Abstract
Purpose
To aid couples wishing to conceive children who are HLA matched to a sibling in need of a hematopoietic progenitor cell transplant, we developed a preimplantation HLA haplotype analysis of embryos that utilizes tri-, tetra-, and pentanucleotide STR markers.
Methods
For preimplantation HLA genotyping, we use polymorphic STR markers located across the HLA and flanking regions, selecting exclusively tri-, tetra-, and pentanucleotide repeats. These markers can be resolved using either capillary electrophoresis (CE) or polyacrylamide gels.
Results
We have developed 43 reliable STR markers for preimplantation HLA matching. Selected STR markers enabled unambiguous identification of embryos whose HLA haplotypes were matched with the affected patient using polyacrylamide gel or capillary electrophoresis.
Conclusions
The use of tri-, tetra-, and pentanucleotide repeat markers and polyacrylamide gels for STR genotyping in HLA matching is a simple and cost effective approach to clinical testing.
Electronic supplementary material
The online version of this article (doi:10.1007/s10815-008-9233-2) contains supplementary material, which is available to authorized users.
Keywords: Haplotype analysis, Preimplantation genetic diagnosis, Preimplantation HLA matching, Short tandem repeats, Human leukocyte antigen
Introduction
Preimplantation genetic diagnosis (PGD) is a technique to analyze embryos for chromosome abnormalities, genetic mutations and HLA haplotypes. Analysis of embryos for single gene disorders allows selection of unaffected embryos for transfer, while analysis of HLA haplotypes of embryos allows selection of embryos that are HLA matched with a sibling in need of hematopoietic progenitor cell (HPC) transplantation [1]. Preimplantation HLA haplotype analysis of embryos allows couples to conceive children who are HLA matched to the affected sibling, and to collect cord blood from the HLA-matched newborn sibling for use in a HPC transplant. In the first case, reported in 2001, a child with Fanconi anemia required an HLA-matched cord blood transplantation. With the aid of PGD the couple gave birth to a child who was unaffected and HLA matched [2]. Cord blood collected at birth from that child was used successfully to reconstitute the bone marrow of the affected child [3]. Since that time, HLA testing of embryos has been used in a variety of settings for families with both sporadic [4] and inherited genetic diseases [5–7].
Three different methods have previously been developed to identify HLA-matched embryos. Both Verlinsky et al. [2] and Fiorentino et al. [8] determined the genotypes of several classical HLA loci. Verlinsky et al. used allele-specific PCR primers for the HLA loci [8], whereas Fiorentino et al. used minisequencing [2]. The HLA complex is highly polymorphic and therefore each family tested required a unique PCR primer design. Because of the need for family-specific PCR protocols, these methods are less efficient. A third method involves the use of short tandem repeats (STRs), also known as microsatellites, across the HLA region [5, 6, 8]. This method provides a general approach for HLA matching, that disregards the genotypes involved and thus avoids the need to develop specific protocols for HLA genotype analysis.
We used the method of Van de Velde et al. [5] and Fiorentino et al. [6, 8] for HLA haplotype analysis and matching, that employs polymorphic STRs located across the HLA and flanking regions, but restricted to tri-, tetra-, and pentanucleotide repeats to identify HLA-matched embryos. Avoiding the shadow bands that can be seen with dinucleotide microsatellites [9] should aid in the unambiguous identification of HLA alleles using either polyacrylamide gel or capillary electrophoresis (CE) methods. For laboratories that do not have ready access to CE, the use of polyacrylamide gels for STR genotyping and HLA matching is a simple and cost-effective alternative to CE for clinical testing.
Methods and materials
Identification of STRs for HLA and flanking regions
STR markers in this paper were chosen from the published literature [10] or identified from human genomic sequence using the online program Tandem Repeats Finder [11]. PCR primers for the selected STR markers were either taken from the published literature [10] or designed using Oligo™ (Molecular Biology Insights, Cascade, CO) software. 6-FAM labeled and unlabeled PCR primers were synthesized and HPLC purified by Invitrogen (Carlsbad, CA). The PCR primers used in clinical assays were also characterized by mass spectrometry. A 2X Multiplex PCR PreMix (Qiagen Multiplex PCR system, Valencia, CA) was used in all PCR, which were carried out in GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA).
For STR genotyping of individuals, a 20-μL PCR reaction was carried out for each STR marker by combining 10 ng of genomic DNA sample with 10 μL of Qiagen Multiplex PCR PreMix and 20 pmol of each PCR primer. These STR primer pairs were optimized at one of two annealing temperatures, either 56°C or 61°C (Supplementary Information I). For this reason, two different PCR amplification protocols were established. For primers optimized at 56°C, the initial denaturation lasted 14 min at 95°C followed by 29 PCR cycles. Cycles 1–10 consisted of 1 min at 96°C, 30 s at 56°C, and 30 s at 72°C. Cycles 11–29 consisted of 1 min at 94°C, 30 s at 56°C, and 25 s at 72°C. The cycles were followed by a final extension of 45 min at 60°C. For PCR primers optimized at 61°C, the same thermal profile was used except that the annealing temperature for cycles 1–29 was changed to 61°C.
For preliminary screening of potential STR markers, PCR products were separated on 8% or 10% (w/v) Novex 1X TBE polyacrylamide gels that were 8 cm long and 1 mm thick (Invitrogen, Carlsbad, CA) by electrophoresing at 200 V for 45 min to 1 h 30 min. Variable volumes of PCR products were needed for successful analysis of STRs on the polyacrylamide gels. The gels were post-stained with 1X SYBR Green I Nucleic Acid Gel Stain (Lonza, Basel, Switzerland) for 20 min or longer on plastic trays. Gel images were captured by AlphaImager 3400 (Alpha Innotech, San Leandro, CA) using a camera equipped with SYBR Green I filter (Alpha Innotech) under UV 302 nm trans- and epi-illumination.
Our initial collection of polymorphic STR markers were evaluated and selected based on linkage analysis in anonymous families. These primers were further evaluated and PCR conditions optimized for single cell application by testing lymphoblastoid cell lines. Finally, 43 STR loci were selected for future preimplantation HLA haplotype matching application. The allele drop-out (ADO) rates were determined for sixteen of the most commonly used STRs using the following lymphoblastoid cell lines: L.060W.5, L.061W.11, 653 M, and 179 M.
HLA haplotype analysis of patient families
All specimens were collected following institutional review board approval and after informed consent was obtained. DNA was isolated from patient peripheral whole blood specimens or control lymphoblastoid cell lines by the salting-out method (Roche DNA Isolation Kit for Mammalian Blood, Indianapolis, IN), and quantified using PicoGreen dye (Invitrogen Quant-iT PicoGreen dsDNA Assay Kit).
STR analysis was performed for family members consisting of mother, father, affected child, and other siblings where possible, in order to identify informative STR markers across the HLA and flanking sequences to establish the haplotypes of family members. An STR marker was considered to be informative when both parents were heterozygous for that marker, and when both parents showed a difference in allelic sizes of one or both STRs, such that one could distinguish paternal and/or maternal HLA alleles in the children. A sufficient number of HLA linked STRs from Table 1 were studied to fulfill the following criteria for each family. For each family, eight informative STR markers were identified. At least two of the eight STRs were located beyond the centromeric end of HLA-DPB2 gene, and at least two STRs were located beyond the telomeric end of HLA-A gene, with the remaining STRs located between HLA-DPB2 and HLA-A genes. This permits the determination of the HLA haplotype that was passed from each parent to the affected offspring.
Table 1.
Polymorphic STR markers for preimplantation HLA genotyping
| STR name | STR amplicon location | Forward primer (5′ --> 3′) | Reverse primer (5′ --> 3′) |
|---|---|---|---|
| Beyond the telomeric end of HLA-A gene | |||
| #31C | 29,644,080–29,644,266 | TGATCATGCCACTGCACAAC | GAACGAAGAACCATATACAACACAA |
| #115 | 29,719,377–29,719,795 | CAGGAAGAGGGCCTTACTTCT | AGATTGCGCCACTACACTCG |
| #32 | 29,747,874–29,748,272 | GGGGTTAGAAGTGTGCTTATG | AGCCAGAGTGACAGAGTAAG |
| #113 | 29,765,439–29,765,830 | TCACACCTTTTGCTCAATAGG | CAGCGCAACTGCACTTAAA |
| #36 | 29,806,777–29,807,098 | TCATGGATCTTATCAGCCTC | TCCCATGGTCAAGTTCTCAG |
| #66 | 29,830,699–29,831,034 | ATGAACTTGTCCTGAGAATGAAG | CTGTCCTATTTCATATGCTCAGG |
| #37 | 29,895,060–29,896,580 | AAAGCCCGCAGAGTGTATG | GCAATCAGCCAAGATAGCAC |
| #38 | 29,898,560–29,900,060 | TGCACCTCTGAAAGAGAGC | AACAAACCTGCACATCCTGC |
| #39B | 29,914,355–29,914,530 | CCAAGAAAGAAAGAACCAATAGCA | GAGCCAATTAGCCCAATAAATCAC |
| STRs within the HLA complex | |||
| Telomeric end of HLA-A gene | 30,018,310 | ||
| #43 | 30,056,037–30,056,525 | AGAGCTGGATTCTTAGAGCG | GTTAATGGATGCAGCACACC |
| #134A | 30,173,660–30,176,170 | GATAGAGCCAGACCTTGTC | CTGAACTTGATGAAAAGACCCTA |
| #150 | 30,458,074–30,458,713 | CCCCACAGTCATAAGTAAGTT | AAGATCGCACCACTGCTCT |
| #124 | 30,954,652–30,955,083 | TTGCAGTGAGCTGAGAATGC | CTTTCATAGCTCTCGCTGTC |
| #123 | 30,971,034–30,971,752 | CAGCAGACCTACCCTAAAAG | AGGAGTTTGAGAGTAGCCTG |
| #125 | 30,980,010–30,982,250 | TCCTGAGTAGCTAGGACTTC | TTGTTTGGACCCAGGAGGTC |
| #65 | 31,145,170–31,146,570 | GCTTGACTTGAAACTCAGAG | GTAGCTGTGGAAACAGTGTC |
| #62 | 31,312,809–31,313,090 | TTGCAGTGAGCCAAGATCGC | TCTCTGTTATCTCTGGGTATAG |
| #63 | 31,316,969–31,317,187 | GCAGTCACTTTCTCAGGTAC | TGACAGAACAAGGCTCCATC |
| #47A | 31,536,275–31,536,395 | TCTAGTGTCTTCTGGCCTTG | ATCAGGGAAAGGTGCTGGT |
| #153 | 31,830,794–31,831,424 | GCCATCACACCGAACTAAGT | GGGCTACAAGAGCAAAATCC |
| #101 | 31,944,353–31,944,793 | GGACATTGCTCTGACTTGAG | AGCTGAGATTGCACTGCTG |
| #103 | 32,142,664–32,143,116 | CCACTTCCTCCACTAGAATC | CGACAGAGTGAGACCTTGTC |
| #104A | 32,153,630–32,153,815 | ATTCCAGCCTGGGCAACAG | ACCACGCCTGACCACAAAG |
| #107 | 32,309,633–32,309,823 | AGGAGTTCATGACCAGCAAC | CAAGTAGCTGGTACCACAAG |
| #16A | 32,318,105–32,318,457 | GGCCTTTTTCACTTGTTTTTCTA | TCTTCTGCTCTACCCACCA |
| #109 | 32,377,514–32,378,034 | ACTCAACCCTGCTGTTGTAG | TGCATGTCCTGTGAGGTAAG |
| #72 | 32,433,900–32,435,440 | CTAGATGACGAGTTAGTGGG | CCGGTGTATCATTGATGGAC |
| #148 | 32,871,680–32,872,160 | TGGTGTGCGACTGTAATCC | TATCTCATGTTGAGAGCAGAC |
| #54 | 33,039,460–33,039,674 | GCTCAGTTCCAGTTGCTTG | GCAGTGAGCCAAGATTGCAC |
| #149 | 33,160,363–33,160,890 | TGCATTCCAGACTTGGCAAC | TCTTGGAGGGGGAAACATTC |
| Centromeric end of HLA-DPB2 gene | 33,204,869 | ||
| Beyond the centromeric end of HLA-DPB2 gene | |||
| #59A | 33,306,131–33,306,834 | AGCCTGGGTGACACAGCAA | CCTATCGCAATGTAAATGCTGTA |
| #78A | 33,309,804–33,310,002 | ATACTCAAATATTCCCCAGTTAG | TCTAGCCTGGGTGACAGT |
| #79 | 33,318,712–33,319,020 | ACAAGAGTGAAACTCCGTCTCAA | CATCACTGGCTCTCATGCAGA |
| #81 | 33,441,780–33,444,190 | CCACATAGCAAGTCTCCGTC | AGTTCAGTTCAAGCTAGGCTCT |
| #82A | 33,453,691–33,454,132 | TAGCCAAGATCGCACCACT | GGCAATATAGCAGAAACTCTGA |
| #84 | 33,556,390–33,560,840 | GGCTGTTGAATTGTGAGAGTTC | GGCAACAAGAGTGAAACTCTG |
| #85B | 33,564,553–33,564,762 | GAATCACTTGAACCTGGGAG | TTTACCGCAACCAAGCAAATG |
| #138 | 33,569,405–33,569,680 | GAGAGAGCGAGATTCTGC | GAATGCTTAGAAGATGAAGGTG |
| #86 | 33,571,119–33,571,243 | TGGAATGATGGGAAAATTATACT | AGCCAAGATCGTGCCATA |
| #92 | 33,641,898–33,642,580 | TTCTATCTGTTTTGACCTAGTGC | GAATGAGACAAATGGTGGAAG |
| #133 | 33,901,277–33,901,669 | CCCAGTGGAGACATTTTTGC | AAAATGACTGGAGGAGGTGC |
| #141 | 34,040,934–34,041,456 | GTCTCACCTCCAAACCATTG | ACAGAGTCAGACTCCGTCT |
| #144 | 34,198,287–34,198,679 | GGCACCATCCTGTTCCTCAGT | CCCTACTTCCAGTGCCTG |
In order to ensure that the PCR primers of eight STRs selected for an affected family were compatible in a single-cell multiplex PCR, they were validated with single lymphoblastoid cells prior to PGD from one of the cell lines described above using the protocol described below.
Preimplantation genetic diagnosis
Cleavage stage embryos were obtained using standard procedures of in vitro fertilization (IVF) and intracytoplasmic sperm injection (ICSI), and embryo biopsy was performed on day 3 post-injection [12]. One blastomere was removed from embryos with five to six cells and two blastomeres were biopsied from embryos with seven to eight cells. Individual blastomeres from each embryo were washed twice in fresh droplets of HEPES-buffered, modified HTF medium, pH 7.4 (Irvine Scientific, Santa Ana, CA) and 5 μL of the last wash droplets were used as blank controls of the assay. Each blastomere was digested in 5 μL of embryo-lysis buffer containing proteinase K by incubating at 61°C for 60–75 min as previously described [13]. Immediately prior to PCR, the cell lysate was heated at 96°C for 13 min and chilled immediately on ice.
First round PCR were 50-μL multiplex reactions prepared by adding 10 pmol each of the eight pairs of family-specific PCR primers for the HLA linked STRs, and 25 μL of Qiagen Multiplex PCR PreMix to each sample. The PCR profile consisted of an initial 14 min denaturation at 95°C followed by 20 PCR cycles: Cycles 1 and 2 consisted of 1 min at 96°C, 1 min 30 s at 61°C, and 1 min at 72°C; while cycles 3 and 4 consisted of 1 min at 96°C, 1 min 30 s at 56°C, and 1 min at 72°C. Cycles 1–4 were then repeated four more times.
The products of the first round multiplex PCR were then diluted 125 fold with water, and followed by separate second round PCR for each STR. Second round PCR were 20-μL reaction for each of the eight selected STRs prepared by adding 20 pmol of each PCR primer and 10 μL of Qiagen Multiplex PCR PreMix. STR primer pairs were optimized at one of two annealing temperatures, either 56°C or 61°C (Supplementary Information I). For this reason, two different protocols of second round PCR were established. For primers optimized at 56°C, the initial denaturation was 95°C for 14 min followed by 38 PCR cycles. Cycles 1–10 consisted of 1 min at 96°C, 30 s at 56°C, and 30 s at 72°C. Cycles 11–38 consisted of 1 min at 94°C, 30 s at 56°C, and 25 s at 72°C. These 38 cycles were followed by a final extension for 45 min at 60°C. The same thermal profile was used for PCR primers optimized at 61°C, except that the annealing temperature for cycles 1–38 was changed to 61°C.
The PCR products were separated on 8% or 10% (w/v) Novex 1X TBE polyacrylamide gels that were 8 cm long and 1 mm thick (Invitrogen, Carlsbad, CA) by electrophoresing at 200 V for 45 min to 1 h 30 min, or on home-made 10% (w/v) polyacrylamide 1X TBE gels that were 20 cm long and 1.5 mm thick (poured the day before testing) by electrophoresing at 400 V for 3–4 h. Detection of PCR products after post-staining with SYBR Green I dye has been described above.
STR analysis of genomic DNA from patient families and controls using fluorescent PCR
The PCR protocol and thermal profiles for fluorescent PCR were the same as those used for STR genotyping of genomic DNA specimens with unlabeled PCR primers (described in “Identification of STRs for HLA and Flanking Regions” section), except that the forward primer was 5′-end labeled with 6-FAM. Prior to CE analysis, each PCR product was purified by first adding 80 μL of Tris–EDTA buffer, pH 8.0 (TE) and 20 μL of PCR product into a well of a Montage* PCR μ96 Filter Plate (Millipore, Billerica, MA) until all of the solution passes through the filter. Filtration took place on a Millipore MultiScreen*384 Vacuum Manifold. The purified DNA samples were eluted from the filters by incubating in 20 μL of TE for 30 min at room temperature, and transferred to a 96-well PCR reaction plate (Applied Biosystems, Foster City, CA). 3 μL of purified DNA sample was then added to 15 μL of sample loading buffer, which was prepared by mixing 200 μL of GeneScan 600 LIZ Size Standard (Applied Biosystems) with 4.8 mL of Hi-Di Formamide (Applied Biosystems). The samples were denatured for 5 min at 95°C, followed by rapid cooling in a -20°C StrataCooler (Stratagene-Agilant Technologies, La Jolla, CA) for 10 min, and then applied to an 3730xl DNA Analyzer (Applied Biosystems). From the peak signals the fragment sizes were determined using GeneMapper Analysis Software Version 4.0 (Applied Biosystems).
STR analysis of PCR products from single blastomere using fluorescent PCR
The first round multiplex PCR of single blastomeres, performed using unlabeled PCR primers (described in “Preimplantation Genetic Diagnosis” section), was followed by separate second round 20-μL PCR reactions for each STR. The first round PCR products were diluted 125 fold with water, and 2 μL of the diluted products were added to a PCR tube containing 10 μL of Qiagen Multiplex PCR PreMix, and 20 pmol of each PCR primer. The forward primer was labeled with 6-FAM. The thermal profiles for fluorescent PCR are the same as described above for the STR genotyping of single blastomeres with unlabeled PCR primers. Clean up of PCR products and CE analysis were performed as described in the previous paragraph.
Results
In the HLA and flanking regions, 147 loci were screened to identify 43 polymorphic tri-, tetra-, and pentanucleotide repeats that were either fully or partially informative for at least one of ten arbitrarily selected anonymous Wisconsin families consisting of father, mother and child (Supplementary Information I). E.g. STR #82 was fully informative in one of ten families, informative for a paternal allele in two additional families, and informative for a maternal allele in one additional family. In each of the ten families, two informative STRs were identified beyond the telomeric end of HLA. In nine of ten families, there were two informative STRs beyond the centromeric end of HLA. In the remaining family, one informative STR and one partially informative STR (for the father) were identified. In each of the ten families, at least two informative STRs were identified within the HLA locus. Sixteen of the STRs were informative in at least four families tested. The nine STRs that were located beyond the telomeric end of HLA-A gene were within 375 kb, and the thirteen STRs located beyond the centromeric end of HLA-A gene were within 994 kb. The ADO rates of PCR performed on single cells varied between 1% and 9% for sixteen commonly used STRs studied (Supplementary Information II). The PCR failure rates were not greater than 1.6%.
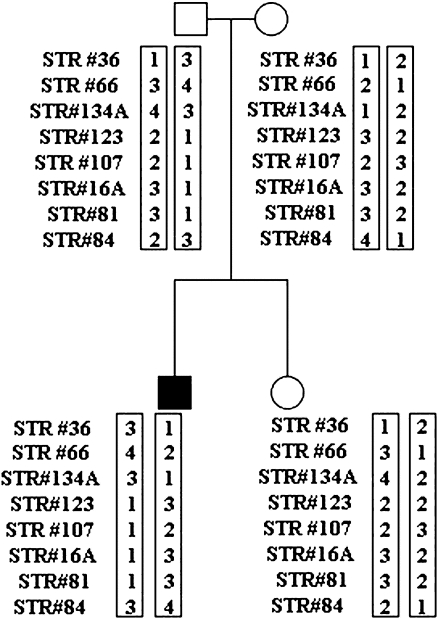
Use of our strategy was illustrated by a clinical case in which PGD for HLA matching was applied to a patient diagnosed with aplastic anemia [14]. We selected eight HLA linked STR markers that were informative for that family. STRs #36 and #66 were located beyond the telomeric end of the HLA locus; STRs #134A, #123, #107, and #16A were within the HLA locus; and STRs #81 and #84 were located beyond the centromeric end of the HLA locus. As illustrated in Fig. 1, STR analysis was carried out on the genomic DNA of the patient and nuclear family in order to establish the HLA haplotypes.
Fig. 1.
STR analysis of father, mother, sibling, and affected child (black square) and resulting HLA haplotypes. Informative STRs are ordered from telomere (top) to centromere (bottom). The numbers within the boxes next to each STR represent allele sizes arbitrarily assigned a number from largest (1) to smallest (4) size. Haplotypes are enclosed in boxes
The patient’s parents undertook an IVF cycle in which 12 eggs were retrieved. Nine embryos were biopsied and tested for HLA matching to the affected child, resulting in three HLA matched embryos. One embryo failed to develop on day 5 post-injection and was not transferred. One embryo did not result in a pregnancy after transfer. A third embryo was frozen and transferred in a subsequent cycle that did not result in a pregnancy.
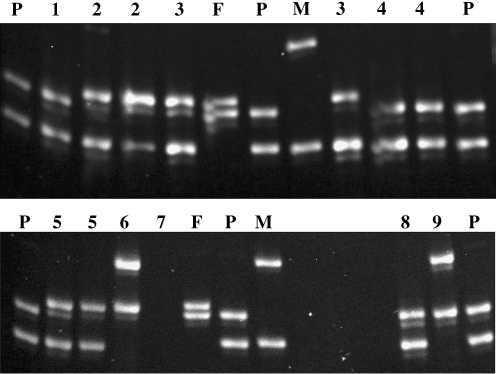
Figure 2 shows electrophoretic analysis for STR #84 of the nine embryos on a polyacrylamide gel. The gel shows that the genotypes of embryos 1, 4, and 8 were identical to that of the affected child. Table 2 summarizes the results of all eight HLA-linked STR markers separated by polyacrylamide gel electrophoresis (PAGE) and the HLA haplotypes deduced from these STR genotypes, revealing that three embryos were HLA matched to the affect child. There was no evidence of meiotic recombination. No contamination was detected in the last wash droplets of the blastomere cells.
Fig. 2.
Polyacrylamide gel analysis of STR #84 for the father, mother, affected child, and blastomeres from the clinical case. The lane labeled P is the affected child, M is the mother, and F is the father. The blastomere lanes are numbered 1 through 9. When a number is used twice, each represents a separate cell from the same embryo. Embryos 1, 4, and 8 were haplo-identical to that of the affected child
Table 2.
Preimplantation HLA haplotyping by PCR using STR markers and detected by polyacrylamide gel electrophoresis
| STRs | Cells | Embryo no. | Individuals | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | F | M | P | ||
| #36 | −1 | 3,1 | 1,1 | 1,1 | 3,1 | 1,1 | 1,2 | FA | FA | 3,2 | 3,1 | 1,2 | 3,1 |
| −2 | 1,1 | 1,1 | 3,1 | * | 3,1 | ||||||||
| #66 | −1 | 4,2 | 3,2 | 3,2 | 4,2 | 3,2 | 3,1 | FA | FA | 4,1 | 4,3 | 2,1 | 4,2 |
| −2 | 3,2 | 3,2 | 4,2 | 3,2 | 4,2 | ||||||||
| #134A | −1 | 3,_ | 4,1 | 4,1 | 3,1 | 4,1 | 4,2 | * | * | 3,2 | 3,4 | 1,2 | 3,1 |
| −2 | 4,1 | 4,1 | 3,1 | 4,1 | 3,1 | ||||||||
| #123 | −1 | 1,3 | 2,3 | 2,3 | 1,3 | 2,3 | 2,2 | FA | FA | 1,2 | 1,2 | 3,2 | 1,3 |
| −2 | 2,3 | 2,3 | 1,3 | 2,3 | 1,3 | ||||||||
| #107 | −1 | * | * | * | * | * | 2,3 | FA | FA | FA | 1,2 | 2,3 | 1,2 |
| −2 | * | * | * | * | FA | ||||||||
| #16A | −1 | 1,3 | 3,3 | 3,3 | 1,3 | 3,3 | 3,2 | FA | FA | 1,2 | 1,3 | 3,2 | 1,3 |
| −2 | 3,3 | 3,3 | 1,3 | 3,3 | 1,3 | ||||||||
| #81 | −1 | 1,3 | 3,3 | 3,3 | 1,3 | 3,3 | * | 1,3 | * | 1,_ | 1,3 | 3,2 | 1,3 |
| −2 | 3,3 | 3,3 | 1,3 | 3,3 | 1,3 | ||||||||
| #84 | −1 | 3,4 | 2,4 | 2,4 | 3,4 | 2,4 | 2,1 | FA | FA | 3,1 | 3,2 | 4,1 | 3,4 |
| −2 | 2,4 | 2,4 | 3,4 | 2,4 | 3,4 | ||||||||
| Predicted genotype | m | m | m | N/A | N/A | N/A | |||||||
Paternally derived markers in each cell are shown on the left and maternally derived markers on the right (not applicable to father and mother). The affected child’s genotypes are shown in boldface.
F Father, M mother, P patient (affected child), STR short tandem repeat, _ allele drop-out, FA failed amplification, N/A not applicable, m HLA matched
*Difficult to interpret
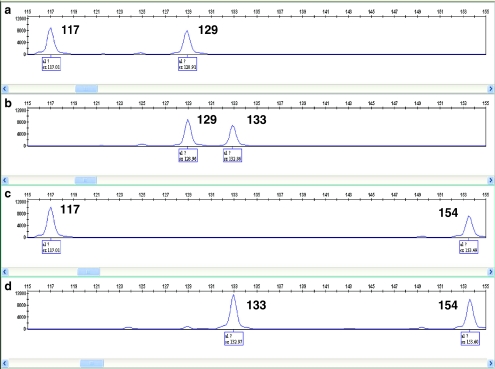
We have also confirmed our genotyping results of the eight STRs by CE analysis. Genotypes of family members were determined by CE from genomic DNA templates using fluorescent PCR. Figure 3 illustrates STR #84 genotyping of family members. In addition, aliquots of first round PCR products from each blastomere were subjected to a second round PCR using fluorescently labeled primers and detection by CE. CE analysis of the eight linked STRs confirmed the accuracy of gel analysis, showing that the genotypes of embryos 1, 4, and 8 were identical to that of the affected child (Supplementary Information III). PCR products for STR #107 were successfully detected by CE, whereas corresponding data were difficult to interpret on the polyacrylamide gels.
Fig. 3.
Capillary electrophoresis (CE) analysis of STR #84 for the affected child (a), father (b), mother (c), and sibling (d). The numbers represent the allele sizes in base pairs (bp). The affected child inherited the 117 bp allele from the mother and the 129 bp allele from the father
Discussion
All but a few of the STR markers used in previous publications for preimplantation HLA matching are dinucleotide repeats [5–8]. The use of tri-, tetra-, and pentanucleotide STRs in our present approach may have several advantages over dinucleotide microsatellites in PGD. It is known that shadow bands are more common for the PCR products for dinucleotide microsatellites, especially for large numbers of PCR cycles required for PGD [9]. In addition, the mutation rate for dinucleotide microsatellites is greater than that of tri-, tetra-, and pentanucleotide STRs [15]. Our approach could minimize the ambiguity that may arise through the use of dinucleotide microsatellites.
One hundred forty seven polymorphic STRs in the HLA and flanking regions were screened in ten arbitrarily chosen families. Screening for polymorphic STRs was carried out on polyacrylamide gels that were 8 cm long, because it was our aim to select STRs that can readily be detected by PAGE. As a result, a number of polymorphic STRs may have been overlooked, as this gel system would fail to clearly resolve alleles that differ by <3% in size. For clinical assays, we also used 10% polyacrylamide gels that were 20 cm long to resolve some alleles that were close in size. Using our collection of 43 resulting STRs, the majority of families were informative for at least two loci both within and flanking the HLA.
Another advantage of tri-, tetra-, and pentanucleotide STRs is that the PCR products may be detected either by CE or PAGE analysis. Gel electrophoresis is more cost effective than CE analysis, and would therefore permit laboratories with limited resources to pursue PGD for HLA matching [16]. An effort to resolve polymorphic dinucleotide repeats for HLA on polyacrylamide gels has been reported by Korzebor et al. [16]. Nevertheless, CE analysis provides better resolution of alleles that are similar in size which can be useful when selecting informative STRs for a given family. In addition, CE detection system equipped with laser excitation is more sensitive than SYBR Green I gel staining, as illustrated by STR #107 (Tables 2 and Supplementary Information III) in which the gel system yielded ambiguous results, whereas the CE analysis yielded clear results.
It is critical to determine whether a meiotic recombination event has occurred within the 3.7-Mb HLA region for an embryo that may be a patient’s potential HLA match. At least two STRs are analyzed on each end of the HLA region to detect possible recombination. This ensures successful assessment of embryos without misinterpretation due to potential amplification failure of a given STR. In addition, STRs for four different loci within the HLA region are studied in order to detect double recombination events. In general, PCR failure for a given STR is less than 2% for the STRs studied in this paper (Supplementary Information II). The probability of failures in two flanking STRs for a given embryo would, therefore, be very low.
The STR markers that we selected from the flanking regions of HLA are not more than 374 kb beyond the telomeric end of HLA, and not more than 994 kb beyond the centromeric end. Therefore, the probability that a recombination occurs between a flanking STR and the HLA region is also low. This significantly reduces the probability that a suitable embryo is discarded.
Based on nested PCR performed on single lymphoblastoid cells, the ADO rates of sixteen selected STR markers varied between 1% and 9% for the STRs. This is comparable to the previous findings of Fiorentino et al. [8] who found ADO rates up to 12.5% for lymphocytes. In another study by Fiorentino et al., the ADO rate for blastomeres ranged from 0% to 10.8% [6].
Selected STR markers described in this paper were recently used to screen for an HLA match among siblings of patients in need of HPC transplantation [Hopp KA, Lau EC, Bick DP, Pietz B and Ellis TM, submitted for publication]. Linkage analysis using STR markers covering the HLA and flanking regions provided a more rapid and less expensive approach to HLA matching when compared to current molecular methods used to screen for HLA-identical siblings [17] and is amenable to automated detection platforms.
Conclusions
The use of polymorphic tri-, tetra-, and pentanucleotide markers may enhance the reliability, accuracy and efficiency of preimplantation HLA matching thereby helping families successfully identify a potential HPC donor for a sibling in need of a transplant. By avoiding dinucleotide microsatellites and their associated shadow bands, there may be improvement of HLA allele resolution using either CE or PAGE systems.
Electronic supplementary materials
Below is the link to electronic supplementary materials.
Polymorphic STR markers for preimplantation HLA genotyping (XLS 26.0 kb)
Validation of common STRs used in preimplantation HLA genotyping using single lymphoblastoid cells (XLS 14.5 kb)
Preimplantation HLA haplotyping by PCR using STR markers and detection by capillary electrophoresis (XLS 18.0 kb)
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s10815-008-9233-2) contains supplementary material, which is available to authorized users.
Capsule Preimplantation HLA matching using tri-, tetra-, and pentanucleotide short tandem repeat (STR) markers in the HLA and flanking sequences for HLA haplotype analysis.
References
- 1.Bick DP, Lau EC. Preimplantation genetic diagnosis. Pediatr Clin North Am 2006;53(4):559–77. [DOI] [PubMed]
- 2.Verlinsky Y, Rechitsky S, Schoolcraft W, Strom C, Kuliev A. Preimplantation diagnosis for Fanconi anemia combined with HLA matching. JAMA 2001;285(24):3130–3. [DOI] [PubMed]
- 3.Grewal SS, Kahn JP, MacMillan ML, Ramsay NK, Wagner JE. Successful hematopoietic stem cell transplantation for Fanconi anemia from an unaffected HLA-genotype-identical sibling selected using preimplantation genetic diagnosis. Blood 2004;103(3):1147–51. [DOI] [PubMed]
- 4.Kuliev A, Rechitsky S, Tur-Kaspa I, Verlinsky Y. Preimplantation genetics: improving access to stem cell therapy. Ann N Y Acad Sci 2005;1054:223–7. [DOI] [PubMed]
- 5.Van de Velde H, Georgiou I, De Rycke M, Schots R, Sermon K, Lissens W, et al. Novel universal approach for preimplantation genetic diagnosis of β-thalassaemia in combination with HLA matching of embryos. Hum Reprod 2004;19(3):700–8. [DOI] [PubMed]
- 6.Fiorentino F, Kahraman S, Karadayi H, Biricik A, Sertyel S, Karlikaya G, et al. Short tandem repeats haplotyping of the HLA region in preimplantation HLA matching. Eur J Hum Genet 2005;13(8):953–8. [DOI] [PubMed]
- 7.Verlinsky Y, Rechitsky S, Sharapova T, Laziuk K, Barsky I, Verlinsky O, et al. Preimplantation diagnosis for immunodeficiencies. Reprod Biomed Online 2007;14(2):214–23. [DOI] [PubMed]
- 8.Fiorentino F, Biricik A, Karadayi H, Berkil H, Karlikaya G, Sertyel S, et al. Development and clinical application of a strategy for preimplantation genetic diagnosis of single gene disorders combined with HLA matching. Mol Hum Reprod 2004;10(6):445–60. [DOI] [PubMed]
- 9.Olejniczak M, Krzyzosiak WJ. Genotyping of simple sequence repeats—factors implicated in shadow band generation revisited. Electrophoresis 2006;27(19):3724–34. [DOI] [PubMed]
- 10.Cullen M, Malasky M, Harding A, Carrington M. High-density map of short tandem repeats across the human major histocompatibility complex. Immunogenetics 2003;54(12):900–10. [DOI] [PubMed]
- 11.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acid Res 1999;27(2):573–80. [DOI] [PMC free article] [PubMed]
- 12.Swanson A, Strawn E, Lau E, Bick D. Preimplantation genetic diagnosis: technology and clinical applications. WMJ 2007;106(3):145–51. [PubMed]
- 13.Krisher RL, Gibbons JR, Gwazdauskas FC. Nuclear transfer in the bovine using microinjected donor embryos: assessment of development and deoxyribonucleic acid detection frequency. J Dairy Sci 1995;78:1282–88. [DOI] [PubMed]
- 14.Davies JK, Guinan EC. An update on the management of severe idiopathic aplastic anaemia in children. Br J Haematol 2007;136(4):549–64. [DOI] [PubMed]
- 15.Chakraborty R, Kimmel M, Stivers D, Davison L, Deka R. Relative mutation rates at di-, tri-, and tetranucleotide microsatellite loci. Proc Natl Acad Sci USA 1997;94:1041–46. [DOI] [PMC free article] [PubMed]
- 16.Korzebor A, Zamani M, Nouri K, Modarressi MH. Statistical analysis of six STR loci located in MHC region in Iranian population for preimplantation genetic diagnosis. Int J Immunogenet 2007;34(6):441–3. [DOI] [PubMed]
- 17.Perz JB, Szydlo R, Sergeant R, Sanz J, O’Shea D, Khan T, et al. Impact of HLA class I and class II DNA high-resolution HLA typing on clinical outcome in adult unrelated stem cell transplantation after in vivo T-cell depletion with Alemtuzumab. Transpl Immunol 2007;18:179–85. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to electronic supplementary materials.
Polymorphic STR markers for preimplantation HLA genotyping (XLS 26.0 kb)
Validation of common STRs used in preimplantation HLA genotyping using single lymphoblastoid cells (XLS 14.5 kb)
Preimplantation HLA haplotyping by PCR using STR markers and detection by capillary electrophoresis (XLS 18.0 kb)