Abstract
BAX is a pro-apoptotic protein of the BCL-2 family stationed in the cytosol until activated by a diversity of stress stimuli to induce cell death. Anti-apoptotic proteins such as BCL-2 counteract BAX-mediated cell death. Although an interaction site that confers survival functionality has been defined for anti-apoptotic proteins, an activation site has not been identified for BAX, rendering its explicit trigger mechanism unknown. We previously developed Stabilized Alpha-Helix of BCL-2 domains (SAHBs) that directly initiate BAX-mediated mitochondrial apoptosis. Here we demonstrate by NMR analysis that BIM SAHB binds BAX at an interaction site that is distinct from the canonical binding groove characterized for anti-apoptotic proteins. The specificity of the BIM SAHB-BAX interaction is highlighted by point mutagenesis that abrogates functional activity, confirming that BAX activation is initiated at this novel structural location. Thus, we have now defined a BAX interaction site for direct activation, establishing a new target for therapeutic modulation of apoptosis.
The BCL-2 family comprises a network of pro-apoptotic and anti-apoptotic proteins whose interactions regulate the critical balance between cellular life and death1-3. The family is structurally defined by the presence of up to four conserved “BCL-2 homology’ (BH) domains, all of which include α-helical segments4. Anti-apoptotic proteins such as BCL-2 and BCL-XL display sequence conservation in three to four BH domains (BH1-4), whereas pro-apoptotic proteins are divided into multi-BH domain members such as BAX and BAK that contain three conserved domains (BH1-3) and “BH3-only” members such as BIM and BID that display homology only to the α-helical BH3 domain. The BH3-only subgroup is diverse and transmits pro-death signals arising from disparate stimuli to the core apoptotic machinery located at the mitochondrion5,6. Once deployed, the BH3 death signal is either neutralized by anti-apoptotic proteins or delivered to the mitochondrial executioners BAX and BAK, which upon activation permeabilize the outer mitochondrial membrane7,8. Released mitochondrial factors activate caspases that then implement the death program9.
Structural studies have established a survival paradigm for BCL-2 family interactions based on sequestration of the α-helical BH3 death domain by a hydrophobic groove formed by the juxtaposition of the BH1-3 domains (BH1: portions of helices α4-α5; BH2: α7-α8; BH3: α2) of anti-apoptotic members10. The selective binding interactions of pro-apoptotic BH3 peptides and multi-BH domain pro-apoptotic proteins with anti-apoptotic BCL-2 family members have been well documented11-16. The small molecules ABT-737 and ABT-263, constructed to selectively inhibit anti-apoptotic BCL-2/BCL-XL by targeting the hydrophobic groove, reactivate apoptosis in select tumors17,18. Thus, there is general agreement that one level of apoptosis regulation is mediated by the competitive interactions between pro- and anti-apoptotic members at the structurally-defined anti-apoptotic groove. However, what directly triggers BAX/BAK activation during apoptotic stress remains a matter of active debate11,13,19. Indeed, this lingering question concerning the biochemical mechanism that initiates BAX/BAK activation has recently been characterized by Youle and Strasser as the “holy grail of apoptosis research”3.
BAX activation is believed to be a highly regulated, multi-step process involving an interaction-triggered conformational change, mitochondrial translocation, and oligomerization that ultimately leads to mitochondrial dysfunction and apoptosis20-23. A diversity of stimuli have been implicated in initiating BAX/BAK activation24-26, including direct engagement by select BH3 domains and BH3-only proteins13,27-34. To investigate the elusive initiating event for BAX activation, we studied the interaction of BAX with the BH3 ligand BIM SAHB, which we previously demonstrated recapitulates the α-helical character of native death domains and binds directly to BAX33. We report the first structural analysis of a BH3 α-helix bound to a full length pro-apoptotic multi-BH domain protein, unveiling a novel site of protein interaction for functional activation and pharmacologic modulation of BAX (Supplementary Fig. 1).
Identification of a Novel BH3 Interaction Site on BAX
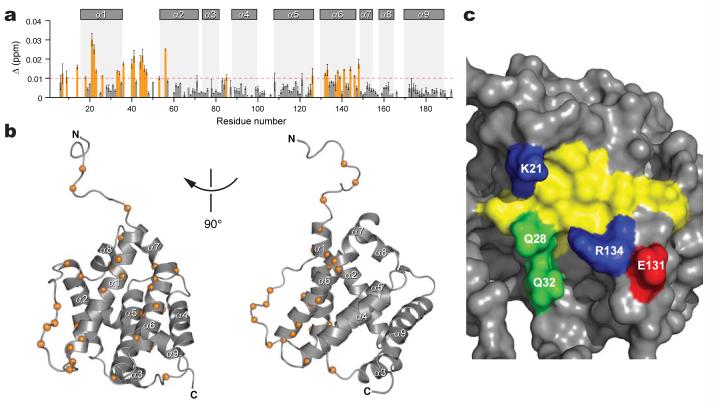
BIM SAHB binding to BAX was monitored using Nuclear Magnetic Resonance (NMR) spectroscopy. Compared to the 1H-15N correlation spectrum of BAX, the addition of BIM SAHB broadened and shifted select NMR cross-peaks, indicating fast exchange between the bound and unbound conformations of BAX. The overall features of the NMR spectra are quite similar except for significant changes in the loop residues between α1 and α2 upon BIM SAHB binding. Chemical shift perturbation mapping of BAX with BIM SAHB titration revealed interactions at a discrete subset of BAX residues. The degree of change in the 1H-15N cross-peak positions for backbone amides is shown in Figure 1a. The largest changes were observed for residues localized in the α1 and α6 helices, as well as residues in the flexible loop between α1 and α2. Significant changes were also observed for the side-chain NH2 of Q28, Q32, and Q52 (Supplementary Fig. 2). In the BAX structure35, the α1 and α6 helices are positioned adjacent to one another, and the residues impacted by BIM SAHB binding localize to a discrete site at the juxtaposition of these α-helices on one side of the protein structure (Fig. 1b). Of note, no residues on the carboxy terminal side of the protein are affected by BIM SAHB titration under these conditions, thus placing the novel binding site on the completely opposite face of the protein from the canonical BH3 binding site of anti-apoptotic proteins (Supplementary Fig. 1). The binding site of BIM SAHB on BAX is thus defined by the two helices α1 and α6, with the interhelical junction forming a hydrophobic cleft surrounded by a perimeter of hydrophilic and charged residues (Fig. 1c).
Figure 1. NMR analysis of BAX upon BIM SAHB titration.
a, Chemical shift changes are plotted as a function of the residue number of BAX. Residues with significant backbone amide chemical shift change (>0.01 ppm) are colored orange. The absence of a bar indicates the presence of a proline or a residue that is overlapped or below detection threshold. Error bars, mean ± s.d. b, The Cα atoms of BAX residues affected by BIM SAHB binding are shown as orange spheres. c, Surface diagram illustrating the BAX binding site. Side chains of hydrophobic, positively charged, negatively charged, and hydrophilic residues are colored yellow, blue, red, and green, respectively.
Orientation of BIM SAHB at the BAX Site
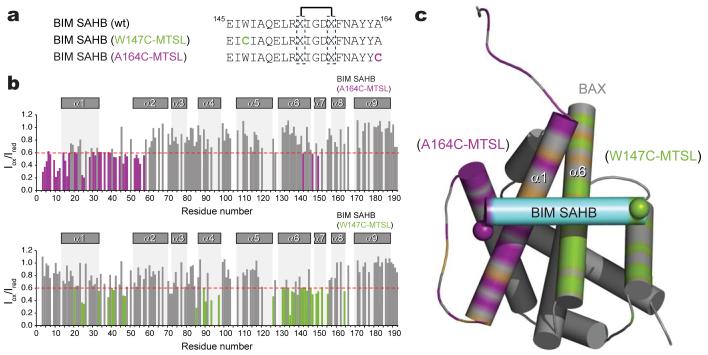
One of the challenges of capturing a BAX-activating interaction by NMR is time- and dose-dependent BAX aggregation due to BIM SAHB addition to the NMR sample. With time, the size of the protein aggregate formed leads to line broadening of the NMR resonances beyond detection, thereby precluding acquisitions of data for the duration required to resolve the initial complex by conventional NMR. Nevertheless, to more precisely orient BIM SAHB at the BAX interaction site, we conducted paramagnetic relaxation enhancement (PRE) NMR experiments, which can be performed on a time-scale that is compatible with the lifespan of the complex under NMR conditions. 1H-15N-BAX HSQC spectra were acquired with methane thiosulphonate (MTSL)-derivatized BIM SAHB compounds (Fig. 2a) in the oxidized state and then repeated after nitroxide reduction. Of note, the chemical shift perturbation maps of BAX in complex with BIM SAHB and the MTSL-labeled derivatives demonstrate consistent changes in BAX α-helices 1 and 6, and in the α1-α2 loop (Supplementary Fig. 3). Whereas the intensity of BAX α1 residues are predominantly reduced in the presence of oxidized C-terminal label, the intensity of BAX α6 residues are predominantly decreased by the presence of oxidized N-terminal label (Fig. 2b). As an example, BIM SAHB(A164C-MTSL) caused marked signal suppression of Ser16 (α1) but had essentially no effect on Asp142 (α6) (Supplementary Fig. 4a), whereas BIM SAHB(W147C-MTSL) had no effect on Ser16 (α1) but reduced the Asp142 (α6) signal (Supplementary Fig. 4b). Because the degree of PRE correlates with the distance between the nitroxide label and the affected BAX residues36, structure calculations using restraints derived from PRE and chemical shift perturbation data were performed to define the orientation of BIM SAHB at the novel BAX site. The calculated model structures converged to place BIM SAHB perpendicular to BAX helices α1 and α6, with the N- to C-terminus directionality of the peptide disposed right to left (Fig. 2c, Supplementary Table 1). Whereas the location of the novel BAX binding site is geographically distinct, the calculated model structure of the BIM BH3-BAX complex reveals an interaction topography that is strikingly similar to that of BIM BH3 with anti-apoptotic BCL-XL, MCL-1, and BFL1/A1, as previously determined by X-ray crystallography (Supplementary Fig. 5).
Figure 2. Orientation of BIM SAHB at the novel BAX binding site.
a,Paramagnetically-labeled derivatives of BIM SAHB. b, Ratios of BAX crosspeak intensities in the presence of oxidized or reduced BIM SAHBMTSL (Iox/Ired) versus residue number. BAX residue intensities reduced below 0.6 are colored. c, Orientation of BIM SAHB at the BAX site. Based on an Iox/Ired threshold of 0.6, BAX residues impacted by BIM SAHB(A164C-MTSL), BIM SAHB(W147C-MTSL), or both are colored in purple, green, or brown, respectively.
Initiation of BAX Activation by BIM SAHB
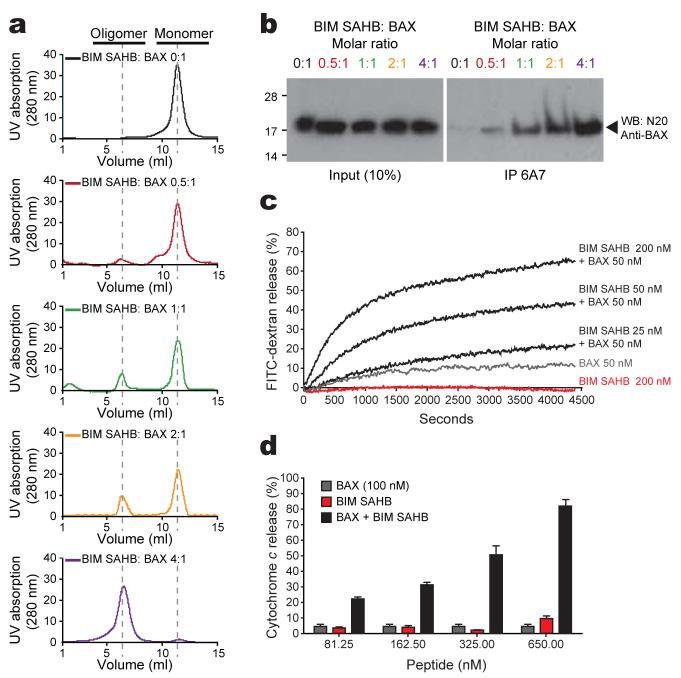
In contrast to the stable inhibitory interactions of BIM BH3 with anti-apoptotic proteins, the BAX-activating interaction triggers a dynamic continuum of events that includes BAX conformational change and oligomerization (Supplementary Fig. 6). Of particular interest, NMR resonances of residues in the α1-α2 loop of BAX shifted significantly upon BIM SAHB binding (Fig. 1a, Supplementary Fig. 3). In monomeric BAX, the center of the loop weakly associates with residues Ile133 and Met137 of the α6 helix35. Changes observed in the loop residues can readily be explained by the shift of the loop conformation into an open form upon BIM SAHB binding. Because loop displacement upon ligand engagement may initiate a conformational change of BAX, we investigated whether BIM SAHB binding could directly activate BAX in solution. We monitored both the conversion of BAX from monomer to oligomer by size-exclusion chromatography (SEC) and the exposure of its buried N-terminal residues using the 6A7 antibody, which selectively detects the conformational change associated with BAX activation37. Indeed, BIM SAHB triggered dose-dependent oligomerization of BAX (Fig. 3a). This BIM SAHB-induced oligomerization correlated with exposure of the N-terminal epitope of BAX as recognized by the 6A7 antibody (Fig. 3b). Thus, the transient stability of the BIM SAHB-BAX complex we observe by NMR correlates with the interaction-triggered BAX conformational change and oligomerization that we detect biochemically.
Figure 3. BIM SAHB directly initiates BAX activation in vitro.
a, Oligomerization (a) and 6A7 immunoprecipitation (b) of BAX after BIM SAHB treatment. c, Liposomal FITC-dextran release in the presence of BAX and BIM SAHB. d, Cytochrome c release from BAX/BAK-null mitochondria upon incubation with BAX and BIM SAHB. Error bars, mean ± s.d.
To confirm that the SEC-based detection of BIM SAHB-induced BAX oligomerization reflects functional activation of BAX for its release activity, we performed correlative liposomal and mitochondrial assays. In liposomal assays that explicitly explore the capacity of BIM SAHB to directly trigger functional BAX activation in the absence of other factors, the combination of BIM SAHB and BAX yielded dose-responsive liposomal release of entrapped FITC-dextran (Fig. 3c). Likewise, in mitochondrial release assays that employed BAX/BAK doubly-deficient mouse liver mitochondria prepared from Alb-creposBaxflox/-Bak-/- mice, BIM SAHB induced dose-responsive BAX-mediated cytochrome c release (Fig. 3d). Thus, in four distinct in vitro assays that measure ligand-induced BAX activation, BIM SAHB directly and dose-responsively triggered BAX.
Specificity of BIM SAHB-Induced BAX Activation
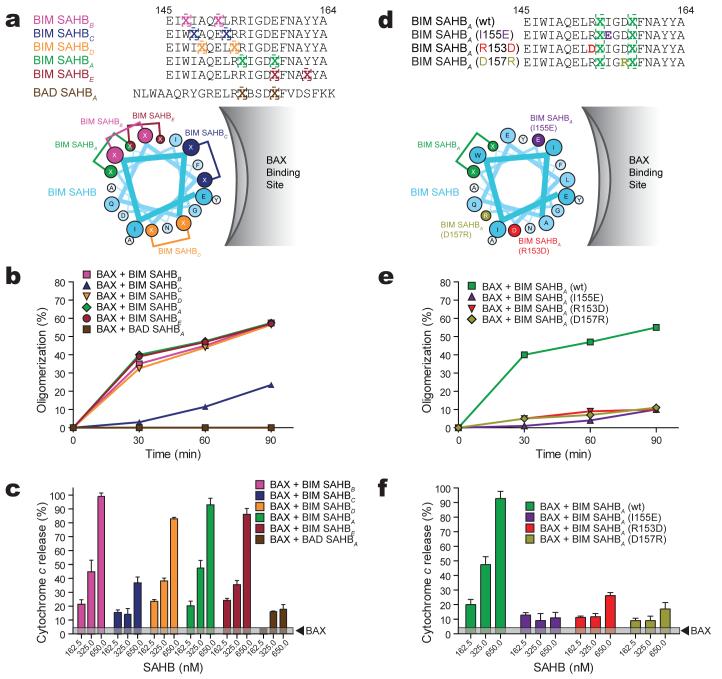
We next performed a “staple scan” and mutagenesis studies to demonstrate and link the specificity of BIM SAHB-induced BAX activation to interaction at the novel site. By performing a staple scan that effectively replaces pairs of amino acid residues within the core BH3 sequence with crosslinked norleucine-like side chains, we simultaneously address which residues are essential to the functional interaction between BIM BH3 and BAX, and further confirm which surface along the BIM BH3 α-helix is essential to BAX engagement (Fig. 4a). Substituting the non-natural amino acid at positions W147, A149, E151, R153, R154, E158, or Y162 did not disrupt BIM SAHB-induced BAX oligomerization (Fig. 4b) or BAX-mediated cytochrome c release (Fig. 4c, Supplementary Fig. 7a). Indeed, hydrocarbon staples located along the length of the α-helix at surfaces that do not face the hydrophobic contact site on BAX did not disrupt the functional interactions between the BIM SAHB analogs and BAX. However, a staple position that replaced I148 and the highly conserved L152, and localizes to the hydrophobic contact surface for BAX on the BIM BH3 α-helix (Fig. 4a), markedly decreased BAX oligomerization (Fig. 4b) and BAX-mediated cytochrome c release (Fig. 4c, Supplementary Fig. 7a), findings consistent with the near abrogation of BIM SAHB-induced chemical shift perturbations of 15N-BAX by BIM SAHBC (Supplementary Fig. 8a). In addition, BAD SAHBA, which has the identical staple position as BIM SAHBA but otherwise contains distinct amino acid sequence, did not bind (Supplementary Fig. 8b) or oligomerize (Fig. 4b) BAX, or induce BAX-mediated cytochrome c release (Fig. 4c, Supplementary Fig. 7a).
Figure 4. Sequence specificity of BIM SAHB-induced BAX activation.
a, BIM SAHBs with differential staple positions and their activities in BAX oligomerization (b) and BAX-mediated cytochrome c release assays (c). d, BIM SAHBA point mutants and their activities in BAX oligomerization (e) and BAX-mediated cytochrome c release assays (f). Error bars, mean ± s.d.
To further examine ligand specificity, we generated and screened a panel of BIM SAHB compounds bearing single point mutations within the core BH3 consensus sequence, including I131E, R153D and D157R (Fig. 4d, Supplementary Fig. 5a). In each case, point mutagenesis abolished BIM SAHB-induced BAX oligomerization (Fig. 4e) and BAX-mediated cytochrome c release (Fig. 4f, Supplementary Fig. 7b), indicating that interference with key hydrophobic or charge interactions specifically disrupts BIM SAHB engagement of the novel BAX site.
Mutagenesis of the BAX Site Impairs Activation
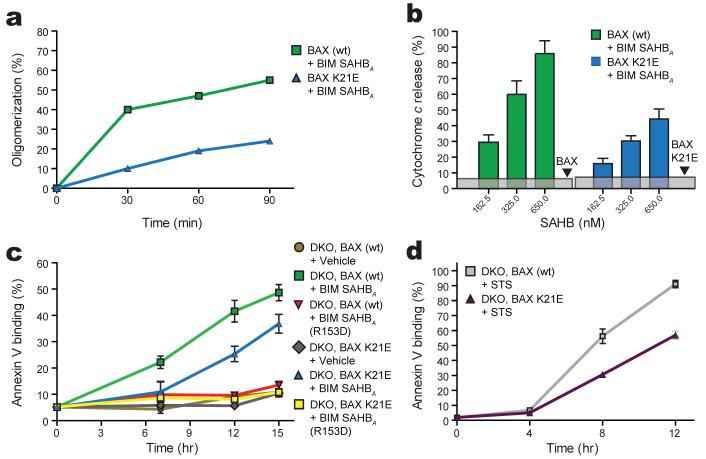
To examine the impact of BAX mutagenesis at the α1-α6 interaction site, we mutated residue K21, which exhibited the most pronounced chemical shift upon BIM SAHB binding (Fig. 1a). We generated recombinant BAX bearing a K21E point mutation (Supplementary Fig. 9) and observed that BIM SAHB-induced BAXK21E activation was significantly reduced compared to wild type BAX as measured by the oligomerization assay (Fig. 5a). BAXK21E-mediated cytochrome c release in response to BIM SAHB was likewise blunted (Fig. 5b). To determine if this single point mutation within the novel BAX binding site impacted the capacity to activate BAX in a cellular context, we retrovirally reconstituted Bax-/-Bak-/- mouse embryonic fibroblasts (DKO MEFs) with either wild type or BAXK21E and monitored apoptosis induction in response to BIM SAHB. Whereas BIM SAHB induced time-dependent apoptosis of BAX-reconstituted MEFs, as quantitated by annexin-V binding, single K21E point mutation within the new BAX binding site reduced BIM SAHB-triggered apoptosis (Fig. 5c). This decrease in BAXK21E-mediated apoptosis correlates with impaired activation of BAXK21E by BIM SAHB in both oligomerization and cytochrome c release assays (Fig. 5a, 5b). As a further measure of specificity, we found that R153D mutagenesis of BIM SAHB, which eliminated its BAX activating capacity in the oligomerization and cytochrome c release assays (Fig. 4e, 4f), likewise abolished BIM SAHB-induced activation of BAX and BAXK21E in a cellular context (Fig. 5c).
Figure 5. Mutagenesis of the BAX interaction site impairs activation and BAX-mediated apoptosis.
a, Impact of BAX K21E mutagenesis in oligomerization and b, cytochrome c release assays. c, Apoptotic response of Bax-/-Bak-/- MEFs reconstituted with BAX or BAXK21E to treatment with BIM SAHBA or BIM SAHBA(R153D). d, Impact of BAX K21E mutagenesis on staurosporine (STS)-induced apoptosis of BAX-reconstituted DKO MEFs. Error bars, mean ± s.d.
To probe the broader physiologic impact of the novel BAX activation site, we examined the apoptotic response of DKO MEFs reconstituted with BAX and BAXK21E to staurosporine (STS), a general stimulus known to operate through endogenous BH3-only proteins13,38, including BIM39. K21E mutagenesis impaired STS-induced apoptosis, as monitored by annexin-V binding over time (Fig. 5d). The reduced activity of BAXK21E was also reflected by impaired cytochrome c release, as detected by subcellular fractionation Western analysis and indirect immunofluorescence (Supplementary Fig. 10). The blunted response of BAXK21E-reconstituted DKO MEFs to STS is uniformly consistent with impaired activation of BAXK21E by BIM SAHB in oligomerization (Fig. 5a), cytochrome c release (Fig. 5b), and cell-based apoptosis assays (Fig. 5c). Thus, the MEF studies extend the mechanistic relevance of direct BAX activation to a cellular context in which BAX-mediated apoptosis is impaired by single amino acid mutagenesis at the novel BH3 interaction site. Taken together, the in vitro and cell-based mutagenesis experiments highlight the exquisite specificity of the BIM BH3-BAX interaction and implicate engagement of the novel binding site as a trigger mechanism for initiating BAX activation.
Discussion
Our data now define an explicit binding site on BAX for BH3 engagement that directly initiates BAX activation. In contrast to the constitutive mitochondrial localization of pro-apoptotic BAK, BAX is predominantly cytosolic, thereby necessitating a trigger mechanism for its activation and translocation to the mitochondria to induce mitochondrial outer membrane permeabilization40. BAX’s C-terminal α-helix folds back into its hydrophobic cleft35, which is homologous to the BH3-binding anti-apoptotic groove10,41 (Supplementary Fig. 1). The amphipathic nature of BAX’s C-terminal α-helix enables the hydrophobic face to closely interact with the canonical hydrophobic groove, while the hydrophilic face promotes solubility by readily engaging the aqueous environment. In order for BAX to insert into the outer mitochondrial membrane during targeting, the C-terminal α-helix must be dislodged, presumably coinciding with an induced global protein conformational change35 (Supplementary Fig. 6). Although the structural characteristics of this conformational change are unknown, the N- and C-termini of BAX have been implicated in locking down cytosolic BAX until structural reorganization and mitochondrial targeting is initiated21,37,42-44. Here, we report the first structural identification of a distinct BH3 binding site on BAX comprised of α-helices 1 and 6. Indeed, our findings support a previous observation that the first α-helix of BAX participates in an interaction with the BH3 peptides of BID and PUMA, as demonstrated by yeast two-hybrid and co-immunoprecipitation analyses27. We find that BIM SAHB engagement directly triggers the functional activation of BAX. Further, BAX activation is specifically disrupted by mutagenesis of the novel interface. These data indicate that BAX can be directly targeted by a BH3 domain at a site that is geographically distinct from but strikingly similar in topography to the canonical anti-apoptotic groove, and thereby unleash the pro-apoptotic activity of BAX.
Structural reorganization to achieve functional activity is a unifying feature of BCL-2 family proteins, and for a subset of pro- and anti-apoptotic members, translocation from cytosol to organelle compartments is also essential21,23,35,40,45. Our identification of a novel BH3 ligand binding site on BAX suggests that engagement of alternate BCL-2 family binding sites may play important roles in regulating apoptotic protein structure, intracellular localization, and functional activation. Whereas blockade of the novel site may effectively repress BAX-induced cell death, ligand engagement may trigger BAX-mediated apoptosis. Thus, our identification of a novel BAX activation site has important implications for the development of pharmacologic agents to respectively activate or inhibit apoptosis in human diseases characterized by unrestrained cell survival or pathologic cell death.
METHODS SUMMARY
SAHBs were synthesized using our established method46,47 and recombinant BAX for NMR and biochemical analyses was generated as previously described33,35. Samples for HSQC and PRE NMR contained uniformly 15N-labeled BAX at 0.2 mM prepared in 10 mM sodium acetate solution at pH 6.0 with up to a 1:1 molar ratio of SAHB. NMR spectra were acquired at 32°C on Bruker 600 and 800 MHz spectrometers, and then processed and analyzed as described in the Full Methods. To evaluate BIM SAHB-induced BAX activation, four in vitro assays were performed. The oligomerization assay employed freshly purified monomeric BAX in combination with BIM SAHB at the indicated ratios and incubation durations followed by size-exclusion chromatography to quantify monomeric vs. oligomeric BAX. The BAX conformational change assay also employed the indicated BIM SAHB:BAX mixtures, which were exposed to the conformation-specific 6A7 anti-BAX antibody, followed by immunoprecipitation and BAX Western analysis to monitor the proportion of activated conformer of BAX upon BIM SAHB exposure. To determine if the BIM SAHB-induced BAX conformational change reflected functional activation of its release activity, we conducted liposomal and mitochondrial release assays as previously described33,48 and using the indicated doses and constructs of BIM SAHB and BAX. For cellular studies, DKO MEFs were reconstituted with BAX by retroviral transduction of BAX-IRES-GFP as previously reported7,13 and as described in the Full Methods. BAX or BAXK21E-reconstituted DKO MEFs were exposed to either BIM SAHBs or staurosporine, and cell death quantified over time by annexin-V-Cy3 staining followed by flow cytometric analysis.
METHODS
SAHB synthesis and characterization
Hydrocarbon-stapled peptides corresponding to the BH3 domain of BIM and its point mutants were synthesized, purified, and characterized using methodologies previously described33,46,47. BIM SAHBA used in NMR and in vitro studies is an N-acetylated, C-amidated 20-mer: Ac-145EIWIAQELRXIGDXFNAYYA164-CONH2, where X represents the non-natural amino acid inserted for olefin metathesis. To generate paramagnetically-labeled BIM SAHB compounds for PRE-NMR studies, cysteine was substituted at position 147 or 164 and then reacted with 5 molar excess methane thiosulphonate (MTSL) reagent in DMSO for 30 minutes at room temperature. Once the reaction reached completion, as confirmed by LC/MS, excess MTSL was removed by lyophilization, the residual powder resuspended in methanol, and then pure MTSL-labeled SAHB precipitated with ether/hexanes. For MEF studies, the cell-permeable N-acetylated, C-amidated 21-mer Ac-146IWIAQELRXIGDXFNAYYARR166-CONH 332 and its R153D mutant were employed.
BAX preparation
Recombinant BAX was produced as previously described33,35. Mutant BAX was generated by PCR-based site-directed mutagenesis followed by DNA sequencing to verify the construct.
NMR sample and spectroscopy
Uniformly 15N-labeled or 13C/15N-labeled full-length human BAX was generated as previously described35. SAHB (1 mM stock) was added to a solution of 0.2 mM Bax to achieve a 1:1 molar ratio. Both peptide stock and protein samples were prepared in 10 mM sodium acetate solution at pH 6.0. Spectra were acquired at 32°C on a Bruker 800 MHz NMR spectrometer, processed using NMRPipe and analyzed with PIPP (see Supplementary Notes for software references). Two independent NMR titrations were performed. The pulse program used to acquire the correlation spectra was 1H-15N HSQC49. The weighted average chemical shift difference Δ at the molar ratio of 1:1 was calculated as in ppm. Ribbon diagrams and molecular models were rendered using PYMOL50.
Paramagnetic relaxation enhancement
Samples for PRE NMR contained uniformly 15N-labeled human BAX at 0.2 mM prepared in 10 mM sodium acetate solution at pH 6.0. NMR spectra were acquired at 32°C on a Bruker 600 MHz NMR spectrometer, processed using NMRPipe and analyzed with NMRView (see Supplementary Notes for software references). Two 1H-15N HSQC49 spectra for PRE NMR were acquired using a 1:1 ratio of BAX to MTSL-labeled SAHB in the oxidized and reduced state. The reduced compound was generated by exposure to five molar excess ascorbic acid for one hour. PRE effects were measured from the ratio of integrated peak intensities of the oxidized and reduced HSQC spectra. Peak intensities were normalized to account for small differences between the paramagnetic and diamagnetic BAX samples using cross peaks arising from unaffected BAX residues that are distant from the SAHB binding site. To evaluate the paramagnetic broadening effects of each BIM SAHBMTSL, the calculated ratios from the peak intensities of the oxidized and reduced spectra (Iox/Ired) were determined. BAX residues closest to a paramagnetic label have calculated values of Iox/Ired <0.6. Measurements for proline or overlapped residues are absent from the intensity ratio plot.
Structure calculations
Structure calculations using restraints derived from the chemical shift perturbation and PRE data were performed as described in the Supplementary Methods.
Oligomerization assay
BIM SAHB was added to a 200 μL solution (20 mM Hepes/KOH pH 7.2-7.4, 150 mM KCl) containing monomeric BAX (38 μM) at a ratio of 0.5:1, 1:1, 2:1 and 4:1 BIM SAHB:BAX. The mixtures and a sample of BAX monomer alone were incubated at 22°C for 15 minutes and then subjected to analysis by size exclusion chromatography (SEC) using an SD75 column. The chromatogram demonstrates the monomeric and oligomeric peaks at ∼11.5 min and ∼6.5 min, respectively. Protein standards (GE Healthcare) were used to calibrate the molecular weights of gel filtration peaks. For time-dependent analysis, BIM SAHB was added to monomeric BAX at a ratio of 1:1, and the mixtures were analyzed by SEC after incubation for 30, 60, and 90 minutes at 22°C. As a baseline for comparison, Bax monomer alone was analyzed at time 0 and the above time points. Replicates were performed using at least two independent preparations of freshly SEC-purified monomeric BAX.
Conformational change assay
BIM SAHB was added to a 20 μL PBS solution containing monomeric BAX (9 μM) at a ratio of 0.5:1, 1:1, 2:1 and 4:1 BIM SAHB:BAX. The mixtures (10 μL) and a BAX monomer sample (10 μL) were incubated at 22°C for 15 minutes and then added to a 3% BSA in PBS solution (250 μL) containing 15 μL of 6A7 anti-BAX antibody for 1 hour incubation at 4°C. Additionally, 1 μL of each input sample (10%) was mixed with 50 μL of SDS-sample buffer to measure baseline BAX levels across specimens. Preclarified sepharose beads (50 μL) were added to the BIM SAHB:BAX and BAX monomer solutions for an additional 2 hour incubation at 4°C. The sepharose beads were spun down, washed 3 times with 1 mL of 3% BSA in PBS solution, resuspended in 50 μL of SDS-sample buffer and boiled at 95 °C for 2 minutes. Ten microliters each of inputs and immunoprecipitation samples were used for analysis. Samples were separated on 12% SDS-PAGE Bis-Tris gel, blotted on a PVDF membrane, and Western analysis performed using the rabbit polyclonal N20 anti-BAX antibody (Santa Cruz Biotechnology) and chemiluminescence-based detection (PerkinElmer).
Release assays
Mitochondrial cytochrome c release assays33,48 were performed in triplicate on Alb-creposBaxflox/-Bak-/- mitochondria, treated with BAX and BIM SAHBs at the indicated doses. Analogous treatments were employed in liposomal assays, in which BAX-induced release of FITC-labeled dextran-10 kD from large unilamellar vesicles was monitored using a FluoroMax-2 spectrofluorometer (SPEX) as previously reported33,48.
Plasmid construction and retrovirus production
BAX was cloned into the retroviral expression vector MSCV-IRES-GFP (pMIG)13. The mutant BaxK21E construct was generated by PCR-based site-directed mutagenesis and confirmed by DNA sequencing. The production of retroviruses was performed as described previously13. Retroviral transduction of BAX constructs were confirmed by Western analysis.
Cell culture, cell transfection and apoptosis assay
Bax-/-Bak-/- DKO MEFs are SV40 transformed and maintained in Dulbecco’s modified Eagles medium supplemented with 10% fetal bovine serum following standard culture conditions and procedures. Reconstitution of BAX and the indicated mutant into DKO cells was achieved by retroviral transduction of BAX-IRES-GFP or BAXK21E-IRES-GFP, followed by MoFlo sorting for GFP positive cells. Comparable expression of wild-type or mutant BAX protein was confirmed by anti-BAX Western analysis. For BIM SAHB treatment, MEFs were trypsinized, collected, and then washed twice in serum-free media followed by plating of cells (3-5x104/well) in 50 μL, exposure to BIM SAHB (20 μM) or vehicle in serum-free media for 2 hours, and serum replacement (2x serum in 50 μL media) for an overall treatment duration as indicated. Cell death of triplicate samples was quantitated by annexin-V-Cy3 (BioVision, Mountain View, CA) staining according to the manufacturer’s protocol, followed by flow cytometric analysis using a FACS Caliber (BD Bioscience, San Jose, CA) and CellQuest software. For staurosporine treatment, plated MEFs in serum-containing media were exposed to drug (1 μM) and cell death of triplicate samples quantitated at the indicated time points by annexin-V binding as above.
Supplementary Material
Acknowledgements
We thank E. Smith for editorial and graphics assistance, W. Beavers for amino acid analyses, A. Perry for technical assistance, C. Turner and A. Bielecki of the MIT/Harvard Center for Magnetic Resonance for NMR technical advice, and R. Youle for feedback on the manuscript. We acknowledge the indelible contributions of the late Stanley J. Korsmeyer, who inspired this work. L.D.W. is supported by National Cancer Institute (NCI) grant 5P01CA92625, a Burroughs Wellcome Fund Career Award in the Biomedical Sciences, a Culpeper Scholarship in Medical Science from the Goldman Philanthropic Partnerships, an American Society of Hematology Junior Faculty Scholar Award, and a grant from the William Lawrence Children’s Foundation. This research was also supported by NCI grant 5RO1CA50239. N.T. is supported by the Intramural Research Program of the National Heart, Lung and Blood Institute, NIH. E.H.-Y.C. is supported by the Searle Scholars Program and NCI grant 5RO1CA125562.
References
- 1.Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 2.Leber B, Lin J, Andrews DW. Embedded together: the life and death consequences of interaction of the Bcl-2 family with membranes. Apoptosis. 2007;12:897–911. doi: 10.1007/s10495-007-0746-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 4.Petros AM, Olejniczak ET, Fesik SW. Structural biology of the Bcl-2 family of proteins. Biochim Biophys Acta. 2004;1644:83–94. doi: 10.1016/j.bbamcr.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 5.Huang DC, Strasser A. BH3-Only proteins-essential initiators of apoptotic cell death. Cell. 2000;103:839–842. doi: 10.1016/s0092-8674(00)00187-2. [DOI] [PubMed] [Google Scholar]
- 6.Walensky LD. BCL-2 in the crosshairs: tipping the balance of life and death. Cell Death Differ. 2006;13:1339–1350. doi: 10.1038/sj.cdd.4401992. [DOI] [PubMed] [Google Scholar]
- 7.Cheng EH, Wei MC, Weiler S, Flavell RA, Mak TW, et al. BCL-2, BCL-X(L) sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol Cell. 2001;8:705–711. doi: 10.1016/s1097-2765(01)00320-3. [DOI] [PubMed] [Google Scholar]
- 8.Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, et al. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, et al. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 10.Sattler M, Liang H, Nettesheim D, Meadows RP, Harlan JE, et al. Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science. 1997;275:983–986. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- 11.Certo M, Del Gaizo Moore V, Nishino M, Wei G, Korsmeyer S, et al. Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell. 2006;9:351–365. doi: 10.1016/j.ccr.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 12.Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 13.Kim H, Rafiuddin-Shah M, Tu HC, Jeffers JR, Zambetti GP, et al. Hierarchical regulation of mitochondrion-dependent apoptosis by BCL-2 subfamilies. Nat Cell Biol. 2006;8:1348–1358. doi: 10.1038/ncb1499. [DOI] [PubMed] [Google Scholar]
- 14.Oltvai ZN, Milliman CL, Korsmeyer SJ. Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell. 1993;74:609–619. doi: 10.1016/0092-8674(93)90509-o. [DOI] [PubMed] [Google Scholar]
- 15.Willis SN, Chen L, Dewson G, Wei A, Naik E, et al. Proapoptotic Bak is sequestered by Mcl-1 and Bcl-xL, but not Bcl-2, until displaced by BH3-only proteins. Genes Dev. 2005;19:1294–1305. doi: 10.1101/gad.1304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhai D, Jin C, Huang Z, Satterthwait AC, Reed JC. Differential regulation of Bax and Bak by anti-apoptotic Bcl-2 family proteins Bcl-B and Mcl-1. J Biol Chem. 2008;283:9580–9586. doi: 10.1074/jbc.M708426200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 18.Tse C, Shoemaker AR, Adickes J, Anderson MG, Chen J, et al. ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res. 2008;68:3421–3428. doi: 10.1158/0008-5472.CAN-07-5836. [DOI] [PubMed] [Google Scholar]
- 19.Willis SN, Fletcher JI, Kaufmann T, van Delft MF, Chen L, et al. Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science. 2007;315:856–859. doi: 10.1126/science.1133289. [DOI] [PubMed] [Google Scholar]
- 20.Annis MG, Soucie EL, Dlugosz PJ, Cruz-Aguado JA, Penn LZ, et al. Bax forms multispanning monomers that oligomerize to permeabilize membranes during apoptosis. Embo J. 2005;24:2096–2103. doi: 10.1038/sj.emboj.7600675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goping IS, Gross A, Lavoie JN, Nguyen M, Jemmerson R, et al. Regulated targeting of BAX to mitochondria. J Cell Biol. 1998;143:207–215. doi: 10.1083/jcb.143.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gross A, Jockel J, Wei MC, Korsmeyer SJ. Enforced dimerization of BAX results in its translocation, mitochondrial dysfunction and apoptosis. Embo J. 1998;17:3878–3885. doi: 10.1093/emboj/17.14.3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolter KG, Hsu YT, Smith CL, Nechushtan A, Xi XG, et al. Movement of Bax from the cytosol to mitochondria during apoptosis. J Cell Biol. 1997;139:1281–1292. doi: 10.1083/jcb.139.5.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, et al. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science. 2004;303:1010–1014. doi: 10.1126/science.1092734. [DOI] [PubMed] [Google Scholar]
- 25.Nie C, Tian C, Zhao L, Petit PX, Mehrpour M, et al. Cysteine 62 of Bax is critical for its conformational activation and its proapoptotic activity in response to H2O2-induced apoptosis. J Biol Chem. 2008;283:15359–15369. doi: 10.1074/jbc.M800847200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pagliari LJ, Kuwana T, Bonzon C, Newmeyer DD, Tu S, et al. The multidomain proapoptotic molecules Bax and Bak are directly activated by heat. Proc Natl Acad Sci U S A. 2005;102:17975–17980. doi: 10.1073/pnas.0506712102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cartron PF, Gallenne T, Bougras G, Gautier F, Manero F, et al. The first alpha helix of Bax plays a necessary role in its ligand-induced activation by the BH3-only proteins Bid and PUMA. Mol Cell. 2004;16:807–818. doi: 10.1016/j.molcel.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 28.Harada H, Quearry B, Ruiz-Vela A, Korsmeyer SJ. Survival factor-induced extracellular signal-regulated kinase phosphorylates BIM, inhibiting its association with BAX and proapoptotic activity. Proc Natl Acad Sci U S A. 2004;101:15313–15317. doi: 10.1073/pnas.0406837101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuwana T, Bouchier-Hayes L, Chipuk JE, Bonzon C, Sullivan BA, et al. BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol Cell. 2005;17:525–535. doi: 10.1016/j.molcel.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 30.Kuwana T, Mackey MR, Perkins G, Ellisman MH, Latterich M, et al. Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell. 2002;111:331–342. doi: 10.1016/s0092-8674(02)01036-x. [DOI] [PubMed] [Google Scholar]
- 31.Letai A, Bassik MC, Walensky LD, Sorcinelli MD, Weiler S, et al. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell. 2002;2:183–192. doi: 10.1016/s1535-6108(02)00127-7. [DOI] [PubMed] [Google Scholar]
- 32.Marani M, Tenev T, Hancock D, Downward J, Lemoine NR. Identification of novel isoforms of the BH3 domain protein Bim which directly activate Bax to trigger apoptosis. Mol Cell Biol. 2002;22:3577–3589. doi: 10.1128/MCB.22.11.3577-3589.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walensky LD, Pitter K, Morash J, Oh KJ, Barbuto S, et al. A stapled BID BH3 helix directly binds and activates BAX. Mol Cell. 2006;24:199–210. doi: 10.1016/j.molcel.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 34.Wang K, Yin XM, Chao DT, Milliman CL, Korsmeyer SJ. BID: a novel BH3 domain-only death agonist. Genes Dev. 1996;10:2859–2869. doi: 10.1101/gad.10.22.2859. [DOI] [PubMed] [Google Scholar]
- 35.Suzuki M, Youle RJ, Tjandra N. Structure of Bax: coregulation of dimer formation and intracellular localization. Cell. 2000;103:645–654. doi: 10.1016/s0092-8674(00)00167-7. [DOI] [PubMed] [Google Scholar]
- 36.Battiste JL, Wagner G. Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear overhauser effect data. Biochemistry. 2000;39:5355–5365. doi: 10.1021/bi000060h. [DOI] [PubMed] [Google Scholar]
- 37.Hsu YT, Youle RJ. Nonionic detergents induce dimerization among members of the Bcl-2 family. J Biol Chem. 1997;272:13829–13834. doi: 10.1074/jbc.272.21.13829. [DOI] [PubMed] [Google Scholar]
- 38.Villunger A, Michalak EM, Coultas L, Mullauer F, Bock G, et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science. 2003;302:1036–1038. doi: 10.1126/science.1090072. [DOI] [PubMed] [Google Scholar]
- 39.Chen D, Zhou Q. Caspase cleavage of BimEL triggers a positive feedback amplification of apoptotic signaling. Proc Natl Acad Sci U S A. 2004;101:1235–1240. doi: 10.1073/pnas.0308050100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsu YT, Wolter KG, Youle RJ. Cytosol-to-membrane redistribution of Bax and Bcl-X(L) during apoptosis. Proc Natl Acad Sci U S A. 1997;94:3668–3672. doi: 10.1073/pnas.94.8.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muchmore SW, Sattler M, Liang H, Meadows RP, Harlan JE, et al. X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death. Nature. 1996;381:335–341. doi: 10.1038/381335a0. [DOI] [PubMed] [Google Scholar]
- 42.Cartron PF, Moreau C, Oliver L, Mayat E, Meflah K, et al. Involvement of the N-terminus of Bax in its intracellular localization and function. FEBS Lett. 2002;512:95–100. doi: 10.1016/s0014-5793(02)02227-5. [DOI] [PubMed] [Google Scholar]
- 43.Nechushtan A, Smith CL, Hsu YT, Youle RJ. Conformation of the Bax C-terminus regulates subcellular location and cell death. Embo J. 1999;18:2330–2341. doi: 10.1093/emboj/18.9.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schinzel A, Kaufmann T, Schuler M, Martinalbo J, Grubb D, et al. Conformational control of Bax localization and apoptotic activity by Pro168. J Cell Biol. 2004;164:1021–1032. doi: 10.1083/jcb.200309013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dlugosz PJ, Billen LP, Annis MG, Zhu W, Zhang Z, et al. Bcl-2 changes conformation to inhibit Bax oligomerization. Embo J. 2006;25:2287–2296. doi: 10.1038/sj.emboj.7601126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bird GH, Bernal F, Pitter K, Walensky LD. Chapter 22 Synthesis and Biophysical Characterization of Stabilized alpha-Helices of BCL-2 Domains. Methods Enzymol. 2008;446:369–386. doi: 10.1016/S0076-6879(08)01622-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walensky LD, Kung AL, Escher I, Malia TJ, Barbuto S, et al. Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science. 2004;305:1466–1470. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pitter K, Bernal F, Labelle J, Walensky LD. Chapter 23 Dissection of the BCL-2 Family Signaling Network with Stabilized alpha-Helices of BCL-2 Domains. Methods Enzymol. 2008;446:387–408. doi: 10.1016/S0076-6879(08)01623-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grzesiek S, BAX A. The importance of not saturating water in protein NMR. Application to sensitivity enhancement and NOE measurements. J Am Chem Soc. 1993;115:12593–12594. [Google Scholar]
- 50.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos: 2002. http://www.pymol.org. ed [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.