Abstract
Estimating detailed transmission trees that reflect the relationships between infected individuals or populations during a disease outbreak often provides valuable insights into both the nature of disease transmission and the overall dynamics of the underlying epidemiological process. These trees may be based on epidemiological data that relate to the timing of infection and infectiousness, or genetic data that show the genetic relatedness of pathogens isolated from infected individuals. Genetic data are becoming increasingly important in the estimation of transmission trees of viral pathogens due to their inherently high mutation rate. Here, we propose a maximum-likelihood approach that allows epidemiological and genetic data to be combined within the same analysis to infer probable transmission trees. We apply this approach to data from 20 farms infected during the 2001 UK foot-and-mouth disease outbreak, using complete viral genome sequences from each infected farm and information on when farms were first estimated to have developed clinical disease and when livestock on these farms were culled. Incorporating known infection links due to animal movement prior to imposition of the national movement ban results in the reduction of the number of trees from 41 472 that are consistent with the genetic data to 1728, of which just 4 represent more than 95% of the total likelihood calculated using a model that accounts for the epidemiological data. These trees differ in several ways from those constructed prior to the availability of genetic data.
Keywords: foot-and-mouth disease virus, transmission trees, contact tracing, complete genome sequencing
1. Introduction
Genetic data from RNA viruses have been used increasingly for tracing disease transmission pathways, taking advantage of their inherent capacity to evolve quickly. Such studies have been carried out with viruses such as HIV (Zhang et al. 1997; Leitner & Albert 1999), hepatitis C virus (Spada et al. 2004; Bracho et al. 2005), SARS coronavirus (Wong et al. 2004; Liu et al. 2005), Ebolavirus (Walsh et al. 2005), Rhinovirus (Savolainen et al. 2002) and noroviruses (Dowell et al. 1995). These genetic data can be used to complement field epidemiological studies that use traditional contact-tracing information and the relative timing and spatial proximity of infection events to each other.
Analysis of the overlapping periods within which individuals or groups are infected and/or infectious can help to decide the most likely direction of transmission, while pathogen genetic data can identify which infected individuals or groups are most closely epidemiologically linked. Although there may be numerous combinations of transmission pathways that are consistent with the genetic or epidemiological data alone, an analysis combining both types of data can lead to the identification of a much smaller set of plausible transmission pathways. Analytical methods to integrate these two types of datasets are beginning to be developed (Wallace et al. 2007), but the increasing speed and economy with which viral genetic data can be generated during epidemics require the development of a wider range of approaches, if full advantage of these data sources is to be taken in improving the tracing of transmission pathways.
Recently, it was shown that foot-and-mouth disease virus (FMDV) transmission can be traced from farm to farm using complete genome sequencing (Cottam et al. 2006). Viruses were sequenced from farms infected at the beginning of the 2001 UK FMDV outbreak, and the genetic data were shown to be consistent with the transmission pathways established from contact-tracing studies. However, for the remainder of the outbreak, and following the national ban on animal movement (NMB), the spread of virus is much less well understood with infection assumed to be transmitted by either airborne spread or mechanical transfer on people or inanimate objects (fomites). Indeed, the precise source and route of infection for the vast majority of the 2030 premises infected during the 2001 UK epidemic remain unknown.
During the epidemic, substantial amounts of epidemiological data were recorded for the farms involved. Although the contact-tracing data are difficult to use to pinpoint the precise origin of infection with high confidence, the information concerning the timing of infection is clear. For every farm involved, the time at which clinical disease began on a farm (estimated from lesion ageing) and the date on which animals were culled were recorded. This information can be used to estimate the most likely date on which a farm was infected and thereby the most likely period over which a farm would have been infectious. These temporal data have been used previously to estimate the sequences of transmission events (Haydon et al. 2003), henceforth referred to as transmission trees. While many of the overall characteristics of these transmission trees (e.g. estimates of the average number of susceptible farms infected by an infectious farm) are robust to variations in their precise structure, the very large numbers of different transmission trees that are consistent with these temporal data render them mostly unhelpful with respect to identifying particular transmission events with confidence.
An integrated analysis, combining both data on the timing of infection and genetic sequence data from each infected farm should improve the resolution and confidence with which virus transmission routes can be identified. It may also highlight anomalous epidemiological information, indicative of unidentified intermediate infected premises missing from chains of transmission.
This study focuses on a set of farms in County Durham that were infected early in the 2001 UK FMDV epidemic. For each of the premises included in the study, we used data on the timing of infection and animal culling to establish distributions describing probable infection dates and infectious periods, together with complete genome sequences acquired from viruses sampled from these infected premises that provided information on their relative relatedness. We have then combined the inferences from these two types of data to determine statistically a set of most likely transmission trees, which we show to be a much smaller set than obtained through analysis of either data type alone. The increasing use of genetic data for forensic epidemiological purposes will require the integration of genetic and epidemiological data, and this study proposes an initial approach to this problem.
2. Material and methods
(a) Infected premises included in the study
The 15 infected premises in the Durham area (established to be FMDV positive through laboratory testing) included in this study were identified from Defra's Animal Health and Welfare FMD Data Archive (http://footandmouth.csl.gov.uk/) that also provided epidemiological data (including lesion age and date of cull). The relative locations of these premises are shown in figure 1. There had been no recorded movements of animals between any of the infected premises; the movements of FMD susceptible livestock having been prohibited throughout the UK since 23 February 2001. The details of the clinical samples included in this study are described in table 1, including source animal, farm type and details of livestock holdings. For farm D, the epithelium was cut in half, and both halves were sequenced independently to investigate the repeatability of the amplification and sequencing method used.
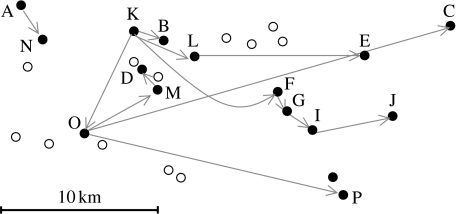
Figure 1.
Map showing the spatial relationship of 15 infected premises confirmed by laboratory testing (filled circles) and 12 infected premises (determined by clinical observations) that were subsequently found to be negative for virus by laboratory testing (open circles). A–P indicate the infected premises from which virus has been sequenced. The direction of most likely transmission events as determined by this study is shown by the grey arrows.
Table 1.
Farm and animal source details for each of the 22 FMDV consensus genomes included.
| virus sample | accession no. | animala | date of examination | date of cull | oldest lesion age (days)c | no. and type of susceptible animals present on farma | no. and type of infected animals reporteda,e |
|---|---|---|---|---|---|---|---|
| 1ab | DQ404179 | P | 24 Feb 2001 | 25 Feb 2001 | 10 | 571P | 450P |
| 1bb | DQ404178 | P | 24 Feb 2001 | 25 Feb 2001 | 10 | 571P | 400P |
| 1cb | DQ404177 | P | 24 Feb 2001 | 25 Feb 2001 | 10 | 571P | 400P |
| 2b | DQ404176 | C | 25 Feb 2001 | 25 Feb 2001 | 7d | 101BC, 366S | 51BC |
| 3b | DQ404175 | C | 24 Feb 2001 | 27 Feb 2001 | 4d | 195BC, 787S,1G | 50BC, 17S |
| 4b | DQ404173 | C | 26 Feb 2001 | 28 Feb 2001 | 3 | 558S | 6S |
| 5b | DQ404172 | C | 1 Mar 2001 | 3 Mar 2001 | 2 | 467BC,12S | 1BC |
| A | EF552688 | S | 31 Mar 2001 | 2 Apr 2001 | 2 | 167DC, 1600S | 3S |
| B | EF552689 | S | 2 Apr 2001 | 3 Apr 2001 | 2 | 113BC, 130S | 104S |
| C | EF552690 | C | 20 Apr 2001 | 20 Apr 2001 | 3 | 8BC, 400S | 3BC |
| D | EF552691 | C | 11 May 2001 | 11 May 2001 | 1 | 188DC | 2DC |
| E | EF552692 | S | 22 Apr 2001 | 24 Apr 2001 | 7 | 110BC, 152S | 6S |
| Fb | DQ404170 | S | 23 Apr 2001 | 24 Apr 2001 | 4 | 234BC, 330S | 264S |
| Gb | DQ404167 | S | 14 May 2001 | 14 May 2001 | 4 | 5BC, 682S, 2P | 545S |
| Ib | DQ404166 | C | 28 May 2001 | 28 May 2001 | 3 | 186BC, 3909S, 27P | 3BC, 2S |
| Jb | DQ404165 | C | 3 Jun 2001 | 4 Jun 2001 | 2 | 456DC | 2DC |
| K | EF552693 | C | 1 Apr 2001 | 4 Apr 2001 | 1 | 215BC, 383S | 1BC |
| L | EF552694 | C | 10 Apr 2001 | 10 Apr 2001 | 2 | 107BC, 124S | 6BC |
| M | EF552695 | S | 17 May 2001 | 18 May 2001 | 5 | 46BC, 188S | 14S |
| N | EF552696 | C | 12 Apr 2001 | 14 Apr 2001 | 1 | 6BC | 1BC |
| O | EF552697 | S | 15 Apr 2001 | 15 Apr 2001 | 2 | 39DC, 47BC, 197S | 3S |
| P | EU214601 | S | 30 Apr 2001 | 1 May 2001 | 4 | 88BC, 530S | 150S |
Animals described by one letter coding; P, pig; S, sheep; C, cattle; DC, dairy cattle; BC, beef cattle; G, goat.
Virus genomic sequences published previously.
Lesion ages according to Defra data warehouse (apart from where indicated), which differ from some original field data, but analysis using both datasets generates similar results.
Lesion ages according to Alexandersen (2003c).
Data from original field records.
(b) Complete genome sequences of foot-and-mouth disease viruses
The full genome sequences of the viruses recovered from four of the infected premises (F, G, I and J) in this cluster had been determined previously together with seven genomes from five other infected premises from the start of the outbreak included in this study (DQ404172, DQ404173, DQ404175–DQ404180, DQ404165, DQ404166, DQ404167, DQ404169, DQ404170; Cottam et al. 2006). Following the method described previously, 11 more viral genomes were sequenced for this study (Cottam et al. 2006; EF552688–EF552697 and EU214601; table 1). The genealogical relationships were based on statistical parsimony as implemented in the software package TCS (Clement et al. 2000). The tree was rooted to the closest FMDV strain SAR/19/2000 (Mason et al. 2003).
(c) Enumeration of transmission trees consistent with the sequence data
Rooting the sequence genealogy with SAR/19/2000 indicated the general direction of the transmission events (away from the node between farms 1 and 2). We took into account the known transmission history between farms 1 and 5 and restricted the tree configurations to those that included these four transmission events as a fixed link. Prior to the movement ban, infected animals could be transferred between farms and we allowed for the possibility of virus transmission without mutation. After the movement ban, farm-to-farm transmission probably required more infection cycles and, given the rate of molecular evolution of FMDV, the possibility of farm-to-farm transfer without mutation was considered to be negligible. The overall strategy was to construct a list of farms on which each putative virus haplotype could have been located, assuming no back mutation and only a single lineage present on each farm, and then to enumerate all transmission trees consistent with all combinations of possible locations of haplotypes. Specifically, the algorithm worked backwards from the tips of the tree derived using TCS, identifying the most recent common ancestors (MRCAs) of viral haplotypes found on pairs of farms, assigning these MRCAs to having been located on one farm or the other, defining the MRCA as a ‘new tip’ and proceeding further back down the tree to assign further MRCAs. Applying this algorithm recursively until each haplotype has a unique farm assigned to it leads directly to one possible transmission tree (an example of this procedure is described in figure 2). The likelihood of each tree was then estimated based on the available epidemiological data as described below.
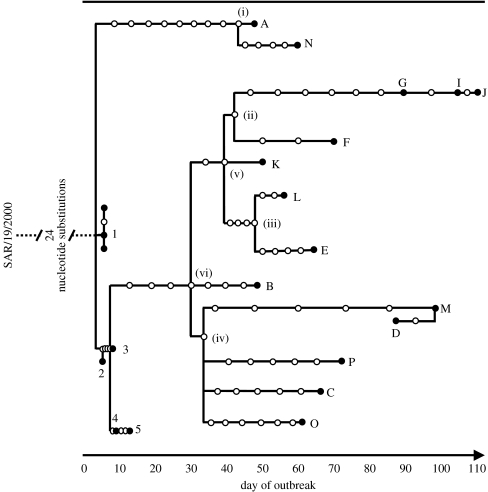
Figure 2.
Statistical parsimony analysis of 22 UK PanAsia O FMDV complete genome sequences rooted to the closest relative SAR/19/2000 by TCS; each connecting branch line represents a nucleotide substitution, with each circle representing a putative ancestral virus haplotype (filled circles indicate sequenced haplotypes). The farms infected through movement of infected livestock are represented by the numbers 1–5 (representing Defra infected premises numbers 4, 6, 7, 16 and 38, respectively), and those from a cluster in Durham are represented by the letters A–P. Day of outbreak corresponds to the number of days since 26 January 2001, the earliest plausible date since the outbreak began. In the most likely tree, it is assumed that the MRCA was present on (i) A, (ii) F, (iii) L, (iv) O, (v) K and (vi) K (and led to the transmission tree in figure 4). Assigning these MRCAs to other farms leads to less likely transmission trees.
(d) Epidemiological data
The first part of the analysis required estimating distributions describing both the likelihood that an individual farm was infected on a particular date and the likelihood that a farm was a source of infection on a particular date (this has been described previously as the temporal risk window; Taylor et al. 2004; Thrusfield et al. 2005). This analysis required two functions: Ii(t), describing the probability that the ith farm was first infected at time t and L(k), the probability that the first infected individual on a given farm incubates virus for k days prior to becoming infectious (and here we assume this function to apply to all infected farms). From these two functions it is possible to estimate a further function, Fi(t), describing the probability that the ith farm is a source of infection at time t.
The mean incubation period was chosen to be 5 days in common with other studies of the 2001 UK outbreak (Keeling et al. 2001), and the distribution of incubation periods, L(k), was assumed to follow a discrete form of the gamma distribution with scale and shape parameters of 3.00 and 1.67, respectively (this results in a 95% probability of incubation periods between 2 and 12 days, in accord with the previous estimates, Gibbens & Wilesmith 2002). The farms were assumed to be a source of infection immediately after the incubation period and up to and including the day the last animal on the farm was culled. This assumption is justified by the imposition of intense farm biosecurity, cleansing and disinfection following livestock culling.
The most likely date of infection for each farm was estimated to be the date on which disease was reported (here termed the examination date) to be present on the farm, minus the age of the oldest lesion on the farm, −5 days for virus incubation. We represented the uncertainty around the most likely date of infection arising from (unknown) error in the lesion dating, and possible variation in the incubation period by Ii(t), a discrete form of a beta distribution. Three pieces of information were used to inform the shape of Ii(t): the estimated most likely date of infection of farm i, which determined the mode of Ii(t); the most likely infection date of the primary case, which determined the earliest possible infection time; and the examination date of the ith farm (less 2 days to allow for a minimum incubation period), which determined the very latest possible infection time.
Fi(t), the probability that the ith farm is infectious at time t, was then calculated from Ii(t) and L(k) as follows:
where Ci was the time at which the ith farm was culled (time in this study is measured in days since 26 January 2001). This expression sums over the probability of farm i becoming infected on day τ, and completing the incubation period in not more than t−τ days. Now it is possible to calculate the likelihood that farm j infected farm i, assuming that farms cannot be multiply infected and that there are only n possible sources of infection
This equation sums the product of the likelihoods that farm i was infected, and farm j infectious over all possible days that transmission to i could have occurred. The denominator is required to ensure the λij sum to 1. These λij can be calculated for all possible pairs of farms, and used to compute the overall log likelihood of any single transmission tree. We proceeded by computing the likelihoods of all transmission trees consistent with the sequence data and to identify which of these trees was the most likely based on the epidemiological data. A subset of trees comprising the top 95% of the distribution of tree likelihoods was identified. For each transmission link in each tree in this top 95%, we also computed the ratio of the likelihood of the tree to the likelihood of the tree that included the next most likely alternative source of infection for the link. This ratio reflects the confidence in the inferred source of infection relative to other possible sources.
(e) Rate of nucleotide substitutions per nucleotide per day
A molecular clock was fitted to the virus sequence data from farms A–P using Markov Chain Monte Carlo techniques implemented in the software package Beast (Bayesian evolutionary analysis sampling trees; Drummond et al. 2002). A relaxed clock was fitted with exponentially distributed rates and fixed population size with the HKY model of base substitution (Hasegawa et al. 1985) and rate heterogeneity assumed.
(f) Number of nucleotide substitutions detectable between consecutive farm infections
The distribution of the number of nucleotide substitutions detected between consecutive farm infections was estimated using the most likely transmission tree determined as described previously. The MRCA of viral genotypes sampled from the source and recipient of infection was assumed to have been present on the source farm. Hence, the number of nucleotide substitutions per farm transfer was estimated as the number of nucleotide substitutions that arose between this common ancestor and the virus sampled on the recipient farm. We included 17 of the 19 transmission events comprising the most likely transmission tree in the analysis (the two transmission events of virus to farms A and K were excluded from the distribution as it is very likely that they were infected from farms outside the study area).
(g) Tests for spatial dependence in the transmission events
Two Monte Carlo tests were conducted to assess whether the 13 deduced transmission events in the County Durham group happened at random among the infected farms or the disease preferentially spread at short distance or in a given direction. To take into account the branching structure of the most likely transmission tree, the expected distributions for the test statistics were generated by randomly relabelling the infected farms (Diggle 1983). The test statistic used to detect potential clustering was the mean transmission distance. To detect directional spread, we used ρ, the norm of the mean of the unit vectors representing the transmission events (known as Rayleigh's test statistic for uniformity of circular data (Batschelet 1981)). The one-tailed p-values were computed from the position of the observed test statistic relative to 100 000 randomizations.
3. Results
(a) Complete genome sequencing of virus isolates and genetic transmission tracing
Epidemiological data from 20 infected premises were included in this study. For five of these infected premises, the transmission tree was determined previously by tracing direct animal movements (farms 1–5); for the other 15 the transmission routes were unknown (A–P). The 11 new sequences (A, B, C, D, E, K, L, M, N, O and P) were between 8193 and 8195 nt in length, with no ambiguous nucleotides, determined with an average of 4.8 times coverage of each nucleotide site. Between all 15 sequences (A–P) there were 85 variant nucleotides, of which 24 resulted in non-synonymous changes. Of the 85 variant nucleotides, five occurred within the VP1 capsid gene. The epithelium sample from farm D (which was sequenced twice independently) yielded identical sequences. A statistical parsimony tree depicting the genetic relationship between the viruses sequenced was generated for the 23 sequences, rooted using the genome sequence of the closest previously sequenced virus (from South Africa) as shown in figure 2.
(b) Temporal infection profiles of infected premises and transmission tracing
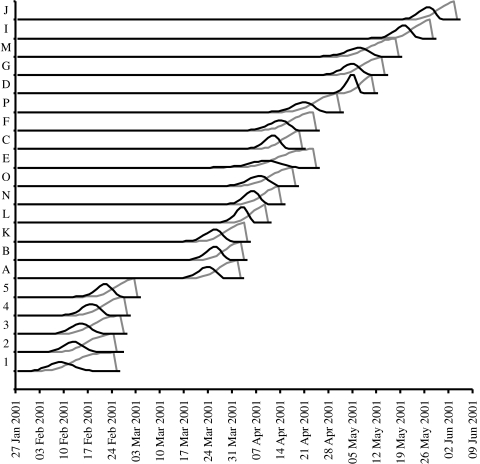
For each farm the distributions of likely date of infection and the most likely period of infectiousness were calculated and are shown in figure 3. The overlaps of infection and infectious periods can be clearly noted, providing a low-resolution impression of possible transmission pathways, together with some obvious breaks in the chain of transmission suggesting the presence of additional infected farms or inaccuracies in the estimates of time of infection.
Figure 3.
Temporal infection profiles of 20 farms infected with FMDV in 2001. Black lines indicate the likelihood of infection on any particular day, Ii(t) and grey lines the likelihood that a farm was infectious on a particular date, Fi(t), prior to culling.
(c) Likelihood analysis of integrated epidemiological and genetic datasets
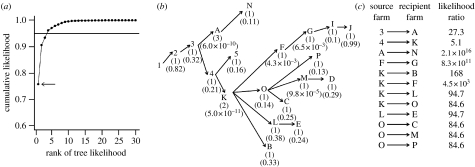
With no knowledge of any transmission routes the number of alternative trees consistent with the genetic data is 41 472. This number reduces to 1728 when the known transmission links between farms 1 to 5 are fixed. Of these 1728, 4 trees account for 95% of the likelihood (figure 4a). For each farm, a maximum of three alternative sources of infection can be identified among these four most likely trees (figure 4b), but the most likely source of infection is always at least 80 times more likely than any other source (except for farms A and K; figure 4c).
Figure 4.
Most likely transmission trees for the sequenced farms. (a) Cumulative likelihood distribution of the transmission trees, where the four most likely trees comprise more than 95% of the sum of all tree likelihoods (arrow, the most likely tree). (b) Most likely tree with, for each farm, the number of different sources of infection among the top 95% set of trees (in parentheses), and λij the likelihood that the recipient farm was infected by the indicated source farm (below). (c) The likelihood ratio between the most likely tree and the next most likely tree with a different source farm for a given infected (recipient) farm, when the source of infection is not assumed.
(d) Rate of substitutions per nucleotide site per day
A relaxed molecular clock for the rate of substitution of all nucleotide changes from farms A to P was estimated to be 2.076×10−5 per site per day (95% CIs 5.739×10−6 to 3.509×10−5).
(e) Number of nucleotide changes upon farm-to-farm transfer
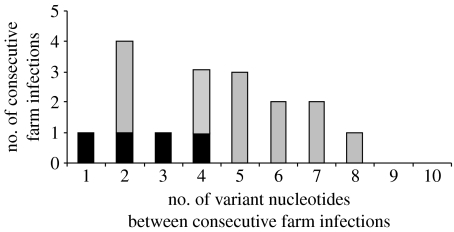
The distribution of the number of nucleotide changes per farm transfer is shown in figure 5. The distribution has a mean of 4.3 nucleotide substitutions, with s.d.=2.1. If this distribution is partitioned into transmission events preceding and proceeding the NMB, then the mean number of substitutions per transmission link proceeding the NMB (farms A–P) is 4.85, which is significantly higher than that preceding the NMB, represented by farms 1–5, of 2.5 substitutions (t-test; t=2.19, p=0.045).
Figure 5.
Distribution of the number of variant nucleotides between viruses recovered from consecutively infected farms. Those that represent transmissions that occurred before the NMB are shown in black. The number of variant nucleotides was determined from the common ancestor of source and daughter farm, as described in §2, using the transmission events shown in figure 4b. The distribution has a mean of 4.3, s.d.=2.1.
(f) Tests for spatial dependence in the transmission events
The Monte Carlo tests demonstrate that transmission did not occur in a spatially random way between pairs of farms. There is a clear preferential transmission towards an easterly and southeasterly direction (p=5.8×10−3), and the mean distance of the transmission events (4.8 km) is significantly less than expected when the transmission between farms occurred randomly with respect to distance (7.5 km, p=1.2×10−3).
4. Discussion
The method described in this study makes a preliminary attempt at integrating information from genetic data with the relative timing of infection events to compute the most likely transmission tree that reflects the spread of infection between farms. The study has demonstrated the power of forensic genetic tracing for FMDV, and highlights the future challenges to analyses of this sort.
Using only the known transmission events relating farms 1–5, 1728 possible transmission trees remain consistent with the rooted parsimony tree identified using the genetic data. This is because while it is possible to identify viruses that share a common ancestor, it is often not possible to identify the farm on which the ancestors were present, and therefore the exact direction of transmission events. However, the difficulties in interpreting the genetic data can be overcome in part by the addition of epidemiological data: here, the most likely time of infection and infectiousness of a farm, which enables the likelihood of particular transmission events to be estimated. This integration of information from the two datasets enables the identification of the most likely trees at a resolution far greater than that possible using either dataset alone.
In this study, we have generated complete genome sequences from all infected farms from a geographical area of approximately 100 km2 found to be FMDV positive by laboratory analysis. Interpretation of the results is complicated because both the epidemiological timing and the genetic data suggest that there may have been more than a single disease introduction event in this area.
Our analysis indicates particularly low probabilities of infection between some pairs of infected farms linked in the most likely transmission tree (figure 4b). These include the links between farms 3 and A, 4 and K, O and M, K and F, and F and G. In some cases, we suspect that additional intermediary infectious farms must exist that are either: (i) outside the geographical area of our study or (ii) located within the study area but from which infected tissue samples were not collected (within the study area there were 108 farms identified as ‘dangerous contacts’ or ‘contiguous premises’ and on which livestock were culled prior to the 28 May; but even if these farms were infected, they may not have been very infectious). Based on the epidemiological activity to the north of our study area throughout March, we believe the former may be a reasonable explanation for how farms A and K became infected.
Explanations for the source of infections to farms M, F and G are more problematic as there are almost no alternative plausible local sources of infection. Furthermore, the possibility of airborne (and potentially longer range) spread occurring among farms C, E, F, G, I and J was considered improbable on the basis of wind strength and direction in relation to the potential sources of infection, and is not thought to have played a role in dissemination of virus to these premises. This forces us to question the accuracy of the data relating to the timing of infection. Lesion dating is imperfect with an unknown amount of error associated with it. However, the dominant source of error associated with using lesions to estimate the date of infection is that the oldest lesions may be overlooked entirely (and it is for these reasons that the function describing the likelihood that a farm was infected on a particular day, Ii(t), was only minimally constrained). This is particularly likely on farms with livestock that include sheep where the clinical symptoms of disease are often mild. The farms M, F and G all maintained substantial sheep herds in which disease was eventually confirmed (table 1) and it is possible that infection was present on these farms considerably earlier than estimated, but went unreported.
From the data generated during this study, we can estimate the distribution of the number of nucleotide changes that arose between consensus sequences recovered from source and recipient infected farms. When this number is unusually large it suggests that virus has been replicating in some other livestock population, either on unidentified intermediate farms, or on the recipient farm but in a population in which disease had previously been overlooked. For example, the large number of changes observed on farm A (more than double the average expected for a single farm–farm transmission event) is suggestive of such an intermediate. It is important to recognize that the number of changes detectable between consecutively infected farms will probably depend on the mode of infection. In this study, the number of nucleotide substitutions per infection generation interval is lower prior to the imposition of the national movement ban, compared with after, probably because there are less viral replication cycles associated with the infections arising as a result of movement of infected animals, compared with fomite-associated transmission. Data of this sort could be used in future investigations to infer the presence of undetected sources of infection or to determine the most likely number of infected premises in a transmission chain. However, for this to be feasible it is important that this distribution is characterized in more detail.
The most likely transmission tree shown in figure 4b raises some interesting points relating to this particular case study. This tree differs from the original contact-tracing tree proposed by Defra (which is inconsistent with the genetic data). First, the separate lineage of infected premises represented by A and N is anomalous, and when compared with the remainder of the sequences available from the 2001 UK outbreak (Cottam et al. 2006) represents transmission of the virus directly into the area from the source of the outbreak, and not via Longtown and Hexham markets (as is thought to be the case for all other infected premises throughout the UK; Gibbens et al. 2001). This finding suggests that further study of the outbreak is necessary to determine the origins of this previously unidentified chain of transmission events. Second, when the layout of the farms is considered (figure 1), it appears that transmission events are significantly clustered with the distance between farms playing an important epidemiological role. This supports the previous interpretations that virus was spreading to nearby farms with a greater likelihood than distant farms. The significant directionality in transmissions is interesting, but could arise for a number of different reasons. It may be a consequence of the road network that connects the farms or may result from infection having been introduced on the westward edge of a cluster of farms (although the existence of susceptible farms to the east, north and south suggests this was not the case).
The rate of nucleotide substitution noted in this study for virus from farms A to P over this short time period is 2.076×10−5 per site per day (95% CIs 5.739×10−6 to 3.509×10−5), which is very close to that estimated previously (2.26×10−5, 95% CIs 1.75×10−5 to 2.80×10−5) for the whole of the 2001 UK outbreak (Cottam et al. 2006). In principle, it would be possible to evaluate the likelihood of the transmission trees based on the genetic data after the imposition of a molecular clock. However, until more is known about the extent of nucleotide variation from single animals, herds and different types of livestock on a single farm, we are inclined to view a more detailed and complex quantitative analysis of ‘one genotype per farm’ data with some caution.
Here, we have presented a simple method by which the transmission tree space is qualitatively restricted through the use of the genetic data, and the likelihood of trees remaining in this space quantitatively evaluated by the use of epidemiological data. This likelihood, estimated from the epidemiological data, could be made more sophisticated by accounting for the mode of transmission (e.g. incorporating information about the possible movement of animals between farms) or the livestock composition on particular farms (for example, pigs are generally regarded as more infectious than cattle or sheep, whereas cattle are more susceptible than sheep or pigs; Alexandersen et al. 2002, 2003a,b). The farm infectiousness is not explicitly quantified in this analysis. While infectiousness is unlikely to remain constant over the course of an infection, available data do not enable a parametrization of possible change (Savill et al. 2007). Here, we have simply assumed that the probability of a farm being a source of infection varies over time in a way dictated by the timing of infection. It is plausible that susceptibility varies between farms, and if this were quantified it could also be incorporated into future models.
Ultimately, the analysis of these sorts of data will be best conducted by development of a single likelihood model that accounts for both the evolutionary and the epidemiological dynamics at the various different scales at which they occur. Developing models that integrate different data types in this way will be a difficult but exciting future research challenge.
Acknowledgments
E.M.C. is the recipient of a BBSRC PhD Studentship, and funding for the laboratory consumables was provided by the Department of Environment, Food and Rural Affairs project SE2936. The collection and archiving of UK 2001 samples was undertaken by the Food and Agricultural Organization World Reference Laboratory for Foot-and-Mouth Disease. We are grateful to Joël Chadœuf for statistical advice.
References
- Alexandersen S, Zhang Z, Reid S.M, Hutchings G.H, Donaldson A.I. Quantities of infectious virus and viral RNA recovered from sheep and cattle experimentally infected with foot-and-mouth disease virus O UK 2001. J. Gen. Virol. 2002;83:1915–1923. doi: 10.1099/0022-1317-83-8-1915. [DOI] [PubMed] [Google Scholar]
- Alexandersen S, Quan M, Murphy C, Knight J, Zhang Z. Studies of quantitative parameters of virus excretion and transmission in pigs and cattle experimentally infected with foot-and-mouth disease virus. J. Comp. Pathol. 2003a;129:268–282. doi: 10.1016/s0021-9975(03)00045-8. doi:10.1016/S0021-9975(03)00045-8 [DOI] [PubMed] [Google Scholar]
- Alexandersen S, Zhang Z, Donaldson A.I, Garland A.J. The pathogenesis and diagnosis of foot-and-mouth disease. J. Comp. Pathol. 2003b;129:1–36. doi: 10.1016/s0021-9975(03)00041-0. doi:10.1016/S0021-9975(03)00041-0 [DOI] [PubMed] [Google Scholar]
- Alexandersen S, Kitching R.P, Mansley L.M, Donaldson A.I. Clinical and laboratory investigations of five outbreaks during the early stages of the 2001 foot-and-mouth disease epidemic in the United Kingdom. Vet. Rec. 2003c;152:489–496. doi: 10.1136/vr.152.16.489. [DOI] [PubMed] [Google Scholar]
- Batschelet E. Academic Press; London, UK: 1981. Circular statistics in biology. [Google Scholar]
- Bracho M.A, Gosalbes M.J, Blasco D, Moya A, Gonzalez-Candelas F. Molecular epidemiology of a hepatitis C virus outbreak in a hemodialysis unit. J. Clin. Microbiol. 2005;43:2750–2755. doi: 10.1128/JCM.43.6.2750-2755.2005. doi:10.1128/JCM.43.6.2750-2755.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement M, Posada D, Crandall K.A. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. doi:10.1046/j.1365-294x.2000.01020.x [DOI] [PubMed] [Google Scholar]
- Cottam E.M, Haydon D.T, Paton D.J, Gloster J, Wilesmith J.W, Ferris N.P, Hutchings G.H, King D.P. Molecular epidemiology of the foot-and-mouth disease virus outbreak in the United Kingdom in 2001. J. Virol. 2006;80:11 274–11 282. doi: 10.1128/JVI.01236-06. doi:10.1128/JVI.01236-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggle P.J. Academic Press; London, UK: 1983. Statistical analysis of spatial point patterns. [Google Scholar]
- Dowell S.F, et al. A multistate outbreak of oyster-associated gastroenteritis: implications for interstate tracing of contaminated shellfish. J. Infect. Dis. 1995;171:1497–1503. doi: 10.1093/infdis/171.6.1497. [DOI] [PubMed] [Google Scholar]
- Drummond A.J, Nicholls G.K, Rodrigo A.G, Solomon W. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics. 2002;161:1307–1320. doi: 10.1093/genetics/161.3.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbens J.C, Wilesmith J.W. Temporal and geographical distribution of cases of foot-and-mouth disease during the early weeks of the 2001 epidemic in Great Britain. Vet. Rec. 2002;151:407–412. doi: 10.1136/vr.151.14.407. [DOI] [PubMed] [Google Scholar]
- Gibbens J.C, Sharpe C.E, Wilesmith J.W, Mansley L.M, Michalopoulou E, Ryan J.B.M, Hudson M. Descriptive epidemiology of the 2001 foot-and-mouth disease epidemic in Great Britain: the first five months. Vet. Rec. 2001;149:729–743. [PubMed] [Google Scholar]
- Hasegawa M, Kishino H, Yano T.-a. Dating of the human–ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 1985;22:160–174. doi: 10.1007/BF02101694. doi:10.1007/BF02101694 [DOI] [PubMed] [Google Scholar]
- Haydon D.T, Chase-Topping M, Shaw D.J, Matthews L, Friar J.K, Wilesmith J, Woolhouse M.E. The construction and analysis of epidemic trees with reference to the 2001 UK foot-and-mouth outbreak. Proc. R. Soc. B. 2003;270:121–127. doi: 10.1098/rspb.2002.2191. doi:10.1098/rspb.2002.2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling M.J, et al. Dynamics of the 2001 UK foot and mouth epidemic: stochastic dispersal in a heterogeneous landscape. Science. 2001;294:813–817. doi: 10.1126/science.1065973. doi:10.1126/science.1065973 [DOI] [PubMed] [Google Scholar]
- Leitner T, Albert J. The molecular clock of HIV-1 unveiled through analysis of a known transmission history. Proc. Natl Acad. Sci. USA. 1999;96:10 752–10 757. doi: 10.1073/pnas.96.19.10752. doi:10.1073/pnas.96.19.10752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lim S.L, Ruan Y, Ling A.E, Ng L.F.P, Drosten C, Liu E.T, Stanton L.W, Hibberd M.L. SARS transmission pattern in Singapore reassessed by viral sequence variation analysis. PLoS Med. 2005;2:e43. doi: 10.1371/journal.pmed.0020043. doi:10.1371/journal.pmed.0020043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason P.W, Pacheco J.M, Zhao Q.-Z, Knowles N.J. Comparisons of the complete genomes of Asian, African and European isolates of a recent foot-and-mouth disease virus type O pandemic strain (PanAsia) J. Gen. Virol. 2003;84:1583–1593. doi: 10.1099/vir.0.18669-0. doi:10.1099/vir.0.18669-0 [DOI] [PubMed] [Google Scholar]
- Savill N.J, Shaw D.J, Deardon R, Tildesley M.J, Keeling M.J, Woolhouse M.E, Brooks S.P, Grenfell B.T. Effect of data quality on estimates of farm infectiousness trends in the UK 2001 foot-and-mouth disease epidemic. J. R. Soc. Interface. 2007;4:235–241. doi: 10.1098/rsif.2006.0178. doi:10.1098/rsif.2006.0178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savolainen C, Mulders M.N, Hovi T. Phylogenetic analysis of rhinovirus isolates collected during successive epidemic seasons. Virus Res. 2002;85:41–46. doi: 10.1016/s0168-1702(02)00016-3. doi:10.1016/S0168-1702(02)00016-3 [DOI] [PubMed] [Google Scholar]
- Spada E, Sagliocca L, Sourdis J, Garbuglia A.R, Poggi V, De Fusco C, Mele A. Use of the minimum spanning tree model for molecular epidemiological investigation of a nosocomial outbreak of hepatitis C virus infection. J. Clin. Microbiol. 2004;42:4230–4236. doi: 10.1128/JCM.42.9.4230-4236.2004. doi:10.1128/JCM.42.9.4230-4236.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor N.M, Honhold N, Paterson A.D, Mansley L.M. Risk of foot-and-mouth disease associated with proximity in space and time to infected premises and the implications for control policy during the 2001 epidemic in Cumbria. Vet. Rec. 2004;154:617–626. doi: 10.1136/vr.154.20.617. [DOI] [PubMed] [Google Scholar]
- Thrusfield M, Mansley L, Dunlop P, Taylor J, Pawson A, Stringer L. The foot-and-mouth disease epidemic in Dumfries and Galloway, 2001. 1: characteristics and control. Vet. Rec. 2005;156:229–252. doi: 10.1136/vr.156.8.229. [DOI] [PubMed] [Google Scholar]
- Wallace R.G, Hodac H, Lathrop R.H, Fitch W.M. A statistical phylogeography of influenza A H5N1. Proc. Natl Acad. Sci. USA. 2007;104:4473–4478. doi: 10.1073/pnas.0700435104. doi:10.1073/pnas.0700435104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh P.D, Biek R, Real L.A. Wave-like spread of Ebola Zaire. PLoS Biol. 2005;3:e371. doi: 10.1371/journal.pbio.0030371. doi:10.1371/journal.pbio.0030371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C.W, Albert T.J, Vega V.B, Norton J.E, Cutler D.J, Richmond T.A, Stanton L.W, Liu E.T, Miller L.D. Tracking the evolution of the SARS coronavirus using high-throughput, high-density resequencing arrays. Genome Res. 2004;14:398–405. doi: 10.1101/gr.2141004. doi:10.1101/gr.2141004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Diaz R.S, Ho D.D, Mosley J.W, Busch M.P, Mayer A. Host-specific driving force in human immunodeficiency virus type 1 evolution in vivo. J. Virol. 1997;71:2555–2561. doi: 10.1128/jvi.71.3.2555-2561.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]