Abstract
Orderly progression through the cell cycle requires the transcriptional activation of histone genes to support packaging of newly replicated DNA. Induction of human histone gene expression is mediated by a co-activation complex containing transcription factor HiNF-P and its co-factor p220NPAT. Here, using cells synchronized in S-phase and in mitosis, as well as serum stimulated cells, we have investigated how HiNF-P is regulated during the cell cycle and examined its stability relative to p220NPAT. We find that while HiNF-P is maintained at steady state levels throughout the cell cycle, both HiNF-P and p220NPAT are actively degraded by the proteasome pathway. Importantly, elevation of HiNF-P levels enhances the stability of its co-activator p220NPAT. The HiNFP-dependent stabilization of p220NPAT may reinforce signaling through the cyclin E/CDK2/ p220NPAT pathway and contribute to coordinate control of histone gene expression.
Keywords: HiNF-P, histone, cell cycle, proteasome, ubiquitin, MG132
The exquisite coupling of histone protein synthesis and DNA replication ensures that newly replicated genomic DNA is properly assembled into chromatin. Coordination of the expression of all histone genes located at different loci is necessary to accommodate the enormous quantity of proteins required to package nascent DNA. Analysis of the promoter sequences of all 15 histone H4 genes has revealed some variability in the promoter organization, consistent with our findings that expression levels of individual H4 genes differ significantly (1-3). Similar observations have been made by Doenecke and others for H3 and H2B histone genes (4-6). Differential expression of individual histone genes suggests flexibility in the regulatory mechanisms by which cells produce histone proteins.
Despite copy-specific differences in H4 gene expression, we have identified a highly conserved sequence (site II) that is present in the majority of mammalian histone H4 genes, including those that are most highly expressed (1;7-9). Site II is responsible for the coordinate cell cycle control of histone H4 gene transcription and interacts with the cell cycle-regulatory transcription factor HiNF-P, as well as IRF-2 and CDP/cut (10-15). Studies from our laboratory have shown that induction of HiNF-P deficiency delays S phase progression and decreases total histone H4 mRNA levels (9).
HiNF-P physically interacts with p220NPAT (9), a nuclear protein that is a substrate of the cyclin E/CDK2 kinase complex (16;17), to co-activate H4 histone genes during S-phase. It has been shown that p220NPAT also activates histone H2B and H3 gene transcription. This activation requires the promoter elements previously reported to mediate cell cycle-dependent transcription of histone genes (i.e., HiNF-P binding site for H4; Oct-1 for H2B) (7-9;16). In normal diploid human fibroblasts during G1, p220NPAT is co-localized at Cajal bodies, a nuclear organelle that is associated with histone gene clusters located at 6p21 (17-19). During the G1/S phase transition, p220NPAT expression increases and the protein accumulates in a new set of foci localized at the 1q21 histone gene cluster, while association with 6p21 is maintained (9;16-20). The increase in p220NPAT levels corresponds with a duplication of the number of p220NPAT foci from two to four. At the beginning of S-phase p220NPAT is phosphorylated by cyclin E/CDK2, and this modification is required for histone gene co-activation (10;17). HiNF-P recruits p220NPAT and increases association of RNA polymerase II at the histone H4/n locus (FO108; HIST2H4 localized at Chr1q21-22, Accession No. M16707), to enhance H4 gene transcription at the G1/S transition (9).
Because HiNF-P is a critical regulatory module that links histone H4 gene expression through p220NPAT with the cyclin E/CDK2 signaling pathway, we examined the temporal regulation of HiNF-P during the cell cycle. We find that HiNF-P protein levels are maintained at a steady state throughout the cell cycle by active proteasome/ubiquitin-mediated degradation. Remarkably, elevation of HiNF-P protein levels increases the half life of its partner p220NPAT. Our data suggest that the dynamic interaction between HiNF-P and p220NPAT may promote histone gene co-activation and the physiological functions of these proteins in cell cycle regulation.
MATERIALS AND METHODS
Cell culture and transfections
Human HeLa S3 cervical adenocarcinoma cells, and human T98G glioblastoma cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco/Invitrogen) supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, and 100 U/ml penicillin G, 100 μg/ml streptomycin. Human SaOS-2 osteosarcoma cells were cultured in McCoy's 5A medium (Gibco/Invitrogen) supplemented with 15% FBS, 2 mM L-glutamine, and 100 U/ml penicillin G, 100 μg/ml streptomycin. For transfections, HeLa cells were seeded at a density of 0.1×106 cells per well in 6-well plates or 1×106 cells per 100 mm plates and transfected with 0.5 μg or 4 μg of pFLAG-HiNF-P respectively, using FuGENE6 (Roche), accordingly to the manufacture's instructions.
Cell cycle and cell proliferation analyses
HeLa cells were synchronized by the double-thymidine-block method as previously described (21). SaOS-2 cells were synchronized by nocodazole block and release. Briefly, cells were incubated 24 hours in medium containing 50 ng of nocodazole (Sigma) per ml. Cells were gently washed twice with PBS and fresh media was added. T98G cells were synchronized by serum starvation for 72 hr, followed by stimulation from quiescence with 20% FBS. Cell cycle distribution was monitored by propidium iodide-stained cells subjected to fluorescence-activated cell sorting (FACS, UMass Medical Core Facility) at different time-points after release.
Protein extracts preparation
Total protein extracts from mammalian cell lines were made in lysis buffer [20 mM Tris HCl pH 7.6, 0.15 M NaCl, 1% NP-40, 1 mM EDTA, 1 mM EGTA, 2.5 mM sodium pyrophosphate, 1 mM ß-glycerol phosphate,0.1 mM ZnCl2, 1X Complete (Roche) , 25 μM MG132, 1X phosphatase inhibitor cocktails I and II (Calbiochem)]. Cells were incubated 5 min on ice and sonicated with a 550 Sonic Dismembrator (Fisher Scientific) set at 10% for 5 sec four times. After centrifugation at 16,100g for 15 min the supernatant was collected and protein concentration was determined by Bradford assay (Bio-Rad).
Western and northern blot analyses
Between 25 to 40 μg of total protein were loaded onto 10% or 4-15% linear gradient sodium dodecyl sulfate (SDS)-polyacrylamide gels and transferred to a polyvinylidene fluoride (PVDF) Immobilon-P membrane (Millipore) for 1 hr 40 min (2 hr 30 min for p220NPAT) at 200 mA at 4°C. Immunodetection was performed using an appropriated dilution of specific antibodies and by using the western Lightning Chemiluminescence Reagent Plus (Perkin Elmer life sciences). Total RNA from synchronized HeLa or T98G cells were prepared using TRIzol reagent following the manufacture's instructions (GIBCO BRL). Total RNA (20 μg) was fractionated in a formaldehyde-containing 1.2% agarose gel, transferred to a Zeta-Probe GT membrane (Bio-Rad), and hybridized with DNA probes labeled by random primer method (Redi Prime II Random Primer Labeling System, Amersham). Band intensities were quantified with a PhosphoImager using ImageQuant 5.0 software (Storm 860, Molecular Dynamics) followed by exposure to X-ray film (Kodak), and expressed as fold-increase/decrease of the specific mRNA to 18S RNA.
Ubiquitination
Exponentially growing HeLa cells were transfected with pFLAG-HiNF-P in 100 mm plates and treated with 25 μM MG132 or vehicle DMSO (control) for 12 hr. Cells were lysed as described above and subjected to immunoprecipitation with rabbit polyclonal anti-FLAG antibody (Sigma) and analyzed by western blot using a mouse monoclonal anti-ubiquitin (clone P4D1) antibody (Santa Cruz). The membrane was treated with Re-blot strong (Chemicon) and re-probed with a mouse monoclonal anti-FLAG antibody (Sigma).
Protein stability
To inhibit protein synthesis, HeLa cells were cultured in the presence of 50 μg/ml cycloheximide (Calbiochem) for the specified times. Cells were directly lysed in SDS-PAGE gel loading buffer and western blots analyses were performed as described above. Band intensities were quantified with an AlphaImager 2200 (Alpha Innotech Corp), and expressed as percentage remaining. Inhibition of the proteasome and other proteases was carried out by culturing HeLa cells for up to 12 hr in presence of 25 μM MG132 (Calbiochem), 10 μM ALLM (Calbiochem), or 10 μM Lactacystin (Calbiochem).
RESULTS AND DISCUSSION
Steady state levels of HiNF-P mRNA and protein during the cell cycle
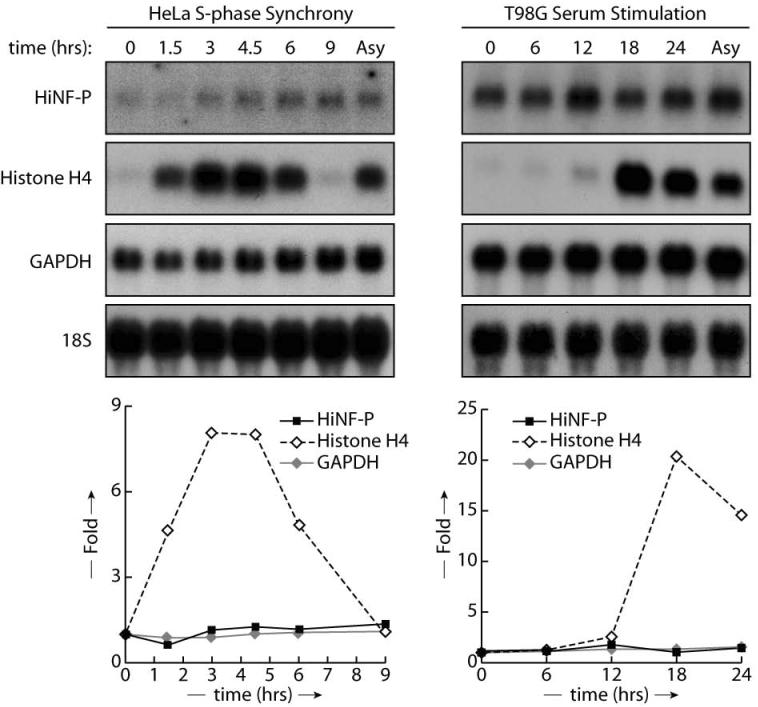
To understand how principal activators of histone gene expression are regulated, we examined the mRNA and protein levels of HiNF-P, a key regulator of histone H4 gene transcription. HiNF-P mRNA levels were quantified during S-phase progression of synchronized HeLa cells and during G1/S phase transit of serum stimulated T98G cells using northern blot analysis (Fig. 1). As expected, histone H4 mRNA levels are transiently up-regulated between 1.5 and 6 hours during S-phase in HeLa cells, and activated at the G1/S transition between 12 and 18 hours in serum-stimulated T98G cells. In contrast, HiNF-P mRNA levels remain relatively constant, with a modest modulation (less than 2-fold) that is comparable to GAPDH mRNA and 18S ribosomal RNA, as cells progress toward mitotic division in HeLa and T98G cells. Thus, expression of HiNF-P mRNA during the cell cycle is temporally uncoupled from histone H4 gene activation.
Figure 1. HiNF-P mRNA levels during the cell cycle and cell proliferation.
HiNF-P mRNA levels were analyzed in either HeLa cells synchronized by the double thymidine block method (left panel) or in T98G released from quiescence by serum stimulation (right panel). Total RNA was isolated from samples taken at the indicated time points after release and analyzed by northern blot using specific probes shown on the left. Lower panels' shows band quantification by PhosphoImager and are expressed as fold change compared to 18S RNA (Asy: asynchronous).
Histone H4 gene transcription is transiently activated by 3-fold in a HiNF-P-dependent manner at the G1/S phase transition (10). Therefore, we addressed whether HiNF-P protein levels are rate-limiting for cell cycle-dependent activation of histone H4 genes. Levels of cyclins E, A, and B1 exhibit characteristic cell cycle stage-specific fluctuations in HeLa cells synchronized in early S phase by double thymidine block (Fig. 2A), SaOS-2 cells released from nocodazole-induced mitotic arrest (Fig. 2B), and serum-stimulated T98G cells (Fig. 2C). The temporal appearance and disappearance of cyclins is consistent with the FACS analysis (lower panels in Fig. 2), and both data sets together establish orderly progression through the cell cycle as expected for synchronized cells.
Figure 2. HiNF-P protein levels during the cell cycle and cell proliferation.
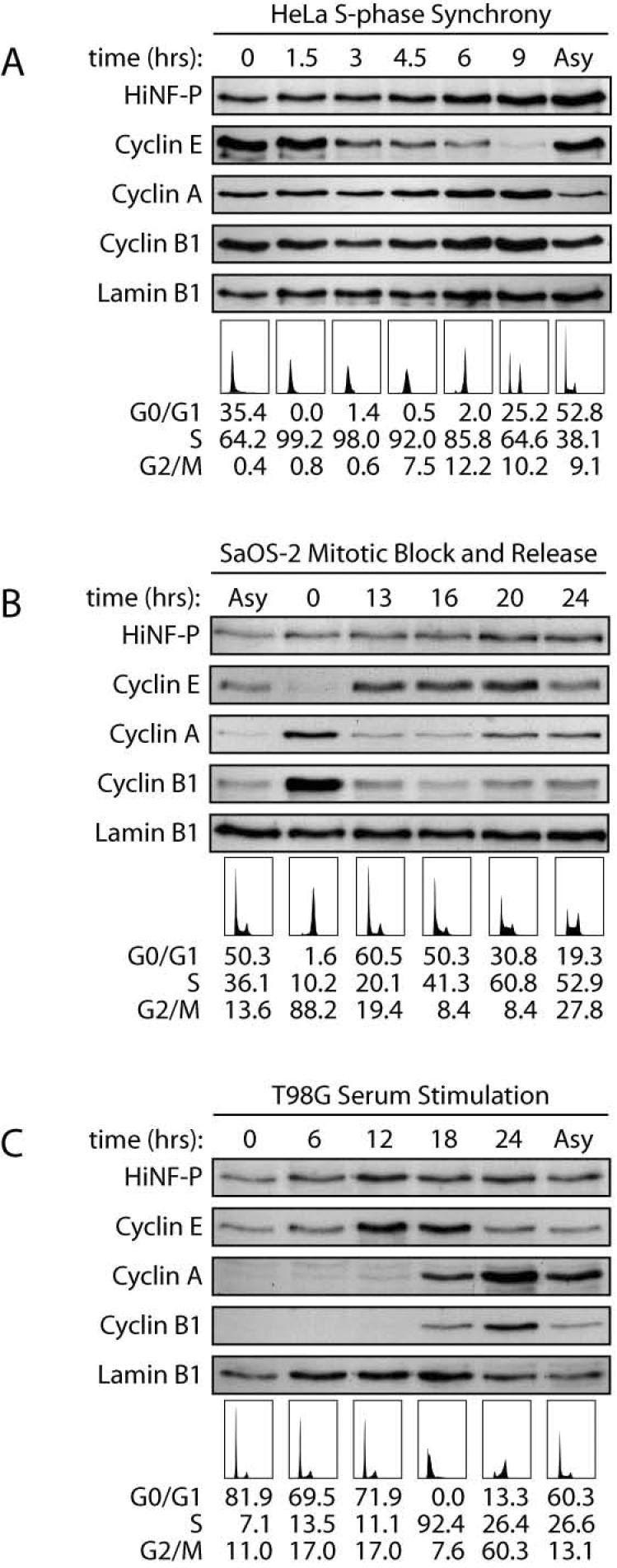
HiNF-P protein levels were analyzed for cell cycle distribution in HeLa cells synchronized by the double thymidine block (A), and SaOS cells blocked by nocodazole treatment (B), and for cell proliferation in T98G released from quiescence by serum stimulation (C). Whole cell extract was prepared from samples taken at the indicated time points after release from blockage and analyzed by western blot using specific antibodies shown on the left. To confirm synchrony in each cell line, cell cycle distribution was analyzed by FACS sorting and profiles along with the percentage in each cell cycle stage are shown underneath.
Importantly, in all three cell types and during both cell cycle progression (Figs. 2A and 2B) and cell growth stimulation (Fig. 2C), HiNF-P protein levels exhibit only minor changes relative to Lamin B1. Combined with results presented in Figure 1, our findings indicate that HiNF-P protein and mRNA levels remain essentially invariable during the cell cycle. The constitutive levels of HiNF-P protein are in agreement with our previous observation that HiNF-P associates with histone H4 gene loci throughout the cell cycle (9).
Destabilization of HiNF-P by the Ubiquitin-proteasome pathway
Many cell cycle regulators are rapidly turned over at specific cell cycle stages to achieve the directionality of cell cycle progression. The constitutive steady state levels of HiNF-P may be achieved by compensatory changes in de novo synthesis of HiNF-P protein and its degradation. We therefore investigated whether HiNF-P is targeted by proteasomal mechanisms that are known to support degradation of cell cycle regulators. HeLa cells were treated for 8 or 12 hours with the protease inhibitor MG132. As expected for proteins known to be degraded by MG132 sensitive mechanisms, the labile proteins cyclin D1 and the p53 tumor suppressor (22-24) are both stabilized in the presence of MG132. There was no effect on the licensing factor MCM6 or total protein levels (Fig. 3A). Our results clearly demonstrate that MG132 increases HiNF-P levels. Thus, the steady state level of HiNF-P is maintained by a MG132-sensitive protease pathway.
Figure 3. Degradation of HiNF-P in HeLa cells.
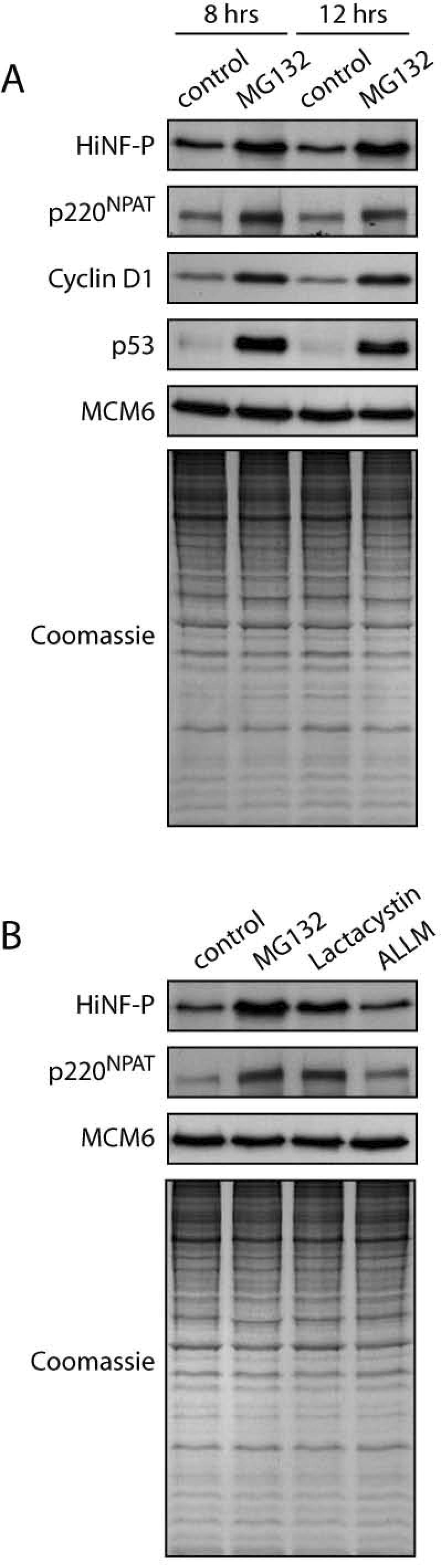
Cells were incubated in presence of DMSO (control) or the protease inhibitor MG132 for 8 or 12 hr (A). Whole cell extracts were analyzed by western blot using specific antibodies shown on the left. MCM6 is shown to confirm equal loading as well as the Coomassie staining underneath. Specificity of the proteasome pathway was addressed by using a combination of protease inhibitors (B). HeLa cells were incubated in presence of DMSO (control), MG132 (proteasome and calpain inhibitor), Lactacystin (proteasome inhibitor) or ALLM (calpain inhibitor) for 12 hr. Whole cell extracts were analyzed by western blot using specific antibodies shown on the left. MCM6 is shown to confirm equal loading as well as the Coomassie staining underneath.
Cell treated with MG132 also exhibit elevated levels of the co-activator p220NPAT, suggesting that this protein may be regulated by a similar mechanism. To demonstrate that the proteasome pathway mediates the turnover of HiNF-P and p220NPAT, we treated cells with the proteasome-specific compound Lactacystin and the calpain protease inhibitor ALLM (25). Both MG132 and Lactacystin, but not ALLM, stabilized HiNF-P and p220NPAT (Fig. 3B). Thus, HiNF-P and p220NPAT are both specifically targeted for degradation via the proteasome.
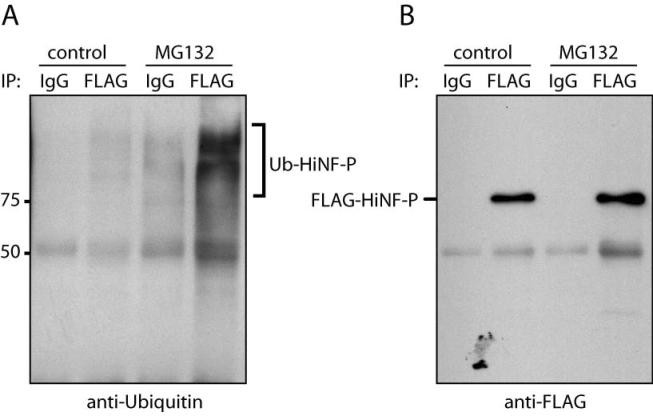
To provide additional evidence of proteasome involvement in HiNF-P degradation, we tested if HiNF-P is poly-ubiquitinated in vivo. We carried out specific immunoprecipitation of exogenous HiNF-P from DMSO (control) and MG132-treated HeLa cells using FLAG antibodies, and assessed the presence of ubiquitinated-HiNF-P by western blot analysis with antibodies against ubiquitin. We detected high molecular weight forms of HiNF-P protein that were immunoreactive with ubiquitin specific antibodies in the control samples and the levels were dramatically enhanced upon MG132 treatment (Fig. 4). Our results demonstrate that HiNF-P is poly-ubiquitinated in vivo. Taken together, our results establish that degradation of HiNF-P is mediated by the ubiquitin-proteasome pathway.
Figure 4. Ubiquitination of HiNF-P in vivo.
Exponentially growing HeLa cells were transfected with pFLAG-HiNF-P. Whole cell extracts were prepared from cells treated with DMSO (control) or MG132 for 12 hr. HiNF-P was immunoprecipitated using a polyclonal anti-FLAG antibody and analyzed by western blot with a monoclonal anti-ubiquitin (clone P4D1) antibody (A). As a control for immunoprecipitations normal rabbit IgG was used. To confirm the presence of HiNF-P in the immunoprecipitated, the membrane was re-blotted using a monoclonal anti-FLAG antibody (B).
Stabilization of the co-activator p220NPAT by elevation of HiNF-P levels
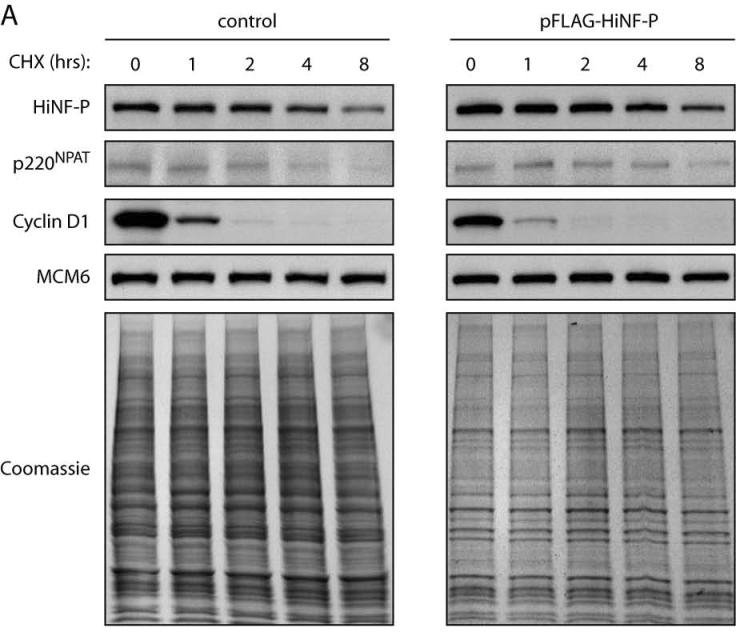
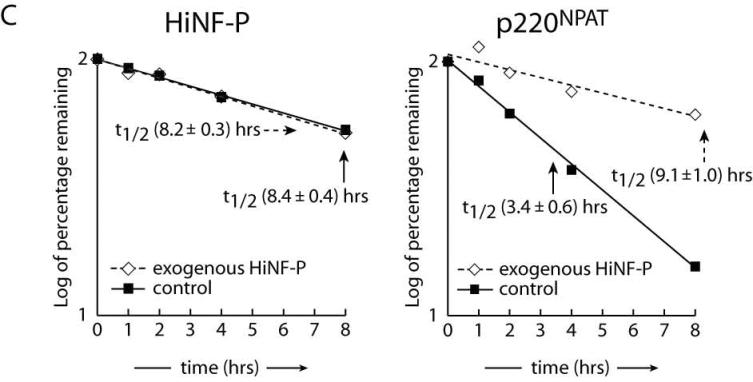
Our findings show that HiNF-P and p220NPAT are both actively degraded by the proteasome pathway during the cell cycle. Because these proteins form a transcriptional co-activation complex, each may influence the stability of the other. To begin addressing this possibility, we examined the half lives of HiNF-P and p220NPAT in asynchronously proliferating HeLa cells. New protein synthesis was prevented by treatment with the translational inhibitor cycloheximide (Fig. 5A). We monitored the decay of pre-existing HiNF-P and p220NPAT as a function of time after cycloheximide treatment. FACS analysis showed that this brief cycloheximide treatment did not alter the cell cycle distribution (data not shown). As expected, the levels of the highly labile Cyclin D1 protein decrease dramatically in the presence of cycloheximide (t1/2 ∼30 min), but no appreciable effect is evident for MCM6 or total protein within this time frame (up to 8 hr). Endogenous HiNF-P levels decay slowly with a t1/2 of 8.4 hours, but p220NPAT levels are much less stable (Fig. 5A, left panel and Fig. 5C control samples). These findings establish that the turnover of the two components of the HiNF-P/p220NPAT co-activation complex occurs with different kinetics. The instability of p220NPAT and the relative stability of HiNF-P are consistent with constitutive association of HiNF-P with histone H4 gene loci, while the interaction of p220NPAT is cell cycle-dependent (9;16).
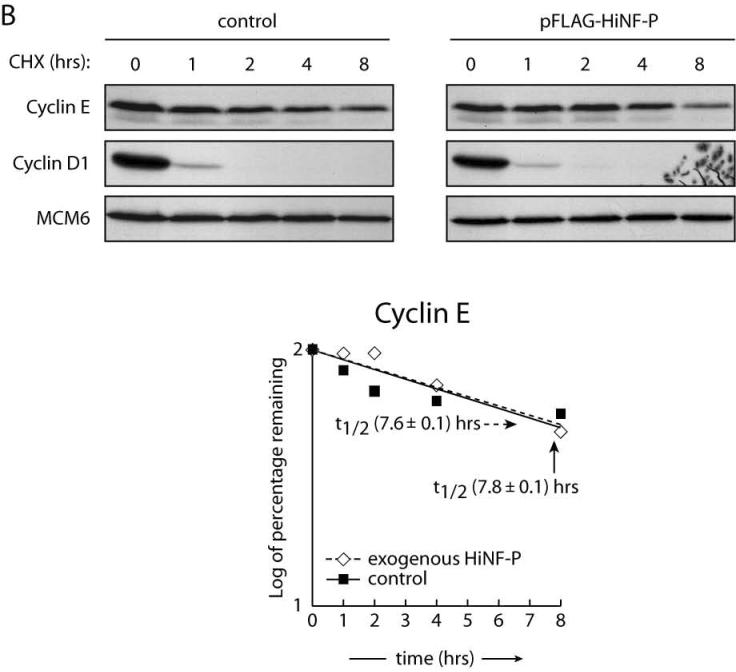
Figure 5. Stabilization of p220NPAT by HiNF-P.
Endogenous (control) or exogenously expressed HiNF-P turnover was analyzed in HeLa cells after treatment with 50 μg/ml cycloheximide (CHX) for the indicated times (A). Cells were harvested and whole cells extracts prepared for western blotting with the indicated antibodies on the left. MCM6 is shown to confirm equal loading as well as the Coomassie staining underneath. Cyclin E turnover was analyzed under conditions described in panel A. The lower panel shows quantification of cyclin E in control cells or cells expressing FLAG-HiNF-P. Results are plotted as the log of the percentage of protein remaining at each time point (B). Panel C shows quantification of the western blots shown in panel A and values are expressed as “log of percentage remaining” as described above. HiNF-P and FLAG-HiNF-P (left) or p220NPAT (right) were analyzed in untransfected cells (control) or cells transfected with pFLAG-HiNF-P (exogenous).
We directly tested whether HiNF-P influences the stability of p220NPAT and vice versa by exogenously expressing either HiNF-P or p220NPAT (Fig. 5 and data not shown). Forced expression of HiNF-P does not affect its intrinsic half life (t1/2 of exogenous HiNF-P = 8.2 hr) (Fig 5A, right panel and Fig. 5C left panel), but significantly stabilizes p220NPAT as reflected by an increased half life, from 3.4 to 9.1 hours (Fig. 5A and 5C right panels). As a control for indirect effects on protein stability, we measured cyclin E protein levels by western blot (Fig. 5B). The results indicate that forced expression of HiNF-P has no effect on the half life of cyclin E (Fig. 5B). As expected, exogenous expression of p220NPAT does not appreciably alter the half life of HiNF-P (t1/2 = 8.6 hr; data not shown), the more stable component of the complex. We propose that elevation of HiNF-P levels increases formation of HiNF-P/p220NPAT co-activation complexes resulting in enhanced p220NPAT stability.
CONCLUSIONS
We have shown here that cellular levels of the human histone H4 transcription factor HiNF-P are constitutive during cell cycle progression through mitosis into S phase, as well as during serum stimulation of quiescent cells. Both HiNF-P and its co-activator p220NPAT are degraded by proteasomal pathways, and p220NPAT is more labile than HiNF-P. Steady state levels of HiNF-P during the cell cycle are consistent with its stable association with H4 genes (9), while the limited half life of p220NPAT supports transient interactions with HiNF-P to modulate histone H4 gene expression at the G1/S phase transition. Because p220NPAT has been shown to be a general co-regulator of histone gene transcription (16;17), our finding that elevation of HiNF-P stabilizes p220NPAT raises the exciting possibility that HiNF-P may coordinate expression of other histone gene subtypes by prolonging the transcriptional activity of p220NPAT.
ACKNOWLEDGMENTS
We thank UMass Medical School FACS facility for Flow cytometry and Judy Rask for editorial assistance. We also thank all members of our research group, especially Hayk Hovhannisyan, Sayyed K. Zaidi, Jitesh Pratap, and Timothy Buck for stimulating discussions.
Grant Acknowledgment: These studies were supported by NIH grants GM32010 and DK32520 DERC.
Abbreviations
- CDK2
cyclin-dependent kinase 2
References
- 1.Holmes WF, Braastad CD, Mitra P, Hampe C, Doenecke D, Albig W, Stein JL, van Wijnen AJ, Stein GS. Coordinate control and selective expression of the full complement of replication-dependent histone H4 genes in normal and cancer cells. J. Biol. Chem. 2005;280:37400–37407. doi: 10.1074/jbc.M506995200. [DOI] [PubMed] [Google Scholar]
- 2.van der Meijden CMJ, Vaughan PS, Staal A, Albig W, Doenecke D, Stein JL, Stein GS, van Wijnen AJ. Selective expression of specific histone H4 genes reflects distinctions in transcription factor interactions with divergent H4 promoter elements. Biochim. Biophys. Acta. 1998;1442:82–100. doi: 10.1016/s0167-4781(98)00147-x. [DOI] [PubMed] [Google Scholar]
- 3.Lichtler AC, Sierra F, Clark S, Wells JR, Stein JL, Stein GS. Multiple H4 histone mRNAs of HeLa cells are encoded in different genes. Nature. 1982;298:195–198. doi: 10.1038/298195a0. [DOI] [PubMed] [Google Scholar]
- 4.Koessler H, Kahle J, Bode C, Doenecke D, Albig W. Human replication-dependent histone H3 genes are activated by a tandemly arranged pair of two CCAAT boxes. Biochem. J. 2004;384:317–326. doi: 10.1042/BJ20040502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trappe R, Doenecke D, Albig W. The expression of human H2A-H2B histone gene pairs is regulated by multiple sequence elements in their joint promoters. Biochim. Biophys. Acta. 1999;1446:341–351. doi: 10.1016/s0167-4781(99)00107-4. [DOI] [PubMed] [Google Scholar]
- 6.Albig W, Trappe R, Kardalinou E, Eick S, Doenecke D. The human H2A and H2B histone gene complement. Biol. Chem. 1999;380:7–18. doi: 10.1515/BC.1999.002. [DOI] [PubMed] [Google Scholar]
- 7.Pauli U, Chrysogelos S, Stein G, Stein J, Nick H. Protein-DNA interactions in vivo upstream of a cell cycle- regulated human H4 histone gene. Science. 1987;236:1308–1311. doi: 10.1126/science.3035717. [DOI] [PubMed] [Google Scholar]
- 8.van Wijnen AJ, van den Ent FM, Lian JB, Stein JL, Stein GS. Overlapping and CpG methylation-sensitive protein-DNA interactions at the histone H4 transcriptional cell cycle domain: distinctions between two human H4 gene promoters. Mol. Cell. Biol. 1992;12:3273–3287. doi: 10.1128/mcb.12.7.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miele A, Braastad CD, Holmes WF, Mitra P, Medina R, Xie R, Zaidi SK, Ye X, Wei Y, Harper JW, van Wijnen AJ, Stein JL, Stein GS. HiNF-P directly links the cyclin E/CDK1/p220NPAT pathway to histone H4 gene regulation at the G1/S phase cell cycle transition. Mol. Cell. Biol. 2005;25:6140–6153. doi: 10.1128/MCB.25.14.6140-6153.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitra P, Xie RL, Medina R, Hovhannisyan H, Zaidi SK, Wei Y, Harper JW, Stein JL, van Wijnen AJ, Stein GS. Identification of HiNF-P, a key activator of cell cycle controlled histone H4 genes at the onset of S phase. Mol. Cell. Biol. 2003;23:8110–8123. doi: 10.1128/MCB.23.22.8110-8123.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vaughan PS, Aziz F, van Wijnen AJ, Wu S, Harada H, Taniguchi T, Soprano KJ, Stein JL, Stein GS. Activation of a cell-cycle-regulated histone gene by the oncogenic transcription factor IRF-2. Nature. 1995;377:362–365. doi: 10.1038/377362a0. [DOI] [PubMed] [Google Scholar]
- 12.Gupta S, Luong MX, Bleuming SA, Miele A, Luong M, Young D, Knudsen ES, van Wijnen AJ, Stein JL, Stein GS. The tumor suppressor pRB functions as a co-repressor of the CCAAT displacement protein (CDP/cut) to regulate cell cycle controlled histone H4 transcription. J. Cell Physiol. 2003;196:541–546. doi: 10.1002/jcp.10335. [DOI] [PubMed] [Google Scholar]
- 13.van Wijnen AJ, van Gurp MF, de Ridder MC, Tufarelli C, Last TJ, Birnbaum M, Vaughan PS, Giordano A, Krek W, Neufeld EJ, Stein JL, Stein GS. CDP/cut is the DNA-binding subunit of histone gene transcription factor HiNF-D: a mechanism for gene regulation at the G1/S phase cell cycle transition point independent of transcription factor E2F. Proc. Natl. Acad. Sci. USA. 1996;93:11516–11521. doi: 10.1073/pnas.93.21.11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramsey-Ewing A, van Wijnen AJ, Stein GS, Stein JL. Delineation of a human histone H4 cell cycle element in vivo: the master switch for H4 gene transcription. Proc. Natl. Acad. Sci. U. S. A. 1994;91:4475–4479. doi: 10.1073/pnas.91.10.4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aziz F, van Wijnen AJ, Stein JL, Stein GS. HiNF-D (CDP-cut/CDC2/cyclin A/pRB-complex) influences the timing of IRF-2 dependent cell cycle activation of human histone H4 gene transcription at the G1/S phase transition. J. Cell. Physiol. 1998;177:453–464. doi: 10.1002/(SICI)1097-4652(199812)177:3<453::AID-JCP8>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 16.Zhao J, Kennedy BK, Lawrence BD, Barbie DA, Matera AG, Fletcher JA, Harlow E. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 2000;14:2283–2297. [PMC free article] [PubMed] [Google Scholar]
- 17.Ma T, Van Tine BA, Wei Y, Garrett MD, Nelson D, Adams PD, Wang J, Qin J, Chow LT, Harper JW. Cell cycle-regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 2000;14:2298–2313. doi: 10.1101/gad.829500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shopland LS, Byron M, Stein JL, Lian JB, Stein GS, Lawrence JB. Replication-dependent histone gene expression is related to Cajal body (CB) association but does not require sustained CB contact. Mol. Biol. Cell. 2001;12:565–576. doi: 10.1091/mbc.12.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frey MR, Matera G. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc. Natl. Acad. Sci. USA. 1995;92:5915–5919. doi: 10.1073/pnas.92.13.5915. erratum in: Proc Natl Acad Sci U S A 1995 Aug 29;92(18):8532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao G, Bracken AP, Burkard K, Pasini D, Classon M, Attwooll C, Sagara M, Imai T, Helin K, Zhao J. NPAT expression is regulated by E2F and is essential for cell cycle progression. Mol. Cell Biol. 2003;23:2821–2833. doi: 10.1128/MCB.23.8.2821-2833.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stein GS, Stein JL, Lian JB, Last TJ, Owen T, McCabe L. Synchronization of normal diploid and transformed mammalian cells. In: Celis JE, editor. Cell Biology: A laboratory Handbook. Academic Press, Inc.; San Diego, CA: 1994. pp. 282–287. [Google Scholar]
- 22.Diehl JA, Zindy F, Sherr CJ. Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev. 1997;11:957–972. doi: 10.1101/gad.11.8.957. [DOI] [PubMed] [Google Scholar]
- 23.Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 24.Maki CG, Huibregtse JM, Howley PM. In vivo ubiquitination and proteasome-mediated degradation of p53(1) Cancer Res. 1996;56:2649–2654. [PubMed] [Google Scholar]
- 25.Lee DH, Goldberg AL. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 1998;8:397–403. doi: 10.1016/s0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]