Abstract
The Nun protein of coliphage HK022 excludes superinfecting λ phage. Nun recognizes and binds to the N utilization (nut) sites on phage λ nascent RNA and induces transcription termination. Over-expression of Nun from a high-copy plasmid is toxic for E.coli, despite the fact that nut sites are not encoded in the E.coli genome. Cells expressing Nun cannot exit stationary phase. Toxicity is related to transcription termination, since host and nun mutations that block termination also suppress cell killing. Nun inhibits expression of wild-type lacZ, but not lacZ expressed from the Crp/cAMP–independent lacUV5 promoter. Microarray and proteomics analyses show Nun down-regulates crp and tnaA. Crp over-expression and high indole concentrations partially reverse Nun-mediated toxicity and restore lacZ expression.
Keywords: phage HK022 Nun, transcription termination, E. coli stationary phase toxicity, fis, indole, Crp
Introduction
The relationships between bacteria and bacteriophages are dynamic. Interactions among phage and host factors are essential for phage development. All phage require and utilize the cellular machinery of the host to duplicate their genomes, synthesize viral proteins, and produce progeny. Bacteriophages compete for a given host, and several molecular mechanisms have evolved that allow a lysogenic phage to exclude superinfecting phages.1 One of the best characterized mechanisms of exclusion occurs between the HK022 prophage and superinfecting λ phage. Exclusion of superinfecting λ phage by HK022 is mediated by the HK022-encoded Nun protein, which acts as a transcription terminator of λ early genes.2 The mechanism entails the binding of the N-terminal arginine rich motif (ARM) of Nun to the boxB RNA sequences within in the nut regions of nascent transcripts produced from the λpL and λpR promoters.3 The Nun C terminal domain (CTD) contacts RNA polymerase (RNAP),4 and these interactions between Nun, the RNA boxB sites and RNAP promote the formation of a stalled transcription elongation complex. In addition, Nun CTD residue W108 interacts with λ DNA template ahead of the transcription elongation complex. Mutation of this amino acid to alanine abrogates Nun termination function. Nun CTD residues K106 and K107 facilitate interaction with template by electrostatic interactions with the phosphate groups of the DNA.5, 6
Nun-mediated transcription termination is stimulated by the E. coli factors NusA, NusB, NusE (ribosomal protein S10), and NusG. Mutations in Nus factors that abrogate Nun-mediated termination have been isolated.7, 8 NusA is a 65kDA protein that binds core RNAP after transcription initiation and slows the elongation rate by stimulating pausing of the transcription elongation complex.9 A NusA point mutant, NusAE136K, specifically blocks Nun-mediated termination in the λpR operon, whereas termination of the λpL transcript is unaffected. NusAE136K has been shown to bind more tightly to Nun than wild-type NusA.7, 10
In this work we show that Nun also affects E. coli. Nun overproduction blocks transition from stationary to logarithmic growth. We characterize the mechanisms underlying Nun-mediated toxicity, and identify several elements that play a critical role. We present evidence that the transcription termination function of Nun is responsible for toxicity, even though E. coli does not carry a boxB sequence. Mutations in NusA and Nun that compromise Nun function in transcription termination also inhibit Nun toxicity. We also show that high indole concentrations and Crp overproduction reduce toxicity. The mechanism of Nun toxicity appears to be mediated by global changes in transcription, and may not involve specific targets.
Results
Nun overexpression inhibits E. coli growth
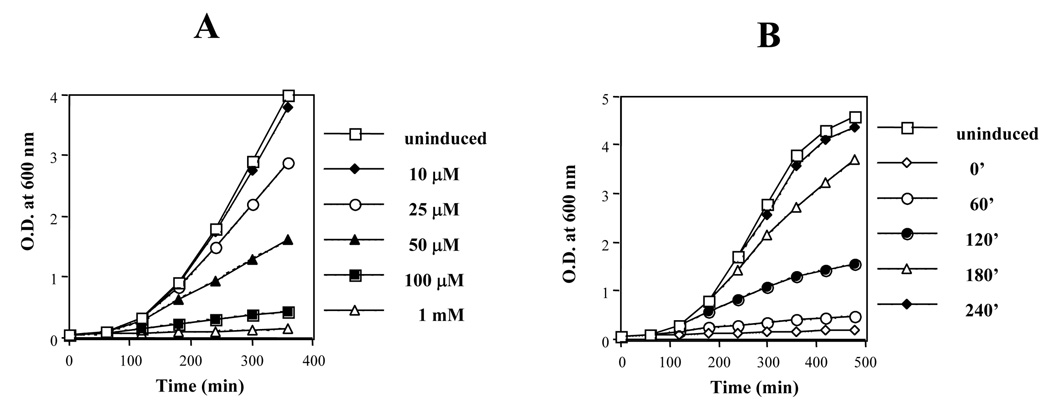
Phage HK022 Nun protein excludes superinfecting phage λ by promoting premature termination of the λpL and λpR early transcripts. Nun is expressed from a dedicated weak promoter in HK022 E. coli lysogens, and is present at a concentration of 120–360 molecules per cell.11 HK022 prophage do not appear to affect cell growth or any other aspect of cell physiology tested. However, Nun expressed from a multicopy plasmid under the control of the strong IPTG-induced trc promoter (AU222; see Table 1) severely inhibited E. coli growth (Fig. 1A). Strain AU221, carrying the empty vector, or uninduced AU222 had no effect (Fig. 1A; data not shown). Nun toxicity depends on two factors: Nun concentration and cell culture phase at the time of Nun overexpression. To determine the effect of Nun concentration, AU222 was grown overnight at 37°C, diluted 1:100, and treated with different IPTG concentrations (10 µM, 25 µM, 50 µM, 100µM and 1 mM). Figure 1A shows that cell growth, as determined by OD600, negatively correlated with increasing inducer concentration. Figure 1B shows that Nun-sensitivity was growth-phase dependent. AU222 was diluted from an overnight culture and treated at different times with 1mM IPTG. We found that Nun blocked cells from exiting stationary phase, and was considerably less toxic to cells in early or late log phase.
TABLE 1.
Strains, plasmids and phages used in this study
| Strains, plasmids, and phages | Genotype | Reference or source |
|---|---|---|
| Strains | ||
| N99 | W3102; Wild-type | NIH collection |
| RJ1800 | MG1655 fis::767 | 16 |
| CA8224 | HfrH placUV5- lacZ | Beckwith J. collection |
| AU221 | N99/pAUM | This work |
| AU222 | N99/pAUMNun | “ |
| AU223 | N99/pAUMNunP9A | “ |
| AU227 | N99 lacZXA21(λ 423)/pAUM | “ |
| AU228 | N99 lacZXA21(λ 423)/pAUMNun | “ |
| AU230 | N99 nusAE136K/pAUM | “ |
| AU231 | N99 nusAE136K/pAUMNun | “ |
| AU246 | N99/pAUMNunW108A | “ |
| AU251 | N99 lacZXA21(λ 4253)/pAUM | “ |
| AU252 | N99 lacZXA21(λ 4253)/pAUM Nun | “ |
| AU260 | N99 lacZXA21 lacUV5-lacZ+/pAUM | “ |
| AU261 | N99 lacZXA21 lacUV5-lacZ+/pAUMNun | “ |
| AU265 | N99 lacZXA21(λ 423), fis:767/pAUM | “ |
| AU266 | N99 lacZXA21(λ 423), fis:767/pAUMNun | “ |
| AU269 | N99 lacZXA21(λ HS3120)/pAUM | “ |
| AU270 | N99 lacZXA21(λ HS3120)/pAUMNun | “ |
| AU271 | N99 lacZXA21(λ HS3134)/pAUM | “ |
| AU272 | N99 lacZXA21(λ HS3134)/pAUMNun | “ |
| AU278 | AU221/pBAD30 | “ |
| AU297 | AU221/pBWCRP | “ |
| AU284 | AU223/pBAD30 | “ |
| AU299 | AU223/pBWCRP | “ |
| Plasmids | ||
| pTrc99 | ColE1ori ampR | Amersham Biosciense |
| pTrcNun | pTrc99-nun+ | Our lab collecton |
| pBAD30 | pACYC184 ori ampR | 25 |
| pBAD18 | ColEIori camR | 25 |
| pHY0 | pEXT20-crp | 26 |
| pAUM | pTrc99, ampS, camR | This work |
| pAUMNun | pAUM-nun+ | “ |
| pAUMNunP9A | pAUM-nunP9A | “ |
| pAUMNunW108A | pAUM-nunW108A | “ |
| pBWCRP | pBAD30-crp | “ |
| Phages | ||
| λ 423 | λcI857 rrnEP1P2 | 27 |
| λ4253 | λcI857 rrnBP2 | 28 |
| λHS3120 | λ pmalE | H. Shuman collection |
| λHS3134 | λ pmalP | H. Shuman collection |
Figure 1. E. coli outgrowth from stationary phase is inhibited by Nun overproduction.
Strain AU222 was grown overnight at 37 °C in LB + chloramphenicol (50µg/ml) and diluted 1:100. A) IPTG was added at time 0 at the indicated concentrations to induce Nun; B) Nun was induced with 1 mM IPTG at the indicated times following dilution. All experiments were at 37 °C.
Mutations that suppress Nun-mediated termination also reduce Nun toxicity
E. coli NusA protein is required for Nun-mediated termination in the λpR operon. A nusA point mutant, nusAE136K, blocks Nun-mediated termination at λnutR but not at λnutL.7, 10 We asked if Nun toxicity was also abrogated by nusAE136K. Overnight cultures were diluted 1:100 and grown for 6 hr in the presence or absence of 50 µM IPTG. As shown in Table 2, Nun reduced the growth rate in wild-type cells by 58% (rows 1 and 2). In contrast, Nun reduced growth by 24% in the nusAE136K background (rows 3 and 4).
Table 2.
nusAE136K and nunW108A suppress Nun toxicity
| Strain | Host mutation | Plasmid | Growth rate(hr−1)a |
|---|---|---|---|
| AU221 | nusA+ | vector | .71 +/− .01 |
| AU222 | nusA+ | nun + | .30 +/− .01 |
| AU230 | nusAE136K | vector | .70 +/− .01 |
| AU231 | nusAE136K | nun + | .53 +/− .04 |
| AU246 | nusA+ | nunW108A | .68 +/− .02 |
Strains were grown overnight and diluted 1:100 into 50µM IPTG.
The initial OD600 was subtracted from the OD600 at 6 hr and divided by 6.
We further tested the idea that Nun-induced transcription termination was lethal in E. coli by asking if the termination-deficient NunW108A mutant was toxic.5 Over-expression of NunW108A had no significant effect on cell growth (Table 2, rows 1 and 5). Taken together, these results suggest that transcription termination is at least partially responsible for Nun toxicity.
Nun inhibits cAMP/Crp – dependent promoters
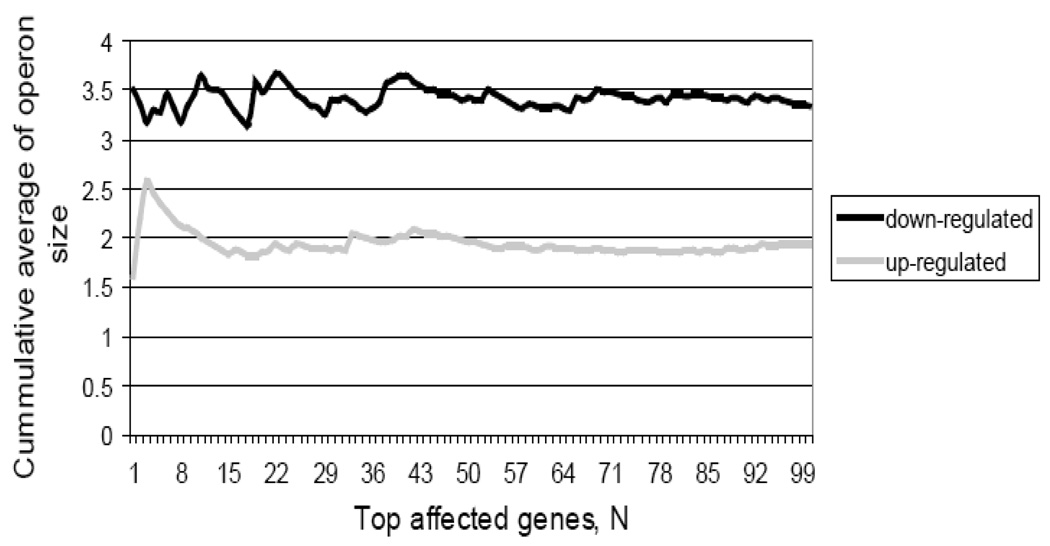
Our results suggest that transcription termination accounts for Nun toxicity. In an attempt to discern general characteristics of the Nun effect on the physiology, we measured relative transcript and protein levels in cells overproducing Nun protein (see Supplementary file for microarray procedures and data analysis). Nun excess affected transcript levels of more than 450 genes: 184 down-regulated and 271 up-regulated (differentially expressed genes are listed in Supplementary Table S1). Further analysis revealed that the down-regulated genes were organized in longer operons than the upregulated ones (Fig.2), supporting an effect of Nun over-expression on transcription elongation. The set of down-regulated proteins in Nun overexpressing cells (See Supplementary Table S2 see Suppl. File for details of proteomics experiments) was enriched for CRP targets, 12 out of 28 proteins whose abundance decreased at least 2-fold (adjusted p<10−2). The set of proteins whose abundance was increased more than 2-fold was not enriched for representatives of any functional class.
Figure 2. Length of operons affected by Nun.
The average length of down- and up-regulated operons was calculated by applying a sliding window of size 5 to the lists of significantly affected genes sorted (from most to least affected) according to the strength of the transcriptional effect. The average size of an unaffected operon with similar signal intensity was 1.9 ± 0.1 genes.
To investigate the role of CRP in mediating the Nun effect, we examined the activity of several CRP- dependent and CRP-independent E. coli operons after Nun induction. Table 3 shows the effect of Nun over-expression on lacZ transcription. β-galactosidase levels were determined 4 hr following induction of Nun and chromosomal lacZ with 100 µM IPTG. LacZ activity was reduced 88% compared to the empty vector control (Table 3: rows 1 and 2). Since the lac operon does not carry a λ nut site, we hypothesized that inhibition was mediated by a general transcription regulator(s). cAMP/Crp controls more than 100 E. coli operons, including lac.12 To ask if Nun acted through cAMP/Crp, we determined the effect of Nun on a lacZ operon driven by the lacUV5 promoter, which is independent of cAMP/Crp.13 Our results show lacUV5 partially relieved Nun inhibition (rows 3 and 4; 47% inhibition compared to the empty vector control).
Table 3.
Nun inhibits lac operon induction.
| Strain | Promoter | Host mutation | Nun | β-galactosidase | % inhibition |
|---|---|---|---|---|---|
| AU221 | plac+ | nusA+ | − | 522 +/− 44 | - |
| AU222 | plac+ | nusA+ | + | 62 +/− 6 | 88 |
| AU260 | placUV5 | nusA+ | − | 1248 +/− 7 | - |
| AU261 | placUV5 | nusA+ | + | 660 +/− 45 | 47 |
| AU230 | plac+ | nusAE136K | − | 606 +/− 42 | - |
| AU231 | plac+ | nusAE136K | + | 649 +/− 45 | < 1 |
| AU246 | plac+ | nusA+ | nunW108A | 505 +/− 15 | 3 |
Overnight cultures were diluted 1:100, grown for 1 hr at 37 °C, and induced with 100 µM IPTG. Cultures were then grown at 37 °C for 4 hr and β-galactosidase units (Miller units) were determined. The above data are an average of three independent experiments.
We then asked whether Nun inhibition of catabolite-repressible gene expression was related to Nun termination activity. Accordingly, we measured lac operon induction in host and phage mutants that abrogate or reduce Nun-mediated termination. Table 3, rows 5 – 7, shows that lacZ expression is not reduced in nunW108A nusA+ or nusAE136K nun+ strains. Thus, Nun down-regulation of the lac operon correlates with Nun termination activity.
We extended our analysis to other catabolite-repressible operons. We tested the effect of Nun on pmalE, which is directly controlled by cAMP/Crp or on pmalP, which is under the control of MalT, whose expression is cAMP/Crp – dependent.14 Overnight cultures carrying pmalE or pmalP fused to lacZ were diluted 1:100, grown for 1 hr, and treated with 100µM IPTG and 0.2% maltose for 3 hr. As shown in Table 4, Nun reduced malE and malP promoter activity 85% and 99%, respectively compared to Nun− controls. We conclude that Nun significantly represses cAMP/Crp – dependent promoters.
Table 4.
Nun inhibits malE and malP promoters
| Strain | lacZ fusion | nun | β-galactosidase | % inhibition |
|---|---|---|---|---|
| AU269 | pmalE | − | 2933 +/− 106 | - |
| AU269 | pmalE | + | 565 +/− 88 | 85 |
| AU270 | pmalP | − | 662 +/− 72 | - |
| AU270 | pmalP | + | < 1 | >99 |
Overnight cultures were diluted 1:100, grown for 1 hr at 37 °C, and then brought to 100µM IPTG and 0.2% maltose to induce nun and mal promoters, respectively. β-galactosidase activity (MU) was determined after an additional 3 hr at 37 °C.
The above data are an average of two independent experiments.
Crp over-expression reverses Nun inhibition of lacZ and partially overcomes toxicity
To determine whether Nun reduced cAMP levels or inhibited Crp, we induced lacZ in a strain carrying a multicopy, arabinose-inducible, plasmid that expressed Crp (pBWCRP; see Table 1). For unknown reasons, we were unable to introduce our Nun+ plasmid into this strain, and therefore substituted a plasmid that expresses a nun derivative, nunP9A, isolated by alanine-scanning mutagenesis. The P9A mutation has no other evident phenotype; it did not reduce Nun toxicity or Nun inhibition of lacZ expression (Table 5). Table 5, column 4, rows 3 and 4, shows that over-expression of Crp completely reversed Nun inhibition of lacZ induction. These results indicate that Nun reduces the concentration of Crp or its activity. Microarray analyses confirmed that Nun reduced crp expression 2.3-fold. Efforts to determine if Nun also reduced cAMP levels were inconclusive.
Table 5.
Crp overproduction suppresses Nun toxicity and restores lac transcription.
| Strain | Nun | Crp | β-galactosidase | Growth rate(hr−1)a |
|---|---|---|---|---|
| AU278 | − | − | 425 +/− 70 | .68 +/− .01 |
| AU297 | − | + | 505 +/− 88 | .67 +/− .02 |
| AU284 | + | − | 146 +/− 10 | .11 +/− .01 |
| AU299 | + | + | 502 +/− 90 | .39 +/− .02 |
Overnight cultures were diluted 1:100 in LB +ampicillin (50µg/ml) + chloramphenicol (50µg/ml), grown for 1 hr at 37 °C, and treated with 100 µM IPTG and 0.02%arabinose to induce nun and crp, respectively. β-galactosidase activity (MU)) was determined after an additional 5 hr at 37 °C. The above data are an average of two independent experiments.
Growth rate was determined as in Table 2.
Crp over-production also partially overcame Nun toxicity, increasing the growth rate of Nun+ cells more than 4-fold (Table 5, column 5, rows 3 and 4).
Nun up-regulates the rrnE operon
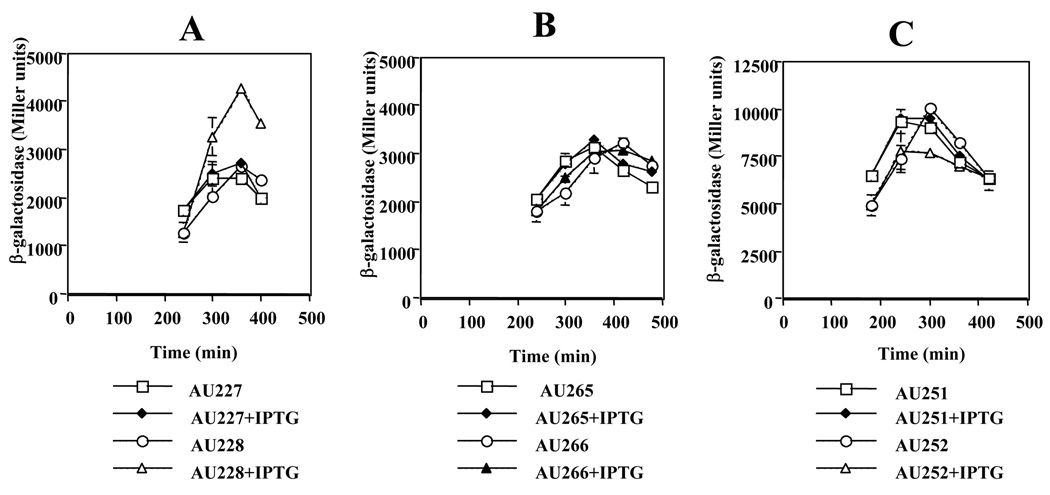
We showed above that Nun was most toxic to cells attempting to exit stationary phase, and had less effect on cells in logarithmic growth. We wondered if Nun might down-regulate ribosomal RNA synthesis, thus preventing entry into log phase. Additionally, rrn operons carry boxA sequences, which are also found in the λnut sites. To measure the effect of Nun on rrn activation, we used fusions carrying various portions of the rrnE promoter region to lacZ. The results of these experiments are shown in Figure 3. Contrary to our expectations, Nun significantly increased rrnE expression (Figure 3A).
Figure 3. Nun induces ribosomal operon transcription in a Fis-dependent fashion.
Strains grown overnight at 30 °C were diluted 1:100 in LB and grown at 30 °C. Four hr later 100 µM IPTG was added where indicated and β-galactosidase (MU) were determined hourly. Strains in panels A and B carry the p1-p2 promoters of rrnE fused to lacZ (Table 1). A) fis+; B) fis::767; C) fis+, the p2 promoter of rrnB is fused to lacZ - Fis binding sites are deleted.
The above data are an average of two independent experiments.
The reason for this increase was suggested by microarray analysis. We found that fis was up-regulated 2.6-fold by Nun (Supplementary Table S1). Fis positively regulates transcription of rrn operons.15 To test the possibility that Nun enhancement of rrnE expression was mediated by Fis, we introduced the non-functional fis::767 mutation into the strains shown in Figure 3A.16 Figure 3B shows that fis::767 completely reversed Nun up-regulation of rrnE transcription. Although the mutation blocked rrnE induction, it had no effect on Nun toxicity (data not shown). The role of Fis in Nun up-regulation of ribosomal RNA synthesis was also demonstrated using rrnB-lacZ fusions that lack the UP and Fis-binding regions. Nun had no effect on this mutant promoter (Figure 3C). Together, these data indicate that Nun induces ribosomal RNA transcription via an increase in Fis concentrations.
Nun down-regulates indole synthesis
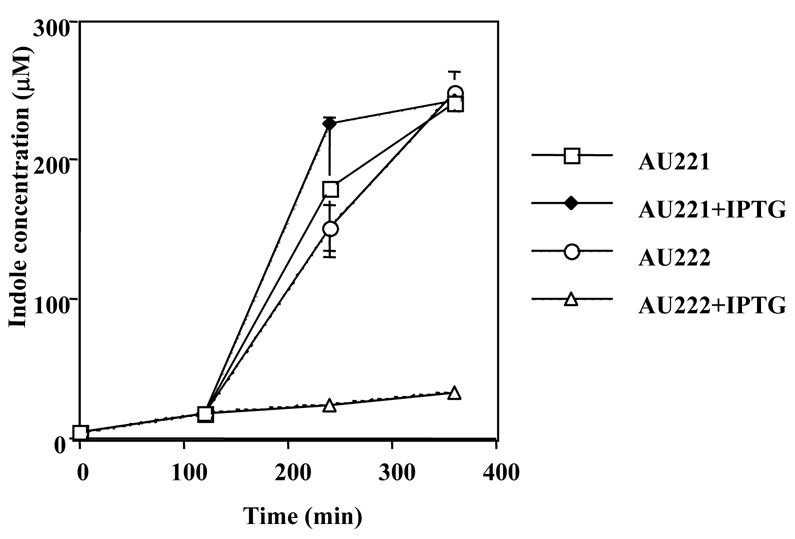
Proteomic analyses of cells exposed to high Nun concentrations showed that the levels of tryptophanase, the enzyme responsible for indole production, were reduced 5.8-fold in response to Nun over-expression (see Supplementary Fig. S1). This is of particular interest, since indole synthesis increases as cells enter stationary phase and may promote this transition.17 Figure 4 shows that Nun repressed indole synthesis 7-fold. Media from cultures carrying the empty vector contained 231µM indole, whereas the media indole concentration in cells expressing Nun was 31µM.
Figure 4. Nun inhibits indole production.
Overnight cultures grown at 37 °C were diluted 1:100 and 100µM IPTG was added where indicated. Samples were collected at 2 hr intervals, and the concentration of indole in the supernatant was determined. The above data are an average of two independent experiments. AU221, vector; AU222, pAUM-nun
Our next experiment was to add indole to the medium and measure the effects of Nun on cell growth and lacZ induction. No effect was seen when we adjusted the indole concentration of the medium to 231µM (data not shown). However, indole at 1.15 mM partially reversed Nun toxicity (Table 6, column 5, compare rows 1 & 2, and 3 & 4). Note that at this concentration, indole inhibited cell growth by ~50%. In addition, indole restored β-galactosidase synthesis in Nun+ cells to ~30% of Nun− levels (Table 6, column 4, compare rows 1 & 2, and 3 & 4).
Table 6.
Indole suppresses Nun.
| Strains | Nun | Indole | β-galactosidase | Growth rate (h−1) |
|---|---|---|---|---|
| AU221 | − | − | 463 +/− 30 | .69 +/− 0.01 |
| AU222 | + | − | 25 +/− 2 | .11 +/− 0.01 |
| AU221 | − | + | 164 +/− 11 | .34 +/− 0.01 |
| AU222 | + | + | 142 +/− 29 | .19 +/− 0.02 |
Overnight cultures grown at 37°C were diluted in fresh LB supplemented with 1.15 mM indole and 100µM IPTG. β-galactosidase (MU)) was determined after 4 hr at 37°C. Growth rate was determined as described in Table 2. The above data are an average of two independent experiments.
Discussion
The known role of phage HK022 Nun protein is to exclude superinfecting λ by terminating transcription prematurely on the λ chromosome. Exclusion is highly specific, affecting only phages that express λ nut RNA. We demonstrate in this report that over-expression of Nun blocked exit of E. coli from stationary phase. Surprisingly, since E. coli does not encode λ nut, the transcription termination activity of Nun was required for this toxicity. In HK022 lysogens, Nun is expressed from a dedicated promoter at very low levels, 120–360 molecules per cell. Nun concentrations increase as lysogens enter stationary phase.11 However, HK022 lysogens grow normally and have no defect in exiting stationary phase (data not shown). No difference between HK022 and HK022 nun mutants in terms of lysogeny or prophage induction have been detected. We suggest that although the drastic effects of Nun over-production on cell growth and gene expression reported here may not have a counterpart in normal cell growth, it is nevertheless possible that subtle alterations of cellular metabolism are induced by physiological concentrations of Nun, and that these favor the propagation of HK022 under conditions that we have not reproduced in the laboratory.
Microarray and proteomic analyses reveal numerous changes induced by Nun in E. coli physiology (see Supplemental Table S1, S2, and Fig. S1). Microarray analysis indicates that genes encoding the tricarboxylic acid (TCA) cycle and chemotaxis are down-regulated, whereas some tRNA ligases, DNA damage repair genes and resident transposon-related genes are up-regulated. Certain genes related to oxidative damage, including soxS and sodB, are also up-regulated. It is plausible that cells exposed to Nun undergo an RpoE-associated stress response.18, 19
Nun over-expression also upregulates fis and down-regulates Crp and tryptophanase. These changes affect many cellular processes. Fis represses transcription of rpoS, which encodes the stationary-phase sigma factor. Nun induction of Fis, although it leads to increased rrn transcription, does not, however, account for Nun toxicity.
Down-regulation of Crp and tryptophanase together do account for Nun inhibition of stationary phase exit. Over-expression of Crp, or addition of indole to the medium, suppressed Nun toxicity. Note, however, that crp or tnaA mutants can exit stationary phase. We assume that down-regulation of Crp or TnaA is only toxic in the context of other changes in cellular physiology induced by Nun. Crp also fully restores induction of lacZ in Nun-inhibited cells and indole has a partial effect. Interestingly, phage λ encodes a gene, hin, whose product blocks lacZ induction. In contrast to Nun, however, Hin inhibition of lacZ expression is due to cAMP depletion subsequent to phage induction.20 With respect to phage development, down-regulation of Crp or cAMP diverts temperate phage from the lysogenic into the lytic pathway.
Indole is synthesized as E. coli enters stationary phase and is also a quorum sensing signal that promotes this transition.17 Indole stimulates expression of Crl, a positive regulator of RpoS.21 Tryptophan is converted to indole by tryptophanase (TnaA). Since transcription of tnaA is controlled by RpoS, a positive regulatory loop amplifies indole secretion as cells transit to stationary phase. Interestingly expression of E. coli cryptic prophage genes are strongly down-regulated by RpoS.22 It is tempting to speculate that accumulation of Nun in HK022 lysogens entering stationary phase reduces indole concentrations down-regulating RpoS activity and thus inducing prophage gene expression.
The pattern of gene deregulation induced by Nun, however, does not indicate a single target. At physiological concentrations, Nun binding to RNA is highly specific; only the boxB RNA sequence of the λ nut region is recognized. Although boxB is not found in the chromosome, boxA, another component of λ nut, is present at several sites. The tnaA operon, which is down-regulated by Nun, includes a boxA sequence, as do the rrn operons, which are up-regulated.
It is possible that at elevated levels, Nun binds RNA non-specifically. This could directly affect translation, and may explain why, despite overall consistency between transcriptomic and proteomic data (see Supplementary Discussion), some genes down-regulated by Nun are seen in proteomic but not in microarray analysis (e.g. TnaA).
Materials and Methods
Bacterial strains, plasmids, and phages
E. coli strains, bacteriophages, and plasmids used in this work are listed in the Table 1. Plasmid pAUM is a derivative of pTrc99 in which chloramphenicol resistance (cat) replaces ampicillin resistance (bla). Two EagI sites were introduced in pTrc99 with the Quickchange site-directed mutagenesis technique (Stratagene). The new EagI sites flank bla. The pair of primers used for the 5’ region of bla were 99EagIF=CGC CCT TAT TCC CTT TTT TGC GGC CGT TTG CCT TCC TGT TTT TGC, and 99EagIR=GAG CAA AAA CAG GAA GGC AAA CGG CCG CAA AAA AGG GAA TAA GGG. The pair of primers used for the 3’ region were 99EagI3F= CGT TCC ACT GAG CGT CAG CGG CCG TAG AAA AGA TCA AAG G, and 99EagI3R= CCT TTG ATC TTT TCT ACG GCC GCT GAC GCT CAG TGG AAC G. The cat gene from pBAD18 (see Table 1) was mutated changing codon 73 (GAA) to disrupt an EcoRI site. The primers used were Cm-EF= CTG ATG AAT GCT CAT CCG GAG TTC CGT ATG GCA ATG AAA GAC, and Cm-ER= GTC TTT CAT TGC CAT ACG GAA CTC CGG ATG AGC ATT CAT CAG. Then this pBAD18 derived plasmid was used as template to amplify by Polymerase Chain Reaction (PCR) a fragment of 930pb containing cat; the primers used were CmEaf= CCA GTC TAT TAA TTG CGG CCG GGA AGC, and CmEar= CAA CGC CAT GAG CGG CCG CAT TTC TTA. The PCR product containing cat and the pTrc99-derived plasmid were digested with EagI (New England Biolabs) and purified with QIAquick PCR purification kit and QIAquick Gel extraction Kit (QIAGEN) respectively. The PCR fragment and pTrc99-derived DNA fragment obtained were ligated by incubation with T4 DNA ligase (New England Biolabs) at 16°C overnight. The ligated mixture, containing plasmid pAUM, was used to transform XL1-Blue cells.
The plasmid pTrc-Nun was digested with EcoRI and HindIII (New England Biolabs) to release a fragment containing nun. pAUM was digested with EcoRI and HindIII, and the resulting pAUM-digested fragment was purified with the QIAquick Gel extraction Kit (QIAGEN). The nun and pAUM fragments were ligated by incubation with T4 DNA ligase (New England Biolabs). The ligation mixture containing plasmid pAUM-Nun was used to transform XL1-Blue cells.
Plasmid pAUM-NunP9A was constructed by the introduction of a mutation in codon nine in nun gene using the Quickchange site-directed mutagenesis technique (Stratagene). The primers used were P9Af= GAC TAT TTA TGT TAA TGC TGA CAG CGG ACA AAA CAG, and P9Ar= CTG TTT TGT CCG CTG TCA GCA TTA ACA TAA ATA GTC.
Plasmid pHY0 (see Table 1) was digested with EcoRI and HindIII to release a fragment containing crp which was ligated to pBAD30 digested with the same nucleases, yielding pBWCRP.
Determination of growth rate
Fresh overnight cultures were diluted 1:100 in LB+ 50 µg/ml chloramphenicol. To induce samples, IPTG was added at time 0 at 50µM final concentration, Growth rate was determined by measuring OD600 at time 0, and 6 hr later. The difference in OD600 was divided by 6 to yield growth rate hr−1. Note that what is being determined is, in fact, the lag time for stationary phase exit.
Transcription from chromosomal promoters
lacZ fusions were used to measure transcription from chromosomal promoters. Overnight cultures were diluted 1:100 in LB supplemented with the appropriate antibiotic chloramphenicol or chloramphenicol + ampicillin, each at 50 µg/ml. Cultures were grown 1 hr at 37°C, and then the inducers were added simultaneously. The inducers used were IPTG (100µM), maltose (0.2%), and arabinose (0.02%). In the case of the determination for the ribosomal fusions the addition of the inducer IPTG (100µM) was done 4 hr after dilution, and the cells were grown at 30°C. β-galactosidase activity is reported as Miller Units.23
Indole assays
Indole was determined in E. coli culture supernatants as described.24 Briefly, 400µl aliquots of growing cultures were removed every 2 hr and centrifuged to remove cells. 200µl of supernatant were added to 200µl of 0.5 M perchloric acid. After centrifugation to remove precipitated proteins, 400µl of Ehrlich’s reagent was added. The samples were incubated for 30 min at 37°C, after which absorbance at 571 nm was determined.
Supplementary Material
Acknowledgements
We thank C. Yanofsky, R. Gourse, and G. Roberts for strains. We also thank R. Washburn, L. Duarte, and C. Vitiello for helpful discussions. MEG was supported by NIH grant GM37219. AK was supported by NIH grant GM066098.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Duckworth DH, Glenn J, McCorquodale DJ. Inhibition of bacteriophage replication by extrachromosomal genetic elements. Microbiol. Rev. 1981;45:52–71. doi: 10.1128/mr.45.1.52-71.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robert J, Sloan SB, Weisberg RA, Gottesman ME, Robledo R, Harbrecht D. The remarkable specificity of a new transcription termination factor suggests that the mechanisms of termination and antitermination are similar. Cell. 1987;51:483–492. doi: 10.1016/0092-8674(87)90644-1. [DOI] [PubMed] [Google Scholar]
- 3.Chattopadhyay S, Hung SC, Stuart AC, Palmer AG, III, Garcia-Mena J, Das A, Gottesman ME. Interaction between the phage HK022 Nun protein and the nut RNA of phage λ. Proc. Nat. Acad. Sci. USA. 1995;92:12131–12135. doi: 10.1073/pnas.92.26.12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watnick R, Gottesman ME. Escherichia coli NusA is required for efficient RNA binding by phage HK022 Nun protein. Proc. Nat. Acad. Sci. USA. 1998;95:1546–1551. doi: 10.1073/pnas.95.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watnick R, Gottesman ME. Binding of transcription termination protein Nun to nascent RNA and template DNA. Science. 1999;286:2337–2339. doi: 10.1126/science.286.5448.2337. [DOI] [PubMed] [Google Scholar]
- 6.Kim HC, Gottesman ME. Transcription termination by phage HK022 Nun is facilitated by COOH-terminal lysine residues. J. Biol. Chem. 2004;279:13412–13417. doi: 10.1074/jbc.M313206200. [DOI] [PubMed] [Google Scholar]
- 7.Robledo R, Atkinson BL, Gottesman ME. Escherichia coli mutations that block transcription termination by phage HK022 Nun protein. J Mol. Biol. 1991;220:613–619. doi: 10.1016/0022-2836(91)90104-e. [DOI] [PubMed] [Google Scholar]
- 8.Burova E, Hung SC, Chen J, Court DL, Zhou JG, Mogilnitskiy G, Gottesman ME. Escherichia coli nusG mutations that block transcription termination by coliphage HK022 Nun protein. Mol. Microbiol. 1999;31:1783–1793. doi: 10.1046/j.1365-2958.1999.01315.x. [DOI] [PubMed] [Google Scholar]
- 9.Greenblatt J, Li J. Interaction of the sigma factor and the nusA gene protein of E. coli with RNA polymerase in the initiation-termination cycle of transcription. Cell. 1981;24:421–428. doi: 10.1016/0092-8674(81)90332-9. [DOI] [PubMed] [Google Scholar]
- 10.Kim HC, Washburn RS, Gottesman ME. Role of E. coli NusA in phage HK022 Nun-mediated transcription termination. J. Mol. Biol. 2006;359:10–21. doi: 10.1016/j.jmb.2006.02.081. [DOI] [PubMed] [Google Scholar]
- 11.King RA, Madsen Pl, Weisberg RA. Constitutive expression of a transcription termination factor by a repressed prophage: promoters for transcribing the phage HK022 nun gene. J. Bacteriol. 2000;182:456–462. doi: 10.1128/jb.182.2.456-462.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng D, Constantinidou C, Hobman JL, Minchin ST. Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res. 2004;32:5874–5893. doi: 10.1093/nar/gkh908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silverstone AE, Arditti RR, Magasanik B. Catabolite-insensitive revertants of lac promoter mutants. Proc. Nat. Acad. Sci. USA. 1970;66:773–779. doi: 10.1073/pnas.66.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Débarbouillé M, Shuman HA, Silhavy TJ, Schwartz M. Dominant constitutive mutations in malT, the positive regulator of the maltose regulon in Escherichia coli. J. Mol. Biol. 1978;124:359–371. doi: 10.1016/0022-2836(78)90304-2. [DOI] [PubMed] [Google Scholar]
- 15.Ross W, Thompson JF, Newlands jT, Gourse RL. E. coli Fis protein activates ribosomal RNA transcription in vitro and in vivo. EMBO. J. 1990;9:3733–3742. doi: 10.1002/j.1460-2075.1990.tb07586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson RC, Bal CA, Pfeffer D, Simon MI. Isolation of the gene encoding the Hin recombinational enhancer binding protein. Proc. Nat. Acad. Sci. USA. 1988;85:3484–3488. doi: 10.1073/pnas.85.10.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang D, Ding X, Rather PN. Indole can act as an extracellular signal in Escherichia coli. J. Bacteriol. 2001;183:4210–4216. doi: 10.1128/JB.183.14.4210-4216.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bauer CE, Elsen S, Bird TH. Mechanisms for redox control of gene expression. Annu. Rev. Microbiol. 1999;53:495–523. doi: 10.1146/annurev.micro.53.1.495. [DOI] [PubMed] [Google Scholar]
- 19.Storz G, Imlay JA. Oxidative stress. Curr. Opin. Microbiol. 1999;2:188–194. doi: 10.1016/s1369-5274(99)80033-2. [DOI] [PubMed] [Google Scholar]
- 20.Court D, Gottesman M, Gallo M. Bacteriophage lambda Hin function I. Pleitropic alteration in host physiology. J. Mol. Biol. 1980;138:715–729. doi: 10.1016/0022-2836(80)90061-3. [DOI] [PubMed] [Google Scholar]
- 21.Pratt LA, Silhavy TJ. Crl stimulates RpoS activity during stationary phase. Mol. Microbiol. 1998;29:1225–1236. doi: 10.1046/j.1365-2958.1998.01007.x. [DOI] [PubMed] [Google Scholar]
- 22.Patten CL, Kirchhof MG, Schertzberg MR, Morton RA, Schellhorn HE. Microarray analisys of RpoS-mediated gene expression in Escherichia coli K-12. Mol Gen. Genomics. 2004;272:580–591. doi: 10.1007/s00438-004-1089-2. [DOI] [PubMed] [Google Scholar]
- 23.Miller JH. A short course in bacterial genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 24.Lelong C, Aguiluz K, Luche S, Kuhn L, Garin J, Rabilloud T, Geiselmann J. The Crl- RpoS regulon of Escherichia coli. Mol. Cell. Proteomics. 2007;6:648–659. doi: 10.1074/mcp.M600191-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Guzman L, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Youn H, Kerby RL, Conrad M, Roberts GP. Study of highly constitutively active mutants suggests how cAMP activates cAMP receptor protein. J. Biol. Chem. 2006;281:1119–1127. doi: 10.1074/jbc.M509421200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gourse RL, de Boer HA, Nomura A. DNA determinants of rRNA synthesis in E. coli: Growth rate dependent regulation, feedback inhibition , upstream activation, antitermination. Cell. 1986;44:197–205. doi: 10.1016/0092-8674(86)90498-8. [DOI] [PubMed] [Google Scholar]
- 28.Ross W, Aiyar SE, Salomon J, Gourse RL. Escherichia coli promoters with UP elements of different strengths: Modular structure of bacterial promoters. J. Bacteriol. 1998;180:5375–5383. doi: 10.1128/jb.180.20.5375-5383.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.