protein 13 with brefeldin A-inhibited guanine nucleotide-exchange protein 1 (BIG1): Effects of FK506. Proc Natl Acad Sci USA. 2003;100:2322–2327. doi: 10.1073/pnas.2628047100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Adamik R, Pacheco-Rodriguez G, Moss J, Vaughan M. Protein kinase A-anchoring (AKAP) domains in brefeldin A-inhibited guanine nucleotide-exchange protein 2 (BIG2) Proc Natl Acad Sci USA. 2003;100:1627–1632. doi: 10.1073/pnas.0337678100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuroda F, Moss J, Vaughan M. Regulation of brefeldin A-inhibited guanine nucleotide-exchange protein 1 (BIG1) and BIG2 activity via PKA and protein phosphatase 1gamma. Proc Natl Acad Sci USA. 2007;104:3201–3206. doi: 10.1073/pnas.0611696104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Padilla PI, et al. Association of guanine nucleotide-exchange protein BIG1 in HepG2 cell nuclei with nucleolin, U3 snoRNA, and fibrillarin. Proc Natl Acad Sci USA. 2008;105:3357–3361. doi: 10.1073/pnas.0712387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saeki N, Tokuo H, Ikebe M. BIG1 is a binding partner of myosin IXb and regulates its Rho GTPase-activating protein activity. J Biol Chem. 2005;280:10128–10134. doi: 10.1074/jbc.M413415200. [DOI] [PubMed] [Google Scholar]
- 15.Hirokawa N, Noda Y. Intracellular transport and kinesin superfamily proteins, KIFs: Structure, function, and dynamics. Physiol Rev. 2008;88:1089–1118. doi: 10.1152/physrev.00023.2007. [DOI] [PubMed] [Google Scholar]
- 16.Miki H, Setou M, Kaneshiro K, Hirokawa N. All kinesin superfamily protein, KIF, genes in mouse and human. Proc Natl Acad Sci USA. 2001;98:7004–7011. doi: 10.1073/pnas.111145398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inoue A, Saito J, Ikebe R, Ikebe M. Myosin IXb is a single-headed minus-end-directed processive motor. Nat Cell Biol. 2002;4:302–306. doi: 10.1038/ncb774. [DOI] [PubMed] [Google Scholar]
- 18.Caviston JP, Holzbaur EL. Microtubule motors at the intersection of trafficking and transport. Trends Cell Biol. 2006;16:530–537. doi: 10.1016/j.tcb.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 19.Egea G, Lázaro-Diéguez F, Vilella M. Actin dynamics at the Golgi complex in mammalian cells. Curr Opin Cell Biol. 2006;18:168–178. doi: 10.1016/j.ceb.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 20.Allan VJ, Thompson HM, McNiven MA. Motoring around the Golgi. Nat Cell Biol. 2002;4:E236–E242. doi: 10.1038/ncb1002-e236. [DOI] [PubMed] [Google Scholar]
- 21.Palmer KJ, Watson P, Stephens DJ. The role of microtubules in transport between the endoplasmic reticulum and Golgi apparatus in mammalian cells. Biochem Soc Symp. 2005;72:1–13. doi: 10.1042/bss0720001. [DOI] [PubMed] [Google Scholar]
- 22.Gross SP, et al. Interactions and regulation of molecular motors in Xenopus melanophores. J Cell Biol. 2002;156:855–865. doi: 10.1083/jcb.200105055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ali MY, et al. Myosin Va maneuvers through actin intersections and diffuses along microtubules. Proc Natl Acad Sci USA. 2007;104:4332–4336. doi: 10.1073/pnas.0611471104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammer JA, 3rd, Wu X. Slip sliding away with myosin V. Proc Natl Acad Sci USA. 2007;104:4332–4336. doi: 10.1073/pnas.0701071104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marszalek JR, Weiner JA, Farlow SJ, Chun J, Goldstein LS. Novel dendritic kinesin sorting identified by different process targeting of two related kinesins: KIF21A and KIF21B. J Cell Biol. 1999;145:469–479. doi: 10.1083/jcb.145.3.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu S, Zhao C, Zhao K, Li N, Larsson C. Novel and recurrent KIF21A mutations in congenital fibrosis of the extraocular muscles type 1 and 3. Archives of Ophthalmology. 2008;126:388–394. doi: 10.1001/archopht.126.3.388. [DOI] [PubMed] [Google Scholar]
- 27.Chan WM, et al. Three novel mutations in KIF21A highlight the importance of the third coiled-coil stalk domain in the etiology of CFEOM1. BMC Genetics. 2007;8:26. doi: 10.1186/1471-2156-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ali M, Venkatesh C, Ragunath A, Kumar A. Mutation analysis of the KIF21A gene in an Indian family with CFEOM1: Implication of CpG methylation for most frequent mutations. Ophthalmic Genetics. 2004;25:247–255. doi: 10.1080/13816810490498198. [DOI] [PubMed] [Google Scholar]
- 29.Simon MI, Strathmann MP, Gautam N. Diversity of G proteins in signal transduction. Science. 1991;252:802–808. doi: 10.1126/science.1902986. [DOI] [PubMed] [Google Scholar]
- 30.van der Voorn L, Ploegh HL. The WD-40 repeat. FEBS Lett. 1992;307:131–134. doi: 10.1016/0014-5793(92)80751-2. [DOI] [PubMed] [Google Scholar]
- 31.Neer EJ, Schmidt CJ, Nambudripad R, Smith TF. The ancient regulatory protein family of WD-repeat proteins. Nature. 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- 32.Manolea F, Claude A, Chun J, Rosas J, Melançon P. Distinct functions for Arf guanine nucleotide exchange factors at the Golgi complex: GBF1 and BIGs are required for assembly and maintenance of the Golgi Stack and trans-Golgi network, respectively. Mol Biol Cell. 2008;19:523–535. doi: 10.1091/mbc.E07-04-0394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoepfner S, et al. Modulation of receptor recycling and degradation by the endosomal kinesin KIF16B. Cell. 2005;121:437–450. doi: 10.1016/j.cell.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 34.Guo X, et al. The GTPase dMiro is required for axonal transport of mitochondria to Drosophila synapses. Neuron. 2005;47:379–393. doi: 10.1016/j.neuron.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 35.Glater EE, Megeath LJ, Stowers RS, Schwarz TL. Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J Cell Biol. 2006;173:545–557. doi: 10.1083/jcb.200601067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Venkateswarlu K, Hanada T, Chishti AH. Centaurin-alpha1 interacts directly with kinesin motor protein KIF13B. J Cell Sci. 2005;118:2471–2484. doi: 10.1242/jcs.02369. [DOI] [PubMed] [Google Scholar]
- 37.De Matteis MA, Luini A. Exiting the Golgi complex. Nat Rev Mol Cell Biol. 2008;9:273–284. doi: 10.1038/nrm2378. [DOI] [PubMed] [Google Scholar]
- 38.Wang G, Wu WW, Zeng W, Chou CL, Shen RF. Label-free protein quantification using LC-coupled ion trap or FT mass spectrometry: Reproducibility, linearity, and application with complex proteomes. Journal of Proteome Research. 2006;5:1214–1223. doi: 10.1021/pr050406g. [DOI] [PubMed] [Google Scholar]
- 39.Shen X, et al. Association of brefeldin A-inhibited guanine nucleotide-exchange protein 2 (BIG2) with recycling endosomes during transferrin uptake. Proc Natl Acad Sci USA. 2006;103:2635–2640. doi: 10.1073/pnas.0510599103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.