Summary
Human immunodeficiency virus (HIV-1) interaction with the primary receptor, CD4, induces conformational changes in the viral envelope glycoproteins that allow binding to the CCR5 second receptor and virus entry into the host cell. The small molecule NBD-556 mimics CD4 by binding the gp120 exterior envelope glycoprotein, moderately inhibiting virus entry into CD4-expressing target cells, and enhancing CCR5 binding and virus entry into CCR5-expressing cells lacking CD4. Studies of NBD-556 analogues and gp120 mutants suggest that: 1) NBD-556 binds within the Phe 43 cavity, a highly conserved, functionally important pocket formed as gp120 assumes the CD4-bound conformation; 2) the NBD-556 phenyl ring projects into the Phe 43 cavity; 3) enhancement of CD4-independent infection by NBD-556 requires the induction of conformational changes in gp120; and 4) increased affinity of NBD-556 analogues for gp120 improves antiviral potency during infection of CD4-expressing cells.
Introduction
Prevention of human immunodeficiency virus (HIV-1) transmission requires approaches that interrupt the early phase of retrovirus infection, before provirus formation. One early event, HIV-1 entry into target cells, involves the viral envelope glycoproteins, gp120 and gp41, and host cell receptors, CD4 and the chemokine receptors (either CCR5 or CXCR4) (Wyatt and Sodroski, 1998). Binding to CD4 induces conformational changes in the exterior envelope glycoprotein gp120 that allow CCR5/CXCR4 engagement and that expose elements of the gp41 transmembrane glycoprotein (Furuta et al., 1998; Trkola et al., 1996; Wu et al., 1996). Further conformational changes in gp41 lead to fusion of the viral and host cell membranes, allowing virus entry. Each of these steps represents a potential target for intervention.
To evade host antibody responses, HIV-1 has evolved envelope glycoproteins with surface variability, dense glycosylation and conformational flexibility (Kwong et al., 2002; Wyatt and Sodroski, 1998). The unliganded HIV-1 gp120 glycoprotein is unusually flexible and partially unstructured; binding CD4 locks gp120 into a rigid conformation (Myszka et al., 2000). This binding event is characterized by an unusually large and favorable enthalpy change that is partially countered by a large unfavorable entropy change. High-resolution structures of CD4-bound HIV-1 gp120 have provided insights into these conformational transitions (Kwong et al., 1998). The conserved gp120 core consists of an inner domain that interacts with gp41, an outer domain with a heavily glycosylated surface, and a bridging sheet that connects these two domains. On the unliganded HIV-1 gp120, the inner and outer gp120 domains are thought to move with respect to each other, with the bridging sheet assuming a conformation different from that seen in the CD4-bound state (Chen et al., 2005; Kwong et al., 1998).
CD4 primarily contacts the gp120 outer domain and bridging sheet (Kwong et al., 1998). CD4 binding creates a 153-Å3 cavity (“the Phe 43 cavity”) at the interface between gp120 and CD4; the Phe 43 cavity is bounded by highly conserved residues from all three gp120 domains and by a single CD4 residue, Phe 43 (Kwong et al., 1998). The contacts made by phenylalanine 43 and arginine 59 of CD4 with gp120 residues in the vestibule of this virtual pocket contribute significantly to gp120-CD4 affinity (Arthos et al., 1990; Ashkenazi et al., 1990; Brodsky et al., 1990; Fontenot et al., 2007; Kwong et al., 1998; Landau et al., 1988; Olshevsky et al., 1990). Thus, the Phe 43 cavity has been suggested as a desirable target for compounds that could disrupt gp120-CD4 interactions (Kwong et al., 1998).
NBD-556 is a 337-dalton compound identified in a screen for inhibitors of the gp120-CD4 interaction (Zhao et al., 2005)s. Remarkably, NBD-556 can also act as a CD4 agonist, inducing thermodynamic changes in gp120 similar to those observed upon CD4 binding (Schön et al., 2006). NBD-556 promotes CCR5 binding and can enhance HIV-1 entry into CD4-negative (CD4−) target cells expressing CCR5 (Schön et al., 2006). Here, we utilize gp120 mutants, molecular modeling, and NBD-556 analogues to test the hypothesis that NBD-556 interacts with the gp120 Phe 43 cavity. The relationship between the ability of NBD-556 analogues to induce changes in gp120 entropy and to stimulate entry-related conformational changes was examined. The potency of inhibition of infection of CD4-expressing target cells was related to the overall affinity of the NBD-556 analogues for gp120. These studies provide a foundation for understanding the interaction of small molecules with the CD4-binding site of HIV-1 gp120.
Results
NBD-556 can replace CD4 in HIV-1 infection
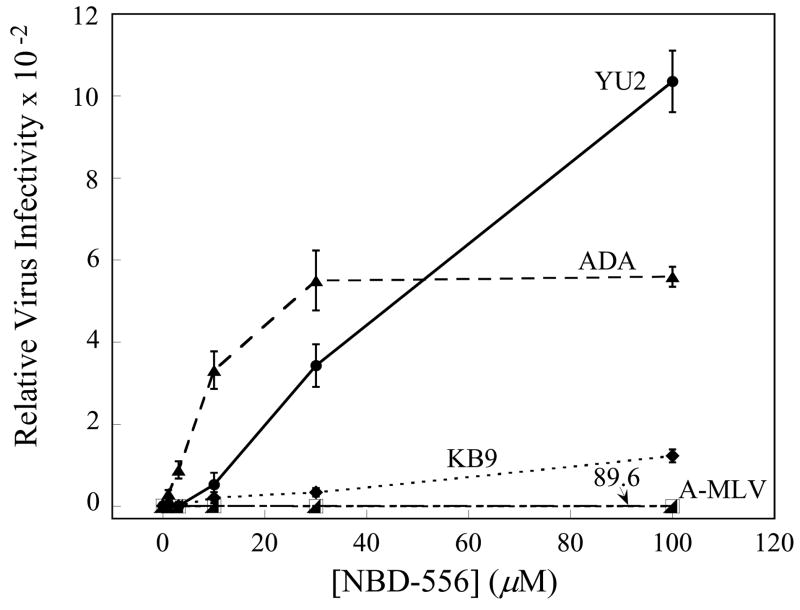
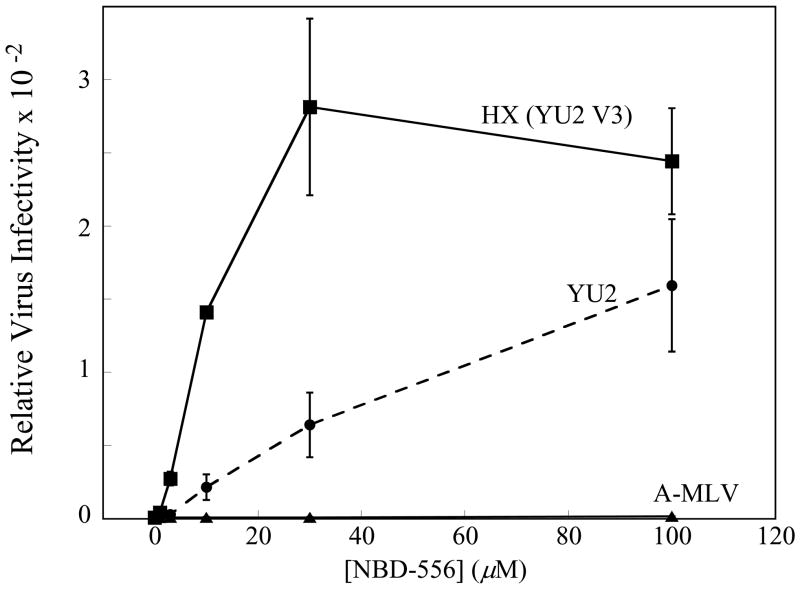
To examine the ability of NBD-556 to replace CD4 during HIV-1 infection, recombinant HIV-1 expressing firefly luciferase was pseudotyped with different envelope glycoproteins and incubated with CD4−CCR5+ Cf2Th-CCR5 cells in the presence of different concentrations of NBD-556. The envelope glycoproteins were derived from CCR5-using (R5) primary HIV-1 isolates (YU2 and ADA), dual-tropic (R5X4) primary isolates (89.6 and KB9) and, as a control, the amphotropic murine leukemia virus (A-MLV). NBD-556 enhanced infection of the Cf2Th-CCR5 cells in the following order of efficiency: YU2, ADA > KB9 > 89.6, A-MLV (Figure 1a). This order corresponds to the affinity of the gp120 glycoproteins from these HIV-1 isolates for CCR5 (Babcock et al., 2001; Karlsson et al., 1998; Staudinger et al., 2003; Wu et al., 1996), and suggests that a high coreceptor-binding affinity is required to achieve CD4-independent infection in this context. Consistent with this interpretation, NBD-556 did not enhance the entry of viruses with the KB9 envelope glycoproteins into CD4−CXCR4+ Cf2Th-CXCR4 cells (data not shown); the HIV-1 gp120 affinity for CXCR4 is significantly lower than that for CCR5 (Babcock et al., 2001). Likewise, the envelope glycoproteins of the laboratory-adapted CXCR4-using (X4) HIV-1 isolate, HXBc2, bound NBD-556 weakly (see below) and were not functionally enhanced by NBD-556 for entry into Cf2Th-CXCR4 cells (data not shown). However, NBD-556 dramatically enhanced infection of Cf2Th-CCR5 cells by viruses containing a chimeric HIV-1 envelope glycoprotein (HX(YU2 V3)) in which the HXBc2 gp120 third variable (V3) loop was replaced by that of the YU2 HIV-1 isolate (Figure 1b); this chimeric envelope glycoprotein exhibits a high affinity for the CCR5 coreceptor (Choe et al., 2003; Sullivan et al., 1998; Xiang et al., 2005). Thus, NBD-556 can replace CD4 during infection of CCR5+ cells by viruses with a variety of HIV-1 envelope glycoproteins, provided that the envelope glycoproteins exhibit sufficient affinity for CCR5.
Figure 1. NBD-556 enhancement of infection of CD4-negative cells by viruses with envelope glycoproteins from different HIV-1 strains.
a. The effect of incubating recombinant, luciferase-expressing HIV-1 bearing the envelope glycoproteins of the indicated HIV-1 strains with increasing concentrations of NBD-556 on infection of CD4-negative Cf2Th-CCR5 cells is shown. Relative virus infectivity represents the amount of infection detected in the presence of the indicated concentration of compound divided by the infection detected in the absence of compound. A recombinant HIV-1 with the A-MLV envelope glycoproteins is included as a control. The values shown are the means+/−SEM from a single experiment (n=3). The experiment was performed three times, with comparable results. b. The effect of incubating recombinant HIV-1 bearing the YU2, HXBc2 or HX(YU2 V3) envelope glycoproteins with increasing concentrations of NBD-556 on infection of Cf2Th-CCR5 cells is shown. The values shown are the means+/−SEM from a single experiment (n=3). The experiment was performed four times, with similar results.
NBD-556 interacts with the conserved core of the HIV-1 gp120 glycoprotein
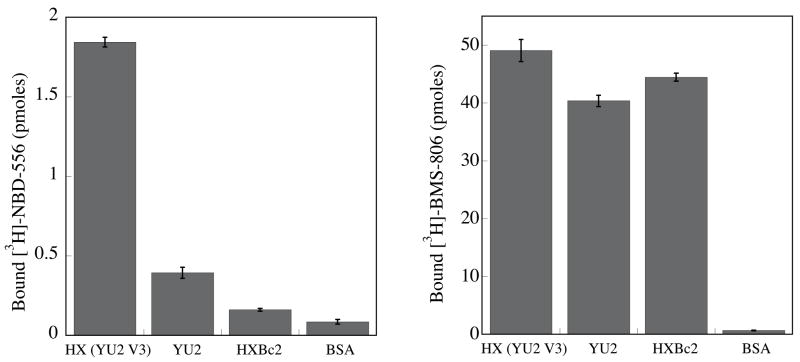
The direct binding of 3H-labeled NBD-556 to gp120 glycoprotein variants was measured (Figure 2a). NBD-556 bound YU2 gp120 efficiently, but HXBc2 gp120 only moderately. The chimeric HXBc2 gp120 with the substitution of the YU2 V3 loop bound NBD-556 with the highest efficiency. All three gp120 glycoproteins bound the control 3H-labeled BMS-806 similarly. These results are consistent with the observed susceptibility of viruses with these envelope glycoproteins to enhancement by NBD-556.
Figure 2. Binding of NBD-556 to the conserved core of HIV-1 gp120.
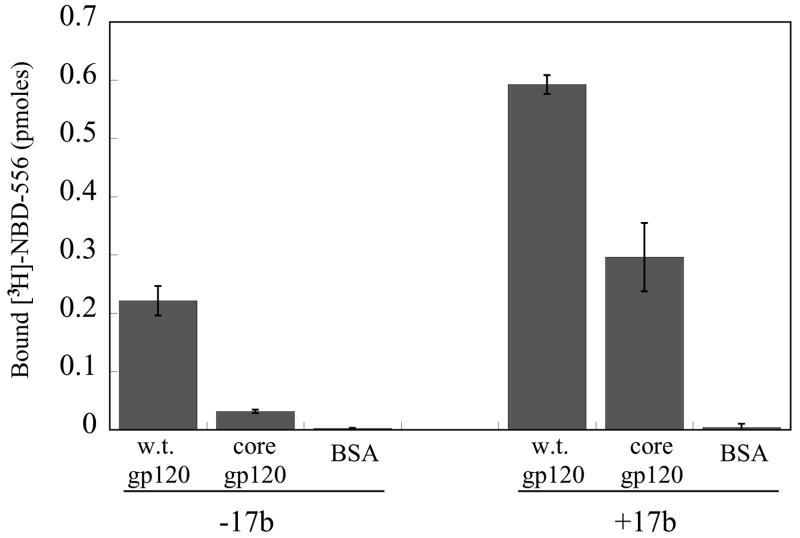
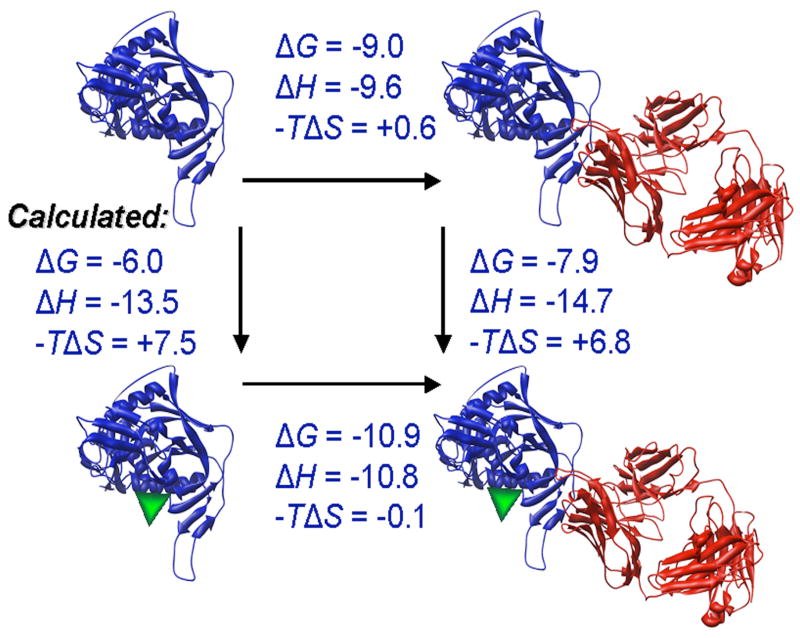
a. The binding of [3H]-NBD-556 to the indicated HIV-1 gp120 envelope glycoprotein variants or to bovine serum albumin (BSA) is shown (left panel). The right panel shows binding of the same gp120 envelope glycoproteins to [3H]-BMS-806 as a control. The values shown are the means+/−SEM from one experiment (n=3). b. The binding of [3H]-NBD-556 to the HIV-1YU2 w.t. gp120 glycoprotein or to the gp120 core protein in the absence or presence of the 17b antibody (1 mg/ml) is shown. Binding of [3H]-NBD-556 to the BSA control protein is shown for comparison. The values shown are the means +/− SEM from a single experiment (n=3). The experiment was performed three times with comparable results. c. The thermodynamic cycle for the binding of NBD-556 and the 17b antibody to the HIV-1YU2 gp120 core was studied by titrating the gp120 core with 17b in the presence of a saturating concentration of NBD-556. The values associated with the direct binding of NBD-556 to the gp120 core were calculated by completing the thermodynamic cycle. The structures of the gp120 core and the 17b Fab fragment (PDB entry 1GC1) are depicted in blue and red, respectively. NBD-556 is represented by the green triangle.
The gp120 core protein lacks the V1, V2 and V3 hypervariable loops and the N and C termini of gp120 (Kwong et al., 1998). The binding of [3H]-NBD-556 to the YU2 gp120 core was tested in the absence and presence of the 17b anti-gp120 antibody; the 17b antibody preferentially recognizes the CD4-bound conformation of gp120 and blocks chemokine receptor binding (Thali et al., 1991; Trkola et al., 1996; Wu et al., 1996). Weak binding of NBD-556 to the YU2 gp120 core was detected in the absence of the 17b antibody; NBD-556 binding to both full-length gp120 and the gp120 core was significantly enhanced by the addition of the 17b antibody (Figure 2b). The thermodynamic cycle of NBD-556 and 17b binding to the HIV-1YU2 gp120 core was determined by isothermal titration calorimetry (Figure 2c). NBD-556 bound weakly to the gp120 core (calculated Kd = 40 μM), but exhibited an approximately 25-fold increase in affinity for the gp120 core bound to the 17b antibody (Kd = 1.5 μM). Conversely, the 17b antibody preferentially recognized the NBD-556-bound form of the gp120 core. BIAcore analysis confirmed that NBD-556 could efficiently recognize the gp120 core-17b complex (Kd = 6.6 μM) (data not shown). Apparently, NBD-556 preferentially recognizes the CD4-bound conformation of HIV-1 gp120. Moreover, NBD-556 binds the relatively conserved portion of gp120 retained in the core molecule.
Modeling the NBD-556-gp120 interaction
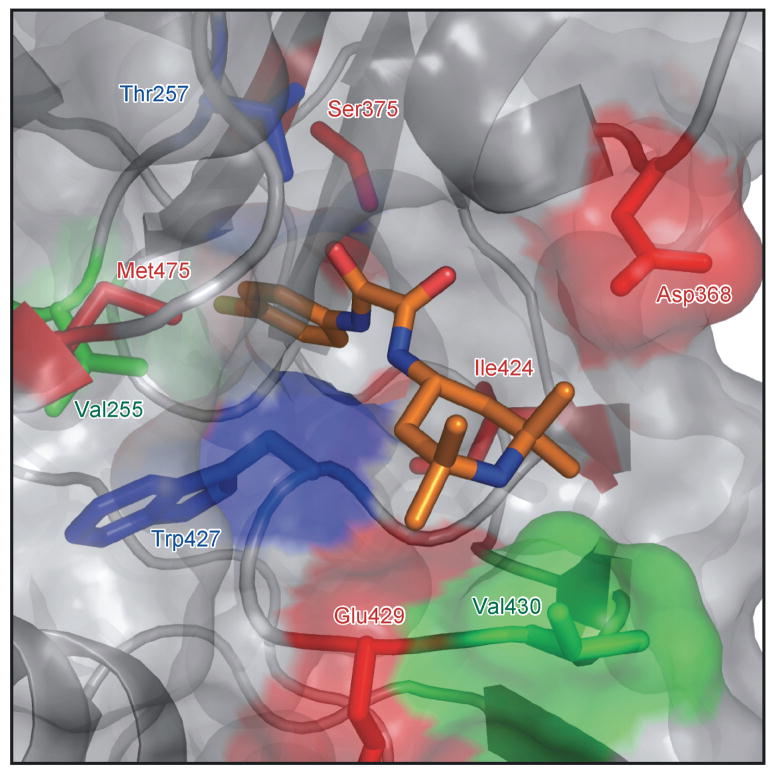
As NBD-556 preferentially recognizes the CD4-bound conformation of the HIV-1 gp120 core, we used the available x-ray crystal structures of gp120-CD4 complexes (Kwong et al., 1998) to model the binding of NBD-556 to HIV-1 gp120 in the CD4-bound state. Models produced independently using Glide (Friesner, 2004; Halgren and L. L.; Pollard, 2004), Gold (Jones, 1997) and Accelrys (Kuntz, 1994; Luty et al., 1995; Stouten et al., 1993) predicted remarkably similar binding modes, with the chloro-phenyl ring of NBD-556 projecting into the Phe 43 cavity of gp120 (Figure 3). Based on the predicted binding mode, the chloro-phenyl ring of NBD-556 sits 6.5 Å deeper in the Phe 43 cavity than the phenyl ring of Phe 43 of CD4. Predicted aromatic-aromatic stacking interactions between the NBD-556 chloro-phenyl ring and Trp 427, Phe 382 and Trp 112 likely stabilize the NBD-566-gp120 complex. Three other gp120 residues, Val 255, Thr 257 and Met 475 are within 4 Å of the NBD-556 chloro-phenyl group situated in the bottom of the gp120 cavity. Docking is also suggestive of hydrogen bonds between one of the NBD-556 oxalamide nitrogens and gp120 backbone carbonyls in the neck of the cavity.
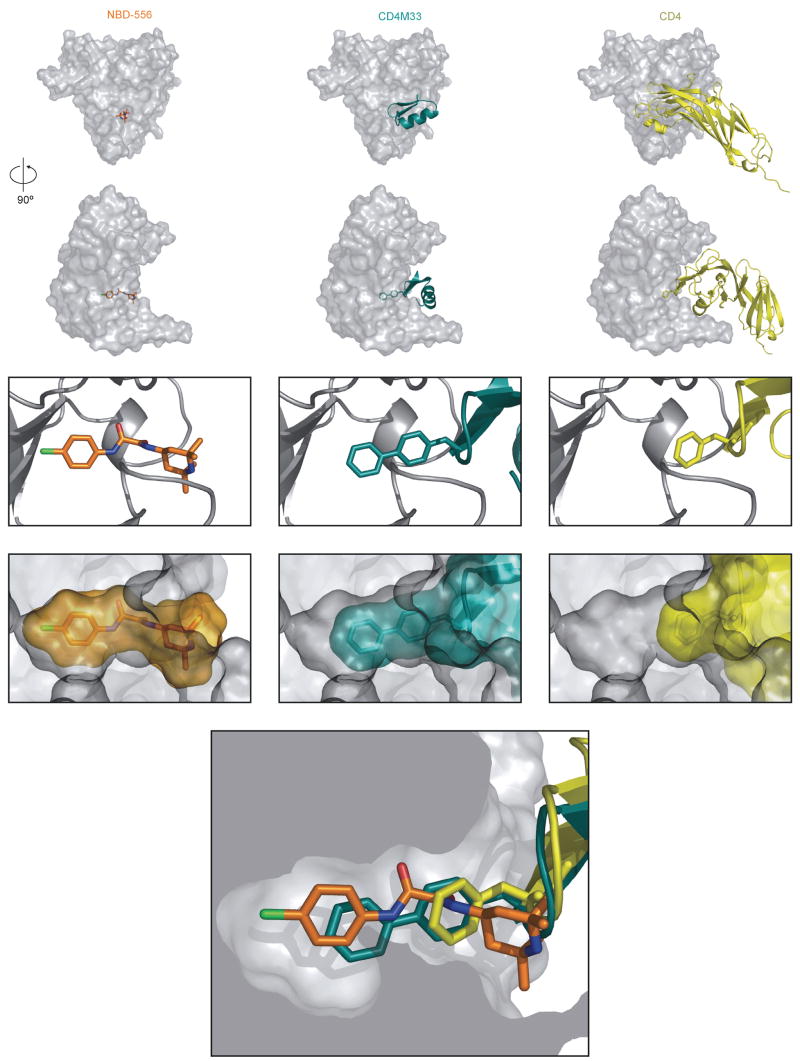
Figure 3. Interaction of NBD-556, CD4M33 and CD4 with the HIV-1 gp120 core.
In the upper row, the molecular surface of the HIV-1 gp120 core (Kwong et al., 1998) is shown, from the perspective of two-domain CD4, which is depicted as a yellow ribbon in the right panels. In the second row, the gp120 core has been rotated 90° around the vertical axis. In the middle column, the CD4-mimetic miniprotein CD4M33 (cyan) is docked onto the gp120 core, based on x-ray crystal structures of the CD4M33:gp120core:17b Fab complex (Huang et al., 2005). In the left column, the bound NBD-556 has been modeled by the Glide program (Friesner, 2004; Halgren et al., 2004). In the third row, a close-up view of the gp120 region surrounding the Phe 43 cavity is shown. The gp120 structure is shown as a gray ribbon. In the right panel, Phe 43 of CD4 is shown (yellow bonds). The biphenyl moiety of CD4M33 (cyan bonds) is shown in the middle panel. In the left panel, NBD-556 is shown, with bonds colored according to the atom type (green = chlorine; orange = carbon; blue = nitrogen; red = oxygen). In the fourth row, the molecular surfaces of NBD-556 (orange), CD4M33 (cyan), and CD4 (yellow) are shown, illustrating the extent to which these ligands fill the Phe 43 cavity (the gp120 surface is shown in gray). In the bottom figure, CD4 Phe 43 (yellow), the CD4M33 biphenyl group (cyan) and the modeled NBD-556 (multicolored) are positioned together in the Phe 43 cavity.
Scyllatoxin or charybdotoxin scaffolds have been used to create CD4-mimetic miniproteins (Vita et al., 1999; Zhang et al., 1999). X-ray crystal structures have demonstrated the similar ways in which Phe 23 of some scyllatoxin-based miniproteins and Phe 43 of CD4 contact gp120 (Huang et al., 2005). In one scyllatoxin derivative, CD4M33, a biphenyl group at residue 23 reaches into the Phe 43 cavity, increasing the affinity for gp120 (Huang et al., 2005; Martin et al., 2003). The predicted position of the chloro-phenyl ring in the bound NBD-556 models is similar to that of the distal phenyl ring of CD4M33 (Figure 3). However, the NBD-556 chloro-phenyl ring projects slightly deeper into the Phe 43 cavity than the CD4M33 biphenyl group.
NBD-556 phenyl ring/oxalamide linker modification
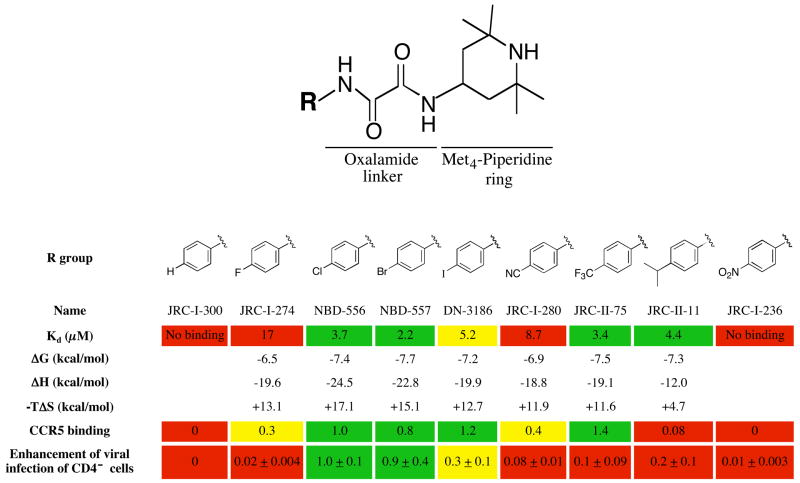
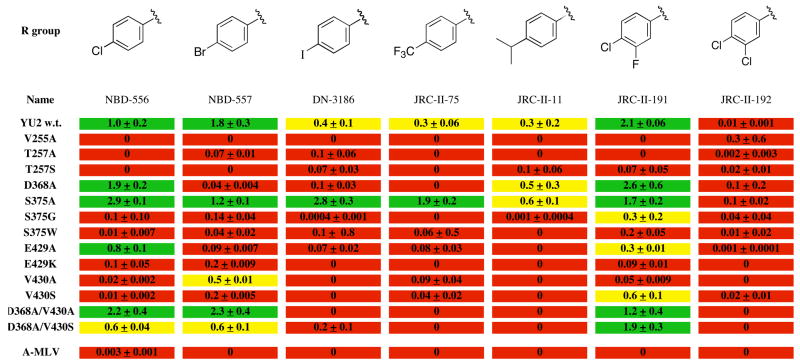
The models predict that changes in the NBD-556 phenyl ring and oxalamide linker will affect gp120 binding and/or functional mimicry of CD4. To test this, NBD-556 analogues were synthesized and tested for the ability to bind gp120 and to enhance CCR5 binding and infection of CCR5+ cells (Figure 4). NBD-557, which has a bromo group at the para position of the phenyl ring, bound gp120 and activated CCR5 binding and entry comparably to NBD-556. Para substitution of the phenyl ring chloro and bromo groups with either larger or smaller groups resulted in decreased enhancement of CCR5 binding and/or HIV-1 entry. The affinity of gp120 binding was sensitive to modifications at the para position of the phenyl ring; for example, the binding of JRC-I-300, having only a hydrogen atom in the para position, was too weak to be determined by isothermal titration calorimetry.
Figure 4. Structure-activity relationships of NBD-556 analogues with different para-phenyl substituents.
The values for Kd, ΔG, ΔH and −TΔS associated with the binding of each compound to the w.t. HIV-1YU2 gp120 glycoprotein were determined by isothermal titration calorimetry (see Methods). The Kd values (in μM) are color-coded as follows: red = no binding or >8; yellow = 5–8; green = <5. CCR5 binding of radiolabeled HIV-1YU2 gp120 was determined after incubation with 10 μM compound (see Methods). The induction of CCR5 binding by each compound was normalized to that observed for NBD-556. The relative induction of CCR5 binding is color-coded as follows: red = <0.25; yellow = 0.25–0.7; green = >0.7.
To study compound enhancement of infection of CD4−CCR5+ cells, recombinant, luciferase-expressing HIV-1 with the wild-type HIV-1YU2 envelope glycoproteins was incubated with increasing concentrations of the compound and then added to Cf2Th-CCR5 cells. Cells were lysed 48h later and luciferase activity measured (see methods). The area under the dose-response curve for each compound was calculated and normalized to the value obtained for NBD-556. The relative enhancement of infection of CCR5-expressing cells is color-coded as follows: red = 0–0.24; yellow = 0.25–0.7; green = >0.7.
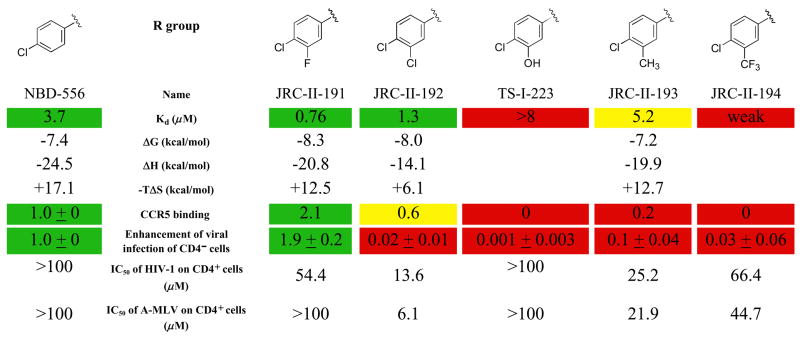
To explore additional options for modifying the interactions of NBD-556 with the Phe 43 cavity of gp120, different groups were substituted at the ortho and meta positions of the NBD-556 phenyl ring. NBD-556 was chosen over NBD-557 for these studies because of better solubility, which resulted in improved reproducibility in the biological assays. The size and nature of the group at the meta position significantly affected the affinity and virus-enhancing ability of the NBD-556 derivatives (Figure 5). JRC-II-191, with a fluoro group at the meta position, exhibited the highest affinity for gp120 and the most potent stimulation of HIV-1 infection of CCR5+ cells. Larger groups at the meta position resulted in decreases in both gp120 binding and viral enhancement (Figure 5 and data not shown). JRC-II-191 specifically inhibited HIV-1YU2 infection of cells expressing CD4 and CCR5 (Figure 5); this contrasts with several of the other meta-substituted analogues, which exhibited only non-specific inhibitory effects on infection of viruses with HIV-1 and A-MLV envelope glycoproteins. Ortho substitutions invariably resulted in loss of gp120 binding and viral enhancement (data not shown). Likewise, replacement of the phenyl ring and/or oxalamide linker resulted in compounds that did not detectably bind gp120 and exhibited no specific enhancement or inhibition of HIV-1 infection (data not shown). These results are consistent with modeling predictions in which the Phe 43 cavity constrains the nature and size of the phenyl and oxalamide substituents that can be tolerated in functionally active compounds.
Figure 5. Structure-activity relationships of NBD-556 analogues with different meta-phenyl substituents.
Values for Kd, thermodynamic parameters, CCR5 binding and enhancement of virus infection of CD4−CCR5+ cells were obtained as described in the Figure 4 legend. In the CCR5-binding experiments, the compounds were used at a concentration of 1 μM. The 50% inhibitory concentration (IC50) is shown for the compound’s inhibition of infection of Cf2Th-CD4/CCR5 cells by recombinant HIV-1 with the w.t. HIV-1YU2 envelope glycoproteins or with the control A-MLV envelope glycoproteins. In the first column, the values for NBD-556 are shown for comparison. The values are color-coded as in Figure 4.
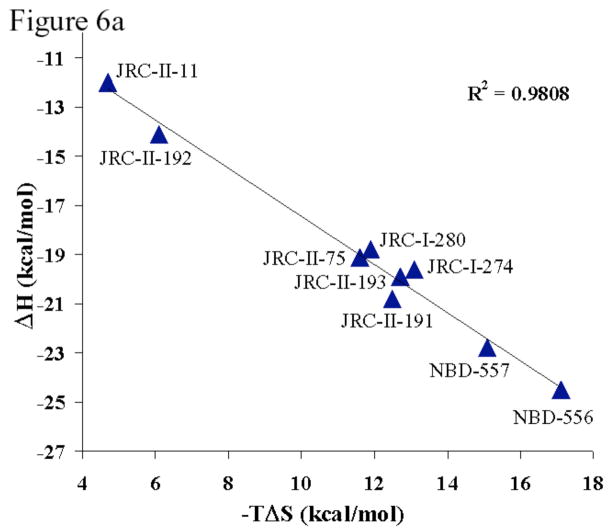
Binding thermodynamics were analyzed for the subset of NBD-556 analogues that exhibited detectable gp120 affinity. As expected for compounds with only modest differences in affinity (i.e., similar ΔG values), a strong correlation between the enthalpy change (ΔH) and the entropy change (−TΔS) associated with gp120 binding was observed (Figure 6a). The enthalpy changes associated with the binding of all of these phenyl ring variants were much larger than those expected from the modeled interactions of the compounds with the gp120 protein. Moreover, differences in −TΔS values of up to 11 kcal/mol were observed for the binding of closely related compounds. The observed entropy changes originate from conformational and solvation changes. Solvation changes are related to the heat capacity change, which is negative (Schön et al., 2006) and consistent with a favorable desolvation entropy (Luque and Freire, 1998). That the overall entropy changes associated with compound-gp120 binding are unfavorable indicates that desolvation does not compensate for the unfavorable conformational entropy changes. It has been estimated that binding of NBD-556 induces the structuring of 67 residues of gp120 on average (Schön et al., 2006). Apparently, the NBD-556 analogues structure the gp120 glycoprotein to different degrees.
Figure 6. Thermodynamic parameters associated with the binding of NBD-556 analogues to HIV-1 gp120.
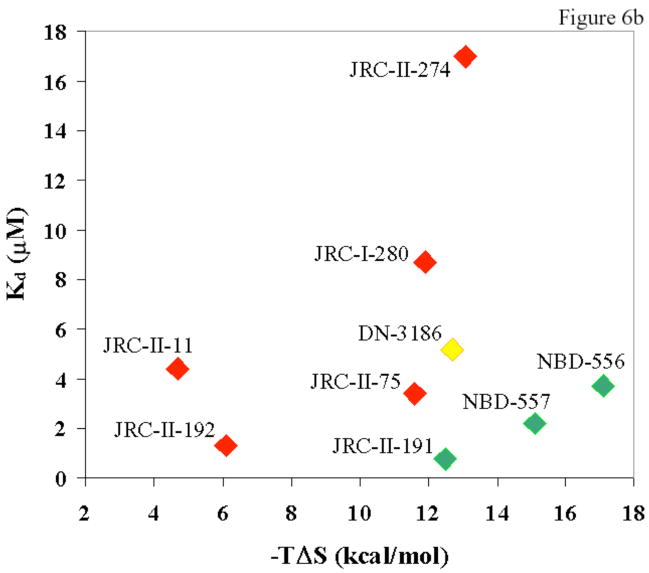
a. The relationship between the entropy change (−TΔS) and enthalpy change (ΔH) associated with the binding of NBD-556 analogues to HIV-1YU2 gp120 is shown. The least-squares regression line is depicted, along with the R2 value. b. The relationship is shown between the ability of an NBD-556 analogue to enhance HIV-1 infection of CCR5+ cells and the dissociation constant (Kd) and entropy change (−TΔS) associated with compound-gp120 binding. The symbols associated with the compounds are colored according to the ability of the compound to promote HIV-1 infection of CD4−CCR5+ cells. The color code is the same as that used in Tables 1 and 2, from red (none/low) to green (high level of enhancement).
The relationship between thermodynamic parameters and the enhancement of HIV-1 entry into CCR5+ cells was examined (Figure 6b). NBD-556 analogues that stimulated HIV-1 infection exhibited both high affinities for gp120 and large unfavorable entropy changes upon gp120 binding. Thus, high affinity for gp120 is necessary but not sufficient for the ability of NBD-556 analogues to replace CD4 in the HIV-1 entry process. The conformational fixation of gp120, reflected in the large entropic change observed during compound-gp120 binding, apparently contributes to CD4 mimicry.
One of the para-substituted analogues (DN-3186), which bound gp120 efficiently but minimally stimulated HIV-1 infection, inhibited the enhancement of HIV-1 infection by NBD-556 (data not shown). This observation, together with the thermodynamic data, supports a model in which these analogues bind, with varying degrees of CD4 mimicry, to the same general region of gp120.
Effects of gp120 changes near the Phe 43 cavity
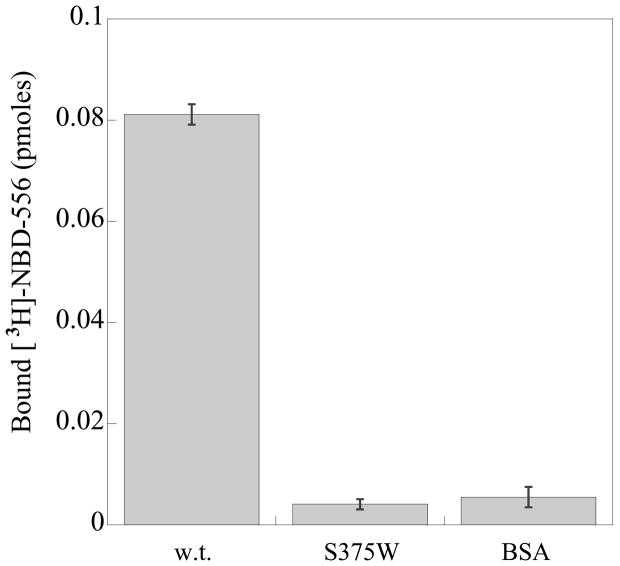
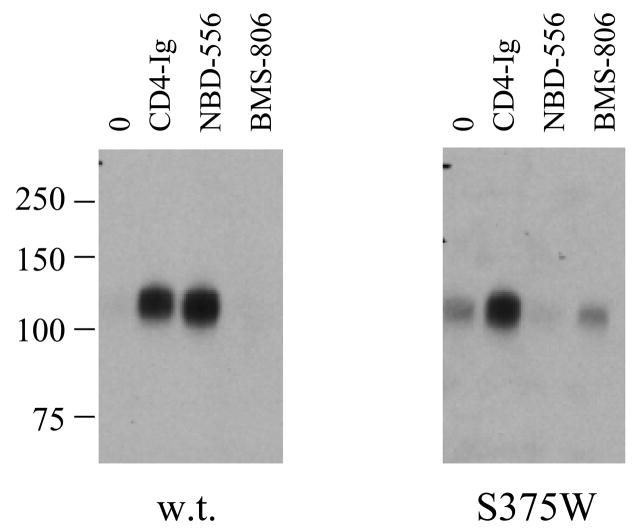
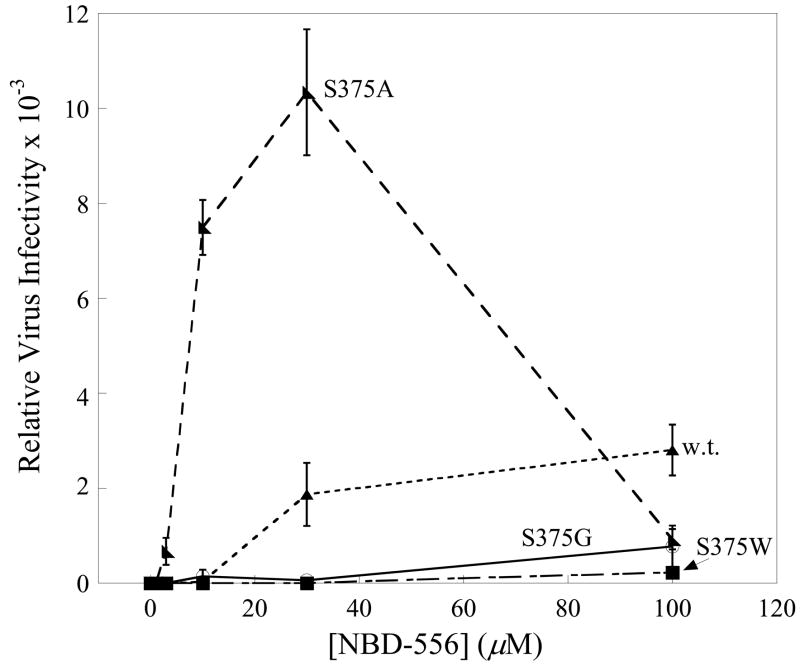
To test the models further, the interaction of mutants of the HIV-1YU2 gp120 glycoprotein with NBD-556 and the analogues was examined. Substitution of gp120 Ser 375 with a tryptophan residue fills the Phe 43 cavity but does not disrupt CD4 binding (Xiang et al., 2002). Compared with the wild-type (w.t.) HIV-1YU2 gp120, the S375W mutant bound radiolabeled NBD-556 inefficiently (Figure 7a). The binding of the S375W gp120 to Cf2Th-CCR5 cells was induced by a soluble form of CD4 but not by NBD-556 (Figure 7b). A control compound, BMS-806, which also binds HIV-1 gp120 (Lin et al., 2003), did not enhance the binding of either w.t. or S375W gp120 to CCR5+ cells. The infection of viruses with the S375W mutant envelope glycoproteins was not enhanced by NBD-556, in contrast to the significant enhancement observed for viruses with the w.t. envelope glycoproteins (Figure 7c). These results suggest that filling the Phe 43 cavity with the indole side chain of tryptophan prevents NBD-556 binding.
Figure 7. Interaction of NBD-556 with HIV-1 gp120 mutants.
a. The binding of [3H]-NBD-556 to equivalent amounts of the w.t. or S375W HIV-1YU2 gp120 glycoprotein or bovine serum albumin (BSA) is indicated. The means from a single experiment, with the standard errors derived from triplicate samples, are shown. The experiment was performed three times with comparable results. b. The amount of radiolabeled w.t. or S375W mutant HIV-1YU2 gp120 bound to Cf2Th-CCR5 cells is shown, after incubation without (0) or with CD4-Ig (1 μg/ml), NBD-556 (100 μM) or BMS-806 (10 μM). Numbers indicate molecular weight (in kD). c. Recombinant viruses bearing the w.t. or indicated mutant HIV-1YU2 envelope glycoproteins were incubated with Cf2Th-CCR5 cells in the presence of increasing concentrations of NBD-556. Luciferase activity in the Cf2Th-CCR5 cells was measured, and the relative virus infectivity, compared with that observed in the absence of added NBD-556, is shown. The mean values and standard errors (from triplicate samples) are shown from a typical experiment. d. The location of HIV-1YU2 gp120 residues in which changes affect NBD-556 binding and/or entry enhancement is shown. The modeled NBD-556, with bonds colored according to the identity of the atoms (see Figure 3 legend), is shown penetrating into the Phe 43 cavity of gp120. The gp120 core is depicted as a Cα ribbon and transparent molecular surface. The HIV-1YU2 gp120 residues that, when changed, affect NBD-556 binding and/or enhancement of HIV-1 entry into Cf2Th-CCR5 cells are labeled and highlighted in arbitrary colors, with side chain bonds shown.
We examined the effect of changes in several gp120 residues that line the Phe 43 cavity on the sensitivity of HIV-1 to enhancement by a panel of NBD-556 analogues (Figure 8). Two analogues, NBD-557 and JRC-II-191, that efficiently enhanced infection by viruses with w.t. HIV-1YU2 envelope glycoproteins did not activate the entry of viruses with the S375W envelope glycoproteins. The replacement of gp120 Ser 375 with glycine dramatically reduced HIV-1 sensitivity to enhancement by any of the NBD-556 analogues (Figure 7c and Figure 8), suggesting that some element of the Ser 375 side chain contributes to NBD-556 efficacy. In some x-ray crystal structures of the CD4-bound gp120 (1G9M) (Kwong et al., 2000), Ser 375 anchors a water molecule in the base of the Phe 43 cavity; this water molecule, which was not displaced in docking NBD-556 with Gold, likely affects the shape and flexibility of the cavity. Viruses bearing envelope glycoproteins with Ser 375 changed to alanine exhibited greater enhancement by NBD-556 than the viruses with w.t. envelope glycoproteins (Figure 7c and Figure 8). Moreover, several compounds (DN-3186, JRC-II-75 and JRC-II-11) stimulated the entry of viruses with the S375A change more efficiently than they enhanced w.t. virus infection (Figure 8). These results suggest that the hydroxyl group of Ser 375 is detrimental to the binding and/or activity of some NBD-556 analogues that contain large para-phenyl substituents.
Figure 8. NBD-556 analogue-mediated enhancement of infection of CD4-negative, CCR5-expressing cells by viruses containing mutant HIV-1YU2 envelope glycoproteins.
As described in the Figure 4 legend, the area under the dose-response curve (in arbitrary units) was determined for each compound incubated with the viruses bearing the indicated HIV-1YU2 envelope glycoprotein mutant (or A-MLV envelope glycoproteins). The value for this area was normalized to the value calculated for NBD-556 and the viruses with w.t. YU2 envelope glycoproteins in the same experiment. The numbers reported represent the mean values and standard deviations obtained from at least three independent experiments. The relative enhancement of infection of Cf2Th-CCR5 cells is color-coded as follows: red = 0–0.24; yellow = 0.25–0.7; green = >0.7.
Alteration of gp120 Asp 368 to alanine reduced the basal level of replication of HIV-1 in cells expressing CD4 and CCR5 (data not shown), consistent with the importance of this residue for CD4 binding (Kwong et al., 1998; Olshevsky et al., 1990; Wyatt and Sodroski, 1998). The infectivity of the D368A mutant viruses in CD4−CCR5+ target cells was enhanced by incubation with NBD-556, JRC-II-191 and JRC-II-11 (Figure 8). In the presence of the first two compounds, the entry of viruses with the D368A envelope glycoproteins into Cf2Th-CCR5 cells was similar to the levels achieved by w.t. viruses. Viruses with the D368A envelope glycoproteins were not enhanced by NBD-557, in contrast to w.t. viruses (Figure 8); this suggests that NBD-556 and NBD-557 differ in their sensitivity to changes in Asp 368 of gp120.
Changes in other gp120 residues lining the Phe 43 cavity or vestibule (Val 255, Thr 257, Glu 429 and Val 430) significantly decreased the enhancement of virus infection by NBD-556 analogues (Figure 8). Although the Glu 429 side chain projects away from the Phe 43 vestibule, it forms a salt bridge with Lys 121 and thus may help to stabilize the CD4-bound conformation of gp120 (Kwong et al., 1998). Replacement of Glu 429 with a lysine residue, which disrupts this salt bridge, eliminated the activation of infection for NBD-556, NBD-557 and JRC-II-191. Substitution of an alanine residue for Glu 429 was less disruptive of NBD-556-induced virus activation, although the virus-enhancing activities of NBD-557 and JRC-II-191 were significantly diminished by this change. Thus, alteration of several gp120 residues that line the Phe 43 pocket or reside in the vestibule leading into the pocket can affect, in either a positive or negative manner, the CD4-mimetic effects of NBD-556 or the analogues on HIV-1 infection.
Binding of NBD-556 to HIV-1 gp120 mutants
The binding of NBD-556 to selected HIV-1YU2 gp120 mutants was studied by isothermal titration calorimetry. Compared with the w.t. gp120 glycoprotein, both the D368A and S375A gp120 mutants bound NBD-556 and JRC-II-191 with higher affinity (Table1). A mutant gp120 glycoprotein, D368A/S375A, that combined both changes bound NBD-556 and JRC-II-191 more tightly than either of the mutants with individual changes. These results agree with measurements of radiolabeled NBD-556 binding to these gp120 glycoproteins (data not shown). Several gp120 mutants (S375W, I424A, W427A and M475A) with changes in residues that contact the Phe 43 cavity did not detectably bind NBD-556 in the isothermal titration calorimetric analyses (Table1). These results support the importance of gp120 residues near the Phe 43 cavity in binding NBD-556 and the analogues, and lend credence to the docked binding mode. Figure 7d illustrates the location of gp120 residues in which changes affect the binding and/or virus-enhancing ability of NBD-556 analogues, underscoring the proximity of these residues to the Phe 43 cavity.
Table 1.
Binding affinities of NBD-556 and JRC-II-191 to different gp120 variantsa
| HIV-1 YU2 gp120 glycoprotein |
Kd (μM)
|
|
|---|---|---|
| NBD-556 | JRC-II-191 | |
| w.t. | 3.7 | 0.80 |
| D368A | 2.2 | 0.72 |
| S375A | 0.31 | 0.14 |
| D368A/S375A | 0.26 | 0.083 |
| S375W | NB | NB |
| I424A | NB | NB |
| W427A | NB | NB |
| V430A | 3.5 | 1.0 |
| M475A | NB | NB |
The binding experiments were carried out by isothermal titration calorimetry at 25°C, as described in Methods. NB - no detectable binding.
Compound-gp120 affinity and HIV-1 inhibition
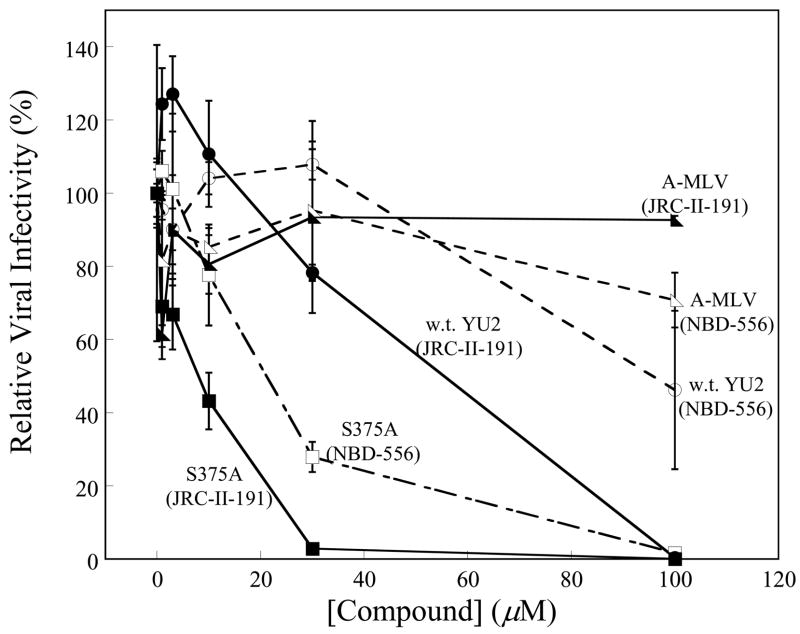
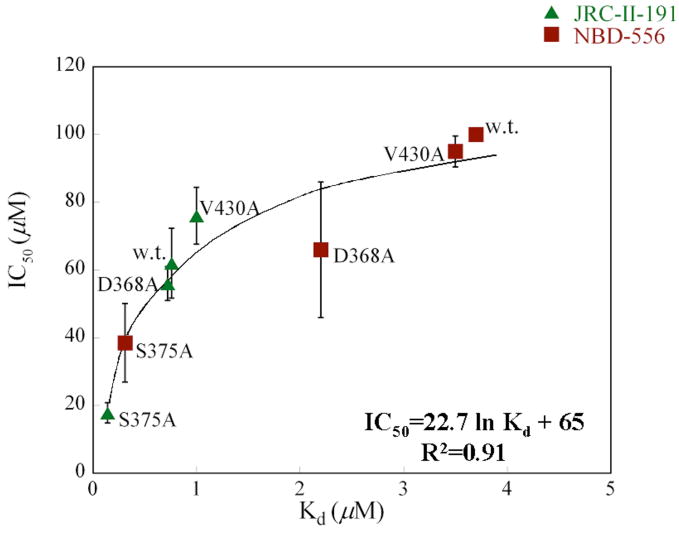
As natural target cells for HIV-1 in vivo all express CD4, future efforts to create antiviral agents directed against the Phe 43 cavity of gp120 would benefit from an understanding of the properties of these compounds that correlate with inhibition of HIV-1 infection of CD4+CCR5+ cells. We used gp120 variants and NBD-556 analogues exhibiting a range of affinities to examine the contribution of this parameter to virus inhibition. Infection of Cf2Th-CD4/CCR5 cells by viruses with the w.t. YU2 envelope glycoproteins was inhibited only by high concentrations of NBD-556 (50% inhibitory concentration (IC50) = 100 μM) (Figure 9a). On the other hand, JRC-II-191 inhibited w.t. virus infection with an IC50 of 54 μM, consistent with the higher affinity of this analogue for the HIV-1YU2 gp120 glycoprotein. Viruses with the S375A envelope glycoproteins, which exhibit a higher affinity for NBD-556 and JRC-II-191, were inhibited by both compounds with IC50 values of 10–20 μM (Figure 9a). There is a close relationship between the Kd of the compound for the particular envelope glycoprotein variant and the IC50 (Figure 9b). The Kd values are less than the IC50 values; this is not surprising, as many factors that can potentially influence the inhibition of virus infection are not expected to affect the binding of the compounds to gp120 monomers. Nonetheless, it is apparent that increasing the binding affinity of a small-molecule CD4 mimic results in improved antiviral effect.
Figure 9. Inhibition of HIV-1 infection of CD4-expressing cells by NBD-556 and JRC-II-191.
a. Recombinant viruses bearing the w.t. or S375A HIV-1YU2 envelope glycoproteins or control viruses with the A-MLV envelope glycoproteins were incubated with increasing concentrations of NBD-556 (open symbols) or JRC-II-191 (filled symbols) and then added to Cf2Th-CD4/CCR5 cells. The level of infection was determined by luciferase assay. The percentage of infection relative to that seen in the absence of added compound is shown. The means and standard errors from a single experiment, performed with triplicate samples, are shown. The experiment was repeated 3–5 times with comparable results. b. The concentrations (IC50) of NBD-556 and JRC-II-191 inhibiting 50% of infection of Cf2Th-CD4/CCR5 cells by viruses bearing the indicated HIV-1 envelope glycoproteins were plotted against the Kd for the binding of the compounds to the indicated HIV-1 gp120 variants. The IC50 values represent the means and standard deviations of values obtained in five independent experiments, with each experiment containing triplicate samples. The equation of the fitted curve is shown.
Discussion
The binding of CD4 and some neutralizing antibodies to the HIV-1 gp120 glycoprotein (or to the gp120 core) is accompanied by unfavorable changes in entropy that are unusually large for protein-protein interactions (Kwong et al., 2002; Myszka et al., 2000). These unfavorable entropic changes, reflecting the introduction of order into the conformationally flexible gp120, result from the formation of intramolecular interactions within gp120, reflected in favorable enthalpic changes. That the binding of a small molecule like NBD-556 to gp120 also involves such dramatic changes in enthalpy and entropy is remarkable (Schön et al., 2006). The observed binding thermodynamics imply that NBD-556, like CD4 and some antibodies (Kwong et al., 2002; Myszka et al., 2000), fixes gp120 into a limited subset of conformations. The NBD-556-induced conformation(s) must resemble the CD4-bound state, given the ability of NBD-556 to stimulate both CCR5 binding and HIV-1 infection of CD4−CCR5+ cells (Schön et al., 2006). The thermodynamic and mechanistic similarities of NBD-556 and CD4 justify the use of the CD4-bound structure of HIV-1 gp120 (Kwong et al., 2000; Kwong et al., 1998) to construct models of the NBD-556-gp120 complex. The consistency of the NBD-556 binding modes predicted by independent modeling approaches attests to the enthalpic benefits of interacting with the Phe 43 pocket. The predicted orientation of the bound NBD-556, with the phenyl ring projecting into the Phe 43 cavity, is necessitated by the steric bulk of the tetramethyl-piperidine ring at the other end of the molecule.
Studies with a panel of HIV-1 gp120 mutants supported the importance of gp120 residues near the Phe 43 cavity to NBD-556 binding. Filling the Phe 43 cavity with the indole ring of tryptophan in the S375W mutant resulted in loss of NBD-556 binding and activity, even though CD4 binding was preserved. These observations support the modeling predictions that NBD-556 inserts more deeply into the Phe 43 pocket than CD4. Alteration of several other gp120 residues near the Phe 43 cavity resulted in increased or decreased NBD-556 binding and/or activity. Notably, substitution of Ser 375 with alanine resulted in a gp120 glycoprotein that bound NBD-556 analogues better than the w.t. glycoprotein. The S375A viruses were also more sensitive to the enhancing effects of NBD-556 analogues (in CD4−CCR5+ cells) and to the inhibitory effects of the compounds (in CD4+CCR5+ cells). Substitution of alanine for Asp 368 reduced CD4 binding and HIV-1 replication, but resulted in tighter NBD-556 binding and increased NBD-556 enhancement of infection of CD4− target cells.
Our results provide insights into the surprising ability of small molecules to structure the conformationally flexible HIV-1 gp120 glycoprotein. The favorable enthalpic changes (up to 24.5 kcal/mol) observed upon binding of a series of NBD-556 analogues are much larger than those values expected from the interaction of the compounds with gp120, as delineated in the docking model. The large enthalpy changes, which are partially compensated by large unfavorable entropy changes, are reminiscent of those changes observed during protein folding (Robertson and Murphy, 1997). The magnitude of the enthalpic change is consistent with the formation of a significant network of interactions within gp120 upon compound binding. Moreover, some NBD-556 analogues that vary in the phenyl ring substituents have similar affinities for gp120, yet exhibit enthalpy changes upon gp120 binding that differ by more than 10 kcal/mol. These observations hint that the formation of new interactions within gp120 accounts for the major portion of the favorable enthalpy changes associated with the binding of some NBD-556 analogues, as has been previously suggested for CD4 binding (Myszka et al., 2000).
NBD-556 contacts with gp120 presumably contribute only a small fraction of the favorable enthalpic changes that occur upon binding. Modeling predicts that these interactions involve aromatic-aromatic stacking interactions in the base of the Phe 43 cavity and hydrogen bonds between the NBD-556 oxalamide and gp120 backbone carbonyls in the neck of the cavity. All known gp120 protein ligands that induce large entropic changes bind to at least two of the three gp120 core domains (Huang et al., 2005; Kwong et al., 2002; Kwong et al., 2000; Myszka et al., 2000; Vita et al., 1999; Zhang et al., 1999), implying that inter-domain flexibility likely contributes to the high entropy of unliganded gp120. Binding in the Phe 43 cavity, at the nexus of all three gp120 domains, provides an appealing explanation for the ability of a small molecule like NBD-556 to decrease gp120 conformational flexibility. Modeling of NBD-556 binding predicts that the para-phenyl substituents are situated adjacent to strand β16 (gp120 outer domain residues 374–379) and loop B (gp120 residues 255–257), which links strands β8 in the inner domain and β9 in the outer domain (Kwong et al., 1998). The differences in the entropy changes associated with the binding of analogues with subtly different para-phenyl substitutions indicate the importance of interactions with these two gp120 elements in inducing or stabilizing the CD4-bound conformation. Studies with chemically modified soluble CD4 or CD4-mimetic miniproteins have suggested that extending ligand interactions deeper into the Phe 43 cavity than those made by the Phe 43 ring of CD4 can increase affinity and antiviral potency (Van Herrewege et al., 2008; Xie et al., 2007; Zhang et al., 1999).
Our analysis of structure-activity relationships for a series of NBD-556-like compounds is consistent with the predicted constraints on the size and polarity of the phenyl ring and oxalamide linker imposed by the nature of the Phe 43 pocket. Although more than 90 NBD-556 analogues with alterations in these moieties were tested, only two related compounds (JRC-II-191 and JRC-II-192) exhibited better gp120 binding than NBD-556 or NBD-557. Both JRC-II-191 and JRC-II-192 have halogen groups at the para position and at one of the meta positions of the phenyl ring, suggesting that asymmetric positioning of the compounds in the Phe 43 cavity is allowed and even favored. Asymmetric binding is supported by docking calculations, which suggest that the meta halogens in JRC-II-191 and JRC-II-192 are oriented towards gp120 loop B and β16, rather than towards the water channel that opens onto the opposing surface of the Phe 43 cavity (Kwong et al., 1998).
Achieving adequate affinity for gp120 is essential but not sufficient for the ability of NBD-556-like compounds to promote CCR5 binding and HIV-1 infection of CD4−CCR5+ cells. A series of NBD-556 analogues with various substituents in the para position of the phenyl ring bound comparably to gp120 yet exhibited a wide range of abilities to activate CCR5 binding or virus infection. Only by taking into account both the binding affinity and entropy changes could functional activation of the HIV-1 envelope glycoproteins be accounted for. Likewise, JRC-II-191 and JRC-II-192, which differ in the meta-phenyl substituent, exhibited similar affinities for gp120 and shared predicted binding modes (see above); nonetheless, only JRC-II-191 activated HIV-1 infection of CD4− target cells. The greater magnitudes of the unfavorable entropy change and favorable enthalpy change associated with JRC-II-191 binding are consistent with the importance of conformational fixation of gp120 to the CD4-mimetic effects of these compounds.
The evolutionary requirement to evade the binding of neutralizing antibodies has resulted in several unusual features of the HIV-1 envelope glycoproteins: surface variability, a high degree of glycosylation and conformational flexibility (Kwong et al., 2002; Wyatt and Sodroski, 1998). Although these features are most effective in abrogating the binding of large molecules like antibodies, they can also influence the interaction of gp120 with small molecules. The variability of the gp120 surface creates a formidable barrier to the development of antiviral agents that are able to inhibit a broad range of natural HIV-1 variants. The Phe 43 pocket and surrounding vestibule represent one of the few well-conserved, accessible surfaces on the HIV-1 envelope glycoprotein trimer (Kwong et al., 1998; Wyatt et al., 1998; Wyatt and Sodroski, 1998). Our data indicate that small molecules binding in the Phe 43 pocket can interact with several different gp120 glycoproteins from distinct HIV-1 strains. Thus, the compounds that most closely mimic CD4 can take advantage of the requirement for HIV-1 to conserve the gp120 region that mediates CD4 binding.
The conformational flexibility of HIV-1 gp120 is thought to protect the receptor-binding sites from neutralizing antibodies (Kwong et al., 2002). Presumably, however, HIV-1 gp120 has also maintained throughout evolution a natural propensity to make the transition into the CD4-bound conformation. The binding of CD4 and CD4 mimics triggers a cascade of cooperative interactions within gp120 that result in the structuring of previously disordered regions (Huang et al., 2005; Kwong et al., 1998; Myszka et al., 2000; Schön et al., 2006; Zhang et al., 1999). These events may involve gp120 structures distant from the binding site, creating the possibility of altering the binding of CD4 mimics by changes outside the gp120 residues that directly contact the ligand. For example, we observed a difference in the efficiency with which NBD-556 bound the gp120 glycoproteins of the YU2 and HXBc2 HIV-1 strains, even though the gp120 residues predicted to contact NBD-556 are identical in these two variants. Presumably, envelope glycoprotein differences outside the NBD-556 binding site influence the affinity of gp120 for the compound in this instance. The propensity of HIV-1 to escape small-molecule CD4 mimics by this mechanism is balanced by the viral requirements to maintain efficient CD4 binding.
The most effective CD4 mimics can allow HIV-1 to circumvent the requirement for CD4 on target cells overexpressing CCR5. The degree to which this activating effect might extend the tropism of HIV-1 to CD4− cells in vivo, which express much lower levels of CCR5, requires further investigation. This activating effect of CD4-mimetic compounds will be balanced by virus-inhibitory effects. The latter include competition for CD4 on natural target cells and the premature triggering of metastable states in the HIV-1 envelope glycoproteins that lead to virus inactivation.
The working model for specific gp120-NBD-556 interactions presented herein should assist interpretation of structure-activity relationships and guide efforts to improve these compounds further. Our results suggest that increasing the affinity of NBD-556 for gp120 should be a high priority in seeking to improve antiviral efficacy. The relatively low molecular weight of JRC-II-191, which is optimized for insertion into the Phe 43 cavity, allows future manipulation of the piperidine ring to achieve greater antiviral potency. CD4-mimetic drugs could eventually be combined with modalities that recognize the highly conserved chemokine receptor-binding surface of gp120 and interfere with CCR5 binding. Further understanding of the HIV-1 receptor-binding regions should assist the development of rational, targeted therapeutic or prophylactic interventions.
Experimental Procedures
Compounds
NBD-556 analogues were synthesized and stored as described previously (Schön et al., 2006). Additional details of synthesis are available from the authors on request.
Cell lines
Cell lines were maintained as described in the supplemental material.
Plasmids expressing HIV-1 envelope glycoproteins
The w.t. and mutant HIV-1 envelope glycoproteins were expressed from the pSVIIIenv vector (Olshevsky et al., 1990; Sullivan et al., 1998). Full description of the preparation of mutant plasmids is provided in the supplemental material.
Effects of compounds on virus infectivity
Recombinant, luciferase-expressing viruses were made as described previously (Schön et al., 2006) and in the supplemental material. Cf2Th-CD4-CCR5 or Cf2Th-CCR5 target cells were seeded at a density of 6 × 103 cells/well in 96-well luminometer-compatible tissue culture plates (Dynex) 24 h before infection. On the day of infection, NBD-556 or its analogues (1 to 100 μM) was added to recombinant viruses (10,000 reverse transcriptase units) in a final volume of 50 μl and incubated at 37°C for 30 min. The medium was removed from the target cells, which were then incubated with the virus-drug mixture for 48 h at 37°C. The medium was removed from each well, and the cells were lysed in 30 μl of passive lysis buffer (Promega) by three freeze-thaw cycles. An EG&G Berthold Microplate Luminometer LB 96V was used to measure luciferase activity in each well after the addition of 100 μl of luciferin buffer (15 mM MgSO4, 15 mM KPO4 [pH 7.8], 1 mM ATP, 1 mM dithiothreitol) and 50 μl of 1 mM D-luciferin potassium salt (BD Pharmingen).
Binding assays
The production of gp120 and assays measuring the binding of gp120 to CCR5 and radiolabeled NBD-556 are described previously (Schön et al., 2006), as well as in the supplemental material.
Isothermal titration calorimetry
Isothermal titration calorimetric experiments were performed as described previously (Schön et al., 2006) and in the supplemental material.
Modeling the binding of NBD-556 and its analogues to HIV-1 gp120
For the docking calculations, small molecule and protein preparation used standard protocols, as described in the supplemental materials. For Glide(4.018) (Friesner, 2004; Halgren et al., 2004) docking, the binding site was defined based on the positions of CD4 Phe 43 and the isopropanol molecule from the 1G9M crystal structure. The Glide grids were computed with a box center at 28.10, −12.35, 81.57 and an inner and outer box range of 14 Å and 36 Å, respectively. Docking calculations were performed in standard sampling mode with maxkeep 5,000 and maxref 1,000. Docking was repeated using Gold (version 3.2). The binding site was defined by using the docked conformation of NBD-556 produced with Glide. Docking calculations were performed with three crystallographic water molecules (HOH 343, HOH 6 and HOH 327) residing in the cavity, as described in the supplemental materials section. Docking calculations were conducted with default parameters with the following exceptions: 100 genetic algorithm (GA) docking runs were performed using an initial_virtual_pt_match_max=3.5, diverse_solutions=1, divsol_cluster_size=1, and divsol_rmsd=1.5 Å.
NBD-556 was independently modeled and docked to the CD4-bound gp120 core using Insight II/Discover Software (Accelrys, San Diego, CA), as described in the supplemental material.
Supplementary Material
Acknowledgments
We thank Drs. Wayne Hendrickson and Irwin Chaiken for valuable discussion. We thank Ms. Yvette McLaughlin and Ms. Elizabeth Carpelan for manuscript preparation, and Jonathan Stuckey for preparation of figures. This study was supported by grants from the National Institutes of Health (GM56550, AI24755 and AI60354), by the International AIDS Vaccine Initiative, and William F. McCarty-Cooper.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arthos J, Deen KC, Shatzman A, Truneh A, Rosenberg M, Sweet RW. The genetic analysis of the HIV envelope binding domain on CD4. Ann N Y Acad Sci. 1990;616:116–124. doi: 10.1111/j.1749-6632.1990.tb17833.x. [DOI] [PubMed] [Google Scholar]
- Ashkenazi A, Presta LG, Marsters SA, Camerato TR, Rosenthal KA, Fendly BM, Capon DJ. Mapping the CD4 binding site for human immunodeficiency virus by alanine-scanning mutagenesis. Proc Natl Acad Sci U S A. 1990;87:7150–7154. doi: 10.1073/pnas.87.18.7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock GJ, Mirzabekov T, Wojtowicz W, Sodroski J. Ligand binding characteristics of CXCR4 incorporated into paramagnetic proteoliposomes. J Biol Chem. 2001;276:38433–38440. doi: 10.1074/jbc.M106229200. [DOI] [PubMed] [Google Scholar]
- Brodsky MH, Warton M, Myers RM, Littman DR. Analysis of the site in CD4 that binds to the HIV envelope glycoprotein. J Immunol. 1990;144:3078–3086. [PubMed] [Google Scholar]
- Chen B, Vogan EM, Gong H, Skehel JJ, Wiley DC, Harrison SC. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature. 2005;433:834–841. doi: 10.1038/nature03327. [DOI] [PubMed] [Google Scholar]
- Choe H, Li W, Wright PL, Vasilieva N, Venturi M, Huang CC, Grundner C, Dorfman T, Zwick MB, Wang L, et al. Tyrosine sulfation of human antibodies contributes to recognition of the CCR5 binding region of HIV-1 gp120. Cell. 2003;114:161–170. doi: 10.1016/s0092-8674(03)00508-7. [DOI] [PubMed] [Google Scholar]
- Fontenot D, Jones JK, Hossain MM, Nehete PN, Vela EM, Dwyer VA, Jagannadha Sastry K. Critical role of Arg59 in the high-affinity gp120-binding region of CD4 for human immunodeficiency virus type 1 infection. Virology. 2007;363:69–78. doi: 10.1016/j.virol.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Friesner RAB, Murphy JL, Halgren RB, Klicic TA, Mainz JJ, Repasky DT, Knoll MP, Shelley EH, Perry M, Shaw JK, Francis DE, Shenkin P, Glide PS. A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- Furuta RA, Wild CT, Weng Y, Weiss CD. Capture of an early fusion-active conformation of HIV-1 gp41. Nat Struct Biol. 1998;5:276–279. doi: 10.1038/nsb0498-276. [DOI] [PubMed] [Google Scholar]
- Halgren TAMRB, Friesner RA, Beard HS, Frye LL, Pollard WTB, Glide JL. A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem. 2004:1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- Huang CC, Stricher F, Martin L, Decker JM, Majeed S, Barthe P, Hendrickson WA, Robinson J, Roumestand C, Sodroski J, et al. Scorpion-toxin mimics of CD4 in complex with human immunodeficiency virus gp120 crystal structures, molecular mimicry, and neutralization breadth. Structure. 2005;13:755–768. doi: 10.1016/j.str.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Jones GW, Glen P, Leach RC, Taylor RAR. Development and validation of a genetic algorithm for flexible docking. J Mol Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- Karlsson GB, Halloran M, Schenten D, Lee J, Racz P, Tenner-Racz K, Manola J, Gelman R, Etemad-Moghadam B, Desjardins E, et al. The envelope glycoprotein ectodomains determine the efficiency of CD4+ T lymphocyte depletion in simian-human immunodeficiency virus-infected macaques. J Exp Med. 1998;188:1159–1171. doi: 10.1084/jem.188.6.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntz ID, Meng EC, Shoichet BK. Structure-based molecular design. Accounts Chem Res. 1994;27:117ff. [Google Scholar]
- Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M, et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Majeed S, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure. 2000;8:1329–1339. doi: 10.1016/s0969-2126(00)00547-5. [DOI] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau NR, Warton M, Littman DR. The envelope glycoprotein of the human immunodeficiency virus binds to the immunoglobulin-like domain of CD4. Nature. 1988;334:159–162. doi: 10.1038/334159a0. [DOI] [PubMed] [Google Scholar]
- Lin PF, Blair W, Wang T, Spicer T, Guo Q, Zhou N, Gong YF, Wang HG, Rose R, Yamanaka G, et al. A small molecule HIV-1 inhibitor that targets the HIV-1 envelope and inhibits CD4 receptor binding. Proc Natl Acad Sci U S A. 2003;100:11013–11018. doi: 10.1073/pnas.1832214100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luque I, Freire E. Structure-based prediction of binding affinities and molecular design of peptide ligands. Methods Enzymol. 1998;295:100–127. doi: 10.1016/s0076-6879(98)95037-6. [DOI] [PubMed] [Google Scholar]
- Luty BA, Wasserman ZR, Stouten PFW, Hodge CN, Zacharias M, McCammon JA. A molecular mechanics/grid method for evaluation of ligand-receptor interactions. J Comp Chem. 1995;16:454–464. [Google Scholar]
- Martin L, Stricher F, Misse D, Sironi F, Pugniere M, Barthe P, Prado-Gotor R, Freulon I, Magne X, Roumestand C, et al. Rational design of a CD4 mimic that inhibits HIV-1 entry and exposes cryptic neutralization epitopes. Nat Biotechnol. 2003;21:71–76. doi: 10.1038/nbt768. [DOI] [PubMed] [Google Scholar]
- Myszka DG, Sweet RW, Hensley P, Brigham-Burke M, Kwong PD, Hendrickson WA, Wyatt R, Sodroski J, Doyle ML. Energetics of the HIV gp120-CD4 binding reaction. Proc Natl Acad Sci U S A. 2000;97:9026–9031. doi: 10.1073/pnas.97.16.9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olshevsky U, Helseth E, Furman C, Li J, Haseltine W, Sodroski J. Identification of individual human immunodeficiency virus type 1 gp120 amino acids important for CD4 receptor binding. J Virol. 1990;64:5701–5707. doi: 10.1128/jvi.64.12.5701-5707.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson AD, Murphy KP. Protein Structure and the Energetics of Protein Stability. Chem Rev. 1997;97:1251–1268. doi: 10.1021/cr960383c. [DOI] [PubMed] [Google Scholar]
- Schön A, Madani N, Klein JC, Hubicki A, Ng D, Yang X, Smith AB, 3rd, Sodroski J, Freire E. Thermodynamics of binding of a low-molecular-weight CD4 mimetic to HIV-1 gp120. Biochemistry. 2006;45:10973–10980. doi: 10.1021/bi061193r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudinger R, Phogat SK, Xiao X, Wang X, Dimitrov DS, Zolla-Pazner S. Evidence for CD4-enchanced signaling through the chemokine receptor CCR5. J Biol Chem. 2003;278:10389–10392. doi: 10.1074/jbc.M212013200. [DOI] [PubMed] [Google Scholar]
- Stouten PFW, Froemmel C, Nakamura H, Sander C. Molecular Stimulation. 1993;10 [Google Scholar]
- Sullivan N, Sun Y, Binley J, Lee J, Barbas CF, 3rd, Parren PW, Burton DR, Sodroski J. Determinants of human immunodeficiency virus type 1 envelope glycoprotein activation by soluble CD4 and monoclonal antibodies. J Virol. 1998;72:6332–6338. doi: 10.1128/jvi.72.8.6332-6338.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thali M, Olshevsky U, Furman C, Gabuzda D, Posner M, Sodroski J. Characterization of a discontinuous human immunodeficiency virus type 1 gp120 epitope recognized by a broadly reactive neutralizing human monoclonal antibody. J Virol. 1991;65:6188–6193. doi: 10.1128/jvi.65.11.6188-6193.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A, Dragic T, Arthos J, Binley JM, Olson WC, Allaway GP, Cheng-Mayer C, Robinson J, Maddon PJ, Moore JP. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- Van Herrewege Y, Morellato L, Descours A, Aerts L, Michiels J, Heyndrickx L, Martin L, Vanham G. CD4 mimetic miniproteins: potent anti-HIV compounds with promising activity as microbicides. J Antimicrob Chemother. 2008;61:818–826. doi: 10.1093/jac/dkn042. [DOI] [PubMed] [Google Scholar]
- Vita C, Drakopoulou E, Vizzavona J, Rochette S, Martin L, Menez A, Roumestand C, Yang YS, Ylisastigui L, Benjouad A, Gluckman JC. Rational engineering of a miniprotein that reproduces the core of the CD4 site interacting with HIV-1 envelope glycoprotein. Proc Natl Acad Sci U S A. 1999;96:13091–13096. doi: 10.1073/pnas.96.23.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Gerard NP, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso AA, Desjardin E, Newman W, et al. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, Sodroski JG. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- Xiang SH, Farzan M, Si Z, Madani N, Wang L, Rosenberg E, Robinson J, Sodroski J. Functional mimicry of a human immunodeficiency virus type 1 coreceptor by a neutralizing monoclonal antibody. J Virol. 2005;79:6068–6077. doi: 10.1128/JVI.79.10.6068-6077.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang SH, Kwong PD, Gupta R, Rizzuto CD, Casper DJ, Wyatt R, Wang L, Hendrickson WA, Doyle ML, Sodroski J. Mutagenic stabilization and/or disruption of a CD4-bound state reveals distinct conformations of the human immunodeficiency virus type 1 gp120 envelope glycoprotein. J Virol. 2002;76:9888–9899. doi: 10.1128/JVI.76.19.9888-9899.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Ng D, Savinov SN, Dey B, Kwong PD, Wyatt R, Smith AB, 3rd, Hendrickson WA. Structure-activity relationships in the binding of chemically derivatized CD4 to gp120 from human immunodeficiency virus. J Med Chem. 2007;50:4898–4908. doi: 10.1021/jm070564e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Canziani G, Plugariu C, Wyatt R, Sodroski J, Sweet R, Kwong P, Hendrickson W, Chaiken I. Conformational changes of gp120 in epitopes near the CCR5 binding site are induced by CD4 and a CD4 miniprotein mimetic. Biochemistry. 1999;38:9405–9416. doi: 10.1021/bi990654o. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Ma L, Jiang S, Lu H, Liu S, He Y, Strick N, Neamati N, Debnath AK. Identification of N-phenyl-N′-(2,2,6,6-tetramethyl-piperidin-4-yl)-oxalamides as a new class of HIV-1 entry inhibitors that prevent gp120 binding to CD4. Virology. 2005;339:213–225. doi: 10.1016/j.virol.2005.06.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.