Abstract
Modification of histone proteins by lysine methylation is a principal chromatin regulatory mechanism (Shi, Y., and Whetstine, J. R. (2007) Mol. Cell 25, 1–14). Recently, lysine methylation has been shown also to play a role in regulating non-histone proteins, including the tumor suppressor protein p53 (Huang, J., and Berger, S. L. (2008) Curr. Opin. Genet. Dev. 18, 152–158). Here, we identify a novel p53 species that is dimethylated at lysine 382 (p53K382me2) and show that the tandem Tudor domain of the DNA damage response mediator 53BP1 acts as an “effector” for this mark. We demonstrate that the 53BP1 tandem Tudor domain recognizes p53K382me2 with a selectivity relative to several other protein lysine methylation sites and saturation states. p53K382me2 levels increase with DNA damage, and recognition of this modification by 53BP1 facilitates an interaction between p53 and 53BP1. The generation of p53K382me2 promotes the accumulation of p53 protein that occurs upon DNA damage, and this increase in p53 levels requires 53BP1. Taken together, our study identifies a novel p53 modification, demonstrates a new effector function for the 53BP1 tandem Tudor domain, and provides insight into how DNA damage signals are transduced to stabilize p53.
Lysine methylation is a principal mechanism involved in chromatin regulation via modification of histone proteins (1). Recently, lysine methylation has been shown to regulate non-histone proteins, including the tumor suppressor p53 (2). p53 plays a central role in directing cellular responses to DNA damage, including the most dangerous DNA lesion, double strand breaks (DSBs)3 (3). A complex network of p53 posttranslational modifications aids in the coordination of these activities (4). Three different lysine residues present within the C-terminal regulatory region of p53 are validated as sites of lysine methylation (5–8). Each of these methylation events either stimulates or represses p53 transcriptional activity, yet with multiple additional lysines in the C terminus of p53 as potential methylation sites, and possible mono-, di-, and trimethylation states, the role of methylation in regulation of p53 and the molecular mechanisms linking different p53 methylation events to biological outcomes are just beginning to be understood.
53BP1 (p53-binding protein 1) is a key mediator of the cell's response to DSBs (9). Upon the induction of DSB lesions, 53BP1 rapidly relocates to the sites of breaks and is believed to promote the stabilization of additional DNA damage response factors at DSBs (9). The recognition of histone H4 dimethylated at lysine 20 (H4K20me2) by the 53BP1(TD) has been shown to be important for 53BP1 localization to DSBs: linking chromatin structure, lysine methylation, and DSB signaling (10). 53BP1 might also have roles in transcription regulation. For example, a recent study reported that 53BP1 recognizes p53 dimethylated at lysine 370 through its Tudor domain and modulates p53 transactivation at several target genes (7).
Here, we identify a number of novel lysine methylated p53 species and provide the first direct evidence of endogenous p53 dimethylated at lysine 382. We show that p53K382me2 is a DNA damage-associated species and that through its recognition by the 53BP1(TD), it is important for regulating a modular and DNA damage-dependent interaction between p53 and 53BP1. This interaction facilitates p53 stabilization in response to DSBs, suggesting that one mechanism by which DSB signals are transduced to activate p53 is via posttranslational modification of p53 by lysine methylation.
MATERIALS AND METHODS
Constructs and Reagents—Human 53BP1 cDNA and hemagglutinin-53BP1 constructs were gifts from Jiri Lukas and Phillip Carpenter, respectively. The 53BP1 tandem Tudor domain and mutants were cloned and generated in pGEX vectors; SET8(Y334F) was generated in pcDNA and pGEX vectors using PCR mutagenesis (Stratagene). Primer sequences are available upon request. The p53K382me2 antibody was generated in rabbits immunized with the peptide 377TSRHKK(me2)LMFKT387, purified over a SulfoLink Coupling Gel (Pierce) coupled to the p53K382me2 peptide, and depleted with recombinant wild-type GST-p53. Other antibodies used in this study include horseradish peroxidase-p53 (R&D Systems); p53 (DO1; Calbiochem); SET8 (Abcam); 53BP1, mouse monoclonal (Upstate) and rabbit polyclonal (Bethyl Laboratories); FLAG (M2); and tubulin (Sigma). p53 peptides bearing different modifications were synthesized at the W. M. Keck Facility at Yale University.
Protein Purification and Mass Spectrometry—Nuclear extracts were prepared from HeLa cells ± doxorubicin treatment (0.5 μg/ml for 4 h) as previously described (8). To immunoprecipitate endogenous p53 proteins, ∼10 mg of nuclear extracts were incubated overnight with 50 μl of DO1-conjugated agarose in buffer containing 20 mm Tris-Cl, pH 8.0, 150 mm NaCl, 0.01% SDS, 1% Triton X-100, 1 mm EDTA, and protease inhibitors with gentle rotation at 4 °C. The beads were washed two times with the same buffer, two times with high salt buffer (20 mm Tris-Cl, pH 8.0, 500 mm NaCl, 0.1% SDS, 1% Triton X-100, 2 mm EDTA), once with LiCl buffer (20 mm Tris, pH 8.0, 500 mm LiCl, 1% Nonidet P-40, 1% deoxycholate, 1 mm EDTA), and once with 10 mm Tris-HCl, pH 8.0, 1 mm EDTA. The p53 protein bound to the beads was subjected to SDS-PAGE, excised from the gels, and incubated with trypsin overnight at 37 °C. Pooled supernatants containing extracted peptides were dried and resuspended in 30% acetonitrile and 0.1% trifluoroacetic acid prior to mass spectrometry analysis. Samples were analyzed on a reflectron time-of-flight mass spectrometer (MALDI TOF instrument, Ultraflex, Bruker Daltonics, Billerica, MA) equipped with a 337-nm nitrogen laser and delayed ion extraction capability (delay times, 30–50 ns). Ion structure information was obtained by post-source decay, using the mass gate feature, to select a specific m/z window for fragmentation. The mass gate resolution was 1% of the precursor mass. Data were recorded in both positive and negative ion modes at 20-kV acceleration, and mass analysis of ions was performed using a dual micro-channel plate detector. Detector output was collected with a 1-GHz digitizer and displayed directly on a Windows NT-based computer. Ten positive ion reflectron time-of-flight mass spectra of 1000 laser shots were accumulated and externally calibrated with commercial peptide mix (Bruker Daltonics). For analysis of in vitro methylated synthetic peptides, the synthetic peptides, untreated and treated with SET8, were equilibrated with 0.1% trifluoroacetic acid and 50% acetonitrile with 0.1% trifluoroacetic acid and applied to the MALDI target plate with equal volumes of the matrix α-cyano-4-hydroxycinnamic acid (Sigma).
Peptide Pulldown Assays—Peptide pulldown assays were performed as previously described (11). Briefly, 1 μg of biotinylated peptides was incubated with 1 μg of protein in binding buffer (50 mm Tris-HCl, pH 7.5, 300 mm NaCl, 0.1% Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitors) for 4 h at 4 °C with rotation. After a 1-h incubation with streptavidin beads (Amersham Biosciences) and extensive washing, bound proteins were analyzed by SDS-PAGE and Western blotting.
Histone Methyltransferase Assay—Methyltransferase assays were performed as previously described (8). Briefly, 2 μgof GST-p53 or 1 μg of p53 peptides was incubated with 1 μgof recombinant histone methyltransferase and 2 μCi of S-adenosylmethionine (Amersham Biosciences) in reaction buffer containing 50 mm Tris-HCl, pH 8.0, 10% glycerol, 20 mm KCl, 5 mm MgCl2, 1 mm dithiothreitol, and 1 mm phenylmethylsulfonyl fluoride at 30 °C for 30 min to 2 h. The reaction mixtures were then subjected to SDS-PAGE, followed by either radiography or Western analysis. The reactions with peptides were subjected to mass spectrometry analysis.
Cell Culture and Transfections—U2OS, H1299, and 293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Cells were transfected with plasmids or siRNA duplexes by TransIT-LT1 (or TransIT-293) (Mirus) or DharmaFECT (Dharmacon), respectively, according to the manufacturers' protocols.
Knockdown of 53BP1—100 nm control or 53BP1 siRNAs was transfected into U2OS cells using DharmaFECT4 transfection reagent (Dharmacon) for 48–96 h before treatments. siRNA target sequences for 53BP1 are 5′-GAGCUGGGAAGUAUAAAUUUU-3′ and 5′-GGACUCCAGUGUUGUCAUUUU-3′. On-target plus siControl siRNA (5′-UGGUUUACAUGUCGACUAA-3′, Dharmacon) and on-target plus SMARTpool siControl siRNA (Dharmacon) were used as controls.
Immunoprecipitation and Western Immunoblotting—Endogenous p53 or ectopically expressed FLAG-tagged p53 were immunoprecipitated with agarose-conjugated p53 or FLAG antibodies from whole cell extracts in cell lysis buffer (50 mm Tris-HCl, pH 7.4, 250 mm NaCl, 0.5% Triton X-100, 10% glycerol, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitors). After an overnight incubation at 4 °C, the beads were washed three times with the same buffer and boiled in 2× Laemmli buffer. The IP was resolved on sodium dodecyl sulfate-polyacrylamide gel and detected by αp53K382me2 and α53BP1 antibodies, or horseradish peroxidase-αp53 to avoid cross-reactivity with IgG heavy chain.
Reverse Transcription-PCR and Real-time PCR—Reverse transcription-PCR and real-time PCR were performed as previously described (8). mRNA was prepared using an RNeasy Plus kit (Qiagen) and reverse-transcribed using a First Strand Synthesis kit (Invitrogen). Quantitative real-time reverse transcription-PCR was performed on the ABI PRISM 7700 Sequence Detection System (Applied Biosystems). Gene expression was calculated and normalized to glyceraldehyde-3-phosphate dehydrogenase levels by the comparative Cycle threshold method.
RESULTS
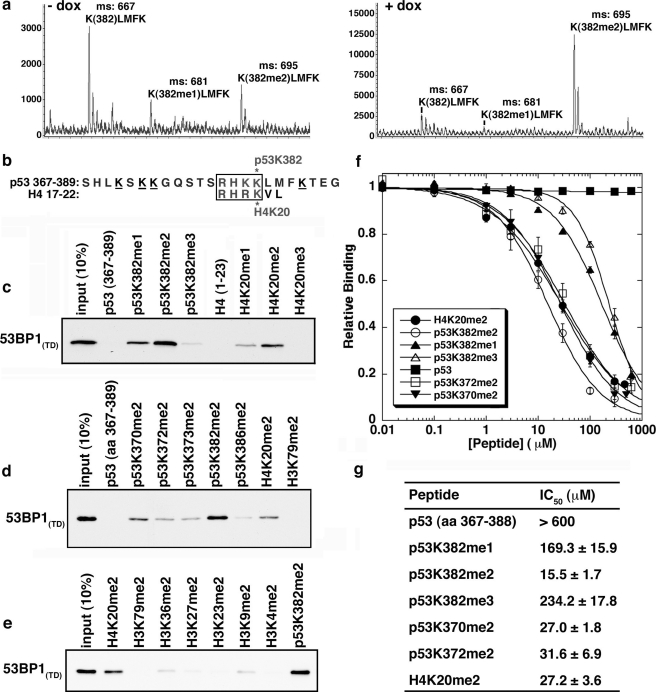
p53 Is Dimethylated at Lysine 382 in Vivo—To search for novel p53 methylation marks, we performed a mass spectrometry analysis of endogenous p53 purified from HeLa nuclear extracts ± DNA damage (Fig. 1a; supplemental Fig. 1; see “Materials and Methods”). In both samples, we identified three peaks representing the digested peptide product containing lysine 382 (K(382)LMFK) that either was unmodified or was shifted by a mass that corresponds to one and two methyl moieties (Fig. 1a). We have previously reported the second peak as the monomethylated species, K(382me1)LMFK (8). The identity of the third peak was confirmed by tandem mass spectrometry to represent dimethylated Lys382 species. These data demonstrate that endogenous p53 is dimethylated at lysine 382 (p53K382me2).
FIGURE 1.
53BP1(TD) binds specifically to p53K382me2 in vitro. a, identification of endogenous p53 dimethylated at lysine 382 by mass spectrometry analysis. Unmethylated, monomethylated, and dimethylated Lys382 within the KLMFK peptide was obtained from trypsinized purified endogenous p53 from HeLa nuclear extract (left, no DNA damage; right,4hof doxorubicin (dox)). b, alignment of amino acid sequences surrounding p53K382 (amino acids 367–389) and H4K20 (amino acids 17–22). Boxed residues, a binding site for 53BP1(TD) (10); underlined residues, other lysines in the p53 C terminus. Asterisks indicate methylation sites. c–e, 53BP1(TD) preferentially binds p53K382me2 peptides. Shown are the results from Western analysis of pulldowns with the indicated biotinylated peptides and GST-53BP1(TD) or GST control. f and g, the binding affinities (IC50) of p53 and H4 peptides for 53BP1(TD) (shown in g), determined by competition assays (with the indicated nonlabeled peptides) using fluorescence anisotropy change of fluorescein-labeled H4K20me2 of known affinity.
We also identified p53 peptides in the digests that contain lysine mono- and dimethylation events (supplemental Fig. 1). Specifically, we have evidence for two novel methylated p53 species, K386me1 and K386me2, as well as potentially confirm by mass spectrometry the previously reported K370me2 species (6, 7).
p53K382me2 Is a High-affinity Ligand for 53BP1 Tudor Domain in Vitro—The amino acid sequence surrounding p53K382 is highly homologous to that of histone H4K20 (Fig. 1b). 53BP1(TD) binds to H4K20me2 via the formation of contacts with the sequence HRKme2 (10). We therefore reasoned that 53BP1(TD) might likewise recognize p53K382me2 through the similar sequence HKKme2. Indeed, as shown in Fig. 1c, in in vitro binding assays, recombinant 53BP1(TD) preferentially bound p53K382me2 peptides versus other p53K382 methylation states. Furthermore, the binding affinity of 53BP1(TD) for p53K382me2 was moderately stronger than that observed for H4K20me2 and p53K370me2 (15.5 μm versus 27.2 and 27.0 μm, respectively), as well as multiple other histone lysine dimethylation sites and potential or reported p53 dimethylation sites (Fig. 1, c–g). Many other methyllysine effector domains, including numerous plant homeodomain fingers and chromodomains, do not bind to p53K382me2 (data not shown). Thus, in vitro, p53K382me2 is a novel and high-affinity binding ligand for the tandem Tudor domain of 53BP1.
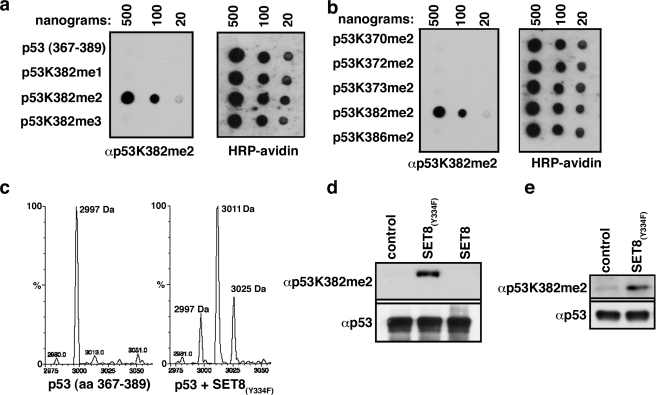
Generation of p53K382me2 in Vitro and in Vivo—Next, a site-specific polyclonal antibody recognizing p53K382me2 was raised. This antibody is specific for dimethylated p53K382me2 and does not recognize unmethylated, monomethylated, or trimethylated p53K382 (Fig. 2a), several additional p53 dimethylation sites (Fig. 2b), and H4K20me2 (supplemental Fig. 2). We have not yet identified the enzyme that generates p53K382me2, despite testing more than 30 lysine methyltransferase enzymes (data not shown). Therefore, to investigate the biological role of p53K382me2, we established a system to generate this species in vitro and in vivo. SET8/PR-Set7 adds a single methyl moiety to H4K20 (12–14) as well as p53K382 (8). Substitution of Tyr334 with Phe within SET8 (SET8(Y334F)) converts its product specificity with respect to H4K20 from a strict monomethyltransferase to a mono- and dimethyltransferase (15). Accordingly, incubation of a p53 peptide encompassing Lys382 (amino acids 367–389) with recombinant SET8(Y334F) generated mono- and dimethylation at p53K382 (Fig. 2c; data not shown). We confirmed that SET8(Y334F) dimethylated p53K382 in the context of recombinant full-length p53 in vitro (Fig. 2d) and validated its activity in cells on endogenous p53 (Fig. 2e). Thus, the SET8(Y334F) enzyme provides a tool for modulation of p53K382 dimethylation levels in vitro and in cells.
FIGURE 2.
In vitro and in vivo generation of p53K382me2. a and b, specific recognition of p53K382me2 by an αp53K382me2 antibody. Dot blot analyses of the indicated biotinylated peptides (a, p53K382me 0–3; b, p53 amino acids (aa) 367–389 with indicated dimethylated lysines) with p53K382me2 antibody are shown. Blots were probed with horseradish peroxidase (HRP)-conjugated streptavidin to control for peptide loading. c, p53 is mono- and dimethylated by SET8(Y334F) at Lys382. Mass spectrometry analysis of p53 peptide (amino acids 367–389) before (left) and after (right) SET8(Y334F) methyltransferase reaction is shown. Masses of peptides are indicated. d, SET8(Y334F) but not wild-type SET8 generates p53K382me2. Western blot analysis with αp53K382me2 of methyltransferase assays on recombinant p53 protein is shown. Total p53 levels detected with DO1 antibody demonstrate equal loading. e, SET8(Y334F) dimethylates endogenous p53 at Lys382 in vivo. Western blot analysis with the indicated antibodies (anti-horseradish peroxidase-p53 for p53 levels) of p53 (DO1) IPs from 293T cells expressing SET8(Y334F) is shown.
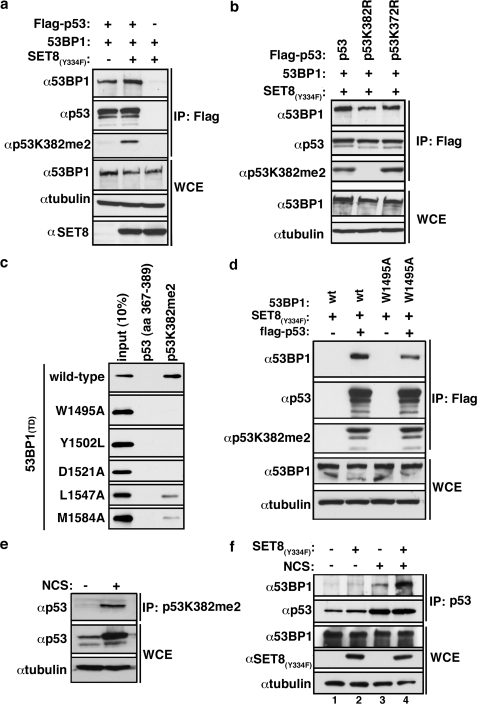
Methylation of p53K382 Modulates Interaction between p53 and 53BP1—53BP1 was originally identified in a two-hybrid screen as a p53-interacting protein (16). Subsequently, the 53BP1 BRCT tandem repeats were shown to bind the DNA-binding domain of p53, although the physiologic context in which this interaction functions remains elusive (9). To determine whether 53BP1(TD) recognition of p53K382me2 can augment the base-line p53-53BP1 interaction, we performed co-immunoprecipitation experiments. As shown in Fig. 3a, the ability of FLAG-p53 to co-immunoprecipitate hemagglutinin-53BP1 was enhanced when levels of p53 dimethylation at p53K382 were increased via co-expression with SET8(Y334F). Substitution of p53K382 with Arg abolished this SET8(Y334F)-mediated increase, whereas the corresponding substitution at p53K372 did not (Fig. 3b). Thus, the enhanced interaction of p53 with 53BP1 following p53 dimethylation by SET8(Y334F) specifically requires Lys382. We note that the affinity of 53BP1(TD) for p53K382me1 is 10-fold lower than for p53K382me2 (Fig. 1g), and therefore it is unlikely that the generation of this species by SET8(Y334F) has a major impact on the p53–53BP1 interaction.
FIGURE 3.
p53K382me2 levels increase with DNA damage to facilitate the interaction between p53 and 53BP1 in vivo. a, ectopic SET8(Y334F) augments 53BP1-p53 interaction. Western blot analysis with the indicated antibodies of whole cell extract (WCE) and FLAG IPs from 293T cells expressing the indicated proteins is shown. Anti-tubulin of whole cell extract is shown to control for loading.b, p53K382 mutation abolishes the methylation-dependent increase of the p53-53BP1 interaction. Western blot analysis as in a in cells expressing FLAG-tagged wild-type or mutant p53 as indicated is shown. c, conservation of the molecular basis of 53BP1(TD) recognition of H4K20me2 and p53K382me2. The indicated GST-53BP1 tandem Tudor domain mutants were tested for binding to p53K382me2 as in Fig. 1c. d, an intact tandem Tudor domain is required for robust 53BP1 recognition of p53 in cells. A Western blot analysis as in a in cells expressing the indicated proteins is shown. e, elevation of endogenous p53K382me2 levels upon DNA damage. Endogenous p53 levels are present in αp53K382me2 IPs from U2OS cells ± 45 nm NCS. Total p53 present in the whole cell extract (input) is shown. f, endogenous association between p53 and 53BP1is augmented by DNA damage and dimethylation at p53K382. Western analysis of p53-bound proteins in co-immunoprecipitates from U2OS cells ± 45 nm NCS and ± SET8(Y334F) expression is shown.
Key Residues within 53BP1 Tudor Domain Are Required for Binding Methylated p53—Due to homology in the amino acid sequence surrounding p53K382 and H4K20, we turned to the crystal structure of the 53BP1(TD)-H4K20me2 peptide complex to obtain insight into the molecular basis of p53K382me2-recognition by the 53BP1(TD) (see Fig. 1b) (10). Mutation of residues within the first Tudor domain of the 53BP1(TD) that constitute the binding cage accommodating the dimethyllysine of H4K20 (W1495A, Y1502L, D1521A), as well as the residues that contact a histidine residue two amino acids N-terminal of the methylated lysine (L1547A, M1584A), abolished or severely compromised the interaction with p53K382me2 (Fig. 3c) (10).
An intact 53BP1(TD) is critical for optimal binding to Lys382-methylated p53 in the context of full-length 53BP1, as introduction of a mutation within the methyllysine binding pocket of 53BP1 (W1495A) impaired its ability to be co-immunoprecipitated with p53 (Fig. 3d). Previous structural analysis of the 53BP1(TD) verified that these mutations do not disrupt folding (10, 17). Taken together, these results indicate that recognition of p53K382me2 by the tandem Tudor domain of 53BP1 can modulate the p53-53BP1 interaction in cells.
Interaction between Endogenous p53 and 53BP1 Increases with DNA Damage—Both p53 and 53BP1 are key mediators of cellular responses to DSBs (9), raising the possibility that p53K382me2 generation, as well as the p53K382me2-53BP1(TD) interaction, might be linked to genotoxic stress. Consistent with this notion, in U2OS cells the endogenous levels of p53K382me2 increased in response to the DSB-inducing drug neocarzinostatin (NCS) relative to control treatment (Fig. 3e). To determine whether the endogenous interaction of p53 and 53BP1 paralleled the DNA damage-dependent increase in p53K382me2, p53 immunoprecipitations were performed from U2OS cells with and without NCS treatment. In the absence of DNA damage, little endogenous 53BP1 was present in the p53 IP, but after DNA damage, 53BP1 was readily detected co-immunoprecipitating with p53 (Fig. 3f, compare lanes 1 and 3). Moreover, this DNA damage-dependent interaction was augmented by expression of SET8(Y334F) (Fig. 3f, compare lanes 3 and 4). Thus, endogenous p53K382me2 levels increase with DNA damage and possibly augment the physiologic interaction between p53 and 53BP1.
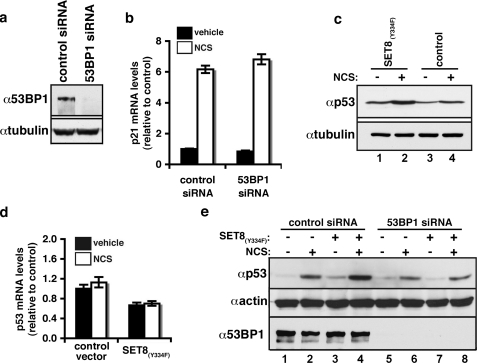
Role for p53K382me2-53BP1 Interaction in Mediating p53 Stability upon DNA Damage—To dissect potential roles for p53K382me2 and p53K382me2-53BP1 interactions in DNA damage responses, we first asked whether 53BP1 acts as a transcriptional co-activator of p53. Treatment of U2OS cells with siRNA targeting 53BP1 (Fig. 4a) did not decrease DNA damage-dependent induction of p21 or a number of additional p53 target genes (Fig. 4b; supplemental Fig. 3; data not shown). These results suggest that the 53BP1-p53K382me2 interaction likely modulates a p53 function distinct from its transactivation activity.
FIGURE 4.
53BP1 recognition of p53K382me2 couples DNA damage to p53 accumulation. a, 53BP1 siRNA treatment knocks down endogenous 53BP1 protein levels. Western analysis of 53BP1 expression in U2OS treated with control or 53BP1 siRNAs is shown. b, 53BP1 knockdown does not impact p53 induction of p21. Real-time PCR analysis of relative mRNA levels of p21 in U2OS cells treated with control or 53BP1 siRNAs as in a is shown, ± 45 nm NCS. c, SET8(Y334F) augments p53 levels at base line and in response to DNA damage. Western analysis of total p53 ± SET8(Y334F) expression and ± 45 nm NCS in U2OS cells is shown. d, SET8(Y334F) expression does not increase p53 mRNA levels. Real-time PCR analysis of relative p53 mRNA levels in U2OS cells treated as in c is shown. e, 53BP1 protein is required for SET8(Y334F)-dependent p53 accumulation. Western analysis of U2OS cells treated with control or 53BP1 siRNA ± SET8(Y334F) and ± 45 nm NCS is shown. Knockdown was confirmed with anti-53BP1 immunoblot.
Regulation of p53 protein levels is critical for proper p53-mediated responses. The stability of p53 was previously shown to be compromised in 53BP1 knockdown cells (18), and 53BP1 was identified in an RNA interference-based screen as a gene necessary for the toxicity associated with the MDM2-inhibitor Nutlin-3 (19). We therefore postulated that recognition of p53K382me2 by the Tudor domain of 53BP1 might positively regulate the accumulation of p53 protein that accompanies DSB lesion formation. To investigate this possibility, total p53 protein levels present in U2OS cells were determined at base line and in response to NCS treatment, under conditions in which p53K382me2 levels were normal or elevated, respectively. We observed that expression of SET8(Y334F) increased p53 protein levels relative to control at both base line and under genotoxic stress conditions (Fig. 4c). This increase was not due to enhanced p53 mRNA synthesis; rather, SET8(Y334F) elicited a small decrease in p53 mRNA levels relative to control (Fig. 4d). Next, we tested the role of 53BP1 in SET8(Y334F)-mediated augmentation of p53 protein levels. Consistent with previous studies, depletion of 53BP1 by siRNA led to a decrease in p53 protein levels (Fig. 4e, compare lanes 2 and 6) (19). Furthermore, although SET8(Y334F) expression increased p53 protein levels in control siRNA-treated cells, it failed to do so in cells lacking 53BP1 (Fig. 4e, compare lanes 4 and 8). Notably, no decrease in p53 mRNA levels was observed in 53BP1 knockdown cells (supplemental Fig. 4). In addition, under conditions in which the proteasome is inhibited, cells lacking 53BP1 have equivalent p53 protein levels relative to control cells (data not shown). Taken together, our data argue that 53BP1 recognition of p53K382me2 plays a role in regulating p53 protein accumulation in response to DNA damage.
DISCUSSION
In summary, we have identified a novel DNA damage-associated p53 species that is dimethylated at Lys382. This modification is recognized by the tandem Tudor domain of 53BP1, and the interaction may promote stabilization of p53. Homo-oligomerization of 53BP1 is important for its DNA repair activity; we propose that this allows 53BP1 to serve as an adaptor at DSB sites, with one 53BP1 molecule bound to H4K20me2 and a paired second 53BP1 molecule free to dock dimethylated p53 or potentially other methylated nuclear proteins (supplemental Fig. 5) (9). The DNA damage-triggered rapid accumulation of 53BP1 at DSB sites, in conjunction with the assembly of additional p53 regulatory factors such as ATM and Chk2, may cooperate to create a high-affinity site for p53. We hypothesize that this can facilitate accumulation of p53 by sequestering it away from proteins that target it for degradation as well as promoting p53-activating posttranslational modifications. In this context, there is evidence that p53 localizes to sites of double strand breaks (20). We have also observed increased occupancy of p53 at a defined DSB (data not shown). The identification of the lysine methyltransferase that generates p53K382me2 will greatly facilitate the uncovering of the molecular function(s) associated with this DNA damage-linked mark.
In our purification of endogenous p53, we identified potentially three p53 methylated species in addition to p53K382me2: p53K370me2, p53K386me1, and p53K386me2 (supplemental Fig. 1). The function of K386 mono- and dimethylation is unknown, and we have not confirmed their existence by independent methods or identified a lysine methyltransferase enzyme that acts on this residue (data not shown). The existence of p53K370me2 has been demonstrated by the use of state-specific antibodies (7). In this study, removal of the p53K370me2 mark by the lysine demethylase LSD1 was shown to repress p53 transactivation by preventing the interaction between p53 and 53BP1, which was shown to be critically dependent on 53BP1(TD) recognition of p53K370me2. Here, we also found that the 53BP1(TD) binds to p53K370me2, albeit with slightly lower affinity than p53K382me2 (see Fig. 1). Indeed, we observe binding of the 53BP1(TD) to several dimethylated p53 peptides, suggesting that many different lysine dimethylation events on p53 can potentially promote a modular protein-protein interaction between p53 and 53BP1. In the case of p53K370me2, recognition of 53BP1 was shown to be required for transcriptional activation by p53 of its target genes (7). In contrast, binding to p53K382me2 by 53BP1 does not appear to play a role in p53 transactivation (Fig. 4). How the recognition of different methylation events on p53 by the same effector protein (53BP1) can result in alternative physiologic outcomes remains an open question. On the basis of the many different potential methylation sites on p53 and the observation that at least two different 53BP1 recognition events can lead to different physiologic outcomes, we propose that lysine methylation-mediated funneling of p53 to particular functions likely depends on additional modifications and binding partners that co-occur with one modification (e.g. K382me2) versus another (e.g. K370me2).
Taken together, our study sheds light on potential molecular mechanisms by which DSB signals are transduced to activate p53 and begins to dissect how lysine dimethylation contributes to a p53 posttranslational code that modulates distinct p53 functions. Last, our study highlights the notion that lysine methylation on non-histone proteins is likely a general molecular paradigm utilized to regulate diverse nuclear processes.
Supplementary Material
Acknowledgments
We thank P. Carpenter for hemagglutinin-53BP1 and J. Lukas for 53BP1 cDNA. We also thank C. Anderson for helpful comments and K. Walters for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant GM079641 (to O. G.) and the Intramural Research Program of the National Institutes of Health (to E. A. and H. Y.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental “Materials and Methods,” Figs. 1–5, and additional references.
Footnotes
The abbreviations used are: DSB, double strand break; 53BP1(TD), 53BP1 tandem Tudor domain; NCS, neocarzinostatin; siRNA, short interfering RNA; IP, immunoprecipitate; GST, glutathione S-transferase.
References
- 1.Shi, Y., and Whetstine, J. R. (2007) Mol. Cell 251 –14 [DOI] [PubMed] [Google Scholar]
- 2.Huang, J., and Berger, S. L. (2008) Curr. Opin. Genet. Dev. 18152 –158 [DOI] [PubMed] [Google Scholar]
- 3.Gottifredi, V., Shieh, S. Y., and Prives, C. (2000) Cold Spring Harbor Symp. Quant. Biol. 65483 –488 [DOI] [PubMed] [Google Scholar]
- 4.Appella, E., and Anderson, C. W. (2001) Eur. J. Biochem. 2682764 –2772 [DOI] [PubMed] [Google Scholar]
- 5.Chuikov, S., Kurash, J. K., Wilson, J. R., Xiao, B., Justin, N., Ivanov, G. S., McKinney, K., Tempst, P., Prives, C., Gamblin, S. J., Barlev, N. A., and Reinberg, D. (2004) Nature 432353 –360 [DOI] [PubMed] [Google Scholar]
- 6.Huang, J., Perez-Burgos, L., Placek, B. J., Sengupta, R., Richter, M., Dorsey, J. A., Kubicek, S., Opravil, S., Jenuwein, T., and Berger, S. L. (2006) Nature 444629 –632 [DOI] [PubMed] [Google Scholar]
- 7.Huang, J., Sengupta, R., Espejo, A. B., Lee, M. G., Dorsey, J. A., Richter, M., Opravil, S., Shiekhattar, R., Bedford, M. T., Jenuwein, T., and Berger, S. L. (2007) Nature 449105 –108 [DOI] [PubMed] [Google Scholar]
- 8.Shi, X., Kachirskaia, I., Yamaguchi, H., West, L. E., Wen, H., Wang, E. W., Dutta, S., Appella, E., and Gozani, O. (2007) Mol. Cell 27636 –646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adams, M. M., and Carpenter, P. B. (2006) Cell Div. 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Botuyan, M. V., Lee, J., Ward, I. M., Kim, J. E., Thompson, J. R., Chen, J., and Mer, G. (2006) Cell 1271361 –1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi, X., Hong, T., Walter, K. L., Ewalt, M., Michishita, E., Hung, T., Carney, D., Pena, P., Lan, F., Kaadige, M. R., Lacoste, N., Cayrou, C., Davrazou, F., Saha, A., Cairns, B. R., Ayer, D. E., Kutateladze, T. G., Shi, Y., Cote, J., Chua, K. F., and Gozani, O. (2006) Nature 44296 –99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang, J., Feng, Q., Ketel, C. S., Wang, H., Cao, R., Xia, L., Erdjument-Bromage, H., Tempst, P., Simon, J. A., and Zhang, Y. (2002) Curr. Biol. 121086 –1099 [DOI] [PubMed] [Google Scholar]
- 13.Nishioka, K., Rice, J. C., Sarma, K., Erdjument-Bromage, H., Werner, J., Wang, Y., Chuikov, S., Valenzuela, P., Tempst, P., Steward, R., Lis, J. T., Allis, C. D., and Reinberg, D. (2002) Mol. Cell 91201 –1213 [DOI] [PubMed] [Google Scholar]
- 14.Rice, J. C., Nishioka, K., Sarma, K., Steward, R., Reinberg, D., and Allis, C. D. (2002) Genes Dev. 162225 –2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Couture, J. F., Collazo, E., Brunzelle, J. S., and Trievel, R. C. (2005) Genes Dev. 191455 –1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwabuchi, K., Bartel, P. L., Li, B., Marraccino, R., and Fields, S. (1994) Proc. Natl. Acad. Sci. U. S. A. 916098 –6102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huyen, Y., Zgheib, O., Ditullio, R. A., Jr., Gorgoulis, V. G., Zacharatos, P., Petty, T. J., Sheston, E. A., Mellert, H. S., Stavridi, E. S., and Halazonetis, T. D. (2004) Nature 432406 –411 [DOI] [PubMed] [Google Scholar]
- 18.Wang, B., Matsuoka, S., Carpenter, P. B., and Elledge, S. J. (2002) Science 2981435 –1438 [DOI] [PubMed] [Google Scholar]
- 19.Brummelkamp, T. R., Fabius, A. W., Mullenders, J., Madiredjo, M., Velds, A., Kerkhoven, R. M., Bernards, R., and Beijersbergen, R. L. (2006) Nat. Chem. Biol. 2202 –206 [DOI] [PubMed] [Google Scholar]
- 20.Al Rashid, S. T., Dellaire, G., Cuddihy, A., Jalali, F., Vaid, M., Coackley, C., Folkard, M., Xu, Y., Chen, B. P., Chen, D. J., Lilge, L., Prise, K. M., Bazett Jones, D. P., and Bristow, R. G. (2005) Cancer Res. 6510810 –10821 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.