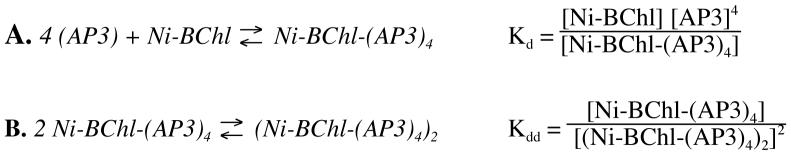Abstract
We demonstrate coordination of the extremely hydrophobic 132-OH-Ni-bacteriochlorophyll (Ni-BChl) to the lipophilic domain of a novel, designed amphiphilic protein maquette (AP3) dispersed in detergent micelles (accompanying manuscript; Discher et al). Sedimentation velocity and equilibrium experiments, and steady state absorption spectra indicate that Ni-BChl-AP3 is a four-helix bundle containing one Ni-BChl axially ligated by one or two histidines. The nature of the ligation was pursued with ultrafast visible spectroscopy. While it is well established that light excitation of axially ligated mono- and bis-imidazole Ni-BChl in solution leads to rapid imidazole dissociation and nanosecond recombination, there is no evidence of axial ligand dissociation in the light-excited Ni-BChl-AP3. This indicates that Ni-BChl is confined within the AP3 protein, ligated to histidines with severely restricted mobility. Dissociation constants show that Ni-BChl binding to AP3 is considerably weaker than the nano-molar range usual for heme and hydrophilic (HP) maquettes; moreover, there is a tendency for the Ni-BChl-AP3 four-helix bundles to dimerize into eight-helix bundles. Nevertheless, the preparation of the Ni-BChl-AP3 four-α-helix maquettes, supported by time-resolved spectroscopic analysis of the nature of the ligation provides a viable new approach to AP maquette designs that address the challenges involved in binding extremely hydrophobic cofactors.
Keywords: Amphiphilic protein maquettes, heme, Ni-bacteriochlorophyll, protein de novo design, analytical ultracentrifugation, time-resolved absorption spectroscopy
The protein maquette concept was introduced about ten years ago with the de-novo design of a four-α-helix bundle protein that ligated hemes (1). Maquettes intend to provide the simplest protein scaffold in which to test and examine the assembly and function of catalytic elements that, in native enzymes, are often obscured by complexity. In many cases, metal ions and clusters, and organic and organometalic molecules are critical parts of such catalytic elements, and usually they are associated within the protein through non-covalent interactions such as coordinative and hydrogen bonds, and/or van der Waals interactions. Understanding the details of these protein-cofactor interactions is essential for developing custom-designed catalysts that will match and expand upon the selectivity, activity and efficiency of natural enzymes.
Most of cofactor binding maquette designs that followed the early prototypes made principal use of metal centers or heme derivatives coordination to the protein (2-14). Binding other organic cofactors to maquettes has proven more challenging and has often resorted to covalent coupling via specific natural or non natural residues that effectively incorporate the desired organic cofactors as a side chain (15, 16). This is because of the great difficulties in achieving the fine balance of hydrophobic and polar interactions between peptides, cofactors, and solvent with organic cofactors unconstrained by any covalent link to the peptide backbone (17, 18). Most importantly, incorporating very hydrophobic cofactors into water-soluble maquette is severely limited by the tendency of these cofactors to self-aggregate in polar solvents.
The preceding paper (19) introduced a new type of amphiphilic (AP) maquettes designed according to the general architecture of natural membrane proteins (20, 21). AP maquettes are based on a construct of hydrophilic (HP) and lipophilic (LP) domains and require hydrophobic environments such as lipid bilayers, or detergent micelles in which to assemble. This work followed effects of the new non-polar environment on heme binding properties of a HP domain of an AP maquette and its implications on heme associated functional elements with respect to the polar environment of a fully hydrophilic HP maquette. The work also demonstrated the first examples of heme and bacteriochlorophyll (BChl) binding to a LP domain of an AP maquette, and drew attention to the challenges faced, particularly the case of the BChl derivatives, in devising strategies to assemble very hydrophobic cofactors within hydrophobic LP domains and, once assembled, determine the nature of the association.
BChls are extremely hydrophobic and have not been incorporated into the hydrophobic interior of previous HP maquettes because of their very high tendency to self-aggregate in water. The only other chlorophyll (Chl) and BChl derivatives successfully incorporated into HP maquettes either had their phytyl chain removed, which significantly increased their solubility in water (22), or were covalently bound (15). Other examples of Chl/BChl binding to synthetic polypeptides include a short randomly coiled polypeptide based on a Chl binding motif from plant light-harvesting complex (23, 24), and the self-assembly of BChls with truncated and modified proteins from purple bacterial light-harvesting complexes (25). In all these cases cofactors were not incorporated into a binding pocket in the protein but rather interacted with several key protein residues and remain exposed to solvent and other cofactors. In fact, BChl-protein assembly in the latter case is driven by pigment self-aggregation. Kashiwada et al. reported on BChl binding to a polypeptide based on a sequence variant of the HP heme-binding maquette in detergent solution (26). However, the CD and absorption spectra of BChls is indicative of aggregates and there is no indication that the peptide retains the four-helix bundle fold as designed.
The intention of this work is to begin to address the unique challenges of non-covalently binding extremely hydrophobic cofactors to de-novo designed proteins. For this purpose, a Ni substituted BChl (Ni-BChl) derivative is especially useful because its rich absorption spectra is well correlated with the type and number of axial ligands, which makes it an excellent reporter for the local coordination and solvation environment (27, 28). We show that Ni-BChl is axially coordinated within the LP domain of the AP3 maquette, most probably by one or two histidine (His) residues, and take advantage of the distinctive excited state dynamics of Ni-BChl derivatives (29-34), especially the well-documented axial ligand photodissociation (29, 31, 33), to examine the physical nature of the axial ligation. Thus, if Ni-BChl is ligated by residues that are confined within the AP3 protein structure, we expect the dynamics as seen by the time-resolved absorption spectra of Ni-BChl-AP3 to be significantly different from those observed in solvents or detergent micelles. Indeed, we show that in contrast to Ni-BChl-Im in micelles there is no indication of axial ligand photodissociation in AP3-bound Ni-BChl. The unique spectroscopic and coordinative properties of Ni-BChl provide a variety of useful probes for the local protein environment which can be monitored by a variety of multi-wavelength spectroscopic measurements and provide useful information about the molecular charcteristics of this new cofactor-maquette assembly. Our success with an extremely hydrophobic cofactor into maquettes demonstrates the viability of the modular design approach for AP maquettes and opens new functional possibilities by incorporating new cofactors previously inaccessible to HP maquettes.
MATERIALS AND METHODS
Peptide and pigment synthesis, and sample preparation
The amphiphilic maquette, AP3, was prepared as described in the preceding paper (19). Ni-BChl was prepared by trans-metallation of native BChl following Hartwich et al. (35). The ratio between 132-OH-Ni-BChl and the 132-OH’-Ni-BChl epimer was not determined and although this ratio might affect the self-aggregation tendency of Ni-BChl as well as specific interactions with AP3, it appears secondary to the strong hydrophobic interactions that drive Ni-BChl self-aggregation. Currently, it is hard to estimate the contribution of specific Ni-BChl-AP3 interactions in the absence of structural information at the molecular level. All aqueous solutions were buffered with 20 mM sodium phosphate at pH 8.0 and contained 100 mM sodium chloride.
The preparation of a stable concentrated sample of Ni-BChl-AP3 has proven to be a significant challenge mostly because of the low solubility and high self-aggregation tendency of Ni-BChl, even in detergent solutions. High detergent concentrations are required in order to prevent such self-aggregation but this lowers the affinity of Ni-BChl for AP3 and hence in turn demands higher concentrations of AP3 in order to obtain significant binding. We tested two different detergents, Zwittergent 3-12 (zwt312, n-Dodecyl-N,N-dimethyl-3-ammonio-1-propanesulfonate) and β-octyl-glucopyranoside (OG). A solution of 1% (w:w, about 100 fold above the critical micelle concentration) zwt312 completely solubilized AP3 but we noticed significant aggregation of Ni-BChl whereas increasing zwt312 concentration to 4% completely solubilized Ni-BChl but prevented association with AP3. Finally, 2% zwt312 solution provided good balance between the opposing requirements for Ni-BChl solubility and AP3 affinity. Nevertheless, the assembly of Ni-BChl-AP3 complex required AP3 concentrations in the order of 0.1 mM. Additionally, we found that heating Ni-BChl/zwt312 samples breaks down some of the aggregates as indicated by narrowing of the Ni-BChl Qx and Qy bands and a blue shift of 2-5 nm in the Qx band (not shown). However, while cooling the sample back to room temperature, re-aggregation occurs quite slowly over several hours. Similar results were obtained with OG, yet Zwt312 was chosen for further experiments because, in contrast to OG, its partial specific volume could be matched by D2O/H2O mixture for sedimentation studies (see below). Thus, to prepare the Ni-BChl-AP3 maquette, Ni-BChl was first dissolved in a minimal volume (5-10 μl) of DMF (Fisher, Fairlawn, NJ) and added to 16% (w:w) zwt312 (Calbiochem, San Diego, CA) solution. Then, the sample was diluted 8 fold and heated to 55°C to dissociate pigment aggregates. After cooling to room temperature, the sample was added to a solution of AP3 in 2% (w:w) zwt312. Similarly, Ni-BChl-Im was prepared by adding solid Im (Sigma, St. Louis, MO) into the Ni-BChl zwt312 solution. All spectroscopic measurements were carried out at room temperature.
Analytical ultracentrifugation
Sedimentation equilibrium and sedimentation velocity measurements were carried out using a Beckman XLI analytical ultracentrifuge. All sample solutions were density matched with D2O (Cambridge isotope labs) to neutralize zwt312 buoyancy. The density match point was determined to be 46.2% D2O by sedimentation equilibrium of 2% zwt312 at various D2O/H2O buffer mixtures. Ni-BChl-AP3 was prepared for ultracentrifugation by mixing 140 μl of 1 mM AP3 solution in 2% zwt312 H2O buffer with D2O and H2O buffers to reach 2% zwt312 in 46.2% D2O. Final sample contained 320 μM AP3 and 35 μM Ni-BChl. Samples were loaded into a standard two-channel analytical ultracentrifuge cell and spun at 50,000 rpm for 72 hrs at 25°C. During this time, the radial absorbance profiles were recorded at 540 and 600 nm. Once sedimentation equilibrium had been reached, a radial-spectral matrix was created by recording absorption spectra from 422 to 800 nm at 17 radial positions along the cell. Then, two additional radial absorbance profiles were recorded at 750 and 790 nm. These were combined with the latest absorbance profiles at 540 and 600 nm and were globally fitted to a sedimentation equilibrium model.
Sedimentation coefficient (S), and molecular weight (M) distributions (c(S) and c(M), respectively), were obtained from the global analysis of sedimentation velocity profiles monitored at 540 and 600 nm by using the multi-signal ck(S) analysis module in the software Sedphat 3.0 (36, 37) (obtained from http://www.analyticalultracentrifugation.com). Sedimentation equilibrium data were analyzed using custom-built programs written in Igor 5.05 (Wavemetrics, Lake Oswego, OR). The model used to describe radial absorbance profiles is similar to that of Schuck et al. (38) given by
| (1) |
where A(r) is the absorbance at a radial position r (in cm); 1.2 cm is the optical path length of the ultracentrifuge cell; p, l, c specify the apoprotein, cofactor, and protein-cofactor complex, respectively; εi(λ) (i = p,l,c) are the respective extinction coefficients (M-1·cm-1) at a wavelength λ; (i = p,l,c) are the respective concentrations (M) at an arbitrary radial position r0 (cm), R is the ideal gas constant (8.315·107 erg·K-1·mole-1), T is the temperature (°K), ω is the angular velocity of the rotor (radians·s-1), and and are the buoyant molecular weight of the protein and cofactor, respectively (g·mol-1) given by
| (2a) |
where i is either p or l, Mwi and are the molecular weights (g·mole-i) and partial specific volume (ml·g-1) of the species, respectively, and ρ is the solvent’s density (g·ml-1). The buoyant molecular weight of the protein-cofactor complex, is given by
| (2b) |
where n is the stoichiometric number of the protein in the complex (we assume one cofactor per n apo-proteins in a complex). The concentrations were calculated by requiring mass conservation for the entire solution column, and the mass action law for complex formation
| (3) |
where Kd is the dissociation constant of the complex in terms of Mn; and are the effective total loading concentrations of protein and cofactor, respectively, and χp, χl and χc are dimensionless factors given by
| (4) |
where b and m are the radial positions of the solution meniscus and the cell bottom, respectively, and is given by Eq. 2a. Eq. 4 can be solved analytically by use of Dawson’s integral (39). Here, we use and as parameters when fitting equation 1 to the experimental data.
Time-resolved absorption spectroscopy
Transient absorption spectra were recorded using a femtoseconds pump-probe absorption spectrometer. Optical pulses (135 fs) centered at 775 nm, were generated using a Ti:Sapphire laser (CPA-2001, Clark-MXR, Dexter, MI), which consisted of a regenerative amplifier seeded by a mode-locked fiber oscillator. A fraction of this output was focused into a 2 mm c-cut sapphire plate to generate a white light continuum that was used as the probe beam, and the rest was passed through optical parametric amplifiers (near-IR and visible OPAs, Clark-MXR) to generate pump excitation pulses tunable in wavelength from the UV through the near-IR region. The pump beam was passed through a computer-controlled delay line and was focused on a 2 mm path length sample cell together with the probe beam. After passing through the sample, the probe light was focused onto the entrance slit of a computer-controlled image spectrometer (SpectraPro-150, Acton Research Corporation, Acton, MA). A CCD array detector (1024 × 128 elements, Roper Scientific, Duluth, GA), interfaced to the spectrometer, recorded the spectrum of the probe light from the UV (∼370 nm) to the near-IR (∼1100 nm), providing spectral resolution better than 0.5 nm. Pairs of consecutive spectra were measured with (Ion(λ)) and without (Ioff(λ)) optical pumping to determine the difference spectrum, ΔA = log(Ioff(λ) / Ion(λ)). All transient spectra reported represent averages obtained over 4-6 scans with each scan consisting of 150-250 points. A more detailed description of the optical setup can be found in the supporting information of (40).
Ni-BChl, Ni-BChl-Im and Ni-BChl-AP3 were excited at 550 nm. Ni-BChl-Im and Ni-BChl-AP3 were also excited at 585 and 610 nm in order to selectively excite mono- and bi-ligated Ni-BChl populations. The resulting kinetic-spectral matrices were fitted to kinetic models by using iterative pseudo-inverse routine described in more details below.
Curve fitting and global analysis
Ni-BChl has rich and informative absorption features over a wide range of the near-UV, visible, and NIR spectrum. In order to take full advantage of these unique features, our experiments were designed to obtain multi-wavelength data. Non-linear global fitting and pseudo-inverse methods provide the most efficient and reliable way to handle this type of data. In global fitting, multiple data-sets are fit simultaneously to functional models for which some parameters are common to several data sets (global), and some are unique for each set (local) (41). We used the built-in non-linear global fitting procedures in the Igor 5.05 software to simultaneously fit radial absorbance curves at four wavelengths. Confidence limits for the non-linear fit parameters were obtained by implementing the linear joint confidence intervals method as described by Johnson and Faunt (40) at 0.95 confidence level. This method provides more reliable error estimates for non-linear fits than the standard routines because it does not assume the confidence regions to be either linear or symmetrical.
Pseudo-inverse methods are useful for analyzing multi-wavelength absorption data. When a sample is comprised of a mixture of species, according to the Beer-Lambert law, its absorption spectrum at each wavelength, A(λ), is the weighted sum of species concentration, Ci, and their respective extinction coefficients at each wavelength εi(λ). If the sample’s composition changes as a function of an experimental parameter, p, then the set of absorption spectra, A(λ, p), is given by
| (5a) |
which may be written as a matrix product
| (5b) |
where A(λk,rj)) is a spectral matrix for which columns and rows correspond to values of the parameter p and wavelengths, respectively, E(λk) is the extinction coefficient matrix and C(pj) is the concentration profile matrix. Using a functional model C(pj, ϕi) where ϕi are a set of parameters, it is possible to obtain E*, a least-squares estimate of E (42) with
| (6) |
where C+ is the pseudo-inverse of C(pj, ϕi). The maximum likelihood set of fitting parameters may be found by any non-linear least squares algorithm that minimizes
The pseudo-inverse method was applied in analyzing the radial-spectral matrix from the sedimentation equilibrium experiments, and the kinetic spectral matrix from time-resolved absorption measurements.
For the radial-spectral matrix A(λk,rj) we used the matrix form of equation 1
| (7) |
where each column in the matrix E(λk) corresponds to the absorption spectra εi(λκ) of apo-AP3, free Ni-BChl, and Ni-BChl-AP3, and each row of radial concentration profile matrix C(rj, , , ω, R, T) contains their respective concentration gradients
For the kinetic-spectral matrix, D(λk,tj) we use
| (8) |
where each column in the matrix S(λk) corresponds to a decay-associated difference spectrum (DADS) of an excited state, and each row of the kinetic profile matrix C contains the respective kinetic profile of each state determined by the set of differential equations
| (9) |
where K is the matrix of rate constants. We assume a simple kinetic model whereby each excited state decays to the next excited state until the final excited state decays to ground state. The matrix elements of K are therefore given by
| (10) |
where τi is the lifetime of the ith excited state, and δ(i,j) is the Kronecker delta function. Additionally, we assume the initial conditions C(t=0) = δ(i,1).
RESULTS
Spectral features of Ni-BChl, Ni-BChl-AP3, and Ni-BChl-Im in zwt312 detergent
Figure 1 compares the absorption spectrum of Ni-BChl in 2% zwt312 solution to its spectra in the presence of either 0.4 mM AP3 or 1 M Im in the same solvent. The effects of both protein and Im on Ni-BChl in the detergent micelle solution are similar to those of nitrogeneous axial ligands in organic solvents. Typically, the Qy band narrows and blue shifts by 10-15 nm whereas the Qx band shifts by 40-70 nm depending on the number and type of axial ligands (27, 28). Here, the Qx band peak shifts from 538 nm in unbound Ni-BChl to 584 nm in Ni-BChl with Im to 609 nm in Ni-BChl with AP3, and the Qy band peak shifts from 783 nm in unbound Ni-BChl to 772 nm in Ni-BChl with Im to 768 nm in Ni-BChl with AP3. These changes are characteristic of Ni-BChl binding one or two axial ligands, respectively. Additionally, the intensity of Ni-BChl-AP3 absorption bands increases significantly compared to Ni-BChl and Ni-BChl-Im, similar to that observed upon binding heme derivatives to both water-soluble maquettes and HP and LP domains of AP maquettes (11, 19). However, in contrast to heme, a significant amount of Ni-BChl remains unligated even at high excess of either Im or AP3 as indicated by the significant shoulder at 538 nm observed in the Qx band of both Ni-BChl-Im and Ni-BChl-AP3 spectra.
Figure 1.
Changes in absorption spectra of 40 μM Ni-BChl (heavy line) in 2% zwt312 solution with the addition of 1M Im (dotted line), or in the presence of 0.4 mM AP3 (solid line).
Within several hours, new features indicative of pigment aggregation appear in the Ni-BChl-Im spectra (not shown) that include 850 nm and 620 nm shoulders at the Qy and Qx bands, respectively. In contrast, the spectra of Ni-BChl and Ni-BChl-AP3 remained unchanged for several days with no indication of either pigment aggregation or degradation.
Ni-BChl-AP3 complex stoichiometry and binding characteristics
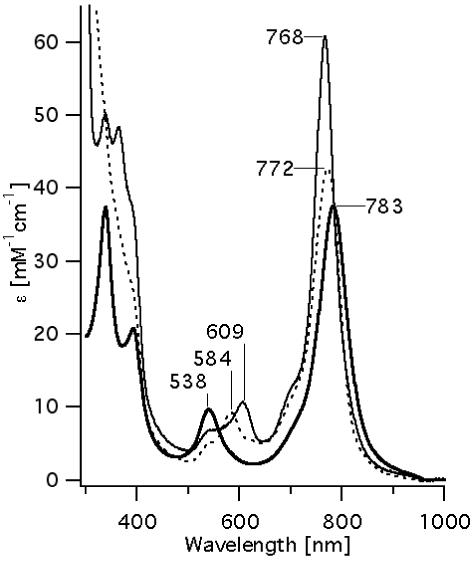
Figure 2 presents a radial-spectral matrix of Ni-BChl-AP3 in density-matched zwt312 once sedimentation equilibrium has been reached. Evidently, the prominent spectral features at longer radii are typical of axially ligated Ni-BChl whereas the spectral features at short radii are almost exclusively those of non-ligated Ni-BChl. This clearly indicates that the high molecular weight Ni-BChl-AP3 complex contains axially coordinated Ni-BChl and strongly suggests that Ni-BChl is bound to AP3 via one or two His ligands. Conversely, non-ligated Ni-BChl is a low molecular weight species and thus it is highly unlikely that this species is bound to AP3. Nonetheless, the spectra at longer radii still feature a significant peak at 540 nm indicative of a low affinity of Ni-BChl to AP3.
Figure 2.
Radial-spectral matrix of Ni-BChl-AP3 sample that have reached sedimentation equilibrium at 50,000 rpm.
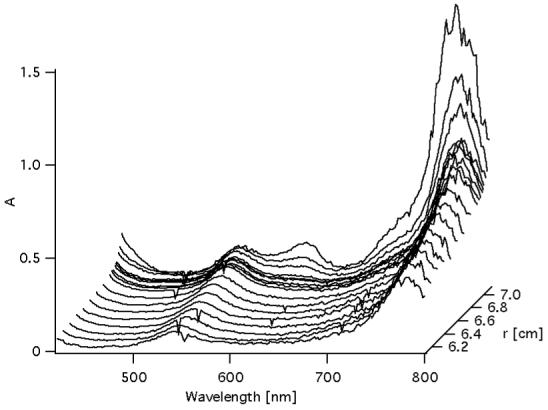
Figure 3 presents two sedimentation coefficients distributions, c(S), of Ni-BChl-AP3 in density matched zwt312 solution obtained by monitoring the same sedimenting sample at 540 nm where free Ni-BChl has the predominant absorbance, and at 600 nm where Ni-BChl-AP3 absorbance predominates. As expected, at 540 nm a major peak (88% of total area) is observed at 0.09 S corresponding to molecular weight of 904 Da and attributed unbound Ni-BChl (actual molecular weight of 944 Da). However, an additional minor peak (12% of total area) at 0.55 S corresponds to a molecular weight of 13,665 Da more typical of a much larger Ni-BChl-AP3 complex. This distribution can be rationalized according to Gilbert-Jenkins theory (43) by assuming rapid reversible interactions between Ni-BChl, AP3, and their complex. As recently shown (36, 44), the sedimentation profile of such a two-component solution gives rise to a bimodal c(S) distribution with a peak appearing at a S-value close to that of the slowly sedimenting free component (Ni-BChl) corresponding to an undisturbed boundary, and a second peak at a position between the S-values of the slowest and fastest sedimenting species corresponding to a reaction boundary comprised of a reaction mixture of interacting species (Ni-BChl, apo-AP3, and Ni-BChl-AP3). Thus, the minor peak is not due to residual absorbance of the Ni-BChl-AP3 at 540 nm but rather to free Ni-BChl in the reaction boundary.
Figure 3.
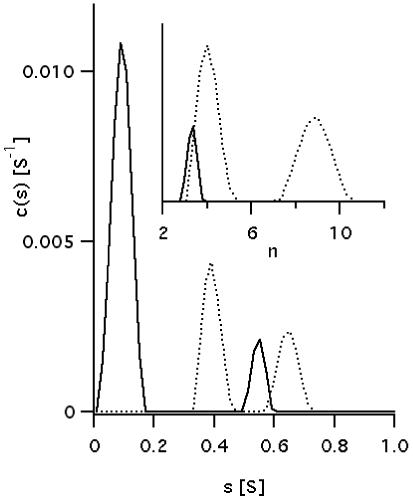
Sedimentation coefficients distribution, c(S), plots of Ni-BChl-AP3 in density matched zwt312 obtained from global fitting of sedimentation profiles monitored at 540 nm (solid line) and 600 nm (dotted line). Distributions were calculated assuming the presence of time invariant and radial invariant noise, an unknown global meniscus position and optimizing the weight-average frictional ratios separately for data sets of each wavelength. Inset: aggregation number distribution, c(n), is plotted for the higher S region. n is the aggregation number in Ni-BChl-(AP3)n.
The c(S) distribution obtained from monitoring sedimentation at 600 nm exhibits two distinct peaks at 0.39 S (61% of total area) and 0.65 S (39% of total area) corresponding molecular weights of 16,091 Da and 34,625 Da, respectively. Assuming a single Ni-BChl per apo-AP3 complex, these peaks correspond to aggregation numbers (n) of 4.0, and 8.9 respectively, as shown in the c(n) distribution graph (Figure 3 inset). Given the assumptions involved in converting sedimentation coefficients to actual molecular weights (45), and the discrete nature of these peaks, it follows that the peaks in c(n) monitored at 600 nm corresponds to the Ni-BChl-AP3 complex comprised of four AP3 monomers, Ni-BChl-(AP3)4, and its dimeric form, (Ni-BChl-(AP3)4)2. Based on the ratio between peak areas, assuming that the AP3:Ni-BChl stoichiometry is maintained in the dimer, and that contributions to the absorbance at 600 nm are additive, we obtained a ratio of one (Ni-BChl-(AP3)4)2 to three Ni-BChl-(AP3)4.
A model consistent with the sedimentation velocity results is given in scheme 1. This model may be used in the analysis of sedimentation equilibrium profiles to provide more quantitative description of Ni-BChl binding to AP3. However, the analysis of a system of such complexity involving association of two components followed by self-assocation of the product is very complicated. In order to simplify the analysis and reduce some degrees of freedom, we recorded sedimentation equilibrium profiles at four characteristic wavelengths: 540 and 790 nm where absorbance is primarily from unbound Ni-BChl, and 600 and 750 nm where absorbance is primarily due to Ni-BChl-AP3. These were globally fitted together with the radial spectral matrix using the equilibrium model in Eq. 1 and scheme 1A. For further simplification we neglect self-association reaction in scheme 1B. The best fit to the experimental data, presented in Figure 4, was obtained by using Mwp = 3,758 Da and n = 4 in Eq. 2a and 2b, respectively, and yielded = 0.7891 ± 0.0007, and = 0.8890 ± 0.0006 ml·g-1 for the partial specific volumes of AP3 and Ni-BChl, respectively, in good agreement with the theoretical values of = 0.7773 and = 0.91. Using Mwp = 15,032 or 7,516 Da in Eq. 2a and n = 1 or 2 in Eq. 2b, respectively, yielded worse fits and unreasonably high values of > 0.8. This supports the stoichiometry in scheme 1A and implies that Ni-BChl-AP3 assembles simultaneously from four apo-AP3 monomers and one Ni-BChl. Sedimentation equilibrium experiments of apo-AP3 under similar conditions (not shown) verified that apo-AP3 is not pre-assembled in four-helical bundles which is not surprising given the high detergent concentration. The resulting dissociation constant Kd = 0.00173 ± 0.00002 mM4 implies that significant dissociation may occur even at AP3 concentration as high as 200 μM. This is much higher than dissociation constants of other heme binding protein maquettes that are typically on the order of 1 μM or lower.
Scheme 1.
Modes of AP3 and Ni-BChl interactions and association equilibrium.
Figure 4.
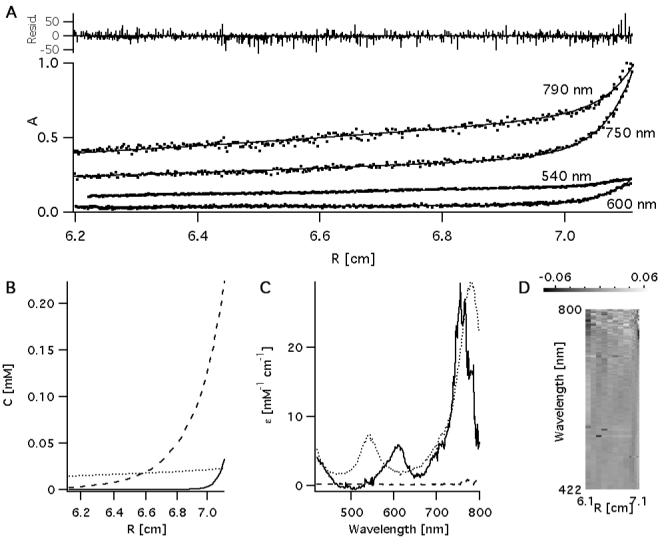
Sedimentation equilibrium traces of Ni-BChl-AP3 at 790, 750, 600, and 540 nm (A) were globally fitted together with the radial-spectral matrix (Figure 2) assuming association of Ni-BChl with 4 AP3 helices. The model yielded a sedimentation profile (B) for apo AP3 (dashed line), unbound Ni-BChl (dotted line) and Ni-BChl-AP3 (solid line). Using this profile and the radial-spectral data it was possible to resolve the corresponding spectra of each component (C). These were used with the radial profiles in (B) to reproduced radial-spectral data and compared to the experimental data. This is presented as an image (D) whereby residual values at each data point are depicted as bright (positive) or dark (negative) pixels.
Time-resolved absorbance spectra
Figure 5 shows the transient absorption spectra of Ni-BChl and Ni-BChl with 1M Im excited at 550 and 610 nm, respectively, and Ni-BChl-AP3 excited at 550 and 610 nm in 2% zwt312. As expected, relaxation of excited Ni-BChl (Figure 5a) is extremely fast and the ground state is completely recovered within 100 ps. This is typical of Ni-BChl and Ni-porphyrins in non-coordinating organic solvents (31, 32). Likewise, the transient absorption spectra of Ni-BChl in the presence of 1 M Im (Figure 5b) resemble those of Ni-BChl in pyridine, a coordinating organic solvent (31). Particularly, a long-lived transient feature with positive amplitude at 780 nm and a negative one at 760 nm becomes obvious after about 100 ps and persists well beyond 1 ns. These bands correspond to the difference between the Qy spectral bands of ligated and non-ligated Ni-BChl and are characteristic of axial ligand photodissociation as has been observed for Ni-BChl in pyridine (31). Similar photodissociation is known to occur in other axially ligated Ni-porphyrins in organic solvents (33). Interestingly, the confined zwt312 micellar environment of Ni-BChl does not prohibit the photodissociation of ligand molecules from Ni-BChl. In contrast, the long-lived photo-dissociative feature is missing in the respective transient spectra of Ni-BChl-AP3 (Figure 5d), yet the short-term spectral features (< 100 ps) are very similar to those of Ni-BChl-Im.
Figure 5.
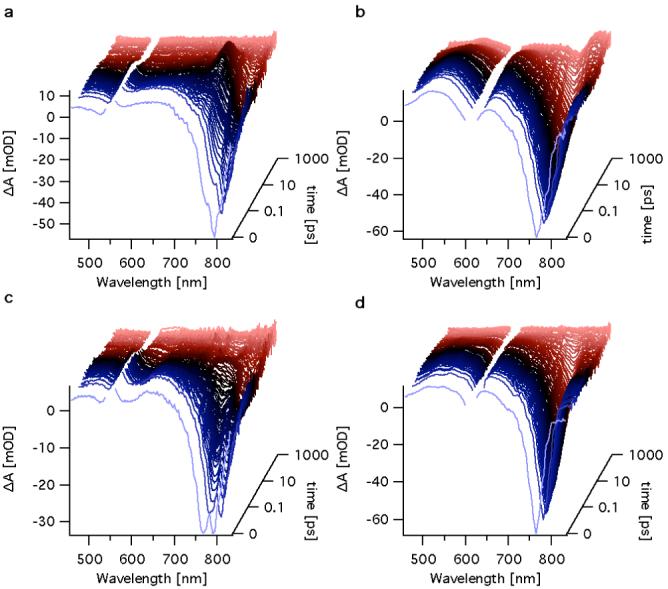
Transient absorption spectra of Ni-BChl in 2% zwt312 excited at 550 nm (a), Ni-BChl in 2% zwt312 containing 1 M Im excited at 610 nm (b), and Ni-BChl-AP3 excited at 550 nm (c) and 610 nm (d).
The transient absorption spectra of both Ni-BChl with Im and Ni-BChl-AP3 following excitation at 585 nm (not shown) are the same as those following excitation at 610 nm. The first wavelength selectively excites mono-ligated Ni-BChl whereas the latter excites bi-ligated Ni-BChl. When the excitation wavelength is at 550 nm we expected to excite mainly non-ligated Ni-BChl but we cannot avoid exciting some mono-ligated Ni-BChls. Indeed, the transient spectra of both Ni-BChl-AP3 (Figure 5c) and Ni-BChl with Im excited at 550 nm are consistent with our expectations. In both cases, the transient spectra did not exhibit any new spectral features but rather a mixture of the typical features of ligated and non-ligated Ni-BChl transient spectra.
The DADS and their associated lifetimes provide more detailed information about the transient states and relaxation processes of photo-excited Ni-BChl in detergent micelles and within the AP3 maquette. Table 1 summarizes the relaxation lifetimes obtained from sets of transient absorption spectra. The resulting excited state lifetimes of Ni-BChl and Ni-BChl-Im in zwt312 are very similar to those reported by Musewald et al. (31) for Ni-BChl in toluene and pyridine, respectively, with the exception of the earlier excited state lifetimes of Ni-BChl. Our analysis indicates two earlier transients with lifetimes of 69.5 fs and 1.0 ps whereas Musewald et al. (31) report on three transients with lifetimes of 0.1, 0.45, and 4 ps whereby the fastest component is only detected by fluorescence up-conversion. While these differences may reflect different excited state dynamics in toluene and zwt312, they may be a result of the different experimental conditions. Particularly, the excitation wavelength used by Mueswald et al. (31) was 780 nm which excites the Qy band whereas we excite the Qx band at 550 nm. Resolving this discrepancy requires a more careful study of the early events of Ni-BChl excitation in both organic solvents and detergent micelles. However, since our primary interest is the excited dynamics of AP3 bound Ni-BChl which is axially ligated by at least one His residue, we chose not to pursue issue any further as it concerns the photo-physics of unbound, non-ligated Ni-BChl which is beyond the scope of this manuscript. This not withstanding, we adopt Musewald et al. (31) suggested assignment of excited states to illustrate the relaxation processes of photoexcited ligated and non-ligated Ni-BChl with proper reservations concerning the earliest transient states of the latter.
Table 1.
Kinetic components of Ni-BChl, Ni-BChl-Im and Ni-BChl-AP3
| λex [nm] | τ1 [fs] | τ2 [ps] | τ3 [ps] | Photodissociation* | |
|---|---|---|---|---|---|
| Ni-BChl | 550 | 69.5 ± 0.4 | 1.01 ± 0.01 | 20.9 ± 0.2 | - |
| Ni-BChl-Im | 550 | 76 ± 1 | 0.83 ± 0.01 | 67.6 ± 0.8 | + |
| 585 | 150 ± 4 | 1.39 ± 0.02 | 85.4 ± 0.5 | + | |
| 610 | 135 ± 3 | 2.40 ± 0.03 | 92.5 ± 0.3 | + | |
| Ni-BChl-AP3 | 550 | 69.4 ± 0.5 | 0.96 ± 0.01 | 16.6 ± 0.2 | - |
| 585 | 130 ± 2 | 1.41 ± 0.01 | 68.5 ± 0.5 | - | |
| 610 | 104 ± 2 | 2.42 ± 0.02 | 79.9 ± 0.4 | - | |
Long-lived transient (τ ≫ 1 ns) with spectral features characteristic of axial ligand photodissociation
The DADS and of Ni-BChl in zwt312 (Figure 6) exhibit transient spectra indicative of a blue shift in the Qy transition. These are associated with an excited state having a high-spin Ni that relaxes to the low-spin Ni ground state.
Figure 6.
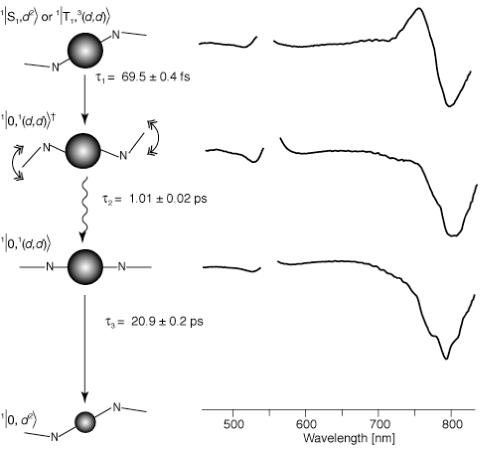
DADS of Ni-BChl in zwt312 excited at 550 nm, and their associated transient states, relaxation processes and lifetimes. State assignments and bracket notations are according to Musewald et al. (31) whereby the first term in the bracket is the macrocycle state (0, S, and T, for ground, singlet and triplet excited states, respectively), the second term is the Ni atom state ((d2), and (d,d) for low and high-spin Ni, respectively), and the superscript numbers to the left are spin multiplicities for each state. In the graphic schemes of each state, Ni-BChl is represented by its central Ni atom and two macrocycle nitrogens. Small and large spheres correspond to low- and high-spin Ni, respectively. Straight and wavy arrows represent electronic and vibrational relaxation, respectively.
The DADS and lifetimes of Ni-BChl in 2% zwt312 with 1M Im following excitation at 610 nm (Figure 7) are in very good agreement with those of Ni-BChl in pure pyridine (31). More specifically, we observe three short-lived transients with lifetimes of 0.135, 2.4, and 92.5 ps, and an additional longer-lived component with a lifetime of more than 1 ns. These correspond to lifetimes of 0.2, 6, and 90 ps for the respective short-lived transients, and more than 2 ns for the longer lived transients in pyridine. The spectral features of the long-lived component in zwt312 and in pyridine are characteristic of ground-state non-ligated Ni-BChl. This long-lived transient is missing in the DADS of Ni-BChl-AP3 in 2% zwt312 following excitation at 610 nm (Figure 8), yet, the three short-lived transients are very similar to the Ni-BChl-Im DADS, with lifetimes of 0.105, 2.5 and 79.9 ps. Thus, as demonstrated schematically in Figures 7 and 8, excited state relaxation of Ni-BChl-AP3 follows the same pathways as Ni-BChl-Im in zwt312 and Ni-BChl in pyridine but non-ligated Ni-BChl ground state is formed only in the latter two cases. This implies that the movement of both Ni-BChl and its ligands within the AP3 maquette is restricted by the protein to an extent that either leads to fast geminate recombination or eliminates ligand photodissociation altogether.
Figure 7.
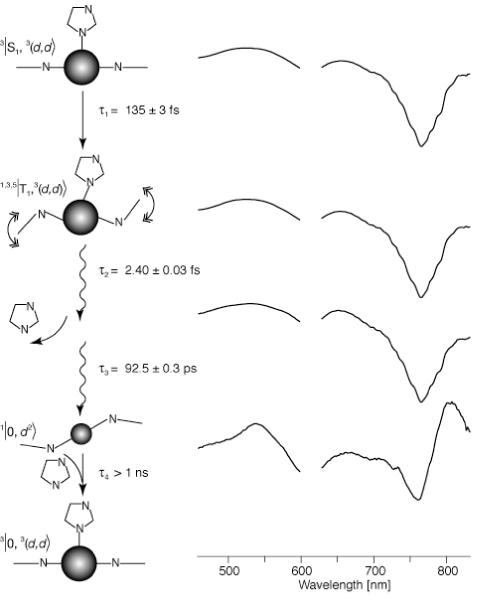
DADS of Ni-BChl in zwt312 with 1 M Im excited at 600 nm, and their associated transient states, relaxation processes and lifetimes. State assignments and bracket notations are according to Musewald et al. (31) whereby the first term in the bracket is the macrocycle state (0, S, and T, for ground, singlet and triplet excited states, respectively), the second term is the Ni atom state ((d2), and (d,d) for low and high-spin Ni, respectively), and the superscript numbers to the left are spin multiplicities for each state. In the graphic schemes of each state, Ni-BChl is represented by its central Ni atom and two macrocycle nitrogens. Small and large spheres correspond to low- and high-spin Ni, respectively. Straight and wavy arrows represent electronic and vibrational relaxation, respectively. For simplicity, we present only one axial Im ligand although the actual process may involve sequential or simultaneous photodissociation of two axial Im molecules.
Figure 8.
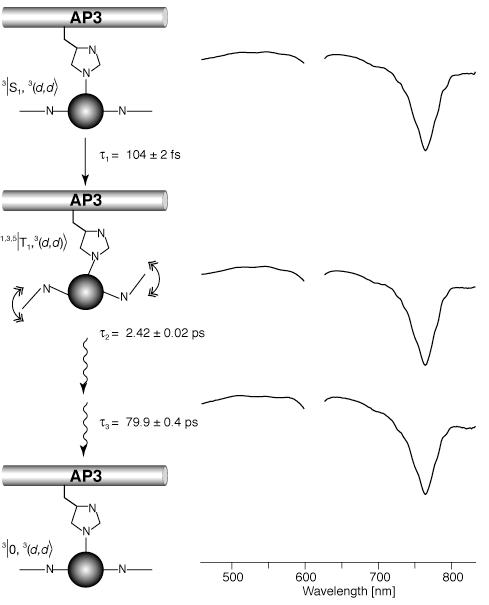
DADS of Ni-BChl-AP3 in zwt312 excited at 610 nm, and their associated transient states, relaxation processes and lifetimes. State assignments and bracket notations are according to Musewald et al. (31) whereby the first term in the bracket is the macrocycle state (0, S, and T, for ground, singlet and triplet excited states, respectively), the second term is the Ni atom state ((d2), and (d,d) for low and high-spin Ni, respectively), and the superscript numbers to the left are spin multiplicities for each state. In the graphic schemes of each state, Ni-BChl is represented by its central Ni atom and two macrocycle nitrogens. Small and large spheres correspond to low- and high-spin Ni, respectively. Straight and wavy arrows represent electronic and vibrational relaxation, respectively. For simplicity, we present only the mono-Histidyl Ni-BChl but the same relaxation scheme applies to bis-Histidyl Ni-BChl as well.
DISCUSSION
Efforts to design proteins that would assemble across hydrophilic-hydrophobic interfaces started some time ago with palmitoyl-terminated HP maquettes (46) culminating in the present AP family (19-21, 47) and novel design creations (48). This series of papers have opened up new avenues for structural and functional analysis of designed protein amphiphiles at air-water interfaces, in detergent-micelle dispersions and most recently across aqueous-membrane interfaces. Assembly across aqueous-membrane bilayers reproduces a very common feature of natural membrane proteins and the possibility of studying selected structural and functional elements of these often large and complex proteins in very much simplified AP maquettes. Indeed, as we have described in the accompanying manuscript, some of the AP maquettes already show promise for incorporating and examining functional elements in de novo designed proteins close to or inside a membrane bilayer (19). Importantly, a large variety of organic and organo-metallic cofactors that have proven to be too hydrophobic to be introduced to hydrophilic, HP, maquettes in aqueous solutions because of their poor solubility become accessible to AP maquettes. The present work has examined the binding of an extremely hydrophobic cofactor, Ni-BChl, to the lipophilic domain of a novel designed amphiphilic protein maquette, AP3. This Ni-BChl derivative was selected for its well-understood properties in solution and its characteristic spectral responses to axial ligation of nitrogenous bases such as imidazole and its amino acid analogue, histidine. The work has addressed in some detail the challenges we face in translating cofactor binding and assembly guidelines from the HP to the AP world, which is not straightforward because HP protein design relies on excluding hydrophobic elements from the polar solvent environment by packing them into the protein core (49). However, in a hydrophobic membrane environment this major driving force for HP peptide and cofactor self-assembly is not a significant factor, hence a finer balance of weaker interactions such as van der Waals packing, hydrogen bonds, metal coordination, and polar interactions is required for successful cofactor incorporation into AP maquettes (50, 51).
Although reconstitution of Chl and BChl derivatives into the heme binding sites of natural proteins and protein maquettes is not unprecedented (22, 52), only hydrophilic analogs such as Zn-chlorin e6, chlorophyllide a, or bacteriochlorophyllide a could be successfully incorporated. The observed Ni-BChl-AP3 dissociation constant of 0.00173 mM4, which is equivalent to 200 μM, is at least three orders of magnitudes higher than the sub-micromolar dissociation constant of AP and HP heme-maquette complexes. Nevertheless, it is still about three order of magnitudes lower than typical dissociation constants of bis- and mono-ligated Ni-BChl with small molecule ligands such as 1-methyl-Im in acetonitrile (25 mM and 709 mM2, respectively) (27). Although the typical dissociation constants of bis-ligated heme are slightly lower than the respective Ni-BChl (Kd = 100 mM2, for heme-Im2) (53), metal coordination alone cannot account for the significantly reduced affinity of AP3 to Ni-BChl as well as Zn-BChl (19). The high self-aggregation tendency of these hydrophobic cofactors is certainly a significant barrier for their incorporation into heme binding proteins and protein maquettes. Another barrier is the bulkiness and conformational flexibility of the reduced Chl and BChl macrocycle make them more disruptive within a protein core than the planar heme macrocycle, which is particularly destabilizing in HP maquettes that rely on good packing of their hydrophobic core. By these criteria, Ni-BChl represent the least favorable BChl derivative to be incorporated into maquettes because of its hydrophobic nature and high tendency to self-aggregate in polar environments, as well as the significant ruffling of its macrocycle (27).
Despite the complications involved in Ni-BChl binding, the properties of Ni-BChl-AP3 are very encouraging. At AP3 concentration of 0.1mM or less, axial coordination by one or two protein residues predominates as indicated by the typical changes in Ni-BChl absorption spectrum upon binding (Figure 1). The shift of the Ni-BChl Qx band from 538 to 605 nm is characteristic of bis-His ligated Ni-BChl, which is similar to bis-His ligation of heme in HP and AP maquettes. Yet, by contrast to the unique bis-His ligation in heme, the small shoulder at ∼585 nm in the Ni-BChl-AP3 spectrum suggests that some of the cofactors are only mono-ligated. Nonetheless, the sedimentation results (Figures 2 and 3) clearly show that non-ligated Ni-BChl is not bound to AP3, indicating that AP3 bound Ni-BChl is incorporated within the designated heme binding site and is either bis- or mono-ligated by His residues.
Sedimentation velocity provides direct measurement of Ni-BChl-AP3 aggregation state (Figure 3) and clearly shows that the aggregation number is primarily 4 similar to other HP and AP maquettes. Yet, about 25% of the Ni-BChl-AP3 appears to be dimers of this four-helical bundle complex. This is once again in contrast to HP binding maquette for which a unique four-helix bundle conformation has been achieved. Given the heterogeneity in coordination and aggregation states, it is tempting to assume correlation between coordination mode and aggregation number and speculate that the minor population of mono-ligated Ni-BChl corresponds to the dimeric form of the Ni-BChl-AP3 complex, yet, since these species are hard to resolve spectroscopically, we could not find any experimental evidence that would validate this speculation.
Time-resolved absorption spectroscopy provides valuable information about the local environment of Ni-BChl within AP3. Clearly, this environment is significantly different from Ni-BChl-Im in either detergent micelles or organic solvents. In Ni-BChl-AP3 we could not observe the long-lived intermediate that is typically observed in organic solvents upon axial ligand photodissociation from Ni-BChl. This is in contrast to Ni-BChl-Im in detergent micelles where such an intermediate was observed upon selective excitation of both mono- and bis-ligated Ni-BChl. While our spectroscopic data are insufficient to determine whether photodissociation in AP3 is followed by fast geminate recombination or is eliminated altogether, the absence of a long-lived photodissociated state indicates that the protein provides a rigid coordination environment which confines axial ligand motion. Furthermore, given the significant changes in macrocycle conformations between ligated and non-ligated Ni-BChl (27), it is likely that the protein environment imposes a significant conformational constraint on photodissociation by restricting macrocycle motion. Thus, it is reasonable to assume that Ni-BChl is incorporated into a binding pocket within the AP3 maquette. Conferring conformational flexibility to the maquette scaffold that will be initiated by axial ligand photodissociation is an aim for future maquette designs. Such a design may allow using Ni-BChl as an ultrafast phototrigger for protein conformational changes.
ACKNOWLEDGMENT
We thank Dr. Roie Yerushalmi from the Plant Sciences dept., Weizmann Institute for useful discussions on Ni-BChl axial ligation.
ABBREVIATIONS
- AP
amphiphilic
- BChl
bacteriochlorophyll
- Chl
chlorophyll
- DADS
decay-associated difference spectrum
- heme
Fe-protoporphyrin IX
- HP
hydrophilic
- HWHM
half-width at half-maximum
- Im
imidazole
- LP
lipophilic
- Ni-BChl
132-OH-Ni-bacteriochlorophyll
- zwt312
Zwittergent 3-12 (n-Dodecyl-N,N-dimethyl-3-ammonio-1-propanesulfonate)
Footnotes
The authors acknowledge financial support from NIH grant GM48130 and NSF grant DMR00-79909 to PLD, NIH grant P41RR003148 to RMH, and Human frontiers science program organization long term fellowship to DN.
REFERENCES
- 1.Robertson DE, Farid RS, Moser CC, Urbauer JL, Mulholland SE, Pidikiti R, Lear JD, Wand AJ, DeGrado WF, Dutton PL. Design and Synthesis of Multi-Heme Proteins. Nature. 1994;368:425–431. doi: 10.1038/368425a0. [DOI] [PubMed] [Google Scholar]
- 2.Benson DE, Haddy AE, Hellinga HW. Converting a maltose receptor into a nascent binuclear copper oxygenase by computational design. Biochemistry. 2002;41:3262–3269. doi: 10.1021/bi011359i. [DOI] [PubMed] [Google Scholar]
- 3.Calhoun JR, Kono H, Lahr S, Wang W, DeGrado WF, Saven JG. Computational design and characterization of a monomeric helical dinuclear metalloprotein. J. Mol. Biol. 2003;334:1101–1115. doi: 10.1016/j.jmb.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Di Costanzo L, Wade H, Geremia S, Randaccio L, Pavone V, DeGrado WF, Lombardi A. Toward the de novo design of a catalytically active helix bundle: A substrate-accessible carboxylate-bridged dinuclear metal center. J. Am. Chem. Soc. 2001;123:12749–12757. doi: 10.1021/ja010506x. [DOI] [PubMed] [Google Scholar]
- 5.Ghirlanda G, Osyczka A, Liu WX, Antolovich M, Smith KM, Dutton PL, Wand AJ, DeGrado WF. De novo design of a D-2-symmetrical protein that reproduces the diheme four-helix bundle in cytochrome bc(1) J. Am. Chem. Soc. 2004;126:8141–8147. doi: 10.1021/ja039935g. [DOI] [PubMed] [Google Scholar]
- 6.Gibney BR, Mulholland SE, Rabanal F, Dutton PL. Ferredoxin and ferredoxin-heme maquettes. Proc. Natl. Acad. Sci. U. S. A. 1996;93:15041–15046. doi: 10.1073/pnas.93.26.15041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibney BR, Isogai Y, Rabanal F, Reddy KS, Grosset AM, Moser CC, Dutton PL. Self-assembly of heme A and heme B in a designed four-helix bundle: Implications for a cytochrome c oxidase maquette. Biochemistry. 2000;39:11041–11049. doi: 10.1021/bi000925r. [DOI] [PubMed] [Google Scholar]
- 8.Kennedy ML, Gibney BR. Metalloprotein and redox protein design. Curr. Opin. Struct. Biol. 2001;11:485–490. doi: 10.1016/s0959-440x(00)00237-2. [DOI] [PubMed] [Google Scholar]
- 9.Lombardi A, Summa CM, Geremia S, Randaccio L, Pavone V, DeGrado WF. Retrostructural analysis of metalloproteins: Application to the design of a minimal model for diiron proteins. Proc. Natl. Acad. Sci. U. S. A. 2000;97:6298–6305. doi: 10.1073/pnas.97.12.6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maglio O, Nastri F, Pavone V, Lombardi A, DeGrado WF. Preorganization of molecular binding sites in designed diiron proteins. Proc. Natl. Acad. Sci. U. S. A. 2003;100:3772–3777. doi: 10.1073/pnas.0730771100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharp RE, Diers JR, Bocian DF, Dutton PL. Differential binding of iron(III) and zinc(II) protoporphyrin IX to synthetic four-helix bundles. J. Am. Chem. Soc. 1998;120:7103–7104. [Google Scholar]
- 12.Wade HV, Stayrook S, DeGrado WF. Structural analysis of a de novo designed metalloprotein. J. Inorg. Biochem. 2003;96:246–246. [Google Scholar]
- 13.Schnepf R, Haehnel W, Wieghardt K, Hildebrandt P. Spectroscopic identification of different types of copper centers generated in synthetic four-helix bundle proteins. J. Am. Chem. Soc. 2004;126:14389–14399. doi: 10.1021/ja0484294. [DOI] [PubMed] [Google Scholar]
- 14.Fahnenschmidt M, Bittl R, Schlodder E, Haehnel W, Lubitz W. Characterization of de novo synthesized four-helix bundle proteins with metalloporphyrin cofactors. Phys. Chem. Chem. Phys. 2001;3:4082–4090. [Google Scholar]
- 15.Rau HK, Snigula H, Struck A, Robert B, Scheer H, Haehnel W. Design, synthesis and properties of synthetic chlorophyll proteins. Eur. J. Biochem. 2001;268:3284–3295. doi: 10.1046/j.1432-1327.2001.02231.x. [DOI] [PubMed] [Google Scholar]
- 16.Sharp RE, Moser CC, Rabanal F, Dutton PL. Design, synthesis, and characterization of a photoactivatable flavocytochrome molecular maquette. Proc. Natl. Acad. Sci. U. S. A. 1998;95:10465–10470. doi: 10.1073/pnas.95.18.10465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barker PD. Designing redox metalloproteins from bottom-up and top-down perspectives. Curr. Opin. Struct. Biol. 2003;13:490–499. doi: 10.1016/s0959-440x(03)00108-8. [DOI] [PubMed] [Google Scholar]
- 18.Cochran FV, Wu SP, Wang W, Nanda V, Saven JG, Therien MJ, DeGrado WF. Computational de novo design and characterization of a four-helix bundle protein that selectively binds a nonbiological cofactor. J. Am. Chem. Soc. 2005;127:1346–1347. doi: 10.1021/ja044129a. [DOI] [PubMed] [Google Scholar]
- 19.Discher BM, Noy D, Ye SX, Moser CC, Lear JD, Blasie JK, Dutton PL. Amphiphilic protein maquettes: design, assembly, membrane insertion, and cofactor interactions. Biochemistry accompanying manuscript. 2005 doi: 10.1021/bi050695m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Discher BM, Koder RL, Moser CC, Dutton PL. Hydrophillic to amphiphilic design in redox protein maquettes. Curr Opin Chem Biol. 2003;7:741–748. doi: 10.1016/j.cbpa.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 21.Ye SX, Strzalka JW, Discher BM, Noy D, Zheng SY, Dutton PL, Blasie JK. Amphiphilic 4-helix bundles designed for biomolecular materials applications. Langmuir. 2004;20:5897–5904. doi: 10.1021/la0363884. [DOI] [PubMed] [Google Scholar]
- 22.Razeghifard AR, Wydrzynski T. Binding of Zn-chlorin to a synthetic four-helix bundle peptide through histidine ligation. Biochemistry. 2003;42:1024–1030. doi: 10.1021/bi026787i. [DOI] [PubMed] [Google Scholar]
- 23.Eggink LL, Hoober JK. Chlorophyll binding to peptide maquettes containing a retention motif. J. Biol. Chem. 2000;275:9087–9090. doi: 10.1074/jbc.275.13.9087. [DOI] [PubMed] [Google Scholar]
- 24.Chen M, Eggink LL, Hoober JK, Larkum AW. Influence of structure on binding of chlorophylls to peptide ligands. J. Am. Chem. Soc. 2005;127:2052–2053. doi: 10.1021/ja043462b. [DOI] [PubMed] [Google Scholar]
- 25.Kashiwada A, Watanabe H, Tanaka T, Nango M. Molecular assembly of zinc bachteriochlorophyll a by synthetic hydrophobic 1 alpha-helix polypeptides. Chem. Lett. 2000:24–25. [Google Scholar]
- 26.Kashiwada A, Nishino N, Wang ZY, Nozawa T, Kobayashi M, Nango M. Molecular assembly of bachteriochlorophyll a and its analogues by synthetic 4 alpha-helix polypeptides. Chem. Lett. 1999:1301–1302. [Google Scholar]
- 27.Yerushalmi R, Noy D, Baldridge KK, Scherz A. Mutual control of axial and equatorial ligands: Model studies with [Ni]-bacteriochlorophyll-a. J. Am. Chem. Soc. 2002;124:8406–8415. doi: 10.1021/ja0121078. [DOI] [PubMed] [Google Scholar]
- 28.Noy D, Yerushalmi R, Brumfeld V, Ashur I, Scheer H, Baldridge KK, Scherz A. Optical absorption and computational studies of [Ni]-bacteriochlorophyll-a. New insight into charge distribution between metal and ligands. J. Am. Chem. Soc. 2000;122:3937–3944. [Google Scholar]
- 29.Eom HS, Jeoung SC, Kim D, Ha JH, Kim YR. Ultrafast vibrational relaxation and ligand photodissociation/photoassociation processes of nickel(II) porphyrins in the condensed phase. J. Phys. Chem. A. 1997;101:3661–3669. [Google Scholar]
- 30.Kobayashi T, Straub D, Rentzepis PM. Energy relaxation mechanism in Ni(II), Pd(II), Pt(II) and Zn(II) porphyrins. Photochem. Photobiol. 1979;29:925–931. [Google Scholar]
- 31.Musewald C, Hartwich G, Lossau H, Gilch P, Pollinger-Dammer F, Scheer H, Michel-Beyerle ME. Ultrafast photophysics and photochemistry of [Ni]- bacteriochlorophyll a. J. Phys. Chem. B. 1999;103:7055–7060. [Google Scholar]
- 32.Rodriguez J, Holten D. Ultrafast vibrational dynamics of a photoexcited metalloporphyrin. J. Phys. Chem. 1989;91:3525–3531. [Google Scholar]
- 33.Rodriguez J, Holten D. Ultrafast photodissociation of a metalloporphyrin in condensed phase. J. Phys. Chem. 1990;92:5944–5950. [Google Scholar]
- 34.Uesugi Y, Mizutani Y, Kitagawa T. Photoinduced solvent ligation to nickel(II) octaethylporphyrin probed by picosecond time-resolved resonance Raman spectroscopy. J. Phys. Chem. A. 1998;102:5809–5815. [Google Scholar]
- 35.Hartwich G, Fiedor L, Simonin I, Cmiel E, Schafer W, Noy D, Scherz A, Scheer H. Metal-substituted bacteriochlorophylls. 1 Preparation and influence of metal and coordination on spectra. J. Am. Chem. Soc. 1998;120:3675–3683. [Google Scholar]
- 36.Dam J, Schuck P. Sedimentation velocity analysis of heterogeneous protein-protein interactions: Sedimentation coefficient distributions c(s) and asymptotic boundary profiles from Gilbert-Jenkins theory. Biophys. J. 2005;89:651–666. doi: 10.1529/biophysj.105.059584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balbo A, Minor KH, Velikovsky CA, Mariuzza RA, Peterson CB, Schuck P. Studying multiprotein complexes by multisignal sedimentation velocity analytical ultracentrifugation. Proc. Natl. Acad. Sci. U. S. A. 2005;102:81–86. doi: 10.1073/pnas.0408399102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vistica J, Dam J, Balbo A, Yikilmaz E, Mariuzza RA, Rouault TA, Schuck P. Sedimentation equilibrium analysis of protein interactions with global implicit mass conservation constraints and systematic noise decomposition. Anal. Biochem. 2004;326:234–256. doi: 10.1016/j.ab.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 39.Weisstein EW. Dawson’s Integral. MathWorld-A Wolfram Web Resource. http://mathworld.wolfram.com/DawsonsIntegral.html.
- 40.Johnson ML, Faunt LM. Parameter-Estimation by Least-Squares Methods. Method. Enzymol. 1992;210:1–37. doi: 10.1016/0076-6879(92)10003-v. [DOI] [PubMed] [Google Scholar]
- 41.Beechem JM. Global Analysis of Biochemical and Biophysical Data. Method. Enzymol. 1992;210:37–54. doi: 10.1016/0076-6879(92)10004-w. [DOI] [PubMed] [Google Scholar]
- 42.Malinowski ER. Factor Analysis in Chemistry. 3rd ed. John Wiley; New York: 2002. [Google Scholar]
- 43.Gilbert GA, Jenkins RC. Boundary problems in the sedimentation and electrophoresis of complex. Nature. 1956;177:853–854. doi: 10.1038/177853a0. [DOI] [PubMed] [Google Scholar]
- 44.Dam J, Velikovsky CA, Mariuzza RA, Urbanke C, Schuck P. Sedimentation velocity analysis of heterogeneous protein-protein interactions: Lamm equation modeling and sedimentation coefficient distributions c(s) Biophys. J. 2005;89:619–634. doi: 10.1529/biophysj.105.059568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dam J, Schuck P. Calculating sedimentation coefficient distributions by direct modeling of sedimentation velocity concentration profiles. Method Enzymol. 2004;384:185–212. doi: 10.1016/S0076-6879(04)84012-6. [DOI] [PubMed] [Google Scholar]
- 46.Chen XX, Moser CC, Pilloud DL, Dutton PL. Molecular orientation of Langmuir-Blodgett films of designed heme protein and lipoprotein maquettes. J. Phys. Chem. B. 1998;102:6425–6432. [Google Scholar]
- 47.Ye SX, Strzalka J, Chen XX, Moser CC, Dutton PL, Blasie JK. Assembly of a vectorially oriented four-helix bundle at the air/wAter interface via directed electrostatic interactions. Langmuir. 2003;19:1515–1521. [Google Scholar]
- 48.Cristian L, Nanda V, Lear JD, DeGrado WF. Synergistic interactions between aqueous and membrane domains of a designed protein determine its fold and stability. J. Mol. Biol. 2005;348:1225–1233. doi: 10.1016/j.jmb.2005.03.053. [DOI] [PubMed] [Google Scholar]
- 49.Huang SS, Koder RL, Lewis M, Wand AJ, Dutton PL. The HP-1 maquette: From an apoprotein structure to a structured hemoprotein designed to promote redox-coupled proton exchange. Proc. Natl. Acad. Sci. U. S. A. 2004;101:5536–5541. doi: 10.1073/pnas.0306676101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chamberlain AK, Faham S, Yohannan S, Bowie JU. Construction of helix-bundle membrane proteins. Advances in Protein Chemistry. 2003;63:19–46. doi: 10.1016/s0065-3233(03)63002-0. [DOI] [PubMed] [Google Scholar]
- 51.Bowie JU. Understanding membrane protein structure by design. Nature Struct. Biol. 2000;7:91–94. doi: 10.1038/72454. [DOI] [PubMed] [Google Scholar]
- 52.Moog RS, Kuki A, Fayer MD, Boxer SG. Excitation Transport and Trapping in a Synthetic Chlorophyllide Substituted Hemoglobin - Orientation of the Chlorophyll-S1 Transition Dipole. Biochemistry. 1984;23:1564–1571. doi: 10.1021/bi00302a034. [DOI] [PubMed] [Google Scholar]
- 53.Hambright P. Chemistry of water soluble porphyrins. In: Kadish KM, Smith KM, Guilard R, editors. The Porphyrin Handbook. Academic Press; San Diego: 2000. pp. 129–210. [Google Scholar]