Summary
Cell type-specific genetic modification using the Cre/loxP system is a powerful tool for genetic analysis of distinct cell lineages. Because of the exquisite specificity of Vasa expression (confined to the germ cell lineage in invertebrate and vertebrate species), we hypothesized that a Vasa promoter-driven transgenic Cre line would prove useful for the germ cell lineage-specific inactivation of genes. Here we describe a transgenic mouse line, Vasa-Cre, where Cre is efficiently and specifically expressed in germ cells. Northern analysis showed that transgene expression was confined to the gonads. Cre-mediated recombination with the Rosa26-lacZ reporter was observed beginning at ~e15, and was >95% efficient in male and female germ cells by birth. There was no ectopic activity in most adults, although some animals showed more widespread lacZ expression. This Vasa-Cre transgenic line should thus prove useful for genetic analysis of diverse aspects of germ cell function and gametogenesis.
Keywords: Cre, vasa, germ cells, gonad
Genetic approaches are well-suited for studies of gametogenesis and reproduction, processes that require complex interactions between germline and somatic cells, and hence are difficult to fully recapitulate in vitro. More recently, conditional gene targeting using the Cre/loxP system has emerged as an especially powerful method in reproductive genetics, particularly in the study of genes whose systemic inactivation is lethal. Critical to the success this approach is the availability of suitable Cre transgenes expressed in a wide range of distinct cell lineages at appropriate developmental timepoints (Kwan, 2002; Lewandoski, 2001). Previous germ cell Cre lines include ZP3-Cre, which is not active in primordial follicles but becomes induced following follicle growth; this line is thus useful for analyses of follicle growth or maternal contributions to the embryo (de Vries et al., 2000). A Cre knock-in into the TNAP (tissue non-specific alkaline phosphatase) is active in primordial germ cells starting as early as e9.5. but the TNAP promoter is also active in other tissues (e.g. placenta and liver, among others) and such widespread expression may complicate interpretation of experiments where germ cell-specific gene inactivation is desired or required (if e.g. the locus is essential) (Lomeli et al., 2000).
Vasa expression is confined to the germ cell lineage in species including Drosophila, zebrafish, mouse, and humans (Castrillon et al., 2000; Lasko and Ashburner, 1988). In the mouse, Vasa (a.k.a. mvh, mouse vasa homolog) expression begins at the time of gonad colonization by primordial germ cells. Vasa expression is detectable by e12.5 in both sexes and continues throughout life (Tanaka et al., 2000). We reasoned that a Vasa-Cre line could prove useful for studies of germ cell function following gonadal colonization, including many aspects of spermatogenesis and oogenesis, such as the assembly, activation, and growth of primordial follicles.
We cloned a 5.6 kb DNA fragment containing mouse Vasa 5’ regulatory sequences, including the transcriptional start site. This fragment contains canonical promoter sequences including a TATA box at −27 and more distant control elements important for germ cell specific expression. A similar homologous fragment has been employed to drive germ cell specific expression of the fluorescent reporter protein GFP in fish (Takeuchi et al., 2002). The 5.6 kb mouse Vasa promoter fragment was ligated to a cassette of the Cre ORF with an engineered nuclear localization sequence (CreN) (Gu et al., 1993). Following linearization and oocyte microinjection of this construct (Fig. 1), we identified eight transgenic founder lines with confirmed transgene integration by Southern analysis. Subsequent screening of these transgenic founder lines was performed by Northern analysis of gonads and somatic tissues, leading to the identification of two lines with the desired gonad-specific pattern of Cre expression. One line (see Fig. 2 for Northern analysis) was selected for subsequent analysis.
FIG. 1.
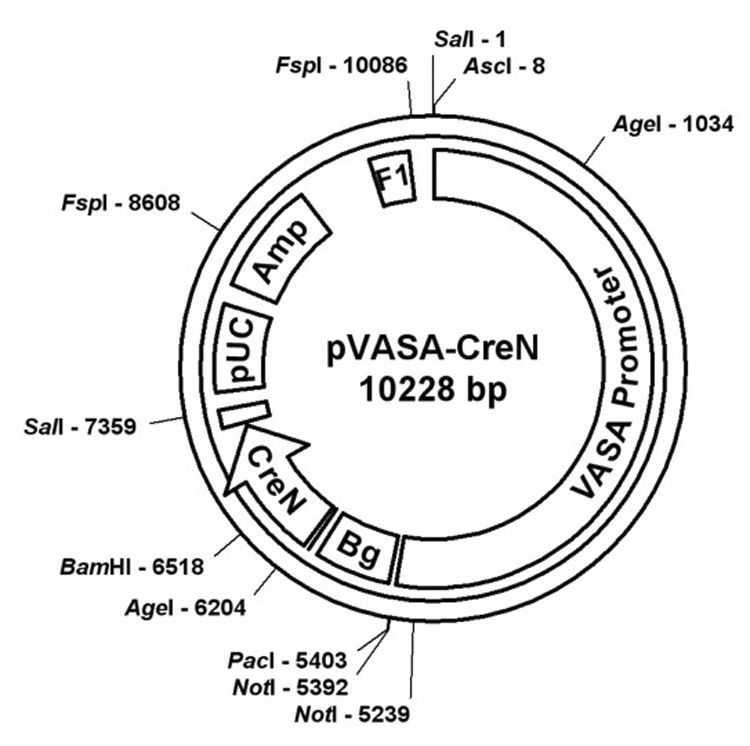
Map of construct used for transgenesis. The construct was linearized with AscI, then injected into oocytes. Vasa promoter=5.6kb genomic fragment, Bg=beta globin intron, CreN=Cre open reading frame with nuclear localization signal, pUC=pUC origin of replication, Amp=ampicillin resistance cassette, F1=F1 origin of single-stranded DNA replication. The unmarked box between pUC and CreN corresponds to the SV40 polyadenylation signal.
FIG. 2.
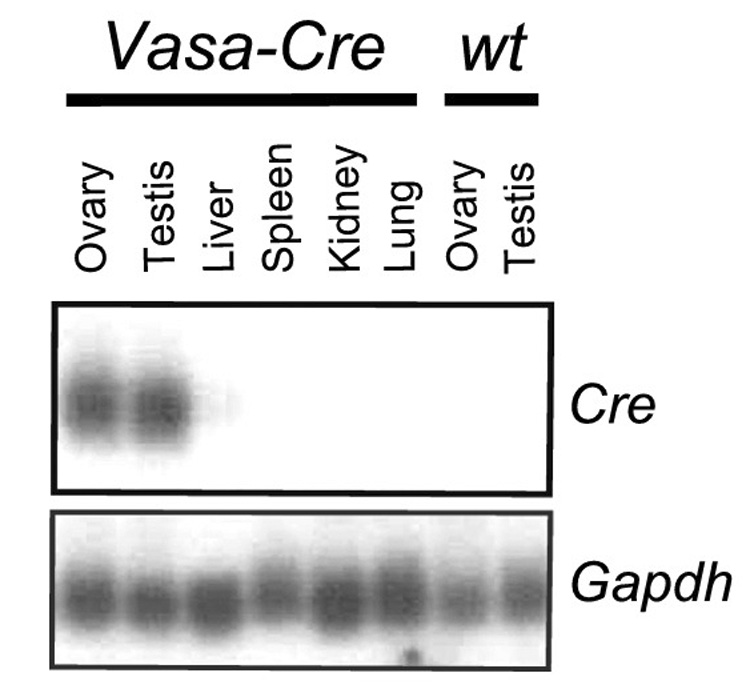
Northern analysis of tissues from Vasa-Cre transgenic and control mice at 3 weeks of age. Each lane corresponds to 10 µg of total RNA. The blot was probed with a Cre cDNA fragment and then stripped and reprobed with Gapdh as a loading control.
To further test the function and specificity of this Vasa-Cre line in vivo, we crossed Vasa-Cre males with female Gt(ROSA)26sor lacZ reporter mice (Soriano, 1999), referred to hereafter as R26R. Gonads from all Vasa-Cre; R26R progeny showed strong, diffuse lacZ expression, but in most animals, skin and all other tissues including the kidney, liver, lung, uterus, were negative (Fig. 3A–F). Occasional animals did express lacZ in a variegated pattern in skin epithelium, both in the ear and other locations such as the tail. Fig. 3G shows an example of high lacZ expression in skin; in this animal, most areas of skin did not express lacZ, and there was no lacZ expression in visceral organs. A minority of animals (<20%) showed more global expression (see explanation below). To assess the timing of Cre induction, we performed X-gal staining of embryonic gonads. At e18, strong recombination was observed in testes, whereas at e15, only very faint blue staining was observed that was distinguishable from controls by eye, but not readily apparent in photographs (Fig. 3 I, J). Therefore, Vasa-Cre activity is strongly induced between e15 and e18. The basis of this apparently later onset of Vasa-Cre expression relative to the endogenous Vasa locus has not been explored, but may relate to the absence of more distant regulatory elements required for earlier expression, or perhaps equally likely, the need for a threshold level of Cre protein to effectively induce recombination.
FIG. 3.
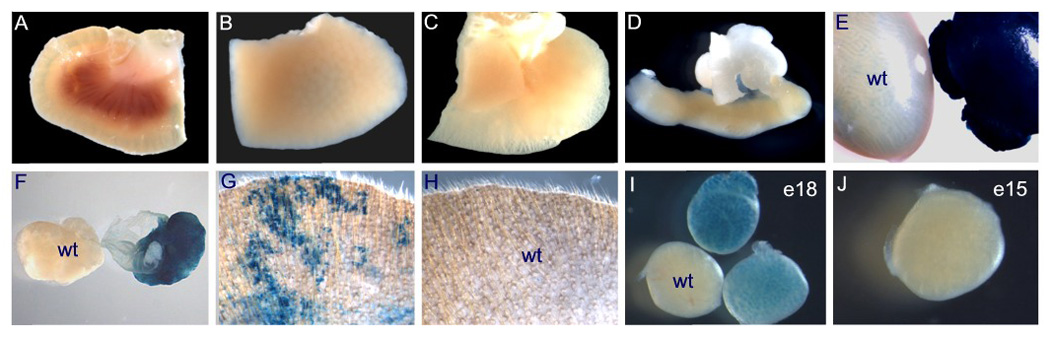
Evaluation of lacZ activity in gonads and other tissues in Vasa-Cre; R26R reporter mice. Vasa-Cre trasgenic males were bred to R26R females. All tissues in this figure were derived from mice carrying the R26R reporter except where noted (wt). A–H, tissues harvested at PND3. A) kidney; B) liver; C) lung; D) uterus and adipose tissue; E) testes (right, Vasa-Cre; left, sibling R26R control); F) ovaries (right, Vasa-Cre; left, R26R control); G) ear from Vasa-Cre mouse; H) ear from sibling control; I) e18 testes; e15 testis shows faint lacZ expression visible by eye but not apparent in photographs. Tissues were fixed in formalin and stained with X-Gal per standard protocols.
Of note, in crosses with R26R or other floxed alleles, we have observed potent parent-of-origin effects that must be taken into account in experimental designs. When the mother is the Vasa-Cre transgene carrier, virtually all progeny undergo global Cre-mediated recombination, even those that do not inherit Vasa-Cre (data not shown). This potent maternal effect is readily explained by the perdurance of Cre protein in the egg. This property should make Vasa-Cre useful as a general germ cell deletor line, in that it permits the efficient conversion of a floxed to a null allele while obviating the need to perform an additional cross to eliminate the Cre transgene. When complete germ line-specific inactivation of a gene is desired (e.g. to study its function in the germline), it is thus necessary to use males as the Vasa-Cre carriers. Even when males are used as carriers, however, occasional Vasa-Cre; R26R progeny (<20%) exhibit global lacZ expression and thus must have undergone recombination very early in zygotic development (perhaps 1 or 2 cell stage). This does not appear to be a paternal effect, as it is dependent on inheritance of the Vasa-Cre transgene. In practice, such animals that have undergone global recombination are readily identifiable by routine genotyping of tail DNA preps. This effect could, however, lower the expected frequency of some genotypes when the floxed allele under investigation is of an essential gene.
To document Cre activity at the cellular level, tissues shown in Fig. 3 were paraffin-embedded, sectioned and counterstained. Activity was observed in a high proportion of germ cells in both the ovary and testis, but not detectable in any somatic cells (Fig. 4A). Unexpectedly, β-galactosidase protein was exclusively confined within germ cells to a cytoplasmic structure that corresponds both in size and localization to the vitelline and chromatoid bodies, germ-cell specific organelles whose function remains poorly understood but are believed to participate in protein and/or RNA sorting (de Smedt et al., 2000). The biological basis of this localization has not been explored, but is presumably mediated by specific amino acid sequences present in the β-galactosidase protein.
FIG. 4.
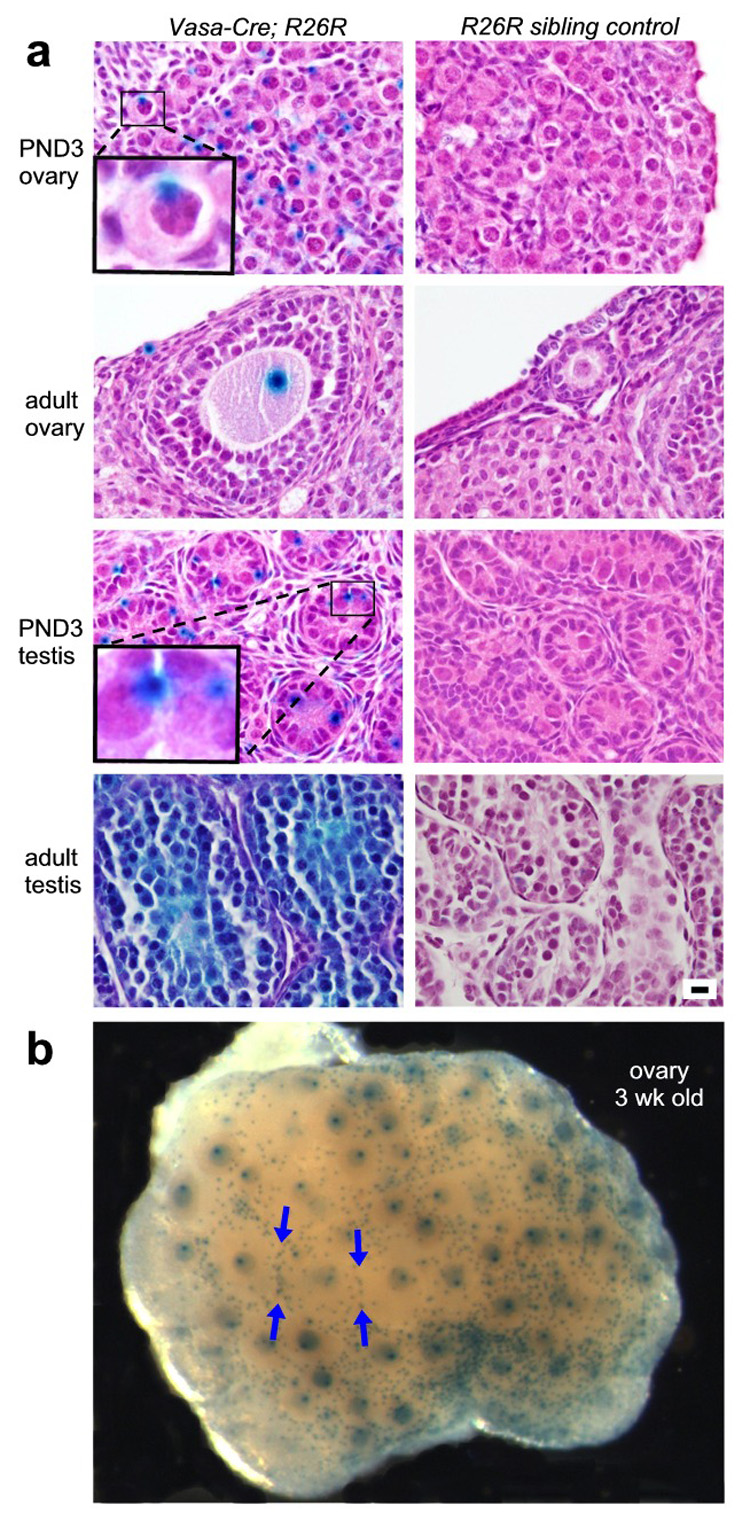
Analysis of X-gal stained tissues from Vasa-Cre; R26R mice and R26R non-transgenic sibling controls. All tissues were formalin-fixed, stained with X-gal, paraffin-embedded, sectioned, and counterstained with hematoxylin and eosin. A) ovaries and testes showing germ-cell specific lacZ expression in a high proportion of germ cells; all panels at same magnification. Insets at higher magnification show distinct localization of β-galactosidase protein to cytoplasmic structures that correspond to the vitelline and chromatoid bodies in oocytes and spermatogonia, respectively. Bar = 10 microns for all panels. B) gross view of intact ovary at 3 weeks of age (more lightly stained than ovary in Figure 3F) shows specific staining of virtually all primordial and growing oocytes. Blue arrows point to the increased concentration of primordial follicles at the interface of larger growing follicles.
At 3 weeks of age, staining of ovaries reveals a strikingly specific pattern wherein virtually all of the primordial oocytes, as well as all of the growing oocytes (but not any somatic cells), are strongly lacZ positive (Fig. 4B). These results suggest that this Vasa-Cre line will prove uniquely useful for studies of primordial follicle assembly, activation, and early follicle growth. Also of note, this procedure permits excellent visualization of the entire cohort of primordial follicles, revealing that primordial follicles are not randomly distributed on the ovarian cortex, but rather are concentrated along the boundaries of growing follicles (Fig. 4B arrows), pointing to unexpectedly complex patterns of follicle movement and/or distribution.
Because β-galactosidase protein is so tightly localized within each germ cell, examination of tissue sections can give the false impression that an individual cell does not express lacZ (i.e. the vitelline or chromatoid body is outside the plane of section). To determine the exact percentage of germ cells undergoing Cre-mediated recombination, we systematically analyzed each cell in serial tissue sections. This analysis showed that Cre-mediated recombination occurred in over 97% of germ cells in both males and females by PND3.
In summary, although care must be taken to account for maternal and early zygotic Vasa-Cre activity, the efficiency and specificity of this Vasa-Cre transgenic line suggest that it will be a useful tool for genetic analysis of diverse aspects of germ cell function, gonadogenesis, and gametogenesis, and in particular, for the genetic analysis of primordial follicle assembly, activation, and early follicle growth.
MATERIALS AND METHODS
Vasa-Cre construct
A 5.6 kb fragment of genomic DNA from strain FVB was amplified using the Takara Long and Accurate PCR system. The primer sequences were om-VASA51–ATAGGCGCGCCTGTGCCACCATGCCTGGCCCAGTTTC and om-VASA53–ATATTAATTAATCTCCGCTCCAGGCTCCCCGGGCTCCT. AscI and NotI restriction sites are engineered into the 5’ ends of these primers. PCR reactions were performed using the LA PCR kit 2.1 (Takara) in 50 µl volumes with tail DNA (0.50 µl), H2O (13 µl), 2× GC Buffer 2 (Takara) (25 µl), 25mM MgCl2, (1.5 µl), 2.5mM dNTP (8 µl), Forward and Reverse primers 10 µm (1 µl each) and LA Taq polymerase (Takara) 5U/µl (0.5 µl). The PCR conditions for long and accurate PCR were 95° 2 min; (95° 30 sec, 68° 10 min) × 35 cycles; 72° 10 min. Following amplification, the PCR product was digested with AscI and NotI, and cloned product directly into AscI/NotI digested pCreN vector that has a CreN open reading frame with a nuclear localization sequence in a pSG5 (Strategene) backbone.
Generation of Vasa-Cre transgenic mice and animal care
Following linearization by AscI, the DNA was purified on an Elutip-D column (Schleicher & Schuell) and microinjected into FVB oocytes by standard protocols (Nagy et al., 2003) at the UT Southwestern Transgenic Core Facility. R26R mice were purchased from Jackson Laboratories. This study was approved by the Institutional Animal Care and Use Committee of UT Southwestern Medical Center.
Initial screening of lines by Southern analysis and subsequent Northern analysis
Genomic DNA was prepared by digesting tail sections overnight in SNET buffer (20mM Tris pH8.0, 5mM EDTA, 400mM NaCl, 1% (w/v) SDS) and 1.5 µl Proteinase K (10mg/ml) at 55C. Samples were purified by phenol:chloroform extraction and ethanol precipitation. DNA was resuspended in 50 µl Tris pH7.5. SalI digests were performed on 10 µg of DNA and electrophoresed on a 0.8% agarose gel. DNA was transferred to Hybond N+ membrane (Amersham) and probed with a Cre cDNA fragment. RNA was prepared from adult tissues using Tripure reagent (Roche) and 10 µg was electrophoresed on a 1% formaldehyde gel. RNA was transferred to Hybond N+, probed with Cre and reprobed with Gapdh as a loading control.
Genotyping
Tail DNA was prepared by digesting overnight at 55° in PBND buffer (50mM KCl, 10mM Tris pH8.3, 2.5mM MgCl2, 0.1 mg/ml gelatin, 0.45% v/v NP40, 0.45% v/v Tween 20) and 1.5 ul Proteinase K 10 mg/ml (Invitrogen). PCR reactions were in 25 µl volumes with tail DNA (0.50 µl), H2O (18 µl), 10× Buffer (2.5 µl), 25mM MgCl2, (1.5 µl), 25mM dNTP (0.2 µl), Forward and Reverse primers 10 µm (1 µml each) and Hotstart Taq polymerase, 5U/µl (0.25 µl). Primer sequences are (For) CACGTGCAGCCGTTTAAGCCGCGT and (Rev) TTCCCATTCTAAACAACACCCTGAA. The PCR conditions were 95 °10 min; (94° 30 sec, 55° 30 sec) × 35 cycles; 72° 30 sec; 72° 7 min. Note: For primer corresponds to the Vasa promoter, Rev to the globin intron of the expression construct.
Histologic analysis and X-gal staining
Wholemount X-gal staining was performed by fixing tissues in 4% paraformaldehyde/PBS (Electron Microscopy Sciences) for one hour at 4° then rinsing twice for twenty minutes each in Buffer I (100mM sodium phosphate pH7.3, 2mM MgCl2, 0.01% sodium deoxycholate, 0.02% NP40) at room temperature. Tissues were then incubated at 50° in Buffer II (100mM HEPES, 5mM DTT, 1mM MgSO4, 2% Triton X-100) for one hour. Tissues were again rinsed in Buffer I and then incubated in Staining solution (5mM potassium ferricyanide, 5mM potassium ferrocyanide, 1 mg/ml X-gal and brought up to volume in Buffer I) for a period of 1.5 hours to overnight. Once staining was complete, tissues were refixed in formalin overnight. Following staining, tissues were embedded in paraffin, cut into 5 micron sections serially, and counterstained with hematoxylin and eosin.
ACKNOWLEDGEMENTS
We thank John Ritter and Bob Hammer of the UTSW Transgenic Core Facility for advice and for performing oocyte microinjections, and Klaus Rajewsky for the Cre cDNA. We acknowledge support for this work through a grant from the Lance Armstrong Foundation.
Grant sponsor: Lance Armstrong Foundation
LITERATURE CITED
- Castrillon DH, Quade BJ, Wang TY, Quigley C, Crum CP. The human VASA gene is specifically expressed in the germ cell lineage. Proc Natl Acad Sci U S A. 2000;97:9585–9590. doi: 10.1073/pnas.160274797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Smedt V, Szollosi D, Kloc M. The balbiani body: asymmetry in the mammalian oocyte. Genesis. 2000;26:208–212. doi: 10.1002/(sici)1526-968x(200003)26:3<208::aid-gene6>3.3.co;2-e. [DOI] [PubMed] [Google Scholar]
- de Vries WN, Binns LT, Fancher KS, Dean J, Moore R, Kemler R, Knowles BB. Expression of Cre recombinase in mouse oocytes: a means to study maternal effect genes. Genesis. 2000;26:110–112. [PubMed] [Google Scholar]
- Gu H, Zou YR, Rajewsky K. Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell. 1993;73:1155–1164. doi: 10.1016/0092-8674(93)90644-6. [DOI] [PubMed] [Google Scholar]
- Kwan KM. Conditional alleles in mice: practical considerations for tissue-specific knockouts. Genesis. 2002;32:49–62. doi: 10.1002/gene.10068. [DOI] [PubMed] [Google Scholar]
- Lasko PF, Ashburner M. The product of the Drosophila gene vasa is very similar to eukaryotic initiation factor-4A. Nature. 1988;335:611–617. doi: 10.1038/335611a0. [DOI] [PubMed] [Google Scholar]
- Lewandoski M. Conditional control of gene expression in the mouse. Nat Rev Genet. 2001;2:743–755. doi: 10.1038/35093537. [DOI] [PubMed] [Google Scholar]
- Lomeli H, Ramos-Mejia V, Gertsenstein M, Lobe CG, Nagy A. Targeted insertion of Cre recombinase into the TNAP gene: excision in primordial germ cells. Genesis. 2000;26:116–117. [PubMed] [Google Scholar]
- Nagy A, Gertsenstein M, Vintersten K, Behringer R. Manipulating the Mouse Embryo: A Laboratory Manual. Third ed. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2003. [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Takeuchi Y, Yoshizaki G, Kobayashi T, Takeuchi T. Mass isolation of primordial germ cells from transgenic rainbow trout carrying the green fluorescent protein gene driven by the vasa gene promoter. Biol Reprod. 2002;67:1087–1092. doi: 10.1095/biolreprod67.4.1087. [DOI] [PubMed] [Google Scholar]
- Tanaka SS, Toyooka Y, Akasu R, Katoh-Fukui Y, Nakahara Y, Suzuki R, Yokoyama M, Noce T. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev. 2000;14:841–853. [PMC free article] [PubMed] [Google Scholar]


