Abstract
Molecular genetic analysis of the fruit fly Drosophila melanogaster has revolutionized our understanding of the transcription/translation loop mechanisms underlying the circadian molecular oscillator. More recently, Drosophila has been used to understand how different neuronal groups within the circadian pacemaker circuit interact to regulate the overall behavior of the fly in response to daily cyclic environmental cues as well as seasonal changes. Our present understanding of circadian timekeeping at the molecular and circuit level is discussed with a critical evaluation of the strengths and weaknesses of present models. Two models for circadian neural circuits are compared: one that posits that two anatomically distinct oscillators control the synchronization to the two major daily morning and evening transitions, versus a distributed network model that posits that many cell-autonomous oscillators are coordinated in a complex fashion and respond via plastic mechanisms to changes in environmental cues.
Keywords: oscillator, entrainment, photoperiod, morning-evening oscillator model
INTRODUCTION
All living organisms are faced with challenges associated with daily environmental changes. Furthermore, most organisms encounter varying degrees of seasonal changes. The question how organisms adjust their behavioral and physiological programs to such recurring environmental cycles has received considerable attention over the past four decades. It has now been established beyond doubt that biological systems use endogenous time-keeping systems called circadian clocks (from ‘circa’ = approximate, ‘dies’ = a day) that modulate a wide range of behavioral and metabolic processes. While we understand a great deal about the various components of the underlying molecular-genetic and neuronal machinery, it is still unclear how these components interact to regulate a wide range of precisely timed molecular and behavioral processes that can be synchronized to daily and annual environmental cycles and persist in the complete absence of environmental time-cues or zeitgebers (time-givers, from ‘zeit’ = time, ‘geber’ = giver).
In the fruit fly Drosophila melanogaster, circadian clocks regulate the timing of a wide range of behavioral and metabolic processes including adult emergence, activity/rest, egg-laying, olfaction, mating, larval photo-responses, axon-caliber of lamina neurons, bouton size of motor neuron terminals, and expression of numerous genes (Konopka and Benzer, 1971; Krishnan et al., 1999; Sakai and Ishida, 2001; Mazzoni et al., 2005; Howlader and Sharma, 2006; Taghert and Shafer, 2006; Mehnert et al., 2007). It is conceivable that separate oscillators control different metabolic and behavioral phenomena and that these oscillators are coupled together such that they influence each other to make up a multi-oscillatory system. Although some of the seminal studies that laid down the canonical features of circadian pacemakers were based on behavioral studies using the fruit fly Drosophila pseudoobscura (Pittendrigh, 1954, 1960), the power of its close relative Drosophila melanogaster as a genetic tool transformed the field of circadian rhythms beginning with the identification of the various underlying molecular components of the circadian machinery, followed by the localization of cells that are the sites of pacemakers of the circadian oscillators. Drosophila melanogaster (henceforth referred to as Drosophila) continues to this day to be the model system of choice to investigate the details of molecular architecture, neural circuitry and interactions with other metabolic and homeostatic processes. Here we present a critical overview of recent developments and the current understanding in the field of Drosophila circadian rhythms and discuss some unanswered questions and theoretical inconsistencies of present empirically based models. Specifically, evidence will be discussed for two current models of how oscillators are coordinated in the Drosophila circadian circuit—a “morning and evening” oscillator model for which it has been posited that individual oscillators are anatomically restricted to two functional sets and “dance” a Pas de Deux, versus a distributed network model for which many cell-autonomous oscillators are coordinated in a more complex, but ultimately flexible fashion, akin to the southern Italian circle-dance, the Tarantella.
MOLECULAR BASIS FOR DROSOPHILA CIRCADIAN OSCILLATORS
A general consensus has emerged about the existence of feedback mechanisms (involving transcription and translation) with a remarkably similar architecture across multiple phylogenetic classes although its position as an essential feature of the circadian machinery has recently come into question (Nakajima et al., 2005; Tomita et al., 2005; Hardin, 2006; Fan et al., 2007; Mori et al., 2007; Rust et al., 2007). In its simplest form, it is believed that the clock consists of at least one transcriptional-translational feedback loop (TTFL), while in more complex organisms such as Drosophila, there are predictions for at least two interlocked-TTFLs (Glossop et al., 1999; Cyran et al., 2003; Yu and Hardin, 2006). In Drosophila, the gene products of period (per) and timeless (tim) form the core of the Drosophila TTFL in pacemaker cells (Figure 1A). Levels of PERIOD (PER) and TIMELESS (TIM) proteins and their mRNAs exhibit cyclic expression in pacemaker cells, and PER and TIM translocate from cytoplasm to nucleus in a time-of-day-dependent manner (Siwicki et al., 1988; Hardin et al., 1990; Sehgal et al., 1994). Starting at noon, the transcription of per and tim is activated by two proteins CLOCK and CYCLE (CLK and CYC) which heterodimerize and bind to E-box sequences in the per and tim promoters. Thus, CLK and CYC form the positive limbs of one of the interlocked-TTFLs (Figure 1A). The rise in per and tim mRNA levels (peaking at dusk) is followed by an increase in PER/TIM protein heteromultimer in the cytoplasm. There is a delay of 6 hours between the rise in PER/TIM multimer formation and their mRNAs production (Hardin et al., 1990; Hunter-Ensor et al., 1996) partly mediated by DOUBLE-TIME (DBT kinase, a casein kinase Iεhomolog; Kloss et al., 1998), which destabilizes PER by phosphorylating and thus facilitating its subsequent degradation. Previous versions of the TTFL models suggested that after PER and TIM have each been phosphorylated by Casein kinase 2 (CK2) and SHAGGY (SGG, a Glycogen Synthase Kinase 3 homolog) respectively the DBT-PER/TIM heteromultimer enters the nucleus around midnight (Martinek et al., 2001; Lin et al., 2002).
FIGURE 1.
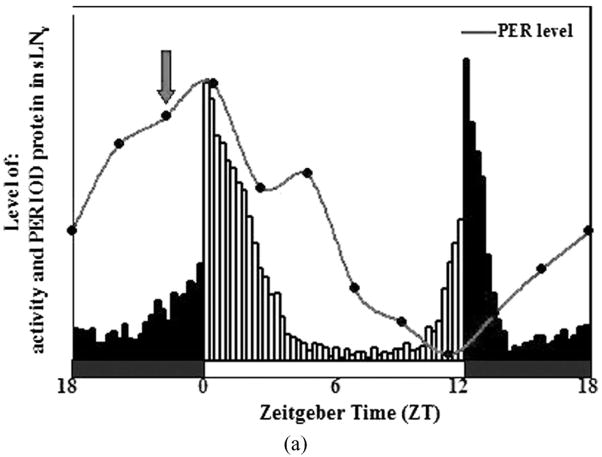
FIGURE 1A The Drosophila molecular circadian clock in pacemaker cells: the essence. The rhythm in activity levels of adult flies shows two peaks, one around dawn and the other around dusk. The pattern of activity rest rhythms is greatly influenced by level and sub cellular localization of PERIOD protein in a group of ventral lateral pacemaker neurons (small LNv and similarly in other pacemaker neurons). Highest levels of PER is seen around dawn (defined as Zeitgeber Time 0, ZT0) and peak nuclear localization occurs shortly before dawn (around ZT 22, Shafer et al., 2002; red arrow). The levels of per mRNA follow a similar pattern with an approximately 6-hour phase advance. Oscillations in mRNA, protein levels and post-translationally modified states of several other genes have been implicated in the generation of rhythmic behavioral and metabolic processes, the intricacies of which are described in Figure1B.
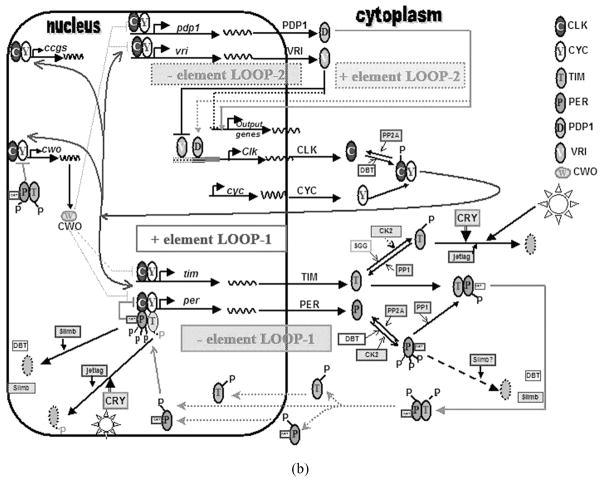
FIGURE 1B The Drosophila molecular circadian clock in pacemaker cells: the gory details. The Drosophila circadian clock consists of interlocked sets of transcription/translation feedback loops (TTFL), which have mRNA and protein components that cycle in abundance and subcellular localization with a near 24-hour period. Dotted or dashed arrows indicate pathways that are not completely resolved or not considered essential for the self-sustained biochemical oscillator function. Lines ending in arrows indicate activation while those ending in a hatched bar indicate repression or inhibition. Degraded proteins are indicated by ovals with dotted walls. Light ordinarily recalibrates the clock at the onset of each day. During the previous night, PER and TIM proteins have accumulated at their highest levels in the nucleus, acting as transcriptional repressors of their own mRNA expression by binding to the transcriptional activators (only per repression is shown for simplicity) CLK-CYC, thus forming the negative limb of one of the TTFLs (loop1). Starting at dawn, light degrades TIM via a CRYPTOCHROME (CRY) mediated pathway, subsequently monomeric PER is phosphorylated and degraded after SLIMB an F-Box protein marks it for proteosomal degradation. By noon, PER and TIM’s degradation releases transcriptional repression and the transcription of period (per) and timeless (tim) genes is activated by CLK-CYC which bind to E-box sequences in per and tim promoters forming the positive limb of loop1 and attaining peak levels of per and tim transcripts at dusk. In the cytoplasm, DOUBLE-TIME (DBT) kinase complexes with and destabilizes PER by phosphorylating and thus facilitating its subsequent degradation. PER and TIM are each also phosphorylated by Casein kinase 2 (CK2) and SHAGGY (SGG), respectively. Since TIM is light sensitive, TIM levels can begin to rise in the cytoplasm only after dusk, after which it complexes with the DBT-PER heteromers. Levels of PER and TIM are maximal by mid-night. The entry of DBT-PER/TIM heteromultimer into the nucleus around midnight is controversial, it is possible that they dissociate such that PER (along with DBT) enters the nucleus at least 3 hours before TIM does. Cytoplasmic PER is stabilized by Protein Phosphatase 2A (PP2A), which dephosphorylates PER while Protein Phosphatase1 (PP1) dephosphorylates and stabilizes TIM thus promoting PER accumulation and hetero-dimerization. Total CLK levels remain constant with circadian oscillation in its phosphorylation state due to the action of the multifunctional DBT and perhaps other kinases and phosphatases including PP2A. CLK heterodimerizes with the constitutively present CYC and this CLK-CYC complex in addition to activation of per and tim transcription also binds to E boxes of at least two other genes vrille (vri) and par domain protein-1ε(pdp-1ε) to activate their transcription. PDP-1 in turn activates transcription of Clk, while VRI represses it by competitively binding to regulatory sequences called VRI/PDP1ε- boxes (V/P-boxes, shown by dotted and hashed line), upstream of Clk, forming a second feedback loop that interlocks with the first via CLK/CYC. Recent studies indicate that the second loop may not be an essential component of the circadian pacemaking machinery as Clk cycling is nonessential for clock function. Instead, the cycling in phosphorylation state of CLK is thought to contribute towards maintaining a robust period. In addition to core-clock genes, CLK regulates mRNA levels of several output genes (and cry via VRI and possibly PDP1ε- not shown). PER-TIM complex is also thought to repress CLK/CYC transcription of vri and pdp-1ε(not shown). PDP1εis now believed to function as an oscillator output component rather than a central oscillator component. The amplitude of the circadian biochemical oscillator may be regulated by Clk-mediated transcriptional activation of various genes including clockwork orange (cwo) which is proposed to contribute to robustness of the amplitude of mRNA oscillations of vri, pdp-1ε, tim, and per. For a depiction of how the amounts and location of these clock molecules vary over time (see Yu and Hardin 2006).
However, two independent studies using very different methods suggest that nuclear entry of PER and TIM could occur independently (Shafer et al., 2002; Meyer et al., 2006). Immunocytochemical assays of PER and TIM levels at high temporal resolution revealed that nuclear entry of PER precedes that of TIM in pacemaker neurons by at least 3 hours (Shafer et al., 2002). In vitro studies using a FRET-based method in S2 cells detected cytoplasmic PER-TIM heteromer formation and disassociation prior to independent nuclear entry, leading the authors to propose that PER-TIM complex formation may act as an interval timer for an event that precedes nuclear entry (Meyer et al., 2006).
If the translocation of PER-TIM-DBT complex is indeed required, it appears to be regulated by phosphorylation which may occur via PER phosphorylation by CK2 (Lin et al., 2002), kinase activity of DBT (Bao et al., 2001; Cyran et al., 2005), concerted action of both DBT and CK2 (Nawathean and Rosbash, 2004; Nawathean et al., 2007) or TIM phosphorylation by SGG (Martinek et al., 2001), while cytoplasmic PER is stabilized by Protein Phosphatase 2A (PP2A) (Sathyanarayanan et al., 2004), which dephosphorylates PER. A recent study (Fang et al., 2007) suggests that another Protein Phosphatase (PP1) dephosphorylates and stabilizes TIM thus promoting PER accumulation and hetero-dimerization. Thus while independent methods confirm the formation of heteromers during the sequence of events that constitute the TTFLs, whether PER-TIM-DBT complex translocation is critical for circadian function remains unresolved. A mathematical model which assumes rapid and cytoplasm-limited PER-TIM heteromers posits that the dissociation of PER-TIM is a means of adjusting the period and phase of molecular oscillations by differential timing of nuclear entry of PER and TIM (Leise and Moin, 2007). This recent model suggests that heteromeric complex translocation is not required for circadian function (Leise and Moin, 2007).
The central feature of the updated TTFL model holds that DBT-PER and TIM complex represses per and tim transcription in the nucleus by binding to CLK and CYC transcription factors and releases CLK/CYC bound to the E-box sequences of the per and tim promoters (for most recent papers on this topic, see Kim and Edery, 2006). Thus, in direct contrast to CLK and CYC, TIM and PER form the negative limb of the TTFLs. TIM is eventually degraded by light through its interaction with the photosensitive CRYP-TOCHROME (CRY) and eventually targeted by proteosomal mechanisms (Naidoo et al., 1999; Busza et al., 2004; Dissel et al., 2004). CRY has also been implicated recently as a potential transcriptional repressor of CLK in peripheral tissues (Collins et al., 2006). Subsequently monomeric PER is phosphorylated and degraded until midnight because SLIMB an F-Box protein marks it for proteosomal degradation (Edery et al., 1994; Zeng et al., 1996; Grima et al., 2002; Ko et al., 2002). In addition to PER, DBT also hyperphosphorylates CLK (Kim and Edery, 2006; Yu et al., 2006). Thus, DBT kinase modulates molecular clock function at multiple stages, components and cellular locations in the circadian cycle.
The loss of PER/TIM mediated repression frees CLK/CYC to begin a new cycle of the molecular clock. A second feedback loop for the Drosophila circadian molecular clock was proposed (Glossop et al., 1999; Cyran et al., 2003) interlocking with the first TTFL. The CLK/CYC complex in addition to transcriptional activation of per and tim also bind to E boxes of two other genes vrille (vri) and par domain protein-1ε(pdp1ε) (Glossop et al., 1999; Cyran et al., 2003). PDP-1 activates transcription of Clk, while VRI represses it by competitively binding to regulatory sequences called VRI/PDP1ε- boxes (V/P-boxes), upstream of Clk thus becoming the positive and negative elements in the second TTFL. VRI levels was shown to peak during the early part of the night, and was believed to repress Clk transcription in the late evening when PDP1εlevels rise to competitively bind V/P-boxes of Clk (shown by dotted and hashed line in Figure 1B) and allow its transcriptional activation (Cyran et al., 2003; Glossop et al., 2003). However, recent studies suggest that the role of VRI as a transcriptional repressor of Clk may not be an essential component of the circadian pacemaking machinery (Kim and Edery, 2006) and that Clk cycling is non-essential for clock function (Kim et al., 2002). More recent work overturns yet another previous assumption of the second feedback loop oscillator model by revealing that PDP1εfunctions as an oscillator output component rather than a central oscillator component (Benito et al., 2007). The authors also propose that an activator of Clk (indicated by a blue bar in Figure 1B) is responsible for the constant high levels of Clk seen in ClkJrk and cyc0 mutants and for rising levels of Clk in wild-type flies during the early day and late night phases when repression by VRI does not occur. In addition, CLK protein levels were revealed to be constant with circadian oscillation in its level of phosphorylation due to the action of the multifunctional DBT and the phosphatase PP2A (Kim and Edery, 2006; Yu et al., 2006). In addition to the core-clock genes, CLK is thought to regulate mRNA levels of several output genes (Cyran et al., 2003; Glossop et al., 2003). This model assumes that post-translational modifications via DBT, CK2, SGG, PP2A, and perhaps yet unknown enzymes are determinants of the period length and the phase of the overt rhythms that the oscillator generates (Bae and Edery, 2006). The amplitude of the rhythm may be regulated by Clk mediated transcriptional activation of various genes (Kim et al., 2002), including the most recent candidate, clockwork orange (cwo) (Kadener et al., 2007; Lim et al., 2007; Matsumoto et al., 2007) which is proposed to contribute to robustness of the amplitude of mRNA oscillations of vri, pdp-1ε, tim and per.
The current TTFL model also does not explain the persistence of overt behavioral rhythms when clk mRNA is expressed using per or tim promoters such that its expression is no longer antiphasic to per and tim (Kim et al., 2002) which according to the model is a critical factor for rhythm generation. Mathematical modeling data also support the hypothesis that robust oscillations can persist in the presence of constant levels of CLK, if total CLK levels remain below that of total PER levels (Leise and Moin, 2007). However, it is possible that the role of the second TTFL is to maintain the stability of the first TTFL (Harms et al., 2004; Lakin-Thomas, 2006). Further, constitutive expression of either tim or per mRNA independently or together does not disrupt oscillations in PER and TIM protein and overt behavioral rhythms, which raises the question as to whether mRNA oscillation is a fundamental feature of the Drosophila circadian oscillator (Frisch et al., 1994; Vosshall and Young, 1995; Cheng and Hardin, 1998; Yang and Sehgal, 2001; Harms et al., 2004)—presenting a long-standing, but still not resolved challenge to the TTFL model. We note that thus far no study has simultaneously blocked mRNA oscillations of all known clock components, and therefore it is likely that there are yet- to be-identified, redundant components of the clock that continue to regulate rhythmicity observed in previous studies.
Can circadian oscillators unequivocally operate without TTFL features? They can. Over the past few years, in vitro studies have shown that a trio of purified cyanobacterial proteins Kai A, B, and C can interact in the absence of transcriptional machinery to function as a self-sustained biochemical oscillator under constant darkness (DD) with a period of ~ 24 hour and can even exhibit a canonical feature of circadian oscillators which is temperature compensation (Nakajima et al., 2005). Studies using mouse fibroblasts have led to the proposal that in fact post-translational modifications are the core oscillator, while transcriptional regulation merely enhances the amplitude and robustness of the oscillation (Kiyohara et al., 2006). Recently, a combination of theoretical and empirical investigations has lead to the proposal that four phosphorylation states (phosphoforms) of the cyanobacterial Kai C protein are generated in an ordered pattern as a result of its intrinsic autokinase and autophosphatase activities and their modulation by Kai A (Rust et al., 2007). Further this study proposes that the negative feedback is achieved by the inhibitory action of one of the Kai C phosphoforms together with Kai B upon Kai A activity. Another study conducted in mouse fibroblasts also challenges the current version of the TTFL model. This was achieved by administering exogenous cell-permeant mCRY1 and mCRY2 proteins modified to include the short hydrophobic sequence AAVLLPVL-LAAP which confers permeability across the plasma membrane, thus the levels of these exogenously administered proteins are not regulated by transcription. Exogenous cell-permeant mCRY1 and mCRY2 proteins can rescue circadian rhythmicity and can act as transcriptional repressors and cause phase shifts in the circadian oscillator (Fan et al., 2007). These results suggest that previous assumption for cycling levels of CRY is non essential for a functional circadian oscillator in mammalian cells. Thus, the TTFL model, while appealing in many ways, has a number of unresolved inconsistencies.
CIRCADIAN CIRCUITS IN DROSOPHILA BRAIN
In the simplest model of organization, all circadian systems can be thought to be composed of input, core-pacemaker, and output components. This organization can be applied to our developing ideas of the Drosophila pacemaker circuit; although it is likely that a particular cell group in the pacemaker circuit has multiple overlapping functions (see below for specific examples).
Core Pacemaker Neurons
In Drosophila, cells that express known ‘clock genes’ occur all over the body. These cells can sustain molecular oscillations under constant conditions and can entrain to environmental zeitgebers such as light/dark and temperature cycles independent of the brain pacemakers (Plautz et al., 1997; Glaser and Stanewsky, 2005). One working definition of a pacemaker is that it should be able to sustain rhythm under long-term DD (Kaneko et al., 2006). In the brain numerous glial cells and approximately 150 neurons express clock genes rhythmically (Kaneko, 1998). Glia have previously been shown to contribute to the generation of activity/rest rhythm (Ewer et al., 1992; Frisch et al., 1994). Recent studies indicate that a biogenic amine synthase coded by the gene ebony is rhythmically expressed in glia with circadian period (Claridge-Chang et al., 2001; Ueda et al., 2002) regulates circadian locomotor activity/rest rhythm by coordinating the action of neurotransmitters such as dopamine and serotonin (Suh and Jackson, 2007). In this review we will focus on the clock pacemaker neurons and their circuits.
Even before the clock genes were discovered and their expression patterns identified, brain pacemakers were considered to be coupled bilaterally between two sides of the brain. This idea was developed from studies in other insects (Helfrich-Forster et al., 1998). Advances in immunocytochemical reagents and reporter constructs allowed for the identification of the pacemaker neurons and their projection patterns and revealed that pacemaker neurons are interconnected both bilaterally and within a brain hemisphere (Helfrich-Forster and Homberg, 1993; Kaneko and Hall, 2000) (Figure 2). More recent studies demonstrate functional coupling among the different pacemaker neuronal subgroups (Peng et al., 2003; Lin et al., 2004; Stoleru et al., 2005; Nitabach et al., 2006).
FIGURE 2.
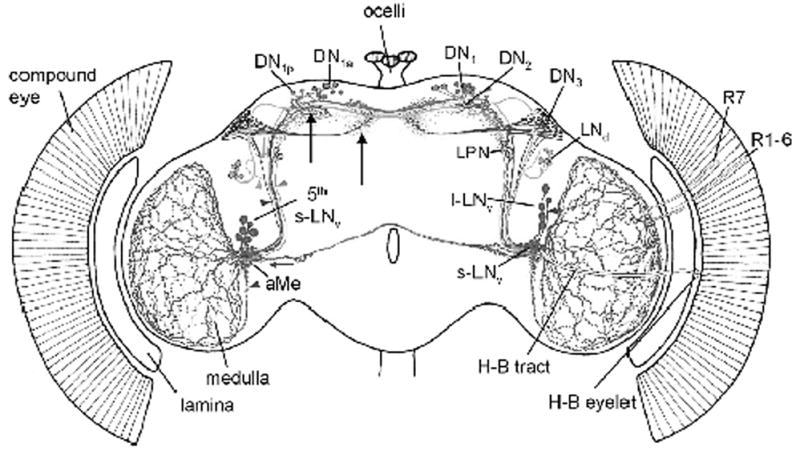
Neuronal network that regulates circadian rhythmicity in the adult Drosophila brain. Locations and putative arborization patterns of Drosophila clock neurons as originally illustrated by Helfrich-Forster et al. (2007). Each neuronal cluster is depicted in distinct color for clarification of neuronal morphology in a frontal view of a brain: Large LNv, brown; PDF-positive small LNv, red; PDF-negative fifth small LNv, dark violet; LNd, orange; DN1a,p, lilac; DN1, blue; DN2, light blue; DN3, navy; LPN cell bodies, green; photoreceptors including H-B eyelet, yellow. Bedsides PDF-positive LNv, fifth small LNv (dark violet arrowhead ), DN1a,p (lilac arrowhead ), LNd (orange arrowhead ), and DN3 (navy arrowhead) invade the AMe ipsilaterally. Large LNv (brown arrow ) and LNd (orange arrow ) send contralateral projections to the AMe. Brown arrowhead points to the ventral elongation of the AMe which receives innervations from large LNv. (Reprinted with permission of Wiley-Liss, Inc., a subsidiary of John Wiley & Sons, Inc.)
Clock pacemaker neurons in adult Drosophila brain can be divided into two groups, lateral neurons (LN) whose cell bodies are located in the anterior lateral cortex of the central brain, and dorsal neurons (DN) with cell bodies in the dorsal cortex. LN are further divided into three groups 4–6 large and 4–6 small ventral LN (LNv), and ~6 dorsal LN (LNd). DN are also divided into at least three subgroups DN1, DN2, and DN3 (Kaneko, 1998). Approximately 15 DN1are located in the pars lateralis, a pair of DN2 are located slightly ventral to DN1 and are located just dorsal to the calyces of the mushroom body, and ~40 small DN3neurons are located more dorsolaterally. In addition to these six clusters, there is another cluster of three per- and tim-expressing neurons in the posterior lateral cortex (LPN) (Kaneko and Hall, 2000; Shafer et al., 2006) (Figure 2).
Projection patterns of most of the pacemaker neurons are shown in Figure 2, and extensively reviewed elsewhere (Helfrich-Forster, 2002, 2003, 2005), the only exception being LPN whose projection pattern is not yet known. The arborization patterns of LNv were first identified by staining for antibody against the crustacean Pigment Dispersing Hormone (PDH) (Helfrich-Forster and Homberg, 1993). For the rest of the pacemaker neurons, per- and tim-GAL4 driven marker gene expression have been used to reveal their morphology (Kaneko and Hall, 2000). Recent attempts from different research groups using various methods such as combination of tim-GAL4 or anti-PDF staining along with anatomical mutations, combinations of GAL4 drivers and GAL80 constructs, new drivers, and cell-filling have revealed detailed and refined patterns of arborizations for DN3 (Veleri et al., 2003), DN1 (Helfrich-Forster, 2003), small and large LNv (Park and Griffith, 2006; Helfrich-Forster et al., 2007), and LNd (Helfrich-Forster et al., 2007).
Most of the clock neurons (small LNv, LNd, DN1, DN2, and DN3) send processes to the dorsal protocerebrum (Helfrich-Forster and Homberg, 1993; Kaneko and Hall, 2000) (Figure 2). The dorsal protocerebrum has connections to many areas of the brain and also contains many neurosecretory cells. Therefore this is a likely location where clock neurons are connected to various behavioral outputs. While processes from these five clusters of neurons extensively overlap in this region, there is no direct evidence of how putative functional coupling might occur—evidence for functional coupling of different neurons in the pacemaker circuit is largely inferential based on projection patterns, and more recently, mutant analysis (Peng et al., 2003; Lin et al., 2004) and genetic perturbation experiments (Nitabach et al., 2006; de la Paz Fernandez et al., 2007). Who directly talks to who in the pacemaker circuit is still an open question.
Where is information in the Drosophila pacemaker circuit integrated? Each large LNv sends contralateral and ipsilateral projections that innervate the medulla and a small neuropil at the inner margin of the medulla called the accessory medulla (AMe) on both sides of the brain (Figure 2) (Park and Griffith, 2006; Helfrich-Forster et al., 2007). This distinct anatomical feature suggests that the large LNv mediate bilateral coupling of the two hemispheres of pacemaker circuit. The large LNv unambiguously sends bilateral projections to the opposite hemisphere of the Drosophila brain (see Park and Griffith, 2006 for high resolution single cell fills). Besides large LNv many other clock neurons (small LNv, LNd, DN1, and DN3) innervate the AMe (Helfrich-Forster, 2003; Veleri et al., 2003; Shafer et al., 2006; Helfrich-Forster et al., 2007). This may be where oscillatory signals as well as light-input signals are integrated to form a coupled pacemaker circuit.
Inputs into Core Pacemaker Neurons
Light is a major Zeitgeber for the circadian clock, and Drosophila has many photoreceptors that participate in the entrainment of circadian rhythms. In addition to light-driven synaptic inputs, CRY a cell autonomous blue-light photoreceptor responsible for circadian entrainment is expressed in many if not all clock cells (Emery et al., 2000; Klarsfeld et al., 2004). Therefore, molecular rhythms in clock cells can be entrained independent of the rest of the circuits through cry, and certainly that is the case for many peripheral oscillators (Stanewsky et al., 1998). CRY interacts with TIM in a light-dependent manner and modifies it, enabling another protein JETLAG (JET) to ubiquitinate TIM thus facilitating its degradation via proteosomal pathways (Koh et al., 2006; Peschel et al., 2006; Van Gelder, 2006). However, locomotor activity rhythms of flies can be entrained without cry (Stanewsky et al., 1998). The compound eyes, ocelli, a structure called the Hofbauer-Buchner eyelet (H-B eyelet), and CRY are all responsible for entraining activity rhythms, and flies lacking one or more of these are partially compromised in their entrainment (Stanewsky et al., 1998; Helfrich-Forster et al., 2001; Rieger et al., 2003; Klarsfeld et al., 2004). Among these photoreceptive structures, the H-B eyelet has a direct input into the AMe and possibly connects with large LNv (Helfrich-Forster et al., 2002a; Malpel et al., 2002; Helfrich-Forster et al., 2007). Exactly how the compound eyes and the ocelli connect to the clock neurons is not yet known. Besides these structures that are responsible for entraining locomotor activity rhythms, clock neurons such as DN1 and DN3 have been shown to be photoreceptive through unknown mechanisms (Veleri et al., 2003; Klarsfeld et al., 2004).
Temperature is another major Zeitgeber of the clock, and can be used to entrain clocks in DD as well as constant light (LL) where flies are normally arrhythmic (Wheeler et al., 1993; Matsumoto et al., 1998; Yoshii et al., 2002; Glaser and Stanewsky, 2005; Yoshii et al., 2005). Temperature-dependent entrainment of the clock seems to occur cell autonomously and temperature-sensing structures such as the antenna are dispensable (Glaser and Stanewsky, 2005). This process needs phospholipase C (Collins et al., 2004; Majercak et al., 2004) and a gene mutated in a novel mutant nocte (Glaser and Stanewsky, 2005). LNv and LNd are dispensable for locomotor activity rhythms under temperature cycles in LL (Yoshii et al., 2005), suggesting that different pacemaker cells may be involved in temperature and light entrainment (Busza et al., 2007; Miyasako et al., 2007). Molecularly, interaction between CRY and TIM-PER complex is seen both after a heat pulse as well as a light pulse, suggesting that this may be a common mechanism for both light- and heat-mediated phase shifts (Kaushik et al., 2007). However, the heat-mediated phase shifts of activity rhythms and accompanying CRY-TIM-PER interaction require rather high temperatures (~37°C) in wild-type flies (Kaushik et al., 2007), whereas locomotor activity rhythms can be entrained by temperature cycles involving much lower temperatures (Wheeler et al., 1993). Furthermore, molecular oscillations can be entrained by temperature cycles in the hypomorphic mutant cryb (Stanewsky et al., 1998; Glaser and Stanewsky, 2005). Therefore, CRY-TIM-PER interaction may not be responsible for entrainment involving moderate temperatures. Preliminary studies in our laboratory suggest that ion channels help modulate temperature sensitivity of the circadian pacemaker circuit within physiologically permissible range of temperatures (Sheeba V, Chou Y, Muirhead, KA, Sharma VK, Holmes TC, unpublished data).
Output from Core Pacemaker Neurons
Large LNv have extensive arborizations with varicosities in the medulla of the optic lobe (Figure 2) (Helfrich-Forster and Homberg, 1993). Therefore, they may underlie communication with the visual system. The visual system also exhibits circadian rhythm in synaptic frequency of photoreceptor cells, screening pigment in photoreceptor terminals, and axon caliber, nuclear size, and dendritic spine of the lamina monopolar cells (Pyza and Meinertzhagen, 1993, 1995, 1997, 1999; Gorska-Andrzejak et al., 2005) suggesting that they are regulated by outputs from large LNv. So what are the molecules that are involved in communication between pacemaker and output structures? Thus far three neuropeptides have been found to be expressed in subsets of clock neurons. PDF is expressed in large LNv and all but one small LNv (Helfrich-Forster, 1995, 1997). Flies lacking either the neuropeptide or cells expressing it cannot sustain robust activity rhythms in constant darkness, and in light/dark cycles (LD), have their evening peak of activity shifted earlier (Renn et al., 1999, Figure 3). The main function of PDF seems to be coupling of molecular oscillations in different clock neurons, because genetic manipulation of PDF-expressing cells and the pdf01 mutation decreases synchrony of molecular oscillations within and among other clock neuronal subgroups (Peng et al., 2003; Lin et al., 2004; Stoleru et al., 2005; Nitabach et al., 2006). A similar role in synchrony of oscillators was previously proposed for a related peptide Pigment Dispersing Hormone (PDH), after injection of PDH into the cockroach brain (Petri and Stengl, 1997). While there is a growing body of functional evidence that PDF is an important neurotransmitter for circadian function, less is known about its receptor, PDFR (a.k.a. GOP, Han). The expression pattern of the receptor for PDF, the best known neuropeptide continues to remain unresolved. While one study, based on immuno-reactivity of a C-terminal sequence, suggests that the receptor is expressed on a pair of DN1, and two- three DN3 in addition to regions around the small LNv, large LNv, LNd (Mertens et al., 2005) another study also based on immuno-reactivity of N-terminus based antibody reveals a larger expression pattern and includes all the large LNv, one of the LNd and five to seven DN1 and one DN3 (PDFR designated as Han, Hyun et al., 2005). Current understanding in the field is that neither of these antibodies accurately depicts the pattern of the receptor distribution. The third study used in situ hybridization and detected signals both in the dorsal as well as lateral brain regions that are known to be regions of pacemaker cells but cannot be confirmed due to lack of colabeling information (PDFR designated as GOP Lear et al., 2005). Thus the final word on its localization awaits further studies.
FIGURE 3.
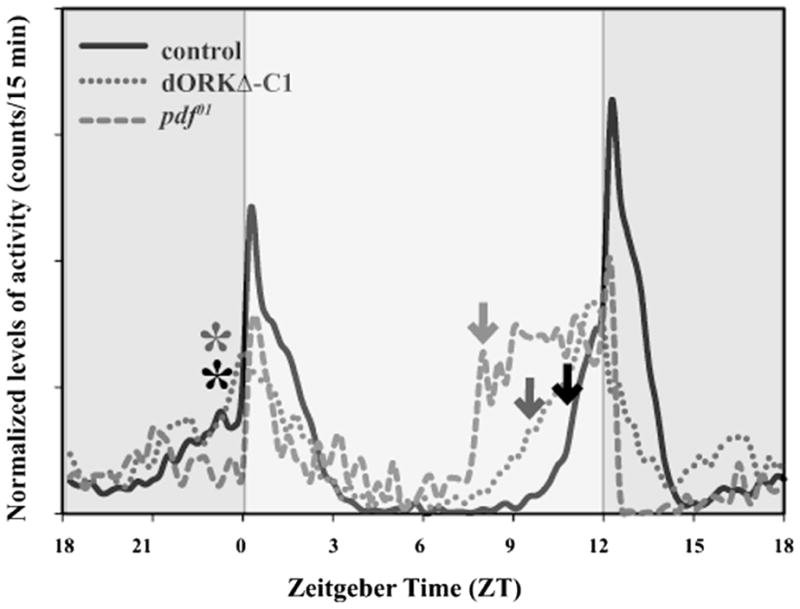
LNv regulation of morning and evening locomotor activity. Representative activity profiles of three genotypes under 12:12 hour light/dark cycles, blue shaded areas represent darkness, and the yellow shaded area represents light. All three genotypes show enhanced activity around dawn and dusk. The controls (solid black curve) show an increase in activity both in anticipation of dawn and dusk as indicated by the black asterisk (morning anticipation) and arrow (evening anticipation). When LNv neurons are electrically silenced by targeted expression of the dORKΔ-C1 channel (red dotted curve) evening anticipation is phase advanced as indicated by the red arrow and occurs around 3 hour before lights OFF while morning anticipation is not significantly altered (red asterisk). A more severe phenotype is obtained with expression of Kir2.1 channel with both a loss of morning anticipation and a shift in evening anticipation (data not shown). The null mutant pdf01 (green dashed curve) shows complete loss of morning anticipation as well as a large phase advance in the evening anticipation. These results indicate a role of the LNv cells in mediating both peaks of the activity rest cycle in Drosophila.
Two-three of ~6 LNd express the Drosophila neuropeptide F (NPF), which is a homologue of mammalian neuropeptide Y (NPY), a neuropeptide expressed in a subset of circadian suprachiasmatic nucleus (SCN) neurons (Lee et al., 2006). Knockout npy−/−mice exhibit defects in their ability to respond to non-photic time cues that ordinarily modulate clock period and these mice show weak entrainment to photoperiods (Harrington et al., 2007). NPF expression is found mostly in males and is regulated by both fruitless and the clock genes Clock and cycle. Two of the DN1 which have a slightly anterior location compared to the rest of this cluster (DN1a) express a neuropeptide called IPNamide (Shafer et al., 2006). These cells along with two other DN1 (DN1p) appear to project back to the AMe and LNv, and thus could mediate a feedback signal within the pacemaker circuit.
FUNCTIONAL OUTPUTS OF THE PACEMAKER
The circadian clock has been shown to control a number of biological processes including cyclic expression of numerous genes. Unfortunately there is poor consensus among the five different studies in which DNA microarray techniques have been used to find cycling gene expressions in the Drosophila genome. The possible reasons behind the heterogeneity of results in the Drosophila circadian genomics studies has been discussed extensively elsewhere (reviewed in Etter and Ramaswami, 2002; Taghert and Shafer, 2006; Wijnen et al., 2006). Briefly, use of different statistical tools, tissues, and experimental paradigms are thought to be the primary reasons behind the lack of consensus among fly micro-array data from different laboratories. In terms of the underlying output mechanism for a behavioral rhythm in Drosophila, the rhythms of adult emergence from the pupal case (eclosion) is perhaps the best understood thanks to endocrinological and developmental studies, and has been comprehensively reviewed elsewhere (Helfrich-Forster, 2005). One important aspect of this rhythm is that it is controlled by the clock in the ecdysteroid secreting prothoracic gland, which in turn appears to be controlled by the PDF-expressing LNv (Myers et al., 2003). This situation is very similar to many peripheral clocks in mammals, which are regulated by the central clock suprachiasmatic nucleus (SCN) (Yamazaki et al., 2000).
Locomotor activity rhythm has been studied extensively as a behavioral readout of the circadian clock in Drosophila (Klarsfeld et al., 2003). For robust free-running activity rhythms in DD, clocks in LN are necessary and sufficient (Ewer et al., 1992; Frisch et al., 1994). Among the LN, PDF-expressing small LNv and their processes in the dorsal protocerebrum are especially important (Helfrich-Forster et al., 1998; Renn et al., 1999). As discussed in detail below several groups have shown data supporting the so-called ‘morning and evening’ oscillator model in which one group of clock neurons is responsible for the fly’s morning peak of activity and another group for evening peak of activity under LD (Grima et al., 2004; Stoleru et al., 2004; Stoleru et al., 2005; Rieger et al., 2006; Stoleru et al., 2007). The circuits downstream of the pacemaker neurons involved in generation of locomotor activity/rest rhythms are yet unknown. Presumably these circuits receive circadian signals from the pacemaker neurons in the dorsal protocerebrum and control arousal level as well as specific behaviors such as mating (Hendricks et al., 2000; Shaw et al., 2000; Sakai and Ishida, 2001). Sleep in flies is believed to be modulated by PKA-CREB pathway and serotonin receptor d5-HT1A in the mushroom body (Joiner et al., 2006; Pitman et al., 2006; Yuan et al., 2006), but sleep regulation is complex and the circadian pacemaker circuit may not control these processes in the mushroom body directly, as activity rhythms are normal in mushroom body-ablated flies (Helfrich-Forster et al., 2002b). Non-PDF-expressing cells regulate some rhythmic outputs such as olfaction and egg-laying rhythms (Krishnan et al., 1999; Tanoue et al., 2004; Howlader et al., 2006).
DEVELOPMENT AND HETEROGENEITY OF THE PACEMAKER CIRCUIT
The expression of neuropeptides revealed heterogeneity among cells within the clusters small LNv, LNd, and DN1. In the case of small LNv, four PDF-expressing ones were the first to be discovered (Helfrich-Forster and Homberg, 1993). Later, it was found that there is a fifth PDF-negative PER- and TIM positive small LNv (Helfrich-Forster, 1995; Kaneko et al., 1997). Recent work suggests that the projection pattern of the fifth PDF-negative small LNv is indistinguishable from those for PDF-expressing small LNv (Helfrich-Forster et al., 2007). Heterogeneity in cell body size and PER cycling amplitude among LNd and DN1was noted even before the neuropeptide expression was discovered (Rieger et al., 2006). The relatively anterior position of two of the DN1 neurons in pupae has been documented (Kaneko and Hall, 2000), and these DN1 (DN1a) have been distinguished as those not expressing GLASS protein and not eliminated by glass60j mutation (Helfrich-Forster et al., 2001; Klarsfeld et al., 2004). In fact, DN1a neurons are the cells that originate from larval DN1 (Klarsfeld et al., 2004), and send projections down to the AMe, while most other DN1 neurons (except DN1p) project proximally and make a commissure in the dorsal protocerebrum (Shafer et al., 2006). In addition to these clusters, heterogeneity in cell body size and projection patterns have been found in DN3 neurons, most of which project proximally to the pars intercerebralis while few project to the AMe (Veleri et al., 2003).
Clock neurons differentiate at different developmental stages. Small LNv are the first to differentiate and have PER and PDF expression from early first instar larval stage onward (Helfrich-Forster, 1997; Kaneko et al., 1997). DN1a and DN2 start expressing PER in late first instar larvae, and these cells persist through metamorphosis (Kaneko et al., 1997; Kaneko and Hall, 2000; Klarsfeld et al., 2004). It has been suggested that LNd and large LNv are present from late third-instar larval stage, because weakly labeled neurons with characteristic projection patterns (POT-like processes for large LNv and projections to the dorsal protocerebrum for LNd) were found in late third instar larvae, and these cells could be observed in early pupae as well (Kaneko and Hall, 2000). These putative larval LNd and large LNv most likely correspond to weakly PER- and TIM-immunoreactive cells near LN in older larvae (Kaneko et al., 1997). Later it was confirmed that these weakly labeled cells near the larval LN are indeed precursors of LNd and large LNv (Helfrich-Forster et al., 2007). PER expression could be observed in majority of DN1 and DN3 from late pupal stage (Kaneko et al., 1997).
SYNCHRONY OF MULTIPLE OSCILLATORS
Considering the fact that many different neuronal subgroups comprise the circadian pacemaker circuit the question arises as to how these neuronal subgroups are orchestrated to generate a single coherent rhythm in overt behavior. In terms of molecular oscillations, the cycling in levels of mRNA and protein for PER and TIM in the different subgroups of cells are synchronous (with one exception) under the influence of LD cycles. In larvae, precursors of the DN2 cells oscillate in anti-phase with the rest of the pacemaker neurons both in LD and DD conditions (Kaneko et al., 1997). In adults, the DN2 oscillation is in-phase with the other pacemaker neurons in LD and for the first two days of DD (Blanchardon et al., 2001) (Figure 4A, E), but on the fifth day of DD, they are out-of-phase leading to the proposition that under the influence of light the DN2 cells are synchronized with the rest of the network (Veleri et al., 2003) (Figure 4I); although its not clear as to what might mediate such synchrony since DN2 are not known to express the photoreceptor CRY (Klarsfeld et al., 2004).
FIGURE 4.
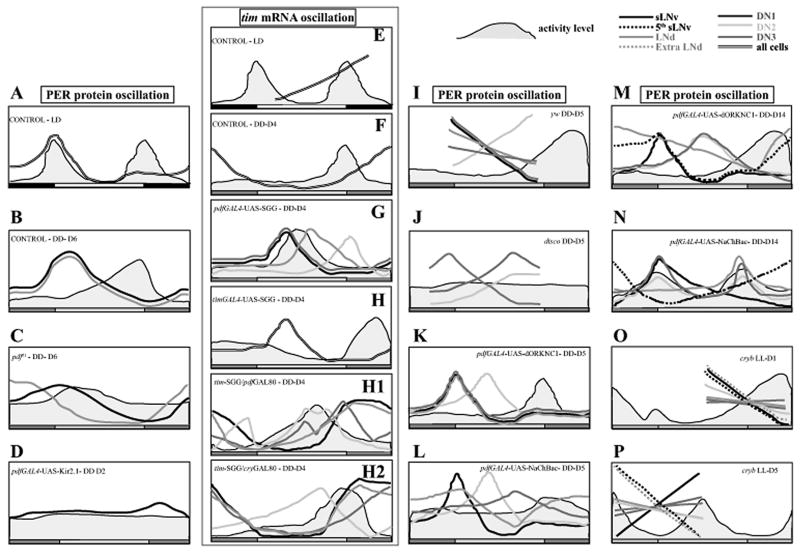
Schematic summary of locomotor activity level and its relationship with molecular oscillation in Drosophila brain circadian pacemaker neuronal subgroups. Horizontal bars at the bottom of each panel depict the light regime. White and black bars indicate the light and dark duration under light/dark (LD) cycles. Constant darkness (DD) is denoted by pale and dark grey bars corresponding to the light and dark durations of previous entrainment regime and similarly constant light (LL) is denoted by white and pale grey bars. Activity level is indicated by the grey filled wave above the LD bars. The neuronal subgroups are color coded in most panels except in cases where the oscillation is synchronous in all the cells assayed, in which case a double lined curve is used. (A) The adult locomotor activity shows two clear peaks in activity under LD 12:12 hr and the level of PER in all cells occur at approximately the same phase and coincides with the morning peak in activity (Bachleitner et al., 2007). Upon transfer to DD, locomotor activity of wild-type flies usually shows only one peak; very often this peak appears to be derived from the evening peak of prior entrainment. (B-C) Lin et al., (2004), showed that molecular oscillations in sLNv and LNd stay tightly synchronized in case of wild-type controls (yw) for up to 6 days in DD, and that in pdf01 flies the dampening in activity rhythm is accompanied by a dampening in molecular oscillations among the sLNv probably due to loss in intercellular communication among sLNv and a phase advance as well as dampening in oscillations among LNd, suggesting that PDF is the agent of synchrony both within and between sLNv and LNd subgroups. (D) Such a dampening and ultimate stop in molecular oscillations was also seen when LNv are electrically silenced using Kir2.1 channels along with arrhythmic locomotor behavior in DD (Nitabach et al., 2002). (E-H) mRNA levels have been used as an indicator of the state of the clock in some studies (Stoleru et al., 2004, 2005, 2007). (E) Under LD 12:12 hr, high level of per mRNA is seen soon after lights OFF and the level is lowest around early morning. (F) Stoleru et al. (2005) report that mRNA level oscillates with a single peak on the fourth day of DD in all the neuronal groups examined and the phase of oscillation remains in close synchrony among the different neuronal subgroups. (G) When SGG is expressed in LNv, the “morning” cells, activity is phase advanced and so is the phase of mRNA oscillation in all cells; with greatest advance in sLNv and DN1 cells, followed by LNd and lastly DN2. (H) Such a phase advance is seen in all the neuronal subgroups to a similar degree when SGG is expressed using the tim-GAL4 driver. (H1) When SGG expression is restricted to non-LNv cells, only DN2 cells (and lLNv, not shown) are phase advanced and other cells are similarly phased as sLNv indicating a dominance of sLNv. (H2) Alternatively when DN2 cells alone express SGG, the advance in molecular oscillation does not alter any of the other cells except lLNv (not shown) (I-J) Veleri et al. (2003) assayed molecular oscillations in control (yw) and disco mutant flies after 5 days in DD and report that oscillation in DN2 cells are now in anti-phase with the rest of the neuronal groups. disco mutants which lack almost all the LN neurons exhibit anti-phase oscillations in both DN1 and DN2 cells compared to DN3 oscillations, which have similar phase as controls. (K) Out-of-phase oscillations in DN2 were also detected in rhythmic control flies expressing dORK-NC1 channel after five days in DD (Nitabach et al., 2006). (L) In contrast, flies expressing NaChBac channel in LNv exhibit arrhythmic behavior and asynchrony in molecular oscillations among the different neuronal subgroups when assayed after 5 days in DD. (M) After 14 days in DD, control flies continue to exhibit robust rhythmic activity with a single peak in activity level. The peak in PER levels in the sLNv occurs at the trough of activity level. The oscillation in DN1 and DN2 are delayed with respect to the sLNv, and LNd shows a dampened oscillation with a clear trough just before activity onset. (N) NaChBac flies show two clear bouts of activity at day 14 DD, one bout exhibits a shorter than 24-hour free running period and the other a longer than 24-hour free running period. The sLNv show peak PER levels coinciding with the long-period activity bout. The DN1 show two peaks each coinciding with one peak of activity. DN2 cells also show higher PER levels coinciding with activity peaks, although they are not significantly different from the other two time points. Under LL, while most wild-type flies are arrhythmic, cryb mutants exhibit two periodicities. This is evident after at least 5 days of LL (Rieger et al., 2006). (O) On the first day of LL only one peak in activity is seen and at that time point, the sLNv, DN2 and a subset of LNd express low levels of PER. At the trough of activity profile, the levels of PER is high in these cells. Other cells do not exhibit significant oscillation. On the fifth day of LL, the activity pattern shows two distinct bouts. DN2, fifth sLNv and one large LNd (extra LNd, dotted orange line) appear synchronous and express high PER coinciding with the activity peak of the short-period bout, and low PER levels corresponding to the time of the long-period bout. PER levels in other four sLNv are in anti-phase with the above cells and have high PER coinciding with the long-period bout. The oscillations in other cell groups as determined by sampling at these two active phases are not statistically significant.
The output molecule PDF is believed to be a synchronizing factor for oscillations among the multiple subgroups by two independent studies using the null mutant pdf01 (Peng et al., 2003; Lin et al., 2004). However these two studies disagree on one important point. Peng et al., 2003 show dampening of cry and tim mRNA oscillations in DD and conclude that behavioral rhythms are generated by the action of PDF coordinating intra-cellular communication, while Lin et al., 2004 show that the oscillations in PER levels in sLNv are in fact not altered by the absence of functional PDF (also discussed in earlier section on molecular oscillators; Figure 4B, C). Instead, the level of synchrony—that is the cycling coherence between the individual cells within a subgroup was significantly reduced. The authors propose that PDF is responsible for maintaining tight synchrony in phase and amplitude of PER oscillation among the small LNv and among the dorsal subgroup LNd. There is good evidence for such synchronizing properties of a related peptide PDH in the cockroach, where injection of PDH in the AMe caused phase shifts in activity/rest behavior in a time dependent manner (Petri and Stengl, 1997).
Studies in our lab using transgenic expression of a voltage-gated Sodium channel NaChBac in the LNv electrically perturbs a small subset of the pacemaker circuit—and these flies exhibit long-term complex locomotor activity/rest rhythms (Nitabach et al., 2006). Further the molecular clock in the different neuronal subgroups in the circuit was desynchronized in flies that express NaChBac in the LNv when assayed during the initial stages of exposure to DD (day 5)—this oscillator desynchrony corresponds with initial short-term arrhythmic locomotor activity (Figure 4L). But upon emergence of stable although complex rhythmicity after ~5–6 days, the molecular oscillations in the different subgroups assume a novel pattern of synchrony that is particularly striking in the dorsal pacemaker cell groups (Sheeba et al., 2008, Figure 4N). We interpret these findings to imply that the pacemaker circuit responds to perturbation by compensatory homeostatic mechanisms that are not cell autonomous, but distributed throughout the neuronal subgroups as reflected by emergence of novel patterns of rhythmicity in behavior and synchrony in molecular oscillations from an initial arrhythmic behavior and asynchronous state of molecular oscillations. These results suggest that oscillator function is not strictly anatomically localized, but plastic throughout the circuit depending on input and cell-cell communication.
ELECTRICAL SIGNALING IN PACEMAKER NEURONS AND CIRCUITS
Before the molecular-genetic characterization of clock genes in Drosophila, circadian rhythms of spontaneous action potential firing was recognized as the key defining feature of pacemaker neurons in vertebrates and invertebrates (Inouye and Kawamura, 1979; Green and Gillette, 1982; Schwartz et al., 1987; Michel et al., 1993). Molecular circadian clocks show conservation both at the operational level and in some cases, the molecular identity of clock components (e.g., alleles of PERIOD are core clock components in both Drosophila and mammals, see Allada et al., 2001). Correspondingly, it appears that there are conserved neurophysiological features of pacemaker neurons. Pacemaker neurons across different invertebrate and vertebrate species tend to fire spontaneous action potentials at high rates during the day and low rates at night (reviewed in Kuhlman and McMahon, 2006). Not surprisingly, neurophysiological recordings from pacemaker neurons in clock gene mutant rats and mice reveal corresponding changes in the phasing of action potential firing patterns (Liu et al., 1997; Herzog et al., 1998; Nakamura et al., 2002). More recent work has shown circadian oscillation of ionic currents in mammalian pacemaker neurons (Colwell, 2000; Pennartz et al., 2002; Ikeda et al., 2003; Itri et al., 2005). Ongoing work is identifying circadian regulated neurophysiological components in Drosophila pacemaker neurons (Rubovszky, Sheeba, Gu, Dahdal, O’Dowd, and Holmes, Soc. Neuroscience Abs. 459.16, 2006). Another recent neurophysiological study shows that Drosophila pacemaker neurons are light-sensitive, based on the finding that the threshold of current-evoked firing is modulated by light levels (Park and Griffith, 2006) although these authors did not report reliable spontaneous firing in these neurons.
Cell-Autonomous Electrical Signaling in Pacemaker Neurons
Isolated pacemaker neurons can retain circadian regulation of membrane electrical properties as shown by cell autonomous circadian rhythms in membrane potential and potassium conductance found in neurons of the mollusk Bulla gouldiana (Michel et al., 1993) and spontaneous action potentials measured in isolated mammalian SCN neurons (Welsh et al., 1995). So is the relationship between electrical signaling in pacemaker neurons and the circadian clock a “one-way street” for which information flows merely as an output function from the circadian clock to regulate electrical activity in the pacemaker neurons? Electrical signaling in pacemaker neurons has been proposed to be a core component of the circadian clock (Njus et al., 1974, 1976; Nitabach et al., 2005a). Experimental support for this idea came initially from the observation that modulation of membrane potential influences both input and output to the circadian clock in Bulla pacemaker neurons (McMahon and Block, 1987). Recent studies have revived the hypothesis that electrical signaling in pacemaker neurons is a core component of circadian clock. In Drosophila, electrical silencing of PDF-expressing LNv by transgenic expression of mutant open rectifier or inward rectifier potassium channels causes run down of the free-running circadian clock in these neurons within few days in DD and abolishes rhythmic circadian locomotor behavior (Nitabach et al., 2002; Nitabach et al., 2005b). PER and TIM levels continue to oscillate in LNv of flies with electrically silenced pacemaker neurons when they are kept in 12:12 hour LD cycles, thus electrical silencing specifically disrupts the free-running circadian clock in DD. Does expression of mutant potassium channels in LNv actually cause electrical silencing? We have recently verified by whole cell patch clamp analysis that expression of the mutant high conductance open rectifier potassium (dORKΔ-C) channel in LNv causes profound membrane hyperpolarization and abolishes spontaneous action potential firing that is normally observed in the LNv (Gu, O’Dowd, and Holmes, unpublished results). While earlier patch clamp recordings of LNv do not reveal spontaneous action potential firing (Park and Griffith, 2006), this study shows clearly that current-injection-evoked action potential firing of LNv is attenuated by dORKΔ-C expression (Park and Griffith, 2006). Thus, all lines of evidence indicate that dORKΔ-C expression causes membrane hyperpolarization and abolishes action potential firing. Based on PER and TIM cycling results, we concluded that electrical silencing of the LNv pacemaker neurons cell-autonomously disrupt circadian molecular oscillations (Nitabach et al., 2002). However, as pointed out by Kuhlman and McMahon (2006), electrical silencing would also likely block output of the LNv pacemaker neurons—and altered cell-cell communication within the pacemaker circuit could potentially feedback to the LNv. We independently considered this as well, and attempted to address this possibility by testing the effects of expressing tetanus toxin light chain throughout the Drosophila pacemaker circuit using the tim-GAL4 driver line (UAS-TeTxLC/tim-GAL4). UAS-TeTxLC/tim-GAL4 flies are behaviorally arrhythmic in constant darkness, but normally phased clock cycling persists in the LNv pacemaker neurons after five days in DD (Nitabach et al., 2005b). This suggests that the effect of electrical silencing on disruption of LNv circadian oscillation is indeed a cell autonomous effect; although we cannot rule out the possibility tetanus toxin light chain expression does not completely block synaptic transmission in the pacemaker circuit. While this question has not been resolved, Nitabach et al. (2002) helped motivate a growing body of work examining circuit-level circadian function in Drosophila.
Circuit-Dependent Electrical Signaling between Components of the Pacemaker Neural Circuit
The circuit-wide effects of altering electrical excitability in a small subset of the Drosophila circadian pacemaker circuit have been tested. As described in a previous section on synchrony of multiple oscillators, targeted expression of the low-threshold voltage-gated sodium channel NaChBac in the LNv causes short-term arrhythmicity, long-term changes in locomotor behavior and short-term molecular clock cycling desynchrony throughout the entire Drosophila pacemaker circuit, particularly in dorsal neurons (Nitabach et al., 2006). Mutations in natively expressed Drosophila channels can also cause circuit-wide perturbation of circadian function as demonstrated by recent studies on SLOWPOKE (a large conductance calcium-activated potassium channel that is expressed widely throughout the Drosophila nervous system). Flies carrying severe mutations in the slowpoke channel gene exhibit weak circadian rhythms that selectively alter clock cycling in the Drosophila pacemaker circuit (de la Paz Fernandez et al., 2007). Future neurophysiological and imaging studies will give us a clearer picture of the functional operation of the Drosophila pacemaker circuit. One note of caution that applies to most of the studies on Drosophila pacemaker circuit described thus far and to those discussed in the following sections is the fact that they do not account for the possibility of plasticity of the wild type neural circuit both during development and in response to environmental changes. Almost all genetic manipulations (and mutants) used in the study of the fly circuit have been chronic, thus pointing to the exciting possibility of unraveling the plasticity of the mature adult as well as the developing larval pacemaker circuit by designing circuit manipulations that are both temporally and spatially restricted.
THE “MORNING” AND “EVENING” OSCILLATOR MODEL
In 1976, Pittendrigh and Daan proposed that circadian clocks simultaneously “measure” daily and seasonal changes in day lengths using two mutually coupled oscillators: the ‘Morning’ (M) oscillator that tracks dawn and the ‘Evening’ (E) oscillator that tracks dusk (Pittendrigh and Daan, 1976). Although the model was originally conceived to explain the peculiar and rare phenomenon of “splitting” and “re-fusion” of morning and evening activity bouts in mammals, it was hoped that it would provide a basis for understanding other characteristic features of circadian pacemakers that could not be explained by other contemporary models based on a single oscillator (Pittendrigh, 1960). Previously a two-oscillator model had been developed to account for the bimodality of circadian rhythms often seen under LD cycles (Aschoff and Wever, 1966)
The M and E oscillator model assigns certain properties to the two oscillators such as (i) the free running period of M and E oscillators are differentially affected by light intensity; the period of morning oscillator is negatively correlated with light intensity and that of the evening oscillator is positively correlated with light intensity, (ii) when mutually coupled, the overall pacemaker period is different from the periods of the individual oscillators, (iii) the relative influence of the oscillators on each other depends upon their phase-relationship, and (iv) the coupling between the two oscillators depends upon the environmental conditions. In many diurnal animals, during long summer days, morning activity occurs earlier and the evening activity occurs later compared to days with near 12:12 hour LD photoperiods, enabling them to avoid the midday heat (Majercak et al., 1999). According to the M and E oscillator model, in the summer (long day conditions, for which LL may act as a proxy) the M oscillator would run with short period while the E oscillator would exhibit a long period. This could, in principle, decouple the two oscillators, resulting in behavioral “splitting.” Indeed, under constant light conditions, behavioral activity components have been seen to split into multiple components in several diurnal and nocturnal rodents, in tree shrews, common marmosets, and in certain species of birds, reptiles, fishes and insects (Rosenwasser and Adler, 1986; Schardt et al., 1989; Smietanko and Engelmann, 1989; Meijer et al., 1990; Hong and Saunders, 1998). Multiple behavioral components that resemble “splitting” have also been observed in certain genetically modified Drosophila strains (Helfrich, 1986; Helfrich-Forster, 2000; Yoshii et al., 2004; Nitabach et al., 2006; Rieger et al., 2006). An important point to note here is that such behavioral decoupling does not usually occur in the wild because constant bright light usually results in behavioral arrhythmicity (Aschoff, 1979; Konopka et al., 1989) and when “splitting” occurs in experimentally manipulated environments such as dim LL (usually up to 100 lux), it is seen after prolonged exposure (as long as two months) to such conditions (Pickard et al., 1993).
Although activity of some diurnal mammals, for example the northern tree shrew (Tupaia belangeri), undergoes splitting under DD, the split components are indistinguishable in terms of their response to light and carbachol perturbations (Meijer et al., 1988; Meijer et al., 1990). Similarly bilaterally distributed identical pacemakers in insects may temporally separate from each other (Koehler and Fleissner, 1978). These results suggest that the left and right SCN or bilaterally distributed pacemakers in insects, which are otherwise indistinguishable structurally, serve as M and E oscillators and show antiphasic coupling in “split” animals. Indeed, partial to complete lesions of one of the SCN lobes in hamsters leads to partial to complete elimination of one of the split activity components (Pickard and Turek, 1982; Davis and Gorski, 1984). Bimodality is also seen at the level of single unit electrical activity of coronal SCN slices of split hamsters (Mason, 1991; Zlomanczuk et al., 1991). Although these studies suggest that the regulation of splitting occurs in the SCN, the left-right distribution of electrical activity is still unclear (Daan et al., 2001). In a relatively recent study in rodents behavioral desynchronization was shown to be coupled with dissociation of clock gene expression in the ventrolateral and dorsomedial region of the SCN (de la Iglesia et al., 2004).
From a purely functional perspective, the M and E oscillator model is appealing because it provides a relatively simple explanation for adjustments of circadian behavior—and thus by inference, circadian oscillator organization—in response to daily and seasonal changes. The M and E oscillator model is particularly attractive for Drosophila melanogaster as it exhibits two distinct bouts of locomotor activity under 12:12 hour LD cycles, one centered at dawn and the other around dusk suggesting that flies anticipate dawn and dusk transitions under natural cyclic environments.
While the M and E oscillator model was originally developed to explain “splitting” behavior in mammals in response to constant light, this model has been applied recently to Drosophila melanogaster using several genetic and behavioral approaches aimed at identifying the putative M and E oscillators in Drosophila circadian system. Helfrich-Forster (2000) suggested that the morning peak of activity is governed by a per-independent clock and is entrained by light signals via photoreceptors, while the evening peak is regulated by the TTFL involving per and is entrained by CRY. Studies by Yoshii and coworkers (2004) with the mutant cryb showed that activity rhythm in these flies has a propensity to dissociate into two components with increasing light intensity, one with a short and the other with a long period (Yoshii et al., 2004). Their investigations led them to the hypothesis that in these flies, the evening oscillator is itself composed of two oscillators, both of which are PER dependent and receive information from photoreceptors (Yoshii et al., 2004). Another approach has been to eliminate different subgroups of clock neurons or to restore clock gene expression in specific neurons in clock mutants (Grima et al., 2004; Stoleru et al., 2004), or to over express core clock genes in different neuronal subgroups to accelerate or alter the molecular oscillations in different neuronal subgroups in an attempt to change the timing of oscillator phase relationships between different oscillator subgroups (Stoleru et al., 2005; Murad et al., 2007; Stoleru et al., 2007). The conclusion from these 2004 studies (Grima et al., 2004; Stoleru et al., 2004, Figure 5) was that the LNv function as the M oscillator in the Drosophila circadian pacemaker circuit, while the LNd functions as the E oscillator (while Stoleru and colleagues acknowledge that they cannot functionally distinguish between various CRY+PDF−cells, they favor the LNd as the E oscillator because of the efferent connections from the LNd to the LNv region, pg. 868 in Stoleru et al., 2004). One of these studies showed that transgenic flies lacking LNv exhibit weak or no anticipation to lights-ON (morning) and altered lights-OFF (evening) anticipation (Figure 2 in Stoleru et al., 2004). Flies that lacked most dorsal neurons exhibit disruptions in evening anticipatory activity. More recently another more detailed study of the behavioral polyrhythmicity and its underlying molecular basis in the cryb mutant was conducted (Rieger et al., 2006). The behavioral read-outs used to interpret the above studies included: 1) anticipatory activities during dawn and dusk and 2) “splitting” of morning and evening activity bouts and rhythmicity in LL (Grima et al., 2004; Stoleru et al., 2004; Yoshii et al., 2004; Rieger et al., 2006; Stoleru et al., 2007).
Although, the claim has been made that the two oscillators are distinct but somehow coupled (Stoleru et al., 2004), the results of this study along with numerous previous studies (Renn et al., 1999; Blanchardon et al., 2001; Nitabach et al., 2002) clearly show a functional contribution by the so-called morning cells (LNv) to the evening bout of activity. Further, cell-specific rescue experiments in circadian clock mutants indicate that absence of LNv molecular oscillation does not modulate LD behavior (Stoleru et al., 2004). While absence of per in the LNv had no effect either on the morning or evening anticipatory activity, its absence in cry expressing cells or in all brain neurons abolished the morning peak and had marginal effect on the evening anticipatory activity (Fig. 4 in Stoleru et al., 2004). Further, while absence of LNv or its output (PDF) is known to abolish morning anticipatory activity it also modifies the evening anticipatory pattern (Figure 2 in Stoleru et al., 2004; Figures 4 & 8 in Renn et al., 1999; Figure 3). In both cases the evening anticipation and the evening peak are phase advanced. These results are consistent with those of previous studies that reported loss of morning anticipatory activity and phase advancement of evening activity peak in flies lacking LNv function by ablation or carrying loss of function mutation for PDF or electrical silencing (Renn et al., 1999; Blanchardon et al., 2001; Nitabach et al., 2002).
In the parallel study which tried to identify the M and E oscillators by restoring clock function in either the LNv or in LNv plus LNd by transgenically expressing PER in per null (per0) genetic background (Grima et al., 2004), it was shown very convincingly that restoring PER expression in the LNv reinstated the morning anticipatory activity alone, while restoring PER expression in both the LNv plus LNd restores both morning and evening anticipatory activity (albeit at weaker amplitudes than wild-type flies). This result is the most striking piece of evidence supporting the role of LNv in regulating morning anticipatory activity via the action of PER protein. Although it was possible to restore the LD activity waveform by expressing PER in the LNv and/or LNd of per0 flies, only 42% of flies showed weak rhythmic activity in DD and the activity profile showed a relatively broader peak in rescued flies compared to wild-type flies suggesting that additional dorsal neurons are important for the normal wild-type DD circadian behavioral pattern (Grima et al., 2004). What is not clear is why disrupting LNv function has such profound effect on the evening activity peak (Figure 3). One possibility is that LNv inhibit the onset of evening activity bout—and that by ablation or electrical silencing this putative inhibition is removed. Alternatively, since PER is a transcription factor that potentially influences the expression of hundreds of genes (Claridge-Chang et al., 2001), the absence of PER could cause defects in receptors or other molecular machinery needed for cell-cell communication between different groups of pacemaker neurons.
In a follow-up to their previous study, Stoleru and coworkers (2005) using a slightly different approach speeded up molecular oscillations in different neuronal subgroups by ectopically expressing SGG (a circadian clock protein described in the section on molecular oscillations) in neuronal subsets of the pacemaker circuit, and examined its effect on activity/rest rhythm during the immediate three days following transfer to DD, and estimated the speed of the molecular oscillators on the fourth day in DD (measured in terms of mRNA levels) in the rest of the circuit (Stoleru et al., 2005) (Figure 4F–H). The speed of the mRNA oscillations in the sLNv was shown to significantly influence those in the LNd, DN1 and DN3 cells, while the DN2 cells remained unaffected. Surprisingly mRNA oscillation in lLNv also remained unaltered despite the over expression of SGG in these cells. Further, when SGG expression is restricted to non-LNv pacemakers, as expected, the molecular oscillation remained unaltered in sLNv, but surprisingly the LNd, DN1 and DN3 also remained in phase with sLNv despite the presence of the accelerator molecule SGG in these cells (Figure 4H). The molecular oscillation was phase advanced in the DN2 and lLNv neurons (even though SGG is not expressed in lLNv). Based on these results the authors proposed that there are two parallel circuits, one which comprises both the “morning” and “evening” oscillators and consists of the sLNv (morning cells) controlling LNd, DN1, and DN3 cells (evening cells) and the second circuit whose function is unknown, comprising cell autonomous DN2 (dominant) oscillator and lLNv. It should be noted that the tim-GAL4 driver regulates expression in many more cells (including neurons and glia throughout the body) of the adult fly than those listed above (Kaneko and Hall, 2000). Further, the authors propose that sLNv neurons provide a daily resetting signal that can function both as a delaying and advancing cue to other members (evening cells) within its circuit. The authors propose that PDF is likely to be the molecule that performs this function. These results are consistent with earlier reported findings that PDF regulates the overall synchrony of multiple oscillators distributed throughout the pacemaker circuit (Petri and Stengl, 1997; Lin et al., 2004). Further studies will be necessary to unveil how both advances and delays in molecular oscillations are achieved by PDF. One unexplained inconsistency between this (Stoleru et al., 2004) and other studies (Veleri et al., 2003; Nitabach et al., 2006) examining molecular oscillations in DD is that this study reports synchronous molecular oscillations on the fourth day in DD in all the pacemaker cell groups, while other studies have shown that DN2 cells have anti-phase oscillations in PER protein by day 5 in DD (Veleri et al., 2003, Nitabach et al., 2006). This inconsistency may have arisen due to the fact that this study examined tim mRNA levels, while others assayed PER protein levels. If one compares activity profile over a 24-hour duration with tim mRNA levels in different neuronal subgroups on the fourth day in DD (Figure 4F–H), it appears that at this stage in DD phase of mRNA oscillation and activity peak has poor correlation with any of the neuronal subgroups. The reliability of mRNA levels as determinants of the state of circadian oscillator is potentially questionable based on the discussions in the earlier section herein on post-transcriptional regulation of molecular oscillator components.
More recent studies by Stoleru and coworkers (2007) and Murad et al. (2007) propose that dorsal neurons act as circadian pacemakers under LL. However, the LNv may not be completely dispensable in LL because these studies also show that activity/rest behavior when SGG is over expressed in all known clock cells in a pdf01 genetic background does not phenocopy the behavior seen when SGG over expression was excluded from LNvin pdf+ genetic background. In the former case, almost all flies were arrhythmic in contrast to the latter where 90% of flies were rhythmic (Stoleru et al., 2007). Although it would be useful to know the phenotype of flies that express SGG in non-LNv cells in a pdf01 genetic background, the results of this study suggests that rhythmic locomotor activity in LL, driven by non-PDF cells is dependent upon the availability of PDF. These results are further complicated by the fact that overexpression of morgue (a gene likely to be involved in the circadian light input pathway) in pdf01 genetic background resulted in rhythmic behavior in about 60% of flies (Murad et al., 2007), suggesting PDF independent mechanism in the generation of rhythmicity in LL.
In a separate study Rieger et al., 2006 examined the “splitting” of behavioral activity/rest rhythm to refine the neural correlates of the M and E oscillators as defined by the studies of Yoshii and coworkers (Yoshii et al., 2004). The activity/rest rhythm of cryb mutant flies in LL predominantly splits into two bouts, each of which free-runs with either a faster or slower than 24-hour rhythm (short-period and long-period activity bouts, respectively) and respond in opposite ways to increase in light intensity, suggesting that they may be behavioral manifestations of M and E oscillators (Rieger et al., 2006). They argue that the designation of LNv and LNd as M and E oscillators respectively is only partly justified. Their studies of cryb mutants in LL examined the level of PER and TIM in different neuronal subgroups on day 1 or day 5 after flies were released into LL at different phases using level of activity as reference (Figure 4O, P). On day 1 of LL cryb flies show only one distinct peak of activity while on day 5 they show two distinct peaks. The authors note that the two bouts do not appear to be derivatives of morning and evening activity bouts of the preceding LD regime, as the faster running bout invariably emerges from the evening activity bout. While all the neuronal subgroups, with oscillating levels of PER appeared to be in-phase on day 1 of LL and have high levels coinciding with the activity trough (Figure 4O), on day 5 sLNv becomes antiphasic with LNd and fifth sLNv and shows high PER levels coinciding with trough of short-period activity bout (Figure 4P). These results led to the conclusion by Rieger and colleagues (2003) that the four PDF expressing small LNv function as both morning and evening oscillators (‘M-E’ oscillators, or ‘Main’ oscillator), while one PDF negative small LNv (the fifth small LNv) along with one of the LNd cells forms the evening oscillator. The differential response of change in speed of the oscillator with changes in light levels supports the original M and E oscillator model, in LL (Wheeler et al., 1993; Helfrich-Forster, 2000). Thus in contrast to the three preceding studies (Grima et al., 2004; Stoleru et al., 2004; Stoleru et al., 2005) the cryb results suggests that the PDF positive small LNv neurons regulate not only the morning activity but also partly the evening activity, indicating that the circadian pacemaker circuit may be far more complex than envisaged in simple labeled-line models (Grima et al., 2004; Stoleru et al., 2004; Stoleru et al., 2005).
The empirical evidence for anatomically restricted neural substrates for the oscillators that regulate the morning and evening anticipatory behavior in Drosophila is equivocal and it appears that many cell groups may jointly regulate activity/rest rhythm whether in LD, LL, or DD. If the LNv and LNd neurons are indeed the M and E oscillators of Drosophila respectively, they should respond differently to DD and LL. This was explored in a recent study (Stoleru et al., 2007) by manipulating different subgroups of pacemaker neurons using ectopic expression of SGG. Over expression of SGG in all known clock cells using tim-GAL4 driver makes the flies rhythmic in LL in contrast to wild-type control flies and those where SGG over expression is restricted to the LNv; both of which are arrhythmic. Stoleru and colleagues (2007) conclude that the so called evening cells and DN2 subgroups of neurons regulate the pacemaker speed in LL based on the interpretation that the DN1 and DN2 are the only cells that exhibit robust cycling in nuclear localization of PER with a pattern similar to that observed in DD in wild-type flies (Shafer et al., 2002). Yet, the authors did not detect significant circadian oscillation in the levels of PER or TIM protein, which has been until now considered as a marker for circadian clock function (Stoleru et al., 2007). It is worth noting in this study that by the use of the tim-GAL4 driver in conjunction with the subtractive pdf-GAL80 driver, the authors essentially extend the definition of “evening” cells to the entire set of clock cells that express TIM with the exception of PDF positive LNv.
Another critical unexplained inconsistency in the assignment of distinct M and E oscillators to the LNv and LNd groups is that despite a near 12-hour phase difference between the observed morning and evening activity peaks, the molecular oscillations (both tim mRNA and PER protein) in the LNv and LNd neuronal groups remain nearly perfectly in phase in LD conditions, a true Pas de Deux (Stoleru et al., 2004; Bachleitner et al., 2007, Figure 4A, E). This raises the possibility that the neural circuits that control morning and evening activity bouts may be downstream from the LNv and LNd circadian pacemakers. The present M and E oscillator model for Drosophila has no explanation for how two sets of oscillators that operate in-phase can control temporally distinct bouts of behavior that are almost completely antiphasic. A comparison of activity levels with the oscillations in the known pacemaker cells (irrespective of whether mRNA or protein levels are examined), suggests that it is highly unlikely that the phase of molecular oscillations and phase of activity have a simple one-to-one correlation (Figure 4).
One possible explanation for the conundrum noted above is that different oscillators can control different outputs at a downstream circuit level. Previous studies in mammals have shown that nocturnal and diurnal rodents that show similar phase of PER oscillation have differences in the brain regions outside the pacemaker center (Smale et al., 2003; Nixon and Smale, 2004; Schwartz et al., 2004; Saper et al., 2005; Schwartz and Smale, 2005). This suggests that molecular and behavioral oscillations may not be linearly correlated and that unimodal rhythm in clock protein cycling might be transformed into a bimodal output. The other possibility is that circadian pacemaker neurons may segregate in a spatio-temporal manner, wherein the same neural subgroup may function as both morning and evening oscillators at different times of the day. Indeed, some of the results of studies described above show that there may be a complex hierarchy among different neuronal subgroups, most of which is still largely unclear, and that circadian outputs are regulated efforts of the entire pacemaker circuit. However, such inferences have not been taken into account, instead it was first proposed that LNv act as the M oscillator and the LNd act as the primary E oscillator (Grima et al., 2004; Stoleru et al., 2004; Stoleru et al., 2005; Stoleru et al., 2007), and later the cell groups LNd, the DNs (with the exception of the DN2, Stoleru et al., 2005) were extended as E oscillators. In the most recent study tim-GAL4/pdf-GAL80 driven SGG expression (which essentially targets ~ 90% LNd; 55% DN1; 75% DN2 and 40% DN3) was implied to target E cells (Stoleru et al., 2007). Thus we have seen a rather flexible assignment of functional identity (morning or evening cells) partly due to constraints due to the lack of availability of more restricted driver lines and the urge to define simple distinct anatomical neural correlates to the two-oscillator model.
Does the M and E Oscillator Model Explain How Circadian Circuits Adapt to Seasonal Change?
One of the original motivations for the development of the M and E model was to explain how the circadian circuit tracks daily and seasonal changes in day and night length. Seasons are marked by gradual changes in day length, spectral properties of light throughout the day, temperature, food availability, and often profound long-term alterations in physical environment. The studies above have led the proponents of the Drosophila M and E oscillator model to test whether pacemaker neuronal subgroups respond differently to environmental manipulations that are proxies for seasonal change—and if so, whether M and E oscillator organization accounts for entrainment to varying photoperiods encountered due to changing seasons. Stoleru and colleagues (2007) show that flies with SGG over expression in all known clock neurons (with the exception of PDF-positive LNv) show diminished responses to light pulses during the early part of the night and conclude that “evening” cells govern entrainability at these phases. In DD, LNv-restricted over-expression of SGG causes shortening of period (τDD = 21.7 hours), close to the shortening achieved when SGG is over expressed in all known pacemakers τDD = 20.8 hours), whereas SGG over expression in the dorsal neurons alone has no observable effect on period (τDD = 23.7 hours) (Stoleru et al., 2005). Previous studies have shown that the LNv are responsible for entraining circadian rhythms especially during the late subjective night (Emery et al., 2000). Since the LNv rely more on the changing levels of clock proteins during the night, it is hypothesized that the active phases during long winter nights would be locked to dusk (Stoleru et al., 2007). In other words, by virtue of having greater sensitivity to light during the early subjective night, the dorsal neurons would play a greater role during the long summer days, while the LNv neurons that serve as the key oscillators in DD, are expected to play a prominent role in winter conditions. The authors tested this hypothesis by comparing the anticipatory activity of flies under different photoperiods which may be comparable to those that flies experience if they live in temperate latitudes during spring/fall (12:12 hours LD), summer (14:10 hours LD) and winter (10:14 hours LD). The phase of anticipatory activity to both the ON and OFF transitions of flies where molecular oscillations in LNv or dorsal neurons were speeded-up was compared to control phase of anticipatory activity. Under spring/fall-like conditions both types of genetic manipulations resulted in phase advances in both transitions, although the magnitude of transition was always greater when dorsal neurons were manipulated. Under “summer-like” photoperiod, control flies show almost no anticipation to lights ON. Under the same light regimen there is a dramatic phase advance in evening anticipatory activity when LNv are speeded up, this advance is even greater with speeding-up of dorsal neurons. But both manipulations barely evoke phase advances for morning anticipation. The authors interpret their results to imply that dorsal neurons by virtue of their more dominant clock under long days have the ability to phase shift both morning and evening anticipatory activity.
Murad et al. (2007) also hypothesize that DN1 are likely to possess a dominant function during “summer-like” long day photoperiods due to the persistence of molecular oscillations in these cells under LL as opposed to the dominance of LNv during “winter-like” conditions due to their role in the persistence of rhythms in DD. Rieger et al. (2006), propose that the extended period of activity seen under long day conditions are in fact due to the period-shortening of the PDF positive sLNv (main oscillators) and the period-lengthening of the fifth sLNv evening oscillator in combination with one of the LNd which is effected by input from the compound eye photoreceptors. They maintain that CRY on the other hand, causes period lengthening in all pacemaker neurons in which it is present (which include the so called morning and evening cells). Thus the above studies suggest a complex interaction among pacemaker neurons rather than a simple anatomically distinct two-oscillator mechanism enabling entrainment to diverse photoperiods in Drosophila. Further, the use of the tim-GAL4 driver, which is known to have a wide pattern of expression including the eyes and other tissues, limits the interpretive power of the results (in terms of oscillator identity) in some of the above studies (Stoleru et al., 2004, 2005, 2007; Murad et al., 2007). A more refined separation of neuronal subgroups along with an experimental paradigm that mimics other environmental factors that are dramatically altered in the different seasons (such as temperature) will probably give us a better understanding of how the neural circuit adjusts to changing seasons.
How faithful are the experimental conditions used in the above studies representing seasonal change? Specifically, can altered day length effectively act as a signifier to the animal for seasonal state? Yes, indeed many studies using mainly multivoltine insect species have shown that they exhibit over-wintering diapause in response to short photoperiods (reviewed in Saunders, 2005). D. melanogaster enters ovarian diapause upon exposure to low temperature and short day lengths characterized by block of haemolymph yolk protein uptake by developing oocytes (Saunders, 1990). One possible mechanism for the induction of diapause has been thought to occur via circadian oscillator whose amplitude dampens with increasing night length (first proposed by Bunning, 1969). Is there a role for any of the known circadian molecular components in entrainment to seasonal changes? Under a range of different photoperiods in combination with different temperatures, the observed alterations in locomotor activity profiles has been demonstrated to be due to alternate splice forms of a regulatory region of the per gene as well as gene phospolipase C (Majercak et al., 1999; Collins et al., 2004; Majercak et al., 2004). A recent collaborative effort by several research groups revealed two alleles of tim gene among populations collected from different latitudes in Europe: one that codes for an ancestral short form and a more recent derivative that codes for both a long and short form (Sandrelli et al., 2007; Tauber et al., 2007). Based on molecular, biochemical and behavioral analysis of seasonal ovarian diapause the authors conclude that light signals are perceived by both the seasonal and circadian timer via TIM degradation pathways (Bradshaw and Holzapfel, 2007; Sandrelli et al., 2007; Tauber et al., 2007). In contrast, an independence of circadian molecule per and the photoperiodic timer was previously proposed by Saunders (1990) based upon studies of ovarian diapause in per mutant flies (Saunders, 1990).
Distributed Oscillator Model: an Alternative to the M and E Oscillator Model
Empirical evidence suggests that circadian pacemaker circuits in both invertebrates and vertebrates function in a distributed spatio-temporal manner. Taken as a whole it appears that different genetic manipulations push the organism’s genetic architecture to different equilibrium steady states, each of which is capable of utilizing the perturbed pacemaker circuit to generate a unique behavioral pattern. While it was quite obvious from the beginning that results of empirical studies testing the M and E oscillator model by eliminating one or other components may not represent how oscillators function in wild-type animals, they have provided us with interesting insights as to how these neuronal subgroups interact to influence behavior when genetically per-turbed.
What is the oscillator organization in mammalian circadian circuits? One of the limitations of studying the circadian circuit in flies is the lack of a highly resolved continuous record of individual oscillators cycling in the large ensemble context of pacemaker circuit. Ideally, we would like to see the precise phase of oscillation between individual cells throughout an entire cycle. Immunocytochemical measurements of clock proteins in fly pacemaker neurons provide only snapshots of dynamic oscillator ensemble activity. While we lack the means to monitor multiple oscillators continuously in Drosophila at present, there is a growing body of such highly resolved data for the mammalian circuit. As noted above, cell autonomous firing of spontaneous action potentials occurs in isolated SCN neurons. Remarkably, spontaneous action potential firing in dispersed SCN neurons occurs with markedly different phases and periods (Welsh et al., 1995). This phase and period dispersal of spontaneous firing does not appear to be an artifact of the isolated culture system, as highly variable circadian phase and period relationships between individual pacemaker neurons are found also in multiunit recordings of SCN in vivo and slice preparations (Meijer et al., 1997; Schaap et al., 2003). Direct comparison of the phase relationship of SCN multiunit firing in vivo and in vitro show that the phase of in vivo firing is actually more variable than in vitro (Meijer et al., 1997). This suggests that greater phase variation between individual oscillators carries more information such as encoding of ultradian components (Meijer et al., 1997).
Since circadian rhythmicity of molecular components of the clock is considered as conditio sine qua non by many investigators for circadian function, what do measurements of the clock itself say about the provocative findings above? Direct support for the electrophysiological findings comes from imaging studies of clock cycling readout in SCN slice cultures. The McMahon group imaged clock cycling in SCN slice cultures prepared from transgenic mice carrying the mPer1 promoter-driven short-lived GFP reporter gene and find that highly variable phased clock cycling in individual neurons occurs across the SCN (Quintero et al., 2003). The Okamura group came to fundamentally the same conclusion by imaging SCN slices prepared from mice carrying the mPer1 promoter-driven luciferase reporter gene (Yamaguchi et al., 2003). They find also that blockage of spontaneous action potentials by bath application of the voltage-gated sodium channel blocker tetrodotoxin (TTX) further disrupts the synchrony and amplitude of individual oscillators. Thus, there is considerable physiological evidence to suggest that the SCN oscillator organization is dispersed as multiple oscillators with widely differing phases and periods. This suggests a more complex organization for the mammalian SCN, akin to the Tarantella (the pell-mell circle dance enjoyed at old-fashioned Southern Italian weddings—as depicted in the film The Godfather). The above results collectively raise questions of whether simple two component models such as the M and E oscillator model, while appealing, can fully account for the functional operations of circadian neural circuits (see also Rohling et al., 2006 for a recent insightful discussion on the SCN). In the case of Drosophila, the limited temporal resolution of current methods precludes assignment of phase relationship between molecular oscillations in the various oscillator subgroups and behavior. The complexity of different oscillator phases in relation to behavioral activity summarized in Figure 4 suggests that a Tarantella model may apply to the Drosophila circadian circuit.
Drosophila has been a fruitful test ground for understanding circadian biology from molecular to behavioral levels. While much of our discussion on the Drosophila TTFL clock model and M and E circuit model is critical, these studies have collectively provided what is arguably our best understanding of circadian mechanisms for any model organism. The noted inconsistencies in present models raise questions for future tests. To what extent are transcriptional loops required for self-sustained clocks? How does clock cycling couple to pacemaker neuronal activity? How does phase distribution of oscillation among different neuronal groups in the pacemaker circuit code for circadian physiological and behavioral output? How do different oscillator groups communicate synaptically? Are oscillator subpopulations anatomically fixed to two functional morning and evening groups, or are they plastically distributed throughout the circuit by recent responses to environmental changes? Can we detect changes in clock and pacemaker circuit function in response to realistic tests of gradual seasonal change? And finally, how does the circadian circuit output couple to circuits that mediate activity versus sleep? Much of what we have learned about circadian biology in flies has informed our understanding of circadian mechanisms in mammals. There is a remarkable conservation of function at the level of the molecules that constitute the circadian clock. The field of Drosophila circadian biology is currently enjoying a shift from inquiry focused on the molecular components of the circadian clock to understanding how circadian information is encoded at the systems level. The advent of more powerful physiological and imaging tools combined with genetics promises to soon reveal whether there are similar levels of conservation for the cellular and circuit physiology between flies and mammals.
Acknowledgments
We gratefully acknowledge the support provided by NSF grants IBN-0323466, IBN-0092753, and NIH grants NS046750 and DA016352 to TCH for our research on circadian biology, and our colleagues Jeff Hall, Jae Park, Leslie Griffith, Michael Young, Justin Blau, Michael Rosbash, Paul Garrity, and Michael Nitabach for providing fly lines and other reagents, and the Bloomington Stock Center for other fly lines.
Footnotes
Publisher's Disclaimer: Full terms and conditions of use: http://www.informaworld.com/terms-and-conditions-of-access.pdf
This article maybe used for research, teaching and private study purposes. Any substantial or systematic reproduction, re-distribution, re-selling, loan or sub-licensing, systematic supply or distribution in any form to anyone is expressly forbidden.
The publisher does not give any warranty express or implied or make any representation that the contents will be complete or accurate or up to date. The accuracy of any instructions, formulae and drug doses should be independently verified with primary sources. The publisher shall not be liable for any loss, actions, claims, proceedings, demand or costs or damages whatsoever or howsoever caused arising directly or indirectly in connection with or arising out of the use of this material.
Contributor Information
Vasu Sheeba, Department of Physiology and Biophysics, University of California, Irvine, Irvine, California, USA.
Maki Kaneko, Department of Physiology and Biophysics, University of California, Irvine, Irvine, California, USA.
Vijay Kumar Sharma, Department of Physiology and Biophysics, University of California, Irvine, Irvine, California, USA; Evolutionary and Organismal Biology Unit, Jawaharlal Nehru Centre For Advanced Scientific Research, Bangalore, India.
Todd C. Holmes, Department of Physiology and Biophysics, University of California, Irvine, Irvine, California, USA
References
- Allada R, Emery P, Takahashi JS, Rosbash M. Stopping time: the genetics of fly and mouse circadian clocks. Annu Rev Neurosci. 2001;24:1091–1119. doi: 10.1146/annurev.neuro.24.1.1091. [DOI] [PubMed] [Google Scholar]
- Aschoff J, Wever R. Circadian period and phase-angle difference in chaffinches (Fringilla coelebs L.) Comp Biochem Physiol. 1966;18:397–404. doi: 10.1016/0010-406x(66)90197-6. [DOI] [PubMed] [Google Scholar]
- Aschoff J. Circadian rhythms: influences of internal and external factors on the period measured in constant conditions. Z Tierpsychol. 1979;49:225–249. doi: 10.1111/j.1439-0310.1979.tb00290.x. [DOI] [PubMed] [Google Scholar]
- Bachleitner W, Kempinger L, Wulbeck C, Rieger D, Helfrich-Forster C. Moonlight shifts the endogenous clock of Drosophila melanogaster. Proc Natl Acad Sci U S A. 2007;104:3538–3543. doi: 10.1073/pnas.0606870104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae K, Edery I. Regulating a circadian clock’s period, phase and amplitude by phosphorylation: insights from Drosophila. J Biochem (Tokyo) 2006;140:609–617. doi: 10.1093/jb/mvj198. [DOI] [PubMed] [Google Scholar]
- Bao S, Rihel J, Bjes E, Fan JY, Price JL. The Drosophila doubletimeS mutation delays the nuclear accumulation of period protein and affects the feedback regulation of period mRNA. J Neurosci. 2001;21:7117–7126. doi: 10.1523/JNEUROSCI.21-18-07117.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito J, Zheng H, Hardin PE. PDP1epsilon functions downstream of the circadian oscillator to mediate behavioral rhythms. J Neurosci. 2007;27:2539–2547. doi: 10.1523/JNEUROSCI.4870-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchardon E, Grima B, Klarsfeld A, Chelot E, Hardin PE, Preat T, Rouyer F. Defining the role of Drosophila lateral neurons in the control of circadian rhythms in motor activity and eclosion by targeted genetic ablation and PERIOD protein overexpression. Eur J Neurosci. 2001;13:871–888. doi: 10.1046/j.0953-816x.2000.01450.x. [DOI] [PubMed] [Google Scholar]
- Bradshaw W, Holzapfel C. Evolution. Tantalizing timeless. Science. 2007;316:1851–1852. doi: 10.1126/science.1145053. [DOI] [PubMed] [Google Scholar]
- Bunning E. Common features of photoperiodism in plants and animals. Photochem Photobiol. 1969;9:219–228. doi: 10.1111/j.1751-1097.1969.tb07286.x. [DOI] [PubMed] [Google Scholar]
- Busza A, Emery-Le M, Rosbash M, Emery P. Roles of the two Drosophila CRYPTOCHROME structural domains in circadian photoreception. Science. 2004;304:1503–1506. doi: 10.1126/science.1096973. [DOI] [PubMed] [Google Scholar]
- Busza A, Murad A, Emery P. Interactions between circadian neurons control temperature synchronization of Drosophila behavior. J Neurosci. 2007;27:10722–10733. doi: 10.1523/JNEUROSCI.2479-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Hardin PE. Drosophila photoreceptors contain an autonomous circadian oscillator that can function without period mRNA cycling. J Neurosci. 1998;18:741–750. doi: 10.1523/JNEUROSCI.18-02-00741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claridge-Chang A, Wijnen H, Naef F, Boothroyd C, Rajewsky N, Young MW. Circadian regulation of gene expression systems in the Drosophila head. Neuron. 2001;32:657–671. doi: 10.1016/s0896-6273(01)00515-3. [DOI] [PubMed] [Google Scholar]
- Collins B, Mazzoni EO, Stanewsky R, Blau J. Drosophila CRYPTOCHROME is a circadian transcriptional repressor. Curr Biol. 2006;16:441–449. doi: 10.1016/j.cub.2006.01.034. [DOI] [PubMed] [Google Scholar]
- Collins BH, Rosato E, Kyriacou CP. Seasonal behavior in Drosophila melanogaster requires the photoreceptors, the circadian clock, and phospholipase C. Proc Natl Acad Sci U S A. 2004;101:1945–1950. doi: 10.1073/pnas.0308240100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell CS. Circadian modulation of calcium levels in cells in the suprachiasmatic nucleus. Eur J Neurosci. 2000;12:571–576. doi: 10.1046/j.1460-9568.2000.00939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyran SA, Buchsbaum AM, Reddy KL, Lin MC, Glossop NR, Hardin PE, Young MW, Storti RV, Blau J. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell. 2003;112:329–341. doi: 10.1016/s0092-8674(03)00074-6. [DOI] [PubMed] [Google Scholar]
- Cyran SA, Yiannoulos G, Buchsbaum AM, Saez L, Young MW, Blau J. The double-time protein kinase regulates the subcellular localization of the Drosophila clock protein period. J Neurosci. 2005;25:5430–5437. doi: 10.1523/JNEUROSCI.0263-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daan S, Albrecht U, van der Horst GT, Illnerova H, Roenneberg T, Wehr TA, Schwartz WJ. Assembling a clock for all seasons: are there M and E oscillators in the genes? J Biol Rhythms. 2001;16:105–116. doi: 10.1177/074873001129001809. [DOI] [PubMed] [Google Scholar]
- Davis FC, Gorski RA. Unilateral lesions of the hamster suprachiasmatic nuclei: Evidence for redundant control of circadian rhythms. J Comp Physiol [A] 1984;154:221–232. [Google Scholar]
- de la Iglesia HO, Cambras T, Schwartz WJ, Diez-Noguera A. Forced desynchronization of dual circadian oscillators within the rat suprachiasmatic nucleus. Curr Biol. 2004;14:796–800. doi: 10.1016/j.cub.2004.04.034. [DOI] [PubMed] [Google Scholar]
- de la Paz Fernandez M, Chu J, Villella A, Atkinson N, Kay SA, Ceriani MF. Impaired clock output by altered connectivity in the circadian network. Proc Natl Acad Sci U S A. 2007;104:5650–5655. doi: 10.1073/pnas.0608260104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissel S, Codd V, Fedic R, Garner KJ, Costa R, Kyriacou CP, Rosato E. A constitutively active cryptochrome in Drosophila melanogaster. Nat Neurosci. 2004;7:834–840. doi: 10.1038/nn1285. [DOI] [PubMed] [Google Scholar]
- Edery I, Zwiebel LJ, Dembinska ME, Rosbash M. Temporal phosphorylation of the Drosophila period protein. Proc Natl Acad Sci U S A. 1994;91:2260–2264. doi: 10.1073/pnas.91.6.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery P, Stanewsky R, Helfrich-Forster C, Emery-Le M, Hall JC, Rosbash M. Drosophila CRY is a deep brain circadian photoreceptor. Neuron. 2000;26:493–504. doi: 10.1016/s0896-6273(00)81181-2. [DOI] [PubMed] [Google Scholar]
- Etter PD, Ramaswami M. The ups and downs of daily life: profiling circadian gene expression in Drosophila. Bioessays. 2002;24:494–498. doi: 10.1002/bies.10109. [DOI] [PubMed] [Google Scholar]
- Ewer J, Frisch B, Hamblen-Coyle MJ, Rosbash M, Hall JC. Expression of the period clock gene within different cell types in the brain of Drosophila adults and mosaic analysis of these cells’ influence on circadian behavioral rhythms. J Neurosci. 1992;12:3321–3349. doi: 10.1523/JNEUROSCI.12-09-03321.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Hida A, Anderson DA, Izumo M, Johnson CH. Cycling of CRYPTOCHROME proteins is not necessary for circadian-clock function in mammalian fibroblasts. Curr Biol. 2007;17:1091–1100. doi: 10.1016/j.cub.2007.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Sathyanarayanan S, Sehgal A. Post-translational regulation of the Drosophila circadian clock requires protein phosphatase 1 (PP1) Genes Dev. 2007;21:1506–1518. doi: 10.1101/gad.1541607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch B, Hardin PE, Hamblen-Coyle MJ, Rosbash M, Hall JC. A promoterless period gene mediates behavioral rhythmicity and cyclical per expression in a restricted subset of the Drosophila nervous system. Neuron. 1994;12:555–570. doi: 10.1016/0896-6273(94)90212-7. [DOI] [PubMed] [Google Scholar]
- Glaser FT, Stanewsky R. Temperature synchronization of the Drosophila circadian clock. Curr Biol. 2005;15:1352–1363. doi: 10.1016/j.cub.2005.06.056. [DOI] [PubMed] [Google Scholar]
- Glossop NR, Lyons LC, Hardin PE. Interlocked feedback loops within the Drosophila circadian oscillator. Science. 1999;286:766–768. doi: 10.1126/science.286.5440.766. [DOI] [PubMed] [Google Scholar]
- Glossop NR, Houl JH, Zheng H, Ng FS, Dudek SM, Hardin PE. VRILLE feeds back to control circadian transcription of Clock in the Drosophila circadian oscillator. Neuron. 2003;37:249–261. doi: 10.1016/s0896-6273(03)00002-3. [DOI] [PubMed] [Google Scholar]
- Gorska-Andrzejak J, Keller A, Raabe T, Kilianek L, Pyza E. Structural daily rhythms in GFP-labelled neurons in the visual system of Drosophila melanogaster. Photochem Photobiol Sci. 2005;4:721–726. doi: 10.1039/b417023g. [DOI] [PubMed] [Google Scholar]
- Green DJ, Gillette R. Circadian rhythm of firing rate recorded from single cells in the rat suprachiasmatic brain slice. Brain Res. 1982;245:198–200. doi: 10.1016/0006-8993(82)90361-4. [DOI] [PubMed] [Google Scholar]
- Grima B, Lamouroux A, Chelot E, Papin C, Limbourg-Bouchon B, Rouyer F. The F-box protein slimb controls the levels of clock proteins period and timeless. Nature. 2002;420:178–182. doi: 10.1038/nature01122. [DOI] [PubMed] [Google Scholar]
- Grima B, Chelot E, Xia R, Rouyer F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature. 2004;431:869–873. doi: 10.1038/nature02935. [DOI] [PubMed] [Google Scholar]
- Hardin PE, Hall JC, Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- Hardin PE. Essential and expendable features of the circadian timekeeping mechanism. Curr Opin Neurobiol. 2006;16:686–692. doi: 10.1016/j.conb.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Harms E, Kivimae S, Young MW, Saez L. Posttranscriptional and posttranslational regulation of clock genes. J Biol Rhythms. 2004;19:361–373. doi: 10.1177/0748730404268111. [DOI] [PubMed] [Google Scholar]
- Harrington M, Molyneux P, Soscia S, Prabakar C, McKinley-Brewer J, Lall G. Behavioral and neurochemical sources of variability of circadian period and phase: studies of circadian rhythms of npy−/− mice. Am J Physiol Regul Integr Comp Physiol. 2007;292:R1306–1314. doi: 10.1152/ajpregu.00383.2006. [DOI] [PubMed] [Google Scholar]
- Helfrich-Forster C, Homberg U. Pigment-dispersing hormone-immunoreactive neurons in the nervous system of wild-type Drosophila melanogaster and of several mutants with altered circadian rhythmicity. J Comp Neurol. 1993;337:177–190. doi: 10.1002/cne.903370202. [DOI] [PubMed] [Google Scholar]
- Helfrich-Forster C. The period clock gene is expressed in central nervous system neurons which also produce a neuropeptide that reveals the projections of circadian pacemaker cells within the brain of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1995;92:612–616. doi: 10.1073/pnas.92.2.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Forster C. Development of pigment-dispersing hormone-immunoreactive neurons in the nervous system of Drosophila melanogaster. J Comp Neurol. 1997;380:335–354. doi: 10.1002/(sici)1096-9861(19970414)380:3<335::aid-cne4>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Helfrich-Forster C, Stengl M, Homberg U. Organization of the circadian system in insects. Chronobiol Int. 1998;15:567–594. doi: 10.3109/07420529808993195. [DOI] [PubMed] [Google Scholar]
- Helfrich-Forster C. Differential control of morning and evening components in the activity rhythm of Drosophila melanogaster—sex-specific differences suggest a different quality of activity. J Biol Rhythms. 2000;15:135–154. doi: 10.1177/074873040001500208. [DOI] [PubMed] [Google Scholar]
- Helfrich-Forster C, Winter C, Hofbauer A, Hall JC, Stanewsky R. The circadian clock of fruit flies is blind after elimination of all known photoreceptors. Neuron. 2001;30:249–261. doi: 10.1016/s0896-6273(01)00277-x. [DOI] [PubMed] [Google Scholar]
- Helfrich-Forster C. The circadian system of Drosophila melanogaster and its light input pathways. Zoology (Jena) 2002;105:297–312. doi: 10.1078/0944-2006-00074. [DOI] [PubMed] [Google Scholar]
- Helfrich-Forster C, Edwards T, Yasuyama K, Wisotzki B, Schneuwly S, Stanewsky R, Meinertzhagen IA, Hofbauer A. The extraretinal eyelet of Drosophila: development, ultrastructure, and putative circadian function. J Neurosci. 2002a;22:9255–9266. doi: 10.1523/JNEUROSCI.22-21-09255.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Forster C, Wulf J, de Belle JS. Mushroom body influence on locomotor activity and circadian rhythms in Drosophila melanogaster. J Neurogenet. 2002b;16:73–109. doi: 10.1080/01677060213158. [DOI] [PubMed] [Google Scholar]
- Helfrich-Forster C. The neuroarchitecture of the circadian clock in the brain of Drosophila melanogaster. Microsc Res Tech. 2003;62:94–102. doi: 10.1002/jemt.10357. [DOI] [PubMed] [Google Scholar]
- Helfrich-Forster C. Neurobiology of the fruit fly’s circadian clock. Genes Brain Behav. 2005;4:65–76. doi: 10.1111/j.1601-183X.2004.00092.x. [DOI] [PubMed] [Google Scholar]
- Helfrich-Forster C, Shafer OT, Wulbeck C, Grieshaber E, Rieger D, Taghert P. Development and morphology of the clock-gene-expressing lateral neurons of Drosophila melanogaster. J Comp Neurol. 2007;500:47–70. doi: 10.1002/cne.21146. [DOI] [PubMed] [Google Scholar]
- Helfrich C. Role of the optic lobes in the regulation of the locomotor activity rhythm of Drosophila melanogaster: behavioral analysis of neural mutants. J Neurogenet. 1986;3:321–343. doi: 10.3109/01677068609106857. [DOI] [PubMed] [Google Scholar]
- Hendricks JC, Finn SM, Panckeri KA, Chavkin J, Williams JA, Sehgal A, Pack AI. Rest in Drosophila is a sleep-like state. Neuron. 2000;25:129–138. doi: 10.1016/s0896-6273(00)80877-6. [DOI] [PubMed] [Google Scholar]
- Herzog ED, Takahashi JS, Block GD. Clock controls circadian period in isolated suprachiasmatic nucleus neurons. Nat Neurosci. 1998;1:708–713. doi: 10.1038/3708. [DOI] [PubMed] [Google Scholar]
- Hong SF, Saunders DS. Internal desynchronization in blow fly (Calliphora vicina) locomotor activity rhythms: Evidence for a complex circadian pacemaker. Biol Rhythm Res. 1998;29:387–396. [Google Scholar]
- Howlader G, Paranjpe DA, Sharma VK. Non-ventral lateral neuron-based, non-PDF-mediated clocks control circadian egg-laying rhythm in Drosophila melanogaster. J Biol Rhythms. 2006;21:13–20. doi: 10.1177/0748730405282882. [DOI] [PubMed] [Google Scholar]
- Howlader G, Sharma VK. Circadian regulation of egg-laying behavior in fruit flies Drosophila melanogaster. J Insect Physiol. 2006;52:779–785. doi: 10.1016/j.jinsphys.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Hunter-Ensor M, Ousley A, Sehgal A. Regulation of the Drosophila protein timeless suggests a mechanism for resetting the circadian clock by light. Cell. 1996;84:677–685. doi: 10.1016/s0092-8674(00)81046-6. [DOI] [PubMed] [Google Scholar]
- Hyun S, Lee Y, Hong ST, et al. Drosophila GPCR Han is a receptor for the circadian clock neuropeptide PDF. Neuron. 2005;48:267–278. doi: 10.1016/j.neuron.2005.08.025. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Sugiyama T, Wallace CS, Gompf HS, Yoshioka T, Miyawaki A, Allen CN. Circadian dynamics of cytosolic and nuclear Ca2+ in ingle suprachiasmatic nucleus neurons. Neuron. 2003;38:253–263. doi: 10.1016/s0896-6273(03)00164-8. [DOI] [PubMed] [Google Scholar]
- Inouye ST, Kawamura H. Persistence of circadian rhythmicity in a mammalian hypothalamic ”island” containing the suprachiasmatic nucleus. Proc Natl Acad Sci U S A. 1979;76:5962–5966. doi: 10.1073/pnas.76.11.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itri JN, Michel S, Vansteensel MJ, Meijer JH, Colwell CS. Fast delayed rectifier potassium current is required for circadian neural activity. Nat Neurosci. 2005;8:650–656. doi: 10.1038/nn1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joiner WJ, Crocker A, White BH, Sehgal A. Sleep in Drosophila is regulated by adult mushroom bodies. Nature. 2006;441:757–760. doi: 10.1038/nature04811. [DOI] [PubMed] [Google Scholar]
- Kadener S, Stoleru D, McDonald M, Nawathean P, Rosbash M. Clockwork Orange is a transcriptional repressor and a new Drosophila circadian pacemaker component. Genes Dev. 2007;21:1675–1686. doi: 10.1101/gad.1552607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M, Helfrich-Forster C, Hall JC. Spatial and temporal expression of the period and timeless genes in the developing nervous system of Drosophila: newly identified pacemaker candidates and novel features of clock gene product cycling. J Neurosci. 1997;17:6745–6760. doi: 10.1523/JNEUROSCI.17-17-06745.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M. Neural substrates of Drosophila rhythms revealed by mutants and molecular manipulations. Curr Opin Neurobiol. 1998;8:652–658. doi: 10.1016/s0959-4388(98)80095-0. [DOI] [PubMed] [Google Scholar]
- Kaneko M, Hall JC. Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J Comp Neurol. 2000;422:66–94. doi: 10.1002/(sici)1096-9861(20000619)422:1<66::aid-cne5>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Kaneko M, Hernandez-Borsetti N, Cahill GM. Diversity of zebrafish peripheral oscillators revealed by luciferase reporting. Proc Natl Acad Sci U S A. 2006;103:14614–14619. doi: 10.1073/pnas.0606563103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik R, Nawathean P, Busza A, Murad A, Emery P, Rosbash M. PER-TIM Interactions with the Photoreceptor Cryptochrome Mediate Circadian Temperature Responses in Drosophila. PLoS Biol. 2007;5:e146. doi: 10.1371/journal.pbio.0050146. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kim EY, Bae K, Ng FS, Glossop NR, Hardin PE, Edery I. Drosophila CLOCK protein is under posttranscriptional control and influences light-induced activity. Neuron. 2002;34:69–81. doi: 10.1016/s0896-6273(02)00639-6. [DOI] [PubMed] [Google Scholar]
- Kim EY, Edery I. Balance between DBT/CKIepsilon kinase and protein phosphatase activities regulate phosphorylation and stability of Drosophila CLOCK protein. Proc Natl Acad Sci U S A. 2006;103:6178–6183. doi: 10.1073/pnas.0511215103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyohara YB, Tagao S, Tamanini F, et al. The BMAL1 C terminus regulates the circadian transcription feedback loop. Proc Natl Acad Sci U S A. 2006;103:10074–10079. doi: 10.1073/pnas.0601416103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klarsfeld A, Leloup JC, Rouyer F. Circadian rhythms of locomotor activity in Drosophila. Behav Processes. 2003;64:161–175. doi: 10.1016/s0376-6357(03)00133-5. [DOI] [PubMed] [Google Scholar]
- Klarsfeld A, Malpel S, Michard-Vanhee C, Picot M, Chelot E, Rouyer F. Novel features of cryptochrome-mediated photoreception in the brain circadian clock of Drosophila. J Neurosci. 2004;24:1468–1477. doi: 10.1523/JNEUROSCI.3661-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloss B, Price JL, Saez L, Blau J, Rothenfluh A, Wesley CS, Young MW. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iepsilon. Cell. 1998;94:97–107. doi: 10.1016/s0092-8674(00)81225-8. [DOI] [PubMed] [Google Scholar]
- Ko HW, Jiang J, Edery I. Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature. 2002;420:673–678. doi: 10.1038/nature01272. [DOI] [PubMed] [Google Scholar]
- Koehler WK, Fleissner G. Internal desynchronisation of bilaterally organised circadian oscillators in the visual system of insects. Nature. 1978;274:708–710. doi: 10.1038/274708a0. [DOI] [PubMed] [Google Scholar]
- Koh K, Zheng X, Sehgal A. JETLAG resets the Drosophila circadian clock by promoting light-induced degradation of TIMELESS. Science. 2006;312:1809–1812. doi: 10.1126/science.1124951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1971;68:2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka RJ, Pittendrigh C, Orr D. Reciprocal behaviour associated with altered homeostasis and photosensitivity of Drosophila clock mutants. J Neurogenet. 1989;6:1–10. doi: 10.3109/01677068909107096. [DOI] [PubMed] [Google Scholar]
- Krishnan B, Dryer SE, Hardin PE. Circadian rhythms in olfactory responses of Drosophila melanogaster. Nature. 1999;400:375–378. doi: 10.1038/22566. [DOI] [PubMed] [Google Scholar]
- Kuhlman SJ, McMahon DG. Encoding the ins and outs of circadian pacemaking. J Biol Rhythms. 2006;21:470–481. doi: 10.1177/0748730406294316. [DOI] [PubMed] [Google Scholar]
- Lakin-Thomas PL. Transcriptional feedback oscillators: maybe, maybe not. J Biol Rhythms. 2006;21:83–92. doi: 10.1177/0748730405286102. [DOI] [PubMed] [Google Scholar]
- Lear BC, Merrill CE, Lin JM, Schroeder A, Zhang L, Allada R. A G protein-coupled receptor, groom-of-PDF, is required for PDF neuron action in circadian behavior. Neuron. 2005;48:221–227. doi: 10.1016/j.neuron.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Lee G, Bahn JH, Park JH. Sex- and clock-controlled expression of the neuropeptide F gene in Drosophila. Proc Natl Acad Sci U S A. 2006;103:12580–12585. doi: 10.1073/pnas.0601171103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leise TL, Moin EE. A mathematical model of the Drosophila circadian clock with emphasis on posttranslational mechanisms. J Theor Biol. 2007;248:48–63. doi: 10.1016/j.jtbi.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Lim C, Chung BY, Pitman JL, McGill JJ, Pradhan S, Lee J, Keegan KP, Choe J, Allada R. Clockwork orange encodes a transcriptional repressor important for circadian-clock amplitude in Drosophila. Curr Biol. 2007;17:1082–1089. doi: 10.1016/j.cub.2007.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JM, Kilman VL, Keegan K, Paddock B, Emery-Le M, Rosbash M, Allada R. A role for casein kinase 2alpha in the Drosophila circadian clock. Nature. 2002;420:816–820. doi: 10.1038/nature01235. [DOI] [PubMed] [Google Scholar]
- Lin Y, Stormo GD, Taghert PH. The neuropeptide pigment-dispersing factor coordinates pacemaker interactions in the Drosophila circadian system. J Neurosci. 2004;24:7951–7957. doi: 10.1523/JNEUROSCI.2370-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Ding JM, Faiman LE, Gillette MU. Coupling of muscarinic cholinergic receptors and cGMP in nocturnal regulation of the suprachiasmatic circadian clock. J Neurosci. 1997;17:659–666. doi: 10.1523/JNEUROSCI.17-02-00659.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majercak J, Sidote D, Hardin PE, Edery I. How a circadian clock adapts to seasonal decreases in temperature and day length. Neuron. 1999;24:219–230. doi: 10.1016/s0896-6273(00)80834-x. [DOI] [PubMed] [Google Scholar]
- Majercak J, Chen WF, Edery I. Splicing of the period gene 3′-terminal intron is regulated by light, circadian clock factors, and phospholipase C. Mol Cell Biol. 2004;24:3359–3372. doi: 10.1128/MCB.24.8.3359-3372.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malpel S, Klarsfeld A, Rouyer F. Larval optic nerve and adult extra-retinal photoreceptors sequentially associate with clock neurons during Drosophila brain development. Development. 2002;129:1443–1453. doi: 10.1242/dev.129.6.1443. [DOI] [PubMed] [Google Scholar]
- Martinek S, Inonog S, Manoukian AS, Young MW. A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell. 2001;105:769–779. doi: 10.1016/s0092-8674(01)00383-x. [DOI] [PubMed] [Google Scholar]
- Mason R. The effects of continuous light exposure on Syrian hamster suprachiasmatic (SCN) neuronal discharge activity in vitro. Neurosci Lett. 1991;123:160–163. doi: 10.1016/0304-3940(91)90920-o. [DOI] [PubMed] [Google Scholar]
- Matsumoto A, Matsumoto N, Harui Y, Sakamoto M, Tomioka K. Light and temperature cooperate to regulate the circadian locomotor rhythm of wild type and period mutants of Drosophila melanogaster. J Insect Physiol. 1998;44:587–596. doi: 10.1016/s0022-1910(98)00046-8. [DOI] [PubMed] [Google Scholar]
- Matsumoto A, Ukai-Tadenuma M, Yamada RG, et al. A functional genomics strategy reveals clockwork orange as a transcriptional regulator in the Drosophila circadian clock. Genes Dev. 2007;21:1687–1700. doi: 10.1101/gad.1552207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoni EO, Desplan C, Blau J. Circadian pacemaker neurons transmit and modulate visual information to control a rapid behavioral response. Neuron. 2005;45:293–300. doi: 10.1016/j.neuron.2004.12.038. [DOI] [PubMed] [Google Scholar]
- McMahon DG, Block GD. The Bulla ocular circadian pacemaker. I. Pacemaker neuron membrane potential controls phase through a calcium-dependent mechanism. J Comp Physiol [A] 1987;161:335–346. doi: 10.1007/BF00603959. [DOI] [PubMed] [Google Scholar]
- Mehnert KI, Beramendi A, Elghazali F, Negro P, Kyriacou CP, Cantera R. Circadian changes in Drosophila motor terminals. Dev Neurobiol. 2007;67:415–421. doi: 10.1002/dneu.20332. [DOI] [PubMed] [Google Scholar]
- Meijer JH, van der Zee E, Dietz M. The effects of intraventricular carbachol injections on the free-running activity rhythm of the hamster. J Biol Rhythms. 1988;3:333–348. doi: 10.1177/074873048800300403. [DOI] [PubMed] [Google Scholar]
- Meijer JH, Daan S, Overkamp GJ, Hermann PM. The two-oscillator circadian system of tree shrews (Tupaia belangeri) and its response to light and dark pulses. J Biol Rhythms. 1990;5:1–16. doi: 10.1177/074873049000500101. [DOI] [PubMed] [Google Scholar]
- Meijer JH, Schaap J, Watanabe K, Albus H. Multiunit activity recordings in the suprachiasmatic nuclei: in vivo versus in vitro models. Brain Res. 1997;753:322–327. doi: 10.1016/s0006-8993(97)00150-9. [DOI] [PubMed] [Google Scholar]
- Mertens I, Vandingenen A, Johnson EC, Shafer OT, Li W, Trigg JS, De Loof A, Schoofs L, Taghert PH. PDF receptor signaling in Drosophila contributes to both circadian and geotactic behaviors. Neuron. 2005;48:213–219. doi: 10.1016/j.neuron.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Meyer P, Saez L, Young MW. PER-TIM interactions in living Drosophila cells: an interval timer for the circadian clock. Science. 2006;311:226–229. doi: 10.1126/science.1118126. [DOI] [PubMed] [Google Scholar]
- Michel S, Geusz ME, Zaritsky JJ, Block GD. Circadian rhythm in membrane conductance expressed in isolated neurons. Science. 1993;259:239–241. doi: 10.1126/science.8421785. [DOI] [PubMed] [Google Scholar]
- Miyasako Y, Umezaki Y, Tomioka K. Separate sets of cerebral clock neurons are responsible for light and temperature entrainment of Drosophila circadian locomotor rhythms. J Biol Rhythms. 2007;22:115–126. doi: 10.1177/0748730407299344. [DOI] [PubMed] [Google Scholar]
- Mori T, Williams DR, Byrne MO, Qin X, Egli M, McHaourab HS, Stewart PL, Johnson CH. Elucidating the Ticking of an In Vitro Circadian Clockwork. PLoS Biol. 2007;5:e93. doi: 10.1371/journal.pbio.0050093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murad A, Emery-Le M, Emery P. A subset of dorsal neurons modulates circadian behavior and light responses in Drosophila. Neuron. 2007;53:689–701. doi: 10.1016/j.neuron.2007.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers EM, Yu J, Sehgal A. Circadian control of eclosion: interaction between a central and peripheral clock in Drosophila melanogaster. Curr Biol. 2003;13:526–533. doi: 10.1016/s0960-9822(03)00167-2. [DOI] [PubMed] [Google Scholar]
- Naidoo N, Song W, Hunter-Ensor M, Sehgal A. A role for the proteasome in the light response of the timeless clock protein. Science. 1999;285:1737–1741. doi: 10.1126/science.285.5434.1737. [DOI] [PubMed] [Google Scholar]
- Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyama T, Kondo T. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- Nakamura W, Honma S, Shirakawa T, Honma K. Clock mutation lengthens the circadian period without damping rhythms in individual SCN neurons. Nat Neurosci. 2002;5:399–400. doi: 10.1038/nn843. [DOI] [PubMed] [Google Scholar]
- Nawathean P, Rosbash M. The doubletime and CKII kinases collaborate to potentiate Drosophila PER transcriptional repressor activity. Mol Cell. 2004;13:213–223. doi: 10.1016/s1097-2765(03)00503-3. [DOI] [PubMed] [Google Scholar]
- Nawathean P, Stoleru D, Rosbash M. A small conserved domain of Drosophila PERIOD is important for circadian phosphorylation, nuclear localization, and transcriptional repressor activity. Mol Cell Biol. 2007;27:5002–5013. doi: 10.1128/MCB.02338-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitabach MN, Blau J, Holmes TC. Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell. 2002;109:485–495. doi: 10.1016/s0092-8674(02)00737-7. [DOI] [PubMed] [Google Scholar]
- Nitabach MN, Holmes TC, Blau J. Membranes, ions, and clocks: testing the Njus-Sulzman-Hastings model of the circadian oscillator. Methods Enzymol. 2005a;393:682–693. doi: 10.1016/S0076-6879(05)93036-X. [DOI] [PubMed] [Google Scholar]
- Nitabach MN, Sheeba V, Vera DA, Blau J, Holmes TC. Membrane electrical excitability is necessary for the free-running larval Drosophila circadian clock. J Neurobiol. 2005b;62:1–13. doi: 10.1002/neu.20053. [DOI] [PubMed] [Google Scholar]
- Nitabach MN, Wu Y, Sheeba V, Lemon WC, Strumbos J, Zelensky PK, White BH, Holmes TC. Electrical hyperexcitation of lateral ventral pacemaker neurons desynchronizes downstream circadian oscillators in the fly circadian circuit and induces multiple behavioral periods. J Neurosci. 2006;26:479–489. doi: 10.1523/JNEUROSCI.3915-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon JP, Smale L. Individual differences in wheel-running rhythms are related to temporal and spatial patterns of activation of orexin A and B cells in a diurnal rodent (Arvicanthis niloticus) Neuroscience. 2004;127:25–34. doi: 10.1016/j.neuroscience.2004.04.052. [DOI] [PubMed] [Google Scholar]
- Njus D, Sulzman FM, Hastings JW. Membrane model for the circadian clock. Nature. 1974;248:116–120. doi: 10.1038/248116a0. [DOI] [PubMed] [Google Scholar]
- Njus D, Gooch VD, Mergenhagen D, Sulzman F, Hastings JW. Membranes and molecules in circadian systems. Fed Proc. 1976;35:2353–2357. [PubMed] [Google Scholar]
- Park D, Griffith LC. Electrophysiological and anatomical characterization of PDF-positive clock neurons in the intact adult Drosophila brain. J Neurophysiol. 2006;95:3955–3960. doi: 10.1152/jn.00117.2006. [DOI] [PubMed] [Google Scholar]
- Peng Y, Stoleru D, Levine JD, Hall JC, Rosbash M. Drosophila free-running rhythms require intercellular communication. PLoS Biol. 2003;1:E13. doi: 10.1371/journal.pbio.0000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennartz CM, de Jeu MT, Bos NP, Schaap J, Geurtsen AM. Diurnal modulation of pacemaker potentials and calcium current in the mammalian circadian clock. Nature. 2002;416:286–290. doi: 10.1038/nature728. [DOI] [PubMed] [Google Scholar]
- Peschel N, Veleri S, Stanewsky R. Veela defines a molecular link between Cryptochrome and Timeless in the light-input pathway to Drosophila’s circadian clock. Proc Natl Acad Sci U S A. 2006;103:17313–17318. doi: 10.1073/pnas.0606675103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petri B, Stengl M. Pigment-dispersing hormone shifts the phase of the circadian pacemaker of the cockroach Leucophaea maderae. J Neurosci. 1997;17:4087–4093. doi: 10.1523/JNEUROSCI.17-11-04087.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard GE, Turek FW. Splitting of the circadian rhythm of activity is abolished by unilateral lesions of the suprachiasmatic nuclei. Science. 1982;215:1119–1121. doi: 10.1126/science.7063843. [DOI] [PubMed] [Google Scholar]
- Pickard GE, Turek FW, Sollars PJ. Light intensity and splitting in the golden hamster. Physiol Behav. 1993;54:1–5. doi: 10.1016/0031-9384(93)90035-e. [DOI] [PubMed] [Google Scholar]
- Pitman JL, McGill JJ, Keegan KP, Allada R. A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature. 2006;441:753–756. doi: 10.1038/nature04739. [DOI] [PubMed] [Google Scholar]
- Pittendrigh C, Daan S. A functional analysis of circadian pacemakers in nocturnal rodents. V. Pacemaker complexity: A clock for all seasons. J Comp Physiol. 1976;106:333–335. [Google Scholar]
- Pittendrigh CS. On temperature independence in the clock system controlling emergence time in Drosophila. Proc Natl Acad Sci U S A. 1954;40:1018–1029. doi: 10.1073/pnas.40.10.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittendrigh CS. Circadian rhythms and the circadian organization of living systems. Cold Spring Harb Symp Quant Biol. 1960;25:159–184. doi: 10.1101/sqb.1960.025.01.015. [DOI] [PubMed] [Google Scholar]
- Plautz JD, Kaneko M, Hall JC, Kay SA. Independent photoreceptive circadian clocks throughout Drosophila. Science. 1997;278:1632–1635. doi: 10.1126/science.278.5343.1632. [DOI] [PubMed] [Google Scholar]
- Pyza E, Meinertzhagen IA. Daily and circadian rhythms of synaptic frequency in the first visual neuropile of the housefly’s (Musca domestica L.) optic lobe. Proc Biol Sci. 1993;254:97–105. doi: 10.1098/rspb.1993.0133. [DOI] [PubMed] [Google Scholar]
- Pyza E, Meinertzhagen IA. Monopolar cell axons in the first optic neuropil of the housefly, Musca domestica L., undergo daily fluctuations in diameter that have a circadian basis. J Neurosci. 1995;15:407–418. doi: 10.1523/JNEUROSCI.15-01-00407.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyza E, Meinertzhagen IA. Circadian rhythms in screening pigment and invaginating organelles in photoreceptor terminals of the housefly’s first optic neuropile. J Neurobiol. 1997;32:517–529. [PubMed] [Google Scholar]
- Pyza E, Meinertzhagen IA. Daily rhythmic changes of cell size and shape in the first optic neuropil in Drosophila melanogaster. J Neurobiol. 1999;40:77–88. doi: 10.1002/(sici)1097-4695(199907)40:1<77::aid-neu7>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Quintero JE, Kuhlman SJ, McMahon DG. The biological clock nucleus: a multiphasic oscillator network regulated by light. J Neurosci. 2003;23:8070–8076. doi: 10.1523/JNEUROSCI.23-22-08070.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- Rieger D, Stanewsky R, Helfrich-Forster C. Cryptochrome, compound eyes, Hofbauer-Buchner eyelets, and ocelli play different roles in the entrainment and masking pathway of the locomotor activity rhythm in the fruit fly Drosophila melanogaster. J Biol Rhythms. 2003;18:377–391. doi: 10.1177/0748730403256997. [DOI] [PubMed] [Google Scholar]
- Rieger D, Shafer OT, Tomioka K, Helfrich-Forster C. Functional analysis of circadian pacemaker neurons in Drosophila melanogaster. J Neurosci. 2006;26:2531–2543. doi: 10.1523/JNEUROSCI.1234-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohling J, Wolters L, Meijer JH. Simulation of day-length encoding in the SCN: from single-cell to tissue-level organization. J Biol Rhythms. 2006;21:301–313. doi: 10.1177/0748730406290317. [DOI] [PubMed] [Google Scholar]
- Rosenwasser AM, Adler NT. Structure and function in circadian timing systems: evidence for multiple coupled circadian oscillators. Neurosci Biobehav Rev. 1986;10:431–448. doi: 10.1016/0149-7634(86)90005-9. [DOI] [PubMed] [Google Scholar]
- Rust MJ, Markson JS, Lane WS, Fisher DS, O’Shea EK. Ordered Phosphorylation Governs Oscillation of a Three-Protein Circadian Clock. Science. 2007;318:809–812. doi: 10.1126/science.1148596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Ishida N. Circadian rhythms of female mating activity governed by clock genes in Drosophila. Proc Natl Acad Sci U S A. 2001;98:9221–9225. doi: 10.1073/pnas.151443298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandrelli F, Tauber E, Pegoraro M, et al. A molecular basis for natural selection at the timeless locus in Drosophila melanogaster. Science. 2007;316:1898–1900. doi: 10.1126/science.1138426. [DOI] [PubMed] [Google Scholar]
- Saper CB, Lu J, Chou TC, Gooley J. The hypothalamic integrator for circadian rhythms. Trends Neurosci. 2005;28:152–157. doi: 10.1016/j.tins.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Sathyanarayanan S, Zheng X, Xiao R, Sehgal A. Posttranslational regulation of Drosophila PERIOD protein by protein phosphatase 2A. Cell. 2004;116:603–615. doi: 10.1016/s0092-8674(04)00128-x. [DOI] [PubMed] [Google Scholar]
- Saunders DS. The circadian basis of ovarian diapause regulation in Drosophila melanogaster: is the period gene causally involved in photoperiodic time measurement? J Biol Rhythms. 1990;5:315–331. doi: 10.1177/074873049000500404. [DOI] [PubMed] [Google Scholar]
- Saunders DS. Erwin Bunning and Tony Lees, two giants of chronobiology, and the problem of time measurement in insect photoperiodism. J Insect Physiol. 2005;51:599–608. doi: 10.1016/j.jinsphys.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Schaap J, Albus H, VanderLeest HT, Eilers PH, Detari L, Meijer JH. Heterogeneity of rhythmic suprachiasmatic nucleus neurons: Implications for circadian waveform and photoperiodic encoding. Proc Natl Acad Sci U S A. 2003;100:15994–15999. doi: 10.1073/pnas.2436298100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schardt U, Wilhelm I, Erkert HG. Splitting of the circadian activity rhythm in common marmosets (Callithrix j. jacchus; primates) Experientia. 1989;45:1112–1115. doi: 10.1007/BF01950173. [DOI] [PubMed] [Google Scholar]
- Schwartz MD, Nunez AA, Smale L. Differences in the suprachiasmatic nucleus and lower subparaventricular zone of diurnal and nocturnal rodents. Neuroscience. 2004;127:13–23. doi: 10.1016/j.neuroscience.2004.04.049. [DOI] [PubMed] [Google Scholar]
- Schwartz MD, Smale L. Individual differences in rhythms of behavioral sleep and its neural substrates in Nile grass rats. J Biol Rhythms. 2005;20:526–537. doi: 10.1177/0748730405280924. [DOI] [PubMed] [Google Scholar]
- Schwartz WJ, Gross RA, Morton MT. The suprachiasmatic nuclei contain a tetrodotoxin-resistant circadian pacemaker. Proc Natl Acad Sci U S A. 1987;84:1694–1698. doi: 10.1073/pnas.84.6.1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal A, Price JL, Man B, Young MW. Loss of circadian behavioral rhythms and per RNA oscillations in the Drosophila mutant timeless. Science. 1994;263:1603–1606. doi: 10.1126/science.8128246. [DOI] [PubMed] [Google Scholar]
- Shafer OT, Rosbash M, Truman JW. Sequential nuclear accumulation of the clock proteins period and timeless in the pacemaker neurons of Drosophila melanogaster. J Neurosci. 2002;22:5946–5954. doi: 10.1523/JNEUROSCI.22-14-05946.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer OT, Helfrich-Forster C, Renn SC, Taghert PH. Reevaluation of Drosophila melanogaster’s neuronal circadian pacemakers reveals new neuronal classes. J Comp Neurol. 2006;498:180–193. doi: 10.1002/cne.21021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287:1834–1837. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- Siwicki KK, Eastman C, Petersen G, Rosbash M, Hall JC. Antibodies to the period gene product of Drosophila reveal diverse tissue distribution and rhythmic changes in the visual system. Neuron. 1988;1:141–150. doi: 10.1016/0896-6273(88)90198-5. [DOI] [PubMed] [Google Scholar]
- Smale L, Lee T, Nunez AA. Mammalian diurnality: some facts and gaps. J Biol Rhythms. 2003;18:356–366. doi: 10.1177/0748730403256651. [DOI] [PubMed] [Google Scholar]
- Smietanko A, Engelmann W. Splitting of circadian rhythm of Musca domestica flies with azadirachtin. J Interdiscipl Cycle Res. 1989;20:71–79. [Google Scholar]
- Stanewsky R, Kaneko M, Emery P, Beretta B, Wager-Smith K, Kay SA, Rosbash M, Hall JC. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell. 1998;95:681–692. doi: 10.1016/s0092-8674(00)81638-4. [DOI] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Agosto J, Rosbash M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature. 2004;431:862–868. doi: 10.1038/nature02926. [DOI] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Nawathean P, Rosbash M. A resetting signal between Drosophila pacemakers synchronizes morning and evening activity. Nature. 2005;438:238–242. doi: 10.1038/nature04192. [DOI] [PubMed] [Google Scholar]
- Stoleru D, Nawathean P, Fernandez Mde L, Menet JS, Ceriani MF, Rosbash M. The Drosophila circadian network is a seasonal timer. Cell. 2007;129:207–219. doi: 10.1016/j.cell.2007.02.038. [DOI] [PubMed] [Google Scholar]
- Suh J, Jackson FR. Drosophila ebony activity is required in glia for the circadian regulation of locomotor activity. Neuron. 2007;55:435–447. doi: 10.1016/j.neuron.2007.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghert PH, Shafer OT. Mechanisms of clock output in the Drosophila circadian pacemaker system. J Biol Rhythms. 2006;21:445–457. doi: 10.1177/0748730406293910. [DOI] [PubMed] [Google Scholar]
- Tanoue S, Krishnan P, Krishnan B, Dryer SE, Hardin PE. Circadian clocks in antennal neurons are necessary and sufficient for olfaction rhythms in Drosophila. Curr Biol. 2004;14:638–649. doi: 10.1016/j.cub.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Tauber E, Zordan M, Sandrelli F, et al. Natural selection favors a newly derived timeless allele in Drosophila melanogaster. Science. 2007;316:1895–1898. doi: 10.1126/science.1138412. [DOI] [PubMed] [Google Scholar]
- Tomita J, Nakajima M, Kondo T, Iwasaki H. No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science. 2005;307:251–254. doi: 10.1126/science.1102540. [DOI] [PubMed] [Google Scholar]
- Ueda HR, Matsumoto A, Kawamura M, Iino M, Tanimura T, Hashimoto S. Genome-wide transcriptional orchestration of circadian rhythms in Drosophila. J Biol Chem. 2002;277:14048–14052. doi: 10.1074/jbc.C100765200. [DOI] [PubMed] [Google Scholar]
- Van Gelder RN. Timeless genes and jetlag. Proc Natl Acad Sci U S A. 2006;103:17583–17584. doi: 10.1073/pnas.0608751103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veleri S, Brandes C, Helfrich-Forster C, Hall JC, Stanewsky R. A self-sustaining, light-entrainable circadian oscillator in the Drosophila brain. Curr Biol. 2003;13:1758–1767. doi: 10.1016/j.cub.2003.09.030. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Young MW. Circadian rhythms in Drosophila can be driven by period expression in a restricted group of central brain cells. Neuron. 1995;15:345–360. doi: 10.1016/0896-6273(95)90039-x. [DOI] [PubMed] [Google Scholar]
- Welsh DK, Logothetis DE, Meister M, Reppert SM. Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron. 1995;14:697–706. doi: 10.1016/0896-6273(95)90214-7. [DOI] [PubMed] [Google Scholar]
- Wheeler DA, Hamblen-Coyle MJ, Dushay MS, Hall JC. Behavior in light-dark cycles of Drosophila mutants that are arrhythmic, blind, or both. J Biol Rhythms. 1993;8:67–94. doi: 10.1177/074873049300800106. [DOI] [PubMed] [Google Scholar]
- Wijnen H, Naef F, Boothroyd C, Claridge-Chang A, Young MW. Control of daily transcript oscillations in Drosophila by light and the circadian clock. PLoS Genet. 2006;2:e39. doi: 10.1371/journal.pgen.0020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Isejima H, Matsuo T, Okura R, Yagita K, Kobayashi M, Okamura H. Synchronization of cellular clocks in the suprachiasmatic nucleus. Science. 2003;302:1408–1412. doi: 10.1126/science.1089287. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- Yang Z, Sehgal A. Role of molecular oscillations in generating behavioral rhythms in Drosophila. Neuron. 2001;29:453–467. doi: 10.1016/s0896-6273(01)00218-5. [DOI] [PubMed] [Google Scholar]
- Yoshii T, Sakamoto M, Tomioka K. A temperature-dependent timing mechanism is involved in the circadian system that drives locomotor rhythms in the fruit fly Drosophila melanogaster. Zoolog Sci. 2002;19:841–850. doi: 10.2108/zsj.19.841. [DOI] [PubMed] [Google Scholar]
- Yoshii T, Funada Y, Ibuki-Ishibashi T, Matsumoto A, Tanimura T, Tomioka K. Drosophila cryb mutation reveals two circadian clocks that drive locomotor rhythm and have different responsiveness to light. J Insect Physiol. 2004;50:479–488. doi: 10.1016/j.jinsphys.2004.02.011. [DOI] [PubMed] [Google Scholar]
- Yoshii T, Heshiki Y, Ibuki-Ishibashi T, Matsumoto A, Tanimura T, Tomioka K. Temperature cycles drive Drosophila circadian oscillation in constant light that otherwise induces behavioural arrhythmicity. Eur J Neurosci. 2005;22:1176–1184. doi: 10.1111/j.1460-9568.2005.04295.x. [DOI] [PubMed] [Google Scholar]
- Yu W, Hardin PE. Circadian oscillators of Drosophila and mammals. J Cell Sci. 2006;119:4793–4795. doi: 10.1242/jcs.03174. [DOI] [PubMed] [Google Scholar]
- Yu W, Zheng H, Houl JH, Dauwalder B, Hardin PE. PER-dependent rhythms in CLK phosphorylation and E-box binding regulate circadian transcription. Genes Dev. 2006;20:723–733. doi: 10.1101/gad.1404406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q, Joiner WJ, Sehgal A. A sleep-promoting role for the Drosophila serotonin receptor 1A. Curr Biol. 2006;16:1051–1062. doi: 10.1016/j.cub.2006.04.032. [DOI] [PubMed] [Google Scholar]
- Zeng H, Qian Z, Myers MP, Rosbash M. A light-entrainment mechanism for the Drosophila circadian clock. Nature. 1996;380:129–135. doi: 10.1038/380129a0. [DOI] [PubMed] [Google Scholar]
- Zlomanczuk P, Margraf RR, Lynch GR. In vitro electrical activity in the suprachiasmatic nucleus following splitting and masking of wheel-running behavior. Levitan Irwin., editor. Brain Res. 1991;559:94–99. doi: 10.1016/0006-8993(91)90291-3. [DOI] [PubMed] [Google Scholar]