Abstract
The loss of Gimap5 (GTPase of the immune-associated protein 5) gene function is the underlying cause of lymphopenia and autoimmune diabetes in the BioBreeding (BB) rat. The in vivo function of murine gimap5 is largely unknown. We show that selective gene ablation of the mouse gimap5 gene impairs the final intrathymic maturation of CD8 and CD4 T cells and compromises the survival of postthymic CD4 and CD8 cells, replicating findings in the BB rat model. In addition, gimap5 deficiency imposes a block of natural killer (NK)- and NKT-cell differentiation. Development of NK/NKT cells is restored on transfer of gimap5−/− bone marrow into a wild-type environment. Mice lacking gimap5 have a median survival of 15 weeks, exhibit chronic hepatic hematopoiesis, and in later stages show pronounced hepatocyte apoptosis, leading to liver failure. This pathology persists in a Rag2-deficient background in the absence of mature B, T, or NK cells and cannot be adoptively transferred by transplanting gimap5−/− bone marrow into wild-type recipients. We conclude that mouse gimap5 is necessary for the survival of peripheral T cells, NK/NKT-cell development, and the maintenance of normal liver function. These functions involve cell-intrinsic as well as cell-extrinsic mechanisms.
Introduction
Gimap5 (GTPase of the immune-associated protein 5; alternate names: imap3,1 iROD,2 IAN4L1,3,4 IAN4,5–7) is a member of a family of Gimap genes that are conserved in mammals, birds, fish, and in higher plants, and are clustered in a single chromosomal locus in each species. Mouse gimap5 mRNA is predominantly expressed in the thymus, spleen, lymph nodes, and lung8 and is present in splenic T and B cells and in all maturation stages of thymic T cells. In the thymus, the highest level of gimap5 mRNA is detected in CD4 single-positive (SP) and CD8SP cells.8 As predicted from its mRNA sequence, mouse gimap5 is a 308-amino acid intracellular molecule that shares a conserved domain structure with other members of the Gimap gene family, ie, an AIG1 domain comprising GTP-binding motifs (residues 34-41, 62-64, 82-84) and 2 functionally undefined regions, the so-called conserved box CB (residues 102-128), and the IAN motif (residues 178-197). The remaining portion of gimap5 comprises a coiled-coil region and a putative carboxyterminal membrane localization signal sequence.9 In mice and humans, but not in rats, a putative duplication and inversion of the Gimap5 gene have occurred, resulting in the presence of a nonfunctional Gimap3 pseudogene in humans and a functional Gimap3 gene in mice
Several independent lines of evidence suggest that gimap5 functions as a negative regulator of apoptosis that modulates intrathymic development and peripheral survival of T lymphocytes. First, in the diabetes-prone BioBreeding (BB) rat, deletion of a nucleotide in the codon for amino acid 85 of rat gimap5 changes the downstream reading frame to encode 19 aberrant amino acids, followed by a premature STOP codon at amino acid position 104.10 The mutation is associated with reduced numbers of peripheral CD4 and CD8 T cells, evidence of spontaneous apoptotic cell death of peripheral T cells, and increased intrathymic frequency of double-negative (DN) thymocytes concomitant with reduced prevalence of double-positive (DP) thymocytes. The development of autoimmune diabetes in the gimap5-deficient BB rat strain could result from a dysregulation of T-cell survival, secondary to a selective survival advantage of autoreactive T cells during thymic selection.11,12 Together with the loss of most naive T cells shortly after thymic exit, such an enrichment of autoreactive T cells would favor autoreactivity of the T-cell repertoire. A functional polymorphism in the polyadenylation signal sequence of the human GIMAP5 gene that reduces the abundance of properly terminated transcripts was recently shown to be associated with the presence of islet autoantibodies in type 1 diabetes patients13 and with an increased risk of systemic lupus erythematosus.14 Introgression of the BB rat Gimap5 mutation into the F344 rat strain reproduces peripheral lymphopenia without causing autoimmune disease,15 and transgenic expression of normal rat gimap5 in F344 rats carrying the defective Gimap5 gene rescues these defects.16 Second, Gimap5 emerged as a candidate in a genome-wide expression profiling approach to detect genes up-regulated during positive selection of mouse thymocytes.8 Knockdown of Gimap5 in immature mouse thymocytes reduced the frequency of DP and CD4SP cells emerging in fetal thymus organ cultures, suggesting that gimap5 is required for optimal generation of DP thymocytes.8 Biochemical evidence suggests that gimap5 physically interacts with positive (ie, Bax, Bak, Bad, and BimEL) and negative (Bcl-2 and Bcl-xL) regulators of the intrinsic mitochondrial pathway of apoptosis.8 Although murine Gimap3 and Gimap5 exhibit a high degree of sequence similarity, these knockdown studies suggest that either gene fulfills different functions in thymocyte maturation.8 Consistent with a predominant role of murine gimap5 in apoptosis regulation, human gimap5 was identified in an expression screen for gene products that protect against radiation-induced apoptosis.2 Of note, in this screen, a fragment of human gimap5 corresponding to residues 230 to 285 within the coiled-coil region of mouse gimap5 was sufficient to confer resistance to radiation-induced cell death. The subcellular localization of gimap5 is still unclear,17 but available data suggest that gimap5 may be associated with intracellular membranes and macromolecular organelles, including endoplasmic reticulum (ER), Golgi, mitochondria, and centrosomes.8
In summary, available evidence suggests that rat, mouse, and human gimap5 regulates through interaction with pro- and/or antiapoptotic genes the mitochondrial pathway of cell death in lymphocytes and that defects in antiapoptotic gimap5 function may alter intrathymic T-cell maturation, survival of peripheral T cells, and bias the repertoire of the residual T-cell population toward autoreactivity. In the current communication, we describe the consequences of gimap5 deficiency in mice. We show that the loss of murine gimap5 function impairs peripheral T-cell survival, imposes a complete block of natural killer (NK) and NKT cell development, and results in lethal postnatal liver failure associated with massive hepatic hematopoiesis.
Methods
Generation of Gimap5−/− mice
BAC clones comprising various portions of the mouse gimap gene cluster were obtained from Invitrogen (Carlsbad, CA). The gene-targeting construct shown in Figure 1 was generated (in 5′-to-3′ order) from a thymidine kinase expression cassette (PGK TK): a 5.4-kb EcoRI/BamHI genomic fragment of Gimap5 intron 1, a neomycin-resistance cassette (PGK-NEOR), and a 1.3-kb fragment composed of the last 713 bp of Gimap5 exon 3 and 597 bp of 3′ flanking region. R1 embryonic stem (ES) cells were electroporated with the linearized targeting vector and selected in medium containing G418 and gancyclovir. ES cell clones having undergone the predicted recombination events where identified by polymerase chain reaction (PCR) amplification of DNA fragments specific for the targeted allele (see Figure 1): primer A, 5′-CTGAAGCAGGTCGGTCTTGACAAAAAG-3′; primer B, 5′-GCCTTCTATCGCCTTCTTGAC-3′; primer C, 5′-GTCTGGTTGCTGGGTTGCAAAC-3′; primer D, 5′-GGACAGGTCGGTCTTGACAAAAAG-3′).
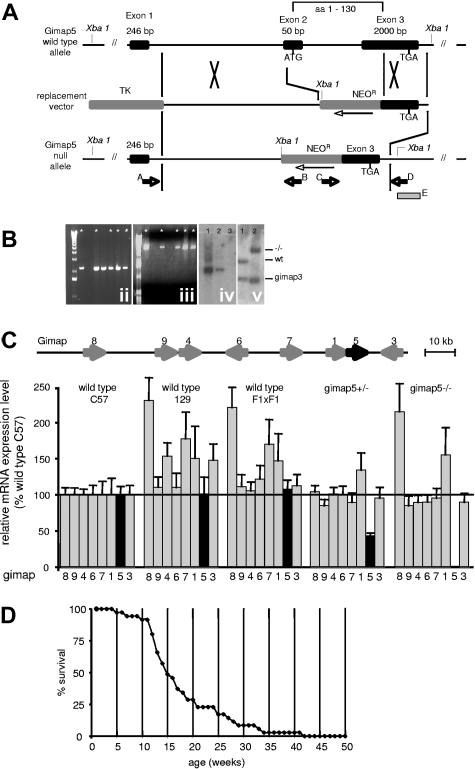
Figure 1.
Generation of gimap5 knockout mice. (A) A replacement-type gene-targeting vector was constructed from a thymidine kinase expression cassette (TK); a 5.4-kb EcoR1-BamH1 fragment (5′-homology) of gimap5 intron 1; a pkg-promoter-neomycin resistance gene cassette (NEOR, orientation opposite to gimap5) and a 1.3-kb fragment comprising the last 713 bp of exon 3 plus 597 bp of 3′-flanking region. Homologous recombination replaces a 2466-bp fragment of the Gimap5 gene containing exon 2 and a fragment of exon 3, thereby deleting the translation start signal within exon 2 and the first 130 amino acids of gimap5 protein. The figure is not drawn to scale. (B) Recombination in the 5′ and 3′ region of the Gimap5 gene was confirmed by amplification of a 4-kb genomic fragment with primers A and B (ii), and a 2.2-kb fragment with primers C and D (iii), respectively. * indicates ES clones having undergone the predicted recombination events in both the 5′ and 3′ homology. (iv) Southern blot hybridization of Xba1-digested genomic DNA from Gimap+/− ES clones (lanes 1,2) or wild-type ES cells (lane 3) with a 32P-labeled neomycin probe detects the predicted 3.1-kb fragment, consistent with a single locus integration of the NEO cassette. (v) Southern blot hybridization analysis of Bgl2-digested genomic DNA from wild-type (lane 1) and Gimap5−/− mice (lane 2) with probe E. The probe detects in wild-type mice a 2.6-kb fragment comprising exon 3 of gimap5 and a 1.8-kb fragment containing a homologous region in exon 5 of Gimap3; in Gimap5−/− DNA, the Gimap3 fragment detected is identical to wild-type, whereas the Gimap5 fragment shifts to 4.1 kb resulting from insertion of the NEO gene. (C) Measurements of gimap gene family mRNA abundance by real-time PCR. The structure of the gimap gene cluster on chromosome 6 is shown on top. Bars indicate the average plus or minus SEM from triplicate determinations from 3 mice each. Data are normalized for gimap abundance in wild-type C57BL/6J mice. Only gimap5 mRNA is significantly reduced in Gimap5+/− mice and is undetectable in Gimap5−/− mice. (D) Survival of Gimap5−/− mice (n = 35) over 42 weeks. The median age of death lies between 14 and 15 weeks. By 42 weeks, no surviving Gimap5−/− mice remained.
Three independent Gimap5+/− ES cells lines were used to generate germline-competent chimeras after injection of ES cells into C57BL6 blastocysts, and transmitted the mutated allele to their offspring after mating of chimeric males to C57BL6 females. The potential existence of an aberrant transcript resulting from splicing of gimap5 exon 1 to the 3′ portion of exon 3 was ruled out by PCR amplification with corresponding primers (forward: 5′-GCTGACAGCCGCTTGG-3′; reverse: 5′-GAACTTTGGGGGACAAGGAT-3′).
Real-time PCR
Total RNA was isolated from the thymus using the RNEasy Minikit (QIAGEN, Valencia, CA). First-strand cDNA synthesis was performed using Superscript III cDNA synthesis kit (Invitrogen) with oligo-dT primers. Real-time PCR was performed using identical amounts of oligo-dT–primed cDNA as a template and primers recognizing specific sequences in 7 members of the mouse Gimap family8 (Gimap1, CGTGGTGTTCACGCGCCAAG; CCCTCACCAAGCACGCAACC; Gimap3, GGATTCCAGTGTATACTACAGAC; TCAGTGGCCTTGACCCTGAG. Gimap4,TCTCTTGCTCACCAGGAAGG; ACCATTTTCCCTCACCATGC; Gimap5, GCTGACAGCCGCTTGG; TGGGCTTCTGGTCCTTGAAC; Gimap6, GACTCTGGACAAGGATTGAC; CAAACTCTCTAGTCCCTTTC; Gimap7, GAAAGAGCGCCACAGCAAAC; GCCCTGGGGTATCAACAACC; Gimap8, GTTCAAAGAAGAGGCAGCC; CCTGCTGTGGGATGCAAAG; Gimap9, CTCACAAAGAAGACCTTGAG; TGTCAGAAAAGTAGGATCCG). Primer sequences (5′ to 3′; forward; reverse) were kindly provided by Drs Nitta and Takahama8 (University of Tokushima, Tokushima, Japan). Real-time PCR was performed with the Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA). PCR thermocycling and fluorescence detection were performed with the Applied Biosystems 7500 Real-Time PCR system. Raw data were analyzed and cycle thresholds determined with the aid of the Applied Biosystems 7500 System SDS Software.
Flow cytometry
Single-cell suspensions were treated with ACK buffer (150 mM ammonium chloride, 10 mM potassium bicarbonate, 1 mM ethylenediaminetetraacetic acid, pH 7.4) to lyse erythrocytes, washed and resuspended in Hank balanced salt solution supplemented with 3% fetal bovine serum (FBS). Nonspecific binding of secondary antibody (Ab) was blocked by preincubation with anti-FcRγII/III (BD Biosciences, San Jose, CA) in 3% FBS. For staining of intracellular antigens (foxP3), cells were fixed and permeabilized with Cytofix/Cytoperm buffer in accordance with the manufacturer's instructions (BD Biosciences). Lineage analysis of cells used the after antibodies (BioLegend, San Diego, CA) included: fluorescein isothiocyanate (FITC)–conjugated anti-CD3e, allophycocyanin (APC)–conjugated anti-CD3e, FITC-conjugated anti-CD4, APC-conjugated anti-CD4, phosphatidylethanolamine (PE)/Cy7-conjugated anti-CD4, FITC-conjugated anti-CD44, APC-conjugated anti-CD44, PE-conjugated anti-CD49b, APC-conjugated anti-CD62l, APC-conjugated anti-CD25, APC-conjugated anti-Gr1, PE/Cy7-conjugated anti-Gr1, APC-conjugated anti-Mac1, PE-conjugated anti-Thy1.2, FITC-conjugated anti-Thy1.2, APC-conjugated anti-Sca1, and PE/Cy7-conjugated anti-cKit. Anti-FcRγII/III, FITC-conjugated anti-CD34, and FITC-conjugated anti-Qa2 were obtained from eBioscience (San Diego, CA). PE-conjugated anti-FcRγII/III, PE-conjugated anti-CD8a, PE-conjugated anti-CD45.1, FITC-conjugated anti-CD45.2, and FITC-conjugated anti-Gr1 were from BD Biosciences. For identification of lin− cells, a pool of biotinylated antibodies recognizing Ter119, CD3e, CD4, CD8, B220 (all from Biolegend), CD19 (eBioscience, San Diego, CA), and Gr1 (BD Biosciences) was used.
Measurement of apoptosis by flow cytometry and TUNEL stain
Identification of apoptotic cells stained with YO-PRO-1 (Invitrogen) and 7-actinomycin D (7-AAD; BD Biosciences) by flow cytometry was conducted as previously described.18,19 TdT-mediated dUTP nick end labeling (TUNEL) assay was performed on paraffin-embedded tissue section as described earlier19 using an In Situ Cell Death Detection Kit (Roche Pharmaceuticals, Nutley, NJ) according to the manufacturer's instructions.
Bone marrow transplantations
Whole bone marrow (BM) was harvested from the femur and tibia of 8- to 12-week-old mice. Cells were treated with ACK buffer to lyse erythrocytes and resuspended in cold phosphate-buffered saline (PBS) supplemented with 2% FBS. On the same day, 2 × 106 of the harvested BM cells were injected in a volume of 0.2 mL into the retro-orbital venous plexus of lethally irradiated (1100 cGy; Gammacell-40 irradiator; MDS Nordion, Ottawa, ON) B6.SJL-PtprcaPepcb/BoyJ (CD45.1 congenic) recipients. Recipients received antibiotic-treated water until death. For reconstitution of immunodeficient Jak3−/− recipients, 2 × 106 cells were transplanted into sublethally irradiated (300 cGy).
Mice
C57BL/6 and 129 mice were obtained from Charles River Breeding Laboratories (Portage, MI). CD45.1 congenic mice (B6.SJL-PtprcaPepcb/BoyJ) and Jak3−/− mice (B6;129S4-Jak3tm1Ljb/J) were obtained from The Jackson Laboratory (Bar Harbor, ME). Rag2−/− mice (C.129S6(B6)-Rag2tm1FwaN12) were obtained from Taconic Farms (Germantown, NY). All procedures involving animals were conducted in accordance with and approved by the Institutional Animal Care and Use Committee of the Medical College of Wisconsin.
Results
Generation of Gimap5-null mice
To selectively disrupt the murine Gimap5 gene, a gene-targeting vector was constructed to replace in ES cells exon 2 and part of exon 3 with a neomycin resistance gene expression cassette (Figure 1A). This replacement deletes the translation initiation signal and amino acids 1 through 130 of the gimap5 gene product, which comprise the putative nucleotide-binding motif. Of 360 ES cell clones analyzed, 5 independent clones were identified by analytical PCR analysis to have undergone the predicted recombination events and to contain a single copy of the neomycin gene cassette (Figure 1B). Three ES clones gave rise to germline-competent, male 129Sv/C57BL6J chimeras that where bred to C57BL6 females and transmitted the mutated allele to their offspring. To confirm the selective disruption of the Gimap5 gene, steady-state mRNA levels of all known mouse gimap genes expressed in the thymus were determined by real-time RT-PCR in 5-week-old wild-type and putative gimap5-null littermates from F1 intercrosses. Only gimap5 mRNA was significantly reduced in heterozygous Gimap5+/− mice and undetectable in Gimap5-null mice (Figure 1C). Reverse transcription (RT)–PCR analysis of Gimap5-null thymus RNA with primers hybridizing to exon 1 and the preserved 3′ region of exon 3 failed to detect potential aberrant transcripts encoding a truncated or frame-shifted gimap5 protein (data not shown).
The genotype distribution of offspring from heterozygous matings conformed to a Mendelian pattern of inheritance, with a sex ratio (male/female) of 0.9 (n = 911; wild-type, 244; Gimap5+/−, 455; Gimap5−/−, 212). Gimap5−/− mice exhibited a reduced life span, with mortality onset as early as 5 weeks, and substantial loss of animals between 10 and 20 weeks. In a selected cohort of 35 Gimap5-null mice, only one female survived beyond 42 weeks (Figure 1D).
These data are consistent with a selective disruption of the Gimap5 gene and show that Gimap5 deficiency does not affect intrauterine embryonic development but significantly reduces the life span of Gimap5−/− animals. All phenotypic abnormalities described in the following were reproduced in at least 2 of 3 independently generated strains of Gimap5−/− mice, in Gimap5−/− animals generated from an intercrosses of 2 independently generated strains, as well as in Gimap5−/− mice backcrossed for 6 generations onto the C57BL6- or BALB/c genetic background (data not shown). None of the phenotypes described below was detected in heterozygous Gimap5+/− mice (not shown).
Gimap5−/− mice exhibit peripheral T-cell lymphopenia
The hematologic profile of Gimap5−/− mice (6-12 weeks old; males and females) differed from that of age- and sex-matched littermates in several respects. Peripheral blood smears exhibited a marked heterogeneity of erythrocyte size (Figure 2A), somewhat reduced hematocrit, erythrocyte counts, and hemoglobin content (Table 1). Mild thrombocytopenia was present in younger mice (< 12 weeks; wild-type, n = 12, 373 ± 58; Gimap5−/−, n = 13, 225 ± 40; P < .05) but was not significant in the subgroup of older animals (> 12 weeks; n = 4; wild-type, n = 5, 526 ± 20; Gimap5−/−, n = 3, 595 ± 136; P = .66). Gimap5−/− mice exhibited peripheral lymphopenia (∼1.6-fold reduction), pronounced granulocytosis (∼ 3.8-fold increase), and unaltered prevalence of monocytes. Fluorescence-activated cell sorter (FACS) analysis of peripheral blood showed a normal representation of B220+ B cells, an approximate 2-fold increased frequency of Gr1+ granulocytes, and an approximate 3-fold reduced abundance of CD90.2 T cells (Table 1).
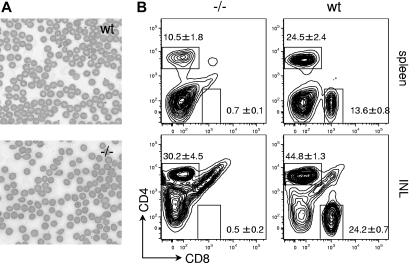
Figure 2.
Erythrocyte abnormalities and peripheral lymphopenia in Gimap5−/− mice. (A) Peripheral blood smear (Wright-Giemsa stain, original magnification ×400). Bars represent 10 μm. wt indicates wild-type; −/−, Gimap5-knockout. Images were acquired on a Nikon Eclipse E600 microscope with 10×, 20×, and 40× PlanFluor objectives and a SPOT Insight 11.2 color mosaic digital camera with Spot software version 4.1 (Diagnostic Instruments, Sterling Heights, MI). (B) Representative FACS analysis of wild-type (wt) and Gimap5-deficient (−/−) splenocytes and inguinal lymph node cells (INL) for expression of CD8 and CD4. Boxes indicate gates used to calculate frequencies presented in Table 1. Gimap5−/− mice exhibit peripheral lymphopenia, with reduced abundance of CD4 T cells and almost complete absence of CD8 T cells from the spleen and inguinal lymph nodes (INL).
Table 1.
Hematologic profile of Gimap5−/− mice
| Units | gimap5−/− | Wild-type | N | P | |
|---|---|---|---|---|---|
| Peripheral blood | |||||
| RBC | 106/μL | 9.0 ± 0.8 | 10.3 ± 0.1 | 15/13 | .13 |
| Hemoglobin | g × 100 mL−1 | 12.9 ± 1.2 | 15.2 ± 0.3 | 15/13 | .08 |
| Hematocrit | % | 42.2 ± 3.9 | 48.2 ± 0.4 | 15/13 | .15 |
| RBC-MCV | fL | 47.3 ± 1.1 | 46.8 ± 0.6 | 15/13 | .68 |
| RBC distribution width | % | 20.2 ± 0.8 | 14.1 ± 0.1 | 15/13 | <.005 |
| Platelets | 103/mL | 271 ± 52 | 399 ± 60 | 15/13 | .12 |
| Leukocytes | 103/μL | 7.0 ± 1.1 | 4.6 ± 0.4 | 13/12 | <.05 |
| Lymphocytes | % | 29.0 ± 3.4 | 63.8 ± 2.4 | 13/12 | <.05 |
| Lymphocytes | 103/μL | 1.8 ± 0.3 | 2.8 ± 0.2 | 13/12 | <.005 |
| Monocytes | % | 8.1 ± 0.7 | 10.8 ± 0.6 | 13/12 | <.05 |
| Monocytes | 103/μL | 0.6 ± 0.1 | 0.5 ± 0.1 | 13/12 | .61 |
| Granulocytes | % | 63.0 ± 3.9 | 25.6 ± 2.1 | 13/12 | <.005 |
| Granulocytes | 103/μL | 4.6 ± 0.8 | 1.2 ± 0.2 | 13/12 | <.005 |
| CD90.2+ | % | 8.6 ± 1.6 | 29.9 ± 2.3 | 6/6 | <.005 |
| Gr1+ | % | 55.4 ± 11.2 | 21.8 ± 2.6 | 6/6 | <.05 |
| B220+ | % | 36.0 ± 11.6 | 48.3 ± 2.3 | 6/6 | .35 |
| Spleen | |||||
| CD4+ plus CD8+ | % | 11.2 ± 1.8 | 38.2 ± 3.0 | 7/5 | <.005 |
| CD4+ | % | 10.5 ± 1.8 | 24.5 ± 2.4 | 7/5 | <.005 |
| CD8+ | % | 0.7 ± 0.1 | 13.6 ± 0.8 | 7/5 | <.005 |
| Gr1+ | % | 32.7 ± 13.1 | 4.9 ± 0.5 | 2/2 | |
| B220+ or CD19+ | % | 16.1 ± 6.3 | 49.6 ± 1.9 | 5/5 | <.005 |
| Inguinal lymph node | |||||
| CD4+ plus CD8+ | % | 30.7 ± 4.6 | 69.0 ± 1.6 | 7/5 | <.005 |
| CD4+ | % | 30.2 ± 4.5 | 44.8 ± 1.3 | 7/5 | <.05 |
| CD8+ | % | 0.5 ± 0.2 | 24.2 ± 0.7 | 7/5 | <.005 |
| Gr1+ | % | 0.9 ± 0.2 | 2.1 ± 01.2 | 5/5 | .38 |
| B220+ or CD19+ | % | 9.0 ± 2.1 | 15.6 ± 4.5 | 5/5 | .24 |
| Pancreatic lymph node | |||||
| CD4+ plus CD8+ | % | 14.8 ± 4.1 | 54.9 ± 2.7 | 3/3 | <.005 |
| CD4+ | % | 13.9 ± 3.7 | 33.3 ± 1.9 | 3/3 | <.05 |
| CD8+ | % | 0.9 ± 0.4 | 21.5 ± 0.9 | 3/3 | <.005 |
| Gr1+ | % | 10.9 ± 7.2 | 6.2 ± 0.2 | 3/3 | .58 |
| B220+ or CD19+ | % | 8.3 ± 3.4 | 29.1 ± 3.3 | 3/3 | <.05 |
In the spleen, the absolute number of T cells (CD4+ plus CD8+) is significantly reduced (wild-type, n = 5: 35.8 ± 5.5; gimap5−/−, n = 7: 4.2 ± 0.6 × 106 cells, ± SEM; P < .005). This approximate 9-fold decrease in splenic T-cell abundance may be attributed to (1) an approximately 20-fold reduction in the frequency of CD8+ T cells (Table 1; Figure 2B), (2) an approximately 2-fold reduction in the frequency of CD4+ T cells (Table 1; Figure 2B), and (3) an approximately 2-fold reduction in total splenic cellularity (wild-type, n = 5, 92.6 ± 9.9; gimap5−/−, n = 7, 42.4 ± 7.1 × 106 cells, ± SEM; P < .005; Table 1; Figure 2B). A significant paucity of B220+ or CD19+ B cells was evident in the spleen of Gimap5−/− mice. Corresponding alterations in the abundance of T and B cells were observed in pancreatic and inguinal lymph nodes (Table 1; Figure 2B).
Compared with wild-type mice, the residual CD8+ and CD4+ T-cell subpopulations in secondary lymphoid organs of Gimap5−/− mice display 2 specific surface markers of activation, ie, increased shedding of CD62L and up-regulation of CD69: The relative abundance of CD4+CD62L− cells and CD8+CD62L− cells, and of CD4+CD69+ and CD8+CD69+ cells is significantly increased in the spleen (Figure 3A) and inguinal lymph nodes (not shown).
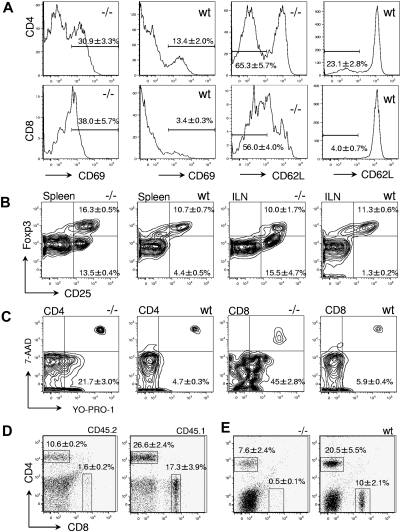
Figure 3.
Characterization of peripheral T cells in Gimap5−/− mice. (A) Representative histograms of CD69 and CD62L expression in CD4+ (top) and CD8+ splenocytes (bottom). Increased expression of CD69 and shedding of CD62L indicate a state of increased activation, compared with wild-type mice. (B) The frequency of CD4+CD25+FoxP3+ regulatory T cells in the spleen or the inguinal lymph node (ILN) is unaltered in Gimap5−/− mice. (C) Apoptosis is increased among peripheral T cells from Gimap5−/− mice. CD4+ (left) and CD8+ (right) splenocytes were stained with the dyes YO-PRO-1 and 7-AAD to identify early apoptotic (YO-PRO-1+7-AAD−) and dead (YO-PRO-1+7-AAD+) T cells. (D) Abundance of CD45.2+ gimap5−/− CD4 and CD8 T-cell populations in lethally irradiated CD45.1+ wild-type recipients 8 weeks after transplantation with unfractionated BM from CD45.2 Gimap5−/− mice. Overall representation of splenic CD45.2 T cells in the example shown was approximately 60%. The relative abundance of donor-derived CD8 and CD4 cells captured by the indicated gates is given as average percentage plus or minus SD (n = 3). The ratio between CD4 and CD8 cells is significantly different between Gimap5−/− donor cells and residual wt recipient cells (P < .05). (E) Jak3−/− mice were sublethally irradiated and transplanted with unfractionated BM from wild-type (wt) or Gimap5−/− mice (−/−). Relative abundance of CD4 and CD8 cells represents average plus or minus SD (n = 3). The ratio between CD4 and CD8 cells is significantly different between Gimap5−/− and wild-type donor cells (P < .05).
In the spleen, the frequency of CD25+FOXP3+ cells among CD4+ cells is somewhat increased in Gimap5−/− mice (Gimap5−/−: 16.3% ± 0.5%, mean ± SEM; n = 3; wild-type, 10.7% ± 0.7%; n = 3; P < .005). However, the frequency of CD25+FOXP3− cells among CD4+ lymphocytes is increased in Gimap5−/− mice (Figure 3B), consistent with a state of partial effector T-cell activation. In the inguinal lymph node, there is a significant increase of CD25+FOXP3− cells among the CD4+ T-cell population but no change in the frequency of CD25+FOXP3+ T cells among CD4+ cells (Figure 3B). Gimap5 deficiency is therefore not associated with a complete loss of putative regulatory T cells but a reduction of the putative FoxP3+ regulatory T (TREG)–cell population proportional to the diminished prevalence of CD4+ T cells and an altered ratio of FoxP3 TREG cells relative to activated CD4+CD25+ T cells. We note that these data are not informative with regard to the functional status of Gimap5−/− putative TREG.
FACS analysis with 7-AAD and YO-PRO-1 was used to distinguish viable (YO-PRO-1−7-AAD−) from apoptotic (YO-PRO-1+7-AAD−) and dead (YO-PRO-1+7-AAD+) lymphocytes in Gimap5−/− animals. No significant difference in the abundance of apoptotic (YO-PRO-1+7-AAD−) cells was observed in blood (data not shown); on the other hand, the prevalence of apoptotic CD8+ and CD4+ splenocytes was increased 4- and 8-fold, respectively (Figure 3C). Similar results were obtained from an analysis of inguinal and pancreatic lymph nodes (data not shown). A corresponding analysis of T cells from blood, spleen, and lymph nodes using specific antibodies to detect the presence of activated caspase 3 showed a nonsignificant trend toward increased apoptosis only in CD8 T cells (data not shown).
Transfer of unfractionated BM from Gimap5−/− mice (N4 backcross to C57BL6; CD45.2) into lethally irradiated CD45.1 congenic wild-type recipients achieved less efficient reconstitution than that achieved in control experiments by transfer of CD45.2 wild-type BM into lethally irradiated CD45.1 wild-type recipients (wild-type donor: 94% ± 2% reconstitution week 8 posttransplant average; range, 90%-96%; Gimap5−/− donor: 76% ± 5%; range, 55%-82%; P = .024). In the spleen of resulting chimeras, Gimap5−/− CD4+ cells were represented at the same level as wild-type CD4+ donor-derived cells (Gimap5−/− donor: 17% ± 2% of splenocytes; wild-type donor: 15% ± 1%; P = 0.49). In contrast, Gimap5−/− CD8 T cells were significantly underrepresented (Gimap5−/− donor: 0.58% ± 0.22% of splenocytes; wild-type donor: 5.3% ± 0.6%; P = .007; Figure 3D). Likewise, transfer of Gimap5−/− marrow into sublethally irradiated Jak3 knockout mice with combined deficiency of T, B, and NK cells20–22 did not rescue the defects in the Gimap5−/− CD4 and CD8 populations (Figure 3E). These results indicate that the loss of peripheral CD8 T cells in Gimap5−/− mice is the result of an intrinsic defect in T cells that persists in a wild-type environment and in the presence of wild-type T, B, and NK cells. We note that these data are not derived from competitive reconstitution experiments, and the analysis of chimeras does therefore not support stringent conclusions about the relative competition between wild-type and Gimap5−/− hematopoietic cells.
Thymic maturation of gimap5−/− T cells
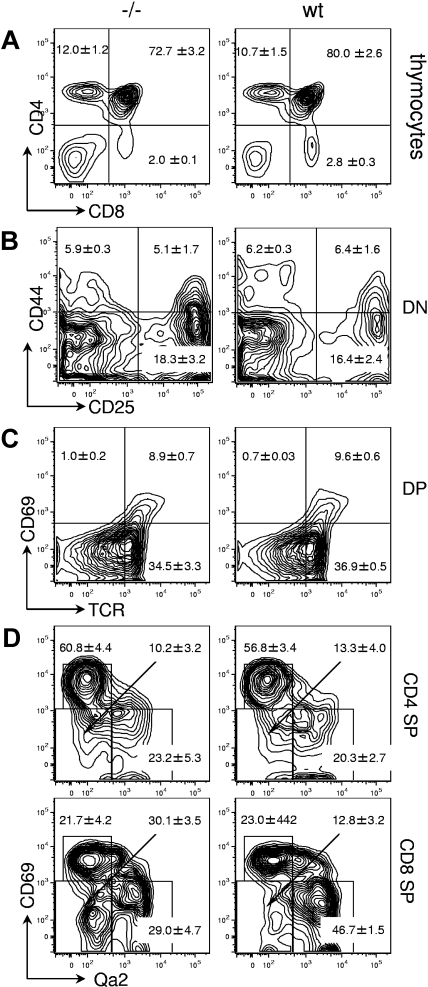
In 6- to 8-week-old Gimap5−/− mice, the grossly observable size and appearance of the thymus were identical to that of wild-type littermates. The proportion of CD4−CD8+ thymocytes was somewhat reduced (Gimap5−/−: 2.0% ± 0.1%; n = 7; wild-type: 2.8% ± 0.3%, ± SEM; n = 5, P = .07), whereas the relative abundance of CD4+CD8− (12% ± 1.2% vs 10.7% ± 1.5%), CD4−CD8− (8.3% ± 2.2% vs 6.5% ± 0.7%), and CD4+CD8+ cells (72.7% ± 3.2% vs 80.0% ± 2.6%) was indistinguishable from that of wild-type animals (Figure 4A). Analysis of the CD4−CD8− subpopulation for expression of CD25/CD44 documented normal progression of T cells through double-negative stages DN1-4 (Figure 4B). Analysis of CD4+CD8+ DP thymocytes for expression of T-cell receptor (TCR) (defined by H57/TCR-β) and CD69 showed a normal abundance of TCR−CD69−, TCR+CD69−, and TCR+CD69+ cells (Figure 4C), consistent with normal maturation CD4+CD8+ thymocytes. Examination of SP thymocytes that have survived selection and constitute the population of prospective thymic emigrants, as defined by up-regulation of Qa2 expression23–26 and concomitant down-modulation of CD69 expression showed a selective deficit of CD4−CD8+CD69LOWQa2HIGH cells (29% ± 4.7% vs 46.7% ± 1.5%, ± SEM; P < .05), and a corresponding accumulation of CD4−CD8+CD69LOWQa2LOW cells (30.1% ± 3.5% vs 12.8% ± 3.2%, ± SEM; P < .05), indicative of a defect in the transition through the final stages of intrathymic maturation of CD8 SP thymocytes (Figure 4D).
Figure 4.
Thymic development in Gimap5−/− mice. Representative FACS plots of wild-type (wt) or Gimap5-deficient (−/−) thymocytes. (A) Expression of CD4 and CD8 in unfractionated thymocytes. Boxes indicate gates used to calculate the average frequencies included in the text. (B) Expression of CD44 and CD25 in DN thymocytes contained in the DN gate of panel A. (C) Expression of CD69 and H57/TCR-β (TCR) in DP thymocytes contained in the DP gate of panel A. (D) Expression of CD69 and Qa2 in CD4SP and CD8SP thymocytes. The relative accumulation of CD69LOWQa2− versus CD69LOWQa2+ cells indicates a defect in the final stages of intrathymic T-cell maturation in Gimap5−/− mice.
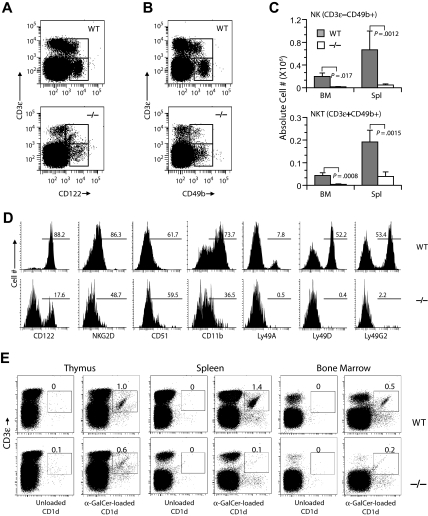
Gimap5−/− mice lack NK and NKT cells
Expression of CD122 marks the commitment of progenitor cells into NK and NKT lineages in the BM and is continued to be expressed on all stages of NK-cell development. Acquisition of CD49b by CD122+ cells marks the second stage of NK maturation.27 Gimap5−/− mice showed an 80% reduction of CD3−CD122+ NK cell abundance among the lymphocytes in the BM and spleen (Figure 5A). Likewise, CD3+CD122+ NKT cells were significantly reduced in BM and spleen. Both the CD3−CD49b+ and the CD3−CD122+ cell populations were similarly diminished, indicating that the few Gimap5−/− NK cells that have committed to NK progenitors are able to transit into the CD3−CD49b+ stage in BM and spleen (Figure 5B). The absolute cell numbers of CD3−CD49b+ NK and CD3+CD49b+ NKT were significantly reduced in BM and spleen (Figure 5C,D) and liver of Gimap5−/− mice. Analysis of Gimap5−/− mice that had been backcrossed to the C57BL/6 background for 7 generations with Cd1–α-galactosylceramide multimers in conjunction with anti-CD3 (Figure 5E), and for expression of the NK-cell marker NK1.1 in BM, spleen, liver, and thymus (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article) yielded corresponding results, documenting the lack of NK cells, and a severe, almost complete lack of NKT cells expressing the invariant Vα14 TCR.
Figure 5.
Gimap5−/− mice lack NK and NKT cells. Development of NK and NKT cells is severely impaired in Gimap5−/− mice. (A) Representative analysis of BM from Gimap5−/− and WT mice stained with anti–CD3-ϵ and anti-CD122; numbers indicate percentage of cells (average ± SD) in gate. CD3-ϵ−CD122+ NK and CD3-ϵ+CD122+ NKT are significantly reduced in Gimap5−/− mice (NK: P = .004; NK-T: P = .002). (B) Representative analysis of BM from Gimap5−/− and WT mice stained with anti–CD3-ϵ and CD49b. (C) The absolute numbers (average ± SD) of CD3-ϵ−CD49b+ NK or CD3-ϵ+CD49b+ NKT cells were significantly reduced in the Gimap5−/− mice. Absolute cell numbers were calculated as percentage cells times percentage lymphocyte times cellularity of spleen (Spl) or BM (n = 6/group). P values calculated by the Student t test. (D) Expression of developmental markers in BM-derived fresh Gimap5−/− CD3-ϵ−CD49b+ NK cells is severely reduced. The frequencies of cells positive for each marker among CD3-ϵ−CD49b+ NK cells are shown along with representative histograms. Gates were set using unstained or nonspecific isotype antibody controls (not shown). Data are representative of 4 mice/genotype. (E) Gimap5−/− mice lack CD1-αGalCer+ NKT cells. Single-cell suspensions prepared from the indicated tissues of wild-type mice (WT) and Gimap5−/− mice (−/−) were stained with anti–CD3-ϵ mAb and α-GalCer–loaded CD1d dimers. Numbers represent percentage Vα14+ CD3-ϵ+ iNKT cells in the gates shown. Anti–CD3-ϵ mAb and unloaded CD1d dimers were used as background controls. Data presented are representative of at least 3 independent analyses.
During the final stages of maturation, NK cells up-regulate integrin CD11b (Mac-1) and CD43 (leukosialin) and acquire one or more Ly49 receptors, whereas CD51 expression declines.28,29 The residual CD3−CD49b+ NK cells in Gimap5 mice exhibited greatly reduced levels of NKG2D and CD11b. However, expression of CD51 was unaffected. The few Gimap5−/− CD3−CD49b+ NK cells lacked expression of either one of the inhibitory Ly49A and L49G or the activating Ly49D receptors, suggesting that the residual NK cells in Gimap5−/− mice fail to undergo this terminal maturation.28 The near absence of NK cells in Gimap5−/− mice, together with potentially reduced responsiveness to interleukin-2 (IL-2; not shown), prevented further analyses, and attempts to derive lymphokine-activated killer cells from spleen or BM were unsuccessful.
Transfer of Gimap5−/− BM into lethally irradiated wild-type recipients (CD45.1 isotype) resulted in partial reconstitution of the splenic NK-cell compartment: following reconstitution with Gimap5−/− BM, donor-derived NK cells (CD45.2+CD49b+) accounted for 0.5% plus or minus 0.1%, plus or minus SEM (n = 4) of total donor cells, in contrast to a CD49b+ NK-cell frequency of 1.3% plus or minus 0.1%, plus or minus SEM (n = 2; P < .05) after transfer of wild-type BM into wild-type recipient. Transfer of Gimap5−/− BM into sublethally irradiated JAK3−/− recipient mice, which completely lack NK cells, resulted in the emergence of Gimap5−/− NK and NKT cells (Figure S2). These data suggest that the defects of NK-cell development in Gimap5−/− mice are at least in part caused by an as yet unknown effect of gimap5 on the environment in which NK cells develop.
Fatal liver failure of Gimap5−/− mice
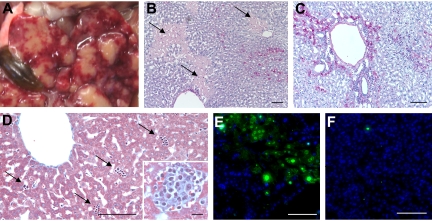
External examination of 10- to 16-week-old gimap5−/− mice was unremarkable. Visual inspection of internal organs and tissues in situ, and examination of histologic sections prepared from brain, lung, heart, kidney, pancreas, gastrointestinal tract (esophagus, stomach, duodenum, colon, cecum, mesenteric membranes), gonads, and bladder did not show any abnormalities or indications of inflammatory changes. Inguinal and mesenteric lymph nodes were of normal size and appearance, whereas pancreatic lymph nodes were small and exhibited gray discoloration. In striking contrast, in every Gimap5−/− mouse of the aforementioned age group analyzed, the liver exhibited marked discoloration, combining pale nodules with scarred, retracted and hyperemic hepatic tissue (Figure 6). Histopathologic alterations included irregular capsular surface, large areas of acute centrilobular and bridging hepatocellular degeneration and necrosis with parenchymal collapse. Large glycogen droplets suggestive of hydropic degeneration were present in many hepatocytes. Some areas with hepatocellular lesions showed evidence of extensive DNA fragmentation, as detected by TUNEL assay (Figure 6E,F).
Figure 6.
Liver pathology of Gimap5−/− mice. (A) In situ appearance of the liver in a 7-week-old Gimap5−/− mouse. The gallbladder (brown, lower left) appears normal. Image was acquired with a Nikon Coolpix digital camera. (B,C) Histologic sections (periodic acid Schiff [PAS] stain). Large areas appear necrotic (pale PAS stain, swollen hepatocytes with finely granular cytoplasm, sparse nuclear staining) but are largely devoid of mononuclear cells. (C) Other areas of the liver are characterized by massive, centrilobular, and periportal accumulation of mononuclear cells (blue nuclear stain, original magnification ×100). Bars represent 100 μm. (D) Liver histology of a 3-week-old gimap5−/− mouse (Trichome stain, original magnification ×200). Bar represents 100 μm. External appearance and histology of the liver parenchyma appear normal; the Gimap5−/− liver contains a large number of hematopoietic foci (→; inset shows high power view; original magnification ×400). Bar represents 10 μm. (B-D) Images were acquired on a Nikon Eclipse E600 microscope with 10×, 20×, and 40× PlanFluor objectives and a SPOT Insight 11.2 color mosaic digital camera with Spot software version 4.1 (Diagnostic Instruments, Sterling Heights, MI). (E) TUNEL stain of liver shown in panel B; areas of TUNEL positivity correlate with the pale-staining areas in PAS-stained sections. (F) DNA fragmentation is not detected in young Gimap5−/− livers shown in panel D. (E,F) TUNEL staining with fluorescein-labeled deoxyuridine triphosphate (dUTP) was recorded in the FITC channel.
A second hallmark of liver pathology was a striking abundance of mononuclear and polymorphonuclear cells, with a conspicuous abundance of hematopoietic precursors, in particular of granulocytic cells, but also including erythropoietic cells and pooled erythrocytes. FACS analysis of liver cell suspensions demonstrated an abundance of hematopoietic progenitor cells, including lin−Sca-1HIc-kitHI hematopoietic stem cells (wild-type, 3.4% ± 0.5%; gimap5−/−, 12.7% ± 1.7%; P < .05), and myeloid progenitors (lin−Sca-1−c-kitHI; wild-type, 3.4% ± 0.5%; Gimap5−/−, 12.7% ± 1.7%; P < .05). The latter population comprised myeloid-erythroid (CD34−FcRγII/IIILO), common-myeloid (CD34+FcRγII/IIILO), and granulocyte-monocytic precursors (CD34+ FcRγII/IIIHI). Gimap5 deficiency increased the abundance of all 3 subpopulations in the liver (common-myeloid Gimap5−/−, 16.6% ± 3.1%; wild-type, 6.7% ± 1.5%, P = .07; granulocyte-monocytic precursors Gimap5−/−, 34.1% ± 3.8%; wild-type, 5.3% ± 2.2%, P < .05; myeloid-erythroid Gimap5−/−, 17.2% ± 2.2%; wild-type, 5.3% ± 2.0%, P < .05). A less pronounced stimulation of hematopoiesis was observed in the spleen of Gimap5−/− mice (data not shown). In contrast, the BM of Gimap5−/− mice contained normal numbers of hematopoietic stem cells (wild-type, 4.6% ± 0.8%; Gimap5−/−, 8.3% ± 2.1%; P = .21) and of lin−Sca-1−c-kitHI myeloid progenitors (wild-type, 62% ± 3.1%; Gimap5−/−, 55% ± 42.7%; P < .05). The abundance of CD4+ and CD8+ T cells in the liver was reduced in a similar manner as shown for secondary lymphoid tissue (not shown).
Analysis of 2 younger Gimap5−/− mice (3 weeks old) showed a normal in situ appearance of the liver. A survey of histologic sections did not show any evidence of hepatocellular lesions, and the extent of DNA fragmentation (TUNEL assay) was indistinguishable from that seen in matched wild-type controls (Figure 6F). On the other hand, Gimap5−/− livers showed numerous small foci of hematopoietic cells (Figure 6D). Such islands were also detected in sections of age-matched wild-type livers but were much less frequent. As in older Gimap5−/− mice, these younger Gimap5−/− mice exhibited T-cell lymphopenia with a selective loss of CD8+ T cells (data not shown).
To examine whether the liver pathology was secondary to altered T- or B-cell function, or caused by accumulation of dying T cells in the liver, Gimap5−/− mice were crossed with T cell– and B cell–deficient Rag2−/− knockout mice. The liver pathology persisted in Gimap5−/−Rag2−/− double knockout mice (n = 3). Likewise, backcrosses of Gimap5−/− mice onto a C57BL6 (N6) or a BALB/c (N4) genetic background did not prevent liver disease, restore NK/NKT cell development, or correct peripheral lymphopenia. Transfer of Gimap5−/− BM into lethally irradiated wild-type recipients did not induce the liver pathology seen in entirely gimap5-deficient knockout mice. As late as 24 weeks after transplantation, the liver of recipient mice appeared morphologically and histologically normal, with no evidence of extramedullary hematopoiesis. Reverse transplants of wild-type marrow into neonatal Gimap5−/− recipients were unsuccessful due to excessive lethality of Gimap5−/− mice, even at low doses of radiation (data not shown). These data suggest that the liver pathology of Gimap5−/− mice is not caused by antibody-mediated autoimmune mechanisms.
Discussion
We show that disruption of the mouse Gimap5 gene is compatible with intrauterine development but results in marked peripheral loss of CD8, and to a lesser extent of CD4 T cells, marked granulocytosis, a complete loss of NK and NKT cells, and severe liver pathology associated with excessive hepatic hematopoiesis. Therefore, the role of gimap5 for T-cell survival, normal function of residual T and TREG cells, and NK/NKT cell development, which were initially documented in the gimap5-deficient lyp/lyp rat model, are conserved in the mouse. Therefore, murine Gimap5, despite the existence of a close related functional gimap3 gene, fulfills a nonredundant role in these contexts. Some differences between the mouse and rat may exist with respect to the role of gimap5 for intrathymic maturation of T cells: short hairpin (sh)RNA-mediated knockdown of Gimap5 in DN thymocytes disturbs the subsequent development of DP T cells in fetal thymic organ culture,9 and similar observations (decreased DP, increased DN) have been made in the Gimap5−/− F344 rat strain.15 In Gimap5−/− mice, we detect only a 2-fold reduction of CD4−CD8+CD69LOWQa2HIGH cells and a corresponding increase in the respective Qa2LOW population, indicative of a block in the very late stages of thymic maturation. A comparative analysis of the global gene expression profile of total thymocytes isolated from Gimap5−/− mice and wild-type littermates detected specific changes in gene expression comprising transcripts with as yet unknown function in thymic maturation of T cells (our unpublished observations, August 2008). Thus, mouse gimap5, like rat lyp/gimap5, also plays a role in intrathymic T-cell development, with minor discrepancies between the rat and mouse probably attributable to differences between the in vivo environment and the fetal thymic organ culture model, and species-specific effects, respectively.
The severe deficiency of NK and NKT cells, or early precursors thereof, dovetails the effects on NK/NKT cells in gimap5-deficient rats, which exhibit reduced numbers of NKT cells in the spleen, as well as γδ T and NK cells in the gut-associated lymphatic system.30–32 Gimap5 deficiency interfered with virtually all stages of mouse NK development and was incompatible with NK formation in any of the anatomic compartments thought to provide a permissive environment for NK maturation, including BM, spleen, lymph nodes, and liver. The presence of a small number of Vα TCR-positive invariant NKT (iNKT) cells (as defined by α-GalCer/CD1d staining) indicates that some of these cells can develop and successfully rearrange their TCR chains. An important novel finding is that Gimap5−/− BM gave rise to CD49b-positive NK when transferred into wild-type recipients, with an efficiency resembling that of BM from wild-type mice. This suggests that the majority of the defects in NK development are not cell intrinsic but are caused at least in part by the loss of gimap5 function in nonhematopoietic cells. Cell-extrinsic effects on NK cell differentiation have been well documented for mice lacking interferon-regulatory factor 1 or lymphotaxin-α/β.33,34 Gimap5 might be similarly required in stromal cells to provide a microenvironment capable of sustaining NK-cell development.
An unexpected finding was that Gimap5 gene disruption was incompatible with long-term survival. The fact that this phenotype was documented in 3 independently generated Gimap5−/− lines of mice as well as in intercrosses between these lines all but rules out inadvertent, secondary mutations as the underlying cause. The cause of death appears to be a progressive loss of liver function, accompanied by excessive hepatic hematopoiesis. This pathology persists in 2 inbred backgrounds (C57BL6/J and BALB/c) and in a Rag2-deficient background in the absence of functional T, B, and NK cells. These experimental findings strongly argue against an autoimmune-based pathogenesis. In addition, infection by environmental pathogens, although theoretically possible as a cause of death, appears doubtful because of the absence of inflammation in all other organs examined and because cohoused immunocompromised Rag2– and Rag2γc–double knockout colonies were not affected. The causal chain of events leading to hepatocyte death and massive hepatic hematopoiesis, and how these pathologic entities are related, is unclear at present. Given that transfer of Gimap5−/− marrow into wild-type recipients does not reproduce the liver pathology, we suspect that the loss of gimap5 expression in resident liver cells, including hepatocytes, may contribute to liver pathology. The peculiar effect of gimap5 deficiency on the liver (ie, apoptosis and persistent hepatic hematopoiesis) is reminiscent of a corresponding phenotype described in double-knockout mice lacking both the nuclear factor-κB (NF-κB) relA/p65 subunit and the tumor necrosis factor-α (TNF-α) receptor-1.35,36 Genetic disruption of NF-κB or of TAK1 compromises postthymic survival of CD8 and, to a lesser extent, of CD4 T cells in a similar manner as the loss of gimap5 function.37–39 The p52/relB alternate NF-κB pathway also appears to control NK development through cell-extrinsic mechanism operating in stromal cells.40 Moreover, gimap5-mediated regulation of T-cell survival and quiescence may involve mitogen-activated protein kinase kinase (MEK)–dependent activation of the upstream regulator of NF-κB, IκB kinase (IKK).41 Together, these observations raise the possibility that gimap5 interacts in a critical manner with the NF-κB signaling pathway. The reported role of NF-κB in the differentiation of marginal zone B cells42 prompted us to examine in detail B-cell differentiation in Gimap5−/− mice, and preliminary results show indeed a severe deficiency of both marginal zone B cells in the spleen, and B1 peritoneal B cells (our unpublished observations, August 2008). Given the substantial complexity of this signaling pathway, a priority for future experiments will be to determine how gimap5 intersects with the classic and alternative NF-κB pathways, define the structural basis of these interactions, and distinguish cell-intrinsic and extrinsic functions of gimap5 on immune cell function, survival, and development.
Supplementary Material
Acknowledgments
This work was supported in part by the American Cancer Society (grant RSG CCG-106204) (D.W.), National Institutes of Health (grants HL073284 [D.W.], AI42380-04 [A.L., A.E.K., J.G., and H.W.], RO1EB001421 [M.J.H.], R01A1064826-01 [S.G.-M.], and R01AI47154 [C.B.W.]), the D. B. and Marjorie Reinhart, Nickolett and Montgomery Family Foundations (C.B.W.), the Max McGee Center Grant from the Children's Research Institute, Children's Hospital of Wisconsin, American Cancer Society Scholar (grant RSG-02-172-LIB) (S.M.), and Roche Organ Transplantation Research Foundation (grant 111662730) (S.M.). H.C. is a recipient of Wisconsin Breast Cancer Showhouse Postdoctoral Fellowship. S.G.-M. is a recipient of American Society for Blood and Marrow Transplantation (ASBMT) Young Investigator Award.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: R.D.S. conducted research and cowrote the paper; X.D., H.C., Y.C., B.E., D.H., S.G.-M., S.J., and E.J.K. conducted experiments; C.B.W. and S.M. analyzed data and cowrote the manuscript; M.J.H. conducted experiments and analyzed data; D.W. analyzed data and cowrote the manuscript; A.E.K. and A.L. contributed critical reagents and analyzed data; S.G. and J.G. designed experiments and analyzed data; and H.W. designed and conducted experiments and cowrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hartmut Weiler, Blood Research Institute, 8727 Watertown Plank Road, Milwaukee, WI 53226; e-mail: hartmut.weiler@bcw.edu.
References
- 1.Stamm O, Krucken J, Schmitt-Wrede HP, Benten WP, Wunderlich F. Human ortholog to mouse gene imap38 encoding an ER-localizable G-protein belongs to a gene family clustered on chromosome 7q32-36. Gene. 2002;282:159–167. doi: 10.1016/s0378-1119(01)00837-x. [DOI] [PubMed] [Google Scholar]
- 2.Sandal T, Aumo L, Hedin L, Gjertsen BT, Doskeland SO. Irod/Ian5: an inhibitor of gamma-radiation- and okadaic acid-induced apoptosis. Mol Biol Cell. 2003;14:3292–3304. doi: 10.1091/mbc.E02-10-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersen UN, Markholst H, Hornum L. The antiapoptotic gene Ian4l1 in the rat: genomic organization and promoter characterization. Gene. 2004;341:141–148. doi: 10.1016/j.gene.2004.06.034. [DOI] [PubMed] [Google Scholar]
- 4.Payne F, Smyth DJ, Pask R, et al. Haplotype tag single nucleotide polymorphism analysis of the human orthologues of the rat type 1 diabetes genes Ian4 (Lyp/Iddm1) and Cblb. Diabetes. 2004;53:505–509. doi: 10.2337/diabetes.53.2.505. [DOI] [PubMed] [Google Scholar]
- 5.Pandarpurkar M, Wilson-Fritch L, Corvera S, et al. Ian4 is required for mitochondrial integrity and T cell survival. Proc Natl Acad Sci U S A. 2003;100:10382–10387. doi: 10.1073/pnas.1832170100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hornum L, Romer J, Markholst H. The diabetes-prone BB rat carries a frameshift mutation in Ian4, a positional candidate of Iddm1. Diabetes. 2002;51:1972–1979. doi: 10.2337/diabetes.51.6.1972. [DOI] [PubMed] [Google Scholar]
- 7.Lang JA, Kominski D, Bellgrau D, Scheinman RI. Partial activation precedes apoptotic death in T cells harboring an IAN gene mutation. Eur J Immunol. 2004;34:2396–2406. doi: 10.1002/eji.200324751. [DOI] [PubMed] [Google Scholar]
- 8.Nitta T, Nasreen M, Seike T, et al. IAN family critically regulates survival and development of T lymphocytes. PLoS Biol. 2006;4:e103. doi: 10.1371/journal.pbio.0040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nitta T, Takahama Y. The lymphocyte guard-IANs: regulation of lymphocyte survival by IAN/GIMAP family proteins. Trends Immunol. 2007;28:58–65. doi: 10.1016/j.it.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 10.MacMurray AJ, Moralejo DH, Kwitek AE, et al. Lymphopenia in the BB rat model of type 1 diabetes is due to a mutation in a novel immune-associated nucleotide (Ian)-related gene. Genome Res. 2002;12:1029–1039. doi: 10.1101/gr.412702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poussier P, Ning T, Murphy T, Dabrowski D, Ramanathan S. Impaired post-thymic development of regulatory CD4+25+ T cells contributes to diabetes pathogenesis in BB rats. J Immunol. 2005;174:4081–4089. doi: 10.4049/jimmunol.174.7.4081. [DOI] [PubMed] [Google Scholar]
- 12.Ramanathan S, Poussier P. BB rat lyp mutation and Type 1 diabetes. Immunol Rev. 2001;184:161–171. doi: 10.1034/j.1600-065x.2001.1840115.x. [DOI] [PubMed] [Google Scholar]
- 13.Shin JH, Janer M, McNeney B, et al. IA-2 autoantibodies in incident type I diabetes patients are associated with a polyadenylation signal polymorphism in GIMAP5. Genes Immun. 2007;8:503–512. doi: 10.1038/sj.gene.6364413. [DOI] [PubMed] [Google Scholar]
- 14.Hellquist A, Zucchelli M, Kivinen K, et al. The human GIMAP5 gene has a common polyadenylation polymorphism increasing risk to systemic lupus erythematosus. J Med Genet. 2007;44:314–321. doi: 10.1136/jmg.2006.046185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moralejo DH, Park HA, Speros SJ, et al. Genetic dissection of lymphopenia from autoimmunity by introgression of mutated Ian5 gene onto the F344 rat. J Autoimmun. 2003;21:315–324. doi: 10.1016/S0896-8411(03)00138-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michalkiewicz M, Michalkiewicz T, Ettinger RA, et al. Transgenic rescue demonstrates involvement of the Ian5 gene in T cell development in the rat. Physiol Genomics. 2004;19:228–232. doi: 10.1152/physiolgenomics.00126.2004. [DOI] [PubMed] [Google Scholar]
- 17.Keita M, Leblanc C, Andrews D, Ramanathan S. GIMAP5 regulates mitochondrial integrity from a distinct subcellular compartment. Biochem Biophys Res Commun. 2007;361:481–486. doi: 10.1016/j.bbrc.2007.07.048. [DOI] [PubMed] [Google Scholar]
- 18.Glisic-Milosavljevic S, Waukau J, Jana S, Jailwala P, Rovensky J, Ghosh S. Comparison of apoptosis and mortality measurements in peripheral blood mononuclear cells (PBMCs) using multiple methods. Cell Prolif. 2005;38:301–311. doi: 10.1111/j.1365-2184.2005.00351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kerschen EJ, Fernandez JA, Cooley BC, et al. Endotoxemia and sepsis mortality reduction by nonanticoagulant activated protein C. J Exp Med. 2007;204:2439–2448. doi: 10.1084/jem.20070404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leonard WJ. Cytokines and immunodeficiency diseases. Nat Rev Immunol. 2001;1:200–208. doi: 10.1038/35105066. [DOI] [PubMed] [Google Scholar]
- 21.Nosaka T, van Deursen JM, Tripp RA, et al. Defective lymphoid development in mice lacking Jak3. Science. 1995;270:800–802. doi: 10.1126/science.270.5237.800. [DOI] [PubMed] [Google Scholar]
- 22.Thomis DC, Berg LJ. The role of Jak3 in lymphoid development, activation, and signaling. Curr Opin Immunol. 1997;9:541–547. doi: 10.1016/s0952-7915(97)80108-2. [DOI] [PubMed] [Google Scholar]
- 23.Egerton M, Scollay R, Shortman K. Kinetics of mature T-cell development in the thymus. Proc Natl Acad Sci U S A. 1990;87:2579–2582. doi: 10.1073/pnas.87.7.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scollay R, Godfrey DI. Thymic emigration: conveyor belts or lucky dips? Immunol Today. 1995;16:268–273. doi: 10.1016/0167-5699(95)80179-0. discussion 273–274. [DOI] [PubMed] [Google Scholar]
- 25.Lucas B, Vasseur F, Penit C. Production, selection, and maturation of thymocytes with high surface density of TCR. J Immunol. 1994;153:53–62. [PubMed] [Google Scholar]
- 26.Gabor MJ, Godfrey DI, Scollay R. Recent thymic emigrants are distinct from most medullary thymocytes. Eur J Immunol. 1997;27:2010–2015. doi: 10.1002/eji.1830270827. [DOI] [PubMed] [Google Scholar]
- 27.Vosshenrich CA, Samson-Villeger SI, Di Santo JP. Distinguishing features of developing natural killer cells. Curr Opin Immunol. 2005;17:151–158. doi: 10.1016/j.coi.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 28.Kim S, Iizuka K, Kang HS, et al. In vivo developmental stages in murine natural killer cell maturation. Nat Immunol. 2002;3:523–528. doi: 10.1038/ni796. [DOI] [PubMed] [Google Scholar]
- 29.Yokoyama WM, Kim S, French AR. The dynamic life of natural killer cells. Annu Rev Immunol. 2004;22:405–429. doi: 10.1146/annurev.immunol.22.012703.104711. [DOI] [PubMed] [Google Scholar]
- 30.Cousins L, Graham M, Tooze R, et al. Eosinophilic bowel disease controlled by the BB rat-derived lymphopenia/Gimap5 gene. Gastroenterology. 2006;131:1475–1485. doi: 10.1053/j.gastro.2006.09.023. [DOI] [PubMed] [Google Scholar]
- 31.Ramanathan S, Marandi L, Poussier P. Evidence for the extrathymic origin of intestinal TCRgammadelta(+) T cells in normal rats and for an impairment of this differentiation pathway in BB rats. J Immunol. 2002;168:2182–2187. doi: 10.4049/jimmunol.168.5.2182. [DOI] [PubMed] [Google Scholar]
- 32.Todd DJ, Forsberg EM, Greiner DL, Mordes JP, Rossini AA, Bortell R. Deficiencies in gut NK cell number and function precede diabetes onset in BB rats. J Immunol. 2004;172:5356–5362. doi: 10.4049/jimmunol.172.9.5356. [DOI] [PubMed] [Google Scholar]
- 33.Ogasawara K, Hida S, Azimi N, et al. Requirement for IRF-1 in the microenvironment supporting development of natural killer cells. Nature. 1998;391:700–703. doi: 10.1038/35636. [DOI] [PubMed] [Google Scholar]
- 34.Wu Q, Sun Y, Wang J, et al. Signal via lymphotoxin-beta R on bone marrow stromal cells is required for an early checkpoint of NK cell development. J Immunol. 2001;166:1684–1689. doi: 10.4049/jimmunol.166.3.1684. [DOI] [PubMed] [Google Scholar]
- 35.Alcamo E, Mizgerd JP, Horwitz BH, et al. Targeted mutation of TNF receptor I rescues the RelA-deficient mouse and reveals a critical role for NF-kappa B in leukocyte recruitment. J Immunol. 2001;167:1592–1600. doi: 10.4049/jimmunol.167.3.1592. [DOI] [PubMed] [Google Scholar]
- 36.Rosenfeld ME, Prichard L, Shiojiri N, Fausto N. Prevention of hepatic apoptosis and embryonic lethality in RelA/TNFR-1 double knockout mice. Am J Pathol. 2000;156:997–1007. doi: 10.1016/S0002-9440(10)64967-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horwitz BH, Scott ML, Cherry SR, Bronson RT, Baltimore D. Failure of lymphopoiesis after adoptive transfer of NF-kappaB-deficient fetal liver cells. Immunity. 1997;6:765–772. doi: 10.1016/s1074-7613(00)80451-3. [DOI] [PubMed] [Google Scholar]
- 38.Liu HH, Xie M, Schneider MD, Chen ZJ. Essential role of TAK1 in thymocyte development and activation. Proc Natl Acad Sci U S A. 2006;103:11677–11682. doi: 10.1073/pnas.0603089103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sato S, Sanjo H, Tsujimura T, et al. TAK1 is indispensable for development of T cells and prevention of colitis by the generation of regulatory T cells. Int Immunol. 2006;18:1405–1411. doi: 10.1093/intimm/dxl082. [DOI] [PubMed] [Google Scholar]
- 40.Sivakumar V, Hammond KJ, Howells N, Pfeffer K, Weih F. Differential requirement for Rel/nuclear factor kappa B family members in natural killer T cell development. J Exp Med. 2003;197:1613–1621. doi: 10.1084/jem.20022234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kupfer R, Lang J, Williams-Skipp C, Nelson M, Bellgrau D, Scheinman RI. Loss of a gimap/ian gene leads to activation of NF-kappaB through a MAPK-dependent pathway. Mol Immunol. 2007;44:479–487. doi: 10.1016/j.molimm.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 42.Cariappa A, Liou HC, Horwitz BH, Pillai S. Nuclear factor kappa B is required for the development of marginal zone B lymphocytes. J Exp Med. 2000;192:1175–1182. doi: 10.1084/jem.192.8.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.