Abstract
A 4.3 Mb duplication of chromosome 21 bands q22.13–q22.2 was diagnosed by interphase fluorescent in‐situ hybridisation (FISH) in a 31‐week gestational age baby with cystic hygroma and hydrops; the duplication was later found in the mother and in her 8‐year‐old daughter by the same method and confirmed by array comparative genomic hybridisation (aCGH). All had the facial gestalt of Down syndrome (DS). This is the smallest accurately defined duplication of chromosome 21 reported with a DS phenotype. The duplication encompasses the gene DYRK1 but not DSCR1 or DSCAM, all of which have previously been implicated in the causation of DS. Previous karyotype analysis and telomere screening of the mother, and karyotype analysis and metaphase FISH of a chorionic villus sample, had all failed to reveal the duplication. The findings in this family add to the identification and delineation of a “critical region” for the DS phenotype on chromosome 21. Cryptic chromosomal abnormalities can be missed on a routine karyotype for investigation of abnormal prenatal ultrasound findings, lending support to the use of aCGH analysis in this setting.
Down syndrome (DS) due to trisomy of chromosome 21 is a condition with a highly recognisable facial phenotype, moderate to severe intellectual disability and characteristic cardiac, gastrointestinal and haematological abnormalities. Cytogenetic and molecular studies of individuals with partial duplications of chromosome 21 and features of DS have sought a “critical region” on this chromosome, the duplication of which is postulated to be both necessary and sufficient to produce the DS phenotype. This has been narrowed down to 21q22, a region named as the Down syndrome critical region (DSCR), approximately 5.4 Mb in length.1,2 A mechanism whereby two of the 30 or so genes in this region, DSCR1 and DYRK1, may produce the DS phenotype when duplicated has recently been described.1 However, reports of individuals with duplications of this region only, without reciprocal deletions of other chromosomes, are extremely rare and the validity of this DSCR has been questioned. We describe a family in which the diagnosis of a 4.3 Mb duplication within 21q22.13–q22.2, encompassing DYRK1 but not DSCR1 or DSCAM, was suggested by the phenotypes of a mother, her daughter and her baby with cystic hygroma and hydrops. Karyotype analysis and metaphase fluorescent in‐situ hybridisation (FISH) were normal in the mother and her baby, but interphase FISH revealed a microduplication within 21q22 in all three individuals, confirmed by array comparative genomic hybridisation (aCGH). The phenotype in this family is of DS with mild learning disability but no major organ malformations.
CLINICAL DETAILS
This family first came to notice when referred to a genetic clinic for investigation of delayed development of the daughter, then aged 3 years. Both mother and daughter had the facial gestalt of DS and mosaicism was suspected, but routine karyotyping and later FISH studies of the subtelomeric regions were normal.
The mother is one of non‐identical twins born at term weighing 1.7 kg compared with 2.7 kg for her twin sister, and requiring nasogastric feeding for some weeks. She walked at 12 months and was not noted to have muscular hypotonia, but lagged in development behind her twin and, despite many years of speech therapy, continues to have articulation difficulties with oromotor dyspraxia. Although she received mainstream schooling until 16 years of age without special assistance, it was described as “a constant struggle”, in contrast to her twin who is professionally trained. She has lived independently since finishing school, but receives a government pension for developmental disability. She presents as a socially mature woman with an open and engaging personality and with a high level of emotional awareness, who stated that she had often been asked if she has DS owing to her appearance.
The daughter, now 8 years, weighed 3 kg at birth with a head circumference of 34.6 cm. She walked at 14 months, but delays in her development were noted by 2.5 years of age. She needed speech and occupational therapy, and now attends a class for children with mild intellectual disability at a mainstream school.
Mother and daughter are similar in appearance (fig 1) but significantly different from other family members. Both have heights on the 10th centile for age, brachycephaly with normal head circumference, round face with malar flush, epicanthic folds, short upslanting palpebral fissures and high arched palate, and the mother has hypotelorism with an interpupillary distance of 50 mm (<3rd centile). The daughter has clinodactyly of the fifth digits, but the mother does not, and both have high palmar triradii, brachydactyly and a wide sandal gap between the 1st and 2nd toes. Neither has Brushfield spots on the iris or single palmar creases. The mother had a normal echocardiogram and the daughter does not have any clinical evidence of cardiac malformation. Neither has hearing loss, nor any other medical problem.
Figure 1 Mother and daughter were found to have microduplication of the Down syndrome critical region (DSCR). Informed consent was obtained for publication of this figure.
The mother next presented at 28 years of age, 11 weeks pregnant in a new relationship. She had had one previous miscarriage with her current partner. An ultrasound revealed a bilateral septated cystic hygroma of 5–6 mm. Chorionic villus sampling at 12 weeks showed a female karyotype with no abnormalities detected on GTG banding. The parents were counselled that the absence of visible chromosomal abnormalities did not predict a normal outcome with certainty but they decided to continue with the pregnancy. Further ultrasounds at 15, 19 and 27 weeks gestation showed persistence of the cystic hygroma with no other fetal abnormalities detected.
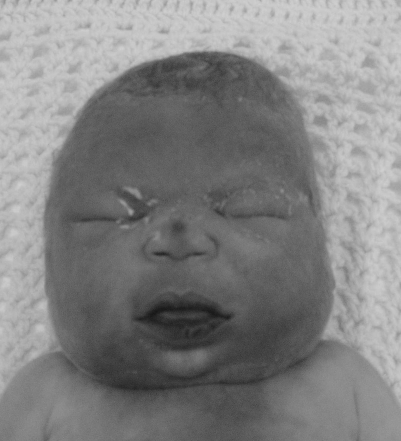
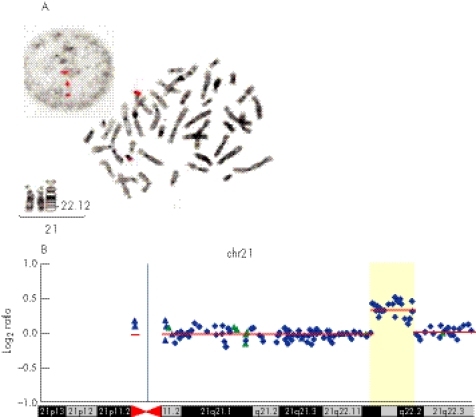
After spontaneous rupture of membranes with chorioamnionitis at 30 weeks gestation, the fetus was found, on repeat ultrasound examination, to have developed generalised hydrops, and died soon after birth 3 days later. External examination of the baby, despite its hydropic appearance, suggested DS, with round face, brachycephaly, upslanting palpebrae and protruding tongue (fig 2). Autopsy showed small size of all internal organs for gestational age, indicating generalised growth failure, but no cardiac or other malformation was found. Re‐examination of the karyotype from Chorionic Villus Sampling including FISH of the DSCR on 21q22 was requested, but no abnormality could be confirmed at the resolution obtained. However, owing to ongoing clinical suspicion of DS in the family, FISH was rescanned on interphase cells. This showed a microduplication with two copies of the DSCR contig probe for D21S259, D21S341 and D21S342 (VysisTM) on one chromosome 21, corresponding to bands 21q22.13–21q22.2 (fig 3A).
Figure 2 Fetus with microduplication of Down syndrome critical region (DSCR), showing round face, protruding tongue and downslanting palpebral fissures. Informed consent was obtained for publication of this figure.
Figure 3 (A) Fluorescent in‐situ hybridisation (FISH) images of the VysisTM locus‐specific probe for chromosome 21 including D21S59, D21S341 and D21S342 on the interphase and metaphase cells of the mother. Partial karyotype of her chromosomes 21 at 550 band resolution. (B) Array comparative genomic hybridization (aCGH) profile of chromosome 21 assayed on the RPCI 19K bacterial artificial chromosome (BAC) array demonstrating duplication at megabase pairs 36–40.3. Average log2 ratios (y axis) were plotted for all clones based on the chromosomal position (x axis), with the blue bar demarcating the centromere. Regions of normal segmental duplications are represented by blue and green triangles, and can be found at http://genome.ucsc.edu/cgi‐bin/hgGateway and http://projects.tcag.ca/variation/, respectively. Horizontal red lines indicate the log2 ratio for each segment as segmented by circular binary segmentation.
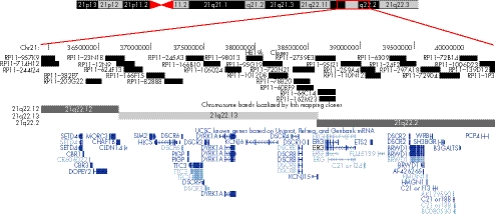
Subsequent to identification of the fetal duplication, repeat FISH using the VysisTM DSCR probe on interphase cell preparations showed both mother and daughter to carry an identical duplication within 21q22. This duplication was excluded in both maternal grandparents. aCGH performed on DNA from the mother using a bacterial artificial chromosome (BAC)‐based array containing 19 000 RPCI‐11 clones demonstrated a 4.3 Mb gain of one copy of the same region (fig 3B), which contains many genes previously identified in the DSCR, but excludes DSCR1 and DSCAM (fig 4). The rest of the genome had an apparently normal aCGH profile with most outlier BAC clones found associated with regions of normal segmental duplications and large‐scale variation in the human genome.3,4,5
Figure 4 University of California Santa Cruz (UCSC) human genome browser screenshot of 4.3 Mb duplicated region of chromosome 21 detailing base‐pair position, BAC clones utilised, cytogenetic band position and known genes (blue).
DISCUSSION
The findings in this family narrow down the critical region of duplication underlying the characteristic features of DS and clarify the contribution of genes previously implicated in the pathogenesis of this condition. Since the identification of trisomy 21 as the cause of DS in 1959, reports of individuals with this condition due to duplication of parts of chromosome 21 have appeared. Attempts were then made to identify the smallest or “critical” region necessary and sufficient in duplication to produce the DS phenotype. This has resulted in some confusion, since many reports included either were too early for accurate chromosome banding, included mosaic individuals6,7,8 or resulted from familial translocations, which may have involved undefined reciprocal deletions.7,9,10,11,12,13,14,15,16
A survey of the literature reveals 29 reported cases of partial trisomy 21 without other reported chromosomal involvement.2,7,13,17,18,19,20,21,22,23 Analysis of these reports has identified the region of duplication common to cases with a recognisable DS phenotype as 21q22, which is not surprising since it contains most of the G‐band‐negative, gene‐rich area of the chromosome. This region has been named the DSCR, but its extent is imprecise, with the smallest estimates being about 5–6 Mb, within 21q22.1–21q22.3.1,2 However, there are only two previous reports of microduplications of the DSCR, which were not visible by routine cytogenetic analysis: one, a 30‐year‐old man with a clinical diagnosis of DS and duplication of a region defined by molecular analysis to be 21q22.11–22.3,24 and the other an 18‐month‐old boy with clinical DS and DNA analysis showing duplication of the superoxide dismutase (SOD1) gene at 21q22.11.25 In the latter, no further delineation of the limit of the duplication was reported. The rarity of such cases is possibly due to the failure of routine metaphase FISH or cytogenetics to identifiy such small duplications, as was our experience. Of interest also in the identification of the DSCR are reports of three individuals who had trisomy 21 with deletion of band 21q22 on one chromosome 21, making them effectively disomic for this area.16,22 These reports clearly indicate a non‐DS phenotype. Another report of a similar individual with trisomy 21 and deletion at 21q22.1–q22.2 does not indicate whether the phenotype was consistent with DS.2
Research into the mechanism whereby duplication of all or part of human chromosome 21 causes DS has not been helped by the limited similarity to mouse models of trisomy 21. However, recent work has linked increased expression of two genes, DYRK1 and DSCR1, to disruption of the NFAT (nuclear factor of activated T cells) regulatory gene pathway, with more specific similarities between mouse models of NFAT dysregulation and human DS (including endocardial cushion defects, aganglionic megacolon, annular pancreas and even increased sociability) than seen in the mouse models of trisomy 21.1 DYRK1 is located within the area duplicated in this family, but DSCR1 and DSCAM are not. Previous reports have linked the genes DSCR1 and DSCAM to cardiac defects in DS, which were not present in this family.23,26
Another issue raised in this case is the possibility of missing the diagnosis of microduplication using karyotype analysis and even metaphase FISH. In the absence of a chromosomal abnormality, published normal live birth rates after diagnosis of ultrasound abnormalities in the first trimester vary enormously, from 4%27 to 80%28 for cystic hygroma and from 46%29 to 82%30 for increased nuchal translucence. It is possible that many of the fetuses included as having a normal karyotype in these figures have microduplications or deletions, which could only be detected by the use of specific probes on interphase cells or high‐resolution aCGH. In the presence of ultrasound abnormalities, more sensitive testing using aCGH to correctly identify cryptic chromosomal abnormalities would assist accurate prenatal diagnosis and counselling.
Key points
A 4.3 Mb duplication of chromosome 21 caused Down syndrome phenotype in three family members.
The microduplication was detected only by interphase FISH and aCGH, not by metaphase FISH.
The duplication involves DYRK1, but not DSCR1 or DSCAM
Acknowledgements
This work was supported by a RPCI Cancer Center Support Grant (P30 CA16056) to NJN. We thank the parents for their interest in and support of this report, and the Maternal Fetal Medicine team at the John Hunter Hospital, Newcastle, New South Wales, Australia.
Abbreviations
aCGH - array comparative genomic hybridisation
BAC - bacterial artificial chromosome
DS - Down syndrome
DSCR1 - Down syndrome critical region 1
FISH - fluorescent in‐situ hybridisation
Footnotes
Competing interests: None declared.
Informed consent was obtained for publication of fig 1 and fig 2.
References
- 1.Arron J R, Winslow M M, Polleri A, Chang C ‐ P, Wu H, Gao X, Neilson J R, Chen L, Heit J J, Kim S K, Yamasaki N, Miyakawa T, Francke U, Graef I A, Crabtree G R. NFAT dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome 21. Nature 2006441595–600. [DOI] [PubMed] [Google Scholar]
- 2.Delabar J, Theophile D, Rahmani Z, Blouin J, Prieur M, Noel B, Sinet P. Molecular mapping of twenty‐four features of Down syndrome on chromosome 21. Eur J Hum Genet 19931114–124. [DOI] [PubMed] [Google Scholar]
- 3.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, Navin N, Lucito R, Healy J, Hicks J, Ye K, Reiner A, Gilliam T C, Trask B, Patterson N, Zetterberg A, Wigler M. Large‐scale copy number polymorphism in the human genome. Science 2004305525–528. [DOI] [PubMed] [Google Scholar]
- 4.Tuzun E, Sharp A J, Bailey J A, Kaul R, Morrison V A, Pertz L M, Haugen E, Hayden H, Albertson D, Pinkel D, Olson M V, Eichler E E. Fine‐scale structural variation of the human genome. Nat Genet 200537727–732. [DOI] [PubMed] [Google Scholar]
- 5.Iafrate A J, Feuk L, Rivera M N, Listewnik M L, Donahoe P K, Qi Y, Scherer S W, Lee C. Detection of large‐scale variation in the human genome. Nat Genet 200436949–951. [DOI] [PubMed] [Google Scholar]
- 6.Cervenka J, Gorlin R, Reza Djavadi G. Down syndrome due to partial trisomy 21q. Clin Genet 197711119–121. [DOI] [PubMed] [Google Scholar]
- 7.Aula P, Leisti J, von Koskull H. Partial trisomy 21. Clin Genet . 1973;4241–251. [DOI] [PubMed]
- 8.Matsumoto N, Mikawa M. Confirmation of Down syndrome critical region by FISH analysis in a patient with add(21)(p11). Am J Med Genet 199559521–522. [DOI] [PubMed] [Google Scholar]
- 9.Korenberg J, Kawashima H, Pulst S, Ikeuchi T, Ogasawara N, Yamamoto K, Schonberg A, West R, Allen L, Magenis E, Ikawa K, Taniguchi N, Epstein C. Molecular definition of a region of chromosome 21 that causes features of the Down syndrome phenotype. Am J Hum Genet 199047236–246. [PMC free article] [PubMed] [Google Scholar]
- 10.Hadebank M, Rodewald A. Moderate Down's syndrome in three siblings having partial trisomy 21q22.2‐ter and therefore no SOD‐1 excess. Hum Genet 19826074–77. [DOI] [PubMed] [Google Scholar]
- 11.Pfeiffer R, Kessel E, Soer K. Partial trisomies of chromosome 21 in man. Two new observations due to translocations 19;21 and 4;21. Clin Genet 197711207–213. [DOI] [PubMed] [Google Scholar]
- 12.Pueschel S, Padre‐Mendoza T, Ellenbogen R. Partial trisomy 21. Clin Genet 198018392–395. [DOI] [PubMed] [Google Scholar]
- 13.Barnicoat A, Bonneau J, Boyd E, Docherty Z, Fennell S, Huret J, King M, Maltby E, McManus S, Pilz D, Shafei‐Benaissa E, Super M, Tolmie J. Down syndrome with partial duplication and del(21) syndrome: study protocol and call for collaboration. Study 1: clinical assessment, Clin Genet 19964920–27. [DOI] [PubMed] [Google Scholar]
- 14.Jenkins E, Duncan C, Wright C, Giordano F, Wilbur L, Wisniewski K, Sklower S, French J, Jones C, Brown W. Atypical Down syndrome and partial trisomy 21. Clin Genet 19832497–102. [DOI] [PubMed] [Google Scholar]
- 15.Williams C, Frias J, Mc Cormick M, Antonarakis S, Cantu E. Clinical, cytogenetic and molecular evaluation of a patient with partial trisomy 21 (21q11–q22) lacking the classical Down syndrome phenotype. Am J Med Genet 19907110–114. [DOI] [PubMed] [Google Scholar]
- 16.Kondo Y, Mizuno S, Ohara K, Nakamura T, Yamada K, Yamamori S, Hayakawa C, Ishii T, Yamada Y, Wakamatsu N. Two cases of partial trisomy 21 (pter‐q22.1) without the major features of Down syndrome. Am J Med Genet 2006140A227–232. [DOI] [PubMed] [Google Scholar]
- 17.Rahmani Z, Blouin J, Creau‐Goldberg N, Watkins P, Mattei J, Poisonnier M, Prieur M, Chettouh Z, Nicole A, Aurias A, Sinet P, Delabar J. Down syndrome critical region around D21S55 on proximal 21q22.3. Am J Med Genet 1990798–103. [DOI] [PubMed] [Google Scholar]
- 18.McCormick M, Schinzel A, Petersen M, Stetten G, Driscoll D, Cantu E, Tranebjaerg L, Mikkelsen M, Watkins P, Antonarakis S. Molecular genetic approach to the characterization of the “Down syndrome region” of chromosome 21. Genomics 19895325–331. [DOI] [PubMed] [Google Scholar]
- 19.Korenberg J, Chen X, Schipper R, Sun Z, Gonsky R, Gerwehr S, Carpenter N, Daumer C, Dignan P, Disteche C, Graham J, Hugdins L, McGillivray B, Miyazaki K, Ogasawara N, Park J, Pagon R, Pueschel S, Sack G, Say B, Schuffenhauer S, Soukup S, Yamanaka T. Down syndrome phenotypes: the consequences of chromosomal imbalance. Proc Natl Acad Sci USA 1994914997–5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leonard C, Gautier M, Sinet P, Selva J, Huret J. Two Down syndrome patients with rec(21), dupq, inv(21)(p11;q2109) from a familial pericentric inversion. Ann Genet 198629181–183. [PubMed] [Google Scholar]
- 21.Petersen M, Tranebjaerg L, Mc Cormick M, Michelsen N, Mikkelsen M, Antonarakis S. Clinical, cytogenetic, and molecular genetic characterization of two unrelated patients with different duplications of 21q. Am J Med Genet 19907104–109. [DOI] [PubMed] [Google Scholar]
- 22.Hagemeijer A, Smit E. Partial trisomy 21. Further evidence that trisomy of band 21q22 is essential for Down's phenotype. Hum Genet 19773815–23. [DOI] [PubMed] [Google Scholar]
- 23.Kosaki R, Kosaki K, Matsushima K, Mitsui N, Matsumoto N, Ohashi H. Refining chromosomal region critical for Down syndrome‐related heart defects with a case of cryptic 21q22.2 duplication. Congenital Anomalies 20054562–64. [DOI] [PubMed] [Google Scholar]
- 24.Delabar J, Sinet P, Chadefaux B, Nicole A, Gegonne A, Stehelin D, Fridlansky F, Creau‐goldberg N, Turleau C, de Grouchy J. Submicroscopic duplication of chromosome 21 and trisomy 21 phenotype (Down syndrome). Hum Genet 198776225–229. [DOI] [PubMed] [Google Scholar]
- 25.Huret J, Delabar J, Marlhens F, Aurias A, Nicole A, Berthier M, Tanzer J, Sinet P. Down syndrome with duplication of a region of chromosome 21 containing the CuZn superoxide dismutase gene without detectable karyotypic abnormality. Hum Genet 198775251–257. [DOI] [PubMed] [Google Scholar]
- 26.Fuentes J J, Genesca L, Kingsbury T J, Cunningham K W, Perez‐Riba M, Estivill X, de la Luna S. DSCR1, overexpressed in Down syndrome, is an inhibitor of calcineurin‐mediated signalling pathways. Hum Mol Genet 200091681–1690. [DOI] [PubMed] [Google Scholar]
- 27.Paoloni‐Giacobino A, Extermann D, Extermann P, Daghoun S. Pregnancy outcome of 30 fetuses with cystic hygroma diagnosed during the first 15 weeks of gestation. Genet Couns 200314413–418. [PubMed] [Google Scholar]
- 28.Trauffer P, Anderson C, Johnson A, Heeger S, Morgan P, Wapner R. The natural history of euploid pregnancies with first‐trimester cystic hygromas. Am J Obstet Gynaecol 19971701279–1284. [DOI] [PubMed] [Google Scholar]
- 29.Senat M, DeKeersmaecker B, Audibert F, Montcharmont G, Frydman R, Ville Y. Pregnancy outcome in fetuses with increased nuchal translucency and normal karyotype. Prenat Diagn 200222345–349. [DOI] [PubMed] [Google Scholar]
- 30.Hyett J. Increased nuchal translucence in fetuses with a normal karyotype. Prenat Diagn 200222864–868. [DOI] [PubMed] [Google Scholar]