Abstract
BACKGROUND
Perineural invasion (PNI) is the dominant pathway for local invasion in prostate cancer. To date, only few studies have investigated the molecular differences between prostate tumors with PNI and those without it.
METHODS
To evaluate the involvement of both microRNAs and protein-coding genes in PNI, we determined their genome-wide expression with a custom microRNA microarray and Affymetrix GeneChips in 50 prostate adenocarcinomas with PNI and 7 without it. In-situ hybridization and immunohistochemistry was used to validate candidate genes.
RESULTS
Unsupervised classification of the 57 adenocarcinomas revealed two clusters of tumors with distinct global microRNA expression. One cluster contained all non-PNI tumors and a subgroup of PNI tumors. Significance analysis of microarray data yielded a list of microRNAs associated with PNI. At a false discovery rate (FDR) < 10%, 19 microRNAs were higher expressed in PNI tumors than in non-PNI tumors. The most differently expressed microRNA was miR-224. In-situ hybridization showed that this microRNA is expressed by perineural cancer cells. The analysis of protein-coding genes identified 34 transcripts that were differently expressed by PNI status (FDR <10%). These transcripts were down-regulated in PNI tumors. Many of those encoded metallothioneins and proteins with mitochondrial localization and involvement in cell metabolism. Consistent with the microarray data, perineural cancer cells tended to have lower metallothionein expression by immunohistochemistry than nonperineural cancer cells.
CONCLUSIONS
Although preliminary, our findings suggest that alterations in microRNA expression, mitochondrial function, and cell metabolism occur at the transition from a non-invasive prostate tumor to a tumor with PNI.
Keywords: Invasion, prostate tumor, gene expression profile, microRNA
INTRODUCTION
Prostate cancer is the most frequently diagnosed malignancy and the second most common cause of cancer mortality in American men [1]. The mortality can be attributed to the spread of cancer cells beyond the prostate. Perineural invasion (PNI) is the dominant pathway for local invasion in prostate cancer and is also a mechanism for extraprostatic spread of the disease [2]. Yet, the prognostic significance of PNI remains controversial [3–5]. Several studies have observed an association of PNI with markers of poor outcome [2,6–8], but others did not find it to be a prognostic factor in prostate cancer [9–12]. The occurrence of PNI is a relatively early event in the development of the clinical disease, and most tumor specimens from radical prostatectomy are PNI-positive [2]. It is this high occurrence rate of PNI in clinical samples (85% to 100%) and the inadequate knowledge of its biology that limit our understanding of PNI’s role in prostate cancer progression and disease outcome.
PNI is the process where cancer cells adhere to and wrap around nerves [13,14]. It occurs in many other types of cancer, including pancreatic and head and neck cancers [15,16]. Prostate cancer cells that have a perineural location acquire a survival and growth advantage and exhibit reduced apoptosis and increased proliferation when compared with cells located away from nerves [17,18]. Altered expression of adhesion molecules in both prostate cancer cells and the adjacent nerves has been observed in PNI, and it has been hypothesized that the changed expression of these molecules allows cancer cells to thrive in the vicinity of nerves [14,19]. Nevertheless, the molecular mechanisms that lead to PNI remain poorly understood. We applied gene expression profiling of both microRNAs and protein-coding genes to identify the gene expression changes associated with PNI in human prostate cancer. We hypothesized that the gene expression signature that differentiates PNI from non-PNI tumors will reveal molecular alterations that take place at the transition from a non-invasive tumor to a tumor with PNI. We assayed microRNAs because a crucial role for them in cancer has been demonstrated [20,21]. Their expression profiles have been shown to classify tumors by developmental lineage and differentiation state [22,23].
MATERIALS AND METHODS
Tissue samples
Frozen tumor specimens were obtained from the NCI Cooperative Prostate Cancer Tissue Resource (CPCTR). The tumors were resected adenocarcinomas that had not received any therapy prior to prostatectomy. The macro-dissected tumor specimens were reviewed by a pathologist, who confirmed the presence of tumor in the frozen specimens. All tissues were collected between 2002 and 2004. Tissue collection was approved by the institutional review boards of the participating institutions.
RNA extraction
Total RNA was isolated using TRIZOL reagent according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA). RNA integrity for each sample was confirmed with the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). Each RNA sample was then split into two aliquots that were either processed for the microRNA microarray or the mRNA microarray.
Gene microarrays
MicroRNA labeling and hybridization were performed as described previously [24]. The microRNA microarray (Ohio State University Comprehensive Cancer Center, Version 2.0) contains probes spotted in quadruplicate for 235 human and 222 mouse microRNAs [24]. The labeling and the hybridization of mRNAs were performed according to Affymetrix standard protocols (Santa Clara, CA). Briefly, 5 µg of total RNA was reverse transcribed with an oligo (dT) primer that has a T7 RNA polymerase promoter at the 5’ end. Second-strand synthesis was followed by cRNA production with incorporation of biotinylated ribonucleotides using the BioArray High Yield RNA Transcript Labeling Kit T3 from Enzo Life Sciences (Farmingdale, NY). The labeled cRNA was fragmented and hybridized to Affymetrix GeneChip HG-U133A 2.0 arrays. This array contains 22,283 probe sets that represent approximately 13,000 human protein-coding genes. Hybridization signals were visualized with phycoerythrin-conjugated streptavidin (Invitrogen) and scanned using a GeneChip Scanner 3000 7G (Affymetrix). In accordance with Minimum Information About a Microarray Experiment (MIAME) guidelines, we deposited the CEL files for the microarray data and additional patient information into the GEO repository (http://www.ncbi.nlm.nih.gov/geo/). The GEO submission accession number for both the microRNA and mRNA profiling data is GSE7055. Additional information about the custom microRNA microarray, Version 2.0, can be found under the ArrayExpress accession number: A-MEXP-258.
Data normalization and statistical analysis
Median-centric normalization was used for the custom microRNA oligonucleotide chips. Affymetrix chips were normalized using the robust multichip analysis (RMA) procedure [25]. To generate lists of significantly differently expressed genes, the resulting data set was subjected to the significance analysis of microarray (SAM) procedure [26]. We generated gene lists based on both P values from two-sided t-tests and intended false discovery rates (FDRs). The FDR calculation followed the method described by Storey and Tibshirani [27]. Unsupervised hierarchical clustering was performed according to principles described by Eisen et al. [28].
Quantitative real-time PCR analysis of microRNA and mRNA
Abundance of mature microRNAs was measured using the stem-loop TaqMan® MicroRNA Assays kit (Applied Biosystems, Foster City, CA) according to a published protocol [29]. Using 10 ng of total RNA, mature microRNA was reverse transcribed into a 5’-extended cDNA with mature microRNA-specific looped RT primers from the TaqMan® MicroRNA Assays kit and reagents from TaqMan® MicroRNA Reverse Transcription kit (Applied Biosystems) following the manufacturer’s directions. Real-time PCR was performed on the cDNA with Applied Biosystems Taqman® 2X Universal PCR Master Mix and the appropriate 5X Taqman® MicroRNA Assay Mix for each microRNA of interest. Triplicate reactions were incubated in an Applied Biosystems 7500 Real-Time PCR system in a 96 well plate for 10 min at 95°C, followed by 40 cycles for 15 s at 95°C and 1 min at 60°C. For each sample, the threshold cycle (Ct) was calculated by the ABI 7500 Sequence Detection System software. Standard curves were used to determine microRNA concentrations in the samples, which were then normalized to U6 RNA. Abundance of mRNA was determined according to a previously described quantitative real-time (qRT) PCR method [30]. Accordingly, 100 ng of total RNA was reverse transcribed using the High-Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA). qRT-PCR was subsequently performed in triplicate using TaqMan Gene Expression Assays (Applied Biosystems), which include pre-optimized probe and primer sets specific for the genes being validated. The assay ID numbers of the validated genes are as follows: Hs00744661_sH for metallothionein 1F and Hs00828387_g1 for metallothionein 1M. Data were collected using the ABI PRISM® 7500 Sequence Detection System. The 18s RNA was used as the internal standard reference. Normalized expression was calculated using the comparative CT method as described and fold changes were derived from the 2−ΔΔCt values for each gene [30].
Immunohistochemistry
Protein expression in perineural and nonperineural cancer cells was assessed immunohistochemically on formalin-fixed, paraffin-embedded tumor sections. The tumors (n = 30) were from prostate patients treated by radical prostatectomy at the Baltimore VA Hospital and the University of Maryland Medical Center. Five micron sections were immunohistochemically stained for S100, a marker for nerve trunks, to visualize areas with PNI. Sections from fourteen tumors were found to contain representative areas with perineural and nonperineural cancer cells. For antigen retrieval, deparaffinized sections were microwaved in 1x Citra buffer (Biogenex, San Ramon, CA). Immunohistochemical staining was performed with the Dako Envision system (DakoCytomation, Carpinteria, CA). The following primary antibodies were used: 1:500 diluted rabbit polyclonal antibody for S100 (Ventana, Tucson, AR); 1:1000 diluted mouse monoclonal antibody for coxsackie adenovirus receptor (CXADR) (Atlas Antibodies, Stockholm, Sweden); and 1:500 diluted mouse monoclonal antibody for metallothionein (DakoCytomation). This antibody (E9) recognizes metallothionein-1 and -2 family members (# M0639). Positive controls: intestine (CXADR) and liver (metallothionein). Omission of the primary antibody was the negative control. A pathologist, who was blinded to the microarray results, evaluated the intensity of the immunostains in perineural and nonperineural cancer cells and categorized immunostaining as less intensive, same, or more intensive in the perineural cancer cells when compared with nonperineural cancer cells. Images of representative areas were taken to document the expression differences.
In-situ Hybridization
In-situ hybridization (ISH) was performed using the GenPoint™ Catalyzed Signal Amplification System (DakoCytomation) following the manufacturer’s protocol. Briefly, slides were incubated at 60°C for 30 minutes and deparaffinized as described. Sections were treated with Proteinase K (DakoCytomation) for 30 minutes at room temperature, rinsed several times with dH2O, and immersed in 95% ethanol for 10 seconds before air-drying. Slides were pre-hybridized at 54°C for 1 hour with in situ hybridization buffer (Enzo Life Sciences, Inc. Farmingdale, NY) before an overnight 54°C incubation in buffer containing either 5'-biotin labeled miR-224 miRCURY™ LNA detection probe (Exiqon, Woburn, MA) or scrambled negative control probe (Exiqon) at 50 nM final concentration. Slides were washed in both TBST and GenPoint™ stringent wash solution (54°C for 30 minutes). Slides were then exposed to H2O2 blocking solution (DakoCytomation) for 20 minutes and further blocked in a blocking buffer (DakoCytomation, X0909) for 30 minutes before being exposed to primary Streptavidin-HRP antibody, biotinyl tyramide, secondary Streptavidin-HRP antibody, and DAB chromogen solutions following the manufacturer’s protocol. Slides were then briefly counterstained in hematoxylin and rinsed with both TBST and water before mounting. A pathologist evaluated the ISH intensity of miR-224 in perineural and nonperineural cancer cells using the same criteria that were used for immunohistochemistry.
Pathway analysis
This analysis was performed with the in-house WPS software [31]. Pathways were annotated according to Gene Ontology Biological Processes (GOBP) (Gene Ontology Consortium: http://www.geneontology.org). Our database had 16,762 human genes annotated for GOBP. Genes were included into the pathway analysis based on the FDR (≤ 30%) of their corresponding probesets on the microarray. If several probesets encoded the same gene, the software recognized this and assured that the gene was counted only once for significance testing at the pathway level. A one-sided Fisher’s exact test was used to determine which biological processes had a statistically significant enrichment of differently expressed genes (P < 0.05). We compiled the Fisher’s exact test results for cluster analyses and displayed the results in color-coded heat maps to reveal the patterns of significantly altered biological processes. The color coding of the heat maps is related to the enrichment of genes in a biological process (−Log(P value)-based) with red indicating a higher enrichment.
RESULTS
Clinical samples and gene expression analysis
We collected macro-dissected tumor specimens from radical prostatectomies of 57 prostate cancer patients (Table 1). Seven (12%) of the tumors were negative for PNI. Consistent with the literature, those tumors had a smaller size and a lower Gleason score than PNI-positive tumors. In addition, all PNI-negative tumors were confined to the prostate. We investigated the gene expression differences between tumors with PNI and those that were negative for PNI. Gene expression profiles from these tumors were generated using both a custom microRNA microarray that represents 235 human microRNAs and the Affymetrix GeneChip HG-U133A 2.0 array that represents approximately 13,000 human protein-coding genes.
TABLE I.
Clinical Characteristics of the Study Population
| All cases (n = 57) |
Perineural Invasion (n = 50) |
No Perineural Invasion (n = 7) |
P value | |
|---|---|---|---|---|
| Age at prostatectomy [median (range)] n = 57 | 61 (46 – 73) | 60 (48 – 77) | 62 (60 – 68) | 0.22b |
| PSA at diagnosis [median (range)] n = 48a | 5.9 (1.3 – 47.7) | 6.1 (3.8 – 47.7) | 5.4 (1.3 – 8.8) | 0.19b |
| Largest individual nodule (grams) median (range) n = 56a | 1.5 (0.2 – 3.0) | 1.6 (0.8 – 3.0) | 1.0 (0.2 – 2.0) | 0.006b |
| Gleason sum score | ||||
| < 7 (5–6) | 13 (23) | 9 (18) | 4 (57) | |
| ≥ 7 (7–9) | 44 (77) | 41 (82) | 3 (43) | 0.04c |
| Extraprostatic extension | ||||
| No | 33 (58) | 26 (52) | 7 (100) | |
| Yes | 24 (42) | 24 (48) | 0 (0) | 0.02c |
| Seminal vesicle invasion | ||||
| No | 47 (82) | 40 (80) | 7 (100) | |
| Yes | 10 (18) | 10 (20) | 0 (0) | 0.33c |
Information was not available for some cases.
Two-sided t-test.
Two-sided Fisher’s exact test.
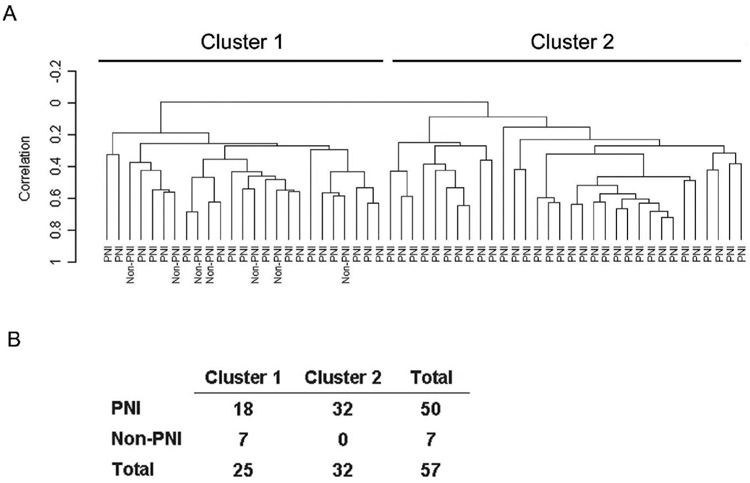
In an initial analysis of our dataset, we applied unsupervised hierarchical clustering to examine whether expression of microRNAs and mRNAs can distinguish between tumors with PNI and those without PNI. Hierarchical clustering based on the global expression of mRNA did not separate PNI cases from non-PNI cases (data not shown). However, the expression patterns of the microRNAs in these samples yielded two prominent clusters with distinct microRNA profiles (Figure 1). Cluster #1 contained all non-PNI tumors and a subgroup of tumors with PNI. Cluster #2 contained PNI tumors that were significantly more likely to have a high Gleason score (≥ 7) and an extraprostatic disease extension than tumors in cluster #1 (P < 0.05, respectively; two-sided Fisher’s exact test).
Figure 1.
Unsupervised hierarchical cluster analysis of 57 prostate tumors based on the expression of 235 microRNAs. A: The microRNA expression yielded two prominent clusters with distinct microRNA profiles. Cluster #1 contained all non-PNI tumors. B: Non-random distribution of tumors by PNI status among the two clusters (P = 0.002; two-sided Fisher’s exact test).
Significance analysis of microarray data revealed that 19 microRNAs and 34 protein-coding genes were significantly differently expressed between PNI and non-PNI tumors at a FDR ≤ 10%. At this threshold, all microRNAs were up-regulated in tumors with PNI (Table 2), while all mRNAs had a lower expression in PNI tumors than in non-PNI tumors (Table 3). This list of differently expressed microRNAs was unique to the comparison between PNI and non-PNI tumors in our dataset. None of these microRNAs were significantly differently expressed by either tumor grade or stage (FDR < 30%). In contrast to the PNI to non-PNI comparison, only very few microRNAs were significantly differently expressed between high (sum score 7–9) and low (sum score 5–6) Gleason score, e.g., miR-1 was down-regulated in tumors with high Gleason score, and between organ-confined and those with extraprostatic extension. Among the protein-coding genes that were differently expressed between PNI and non-PNI tumors, many encoded either metallothioneins (metallothionein 1F, 1G, 1H, 1M, 1X, 2A) or proteins with mitochondrial localization (4-aminobutyrate aminotransferase, ferrochelatase, long chain acyl-coenzyme A dehydrogenase, mitochondrial ribosomal proteins L39/S1). A subset of these genes was also down-regulated in tumors with a high Gleason score when compared with low Gleason score tumors (Figure 3). There was no overlap with genes differently expressed by tumor stage.
TABLE II.
Up-regulated MicroRNAs in Tumors with PNI (FDR ≤ 10%)
| microRNA | Fold Change | Chromosomal Location |
|---|---|---|
| miR-224 | 2.68 | Xq28 |
| miR-21 | 2.65 | 17q23.2 |
| miR-10 (a/b) | 2.63 | 17q21.32/2q31.1 |
| miR-125b (−1/2) | 2.42 | 11q24.1/21q21.1 |
| miR-30a/b/c-2/ | 2.33 | 6q13/8q24.22 |
| miR-100 | 2.24 | 11q24.1 |
| miR-24 (−1/2) | 2.12 | 9q22.32/19p13.12 |
| miR-15a-2 | 2.06 | 13q14.2 |
| miR-191 | 2.04 | 5p21.31 |
| miR-99b | 2.03 | 19q13.41 |
| miR-27a/b | 2.00 | 19p13.12/9q22.32 |
| miR-26a (−1/2) | 1.87 | 3p22.3/12q14.1 |
| miR-126 | 1.68 | 9q34.3 |
| miR-145 | 1.84 | 5q32 |
| miR-195 | 1.67 | 17p13.1 |
| miR-181a-1 | 1.64 | 1p31.1 |
| miR-199b | 1.58 | 9q34.11 |
| miR-151 | 1.55 | 8q24.3 |
| let-7g | 1.47 | 3p21.2 |
Fold change: Expression PNI versus non-PNI (reference)
TABLE III.
Protein-coding RNAs with Differential Expression between PNI and non-PNI Tumors
| Gene | Gene Name | GenBank ID | Fold Change* | FDR (%) | Gleason Score** |
|---|---|---|---|---|---|
| MT1M | metallothionein 1M | R06655 | 0.30 | 0 | 0.52 (0) |
| PTPRM | protein tyrosine phosphatase, receptor type, M | NM_002845 | 0.37 | 0 | ND |
| PTGER4 | prostaglandin E receptor 4 (subtype EP4) | AA897516 | 0.46 | 0 | ND |
| C9orf46 | chromosome 9 open reading frame 46 | NM_018465 | 0.47 | 0 | 0.74 (6) |
| MAN2B2 | mannosidase, alpha, class 2B, member 2 | AW954107 | 0.68 | 0 | ND |
| SLC38A4 | solute carrier family 38, member 4 | NM_018018 | 0.19 | 5 | ND |
| NPR3 | atrionatriuretic peptide receptor C | AI628360 | 0.29 | 5 | ND |
| ODZ1 | odd Oz/ten-m homolog 1 | AL022718 | 0.35 | 5 | ND |
| MT1F | metallothionein 1F | M10943 | 0.52 | 5 | 0.66 (0) |
| ABAT | 4-aminobutyrate aminotransferase | AF237813 | 0.52 | 5 | 0.79 (18) |
| INPP4B | inositol polyphosphate-4-phosphatase, type II | NM_003866 | 0.53 | 5 | ND |
| FECH | Ferrochelatase | NM_000140 | 0.56 | 5 | ND |
| TNFAIP8 | tumor necrosis factor, alpha-induced protein 8 | NM_014350 | 0.58 | 5 | ND |
| TTC12 | tetratricopeptide repeat domain 12 | NM_017868 | 0.58 | 5 | ND |
| MT1H | metallothionein 1H | NM_005951 | 0.59 | 5 | 0.64 (0) |
| MT2A | metallothionein 2A | NM_005953 | 0.64 | 5 | 0.68 (0) |
| AP2S1 | Adaptor-related protein complex 2, S1 subunit | NM_004069 | 0.66 | 5 | ND |
| RAB27A | RAB27A, member RAS gene family | AF125393 | 0.50 | 7 | 0.66 (3) |
| SMS | spermine synthase | NM_004595 | 0.56 | 7 | ND |
| LACTB2 | lactamase, beta 2 | NM_016027 | 0.59 | 7 | ND |
| EIF5B | eukaryotic translation initiation factor 5B | NM_015904 | 0.81 | 7 | ND |
| GPR37 | G protein-coupled receptor 37 | U87460 | 0.29 | 9 | ND |
| MAF | v-maf oncogene homolog | NM_005360 | 0.43 | 9 | 0.69 (2) |
| ACADL | acyl-coenzyme A dehydrogenase, long chain | NM_001608 | 0.46 | 9 | 0.75 (7) |
| MAGEH1 | melanoma antigen family H, 1 | NM_014061 | 0.55 | 9 | 0.72 (2) |
| TBC1D4 | TBC1 domain family, member 4 | NM_014832 | 0.55 | 9 | ND |
| MT1G | metallothionein 1G | NM_005950 | 0.57 | 9 | 0.61 (0) |
| ZNF652 | zinc finger protein 652 | NM_014897 | 0.59 | 9 | ND |
| MT1X | metallothionein 1X | NM_005952 | 0.64 | 9 | 0.67 (0) |
| APXL | apical protein-like (Xenopus laevis) | NM_001649 | 0.64 | 9 | 0.82 (12) |
| CXADR | coxsackie virus and adenovirus receptor | NM_001338 | 0.68 | 9 | ND |
| MRPL39 | mitochondrial ribosomal protein L39 | NM_017446 | 0.72 | 9 | ND |
| MGST3 | microsomal glutathione S-transferase 3 | NM_004528 | 0.73 | 9 | 0.75 (0) |
| MRPS11 | mitochondrial ribosomal protein S1 | AB049944 | 0.80 | 9 | ND |
Fold change: Expression PNI versus non-PNI (reference)
Gleason score: Fold change (FDR %) for high (7–9) versus low (5–6) Gleason score (reference).
ND: no difference (FDR ≤ 30%)
Figure 3.
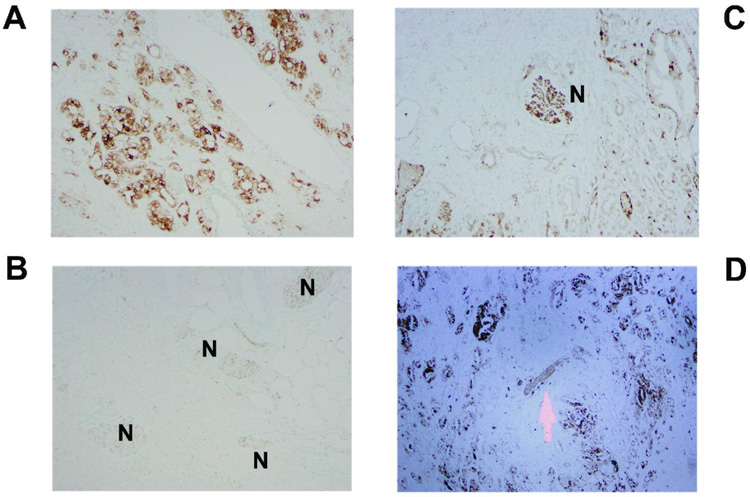
Expression of metallothionein in prostate tumors by immunohistochemistry. The panels show examples of metallothionein expression in the tumor epithelium. Marked cytoplasmic expression of metallothionein in cancer cells distant to neurons (A) and absence of this expression in perineural cancer cells (B) in the same tumor. The expression of metallothionein is decreased as tumor cells approach the nerve (C,D). Arrow and “N” indicate the location of the brown stained nerve trunks. Counterstain: Methyl green.
Validation of microarray data by qRT-PCR
Five microRNAs and two mRNAs were chosen for validation by qRT-PCR (Table 4). Consistent with the microarray data, we found a significantly higher expression of mature miR-224, miR-10, miR-125b, miR-30c, and miR-100 in PNI tumors when compared with non-PNI tumors. The transcript levels of the metallothioneins 1M and 1F were significantly lower in PNI tumors when compared with non-PNI tumors, which is also consistent with our microarray data.
Table IV.
Validation of Microarray Results by qRT-PCR for Selected Genes
| Gene | qRT-PCR Fold changea | Array Fold change |
|---|---|---|
| Metallothionein-1M | 0.26 | 0.30 |
| Metallothionein-1F | 0.31 | 0.52 |
| microRNA | qRT-PCR Fold changeb | Array Fold change |
| miR-224 | 5.72 | 2.68 |
| miR-10b | 2.84 | 2.63 |
| miR-125b | 4.29 | 2.42 |
| miR-30c | 2.15 | 2.44 |
| miR-100 | 2.08 | 2.24 |
Fold change: Expression PNI (n = 36) versus non-PNI (reference; n = 7)
Fold change: Expression PNI (n = 23) versus non-PNI (reference; n = 7)
All fold change differences were statistically significant (P < 0.05; two-sided Mann Whitney U test)
Pathway association of protein-coding genes that are differently expressed by PNI status
We performed a pathway analysis based on those GOBP-annotated genes (n = 62) whose mRNA was differently expressed between PNI and non-PNI tumors at a FDR ≤ 30%. The analysis revealed a number of biological processes that were enriched for differently expressed genes comparing PNI tumors with non-PNI tumors. The most significantly altered biological processes included transport and metabolism of organic (carboxylic) acids/fatty acids, amino acids, and (poly)amines (Table 5). They also included the biological process of “neurogenesis", which is consistent with the known interaction between tumor cells and nerves in PNI.
TABLE V.
Biological Processes Most Significantly Enriched for Differently Expressed Genes Comparing PNI Tumors with non-PNI Tumors
| GOBP Term | PNI List Term hitsa | PNI List Totalb | Population Term hitsc | Population Totald | Fisher's exact test P value |
|---|---|---|---|---|---|
| Organic (carboxylic) acid metabolism | 9 | 62 | 453 | 16762 | 4.0E-05 |
| Amino acid transport | 4 | 62 | 68 | 16762 | 1.2E-04 |
| Amine transport | 4 | 62 | 73 | 16762 | 1.5E-04 |
| Organic (carboxylic) acid transport | 4 | 62 | 83 | 16762 | 2.5E-04 |
| L-phenylalanine metabolism | 2 | 62 | 10 | 16762 | 5.9E-04 |
| Aromatic amino acid family catabolism | 2 | 62 | 12 | 16762 | 8.7E-04 |
| Aromatic compound metabolism | 2 | 62 | 14 | 16762 | 1.2E-03 |
| Fatty acid metabolism | 4 | 62 | 140 | 16762 | 1.8E-03 |
| Neurogenesis | 5 | 62 | 259 | 16762 | 2.7E-03 |
| Aromatic amino acid family metabolism | 2 | 62 | 29 | 16762 | 5.1E-03 |
| Amino acid and derivative metabolism | 5 | 62 | 309 | 16762 | 5.7E-03 |
Number of annotated genes in a GOBP term that are differentially expressed when comparing tumors with PNI versus tumors without PNI
All GOBP-annotated genes that are differentially expressed in this comparison
All annotated genes in a GOBP term
All GOBP-annotated genes
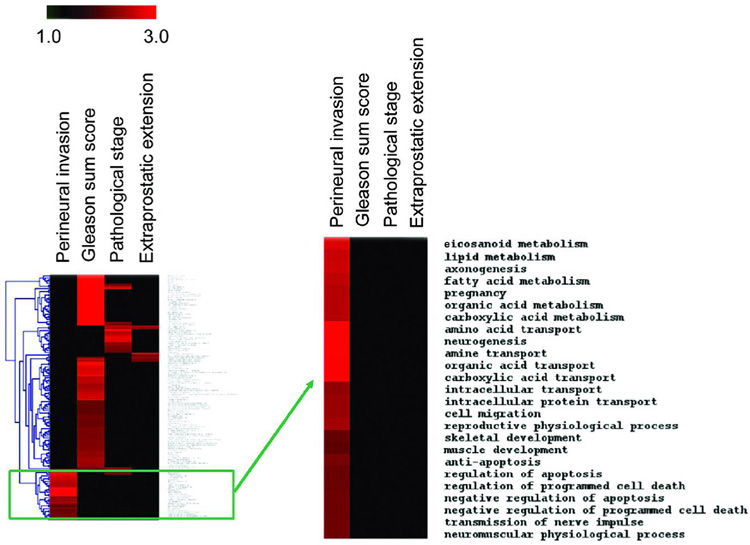
A cluster analysis was performed to identify biological processes that are enriched for differently expressed genes by tumor PNI status (PNI-positive versus PNI-negative), but not by Gleason score (high versus low Gleason score), pathological stage (pT3 versus pT2), or by the presence of extraprostatic extension (yes versus no). As shown by a heatmap, the analysis identified a number of biological processes that were uniquely enriched for differently expressed genes comparing PNI-positive with PNI-negative tumors (Figure 2). These biological processes included metabolism and transport of organic (carboxylic) acids/fatty acids, amino acids, and (poly)amines, as described before, but also processes related to the negative regulation of programmed cell death.
Figure 2.
Cluster analysis of Gene Ontology Biological Processes that are enriched for differently expressed genes comparing PNI tumors with non-PNI tumors. The results of a cluster analysis are displayed in a heatmap with the red color indicating an enrichment of differentially expressed genes in a biological process, e.g., eicosanoid metabolism, for a particular comparison, e.g., PNI tumor versus non-PNI tumor (“Perineural invasion”). The heatmap also shows the cluster analysis for the high (7–9) versus low (5–6) Gleason score comparison (“Gleason sum score”), the pT3 versus pT2 comparison (“Pathological stage”), and the positive versus negative extraprostatic extension comparison (“Extraprostatic extension”). Our analysis revealed that gene expression differences are non-random and create unique patterns of frequently affected biological processes for the four comparisons. The enlarged cluster shows the biological processes that are uniquely enriched for differentially expressed genes comparing PNI tumors with non-PNI tumors. Eicosanoid metabolism, lipid metabolism, and axonogenesis are also enriched for differentially expressed genes comparing pT3 versus pT2.
Expression of metallothionein, coxsackie adenovirus receptor, and miR-224 in perineural cancer cells
Although our microarray-based analysis indicated that PNI and non-PNI tumors differ in their gene expression pattern, this approach is not informative with respect to the expression of these genes in perineural and nonperineural cancer cells. We used immunohistochemistry and in situ hybridization to investigate the relative expression of two protein-coding genes, metallothionein (metallothionein-1 and -2) and coxsackie adenovirus receptor (CXADR), and of miR-224 in perineural and nonperineural cancer cells. Immunohistochemistry was performed on sections from 14 tumors that contained representative areas for perineural and nonperineural cancer cells. In situ hybridization was performed on sections from 11 tumors. Metallothionein, CXADR and miR-224 were found to be expressed in the tumor epithelium (Figure 3–Figure 5). The labeling pattern for metallothionein (epithelial, cytoplasmic, nuclear) and CXADR (epithelial, membranous, cytoplasmic) was consistent with that described by others [32,33]. A lower expression of metallothionein and CXADR was observed in perineural cancer cells of 6 tumors (43%) and 7 tumors (50%), respectively, when compared with nonperineural cancer cells in the same tissues (Figure 3 and Figure 4). No difference was detected in the other tumors with the exception of one (7%) where the expression of metallothionein was scored to be higher in perineural cancer cells than nonperineural cancer cells. A marked increased expression of miR-224 in perineural cancer cells was observed in 4 tumors (36%) (Figure 5). No such difference was seen in the other 7 tumors where miR-224 expression was mostly low to undetectable in the tumor epithelium.
Figure 5.
miR-224 in prostate tumors by in-situ hybridization. Shown are representative examples of cytoplasmic expression of miR-224 in the tumor epithelium. The granular brown staining shows the presence of miR-224. Most tumors showed weak labeling for miR-224 (A). In a subset of tumors, moderate to strong miR-224 labeling was observed in perineural cancer cells (B, C, D). N = nerve trunk. Counterstain: Hematoxylin.
Figure 4.
Expression of the coxsackie adenovirus receptor in prostate tumors by immunohistochemistry. The panels show examples of receptor expression in the tumor epithelium. Membranous and cytoplasmic staining for the receptor in cancer cells distant to neurons (A) and in perineural cancer cells (B) in the same tumor. The expression of the coxsackie adenovirus receptor is decreased in perineural cancer cells (C,D). N: nerve trunk. Counterstain: Methyl green.
DISCUSSION
We investigated the gene expression profiles of PNI and non-PNI tumors and found significant differences in microRNA and mRNA expression between them. Most strikingly, unsupervised hierarchical cluster analysis based on the expression of 235 microRNAs yielded two main tumor clusters, one of which contained all non-PNI tumors. We could not achieve such a classification based on the expression of 13,000 protein-coding transcripts which is in agreement with other studies that could not find an mRNA expression signature associated with local invasion in prostate cancer [34]. Our findings suggest that microRNA expression could be a more distinctive feature of PNI tumors, when compared with non-PNI tumors, than mRNA expression. Although these findings are preliminary, they are consistent with previous reports showing that microRNA expression profiles can be superior to mRNA expression profiles in classifying tumors by developmental lineage and differentiation state [22,23].
Nineteen microRNAs were found to be higher expressed in PNI tumors than non-PNI tumors. Of those, miR-10, miR-21, and miR-125b are candidate oncogenes [35–37]. Furthermore, miR-21 and miR-224 are located in malignancy-associated chromosomal regions that were found to have an increased gene expression in human prostate cancer [38]. A microRNA expression signature common to several human solid cancers, including prostate cancer, has been described [23]. The shared microRNAs between that study and our PNI signature are miR-21, miR-24, and miR-30c. Most notable, however, is the overlap of the PNI signature with other microRNA signatures that were discovered under experimental conditions. Hypoxia has been found to induce miR-24, miR-26, miR-27, and miR-181 [39]. Those microRNAs are also upregulated in PNI tumors. Even more prominent are the similarities between the PNI signature and an inflammation-induced microRNA signature in lungs of LPS-treated mice. Here, LPS induced miR-21, miR-27b, miR-100, and miR-224, among several other microRNAs [40]. Thus, the observed PNI microRNA signature could be partly the result of a pro-inflammatory environment and hypoxia in the cancerous prostate. This interpretation is preliminary. Future studies will have to evaluate whether mediators of inflammation and hypoxia can induce these microRNAs in human prostate cancer cells.
To evaluate the possibility of confounding effects by tumor grade and stage in the PNI signature, we compared the list of differently expressed microRNAs between PNI and non-PNI tumors with the same lists comparing high with low Gleason score tumors and organ-confined tumors with tumors that showed extraprostatic extension. This additional analysis revealed that the PNI signature was not shared by these two contrasts. Instead, only very few microRNAs were found to be significantly differently expressed by tumor grade and stage. Perhaps, the heterogeneous nature of prostate tumors limited our ability to find a microRNA signature associated with these two prognostic factors. Alternatively, the PNI signature could be very distinct and unique to the transition of non-PNI to PNI and may specifically involve the interaction between nerve and cancer cells. This signature could also be a transient phenomenon of cancer cells and disappears when these cells disseminate from their perineural location. We analyzed the expression of miR-224, the most differently expressed microRNA by PNI status, in perineural and nonperineural cancer cells and found an increased expression of it in perineural cancer cells in a subset of the tumors. Although not all tumors showed upregulation of miR-224 in perineural cancer cells, the observation indicates that mechanisms by which cancer cells adhere to nerves could be involved in the induction of miR-224.
Analysis of the mRNA expression profile revealed 34 genes that were down-regulated in PNI tumors at a FDR threshold of ≤ 10%. Even though we observed genes that were higher expressed in PNI tumors than non-PNI tumors, e.g., CRISP3, PSCA, BMP7, or BCL2, their high FDR excluded them from our list of significantly differently expressed genes. Only two other studies, using a co-culture model of DU-145 prostate cancer cells with neuronal cells, examined the expression profile of mRNA associated with PNI [18,19]. Those studies discovered that the genes encoding bystin and Pim-2 are upregulated in PNI. We did not detect an increase of the corresponding mRNAs in PNI tumors. Different methodologies may explain some of the differences among the gene lists generated in the various studies. In addition, our chip did not contain probesets for the gene encoding bystin.
Several of the 34 differently expressed genes were members of the metallothionein gene family. These genes are located in a gene cluster on chromosome 16q13 [41] and have been found to be down-regulated in prostate cancer by promoter hypermethylation and reduced zinc availability [32,42,43]. By immunohistochemistry, we could confirm that metallothionein expression is noticeably lower in perineural cancer cells when compared with nonperineural cancer cells in a subset of the prostate tumors. The down-regulation at the transition from a non-PNI tumor to a PNI tumor may indicate important changes in the metal metabolism of cancer cells that take place at this stage of the disease. Several other genes in our list of differently expressed genes encode proteins with mitochondrial localization, e.g., 4-aminobutyrate aminotransferase, ferrochelatase, and long chain acyl-coenzyme A dehydrogenase, among others. The aminobutyrate aminotransferase and the long chain acyl-coenzyme A dehydrogenase are key genes in the organic (carboxylic) acid metabolism (e.g., ketone body, fatty acid) of cells, whereas the ferrochelatase is involved in the biosynthesis of heme [44]. Alterations in metabolism and in the genome of mitochondria are common events in prostate carcinogenesis [45–47]. Our data suggest that some of these changes may occur at the transition into a PNI-positive tumor.
Other genes that were found to be down-regulated in PNI tumors were those encoding the spermine synthase, the v-MAF oncogene homolog (MAF), and CXADR. Spermine synthase is a key enzyme of the polyamine synthesis pathway that catalyzes the conversion of spermidine into spermine. A transcriptional dysregulation of the polyamine synthesis pathway in prostate cancer has been observed [48]. Spermine is an endogenous inhibitor of prostate carcinoma cell growth [49]. Therefore, down-regulation of the spermine synthase may allow increased growth and survival of prostate cancer cells in a perineural environment. MAF is an oncogene in lymphomas and myelomas, but it was found to be a candidate tumor suppressor gene in prostate cancer [50]. CXADR has a crucial function in the uptake of adenoviruses into human cells [51]. This receptor was found to be down-regulated in locally advanced prostate cancer when compared with normal prostate [33].
Because single gene effects are unlikely to cause PNI, we conducted a pathway analysis for the protein-coding genes that were differently expressed between PNI tumors and non-PNI tumors. This analysis revealed that the most significantly altered biological processes in PNI tumors, when compared to non-PNI tumors, are those that regulate cell and energy metabolism. Other altered biological processes related to neuronal functions, such as neurogenesis and the transmission of nerve impulse, and to the negative regulation of cell death. The latter is consistent with previous findings that prostate cancer cells in a perineural location show decreased apoptosis and increased survival [17,18].
CONCLUSIONS
We observed significant alterations in microRNA and mRNA expression at the transition from a non-PNI tumor to a PNI tumor. Unsupervised hierarchical clustering revealed that non-PNI tumors are more distinct from PNI tumors by their microRNA expression profile than by their mRNA expression profile. Finally, we identified various genes and biological processes related to mitochondrial function and cell metabolism that could be functionally significant in PNI.
ACKNOWLEDGEMENTS
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, and by National Institutes of Health grants CA081534 and CA128609 (C. Croce). The authors thank the Cooperative Prostate Cancer Tissue Resource for providing tissue specimens and supporting data.
Abbreviations
- PNI
perineural invasion
- FDR
false discovery rate
REFERENCES
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Villers A, McNeal JE, Redwine EA, Freiha FS, Stamey TA. The role of perineural space invasion in the local spread of prostatic adenocarcinoma. J Urol. 1989;142:763–768. doi: 10.1016/s0022-5347(17)38881-x. [DOI] [PubMed] [Google Scholar]
- 3.Bostwick DG, Grignon DJ, Hammond ME, Amin MB, Cohen M, Crawford D, Gospadarowicz M, Kaplan RS, Miller DS, Montironi R, Pajak TF, Pollack A, et al. Prognostic factors in prostate cancer. College of American Pathologists Consensus Statement 1999. Arch.Pathol.Lab Med. 2000;124:995–1000. doi: 10.5858/2000-124-0995-PFIPC. [DOI] [PubMed] [Google Scholar]
- 4.Montironi R, Mazzucchelli R, Scarpelli M, Lopez-Beltran A, Mikuz G. Prostate carcinoma I: prognostic factors in radical prostatectomy specimens and pelvic lymph nodes. BJU.Int. 2006;97:485–491. doi: 10.1111/j.1464-410X.2005.05972.x. [DOI] [PubMed] [Google Scholar]
- 5.Montironi R, Mazzucchelli R, Scarpelli M, Lopez-Beltran A, Mikuz G, Algaba F, Boccon-Gibod L. Prostate carcinoma II: prognostic factors in prostate needle biopsies. BJU.Int. 2006;97:492–497. doi: 10.1111/j.1464-410X.2006.05973.x. [DOI] [PubMed] [Google Scholar]
- 6.Beard C, Schultz D, Loffredo M, Cote K, Renshaw AA, Hurwitz MD, D'Amico AV. Perineural invasion associated with increased cancer-specific mortality after external beam radiation therapy for men with low- and intermediate-risk prostate cancer. Int.J Radiat.Oncol.Biol Phys. 2006;66:403–407. doi: 10.1016/j.ijrobp.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 7.Quinn DI, Henshall SM, Brenner PC, Kooner R, Golovsky D, O'Neill GF, Turner JJ, Delprado W, Grygiel JJ, Sutherland RL, Stricker PD. Prognostic significance of preoperative factors in localized prostate carcinoma treated with radical prostatectomy: importance of percentage of biopsies that contain tumor and the presence of biopsy perineural invasion. Cancer. 2003;97:1884–1893. doi: 10.1002/cncr.11263. [DOI] [PubMed] [Google Scholar]
- 8.Harnden P, Shelley MD, Clements H, Coles B, Tyndale-Biscoe RS, Naylor B, Mason MD. The prognostic significance of perineural invasion in prostatic cancer biopsies: a systematic review. Cancer. 2007;109:13–24. doi: 10.1002/cncr.22388. [DOI] [PubMed] [Google Scholar]
- 9.Egan AJ, Bostwick DG. Prediction of extraprostatic extension of prostate cancer based on needle biopsy findings: perineural invasion lacks significance on multivariate analysis. Am J Surg.Pathol. 1997;21:1496–1500. doi: 10.1097/00000478-199712000-00013. [DOI] [PubMed] [Google Scholar]
- 10.Ng JC, Koch MO, Daggy JK, Cheng L. Perineural invasion in radical prostatectomy specimens: lack of prognostic significance. J Urol. 2004;172:2249–2251. doi: 10.1097/01.ju.0000143973.22897.f8. [DOI] [PubMed] [Google Scholar]
- 11.Merrick GS, Butler WM, Wallner KE, Galbreath RW, Allen ZA, Adamovich E. Prognostic significance of perineural invasion on biochemical progression-free survival after prostate brachytherapy. Urology. 2005;66:1048–1053. doi: 10.1016/j.urology.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 12.O'Malley KJ, Pound CR, Walsh PC, Epstein JI, Partin AW. Influence of biopsy perineural invasion on long-term biochemical disease-free survival after radical prostatectomy. Urology. 2002;59:85–90. doi: 10.1016/s0090-4295(01)01486-8. [DOI] [PubMed] [Google Scholar]
- 13.Ayala GE, Wheeler TM, Shine HD, Schmelz M, Frolov A, Chakraborty S, Rowley D. In vitro dorsal root ganglia and human prostate cell line interaction: redefining perineural invasion in prostate cancer. Prostate. 2001;49:213–223. doi: 10.1002/pros.1137. [DOI] [PubMed] [Google Scholar]
- 14.Li R, Wheeler T, Dai H, Ayala G. Neural cell adhesion molecule is upregulated in nerves with prostate cancer invasion. Hum.Pathol. 2003;34:457–461. doi: 10.1016/s0046-8177(03)00084-4. [DOI] [PubMed] [Google Scholar]
- 15.Hyland C, Kheir SM, Kashlan MB. Frozen section diagnosis of pancreatic carcinoma: a prospective study of 64 biopsies. Am J Surg.Pathol. 1981;5:179–191. doi: 10.1097/00000478-198103000-00007. [DOI] [PubMed] [Google Scholar]
- 16.Ballantyne AJ, McCarten AB, Ibanez ML. The extension of cancer of the head and neck through peripheral nerves. Am J Surg. 1963;106:651–667. doi: 10.1016/0002-9610(63)90074-6. [DOI] [PubMed] [Google Scholar]
- 17.Yang G, Wheeler TM, Kattan MW, Scardino PT, Thompson TC. Perineural invasion of prostate carcinoma cells is associated with reduced apoptotic index. Cancer. 1996;78:1267–1271. doi: 10.1002/(SICI)1097-0142(19960915)78:6<1267::AID-CNCR15>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 18.Ayala GE, Dai H, Ittmann M, Li R, Powell M, Frolov A, Wheeler TM, Thompson TC, Rowley D. Growth and survival mechanisms associated with perineural invasion in prostate cancer. Cancer Res. 2004;64:6082–6090. doi: 10.1158/0008-5472.CAN-04-0838. [DOI] [PubMed] [Google Scholar]
- 19.Ayala GE, Dai H, Li R, Ittmann M, Thompson TC, Rowley D, Wheeler TM. Bystin in perineural invasion of prostate cancer. Prostate. 2006;66:266–272. doi: 10.1002/pros.20323. [DOI] [PubMed] [Google Scholar]
- 20.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat.Rev.Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 21.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat.Rev.Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 22.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 23.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc.Natl.Acad.Sci U.S.A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, Dumitru CD, Shimizu M, Zupo S, Dono M, Alder H, Bullrich F, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci U.S.A. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc.Natl.Acad.Sci.U.S.A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc.Natl.Acad.Sci.U.S.A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc.Natl.Acad.Sci.U.S.A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bookout AL, Mangelsdorf DJ. Quantitative real-time PCR protocol for analysis of nuclear receptor signaling pathways. Nucl.Recept.Signal. 2003;1:e012. doi: 10.1621/nrs.01012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yi M, Horton JD, Cohen JC, Hobbs HH, Stephens RM. WholePathwayScope: a comprehensive pathway-based analysis tool for high-throughput data. BMC.Bioinformatics. 2006;7:30. doi: 10.1186/1471-2105-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garrett SH, Sens MA, Shukla D, Flores L, Somji S, Todd JH, Sens DA. Metallothionein isoform 1 and 2 gene expression in the human prostate: downregulation of MT-1X in advanced prostate cancer. Prostate. 2000;43:125–135. doi: 10.1002/(sici)1097-0045(20000501)43:2<125::aid-pros7>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 33.Rauen KA, Sudilovsky D, Le JL, Chew KL, Hann B, Weinberg V, Schmitt LD, McCormick F. Expression of the coxsackie adenovirus receptor in normal prostate and in primary and metastatic prostate carcinoma: potential relevance to gene therapy. Cancer Res. 2002;62:3812–3818. [PubMed] [Google Scholar]
- 34.Singh D, Febbo PG, Ross K, Jackson DG, Manola J, Ladd C, Tamayo P, Renshaw AA, D'Amico AV, Richie JP, Lander ES, Loda M, et al. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell. 2002;1:203–209. doi: 10.1016/s1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- 35.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2006;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 36.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 37.Shi XB, Xue L, Yang J, Ma AH, Zhao J, Xu M, Tepper CG, Evans CP, Kung HJ, deVere White RW. An androgen-regulated miRNA suppresses Bak1 expression and induces androgen-independent growth of prostate cancer cells. Proc Natl Acad Sci U.S.A. 2007;104:19983–19988. doi: 10.1073/pnas.0706641104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Glinsky GV, Krones-Herzig A, Glinskii AB. Malignancy-associated regions of transcriptional activation: gene expression profiling identifies common chromosomal regions of a recurrent transcriptional activation in human prostate, breast, ovarian, and colon cancers. Neoplasia. 2003;5:218–228. doi: 10.1016/S1476-5586(03)80054-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, Davuluri R, Liu CG, Croce CM, Negrini M, Calin GA, Ivan M. A MicroRNA Signature of Hypoxia. Mol Cell Biol. 2007;27:1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moschos SA, Williams AE, Perry MM, Birrell MA, Belvisi MG, Lindsay MA. Expression profiling in vivo demonstrates rapid changes in lung microRNA levels following lipopolysaccharide-induced inflammation but not in the anti-inflammatory action of glucocorticoids. BMC.Genomics. 2007;8:240. doi: 10.1186/1471-2164-8-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.West AK, Stallings R, Hildebrand CE, Chiu R, Karin M, Richards RI. Human metallothionein genes: structure of the functional locus at 16q13. Genomics. 1990;8:513–518. doi: 10.1016/0888-7543(90)90038-v. [DOI] [PubMed] [Google Scholar]
- 42.Henrique R, Jeronimo C, Hoque MO, Nomoto S, Carvalho AL, Costa VL, Oliveira J, Teixeira MR, Lopes C, Sidransky D. MT1G hypermethylation is associated with higher tumor stage in prostate cancer. Cancer Epidemiol.Biomarkers Prev. 2005;14:1274–1278. doi: 10.1158/1055-9965.EPI-04-0659. [DOI] [PubMed] [Google Scholar]
- 43.Wei H, Desouki MM, Lin S, Xiao D, Franklin RB, Feng P. Differential expression of metallothioneins (MTs) 1, 2, and 3 in response to zinc treatment in human prostate normal and malignant cells and tissues. Mol.Cancer. 2008;7:7. doi: 10.1186/1476-4598-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ajioka RS, Phillips JD, Kushner JP. Biosynthesis of heme in mammals. Biochim.Biophys.Acta. 2006;1763:723–736. doi: 10.1016/j.bbamcr.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 45.Dakubo GD, Parr RL, Costello LC, Franklin RB, Thayer RE. Altered metabolism and mitochondrial genome in prostate cancer. J Clin Pathol. 2006;59:10–16. doi: 10.1136/jcp.2005.027664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh KK, Desouki MM, Franklin RB, Costello LC. Mitochondrial aconitase and citrate metabolism in malignant and nonmalignant human prostate tissues. Mol Cancer. 2006;5:14. doi: 10.1186/1476-4598-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petros JA, Baumann AK, Ruiz-Pesini E, Amin MB, Sun CQ, Hall J, Lim S, Issa MM, Flanders WD, Hosseini SH, Marshall FF, Wallace DC. mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci U.S.A. 2005;102:719–724. doi: 10.1073/pnas.0408894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rhodes DR, Barrette TR, Rubin MA, Ghosh D, Chinnaiyan AM. Meta-analysis of microarrays: interstudy validation of gene expression profiles reveals pathway dysregulation in prostate cancer. Cancer Res. 2002;62:4427–4433. [PubMed] [Google Scholar]
- 49.Smith RC, Litwin MS, Lu Y, Zetter BR. Identification of an endogenous inhibitor of prostatic carcinoma cell growth. Nat.Med. 1995;1:1040–1045. doi: 10.1038/nm1095-1040. [DOI] [PubMed] [Google Scholar]
- 50.Watson JE, Doggett NA, Albertson DG, Andaya A, Chinnaiyan A, van Dekken H, Ginzinger D, Haqq C, James K, Kamkar S, Kowbel D, Pinkel D, et al. Integration of high-resolution array comparative genomic hybridization analysis of chromosome 16q with expression array data refines common regions of loss at 16q23-qter and identifies underlying candidate tumor suppressor genes in prostate cancer. Oncogene. 2004;23:3487–3494. doi: 10.1038/sj.onc.1207474. [DOI] [PubMed] [Google Scholar]
- 51.Bruning A, Runnebaum IB. CAR is a cell-cell adhesion protein in human cancer cells and is expressionally modulated by dexamethasone, TNFalpha, and TGFbeta. Gene Ther. 2003;10:198–205. doi: 10.1038/sj.gt.3301887. [DOI] [PubMed] [Google Scholar]