Abstract
Eukaryotic translation elongation factor 1A (eEF1A) is a guanine-nucleotide binding protein, which transports aminoacylated tRNA to the ribosomal A site during protein synthesis. In a yeast two-hybrid screening of a human skeletal muscle cDNA library, a novel eEF1A binding protein, immunoglobulin-like and fibronectin type III domain containing 1 (IGFN1), was discovered, and its interaction with eEF1A was confirmed in vitro. IGFN1 is specifically expressed in skeletal muscle and presents immunoglobulin I and fibronectin III sets of domains characteristic of sarcomeric proteins. IGFN1 shows sequence and structural homology to myosin binding protein-C fast and slow-type skeletal muscle isoforms. IGFN1 is substantially upregulated during muscle denervation. We propose a model in which this increased expression of IGFN1 serves to down-regulate protein synthesis via interaction with eEF1A during denervation.
Keywords: TRANSLATION ELONGATION FACTOR EEF1A, PROTEIN SYNTHESIS, YEAST TWO-HYBRID SYSTEM, SARCOMERIC PROTEIN, MYOSIN-BINDING PROTEIN-C, SKELETAL MUSCLE, DENERVATION
Eukaryotic translation elongation factor 1A (eEF1A), is a guanine-nucleotide binding protein essential for protein biosynthesis. The factor consists of three structural domains of which domain 1 is responsible for binding the guanine nucleotide [Andersen et al., 2003]. In its active, GTP-bound form, eEF1A transports aminoacylated tRNAs and facilitates their binding to the ribosomal A site [Negrutskii and El’skaya, 1998]. During this process, eEF1A cycles from its active, GTP-bound form to its inactive, GDP-bound form. Reactivation of eEF1A is catalyzed by the guanine-nucleotide exchange factor eEF1B.
Besides a canonical role for eEF1A in protein biosynthesis, interactions between eEF1A and proteins involved in other cellular processes, including signal transduction, apoptosis, microtubule severing and actin bundling, have been described. More recently, eEF1A activity has been linked with diseases such as diabetes, cancer and viral infections. Thus, eEF1A appears to be a so-called moonlighting protein (reviewed by Ejiri [2002]); that is, a protein that performs several functions in the cell depending on changes in cellular location, cell type, or concentration of ligands, substrates or cofactors [Jeffery, 1999]. The multitude of functions ascribed to eEF1A may explain the overall abundance of the protein, which amounts to 3–10% of the total cellular protein content [Merrick, 1992].
In mammals, two eEF1A isoforms, eEF1A1 and eEF1A2, exist. The two isoforms display very different patterns of expression: eEF1A1 is the predominant form present in every tissue except for skeletal muscle; eEF1A2 is specifically expressed in differentiated skeletal muscle cells, where it presumably takes over the role of eEF1A1 postnatally, and also in terminally differentiated cells of heart and brain eEF1A2 is found [Kahns et al., 1998; Knudsen et al., 1993; Lee et al., 1993]. Interestingly, the individual eEF1A isoforms are almost fully conserved between species, while eEF1A1 and eEF1A2 within the same organism display approximately 92% identity. This pattern of homology hints that eEF1A1 and eEF1A2 fulfill different roles in the cell apart from their common role in translation (Kahns et al., 1998) or that their activity in translation might be differentially regulated.
The wasted mutation in mice is a deletion in the Eef1a2 gene encoding eEF1A2 that removes the promoter region and first noncoding exon [Chambers et al., 1998]. As a consequence, eEF1A2 is not expressed in these mice, which suffer from severe muscle wasting and degeneration of motor neurons beginning at 3 weeks after birth at precisely the same time when eEF1A1 expression in muscle and neuronal cells declines [Khalyfa et al., 2001]. Since these defects in muscle structure develop only when eEF1A2 expression has declined, eEF1A1 and eEF1A2 appear to have overlapping roles but distinct temporal patterns of expression. Likewise, eEF1A1 and eEF1A2 expression are regulated differently in adult muscle, since eEF1A1 expression increases, while eEF1A2 expression remains constant in long-term denervated muscle [Khalyfa et al., 2003].
In order to elucidate further the cellular functions of eEF1A isoforms and with the aim of finding specific binding partners for eEF1A2, we screened a human skeletal muscle cDNA library using a yeast two-hybrid protocol, and identified a cDNA encoding a novel protein, which was named immunoglobulin-like and fibronectin type III domain containing 1 (IGFN1). IGFN1 presents structural similarity to sarcomeric proteins belonging to the intracellular Ig superfamily [Fürst and Gautel, 1995], including myosin-binding protein C (MyBP-C) fast and slow-type skeletal muscle isoforms [Weber et al., 1993]. We show that IGFN1 is upregulated approximately 100-fold following short-term muscle denervation in rat, which implies a regulatory role of IGFN1 during the modulation of protein synthesis characteristic of muscular atrophy. Our results suggest that the activity of eEF1A may be down-regulated via binding to IGFN1, and the reported interaction between IGFN1 and eEF1A opens the possibility of a direct link between the sarcomeric network of proteins and localized protein synthesis.
MATERIALS AND METHODS
PLASMID CONSTRUCTS
Human eEF1A1 [Brands et al., 1986] and eEF1A2 [Knudsen et al., 1993] cDNAs were PCR amplified with primers containing either 5′ EcoRI and 3′ PstI cloning sites or 5′ EcoRI and 3′ SalI cloning sites, respectively. The tufA gene encoding Escherichia coli EF1A [Knudsen et al., 1992] was PCR amplified with primers containing 5′ EcoRI and 3′ PstI sites. The purified fragments were cloned in pGBDUC2 [James et al., 1996] and sequenced.
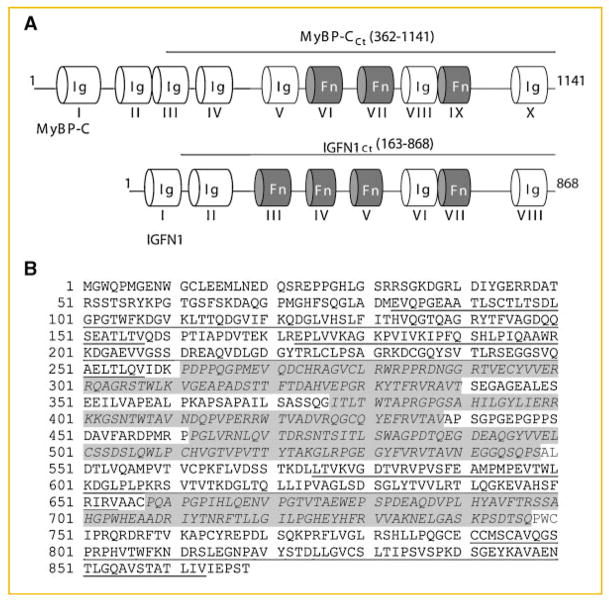
The vector pACT2: hIGFN1Ct encodes a C-terminal fragment of human IGFN1 corresponding to amino acids 163–868, and was directly retrieved from the yeast two-hybrid screening. The vector pACT2: MyBP-CCt encodes a C-terminal fragment (amino acids 362–1,141) of human myosin binding protein C slow-type (Q00872). The vector pACT2: hIGFN1 encodes full-length human IGFN1 (amino acids 1–868). The IGFN1 structural motifs encoded by these constructs are illustrated in Figure 1A.
Fig. 1.
Motifs and primary structure of human IGFN1. A: Schematic representation of the domain distribution of human skeletal muscle myosin binding protein-C slow type (top), and human IGFN1 (bottom). Immunoglobulin I domains (Ig) and fibronectin III domains (Fn) are depicted and numbered with Roman numerals. Lines indicating the extent of the protein sequences present in the MyBP-CCt (amino acids 362–1,141) and hIGFN1Ct (amino acids163–868), constructs used in the yeast two-hybrid analysis summarised in Table II are shown above the appropriate schematic. B: Deduced amino acid sequence for hIGFN1. The Ig domains are underlined and the Fn domains are highlighted and in italics.
The full-length coding sequence of hIGFN1 was PCR amplified from the human skeletal muscle Marathon ready™ cDNA (Clontech Laboratories, Inc., France) adding EcoRI and SalI restriction sites via the PCR primers. The PCR product was purified, cloned in pGBDUC2 and pGBKT7 (Clontech Laboratories) and sequenced.
The human eEF1A1 domain constructs pGBDUC2:eEF1A1-D1 (amino acids 1–239), pGBDUC2:eEF1A1-D2 (amino acids 240–328), pGBDUC2:eEF1A1-D3 (amino acids 329–462), pGBDUC2:eEF1A1-D(1 + 2) (amino acids 1–328), pGBDUC2:eEF1A1-D(2 + 3) (amino acids 240–462) were subcloned into pGBDUC2 from their respective pGADT7 constructs using standard cloning procedures. The plasmids were sequenced to ensure proper open reading frame insertion.
The open reading frames encoding hIGFN1Ct and full-length hIGFN1 were PCR amplified from the skeletal muscle Marathon ready™ cDNA (Clontech Laboratories) with primers containing 5′ EcoRI and 3′ XhoI restriction sites. Both PCR products were purified, cloned in pCMV-myc (Clontech Laboratories) and sequenced. These constructs were named pCMV-myc: hIGFN1Ct and pCMV-myc: hIGFN1, respectively.
YEAST TWO-HYBRID SCREENING
Saccharomyces cerevisiae strain PJ69-4A (MATa, trp1-901, leu2-3, aa2, ura3-522, his3-200, gal4D, gal80Δ, LYS2::GAL1-HIS3, GAL2-ADE2, met2::GAL7-lacZ) [James et al., 1996] was transformed with the bait plasmid pGBDUC2:eEF1A2 and subsequently with a human skeletal muscle cDNA library constructed in the pACT2 vector (Clontech Laboratories). Selection was performed on synthetic drop out (SD) plates lacking uracil, leucine, and adenine (ULA) at 30°C for 10 days until colonies appeared. The selected colonies were plated on SD plates lacking uracil, leucine, and histidine (ULH) added 1 mM 3-aminotriazole (3-AT).
Plasmid DNA isolated from the resulting colonies was rescued in E. coli KC8 strain (Clontech Laboratories) (hsdR, leuB600, trpC9830, pyrF::Tn5, hisB643, lacDX74, strA, galU, K) and sequenced.
Clones of interest were tested against eEF1A1/2 and proper controls including a related protein (pGBDUC2:EF1A) and an empty vector (pGBDUC2) on ULA plates and ULH plates containing increasing concentrations of 3-AT. The lacZ reporter was discarded in these studies, as the background level of self-activation for some constructs was too high. For further mapping of the interaction, PJ69-4A was transformed with each of the eEF1A1 domain constructs pGBDUC2:eEF1A1-D1, pGBDUC2:eEF1A1-D2, pGBDUC2:eEF1A1-D3, pGBDUC2:eEF1A1-D(1 + 2) and pGBDU-C2:eEF1A1-D(2 + 3) and pACT2:hIGFN1Ct. All transformations were performed as described by Clontech Laboratories, Inc. in their ‘‘Yeast Protocols Handbook.’’
RACE, NORTHERN BLOTTING AND RNase PROTECTION ASSAYS
5′ RACE amplification of the gene encoding human IGFN1 was performed using human skeletal muscle Marathon ready™ cDNA (Clontech Laboratories) with Advantage® 2 polymerase mix (Clontech Laboratories) as recommended by the manufacturer. The primers used were AP1 or AP2 (Clontech Laboratories) and an IGFN1-specific antisense primer 5′-AGGGACCATCTAAGCCATTCCAGG-3′. The unique, amplified PCR fragment was cloned into a pCR®4-TOPO® vector using the TOPO TA cloning® kit (Invitrogen) and sequenced.
The cDNA encoding the rat homologue of IGFN1, rIGFN1, was obtained using a PCR-mediated subtractive-hybridization and cloning method. A denervated-minus-innervated rat skeletal muscle cDNA library was screened with a subtractive probe as described [Zhu et al., 1994] (rIGFN1 is referred to as DEN1 in this reference). The 198 bp rat IGFN1 cDNA obtained in this way was used to probe a cDNA library created from 5-day denervated rat skeletal muscle RNA using standard procedures. Adult Sprague-Dawley rats were anesthetized by interperitoneal injection (20 mg/ml ketamine (KetaSet®), 1 mg/ml xylazine (Xylaject®) in 0.9% NaCl), and the lower leg was denervated by removing approximately 2 mm of sciatic nerve. All experiments involving rats were approved by the Animal Care and Use Committee at Massachusetts Institute of Technology. RNA from the denervated (4–5 days) and the contralateral innervated lower leg muscle was isolated as described [Chomczynski and Sacchi, 1987], and stored at −80°C.
Northern blotting was carried out as described in the legends of the relevant figures.
RNase protection assays were performed and quantitated, essentially as described previously [Simon et al., 1992], using a probe of 198 nucleotides from the 3′ untranslated region of rIGFN1. Briefly, the antisense RNA probe was radiolabeled during transcription in vitro from a linearized plasmid containing the corresponding cDNA insert. Subsequently, the labeled probe was hybridized to total RNA in hybridization buffer overnight at 42°C. The reaction was treated with RNases A and T1 to remove unhybridized RNA, digested with proteinase K, and extracted using phenol/chloroform. Protected RNA was precipitated with ethanol, and fractionated in a 5% denaturing polyacrylamide gel. Protected RNAs were quantitated by Phosphoimaging (BioRad).
BIOINFORMATICS
Nucleotide sequences were analyzed using different programs. BLAST at NCBI, herein the human genome resource (http://www.ncbi.nlm.nih.gov/genome/guide/human/) was used for sequence identification. The Genestream resource center (http://www.igh.cnrs.fr/bin/align-guess.cgi) was applied for DNA sequence comparison and GENSCAN (http://genes.mit.edu/genscan.html) for open reading frame detection. Protein sequence alignment was performed using clustalW (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_clustalw.html).
COIMMUNOPRECIPITATION IN C2C12 CELLS
The mouse muscle myoblast cell line C2C12 (American Type Culture Collection) was cultured in Dulbecco’s modified Eagle’s medium (DMEM): high glucose (Bio Whittaker) added 10% fetal calf serum, 2 mM glutamine, 100 units/ml penicillin and 100 μg/ml streptomycin at 37°C and 5% CO2.
C2C12 cells (6 × 105/dish) were transfected with pCMV-myc:hIGFN1 or an empty pCMV-myc vector using Lipofectamine™ 2000 (Invitrogen) following the instructions of the manufacturer, with the exception that the cells were plated in medium containing antibiotic.
Cells were washed after 48 h, harvested and lysed on ice for 1 h in 150 μl modified RIPA buffer (150 mM NaCl, 1 mM EDTA, 10 mM MgCl2, 1 mM DTT, 50 μM GTP, 10% glycerol, 50 μM PMSF, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 50 mM Tris/HCl pH 7.5 and complete EDTA-free inhibitor (Roche Applied Science)). Lysates (100 μl) were mixed with 25 μl anti-c-myc conjugated agarose beads (Santa Cruz Biotechnology, Inc.) and 450 μl CoIp buffer (150 mM NaCl, 20 mM Tris–HCl pH 7.5, 1% NP-40, 10 mM MgCl2, 20% glycerol, 1 mM DTT, 50 μM GTP and complete EDTA-free inhibitor (Roche Applied Science)) and incubated for 2 h at 4°C. The beads were washed six times with PBS containing 50 μM GTP and finally resuspended in 40 μl 5xSDS loading buffer (125 mM Tris/PO4, pH 6.8; 10% SDS; 1% β-mercaptoethanol; 50% glycerol; 0.005% bromphenol blue). Samples were boiled, and 20 μl aliquots were subjected to 12% SDS–PAGE. The gel-separated proteins were transferred to a nylon membrane. eEF1A was detected with scFv eEF1A antibody A8 [Kjær et al., 2001] followed by a rabbit polyclonal anti c-myc antibody (Santa Cruz Biotechnology) and finally a HRP-conjugated swine anti-rabbit antibody (Dako). c-myc-hIGFN1 was detected by incubation with a rabbit polyclonal anti c-myc antibody followed by a HRP-conjugated swine anti-rabbit antibody and visualized using Super Signal™ (Pierce Biotechnolgy).
CONFOCAL LASER SCANNING MICROSCOPY
Semi confluent C2C12 cells seeded in 10 cm2 slide flasks (Nunc) 24 h before transfection, were transfected either with pCMV-myc:hIGFN1 or pCMV-myc using FuGENE6™ transfection reagent (Roche Applied Science). A control with nontransfected cells was also performed. The day after transfection, the medium was changed to differentiation medium (DMEM: high glucose (Bio Whittaker, Inc.), 2% horse serum, 2 mM glutamine, 100 units/ml penicillin and 100 μg/ml streptomycin) were relevant.
In order to analyze the staining pattern of human IGFN1 in both undifferentiated myoblasts and differentiated myotubes, C2C12 cells were immunostained 24 or 96 h after transfection, respectively, essentially as previously described [Corydon et al., 1998]. In brief, the cells were fixed in NBF (Mallinckrodt Baker) for 15 min at room temperature and washed three times in PBS. Then the cells were incubated in ice-cold 70% (v/v) ethanol, followed by three washes in PBS. The cells were then incubated with the primary antibodies polyclonal rabbit-anti-c-myc (Santa Cruz Biotechnology) and monoclonal mouse-anti-eEF1A (Upstate Biotechnology Inc.). Thereafter, cells were washed and incubated with the secondary antibodies rhodamine-conjugated Alexa Fluor® 568 goat anti-rabbit antibody and fluorescein-conjugated Alexa Fluor® 488 goat anti-mouse antibody. Nuclei were stained with Hoescht solution (33258; 1 μg/ml). Confocal analysis was performed using a Leica TCS confocal laser-scanning microscope (Leica Microsystems). Green and red fluorescence was collected using 40× objective (myotubes) or 100× objective (myotubes and myoblasts).
RESULTS
TWO-HYBRID SYSTEM SCREENING
In order to identify novel binding partners to account for noncanonical functions ascribed to eEF1A, the eEF1A2 isoform was used as the bait to screen a human skeletal muscle cDNA library using the yeast two-hybrid system. As eEF1A is a protein with a potential tendency to bind other proteins, the screening was limited to 1.3 × 106 clones, a number that under represents the cDNA library (3.5 × 106 clones). Nevertheless, more than 250 clones were retrieved from the first ULA selection, and of these clones, 45 remained after further selection on ULH plates containing 3-AT. Among these, one particular clone, pACT2:hIGFN1Ct, was retrieved five times, and was subjected to further studies after sequence analysis.
cDNA, DEDUCED PROTEIN SEQUENCE, GENE STRUCTURE AND TISSUE EXPRESSION
The human cDNA library clone pACT2:hIGFN1Ct carries a 2,622 bp long cDNA fragment with a poly A tail at its 3′ end. Sequence analysis identified an open reading frame encoding a potentially 686 amino acid protein. Sequence comparison using BLAST retrieved a nucleotide entry, XM_036558, which is nearly identical to the pACT2:hIGFN1Ct cDNA with one significant difference: a 306 bp region (1,094–1,399) is found in XM_036558, which is not present in the pACT2:hIGFN1Ct cDNA.
Based on the sequence contained in XM_036558, we designed a specific antisense primer to obtain the 5′-UTR of the hIGFN1 cDNA by a standard RACE procedure. In addition, several overlapping PCR fragments were amplified with internal specific primers and sequenced from the same template source. Thus, we could confirm the existence of a complete cDNA sequence comprising 3,951 bp (accession number NM_178275). The corresponding protein was named immunoglobulin-like and fibronectin type III domain containing 1 (IGFN1; accession number NP_840059). The gene encoding IGFN1 is also known as DKFZp434B1231 or EEF1A2BP1, while the protein is also known as eEF1A2 binding protein (eEF1A2BP).
A search of the human genome resource database at NCBI localized our IGFN1 gene to chromosome 1, cytogenetic marker 1q32.1. The general structure of the gene comprises 13 exons and 12 introns of variable length from approximately 100 bp to up to 3.5 kb (Table I).
TABLE I.
Schematic Representation of the Exon–Intron Structure and Splice Junction Sites of the Human IGFN1 Gene
| Exon no | Size (bp) | 5′ splice donor | Int size (bp) | 3′ splice acceptor |
|---|---|---|---|---|
| I | 981 | GATGCCCAAGgtaggtgctt | 554 | gtctctccagGCCCCATGGG |
| II | 125 | TGGCGTCAAGgtactgcctc | 711 | ctgtctccagCTCACCACCC |
| III | 142 | ACTGTCCAGGgtaaggccca | 394 | ttatttctagATTCCCCTAC |
| IV | 300 | CAAGTCATAGgtaccagccc | 1064 | gggtccccagACAAGCCTGA |
| V | 298 | GCTCCTGAGGgtgagagaaa | 534 | ttacccccagCTCTCCCCAA |
| VI | 173 | CCGGTGCCTGgtgagcattg | 1070 | tgttcagaagAGAGGAGGTG |
| VII | 129 | GACCCCATGAgtaagtaggg | 3358 | tgcctggcagGACCCCTGGG |
| VIII | 303 | CCTGTTACTGgtgagtgctg | 991 | tgcgtttcagTCTGTCCCAA |
| IX | 90 | CTCCTTTGAAgtgagtgtac | 1513 | cctttctcagGCCATGCCCA |
| X | 195 | AGGGTGGCAGgtgaggcagg | 947 | tctcctgaagCATGCCCGCA |
| XI | 298 | CGGCAGCGCGgtaagcagcc | 758 | ttcaccacagACAGGTTCAC |
| XII | 327 | ATTGTCATAGgtaatggtgg | 1177 | tgtcttgcagAACCCAGCAC |
| XIII | 972 |
Exon sequences are shown in capital letters, while intron sequences are shown in lower-case letters. Conserved sequences marking the splice site are indicated in bold face. The complete cDNA sequence has accession number NM_178275.
Subsequently, we performed a prediction of the gene structure and the protein amino acid sequence encoded using GENSCAN. For this purpose we used a 24 kb stretch containing up to 8 kb upstream from the IGFN1 5′UTR. The program found a putative initiation site, a termination site and a poly A signal. In particular, the deduced initiation site at position 773 of the complete cDNA presents an ATG in a Kozak sequence context (GGAAGCATGG), which is the most favorable of all the possible initiation sites in the three reading frames found in the first exon. Upstream, at nucleotide position 62 and in the same open reading frame, a second ATG start codon exists (GGCTCTATGC), but the potential Kozak context of this ATG is very weak, as it lacks a G at position +4 and an A at position −3, which are reported to be especially important for initiation of translation [Kozak, 1996]. Therefore, we discarded the ATG at position 62. Using the favored ATG at position 773, the deduced amino acid sequence of IGFN1 is 868 amino acids (Fig. 1B).
The domain structure of human IGFN1 is shown in Figure 1A. It presents four fibronectin III (Fn) [Potts and Campbell, 1996] and four immunoglobulin (Ig) domains [Williams and Barclay, 1988] arranged Ig-Ig-Fn-Fn-Fn-Ig-Fn-Ig. The extension of each domain and their amino acid sequences are shown in Figure 1B. The Fn and Ig domain architecture of IGFN1 resembles those of MyBP-C fast and slow-type isoforms found in skeletal muscle. The sequence identity in humans between IGFN1 and MyBP-C slow and fast type isoforms is 18% and 20%, respectively. Their similarity is 33% for both types. The sequence identity is more pronounced in the C-terminal region of the protein. Interestingly, IGFN1 presents a unique extra Fn domain (Fn IV) between the Ig II and Ig VI domains, which is not found in MyBP-C (Fig. 1A).
In order to determine the tissue expression pattern of human IGFN1 mRNA, we performed a Northern blot with mRNA from eight different human tissues using a 0.4 kb IGFN1-specific probe. A single band of more than 10 kb is detected only in skeletal muscle suggesting that IGFN1 expression is restricted to skeletal muscle (Fig. 2). This expression pattern agrees with the analysis shown by SAGE genie anatomic viewer for tissues only (http://cgap.nci.nih.gov/sage).
Fig. 2.
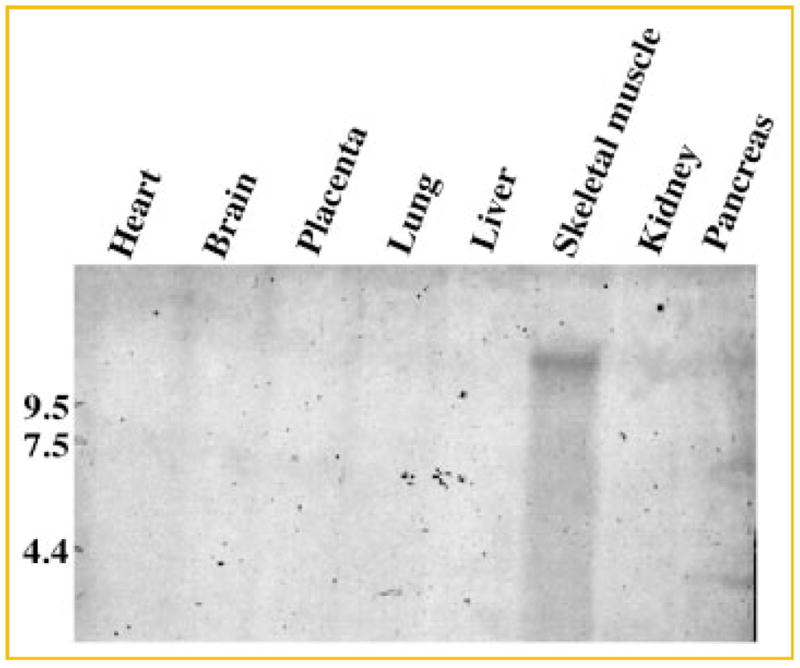
Tissue distribution of human IGFN1. A 0.4-kb fragment that spans the nucleotide region 965–1,350 of IGFN1 was labeled with biotin using a labeling kit (Strip-EZ™, Ambion) and then used to probe a normalized, human 8-lane multiple tissue Northern blot (GE Healthcare). Hybridization took place at 42°C overnight in hybridization buffer (ULTRAhyb™, Ambion). The blot was developed using the BrigthStar™ BioDetect™ kit (Ambion). The probe hybridizes only with skeletal muscle cDNA and is negative for the rest of the tissues analyzed. A RNA size marker is indicated to the left.
In parallel with the identification of IGFN1 from human, a cDNA encoding Igfn1 from rat was also identified. A cDNA library made from 5-day denervated rat skeletal muscle RNA was screened using a 198-bp cDNA isolated by subtractive hybridization cloning [Zhu et al., 1994], and several overlapping cDNAs were isolated. The largest of these, a 3,318 bp cDNA, contained a large open reading frame encoding 929 amino acids. The rat protein, rIgfn1, displays 77% identity and 85% similarity to the human IGFN1 protein. Moreover, the Ig and Fn domain organization in the two proteins is identical. In the rat cDNA, the open reading frame begins at the 5′ end of the 3.3 kb cDNA and extends to a stop codon at nt 2,790. This stop codon is followed by a 3′ untranslated region of 529 bp including a polyadenylation signal and a polyA sequence of 25 bp. This cDNA (from position 152) can be aligned with accession number XM_001063557 (from position 1,227). The two sequences are 99% identical from these positions and throughout the rest of the sequences. The rat gene is localized on chromosome 13, cytogenetic marker 13q13, in the rat genome and has been given the name Igfn1 (the gene is also known as RGD1565384_predicted).
Other putative orthologous sequences of human IGFN1 have been predicted in additional species, including mouse, dog, elephant, hedgehog, chicken and armadillo based on genomic alignment. In general, the homologies of the predicted proteins with human IGFN1 are high indicating a vital role of IGFN1 in the cell.
Innervation differentially regulates the expression of the two eEF1A isoforms [Khalyfa et al., 2003]. Thus, we asked whether innervation also regulates Igfn1 expression. We isolated RNA from innervated and 5-day denervated rat skeletal muscles and probed Northern blots with an Igfn1 probe and a control Gadph probe. In accordance with our data for human IGFN1, rat Igfn1 is encoded by a large, single transcript of approximately 9 kb (Fig. 3A). The transcript appears to be substantially more abundant in denervated than innervated rat muscle. We next quantitated the magnitude and the time-course of rat Igfn1 mRNA induction following skeletal muscle denervation by RNase protection assay. In these studies, induction of rat Igfn1 mRNA is first evident by 3 days and reaches maximal induction (approximately 100-fold) by 5 days following denervation (Fig. 3B). Rat Igfn1 mRNA induction is modestly slower than that observed for the acetylcholine receptor (AChR) delta subunit gene, following denervation [Zhu et al., 1994].
Fig. 3.
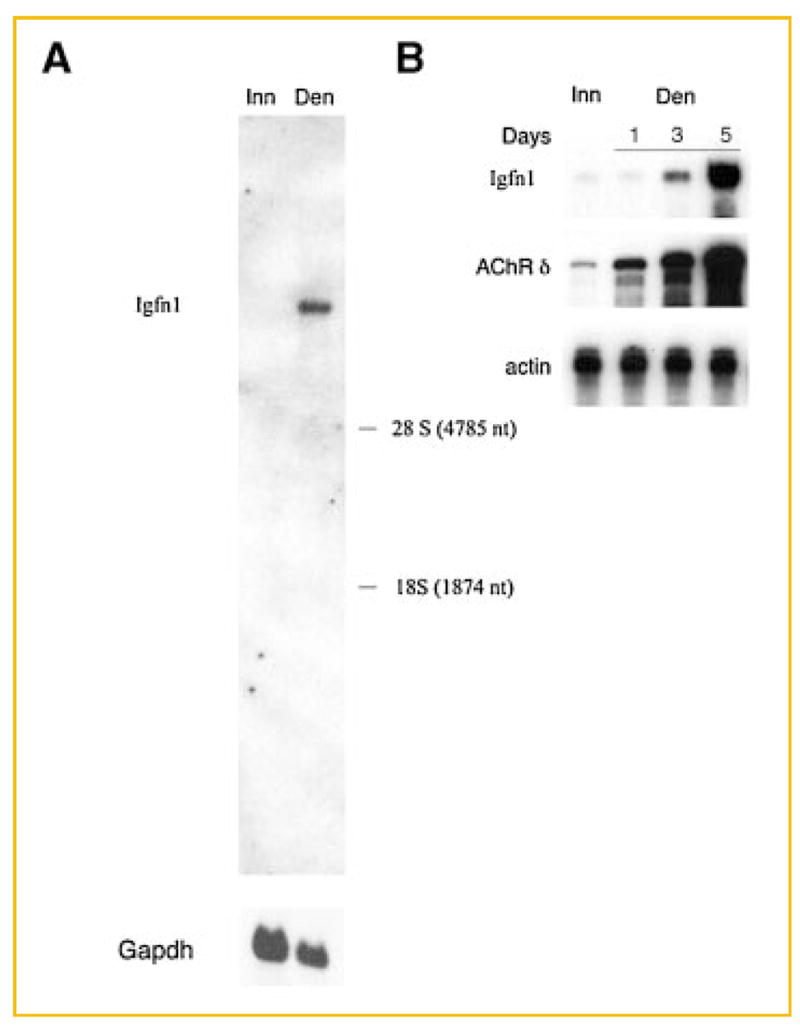
The rat Igfn1 gene encodes a single RNA transcript that is induced 100-fold following denervation. A: Total RNA (15 mg) from innervated (Inn) and 5-day denervated (Den) rat lower leg muscle were separated in 1.1% formaldehyde/agarose gels and transferred to nitrocellulose membranes. The RNA blots were hybridized with a [32P]-labeled 3.4 kb cDNA probe, which includes 2.8 kb of coding sequence and 0.6 kb of the 3′ untranslated region from the eEF12BP mRNA. Subsequently, the blot was washed and exposed to film with an intensifying screen. The blot was stripped and re-hybridized with a Gadph probe. The positions of 28S and 18S ribosomal RNA are indicated. B: Expression of rat Igfn1 in innervated (Inn) and denervated (Den) muscle was measured by an RNase protection assay. Induction of the Igfn1 mRNA is evident by 3 days after denervation and maximal by 5 days after denervation. By 5 days after denervation, the rIgfn1 mRNA is ~100-fold more abundant in denervated than innervated muscle.
YEAST TWO-HYBRID ANALYSIS OF THE INTERACTION BETWEEN HUMAN IGFN1 AND eEF1A
By yeast two-hybrid analysis, both eEF1A isoforms were shown to bind IGFN1Ct (Table IIA). Remarkably, the interaction between IGFN1 and eEF1A2 is the strongest in the system, as demonstrated by yeast growth upon selection on TLH plates containing up to 10 mM 3-AT. The prokaryotic counterpart of eEF1A, EF1A, is unable to activate reporter gene expression in the presence of IGFN1Ct. Thus, IGFN1 binds specifically to the eukaryotic eEF1A isoforms. In our yeast two-hybrid system full-length IGFN1 also binds both eEF1A isoforms, and again with a higher affinity for eEF1A2. Overall, however, the yeast growth observed using the full-length IGFN1 probe is weaker than when IGFN1Ct is applied. It is noteworthy that only eEF1A2 is able to elicit activation of the Ade2 reporter gene upon interaction with full-length IGFN1. In the reporter strain, PJ69-4A, used in this study, the Ade2 reporter gene is controlled by a very tight promoter and its response is normally associated with strong interactions [James et al., 1996]. Thus, also for the most physiologically relevant interaction between native IGFN1 and eEF1A, eEF1A2 appears to have an advantage.
TABLE II.
Yeast Two-Hybrid Analysis of Protein–Protein Interactions
| Cotransformed vector constructs
|
Reporter gene activation
|
||||||
|---|---|---|---|---|---|---|---|
| Ade2
|
His3
|
||||||
| pGBDUC2 | pACT2 | ULA | ULH 0 mM | ULH 1 mM | ULH 3 mM | ULH 5 mM | ULH 10 mM |
| A | |||||||
| eEF1A1 | IGFN1Ct | 10 | 10 | 10 | 2 | 1 | 0 |
| eEF1A2 | IGFN1Ct | 10 | 10 | 10 | 10 | 5 | 3 |
| eEF1A1 | MyBP-CCt | 0 | 1 | 0 | 0 | 0 | 0 |
| eEF1A2 | MyBP-CCt | 0 | 1 | 0 | 0 | 0 | 0 |
| eEF1A1 | IGFN1 | 0 | 5 | 0 | 0 | 0 | 0 |
| eEF1A2 | IGFN1 | 5 | 7 | 0 | 0 | 0 | 0 |
| EF1A | IGFN1Ct | 0 | 2 | 0 | 0 | 0 | 0 |
| eEF1A1-D1 | IGFN1Ct | 0 | 0 | 0 | 0 | 0 | 0 |
| eEF1A1-D2 | IGFN1Ct | 6 | 10 | 6 | 0 | 0 | 0 |
| eEF1A1-D3 | IGFN1Ct | 0 | 0 | 0 | 0 | 0 | 0 |
| eEF1A1-D(1 + 2) | IGFN1Ct | 10 | 10 | 10 | 10 | 10 | 10 |
| eEF1A1-D(2 + 3) | IGFN1Ct | 10 | 7 | 4 | 0 | 0 | 0 |
| Co-transformed vector construct
|
Reporter gene activation
|
||||||
|
Ade2
|
His3
|
||||||
| pGBKT7 | pACT2 | ULA | ULH 0 mM | ULH 1 mM | ULH 10 mM | ULH 20 mM | ULH 40 mM |
|
| |||||||
| B | |||||||
| IGFN1 | Ø | 10 | 10 | 9 | 0 | 0 | 0 |
| IGFN1 | IGFN1 | 10 | 10 | 10 | 10 | 10 | 10 |
Yeast strain PJ69-4A was transformed with the combination of plasmids indicated in the table and plated on SD medium lacking uracil and leucin to select for the presence of the vectors. pGBDUC2 provides a binding domain fusion protein, while pACT2 expresses an activation domain fusion protein. Four independent colonies from each transformation plate were tested for growth on ULA and ULH plates of which the latter contained different concentrations of 3-AT as indicated in mM concentrations. Growth after 6 days is expressed on a scale from 10 for maximum growth to 0 for no growth. (A) Mapping of the interaction between human eEF1A and human IGFN1 was done by testing interactions between full-length eEF1A1, eEF1A1 domains or full-length eEF1A2 and IGFN1Ct (amino acids 183-862), full-length IGFN1 or a C-terminal fragment of myosin binding protein C slow-type (amino acids 362-1,141; MyBP-CCt). Controls with empty vectors were performed and showed no self-activation for any of the constructs shown in A. (B) The ability of human IGFN1 to dimerize was tested by co-transformation of pGBKT7:hIGFN1 and pACT2:hIGFN1 constructs. The pGBKT7:hIGFN1 construct provides a BD fusion protein and was preferred over pGBDUC2:hIGFN1 due to its lower level of self-activation. No self-activation was observed with the pACT2:hIGFN1 construct, which provides an AD fusion protein. The symbol Ø designates an empty vector.
The structural domains of eEF1A responsible for binding to IGFN1Ct were also identified using the yeast two-hybrid system. While domains 1 or 3 of eEF1A1 do not sustain growth, domain 2 of eEF1A1 gives rise to growth on TLA plates and TLH plates added 1 mM 3-AT thus indicating a weak capacity for binding (Table IIA). When domain 2 is combined with either of domains 1 and 3, yeast growth is also supported. The combination of domains 1 and 2 gives rise to the strongest growth signal suggesting that these domains are important for an optimal tuning of the eEF1A1-IGFN1Ct interaction and presumably also for the interaction between eEF1A2 and full-length IGFN1. In contrast, domain 3 appears to have a negative effect on the binding.
We also used our yeast two-hybrid system to test for potential associations between a C-terminal fragment of myosin binding protein C slow type (MyBP-CCt; aa 362-1,141), which resembles IGFN1Ct (aa 163–868), (see Fig. 1A), and eEF1A. Neither eEF1A1 nor eEF1A2 show any binding with MyBP-CCt indicating that despite of a similar Ig and Fn domain structure in MyBP-C and IGFN1, the presence of an extra fibronectin III domain in the latter is critical for eEF1A binding.
The domain structure of IGFN1 contains four immunoglobulin and four fibronectin type 3 domains (Fig. 1A). These domains are known to mediate protein–protein interactions [Cunningham et al., 1987; Yarden and Ullrich, 1988], as well as protein oligomerization as reported for MyBP-C [Moolman-Smook et al., 2002]. We analyzed the possibility of dimerization in the case of human IGFN1. The growth signal induced by an IGFN1:IGFN1 interaction is very strong (Table IIB) suggesting that IGFN1 can, at least, form dimers.
COIMMUNOPRECIPITATION
The interaction between eEF1A and full-length IGFN1 was validated further using a coimmunoprecipitation procedure. Mouse C2C12 cells were transiently transfected with a construct directing the expression of full-length human IGFN1 with an N-terminal c-myc tag. Transfection of the cells with an eEF1A expression plasmid was not necessary, since C2C12 myoblasts express large amounts of endogenous eEF1A1. c-myc-IGFN1 was detected in the supernatant (Fig. 4, lane 4) and bound to anti-c-myc coupled agarose beads (Fig. 4, lane 2). Although eEF1A1 bound nonspecifically to the agarose beads, the amount of eEF1A1 immunoprecipitated from the IGFN1-transfected cells is approximately four-fold higher (Fig. 4, lane 1 vs. 2).
Fig. 4.
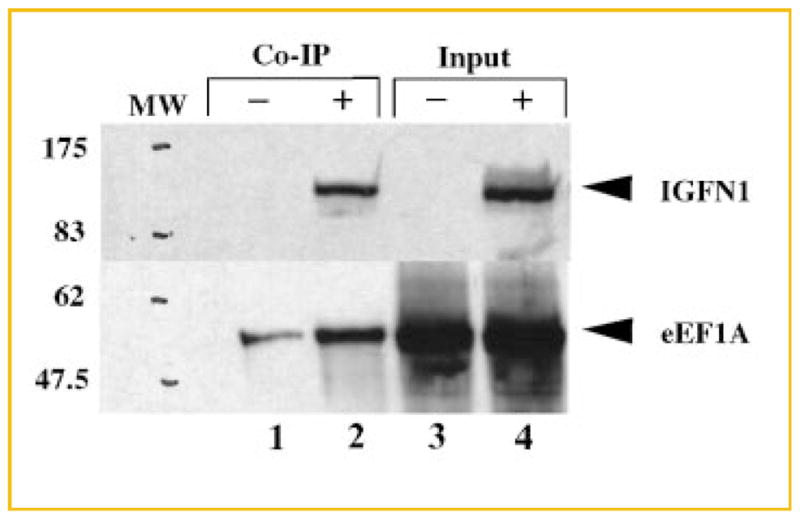
Co-immunoprecipitation of endogenous eEF1A and overexpressed c-myc-IGFN1. Cell lysates containing endogenous eEF1A and recombinant c-myc-tagged IGFN1 were immunoprecipitated using an agarose-coupled antibody directed against c-myc. Immunoprecipitation of eEF1A was detected by Western blotting using scFv eEF1A antibody A8 [Kjær et al., 2001] followed by rabbit polyclonal anti-c-myc antibody and finally swine anti-rabbit HRP conjugated antibody (lower half of blot). The expression of c-myc-IGFN1 was confirmed with a rabbit polyclonal anti-c-myc antibody followed by a swine anti-rabbit HRP conjugated antibody (upper half of blot). Lanes 3 and 4 show samples of the cell extract supernatant prior to coimmunoprecipitation (Input). Lanes 1 and 3 represent negative controls transfected with an empty pCMV-myc vector (denoted ‘‘–’’), and lanes 2 and 4 correspond to cells overexpressing IGFN1 (denoted ‘‘+’’). Lanes 1 and 2 show the results of the coimmunoprecipitation (Co-IP) in the absence and presence of IGFN1, respectively.
The C2C12 cells used in this experiment were undifferentiated, and therefore we assume that only eEF1A1 was present in these cells [Ruest et al., 2002]. If the experiment was carried out with differentiated myotubes that is in the presence of 1/3–2/3 of eEF1A2 [Ruest et al., 2002], the signal of binding presumably would be even stronger.
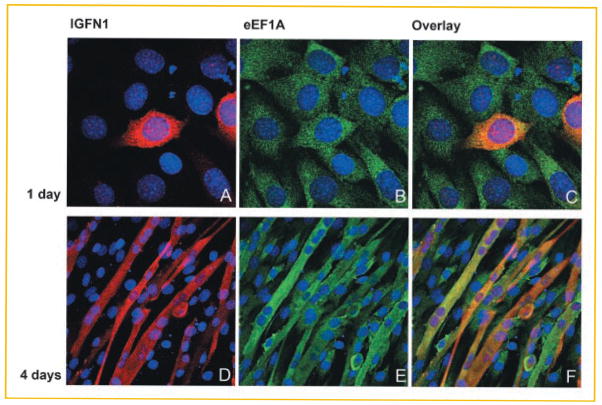
LOCALIZATION STUDIES
Confocal laser scanning microscopy was used to show identical localizations of eEF1A and IGFN1, providing further evidence of interaction between the two proteins. Undifferentiated and differentiated c-myc-IGFN1-transfected C2C12 cells were stained for human myc-tagged IGFN1 (red; Fig. 5A and 5D, respectively) and eEF1A (green; Fig. 5B and 5E, respectively). In the two cell types, both c-myc-IGFN1 and eEF1A1 can be seen in the cytoplasm. The cytoplasmatic location of the two proteins was confirmed in density gradient fractionation experiments (data not shown). The cytoplasmatic localization of eEF1A is well known [Bohnsack et al., 2002; Kjær et al., 2001]. The merged pictures show that IGFN1 colocalizes with eEF1A1 in myoblasts (Fig. 5C) and eEF1A1/eEF1A2 in myotubes (Fig. 5F) as indicated by the yellow colour. It should be noticed that the anti-eEF1A antibody applied recognizes both eEF1A isoforms. Inspection of different confocal sections confirmed that the colocalization was not confined to the specific section shown in Figure 5. Mock transfection with an empty pCMV-myc vector showed no staining in either myoblasts or myotubes (data not shown). Our results suggest that an interaction between eEF1A and IGFN1 can physically take place in the cytosol of myotubes.
Fig. 5.
C2C12 immunostaining and confocal laser scanning microscopy. C2C12 cells were transfected with pCMV-myc:hIGFN1 and maintained for either 1 (undifferentiated) or 4 days (differentiated) after transfection. Overexpressed c-myc-hIGFN1 was detected with an anti c-myc antibody. In parallel, endogenous eEF1A was detected with a monoclonal anti-eEF1A antibody. IGFN1 and eEF1A expression was visualized using Alexa Fluor® 568 (red color) or Alexa Fluor® 488 (green color) labeled antibodies, respectively. Nuclear DNA (blue color) was stained with Hoestch 33258 solution. Colocalized proteins (yellow-orange color) were detected by superimposing pictures of the same focal sections resulting in merging of the red and green color. Pictures A and D show the cellular localization of IGFN1 1 day (undifferentiated cells) or 4 days (differentiated myotubes) post transfection, respectively; Pictures B and E show the cellular localization of the endogenously expressed eEF1A in undifferentiated (day 1) or differentiated (day 4) cells, respectively. Both eEF1A and IGFN1 present a cytosolic distribution. In pictures C and F, pictures from A and B, and D and E, respectively, are superimposed. Cells transfected with empty pCMV-myc vector were analyzed in parallel as a control (data not shown). The results shown are representative of cells examined in several separate experiments. The original magnifications applied are 100x (myoblasts) and 40x (myotubes).
DISCUSSION
The existence of two mammalian translation elongation factor isoforms with very distinct expression patterns raises the question of whether they might have specific binding partners related to specialized functions in the tissue where they are unique.
In this study, we used eEF1A2 as the bait in a yeast two-hybrid screening of a human skeletal muscle cDNA library, and identified a novel MyBP-C-like protein, IGFN1 that binds to both eEF1A isoforms, but with an apparent preference for binding to eEF1A2. Interactions between IGFN1 and the eEF1A isoforms were confirmed by coimmunoprecipitation and were substantiated further by confocal laser scanning microscopy showing colocalization of the two binding partners in the cytosol of both myoblasts and myotubes.
FEATURES OF THE NOVEL PROTEIN
Several features strongly suggest that IGFN1 is a sarcomeric protein. Firstly, the majority of the protein is composed of Ig and Fn repeats, whose sequence and arrangement (Ig-Fn-Fn-Fn-Ig-Fn-Ig) are very similar to sarcomeric structural proteins such as titin, M-protein, myomesin and MyBP-C and H [Fürst and Gautel, 1995]. All these proteins are evolutionarily related, and notably MyBP-C and MyBP-H and their multiple isoforms share an Ig-Fn-Ig C-terminal region. Human IGFN1 shares this Ig-Fn-Ig C-terminal region and 33% homology with human MyBP-C slow and fast-type isoforms suggesting that these proteins may be evolutionarily related. The N-terminal regions of members of the immunoglobulin superfamily appear to be unique, and, indeed, the N-terminal parts of IGFN1 from human and rat display no or only very little homology upstream of the first, conserved Ig domain.
Three adult myosin-binding protein isoforms are known: skeletal muscle slow and fast-type, and the cardiac isoform. All three are encoded by different genes [Gautel et al., 1995; Weber et al., 1993]. MyBP-C is found in the C-zone of the A bands of the sarcomere [Bahler et al., 1985; Bennett et al., 1986; Craig and Offer, 1976] and belongs to the intracellular immunoglobulin superfamily [Carrier et al., 1998]. The modular architecture of MyBP-C is responsible for its binding to myosin [Gruen et al., 1999; Okagaki et al., 1993] and titin [Freiburg and Gautel, 1996] as well as for multimerization [Moolman-Smook et al., 2002].
Besides the presence of Ig and Fn domains, the cytosolic localization and the association with the cytoskeleton are common properties of sarcomeric proteins. Human IGFN1 is found in the cytosol by confocal microscopy and in the pellet of cell extracts not treated with detergents (data not shown), another feature of several proteins associated with the cytoskeleton [Salmikangas et al., 1999]. In addition, IGFN1 is able to form, at least, dimers (Table IIB), which also is a typical characteristic among cytoskeletal and sarcomeric proteins presenting Ig domains [Clark et al., 2002; Moolman-Smook et al., 2002]. Thus, IGFN1 is a strong candidate for being a novel, sarcomeric protein.
There is a discrepancy between the size of the human IGFN1 cDNA detected by RACE and the mRNA detected by Northern blotting of 6–7 kb indicating that the IGFN1 protein might be bigger than reported here. In Northern blots of 5 day denervated muscle from rat, the mRNA encoding rat Igfn1 also is considerably larger than the sequence of the rat Igfn1 predicted from the rat genomic sequence. Searching the human and rat genomes for sufficiently long mRNAs failed. In the human genome, the 5′ UTR of the IGFN1 gene (genome symbol DKFZp434B1231) overlaps with the last exon of another gene, LOC440706, derived by the gene prediction method GNOMON. The corresponding protein is designated ‘‘similar to titin isoform N2-A.’’ Upon fusion of these two genes, the corresponding mRNA reaches a size of approximately 6 kb, which is not sufficient to account for the mRNAs observed upon Northern blotting. When GENSCAN was applied to a 57 kb fragment covering IGFN1 and an upstream region of 41 kb, only the LOC440706 and IGFN1 genes were detected. Apparently, currently available programs for the prediction of genes and intron/exon boundaries are insufficient to account for the size of the IGFN1 mRNA.
The entire 5′-UTR of the hIGFN1 mRNA can be translated in the reading frame identified by the IGFN1 start codon without encountering a stop codon. Thus, the identified cDNA may encode only the C-terminal portion of an even larger protein.
INTERACTION OF IGFN1 WITH eEF1A AND OTHER PROTEINS
In spite of the structural similarity between IGFN1 and MyBP-C, IGFN1 is a specific binding partner for eEF1A, as MyBP-C slow type shows no binding to eEF1A (Table IIA). IGFN1 differs from MyBP-C at the N-terminal region, and presents a unique Ig-Fn-Fn-Fn domain distribution not yet found in other sacomeric proteins. M-protein and myomesin, two proteins known to bind to the C-terminal part of titin in the M-band of the sarcomere, are the closest related proteins structurally, presenting five consecutive Fn domains.
Recently, fragments of IGFN1 cDNA were identified in another yeast two-hybrid screening of a human skeletal muscle cDNA library using the cDNA encoding the muscular dystrophy KY protein as the bait (in that study, IGFN1 is named KYIP1; [Beatham et al., 2004]). In addition, KY was shown to interact with other sarcomeric cytoskeletal proteins including filamin C, titin and MyBP-C-slow. This is in accordance with results obtained by confocal laser scanning microscopy showing partial colocalization between IGFN1 and titin (data not shown). Titin is the largest known protein (approx. 3 MDa). In the sarcomere, titin molecules span the distance from the Z-line to the M-line and acts as a ruler to guide the precise placement of other proteins along muscle fibers. In addition, titin’s elastic properties accounts for the development of passive force in response to sarcomere stretch thereby generating passive tension in striated muscle [Labeit and Kolmerer, 1995].
In a preliminary yeast two-hybrid screening of a human skeletal muscle library using IGFN1 as the bait, a total of 13 clones were retrieved. Of these, five turned out to be myotilin (our unpublished data). Myotilin is a component of the sarcomere Z-disc, where it plays an important role in Z-disc maintenance, and several myotilin point mutations have been described in patients suffering from various muscular dystrophies. Like eEF1A, myotilin binds directly to F-actin and bundles actin filaments (Von Nandelstadh et al., 2005).
Thus, IGFN1 appears to be placed in a network of protein–protein interactions in the sarcomere.
FUNCTIONAL IMPLICATIONS OF THE DISCOVERED INTERACTION
Interestingly, although both eEF1A isoforms are capable of binding to hIGFN1 in the yeast two-hybrid system, eEF1A2 binds more strongly. This correlates with the specific expression of IGFN1 in skeletal muscle, where eEF1A2 is the prevalent eEF1A isoform.
The reason why tissue specific eEF1A isoforms exist in muscle is unknown [Chambers et al., 1998]. The presence of other tissue-specific translation elongation factors [Pizzuti et al., 1993], however, suggests that translation in terminally differentiated cells may be different from that of actively growing cells [Chambers et al., 1998]. We have previously shown that the binding between the canonical guanine-nucleotide exchange factors, eEF1B α/β and eEF1A2 is either very weak or absent [Mansilla et al., 2002]. This indicates that guanine-nucleotide exchange may be regulated differently for eEF1A2 as compared to eEF1A1. IGFN1 may play a role in this regulation pattern.
Northern blotting and RNase protection analysis has demonstrated that rIgfn1 expression is regulated by innervation. It is possible that the increase in rIgfn1 expression observed upon short-term denervation may contribute to the decrease in the rate of translation following denervation. Atrophy of skeletal muscle, caused by denervation, nonweight bearing activity or hind limb unloading, is associated with a short-term increase in the rate of protein degradation and a decrease in the rate of protein synthesis with the purpose of regulating the amount of tissue protein [Goldspink, 1976; Hornberger et al., 2001]. Although the decrease in protein synthesis is caused, at least in part, by a reduction in the rate of translation, the mechanisms by which innervation regulates the rate of translation are not understood. Studies of polysome profiles during nonweight-bearing activity show an increasing number of ribosomes per mRNA after 18 h of muscle disuse. These observations indicate that the initiation of protein synthesis is not rate limiting, while a slowing of the elongation rate may explain the ‘‘traffic jam’’ of ribosomes on the mRNA [Ku and Thomason, 1994]. During the first month after denervation, the levels of eEF1A1 and eEF1A2 remain unchanged. After approximately 1 month, the level of eEF1A1 increases and reaches the level of eEF1A2, which remains unaffected [Khalyfa et al., 2003]. Thus, the rapid reduction in elongation rate cannot be attributed to a change in the absolute amount of eEF1A2, but may be caused by modification of its activity.
Structure analysis of the complex between eEF1A and its guanine-nucleotide exchange factor eEF1Bα from yeast [Andersen et al., 2000] shows that the C-terminal region of eEF1Bα, which is essential for the exchange of GDP by GTP, is buried between domains 1 and 2 of yeast eEF1A upon binding. The fact that hIGFN1 also binds to domain 2 and to a smaller extent also domain 1 of eEF1A (Table IIA) raises the possibility that IGFN1 may reduce the rate of translation elongation by regulating the binding of eEF1A to its guanine-nucleotide exchange factor following delivery of aminoacyl-tRNA to the ribosome, and thereby hamper regeneration of the active, GTP-bound form of eEF1A. In this model, the increase in IGFN1 expression in denervated muscle would lead to a decrease in translation elongation and thereby contribute to a decrease in protein synthesis. This proposal is consistent with data indicating that the translocase, eEF2, which is a common target of elongation regulation via phosphorylation [Carlberg et al., 1990], cannot account for the decrease in translation observed in atrophic rat skeletal muscle [Hornberger et al., 2001]. However, multiple elements related to the translation apparatus probably contribute to the reduced protein synthesis characteristic of short-term denervation. As an example, the heat-shock protein 70 (HSP-70) has been observed to dissociate from polysomes during skeletal muscle atrophy [Ku et al., 1995]. HSP-70 facilitates the treading of the nascent polypeptide chain through the ribosomal polypeptide exit tunnel and thus, the elongation rate of protein synthesis may decrease in its absence.
When skeletal muscle is exercised, the muscle undergoes hypertrophy accompanied by an increase in synthesis of sarcomeric proteins. How muscle activity regulates protein synthesis and whether the synthesis of sarcomeric proteins is coupled to their assembly in sarcomeres is poorly understood. In several cell types, the distribution and translation of mRNAs is finely coordinated and involves transport of mRNA along microtubules or actin filaments, followed by local translation at the final destination [Jansen, 2001]. These findings raise the possibility that granules, containing not only mRNA but also other components of the translational machinery [Knowles et al., 1996], may ensure specific delivery in space and time of relevant sarcomeric proteins in skeletal muscle [Jansen, 1999]. eEF1A is an actin-binding protein [Edmonds et al., 1998; Kurasawa et al., 1996] whose association with the translation apparatus is regulated by its ability to dissociate from actin following changes in pH [Liu et al., 1996]. eEF1A presents two actin-binding sites in domains 1 and 3, respectively [Liu et al., 2002]. Thus, the simultaneous binding of actin and IGFN1 to eEF1A may both regulate the rate of mRNA translation and maintain eEF1A in the sarcomeric vicinity. Translation in muscle, therefore, could be confined in a sarcomeric compartment, in a similar manner to the local translation model described for dendrites [Giustetto et al., 2003; Macchi et al., 2003].
Further studies designed to learn whether IGFN1 regulates the rate of translation elongation, will provide important insights into how IGFN1 may contribute to protein synthesis regulation in skeletal muscle.
Acknowledgments
We thank Dr. Bjarne Bonven for valuable advice on setting up the yeast two-hybrid system and the PJ69-4A yeast strain, Meritxell Orpinell for her vector constructs pACT2:MyBP-CCt and pAC-T2:IGFN1, and Mandana Jadidi for the eEF1A1 domain constructs in pGADT7. We also thank Karen M. Nielsen, Helle Jakobsen, Anja Aagaard and Anja Fjorback for excellent technical assistance.
Grant sponsor: The Novo Nordic Foundation; Grant sponsor: NIH; Grant number: NS36193.
References
- Andersen GR, Pedersen L, Valente L, Chatterjee I, Kinzy TG, Kjeldgaard M, Nyborg J. Structural basis for nucleotide exchange and competition with tRNA in the yeast elongation factor complex eEF1A:eEF1Bα. Mol Cell. 2000;6:1261–1266. doi: 10.1016/s1097-2765(00)00122-2. [DOI] [PubMed] [Google Scholar]
- Andersen GR, Nissen P, Nyborg J. Elongation factors in protein synthesis. Trends Biochem Sci. 2003;28:434–441. doi: 10.1016/S0968-0004(03)00162-2. [DOI] [PubMed] [Google Scholar]
- Bahler M, Eppenberger H, Walliman T. Novel thick filament protein of chicken pectoralis muscle: The 86 kD protein. I. Purification and characterization. J Mol Biol. 1985;186:393–401. doi: 10.1016/0022-2836(85)90113-5. [DOI] [PubMed] [Google Scholar]
- Beatham J, Romero R, Townsend SK, Hacker T, van der Ven PF, Blanco G. Filamin C interacts with the muscular dystrophy KY protein and is abnormally distributed in mouse KY deficient muscle fibres. Hum Mol Genet. 2004;13:2863–2874. doi: 10.1093/hmg/ddh308. [DOI] [PubMed] [Google Scholar]
- Bennett P, Craig R, Starr R, Offer G. The ultrastructural location of C-protein, X-protein and H-protein in rabbit muscle. J Muscle Res Cell. 1986;7:550–567. doi: 10.1007/BF01753571. [DOI] [PubMed] [Google Scholar]
- Bohnsack M, Regener K, Schwappach B, Saffrich R, Paraskeva E, Hartmann E, Görlich D. Exp5 exports eEF1A via tRNA from nuclei and synergyzes with other transport pathways to confine translation to the cytoplasm. EMBO J. 2002;21:6205–6215. doi: 10.1093/emboj/cdf613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brands JH, Maassen JA, van Hemert FJ, Amons R, Möller W. The primary structure of the alpha subunit of human elongation factor 1. Structural aspects of guanine-nucleotide-binding sites. Eur J Biochem. 1986;155:167–171. doi: 10.1111/j.1432-1033.1986.tb09472.x. [DOI] [PubMed] [Google Scholar]
- Carlberg U, Nilsson A, Nygard O. Functional properties of phosphorylated elongation factor 2. Eur J Biochem. 1990;191:639–645. doi: 10.1111/j.1432-1033.1990.tb19169.x. [DOI] [PubMed] [Google Scholar]
- Carrier L, Bonne G, Schwartz K. Cardiac myosin-binding protein C and hypertrophic cardiomyopathy. Trends Cardiovasc Med. 1998;8:151–157. doi: 10.1016/S1050-1738(97)00144-8. [DOI] [PubMed] [Google Scholar]
- Chambers DM, Peters J, Abbott CM. The lethal mutation of the mouse wasted (wst) is a deletion that abolishes expression of a tissue-specific isoform of translation elongation factor 1alpha, encoded by the Eef1a2 gene. Proc Natl Acad Sci USA. 1998;95:4463–4468. doi: 10.1073/pnas.95.8.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Clark KA, McElhinny AS, Beckerle MC, Gregorio CC. Striated muscle cytoarchitecture: An intrincate web of form and function. Cell Dev Biol. 2002;18:637–706. doi: 10.1146/annurev.cellbio.18.012502.105840. [DOI] [PubMed] [Google Scholar]
- Corydon TJ, Bross P, Holst HU, Neve S, Kristiansen K, Gregersen N, Bolund L. A human homologue of Escherichia coli ClpP caseinolytic protease: Recombinant expression, intracellular processing and subcellular localization. Biochem J. 1998;331:309–316. doi: 10.1042/bj3310309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig R, Offer G. Empirical predictions of protein conformation. Annu Rev Biochem. 1976;49:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- Cunningham B, Hemperley J, Murray B, Prediger E, Brackenbury R, Edelman G. Neural cell adhesion molecule: Structure, immunoglobulin-like domains, cell surface modulation, and alternative RNA splicing. Science. 1987;236:799–806. doi: 10.1126/science.3576199. [DOI] [PubMed] [Google Scholar]
- Edmonds BT, Bell A, Wyckoff J, Condeelis J, Leyh TS. The effect of F-actin on the binding and hydrolysis of guanine nucleotide by Dictyostelium elongation factor1A. J Biol Chem. 1998;273:10288–10295. doi: 10.1074/jbc.273.17.10288. [DOI] [PubMed] [Google Scholar]
- Ejiri S. Moonlighting functions of polypeptide elongation factor 1: From actin bundling to zinc finger protein R1-associated nuclear localization. Biosci Biotechnol Biochem. 2002;66:1–21. doi: 10.1271/bbb.66.1. [DOI] [PubMed] [Google Scholar]
- Freiburg A, Gautel M. A molecular map of the interactions between titin and myosin-binding protein C: Implications for sarcomeric assembly in familial hypertrophic cardiomyopathy. Eur J Biochem. 1996;235:317–326. doi: 10.1111/j.1432-1033.1996.00317.x. [DOI] [PubMed] [Google Scholar]
- Fürst DO, Gautel M. The anatomy of a molecular giant: How the sarcomere cytoskeleton is assembled from immunoglobulin superfamily molecules. J Mol Cell Cardiol. 1995;27:951–959. doi: 10.1016/0022-2828(95)90064-0. [DOI] [PubMed] [Google Scholar]
- Gautel M, Zuffardi O, Freiburg A, Labeit S. Phosphorylation switches specific for the cardiac isoform of myosin binding protein-C: A modulator of cardiac contraction? EMBO J. 1995;14:1952–1960. doi: 10.1002/j.1460-2075.1995.tb07187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giustetto M, Hedge AN, Si K, Casadio A, Inokuchi K, Pei W, Kandel ER, Schwartz JH. Axonal transport of eukaryotic translation elongation factor 1alpha mRNA couples transcription in the nucleus to long-term facilitation at the synapse. Proc Natl Acad Sci USA. 2003;100:13680–13685. doi: 10.1073/pnas.1835674100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldspink DF. The effects of denervation on protein turnover of rat skeletal muscle. Biochem J. 1976;156:71–80. doi: 10.1042/bj1560071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruen M, Prinz H, Gautel M. cAPK-phosphorylation controls the interaction of the regulatory domain of cardiac myosin binding protein C with myosin-S2 in an on-off fashion. FEBS Lett. 1999;453:254–259. doi: 10.1016/s0014-5793(99)00727-9. [DOI] [PubMed] [Google Scholar]
- Hornberger TA, Hunter RB, Kandarian SC, Esser KA. Regulation of translation factors during hindlimb unloading and denervation of skeletal muscle in rats. Am J Physiol Cell Physiol. 2001;281:C179–187. doi: 10.1152/ajpcell.2001.281.1.C179. [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R-P. RNA-cytoskeletal associations. FASEB J. 1999;13:455–466. [PubMed] [Google Scholar]
- Jansen R-P. mRNA localization: Message on the move. Nat Rev. 2001;2:247–256. doi: 10.1038/35067016. [DOI] [PubMed] [Google Scholar]
- Jeffery CJ. Moonlighting proteins. Trends Biotechnol Sci. 1999;24:8–11. doi: 10.1016/s0968-0004(98)01335-8. [DOI] [PubMed] [Google Scholar]
- Kahns S, Lund A, Kristensen P, Knudsen CR, Clark BFC, Cavallius J, Merrick WC. The elongation factor 1 A-2 isoform from rabbit: Cloning of the cDNA and characterization of the protein. Nucl Acids Res. 1998;26:1884–1890. doi: 10.1093/nar/26.8.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalyfa A, Bourbeau D, Chen E, Petroulakis E, Pan J, Xu S, Wang E. Characterisation of elongation factor-1A (eEF1A-1) and eEF1A-2/S1 protein expression in normal and wated mice. J Biol Chem. 2001;276:22915–22922. doi: 10.1074/jbc.M101011200. [DOI] [PubMed] [Google Scholar]
- Khalyfa A, Carlson BM, Debkov EI, Wang E. Changes in protein levels of elongation factors, eEF1A-1 and eEF1A-2/S1, in long-term denervated rat muscle. Restor Neurol Neurosci. 2003;21:47–53. [PubMed] [Google Scholar]
- Kjær S, Wind T, Ravn P, Østergaard M, Clark BFC, Nissim A. Generation and epitope mapping of high-affinity scFv to eukaryotic elongation factor 1A by dual application of phage display. Eur J Biochem. 2001;268:3407–3415. doi: 10.1046/j.1432-1327.2001.02240.x. [DOI] [PubMed] [Google Scholar]
- Knowles RB, Sabry JH, Martone MA, Ellisman M, Bassell GJ, Kosik KS. Translocation of RNA granules in living neurons. J Neurosci. 1996;16:7812–7820. doi: 10.1523/JNEUROSCI.16-24-07812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen CR, Clark BFC, Degn B, Wiborg O. One-step purification of E. coli elongation factor. Tu Biochem Int. 1992;28:352–362. [PubMed] [Google Scholar]
- Knudsen SM, Frydenberg J, Clark BFC, Leffers H. Tissue-dependent variation in the expression of elongation factor-1α isoforms: Isolation and characterisation of a cDNA encoding a novel variant of human elongation-factor 1α. Eur J Biochem. 1993;215:549–554. doi: 10.1111/j.1432-1033.1993.tb18064.x. [DOI] [PubMed] [Google Scholar]
- Kozak M. Interpreting cDNA sequences: Some insights from studies on translation. Mamm Genome. 1996;7:563–574. doi: 10.1007/s003359900171. [DOI] [PubMed] [Google Scholar]
- Ku Z, Thomason DB. Soleus muscle nascent polypeptide chain elongation slows protein synthesis rate during non-weight-bearing activity. Am J Physiol. 1994;267:C115–C126. doi: 10.1152/ajpcell.1994.267.1.C115. [DOI] [PubMed] [Google Scholar]
- Ku Z, Yang J, Menon V, Thomason DB. Decreased polysomal HSP-70 may slow polypeptide elongation during skeletal muscle atrophy. Am J Physiol. 1995;268:C1369–C1374. doi: 10.1152/ajpcell.1995.268.6.C1369. [DOI] [PubMed] [Google Scholar]
- Kurasawa Y, Hanyu K, Watanabe Y, Numata O. F-actin bundling activity of Tetrahymena elongation factor 1alpha is regulated by Ca2+/calmodulin. J Biochem. 1996;119:791–798. doi: 10.1093/oxfordjournals.jbchem.a021309. [DOI] [PubMed] [Google Scholar]
- Labeit S, Kolmerer B. Titins: Giant proteins in charge of muscle ultrastructure and elasticity. Science. 1995;270:293–296. doi: 10.1126/science.270.5234.293. [DOI] [PubMed] [Google Scholar]
- Lee S, Wolfraim L, Wang E. Differential expression of S1 and elongation factor-1 alpha during rat development. J Biol Chem. 1993;268:24453–24459. [PubMed] [Google Scholar]
- Liu G, Tang J, Edmonds BT, Murray J, Levin S, Condeelis J. F-actin sequesters elongation factor 1α from interaction with aminoacyl-tRNA in a pH-dependent reaction. J Cell Biol. 1996;135:953–963. doi: 10.1083/jcb.135.4.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Grant WM, Persky D, Latham VM, Jr, Singer RH, Condeelis J. Interactions of elongation factor 1alpha with F-actin and beta-actin mRNA: Implications for anchoring mRNA in cell protrusions. Mol Biol Cell. 2002;13:579–592. doi: 10.1091/mbc.01-03-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macchi P, Hemraj I, Goetze B, Grunewald B, Mallardo M, Kiebler MA. A GFP-based system ot uncouple mRNA tranport and translation in a single living neuron. Mol Biol Cell. 2003;14:1570–1582. doi: 10.1091/mbc.E02-08-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansilla F, Friis I, Jadidi M, Nielsen KM, Clark BFC, Knudsen CR. Mapping the human translation elongation factor eEF1H complex using the yeast two-hybrid system. Biochem J. 2002;365:669–676. doi: 10.1042/BJ20011681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick WC. Mechanism and regulation of eukaryotic protein synthesis. Microbiol Rev. 1992;56:291–315. doi: 10.1128/mr.56.2.291-315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moolman-Smook J, Flashman E, de Lange W, Corfield V, Redwood C, Watkins H. Identification of novel interactions between domains of myosin binding protein-C that are modulated by hypertrophic cardiomyopathy missense mutations. Circ Res. 2002;91:704–711. doi: 10.1161/01.res.0000036750.81083.83. [DOI] [PubMed] [Google Scholar]
- Negrutskii BS, El’skaya AV. Eukaryotic translation elongation factor 1α: Structure, expression, functions, and possible role in aminoacyl-tRNA channeling. Prog Nucl Acid Res Mol Biol. 1998;60:47–78. doi: 10.1016/s0079-6603(08)60889-2. [DOI] [PubMed] [Google Scholar]
- Okagaki T, Weber FE, Fischman DA, Vaughan KT, Mikawa T, Reinach FC. The major myosin-binding domain of skeletal muscle MyBP-C (C protein) resides in the COOH-terminal, immunoglobulin C2 motif. J Cell Biol. 1993;123:619–626. doi: 10.1083/jcb.123.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzuti A, Gennarelli M, Novelli G, Colosimo A, Cicero SL, Caskey CT, Dallapiccola B. Human elongation factor EF-1β: Cloning and characterization of the EF1β5a gene and assignment of EF-1β isoforms to chromosomes 2,5 15 and X. Biochem Biophys Res Com. 1993;197:154–162. doi: 10.1006/bbrc.1993.2454. [DOI] [PubMed] [Google Scholar]
- Potts JR, Campbell ID. Structure and function of fibronectin modules. Matrix Biol. 1996;15:313–320. doi: 10.1016/s0945-053x(96)90133-x. [DOI] [PubMed] [Google Scholar]
- Ruest LB, Marcotte R, Wang E. Peptide elongation factor eEF1A-2/S1 expression in cultured differentiated myotubes and its protective effect against caspase-3-mediated apoptosis. J Biol Chem. 2002;277:5418–5425. doi: 10.1074/jbc.M110685200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmikangas P, Mykkänen OM, Grönholm M, Heiska L, Kere J, Carpen O. Myotilin, a novel sarcomeric protein with two Ig-like domains, is encoded by a candidate gene for limb-girdle muscular dystrophy. Hum Mol Genet. 1999;8:1329–1336. doi: 10.1093/hmg/8.7.1329. [DOI] [PubMed] [Google Scholar]
- Simon AM, Hoppe P, Burden SJ. Spatial restriction of AChR gene expression to subsynaptic nuclei. Development. 1992;120:1799–1804. doi: 10.1242/dev.114.3.545. [DOI] [PubMed] [Google Scholar]
- Von Nandelstadh P, Grönholm M, Moza M, Lamberg A, Savilahti H, Carpén O. Actin-organising properties of the muscular dystrophy protein myotilin. Exp Cell Res. 2005;310:131–139. doi: 10.1016/j.yexcr.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Weber FE, Vaughan KT, Reinach FC, Fischman DA. Complete sequence of human fast-type and slow-type muscle myosin binding protein C (MyBP-C): Differential expression, conserved domain structure and chromosome assignment. Eur J Biochem. 1993;216:661–669. doi: 10.1111/j.1432-1033.1993.tb18186.x. [DOI] [PubMed] [Google Scholar]
- Williams AF, Barclay AN. The immunoglobulin superfamily—Domains for cell surface recognition. Annu Rev Immunol. 1988;6:381–405. doi: 10.1146/annurev.iy.06.040188.002121. [DOI] [PubMed] [Google Scholar]
- Yarden Y, Ullrich A. Growth factor receptor tyrosine kinases. Annu Rev Biochem. 1988;57:443–478. doi: 10.1146/annurev.bi.57.070188.002303. [DOI] [PubMed] [Google Scholar]
- Zhu X, Yeadon JE, Burden SJ. ALM1 is expressed in skeletal muscle and is regulated by innervation. Mol Cell Biol. 1994;14:8051–8057. doi: 10.1128/mcb.14.12.8051. [DOI] [PMC free article] [PubMed] [Google Scholar]