Abstract
A catalase-related allene oxide synthase (cAOS) and true catalases that metabolize hydrogen peroxide have similar structure around the heme. One of the distal heme residues considered to help control catalysis is a highly conserved asparagine. Here we addressed the role of this residue in metabolism of the natural substrate 8R-hydroperoxyeicosatetraenoic acid by cAOS and in H2O2 breakdown by catalase. In cAOS, the mutations N137A, N137Q, N137S, N137D, and N137H drastically reduced the rate of reaction (to 0.8–4% of wild-type), yet the mutants all formed the allene oxide as product. This is remarkable because there are many potential heme-catalyzed transformations of fatty acid hydroperoxides and special enzymatic control must be required. In human catalase, the N148A, N148S, or N148D mutations only reduced rates to ~20% of wild-type. The distal heme Asn is not essential in either catalase or cAOS. Its conservation throughout evolution may relate to a role in optimizing catalysis.
Keywords: Peroxidase, Distal heme, Enzyme kinetics, HPETE, Hydroperoxyeicosatetraenoic acid, Hydrogen peroxide, Allene oxide, Allene oxide synthase, Catalase
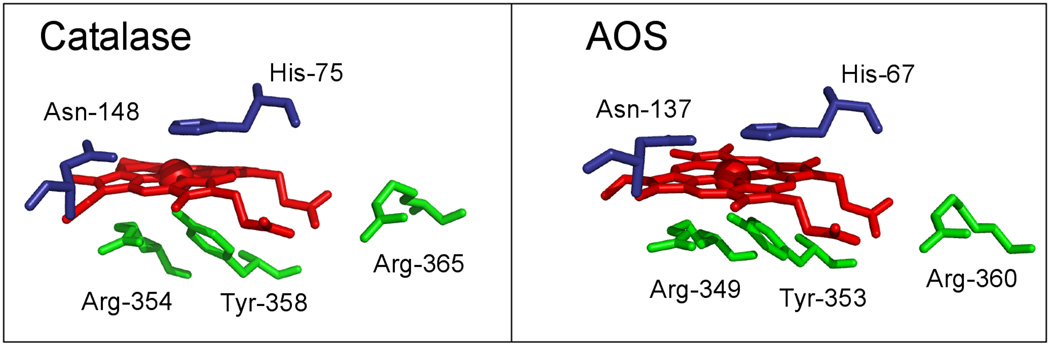
Catalases eliminate hydrogen peroxide by utilizing one H2O2 molecule for heme activation to Compound I and a second H2O2 molecule for heme reduction back to the resting state, in the process converting 2H2O2 to 2H2O plus O2. Currently there are hundreds of catalase sequences in the databases, identified through their sequence similarities [1–4]. Their common structural features comprising the catalase fold include the conserved sequence context of three significant amino acids around the catalytic heme in the active site. In order of appearance in the protein sequence, the first is a conserved histidine (His75 using the numbering for human catalase) that resides over the distal side of the heme and that is viewed as essential to catalysis [5, 6] (Figure 1, left hand side). The distal side of the heme also includes a conserved asparagine (Asn148) positioned over the heme near to His75 [6]. This residue is often implicated along with the distal His in modulating or tuning catalysis, and its role is the focus of this study. In bacterial large (75–84 kDa) subunit monofunctional catalase HPII, mutation of the equivalent distal heme Asn is associated with reduced catalatic activity [7]. The third conserved amino acid in order of appearance in the sequence is the proximal heme ligand, a tyrosine (Tyr388) in the sequence context of Arg-(three hydrophobic residues)-Tyr-(six residues)-Arg. The Arg residues help brace the Tyr on the underside of the heme. Taken together, in the correct sequence context, these three amino acids are conserved as part of the signature sequence of catalases.
Fig. 1.
Key residues in the heme environment in the active sites of human catalase and P. homomalla cAOS.
The heme (red) has a conserved Asn and His residue (blue) on the distal side. The proximal heme ligand, Tyr, is flanked by two conserved Arg residues (green). The structures are from PDB files 1DGF (human catalase) and 1U5U (cAOS).
In addition to the ubiquitous true catalases, there are a few organisms that express an additional catalase-related protein that utilizes a substrate other than hydrogen peroxide. We characterized the first of these a decade ago when we identified the heme domain of a natural fusion protein as a catalase-related allene oxide synthase (cAOS) [8]. cAOS transforms the lipoxygenase product 8R-HPETE (8R-hydroperoxyeicosatetraenoic acid) to the allene epoxide, 8,9-epoxyeicosa-5Z,9,11Z,14Z-tetraenoic acid (Scheme) [8, 9]. Allene oxides are unstable epoxides that are intermediates in the biosynthetic pathways of corals and plants [10]. The fatty acid hydroperoxide 8R-HPETE is synthesized from arachidonic acid by the lipoxygenase that is encoded as the C-terminal domain of the fusion protein with the catalase-related AOS at the N-terminus. The recent characterization of the X-ray crystal structure of this heme domain revealed a remarkable similarity to the central core structure of catalase (Figure 1, right side) [11]. However, quite remarkably the coral catalase-related domain exhibits no reaction with H2O2 [9, 12]. From recent studies we know that this coral catalase-related/LOX fusion protein is not unique. For example, our group and others reported on a cyanobacterial catalase-related/LOX fusion protein that forms allylic epoxides from C18 fatty acid substrates [13–15].
Scheme.
The chemistry of allene oxide synthesis catalyzed by the coral AOS (cAOS) is analogous to the reaction of the plant AOS that convert C18 fatty acid hydroperoxides to allene oxides. The plant AOS are members of the CYP74 family of cytochromes P450 [16, 17]. The CYP74 family includes enzymes that metabolize the same C18 fatty acid hydroperoxide substrates to a number of different products [18]. Thus CYP74A encodes AOS enzymes [17, 19–22], CYP74B are hydroperoxide lyases [23], CYP74C includes both AOS and lyases [24], and CYP74D comprises divinyl ether synthases [25]. All of these CYP74 enzymes share at least 35% amino acid sequence identity. With this knowledge in mind, we questioned whether the coral AOS has the potential to be converted into a hydroperoxide-metabolizing enzyme with a different catalytic specificity. With the understanding that reaction is catalyzed on the distal side of the heme, we hypothesized that changes in the distal heme environment might affect the chemistry of fatty acid hydroperoxide transformation. Accordingly we have explored the influence of the distal heme Asn in cAOS and for comparison with a true catalase, in the distant relative human catalase.
Materials and methods
Materials
Arachidonic acid (AA) was purchased from NuChek Prep Inc (Elysian, MN), and 8R-HPETE was synthesized using 8R-LOX [9]. H2O2 was from Fisher Scientific.
Construction of expression vectors
The construction of allene oxide synthase was described in the previous studies [9]. Human catalase was cloned from tonsil tissue. Firstly, human catalase cDNA was PCR-amplified using the primers 5'CAT ATG GCT GAC AGC CGG GAT CCC GCC 3' and 5' GAT ATC TCA GTG ATG GTG ATG GTG ATG CAG ATT TGC CTT CTC CCT TGC CGC 3', thereby adding a C-terminal His tag before the stop codon and including Nde I and EcoR V restriction sites at N-terminal and C-terminal ends respectively. The DNA sequencing confirmed the correct cloning of human catalase. Then the human catalase gene was sub-cloned into pET17b expression vector (Invitrogen). Plasmids containing the correct insert were transformed into E. coli BL21(DE3) cells for the expression of human catalase.
Site-directed mutagenesis for cAOS and Human catalase
Site-directed mutagenesis for cAOS and human catalase were generated utilizing the QuikChange Site-Directed Mutagenesis Kit (Stratagene, Cedar Creek, TX, USA) following the manufacturer's instructions. Wild type plasmids were used as templates, and point mutation primers designed as in Table 1 were added to the PCR reaction. After the PCR reaction, the amplified products were digested with Dpn I restriction enzyme to remove parental DNA, the mutant constructs were transformed into DH5α cells for the amplification of plasmid DNA. Consequently, plasmids were extracted by Wizard® Plus SV Miniprep DNA purification system (Madison, WI, USA) from a 2 mL overnight growth culture and submitted for sequencing. All point mutations were verified by DNA sequencing (Vanderbilt University). Plasmids containing the correct insert were transformed into BL21(DE3) expression cells for expression of the mutant proteins.
Table 1.
Primers designed for the mutagenesis of conserved Asn in cAOS and human catalase
| N148Q | 5' GGT AAC TGG GAT CTC GTT GGA CAG AAC ACC CCC ATT TTC TTC ATC 3' |
| 5' GAT GAA GAA AAT GGG GGT GTT CTG TCC AAC GAG ATC CCA GTT ACC 3' | |
| N148D | 5' GGT AAC TGG GAT CTC GTT GGA GAT AAC ACC CCC ATT TTC TTC ATC 3' |
| 5' GAT GAA GAA AAT GGG GGT GTT ATC TCC AAC GAG ATC CCA GTT ACC 3' | |
| N148A | 5' GGT AAC TGG GAT CTC GTT GGA GCT AAC ACC CCC ATT TTC TTC ATC 3' |
| 5' GAT GAA GAA AAT GGG GGT GTT AGC TCC AAC GAG ATC CCA GTT ACC 3' | |
| N148S | 5' GGT AAC TGG GAT CTC GTT GGA TCT AAC ACC CCC ATT TTC TTC ATC 3' |
| 5' GAT GAA GAA AAT GGG GGT GTT AGA TCC AAC GAG ATC CCA GTT ACC 3' | |
| N137A | 5' GGT CCA CTG GAT ATC GTC ATG GCT ACG GGT GAA GCC AAT ATA TTT 3' |
| 5' AAA TAT ATT GGC TTC ACC CGT AGC CAT GAC GAT ATC CAG TGG ACC 3' | |
| N137L | 5' GGT CCA CTG GAT ATC GTC ATG CTT ACG GGT GAA GCC AAT ATA TTT 3' |
| 5'AAA TAT ATT GGC TTC ACC CGT AAG CAT GAC GAT ATC CAG TGG ACC 3' | |
| N137H | 5'GGT CCA CTG GAT ATC GTC ATG CAT ACG GGT GAA GCC AAT ATA TTT 3' |
| 5'AAA TAT ATT GGC TTC ACC CGT ATG CAT GAC GAT ATC CAG TGG ACC 3' | |
| N137S | 5' GGT CCA CTG GAT ATC GTC ATG TCT ACG GGT GAA GCC AAT ATA TTT 3' |
| 5' AAA TAT ATT GGC TTC ACC CGT AGA CAT GAC GAT ATC CAG TGG ACC 3' | |
| N137K | 5' GGT CCA CTG GAT ATC GTC ATG AAA ACG GGT GAA GCC AAT ATA TTT 3' |
| 5' AAA TAT ATT GGC TTC ACC CGT TTT CAT GAC GAT ATC CAG TGG ACC 3' | |
| N137Q | 5' GGT CCA CTG GAT ATC GTC ATG CAG ACG GGT GAA GCC AAT ATA TTT 3' |
| 5' AAA TAT ATT GGC TTC ACC CGT CTG CAT GAC GAT ATC CAG TGG ACC 3' | |
| N137V | 5' GGT CCA CTG GAT ATC GTC ATG GTG ACG GGT GAA GCC AAT ATA TTT 3' |
| 5' AAA TAT ATT GGC TTC ACC CGT CAC CAT GAC GAT ATC CAG TGG ACC 3' | |
| N137D | 5' GGT CCA CTG GAT ATC GTC ATG GAT ACG GGT GAA GCC AAT ATA TTT 3' |
| 5' AAA TAT ATT GGC TTC ACC CGT ATC CAT GAC GAT ATC CAG TGG ACC 3' |
Bacterial Expression of cAOS, catalase and their mutants
Protein expression were carried out in BL21(DE3) cells using a modified method described by Hoffman et al [26] and Boutaud et al.[9]. The cells were harvested at 5,000 × g for 15 min in a Sorvall RC-3 centrifuge, washed with 25 ml of 50 mM Tris, pH 7.9, pelleted again at 5,000 × g for 20 min, and resuspended in 10 ml of TSE buffer (100 mM Tris acetate, pH 7.6, 500 mM sucrose, 0.5 mM EDTA) containing 1 mg/ml lysozyme and kept on ice for 30 min. and spun down at 5,000 × g for 20 min. The pellets were frozen at −80 °C for later use.
Purification of His-tagged Proteins
The histidine-tagged cAOS or catalase or their mutants were purified following the protocol of Imai et al. [27] and Boutaud et al. [9] with some modifications. The pellet was resuspended in BugBuster® Protein Extraction Reagent (Novagen) by sonication, and homogenized with glass homogenizer. The 16,000 × g supernatant was loaded on a nickel-NTA column (0.5 ml bed volume, Qiagen) equilibrated with 50 mM potassium phosphate buffer, pH 7.2, 500 mM NaCl at 0.5 ml/min. The column was then washed with the equilibration buffer and the nonspecific bound proteins were eluted with 50 mM potassium phosphate buffer, pH 7.2, 500 mM NaCl, 70 mM glycine. A final wash was performed by 50 mM potassium phosphate buffer, pH 7.2, 500 mM NaCl, 20 mM imidazole. The His-protein was then eluted with 50 mM potassium phosphate buffer, pH 7.2, 500 mM NaCl, 250 mM imidazole. Fractions of 0.5 ml were collected and assayed for the AOS or catalase activity. Up to this point all procedures were the same for cAOS and catalase. The cAOS positive fractions were pooled and dialyzed overnight against 50 mM Tris buffer, pH 7.9, 500 mM NaCl, 20% glycerol and aliquoted and frozen in −80 °C for later use. Catalase fractions were pooled and dialyzed twice against 2 L buffer (20 mM Tris, pH 7.5) to get rid of salt for Mono Q column purification by FPLC. The Mono Q column was pre-equilibrated with 20 mM Tris buffer pH 7.5, and loaded with catalase fractions. The gradient was set 0–250 mM NaCl with flow rate of 1 ml/min. The fractions were monitored by UV detector and confirmed pure by SDS-PAGE.
Incubation and extraction conditions
30 µg 8R-HPETE was incubated with an appropriate amount of cAOS enzyme or its mutants in 3 ml of 50 mM Tris buffer, pH 7.5, 150 mM NaCl for 3 min, then acidified with 0.6 ml of 1M KH2PO4 solution, 30 µl of 1N HCl to reach a pH of 4–5. Products were extracted by 1ml C18 HLB Cartridge (Waters OASIS®). The column was prepared by running methanol first and then with water. Products were loaded onto the column, washed with water and eluted with methanol.
HPLC analysis of cAOS products
Extracts were dried under a stream of nitrogen and redissolved in HPLC solvent. Analyses were carried out by RP-HPLC using a Waters Symmetry C18 column (25 × 0.46 cm), a solvent system of methanol/water/acetic acid in the proportions 80/20/0.01 (v/v/v), a flow rate of 1 ml/min, with UV detection at 205 nm, 220 nm, 235 nm and 270 nm using an Agilent 1100 series diode array detector.
Activity assays
The AOS and its mutants were assayed by following the decrease in absorbance at 235 nm due to the breakage of hydroperoxides in 50 mM Tris buffer, pH 7.5, 150 mM NaCl using a Lambda 35 UV/VIS spectrophotometer (PerkinElmer). The initial rate was expressed as nanomoles of hydroperoxides decomposed per nanomole of heme per second. Catalase and its mutants were assayed by measuring the decrease at 240 nm due to the consumption of H2O2 in 50 mM potassium phosphate buffer, pH 7.0, using a Lambda 35 UV/VIS spectrophotometer (PerkinElmer). Similarly, the initial rate was expressed as nanomoles of H2O2 decomposed per nanomole of heme per second.
Results
Expression and purification of human catalase and cAOS
Human catalase cDNA including a C-terminal His-tag was cloned into the expression vector pET17b and the protein was expressed in E. coli BL21(DE3) with a 36 mg yield of active protein per liter growth under the conditions described in Materials and Methods. cAOS expression yielded 110 mg protein per liter growth using the method described by Boutaud et al [9]. Mutants made by site-directed mutagenesis of the distal heme Asn in human catalase (Asn148) and cAOS (Asn137) were also expressed in E. coli BL21(DE3). In most cases proteins expressed well (20–100 mg/liter). The cAOS was purified using a Ni-NTA affinity column to greater than 90% homogeneity, except for the Asp and Leu mutants which had low levels of expression and gave impure preparations of enzyme (Figure 2). The N137L and the N137V mutants expressed with a low content of heme (<5%). Human catalase and its mutants were isolated using Ni-NTA affinity and further purified using Fast Protein Liquid Chromatography on a Mono-Q column; the proteins recovered from the Mono-Q column showed a single band on SDS-PAGE (Figure 3). UV/VIS spectroscopy indicated a high heme content as evidenced by an absorption ratio for A406/280 of 1.2, better than the value of 1.04 reported for the human catalase used in X-ray crystallography [28]
Fig. 2.
SDS -PAGE with Coomassie staining of wild type cAOS and its mutant proteins after Ni- NTA affinity chromatography. Each lane was loaded with 1–2 µg of protein, except for the Asp and Leu mutants, which were loaded with ~4 µg protein due to their lower levels of cAOS expression in E. coli.
Fig. 3.
SDS-PAGE of wild type human catalase and its mutant proteins recovered from Mono-Q anion exchange chromatography.
Steady-state kinetic analyses of human catalase and its mutants
To identify the role of the distal Asn148, mutations were designed to change or eliminate the hydrogen bonding side chain in the active site. Replacement of Asn148 with Ala, Gln, Asp, and Ser each gave a protein with similar heme content to wild-type. The catalatic activities of wild type human catalase and the mutants were determined by spectrophotometric measurement of H2O2 consumption using the absorbance decrease at 240 nm [29]. The results in Figure 4 illustrate the initial rates measured up to a H2O2 concentration of 50 mM, the highest concentration compatible with this spectroscopic assay. Human catalase, like other true catalases, does not attain Vmax at H2O2 concentrations up to 1–2 M [4]. That Vmax is not attained is evident in the wild-type kinetics (Figure 4), whereas several of the mutants, in addition to exhibiting overall lower activity, also appear to reach close to maximal rates of reaction.
Fig. 4.
Kinetic analysis on wild type human catalase and its mutants. The initial velocity was fitted to the Michaelis-Menten equation using the KaleidaGraph (Synergy software) program.
Table 2 summarizes the kinetic parameters obtained from the steady-state kinetic assay. The wild-type enzyme achieved a kcat of ~4 × 105 per sec per heme moiety, in accord with published values [4]. The N148A, N148S and N148D mutants each gave ~5-fold lower values for kcat, while the N148Q mutant displayed feeble catalytic activity, about 400-fold lower than wild-type. The estimated KM of 72 mM for the wild type catalase is in good agreement with a previous report from human erythrocyte catalase (81 mM) [4]. There is a considerable decrease in KM for the N148A, N148S and N148D mutants compared to wild type catalase (Table 2). This likely reflects the fact that the mutants reach near a maximal rate at 50 mM H2O2 concentration whereas the wild type has not even reached its estimated KM at that point. As a consequence of the lower KM, the catalytic efficiencies (kcat/KM) of the mutants compute to values similar to wild-type catalase.
Table 2.
Kinetic parameters for wild type human catalase and its mutants.
| Enzyme | KM (mM) | kcat (s−1) | kcat/KM (M−1s−1) | R value |
|---|---|---|---|---|
| Catalase | 71.8 | 3.8×105 | 5.3×106 | 0.99 |
| N148A | 12.5 | 7.1×104 | 5.7×106 | 0.99 |
| N148S | 21.9 | 7.9×104 | 3.6×106 | 0.99 |
| N148D | 33.8 | 7.4×104 | 2.2×106 | 0.99 |
| N148Q | 7.5 | 9.6×102 | 1.3×105 | 0.99 |
R value represents the fit to Michaelis-Menten equation.
Product analyses of cAOS and its mutants
To investigate the role of the distal heme residues in cAOS, we first checked the involvement of the distal His67 by its mutagenesis to Val or Asn. Both mutations resulted in protein with a low heme content and with zero cAOS activity (data not shown). To investigate the role of the distal Asn137, we prepared an extensive series of mutants to test the importance of hydrogen bonding, positive or negative charge, and space-filling/hydrophobicity; Asn137 was replaced with Ser, Gln, Asp, His, Lys, Ala, Val, or Leu. These proteins expressed well, with the exception of the N137V and N137L mutants, which exhibited a low content of heme.
Wild-type cAOS converts 8R-HPETE to the unstable 8,9-epoxy allene oxide, which is subject to almost instant non-enzymatic hydrolysis to the α-ketol 8-hydroxy-9-ketoeicosa-5,11,14-trienoic acid and lesser amounts of the γ-ketol 12-hydroxy-9-ketoeicosa-5,10,14-trienoic acid; non-enzymatic cyclization of the allene oxide also occurs and accounts for about 15–20% of the end products as a prostaglandin-like cyclopentenone (Scheme) [30]. These three degradation products of the allene oxide give a distinctive pattern on HPLC analysis with diode array detection (Figure 5A). Also, the transformation of 8R-HPETE is associated with loss of its conjugated diene chromophore, allowing assay of cAOS reaction rates by following the decrease in absorbance at 235 nm. As shown in Table 3, all the Asn137 mutant proteins displayed a severe loss in activity compared to wild-type. The N137A and N137Q retained the most activity (only ~4%), while the N137K (0.2%) and N137L (0.03%) showed the least.
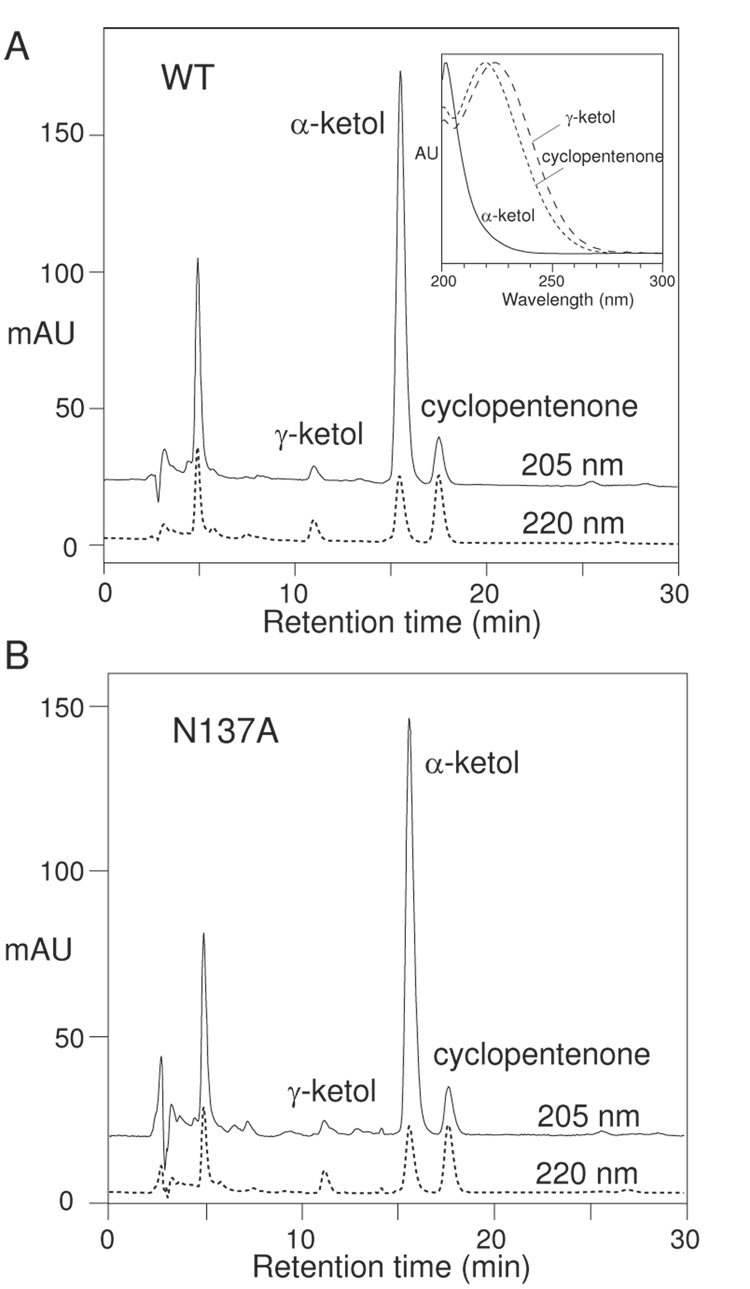
Fig. 5.
RP-HPLC analysis of products from the reaction of 8R-HPETE with (A) wild-type cAOS, and (B) the N137A mutant. Inset: the UV spectra of α-ketol, γ-ketol and cyclopentenone.
Table 3.
Activities for cAOS and its mutants with the substrate 8R-HPETE (40 µM)
| Enzyme | Ratea, s−1 (range) | % of wild type |
|---|---|---|
| cAOS, wild type | 1603 (1309–2045) | 100 |
| N137A | 69 (62–73) | 4.3 |
| N137Q | 70 (50–100) | 4.4 |
| N137D | 52 (41–60) | 3.2 |
| N137S | 28 (22–31) | 1.7 |
| N137H | 23 (21–25) | 1.4 |
| N137V | 14 (13–18) | 0.9 |
| N137K | 2.5 (2–3) | 0.2 |
| N137L | 0.5 (0–1) | 0.03 |
mean of 3 or 4 determinations
Units of rates are nanomoles of 8R-HPETE per nanomole heme moiety per second.
Despite this major impact on activity, the Ser, Gln, Asp, and Ala mutants all showed an identical characteristic pattern of allene oxide-derived products on HPLC analysis, illustrated in Figure 5B for the N137A enzyme. The nature of the products was confirmed both by their distinctive HPLC-UV profiles at specific wavelengths and by their diagnostic UV spectra (Figure 5A, inset). The His mutant formed prominent allene oxide-derived products along with some non-specific reaction products such as 8,15-di(hydro)peroxides with conjugated triene chromophores (cf. ref. [31]) and a trace of a C13 aldehyde cleavage product of 8R-HPETE (cf. ref. [32]). The Val and Leu mutants formed barely detectable levels of allene oxide-derived products along with non-specific by-products similar in type to the His mutant. Only the N137K mutant formed no discernible allene oxide; instead, the HPLC chromatogram showed multiple products (more than 20), many with 205 nm absorbance, likely reflecting the non-specific transformation to mixtures of epoxyalcohol isomers and other rearrangements, a characteristic of non-specific heme-catalyzed breakdown of fatty acid hydroperoxides (data not shown) [33–35]. The important point, to re-state it, is that Asn mutations to Ser, Gln, Asp, Ala, His, Val, and Leu all resulted in enzymes capable of specific allene oxide synthesis from 8R-HPETE, albeit at a greatly reduced rate.
Discussion
The X-ray crystal structure of several catalases was determined in the 1980’s, giving a good picture of the active site environment and the potential amino acid residues participating directly in catalysis [5, 36–38]. The distal heme histidine, a residue conserved in monofunctional catalases, was deduced to be of vital importance and this was confirmed by mutagenesis in the 1990’s [7]. From the time of the first crystal structures the distal heme Asn (N148 in human catalase) was also implicated in catalysis, and even relatively recently the Asn148 was featured prominently in the catalatic cycle [6]. For example, one proposed role for Asn148 was to cooperate with His75 in hydrogen bonding to one oxygen of H2O2 and thus increase affinity for the substrate [6]. Our results tend to show that, in the Asn148 mutants, high concentrations of H2O2 show very markedly reduced reaction rates whereas low concentrations are handled almost as well as in wild-type; the Asn148Ala mutant, for example, shows 70–80% of the reaction rate as wild type at H2O2 concentrations up to 10 mM. Kinetically these effects appear as a much lower Vmax in the mutants, a lower KM, and similar values for kcat/KM, reflecting the near-normal reaction rates with modest concentrations of H2O2. A potential explanation for these atypical kinetics is that the distal Asn helps promote reaction at the higher concentrations of H2O2, perhaps by binding substrate, and substitution of the Asn thus lowers Vmax, as a consequence leading to a computed lower value for KM. As noted in the Introduction, the equivalent Asn in a large subunit bacterial catalase (E. coli HPII) was mutated and shown to diminish but not abolish catalatic activity. We obtained a similar result here using human catalase, which is classified as a typical clade III small subunit catalase by the convention of Klotz and Loewen [2]. In both cases the Asn-to-Ala mutant retained appreciable activity, although distinctly lower than wild-type. With the different mutants, generally if heme was retained in the protein, so was catalytic activity. The idea that this fairly well-conserved residue is not essential to catalysis has emerged recently for a hemoprotein peroxidase. Thus, the peroxidase activity of ovine prostaglandin H synthase-1 was found to be retained in the Gln-to-Val mutant, indicating that the hydrogen bonding capability of distal heme Gln is not required [39].
While it is all but self-evident that allene oxide synthesis occurs in the heme active site of cAOS, direct proof of the reaction pathway is only partly demonstrated. Certainly, the X-ray structure of cAOS shows that the distal side of the heme is comparatively open and accessible compared to catalase [11], although so far there is no structure with a bound substrate analogue. Compound I, a key intermediate in catalysis, is observed in cAOS treated with peracetic acid [12]. Raman spectral data indicate that catalase adopts a 5-coordinate/high spin structure, whereas cAOS has purely a 6-coordinate/high spin heme. Significantly, an active site mutation on the residue adjacent to the distal heme His67 (Thr66Val) confers H2O2-metabolizing capacity on cAOS; this T66V mutant has a 5-coordinate/high spin heme as a minor species, and probably because of this it exhibits some catalatic activity [12]. The transformation of 8R-HPETE to allene oxide is understood in chemical terms (e.g. ref. [40]), but the step-by-step dissection of the reaction in cAOS has yet to be resolved.
In contrast to the breakdown of hydrogen peroxide, the chemical breakdown or rearrangement of fatty acid hydroperoxides has many potential outcomes, so there is a need for strict enzymatic control in the transformation to an allene epoxide. Indeed, in a half century of studies on the degradation of fatty acid hydroperoxides, in the course of which the degradation to multiple epoxides, aldehydes, ethers and alcohols has been documented [35, 41, 42], no evidence of non-enzymatic formation of allene oxides has ever been produced. The only known examples of allene oxide synthesis in biology are the enzymatic transformations catalyzed by the CYP74 cytochrome P450s and the equivalent reactions of the catalase-related AOS [10]. We also know that different members of the CYP74 family of enzymes display distinct catalytic activities, giving different products from the same fatty acid hydroperoxide substrate [18]. For all these reasons we suspected that a marked alteration in the cAOS active site might disrupt the process of allene oxide synthesis and lead to another outcome. Surprisingly, however, the cAOS mutants with substitutions in the distal heme Asn did not form new products. Although their catalytic efficiencies were seriously impaired, retaining no more than 4% of wild type activity, most of mutants were able to catalyze the formation of allene oxide. Even the Asn137Ala mutant produced the allene oxide. Our previous assumption on the catalytic mechanism of allene oxide formation invoked a role for hydrogen bonding from the distal heme Asn137 [11], but our results shown here indicate that this can play no role in the specific transformation to the allene oxide product.
We conclude that the importance of the conserved distal heme Asn in catalase and catalase-related enzymes may have been overestimated, or at least misrepresented. The conserved Asn occupies a conspicuous position, and as the nearest hydrogen-bonding residue to the center of catalytic activity it seems obvious that it should play a crucial role in catalysis. It does function catalytically in speeding up the reaction rate ~5-fold in catalases and ~20 – 100-fold in AOS, although it is not essential in either enzyme. While crucial to an efficient reaction rate in cAOS, it plays no role in directing fatty acid hydroperoxide transformation to the allene oxide. It is clear from our results that the distal Asn optimizes reaction rate. Its conservation throughout evolution may relate to this role in optimization of the inherent catalytic activity.
Acknowledgements
This work was supported by NIH grant GM-74888 to A.R.B..
Abbreviations footnote
- AOS
allene oxide synthase
- cAOS
catalase-related allene oxide synthase
- LOX
lipoxygenase
- H(P)ETE
hydro(pero)xyeicosatetraenoic acid
- RP-HPLC
reversed-phase high pressure liquid chromatography
- SP-HPLC
straight-phase high pressure liquid chromatography
- Ni-NTA
nickel nitrilotriacetic acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chelikani P, Fita I, Loewen PC. Cell Mol Life Sci. 2004;61:192–208. doi: 10.1007/s00018-003-3206-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klotz MG, Loewen PC. Mol Biol Evol. 2003;20:1098–1112. doi: 10.1093/molbev/msg129. [DOI] [PubMed] [Google Scholar]
- 3.Nicholls P, Fita I, Loewen PC. Adv. Inorg. Chem. 2001;51:51–106. [Google Scholar]
- 4.Switala J, Loewen PC. Arch Biochem Biophys. 2002;401:145–154. doi: 10.1016/S0003-9861(02)00049-8. [DOI] [PubMed] [Google Scholar]
- 5.Murthy MR, Reid TJ, 3rd, Sicignano A, Tanaka N, Rossmann MG. J Mol Biol. 1981;152:465–499. doi: 10.1016/0022-2836(81)90254-0. [DOI] [PubMed] [Google Scholar]
- 6.Putnam CD, Arvai AS, Bourne Y, Tainer JA. J Mol Biol. 2000;296:295–309. doi: 10.1006/jmbi.1999.3458. [DOI] [PubMed] [Google Scholar]
- 7.Loewen PC, Switala J, von Ossowski I, Hillar A, Christie A, Tattrie B, Nicholls P. Biochemistry. 1993;32:10159–10164. doi: 10.1021/bi00089a035. [DOI] [PubMed] [Google Scholar]
- 8.Koljak R, Boutaud O, Shieh BH, Samel N, Brash AR. Science. 1997;277:1994–1996. doi: 10.1126/science.277.5334.1994. [DOI] [PubMed] [Google Scholar]
- 9.Boutaud O, Brash AR. J Biol Chem. 1999;274:33764–33770. doi: 10.1074/jbc.274.47.33764. [DOI] [PubMed] [Google Scholar]
- 10.Tijet N, Brash AR. Prostaglandins Other Lipid Mediat. 2002;68–69:423–431. doi: 10.1016/s0090-6980(02)00046-1. [DOI] [PubMed] [Google Scholar]
- 11.Oldham ML, Brash AR, Newcomer ME. Proc. Natl. Acad. Sci. U S A. 2005;102:297–302. doi: 10.1073/pnas.0406352102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tosha T, Uchida T, Brash AR, Kitagawa T. J Biol Chem. 2006;281:12610–12617. doi: 10.1074/jbc.M600061200. [DOI] [PubMed] [Google Scholar]
- 13.Lang I, Gobel C, Porzel A, Heilmann I, Feussner I. Biochem J. 2008;410:347–357. doi: 10.1042/BJ20071277. [DOI] [PubMed] [Google Scholar]
- 14.Schneider C, Niisuke K, Boeglin WE, Voehler M, Stec DF, Porter NA, Brash AR. Proc Natl Acad Sci U S A. 2007;104:18941–18945. doi: 10.1073/pnas.0707148104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng Y, Boeglin WE, Schneider C, Brash AR. J Biol Chem. 2007 [Google Scholar]
- 16.Feussner I, Wasternack C. Annu Rev Plant Biol. 2002;53:275–297. doi: 10.1146/annurev.arplant.53.100301.135248. [DOI] [PubMed] [Google Scholar]
- 17.Song WC, Funk CD, Brash AR. Proc Natl Acad Sci U S A. 1993;90:8519–8523. doi: 10.1073/pnas.90.18.8519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grechkin AN. Prostaglandins Other Lipid Mediat. 2002;68–69:457–470. doi: 10.1016/s0090-6980(02)00048-5. [DOI] [PubMed] [Google Scholar]
- 19.Laudert D, Pfannschmidt U, Lottspeich F, Hollander-Czytko H, Weiler EW. Plant Mol Biol. 1996;31:323–335. doi: 10.1007/BF00021793. [DOI] [PubMed] [Google Scholar]
- 20.Maucher H, Hause B, Feussner I, Ziegler J, Wasternack C. Plant J. 2000;21:199–213. doi: 10.1046/j.1365-313x.2000.00669.x. [DOI] [PubMed] [Google Scholar]
- 21.Pan Z, Durst F, Werck-Reichhart D, Gardner HW, Camara B, Cornish K, Backhaus RA. J Biol Chem. 1995;270:8487–8494. doi: 10.1074/jbc.270.15.8487. [DOI] [PubMed] [Google Scholar]
- 22.Song WC, Brash AR. Science. 1991;253:781–784. doi: 10.1126/science.1876834. [DOI] [PubMed] [Google Scholar]
- 23.Matsui K, Shibutani M, Hase T, Kajiwara T. FEBS Lett. 1996;394:21–24. doi: 10.1016/0014-5793(96)00924-6. [DOI] [PubMed] [Google Scholar]
- 24.Matsui K, Ujita C, Fujimoto S, Wilkinson J, Hiatt B, Knauf V, Kajiwara T, Feussner I. FEBS Lett. 2000;481:183–188. doi: 10.1016/s0014-5793(00)01997-9. [DOI] [PubMed] [Google Scholar]
- 25.Stumpe M, Kandzia R, Gobel C, Rosahl S, Feussner I. FEBS Lett. 2001;507:371–376. doi: 10.1016/s0014-5793(01)03019-8. [DOI] [PubMed] [Google Scholar]
- 26.Hoffman BJ, Broadwater JA, Johnson P, Harper J, Fox BG, Kenealy WR. Protein Expr Purif. 1995;6:646–654. doi: 10.1006/prep.1995.1085. [DOI] [PubMed] [Google Scholar]
- 27.Imai T, Globerman H, Gertner JM, Kagawa N, Waterman MR. J Biol Chem. 1993;268:19681–19689. [PubMed] [Google Scholar]
- 28.Ko TP, Safo MK, Musayev FN, Di Salvo ML, Wang C, Wu SH, Abraham DJ. Acta Crystallogr D Biol Crystallogr. 2000;56:241–245. doi: 10.1107/s0907444999015930. [DOI] [PubMed] [Google Scholar]
- 29.Beers RF, Jr, Sizer IW. J Biol Chem. 1952;195:133–140. [PubMed] [Google Scholar]
- 30.Brash AR, Baertschi SW, Ingram CD, Harris TM. J Biol Chem. 1987;262:15829–15839. [PubMed] [Google Scholar]
- 31.Brash AR, Porter AT, Maas RL. J Biol Chem. 1985;260:4210–4216. [PubMed] [Google Scholar]
- 32.Brash AR, Hughes MA, Hawkins DJ, Boeglin WE, Song WC, Meijer L. J Biol Chem. 1991;266:22926–22931. [PubMed] [Google Scholar]
- 33.Chang MS, Boeglin WE, Guengerich FP, Brash AR. Biochemistry. 1996;35:464–471. doi: 10.1021/bi952081v. [DOI] [PubMed] [Google Scholar]
- 34.Dix TA, Marnett LJ. J Biol Chem. 1985;260:5351–5357. [PubMed] [Google Scholar]
- 35.Gardner HW. Free Radic Biol Med. 1989;7:65–86. doi: 10.1016/0891-5849(89)90102-0. [DOI] [PubMed] [Google Scholar]
- 36.Fita I, Rossmann MG. J Mol Biol. 1985;185:21–37. doi: 10.1016/0022-2836(85)90180-9. [DOI] [PubMed] [Google Scholar]
- 37.Reid TJ, 3rd, Murthy MR, Sicignano A, Tanaka N, Musick WD, Rossmann MG. Proc Natl Acad Sci U S A. 1981;78:4767–4771. doi: 10.1073/pnas.78.8.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vainshtein BK, Melik-Adamyan WR, Barynin VV, Vagin AA, Grebenko AI. Nature. 1981;293:411–412. doi: 10.1038/293411a0. [DOI] [PubMed] [Google Scholar]
- 39.Liu J, Seibold SA, Rieke CJ, Song I, Cukier RI, Smith WL. J Biol Chem. 2007;282:18233–18244. doi: 10.1074/jbc.M701235200. [DOI] [PubMed] [Google Scholar]
- 40.Song WC, Baertschi SW, Boeglin WE, Harris TM, Brash AR. J Biol Chem. 1993;268:6293–6298. [PubMed] [Google Scholar]
- 41.Frankel EN. Chem Phys Lipids. 1987;44:73–85. doi: 10.1016/0009-3084(87)90045-4. [DOI] [PubMed] [Google Scholar]
- 42.Porter NA, Caldwell SE, Mills KA. Lipids. 1995;30:277–290. doi: 10.1007/BF02536034. [DOI] [PubMed] [Google Scholar]