Abstract
Protein misfolding, whether caused by aging, environmental factors, or genetic mutations, is a common basis for neurodegenerative diseases. The misfolding of proteins with abnormally long polyglutamine (polyQ) expansions causes several neurodegenerative disorders, such as Huntington’s disease (HD). Although many cellular pathways have been documented to be impaired in HD, the primary triggers of polyQ toxicity remain elusive. We report that yeast cells and neuron-like PC12 cells expressing polyQ-expanded huntingtin (htt) fragments display a surprisingly specific, immediate, and drastic defect in endoplasmic reticulum (ER)-associated degradation (ERAD). We further decipher the mechanistic basis for this defect in ERAD: the entrapment of the essential ERAD proteins Npl4, Ufd1, and p97 by polyQ-expanded htt fragments. In both yeast and mammalian neuron-like cells, overexpression of Npl4 and Ufd1 ameliorates polyQ toxicity. Our results establish that impaired ER protein homeostasis is a broad and highly conserved contributor to polyQ toxicity in yeast, in PC12 cells, and, importantly, in striatal cells expressing full-length polyQ-expanded huntingtin.
Keywords: Polyglutamine (polyQ), Huntington’s disease (HD), neurodegeneration, endoplasmic reticulum (ER)-associated protein degradation (ERAD), unfolded protein response (UPR), ER stress
Polyglutamine (polyQ) expansions in proteins are the basis for at least nine different neurodegenerative disorders, including Huntington’s disease (HD) (Orr and Zoghbi 2007). The proteins carrying polyQ expansions are broadly expressed, but each disease is characterized by the vulnerability of a particular subset of neurons. Interactions between sequences flanking the polyQ expansion and the proteome unique to distinct neurons must determine the specific character of each disease. However, in virtually every case, toxicity ensues when the expansion reaches ∼40 residues. Further, the age of disease onset decreases and the severity of disease progression increases as the length of the polyQ expansion increases. Thus, even though each disease is distinct, there must be common underlying toxic mechanisms related to polyQ-mediated misfolding.
To investigate polyQ toxicity, we used a combination of yeast, PC12, and striatal cell models. We and others have developed yeast models that express N-terminal fragments of huntingtin (htt exonI) (Krobitsch and Lindquist 2000; Muchowski et al. 2000; Meriin et al. 2002; Duennwald et al. 2006a, b). Our yeast model recapitulates major features of neuronal polyQ pathology, including the hallmark feature of increasing toxicity with increasing polyQ length (Duennwald et al. 2006b). Thus, the yeast model presents the opportunity to identify factors that specifically determine polyQ toxicity in a genetically tractable model organism.
Numerous cellular pathways, such as transcriptional regulation (Riley and Orr 2006), vesicular transport (Gunawardena and Goldstein 2005), and protein turnover (Bence et al. 2001; Bennett et al. 2007) are impaired by polyQ expansion proteins. It remains unclear, however, which of these cellular defects are initial and specific triggers of polyQ toxicity. Here, we focused on how the well-established polyQ-induced defect in the ubiquitin proteasome system (UPS) (Bence et al. 2001; Holmberg et al. 2004; Venkatraman et al. 2004; Bennett et al. 2007) contributes to polyQ toxicity. Specifically, we asked whether the polyQ-induced defect in the UPS is global or whether it affects certain degradation pathways more than others.
We find that polyQ-expanded htt exonI strongly impairs endoplasmic reticulum (ER)-associated protein degradation (ERAD). We provide mechanistic insight into this specific defect: Toxic polyQ-expanded proteins entrap the ERAD proteins Ufd1, Npl4, and p97 (VCP) and thereby inhibit their essential participation in ERAD. Our results explain the molecular basis of the previously reported polyQ-induced unfolded protein response (UPR) and ER stress (Kouroku et al. 2002; Nishitoh et al. 2002; Thomas et al. 2005) and, as noted in the Discussion, suggest that toxicity may be related to the normal function of SCA3 (Zhong and Pittman 2006). Further, we document that the dysfunction in ER protein homeostasis occurs with high specificity and is an early response, the first we detected after the expression of polyQ-expanded htt exonI. The defect in ER protein homeostasis might therefore be an early contributor to polyQ toxicity.
Because it is unclear to what extent overexpressed polyQ-expanded htt fragments mimic HD, we also investigated ER protein homeostasis in striatal cells expressing full-length huntingtin with a polyQ expansion expressed from its endogenous locus (Trettel et al. 2000). In this more accurate cellular model of HD, we also find an induction of the UPR and a specific and strong sensitization to ER stress. Our results therefore mechanistically explain the previously reported polyQ-induced activation of the UPR and define ER stress as a highly conserved event in polyQ toxicity that may be relevant to HD. The nature of these defects suggests relevance to other polyQ expansion diseases as well.
Results
Toxic polyQ expansion proteins impair protein degradation selectively
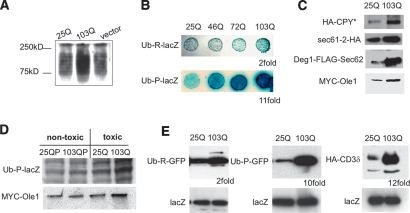
We first investigated whether polyQ-expanded htt exonI impaired the UPS in our yeast model, as reported in other models and in the brains of HD patients (Bence et al. 2001; Holmberg et al. 2004; Bennett et al. 2007). Indeed, the turnover of polyubiquitinated proteins was mildly yet reproducibly reduced in yeast cells expressing 103Q htt exonI for 12 h when compared with yeast cells expressing 25Q htt exonI (Fig. 1A).
Figure 1.
PolyQ-expanded htt exonI selectively impaired UFD and ERAD. (A) Yeast cells expressing polyQ-expanded htt exonI accumulate polyubiquitinated proteins. Protein lysates from yeast cells expressing either 25Q or 103Q htt exonI for 12 h or a vector control were analyzed by Western blotting using an anti-ubiquitin antibody. (B) Longer polyQ expansions caused stronger inhibition of UFD in yeast. The N-end rule reporter protein, Ub-R-lacZ, and the UFD reporter protein Ub-P-lacZ were expressed in yeast cells that expressed htt exonI fragments with 25Q, 46Q, 72Q, and 103Q for 6 h. Turnover of the N-end rule and UFD reporter proteins was visualized by a β-galactosidase overlay assay. (C) 103Q htt exonI impairs ERAD in yeast. Western blot analysis of protein lysates from yeast cells expressing the ERAD reporter proteins HA-CPY*, sec61-2-HA, Deg1-Flag-Sec62, and MYC-Ole1 coexpressing either 25Q or 103Q htt exonI for 6 h. (D) Only toxic polyQ-expanded htt exonI impaired UFD and ERAD. Western blot analysis of protein lysates from yeast cells expressing either the nontoxic htt exonI proteins 25QP, 103QP, and 25Q or the toxic 103Q for 6 h. These cells coexpressed either the UFD reporter (Ub-P-lacZ) or the ERAD reporter (MYC-Ole1). (E) UFD and ERAD were impaired in PC12 cells expressing 103Q htt exonI. PC12 cells harboring inducible versions of 25Q or 103Q htt exonI were transfected with the N-end rule reporter Ub-R-GFP, the UFD-reporter Ub-P-GFP, and the ERAD reporter HA-CD3δ. For all transfections, a plasmid for the expression of lacZ was cotransfected. Two days after transfection, 25Q and 103Q expression was induced for 8 h, cells were lysed, and Western blot analyses were performed. Equal protein loading for the Western blots shown in this figure was documented by the levels of the transfection control lacZ. The numbers below the Western blots indicate the quantification of the fold stabilization of the degradation substrate in cells expressing 103Q compared with cells expressing 25Q (average of three independent experiments).
To determine if some degradation pathways are affected more than others, we compared proteins that are degraded by two distinct and well-characterized pathways: the N-end rule degradation pathway (Varshavsky 1992) and the ubiquitin fusion degradation (UFD) pathway (Johnson et al. 1995). Both types of substrates are polyubiquitinated, and both are then degraded by the proteasome. Yet, they use different modes of substrate recognition and different factors to facilitate ubiquitination prior to degradation (Varshavsky 1992; Johnson et al. 1995).
We expressed an N-end rule substrate and a UFD substrate, each as β-galactosidase fusion proteins, in yeast cells that also expressed htt exonI protein variants, a nontoxic form (25Q), and increasingly toxic forms (46Q, 72Q, and 103Q). The UFD (Ub-P-lacZ) was strongly impaired in a polyQ length-dependent fashion (Fig. 1B). Quantification revealed that 103Q htt exonI caused an 11-fold increase in β-galactosidase stability for the UFD substrate relative to 25Q htt exonI, whereas it caused only a twofold increase in the stability of the N-end rule substrate Ub-R-lacZ (data not shown). Thus, polyQ-expanded htt exonI impairs the degradation of a UFD substrate more strongly than that of an N-end rule substrate. These results were confirmed by pulse-chase analyses. In the presence of 103Q htt exonI, the half life of the N-end rule substrate Ub-R-lacZ was increased less than twofold when compared with 25Q htt exonI-expressing cells. The half-life of the UFD substrate Ub-P-lacZ was significantly increased due to 103Q htt exonI (Supplemental Fig. S2; data not shown). We also tested the degradation of several other proteasome substrates: six additional N-end rule substrates—Ub-I-lacZ, Ub-L-lacZ, Ub-N-lacZ, Ub-Q-lacZ, Ub-Y-lacZ, and Ub-F-lacZ (Varshavsky 1992)—and proteins bearing a well-defined degradation tag from the Mat α2 repressor (Laney et al. 2006). All of these were much less affected than the UFD substrate (data not shown).
The precise role of the UFD pathway is unclear but it shares many features with ERAD (Raasi and Wolf 2007). ERAD is a central component of ER protein quality control. During ERAD, damaged proteins residing in the ER lumen or the ER membrane are transported to the cytosol where they are ubiquitinated by ERAD-specific factors and then degraded by the proteasome (Hampton 2002). We asked whether 103Q htt exonI impaired ERAD by expressing several ERAD substrates of diverse types (HA-CPY*, sec61-2-HA, Deg1-Flag-Sec62, and MYC-Ole1) (Braun et al. 2002; Haynes et al. 2002). All four ERAD substrates were stabilized in cells expressing 103Q htt exonI compared with cells expressing 25Q htt exonI (Fig. 1C). Again, these results were confirmed by pulse-chase analysis and shut-off experiments (Supplemental Fig. S2). Thus, polyQ-expanded htt exonI significantly impaired ERAD. Notably, a defect in ERAD occurred after relatively short induction of 103Q htt exonI in yeast (6 h) and PC12 cells (8 h), whereas the degradation of polyubiquitinated proteins could only be observed after longer induction of the 103Q htt exonI (Supplemental Fig. S1). While cells exhibited a block in ERAD after only 6 h of 103Q induction, they did not stop growing (as determined by the optical density of the culture) until 12 h, and remained fully viable for at least 24 h (M.L. Duennwald and S. Lindquist, in prep.).
We next asked if the defects in UFD and ERAD were specific to toxic forms of polyQ expansion proteins. The endogenous proline-rich region downstream from the Q expansion in htt exonI has a profound effect on toxicity: Proteins that do not contain the proline-rich region (103Q) rapidly arrest growth and eventually kill yeast cells, whereas proteins that contain the proline-rich region (103QP) have no effect on growth (Duennwald et al. 2006b). As shown in Figure 1D, only the toxic htt exonI protein caused an impairment of UFD and ERAD.
To determine if the impairment of UFD and ERAD by toxic 103Q that we had discovered was also characteristic of mammalian neuron-like cells, we used PC12 cells carrying a nontoxic 25Q htt exonI or a toxic 103Q htt exonI under the control of an inducible promoter (Aiken et al. 2004; Apostol et al. 2006). The cells were transfected with an N-end rule substrate (Ub-R-GFP), a UFD substrate (Ub-P-GFP) (Dantuma et al. 2000), or an ERAD substrate (HA-CD3δ) (Tiwari and Weissman 2001), all of which were constitutively expressed. After recovery from transfection, 25Q and 103Q htt exonI were induced and substrate degradation was measured by Western blotting. PC12 cells showed the same strong selective impairment of UFD (10-fold) and ERAD (12-fold) by 103Q htt exonI as the yeast model (Fig. 1E). The degradation of the N-end rule substrate was affected only twofold.
Next, we examined the temporal relationship between the defect in UFD and ERAD that is induced by polyQ and polyQ toxicity. In yeast cells, we simply measured changes in growth rates in liquid media. For PC12 cells, we measured the induction of apoptosis by the activation of caspase3 and caspase7 and reductions in respiratory activity with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (Aiken et al. 2004). In both models, the impairment of UFD and ERAD occurred very early after the induction of the toxic htt proteins: For yeast, a strong ERAD defect was apparent within 6 h, while the very first effects on cell growth were detected only after 8 h (Fig. 1C,E; data not shown). In PC12 cells, UFD and ERAD were strongly affected within 8 h, while the earliest signs of toxicity could be detected only after 12 h by caspase activation and only after 24 h by MTT assays (data not shown). In contrast to the more immediate defect in UFD and ERAD, a general accumulation of polyubiquitinated proteins occurred only after a substantially longer induction (12 h in yeast and up to 48 h in PC12 cells) (Supplemental Fig. S1). Therefore, the selective defect in UFD and ERAD is not simply a downstream consequence of general effects of polyQ-mediated toxicity—rather, it preceded the onset of toxicity in yeast and PC12 cells.
Toxic polyQ expansion proteins elicit the UPR but not the heat-shock response (HSR)
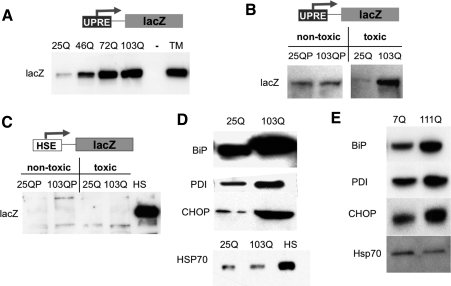
In other systems, a defect in ERAD induces the UPR, a specific reaction to protein folding stress in the ER (Travers et al. 2000). This involves the induction of several genes involved in ER protein homeostasis that are under the transcriptional control of the UPR element (UPRE) (Bernales et al. 2006). To determine if toxic polyQ expansion proteins elicit UPR induction in yeast, we employed a reporter construct, UPRE-lacZ (Patil and Walter 2001). Indeed, the UPR reporter was induced in yeast cells in a manner that correlated with the length of the polyQ expansions (Fig. 2A). Nontoxic htt exonI proteins (25Q, 25QP, and 103QP) did not elicit the UPR (Fig. 2B).
Figure 2.
PolyQ-expanded htt exonI induces the UPR. (A) Longer polyQ expansions caused stronger UPR induction. Yeast cells expressing the UPR reporter UPRE-lacZ were induced for the expression of 25Q, 46Q, 72Q, and 103Q htt exonI for 6 h. Protein lysates were prepared, and lacZ induction was analyzed by Western blotting. TM-treated cells (1 μg/mL for 1 h) served as a positive control and vector-transformed cells as a negative control for UPR induction. (B) Only toxic polyQ expansion proteins induce the UPR. Yeast cells expressing UPRE-lacZ were induced for the expression of either the nontoxic htt exonI fragments 25QP, 103QP, and 25Q or the toxic 103Q for 6 h. Protein lysates were prepared, and lacZ induction was monitored by Western blotting. (C) PolyQ-expanded htt exonI did not induce the HSR. Yeast cells expressing the heat-shock reporter HSE-lacZ were induced for the expression of either the nontoxic (25QP, 103QP, and 25Q) or the toxic 103Q htt exonI fragment for 8 h. Heat-shocked (HS, 2 h 39°C) cells served as a positive control. Protein lysates were prepared, and lacZ induction was monitored by Western blotting. (D) PolyQ-expanded htt exonI induced the UPR in PC12 cells. Protein lysates of PC12 cells induced for the expression of either 25Q or 103Q htt exonI for 8 h were analyzed by Western blot. The UPR-induced proteins BiP, PDI, and CHOP were monitored. The level of Hsp70, a protein induced by the HSR, was not increased by 103Q. Heat-shocked cells (HS, 2 h 39°C) served as a positive control. Equal protein loading for the Western blots shown in this figure was documented by Ponceau-S staining of the blots (Supplemental Fig. S4). (E) Full-length htt bearing a polyQ expansion (111Q) induced the UPR in striatal cells. The UPR-induced proteins BiP, PDI, and CHOP were detected on Western blots prepared with protein lysates from either wild-type (7Q) or mutant (111Q) striatal cells. Hsp70, a marker of the HSR, was not induced in mutant striatal cells. Equal protein loading for the Western blots shown was documented by Ponceau-S staining of the blots (Supplemental Fig. S3).
Is the UPR induced selectively, or do polyQ expansion proteins activate other cellular stress programs? The HSR is a highly conserved cellular program in all living organisms that is activated by the dysfunction of cytosolic protein homeostasis in response to a vast array of conditions; e.g., exposure to high temperatures, anoxia, and the presence of misfolded proteins in the cytosol (Lindquist 1986). Despite the fact that the polyQ expansion proteins were expressed in the cytosol and accumulated there as aggregated species, neither the nontoxic nor the toxic polyQ-expanded htt exonI proteins had induced a HSR at times when the UPR was fully induced. This was true whether the response was measured by the sensitive heat-shock reporter, HSE-lacZ (lacZ under transcriptional control of the heat-shock element, HSE) (Fig. 2C; Kirk and Piper 1991) or by the induction the heat-shock proteins Hsp104 and Hsp26, which are the most sensitive heat-shock indicators in yeast (Supplemental Fig. S6).
The selective induction of the UPR by toxic polyQ-expanded htt exonI proteins was also found in PC12 cells. Here, the UPR was monitored by Western analysis of the UPR-induced proteins (Schroder and Kaufman 2005) BiP, PDI, and CHOP (Fig. 2D). Cells expressing 103Q htt exonI exhibited elevated levels of BiP, PDI, and CHOP compared with cells expressing 25Q htt exonI. The cytosolic stress-specific Hsp70 isoform, a hallmark of the mammalian HSR, was not induced by the 103Q htt exonI (Fig. 2D). The HSR is universally triggered by the presence of misfolded proteins in the cytosol. The fact that misfolded polyQ expansion proteins do not trigger a HSR in neurons (Hay et al. 2004; Tagawa et al. 2007), neuron-like PC12 cells, or, remarkably, even in our yeast model further suggests that the peculiar properties of these misfolded proteins are universal.
Impaired ER protein homeostasis enhances polyQ toxicity
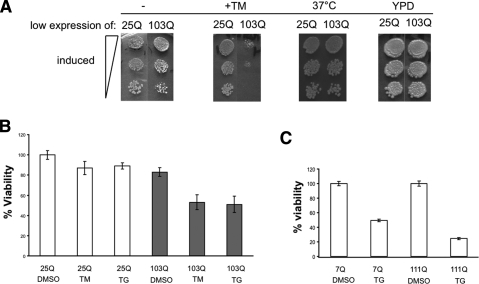
To further probe the interaction between toxic polyQ-expanded htt exonI proteins and perturbations in ER protein homeostasis, we employed chemicals that specifically disturb this process in both yeast and mammalian cells. For yeast we used a sensitized model in which we expressed the toxic 103Q htt exonI protein at a low level (using the MET13 promoter) causing only a very mild retardation of growth (Fig. 3A; Duennwald et al. 2006b). Tunicamycin (TM) inhibits the glycosylation of proteins in the ER. Very low concentrations of TM (0.5 μg/mL), which have no effect on the growth of yeast cells expressing 25Q htt exonI, caused strong synthetic toxicity with low levels of 103Q htt exonI protein (Fig. 3A).
Figure 3.
ER stress enhances polyQ toxicity. (A) TM increased the toxicity of 103Q at low expression levels in yeast. Wild-type yeast cells were spotted in three fivefold dilutions on plates that contained methionine for low expression of 25Q and 103Q and in parallel on plates that induce the low expression of 25Q and 103Q and that additionally contain 0.5 μg/mL TM. Cells expressing low levels of 25Q and 103Q htt exonI were grown at 37°C to induce heat stress (HS). Cells grown at 30°C on YPD plates served as spotting and growth control. (B) TM and TG enhance polyQ toxicity in PC12 cells. PC12 cells were induced for 8 h for the expression of 25Q and 103Q and in parallel treated with TM (0.5 μg/mL) and TG (0.5 μM) or DMSO as a vector control. Cellular viability was monitored 8 h post-polyQ induction by MTT assays. Viability of PC12-expressing 25Q htt exonI in the presence of DMSO was set to 100% and relative viabilities were calculated. The means and standard deviations (error bars) of three independent experiments are shown. (C, top panel) Striatal cells expressing full-length htt with a polyQ expansion (111Q) are sensitized to ER stress induced by TG (0.5 μM) when compared with wild-type (7Q) cells. Luciferase activity of 7Q and 111Q cells treated with DMSO (vector control) were each set as 100%. The relative luciferase activity of 7Q and 111Q cells treated with TG is shown as viability. The means and standard deviations (error bars) of three independent experiments are shown.
In PC12 cells we looked for sensitivity to TM by examining cells very shortly (6 h) after induction of 25Q or 103Q htt exonI. Low concentrations of TM (0.5 μg/mL) enhanced toxicity in PC12 cells that had been expressing 103Q htt exonI (Fig. 3B). Similarly, low concentrations (0.5 μM) of thapsigargin (TG), which induced ER stress by disturbing ER-Ca2+ levels, specifically enhanced 103Q htt exonI toxicity. Heat stress, as induced by growth at higher temperatures did not enhance polyQ toxicity in either the yeast model (Fig. 3A) or PC12 cells (data not shown).
PolyQ expansions in full-length htt induce the UPR and sensitize striatal cells to ER stress
In HD, neurons in the striatum are the most severely damaged by the polyQ-expanded htt (DiFiglia et al. 1997; Trettel et al. 2000). Recently, cell lines have been derived from the striatum of mice that are homozygous for either wild-type (7Q) htt or 111Q expansion gene replacements. We could not directly examine defects in the degradation of UFD or ERAD substrates in these cells because they are very difficult to transfect and because endogenous substrates have not been characterized in them. We did, however, ask if abnormal polyQ expansions elicited a strong UPR in these cells. Indeed, the polyQ expansion protein expressed from its own promoter in its normal chromosomal context was sufficient to cause a strong constitutive UPR. All three of the UPR proteins we examined, BiP, PDI, and CHOP (Fig. 2E), were expressed at high levels. The induction of BiP, PDI, and CHOP were not as drastic as in the PC12 model, which correlates with the less acute nature of polyQ toxicity in this model. In accordance with results in yeast and PC12 cells, the polyQ expansion in the endogenous htt protein did not induce expression of Hsp70 (Fig. 2E).
We also tested the sensitivity of striatal cells to ER stress. As reported previously, the toxicity of polyQ is most sensitively detected by reductions in ATP levels in these cells, as measured by luciferase assays (Trettel et al. 2000). We found that striatal cells were unusually sensitive to TG. Low concentrations (0.5μM) caused a reduction in ATP levels even in wild-type cells (7Q) (Fig. 3C). Cell expressing full-length polyQ-expanded allele (111Q) were about twofold more sensitive (Fig. 3C). Thus, striatal cells, already sensitive to ER stress, are further sensitized when expressing full-length polyQ-expanded huntingtin.
Genetic impairment of ER protein homeostasis enhances polyQ toxicity
Having demonstrated common features of protein homeostasis dysfunction in yeast and mammalian neuronal cells, we took advantage of yeast genetics to explore the genetic interactions of toxic polyQ-expanded htt exonI with other cellular proteins. We did not perform a genome-wide screen, because transformation of the polyQ strains produces problems with spontaneous suppressors. Instead, we took a candidate approach, testing several different cellular protein quality-control systems. Using a series of yeast strains with mutations in different cellular pathways, we searched for synthetic toxic phenotypes that would reveal specific genetic interactions with polyQ toxicity. Cells expressing 25Q htt exonI and the nontoxic 103QP served as reference points because they did not inhibit growth under any experimental conditions tested. To provide a specificity control for the toxicity due to polyQ expansions, we also examined cells expressing α-synuclein, a protein associated with Parkinson’s disease that is also toxic in yeast (Outeiro and Lindquist 2003). This protein was expressed from a single-copy plasmid at a level that caused mild toxicity, equivalent to that caused by 103Q expressed from the MET13 promoter.
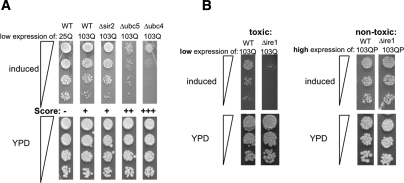
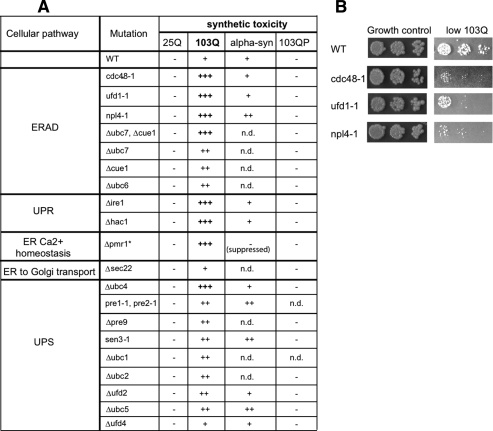
Our scoring system for synthetic toxicity is illustrated in Figure 4A, in cells carrying a deletion of the histone deacetylase SIR2 (Δsir2), the ubiquitin-conjugating enzyme UBC5 (Δubc5), and a deletion of the ubiquitin-conjugating enzyme UBC4 (Δubc4). SIR2 has no known role in protein homeostasis. UBC4 and UBC5 are highly homologous genes involved in the turnover of aberrant proteins, but they have distinct expression profiles. 25Q exhibited no toxicity in wild-type yeast cells and was scored as −. The low toxicity of 103Q htt in wild type was scored as +. The SIR2-deletion did not enhance this toxicity and was hence also scored +. The synthetic toxicity of 103Q in Δubc5 cells was scored as ++, and the more drastic synthetic toxicity of 103Q in Δubc4 was scored as +++.
Figure 4.
Genetic modulation of polyQ toxicity. (A) Wild-type yeast cells expressing 25Q htt exonI and wild type and cells carrying deletions of the genes SIR2, UBC5, and UBC4 expressing low levels of 103Q htt exonI were spotted in four fivefold dilutions on plates that induce htt expression (SD without methionine; top panel) or YPD plates as a growth and spotting control (bottom panel). 25Q exhibited no toxicity (−). The low expression levels of 103Q htt exonI causes only mild toxicity in wild-type (WT) cells (+); the deletion of SIR2 (Δsir2) exhibited the same toxicity. The deletion of UBC5 resulted in synthetic toxicity (++), and the deletion of ubc4 (Δubc4) resulted in drastically enhanced synthetic toxicity (+++). (B) Impaired ERAD and UPR genetically interact only with toxic polyQ-expanded htt exonI. Wild-type and Δire1 cells expressing either low levels of toxic103Q or high levels of the nontoxic 103QP were spotted in three fivefold dilutions on plates that induce the expression of the respective htt exonI fragments (top panel) or YPD plates as a growth and spotting control (bottom panel).
Mutations in, or deletions of, genes involved in ERAD, UPR, ER Ca2+ homeostasis, ER-to-Golgi transport, and the UPS were tested for increased polyQ toxicity. Importantly, except for Δpmr1, none of these mutations had any effect on the growth of wild-type cells under the conditions tested. Further, they did not cause toxicity in cells expressing 25Q or the nontoxic 103QP protein (Figs. 4B, 5A; data not shown). Cells expressing low levels of the toxic 103Q were most strongly affected by perturbations in genes involved in ERAD, ER calcium homeostasis, and UPR (Figs. 4B, 5A). Most perturbations in genes involved in the UPS only caused mild increase in polyQ toxicity and a deletion of a gene involved in ER-to-Golgi transport (Δsec22) did not enhance polyQ toxicity at all. Also, several deletions of cytosolic heat-shock proteins and molecular chaperones did not enhance, or only very mildly enhanced, 103Q htt exonI toxicity (e.g., HSP104, HSP26, and HSP42) (data not shown). This same pattern of synthetic toxicity was observed in cells that expressed higher levels of a less toxic polyQ-expanded htt exonI protein with a shorter expansion (46Q) (data not shown). The α-synuclein protein produced a different pattern of synthetic toxicity (Fig. 5A).
Figure 5.
Impaired ERAD and UPR enhance polyQ toxicity. (A) Wild type (WT) and cells carrying deletions or mutations of genes involved in UFD, ERAD, UPR, ER Ca2+ homeostasis, ER-to-Golgi transport, and the UPS were transformed with a vector for low expression of either 25Q or 103Q htt exonI. All yeast cells carrying deletions or mutations in the table were grown under conditions where they do not exhibit any reduced growth phenotypes (permissive temperature). The only exception (*) is the Δpmr1 strain, which shows reduced growth under all experimental conditions tested. However, the low expression of 103Q htt exonI causes the complete growth arrest of this strain, and hence it was scored (+++). The toxicity of cells expressing the nontoxic 25Q htt exonI, 103QP htt exonI and low levels of α-synuclein (mildly toxic) were listed as controls; n.d. indicates not determined. (B) Wild-type yeast cells and yeast cells carrying temperature-sensitive alleles of the indicated genes expressing low levels of 103Q htt exonI were spotted in three fivefold dilutions.
The ERAD complex p97/Npl4/Ufd1 is entrapped by toxic polyQ expansion proteins
Why is ER protein homeostasis specifically impaired by polyQ-expanded huntingtin exonI? Mutations in CDC48 (the yeast homolog of p97), NPL4, and UFD1 showed very strong synthetic toxicity with 103Q htt exonI (Fig. 5B). p97 is an abundant cytosolic protein remodeling factor of the AAA+ family that, depending on its association with different cofactors, participates in many different biological processes. In complex with Npl4 and Ufd1, p97 facilitates the transport of ERAD substrates from the ER to the cytosol and their delivery to the proteasome for degradation (Meyer et al. 2002).
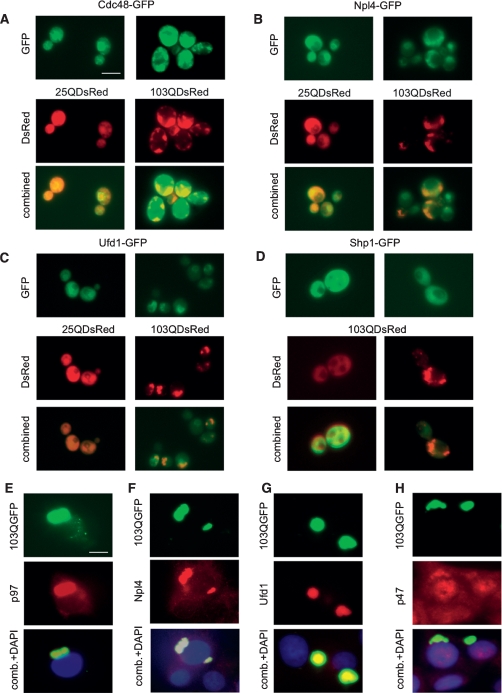
To determine whether polyQ-expanded htt exonI fragments directly interact with this protein complex, we expressed DsRed-tagged 25Q or 103Q htt exonI proteins in yeast strains that also expressed GFP-tagged Cdc48, GFP-tagged Npl4, or GFP-tagged Ufd1. As a specificity control, we examined GFP-tagged Shp1. Like Npl4 and Ufd1, this protein interacts with Cdc48. Yet it acts in vesicular transport in the Golgi and is not involved in ERAD (Ye et al. 2001). Aggregated 103Q htt exonI colocalized with Cdc48-GFP (Fig. 6A) and strongly colocalized with Npl4-GFP (Fig. 6B) and Ufd1-GFP (Fig. 6C). It did not, however, colocalize with Shp1-GFP (Fig. 6D). Cdc48-GFP also colocalized with the nontoxic version of htt exonI (103QP). In contrast, Npl4-GFP and Ufd1-GFP did not strongly colocalize with the nontoxic version of the polyQ-expanded protein, 103QP (Supplemental Fig. S8).
Figure 6.
ERAD proteins Cdc48 (p97), Ufd1, and Npl4 colocalize with polyQ-expanded huntingtin exonI in yeast and PC12 cells. (A–C) 25Q and 103Q huntingtin exonI proteins tagged with DsRed were expressed for 6 h in yeast cells expressing GFP fusion proteins of CDC48 (the yeast homolog of p97) (A), Npl4 (B), and Ufd1 (C). (D) Yeast cells coexpressing a Shp1-GFP fusion and 103QDsRed served as a specificity control. 25Q and 103Q expression was induced for 8 h and then monitored by fluorescence microscopy. p97, Ufd1, and Npl4 colocalize with htt exonI 103Q-GFP in PC12. (E–H) PC12 cells induced for 12 h for the expression of 103Q htt exonI fused to GFP were fixed and immunostained with anti-p97 (E), anti-Npl4 (F), anti-Ufd1 (G), and anti-p47 (H) (the homolog of yeast Shp1 served as specificity control), respectively, followed by fluorescence microcopy. Bars, 10 μm.
To test whether these specific associations also occurred in mammalian neuronal cells, we again used PC12 cells expressing either 25Q or 103Q htt exonI. Here, too, p97, Npl4, and Ufd1 strongly colocalized with aggregated 103Q htt exonI-GFP (Fig. 6E–G). The mammalian homolog of yeast Shp1, p47, only weakly colocalized with 103Q htt exonI-GFP (Fig. 6H) in PC12 cells. Thus, the p97/Npl4/Ufd1 complex seemed to be trapped in aggregates of 103Q htt exonI in both yeast and PC12 cells. To confirm the specific association of p97, Ufd1, and Npl4, we performed coimmunoprecipitation experiments on PC12 cells (Supplemental Fig. S9). All three proteins coimmunoprecipitated with the toxic 103Q htt exonI protein but not with the nontoxic 25Q htt exonI. Further p47, the protein that interacts with p97 yet is not involved in ERAD, did not coimmunoprecipitate with 103Q htt exonI.
The entrapment of the p97/Npl4/Ufd1 complex contributes to toxicity
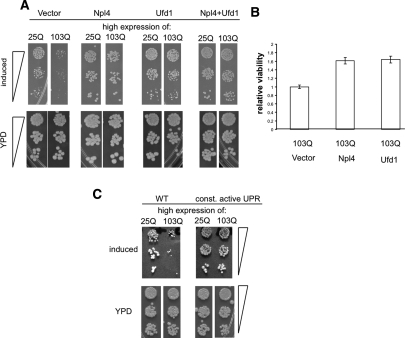
p97 is an essential and very abundant protein in both yeast and mammalian cells. In contrast, the adaptor proteins Npl4 and Ufd1 are essential yet relatively scarce (Ghaemmaghami et al. 2003). Given the importance of these proteins in ERAD and the strong and immediate effects of toxic polyQ expansions on ERAD and ER stress, we speculated that the entrapment of Npl4 and Ufd1 by 103Q htt exonI contributed to its toxicity. To test this, we constitutively overexpressed Npl4 and Ufd1 and a combination of the two in yeast cells expressing the inducible toxic 103Q htt exonI. Ufd1 and Npl4 overexpression individually, or in combination, effectively suppressed polyQ toxicity (Fig. 7A). Importantly, the overexpression of numerous other stress proteins, including ubiquitin and ubiquitin-conjugating enzymes, did not suppress polyQ toxicity (Supplemental Fig. S7; K. Matlack and S. Lindquist, unpubl.).
Figure 7.
Npl4 and Ufd1 and a constitutive active UPR suppressed polyQ toxicity. (A) Yeast cells expressing either high levels of 25Q or 103Q huntingtin exonI and expressing a vector control, or constitutively expressing extra copies of Npl4, Ufd1, and Npl4 and Ufd1 combined were spotted on plates that induce htt exonI or YPD plates as growth and spotting controls. (B) PC12 cells were transfected with a vector control, a plasmid for the constitutive expression of Npl4, and a plasmid for expression of Ufd1. Four days post-transfection 103Q htt exonI expression was induced for 72 h, and polyQ toxicity was monitored by MTT assays. The viability of PC12 cell expressing 103Q htt exonI and a vector control was set as 1 and the relative viabilities were calculated. The means and standard deviations (error bars) of three independent experiments are shown. (C) Constitutively active UPR antagonizes polyQ toxicity. Wild-type yeast cells or yeast cells expressing a constitutively active allele of Hac1 (const. active UPR) were spotted on plates that induce the expression of 25Q or 103Q htt exonI and YPD plates as spotting and growth controls.
Similarly, Npl4 and Ufd1 expression reduced polyQ toxicity in PC12 cells (Fig. 7B). The suppression of toxicity achieved by Npl4 and Ufd1 expression was an underestimate of their efficacy because only ∼80% of the PC12 cells were successfully transfected (data not shown). We were not able to determine the effect of combined overexpression of Ufd1 and Npl4 in PC12 cells because of very low cotransfection efficiencies. Importantly, the overexpression of Npl4 and Ufd1 did not reduce the expression of the polyQ proteins in yeast or in PC12 cells, nor did it change the morphology of aggregates (data not shown). The effects of Ufd1 and Npl4 were specific to polyQ-mediated toxicity, because α-synuclein-induced toxicity was not antagonized by the same proteins (data not shown).
Next, we asked whether the toxicity of polyQ could also be ameliorated by an enhanced UPR. Unlike the more complexly regulated mammalian UPR, the simple signaling cascade of the yeast UPR can be genetically manipulated so that it becomes fully constitutively active using a mutant version of the central UPR transcription factor Hac1(Papa et al. 2003). When the toxic 103Q htt exonI protein was expressed in yeast cells with a constitutively active UPR the morphology of 103Q htt exonI aggregates remained unaltered (data not shown). However, the constitutively active UPR greatly reduced the toxicity of 103Q htt exonI (Fig. 7C).
Discussion
We investigated how polyQ-expanded huntingtin affects protein turnover, a pillar of protein quality control, which is one of the first lines of defense against protein misfolding and its toxic consequences. It is also a system that is severely and broadly challenged by misfolded proteins (Gidalevitz et al. 2006), initiating a vicious cycle of increasing toxicity in protein-misfolding disorders. Thus, the study of protein quality control in neurodegenerative diseases caused by protein misfolding holds great promise to decipher the molecular mechanisms underlying these maladies and also to inform effective therapeutic approaches. Using a combination of yeast and neuronal models we find that polyQ-expanded htt specifically and strongly impairs the turnover of ER proteins (ERAD).
The impairment of ERAD and the ensuing accumulation of ERAD substrates induces stress in the ER, elicits the UPR, and sensitizes cells to other forms of ER stress. Notably, several other studies have documented polyQ-induced UPR and ER stress in the context of different polyQ expansion proteins (Kouroku et al. 2002; Nishitoh et al. 2002; Thomas et al. 2005). The ER, however, remained an unexplained nexus for polyQ toxicity because the misfolding of polyQ-expanded proteins occurs in the cytosol or the nucleus but not in the ER. Our findings elucidate this baffling connection between ER stress and polyQ toxicity: The entrapment of the ERAD complex p97/Npl4/Ufd1 connects polyQ-mediated protein misfolding in the cytosol with toxic defects in protein homeostasis in the ER. Importantly, the fact that overexpression of Npl4 and Ufd1 ameliorate toxicity establishes that this entrapment contributes to the toxicity of the polyQ-expanded protein. Thus, our study provides a molecular mechanism underpinning the cross-talk between different cellular compartments in response to misfolded polyQ proteins. We suggest that it might be fruitful to investigate cellular compartments or pathways that—at least at the first glimpse—have no physical contact with the misfolded protein in the analyses of other types of protein misfolding toxicities. We also found that striatal cells, at least in tissue culture, are especially sensitive to ER stress. If this is true in vivo, it may contribute to the selective vulnerability of this cell type in HD.
The defect in protein homeostasis is highly selective for ERAD and was the earliest cellular response to the expression of toxic polyQ-expanded htt that we detected, earlier than defects in degradation of polyubiquitinated proteins and induction of a HSR. Our genetic data further identify defects in ERAD and ER stress as central events in polyQ toxicity. Moreover, we show that polyQ-induced defects in ER homeostasis are conserved between cells of organisms as diverse as yeast and striatal neurons and occur not only in response to the expression of polyQ-expanded htt fragments but also in response to the expression of full-length polyQ-expanded htt in striatal cells. Importantly, neurons in the striatum are most severely affected in HD. Our results therefore add ER protein homeostasis to other cellular pathways whose dysfunction contributes to htt polyQ toxicity.
How might the polyQ-expanded protein lead to the entrapment of an ERAD protein complex? Recently, polyQ-expanded SCA3 (ataxin3), the protein that causes Machado-Joseph disease, was shown to cause a defect in ERAD (Wang et al. 2006; Zhong and Pittman 2006). In this case, there is a logical connection because the normal function of SCA3 is to regulate the pace of ERAD. Through a beautiful combination of in vitro and in vivo investigations (Boeddrich et al. 2006; Zhong and Pittman 2006), it was determined that SCA3 binds p97 and inhibits its interaction with Npl4 and Ufd1, interfering with the degradation of ERAD substrates by the proteasome. PolyQ-expanded SCA3 competes with Npl4 and Ufd1 in binding to p97 and reduces the pool of the p97/Ufd1/Npl4 complex that functions in ERAD (Zhong and Pittman 2006). In contrast, there is no evidence that huntingtin or huntingtin fragments participate in ERAD as part of their normal physiological function. Yet we observe that polyQ-expanded htt exon1 selectively binds and inhibits the same complex and, moreover, that this is one of the early events in toxicity. This must be more than coincidence.
It has been determined that an arginine/lysine-rich motif in the SCA3 protein that is N-terminal to the polyQ region interacts with p97 and is essential for its normal activity during ERAD (Boeddrich et al. 2006; Zhong and Pittman 2006). It has been speculated that the polyQ expansion of this protein causes a conformational change that presents this motif to p97 more effectively, allowing the polyQ-expanded protein to outcompete the wild-type protein and inhibit ERAD. An alternative explanation, suggested by our findings, is that the short polyQ region in the wild-type protein directly contributes to the interaction with Npl4 and Ufd1 with p97 and that the polyQ-expanded version of SCA3 stabilizes it even further. The toxicity of the htt protein in our experiments is relieved by overexpression of Npl4 and Ufd1. The polyQ expansion in htt might act by entrapping Sca3. If so, it might represent a common (albeit not exclusive) toxic mechanism for toxic polyQ expansion proteins that causes ER stress.
Surprisingly, the HSR, the most general defense against protein misfolding in the cytosol, is not activated early by polyQ-expanded htt exonI or full-length htt in our models. The same finding has been reported in neurons affected in polyQ toxicity (Hay et al. 2004; Tagawa et al. 2007). Yet the expression of heat-shock proteins has been shown to provide effective protection against polyQ toxicity (Warrick et al. 1999; Carmichael et al. 2000; Muchowski et al. 2000; Wyttenbach et al. 2000, 2002; Bao et al. 2002; Barral et al. 2004; Muchowski and Wacker 2005; Vacher et al. 2005). The failure to induce the most important response to defects in cytosolic protein homeostasis might be another contributor to the toxicity of polyQ expansion proteins. We do not know why the misfolding of polyQ-expanded htt fails to promptly activate the HSR. Perhaps cells do not recognize polyQ-expanded htt as a misfolded species because of intrinsic structural features of polyQ expansion proteins. In addition, polyQ can entrap certain transcription factors (Schaffar et al. 2004; Riley and Orr 2006), and might differentially affect particular transcriptional responses, such as the HRR. We and others have found recently that the neurons most severely affected by polyQ expansions have the most reduced levels of heat-shock proteins (Hay et al. 2004; Tagawa et al. 2007; M.L. Duennwald and S. Lindquist, in prep.). Methods for activating both the UPR and the HSR might therefore provide a novel and effective combinatorial therapeutic strategy to combat polyQ expansion disorders.
Materials and methods
Antibodies
The following antibodies were used in this study: anti-p97 and anti-Ufd1 from BD Biosciences; anti-Npl4 and anti-p47 antibodies were a kind gift of Hemmo Meier (ETH, Zurich, Switzerland); anti-BiP, anti-Hsp70, and anti-PDI from Stressgen; anti-CHOP from Affinity BioReagents; anti-β-galactosidase antibody from Cappel (MP Biomedicals), anti-ubiquitin antibody from BabCo, Covance. Secondary antibodies (anti-rabbit and anti-mouse antibodies coupled to horseradish peroxidase) and anti-GFP antibody were purchased from Sigma-Aldrich.
Chemicals
TG and TM were purchased from Sigma-Aldrich.
General yeast methods
Yeast strains expressing the huntingtin exonI GFP/CFP fusion proteins were described in our previous work (Duennwald et al. 2006b). Yeast media were prepared as described previously (Duennwald et al. 2006b). If not stated otherwise, yeast cells were grown in liquid cultures with medium agitation or on plates at 30°C. Transformation of yeast with plasmid DNA was performed by the lithium acetate method (Ito et al. 1983). Proteins extraction from yeast was performed by the glass bead method.
Yeast growth assays
For growth assays, yeast cells were grown overnight in selective media with 2% glucose as a sole carbon source. After the OD600 was determined, cultures were diluted to equal concentrations and then spotted in three or four fivefold dilutions with the most concentrated spot containing 100,000 cells by using a spotter (Frogger; V&P Scientific). Equal spotting was controlled by simultaneously spotting the cells on YPD (yeast/extract/ peptone/dextrose) plates.
Yeast β-galactosidase activity assays
Visualization of β-galactosidase activity was achieved by an overlay assay. Yeast cells were grown overnight in medium containing 2% raffinose as sole carbon source. The cells were then diluted to an OD600 of 0.2 in medium containing 2% galactose as carbon source and then grown for 6 h at 30°C. Cultures were diluted to equal concentrations and spotted onto plates containing glucose as sole carbon source with one spot containing ∼106 cells. Once the spots dried a mixture of a warm (55°C) agarose mix (6% [v/v] DMF, 0.1% [v/v] SDS, 0.1 mg/mL X-Gal, 0.5% [w/v] agarose) were poured on top of the plates. After the agarose mix solidified, the plates were incubated for 8 h at 30°C before pictures where taken. For quantitative β-galactosidase assays cells were grown exactly as described for the overlay assay. β-Galactosidase activity was then determined by permeabilizing yeast cells in a mild lysis buffer and then measuring the luminescence with the Galacto-Light kit (Tropix, Applied Biosciences).
Tissue culture methods
PC12-expressing huntingtin exonI GFP fusions were a kind gift from Eric Schweitzer (University of California at Los Angeles, Los Angeles, CA). Cells were grown and differentiated, and huntingtin exonI GFP expression was induced as described previously (Aiken et al. 2004). Transfection of PC12 cells was carried out using the Nucleofector kit from Amaxa, using buffer V and following the manufacturer’s protocol precisely with the exception that 10% more cells were used per transfection. The plasmids for the expression of Npl4, Ufd1, and p97 under control of the CMV promoter were purchased from Origen.
Striatal cells expressing full-length huntingtin with either seven or 111 glutamines under the control of the endogenous huntingtin promoter were a kind gift from Marcy MacDonald (MGH, Boston, MA). Striatal cells were grown as described previously (Trettel et al. 2000). For all assays presented in this study, cells were grown at 39°C to block cell division.
Protein extraction from PC12 cells and striatal cells
Cells were washed with PBS and then harvested by scraping them off in lysis buffer (150 mM NaCl, 1% NP-40, 10 mM EDTA) containing complete protease inhibitor cocktail (Roche). Cells were then passed through a small syringe 10 times. After a mild clarifying spin (500g for 5 min), the lysates were mixed with equal volumes of 2× SDS sample buffer (100 mM Tris at pH 6.8, 4% [v/v] β-mercaptoethanol, 4% [w/v] SDS, 20% [v/v] glycerol, 0.1% [w/v] bromophenol blue) and boiled for 5 min. In parallel, aliquots of the protein lysates were used to perform BCA protein concentration assays (see below) to guarantee equal loading on SDS-PAGE.
Viability assays
To assay the viability of cells either MTT assays (ATCC) or luciferase assays (CellTiter-Glo Luminescent Cell Viability Assay, Promega) and caspase activity assays (Caspase-Glo 3/7 Assay, Promega) were performed in 96-well plates using 2000 cells per well. Data from viability assays represent at least three independent experiments carried out in triplicate for each experimental condition. The error bars represent standard deviations.
Western blot
For SDS-PAGE, protein concentrations of different samples of one experiment were equalized according to BCA assays (Pierce). Proteins were transferred to PVDF membranes by semi-dry blotting. As a control for equal loading membranes were stained with Ponceau-S after blotting (see Supplemental Figs. S3–S5).
Microscopy
Microscopic experiments were carried out with a Zeiss Axioplan II microscope operating with Openlab software. Pictures were taken using Metamorph. Pictures of yeast cells were taken using a 100× objective. PC12 cells were fixed as described previously for striatal cells (Trettel et al. 2000). Pictures of PC12 cells were taken using a 40× objective. Pictures were processed with Adobe Photoshop.
Acknowledgments
We thank M. Sherman, A. Varshavsky, E. Johnsson, R. Hampton, N. Dantuma, A. Weissman, S. Jentsch, T. Sommer, E. Schweitzer, P. Walter, M. MacDonlad, and A. Cooper for yeast strains, plasmids, or cells, and H. Meyer for antibodies. We are grateful to J. Shorter and K. Allendoerfer for insightful comments on the manuscript. M.L.D. was supported by a post-doctoral fellowship from the Huntington’s Disease Society of America. This work was supported by an Ellison Medical Foundation Senior Scholar Award to S.L. S.L. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1673408.
References
- Aiken C.T., Tobin A.J., Schweitzer E.S. A cell-based screen for drugs to treat Huntington’s disease. Neurobiol. Dis. 2004;16:546–555. doi: 10.1016/j.nbd.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Apostol B.L., Illes K., Pallos J., Bodai L., Wu J., Strand A., Schweitzer E.S., Olson J.M., Kazantsev A., Marsh J.L., et al. Mutant huntingtin alters MAPK signaling pathways in PC12 and striatal cells: ERK1/2 protects against mutant huntingtin-associated toxicity. Hum. Mol. Genet. 2006;15:273–285. doi: 10.1093/hmg/ddi443. [DOI] [PubMed] [Google Scholar]
- Bao Y.P., Cook L.J., O'Donovan D., Uyama E., Rubinsztein D.C. Mammalian, yeast, bacterial, and chemical chaperones reduce aggregate formation and death in a cell model of oculopharyngeal muscular dystrophy. J. Biol. Chem. 2002;277:12263–12269. doi: 10.1074/jbc.M109633200. [DOI] [PubMed] [Google Scholar]
- Barral J.M., Broadley S.A., Schaffar G., Hartl F.U. Roles of molecular chaperones in protein misfolding diseases. Semin. Cell Dev. Biol. 2004;15:17–29. doi: 10.1016/j.semcdb.2003.12.010. [DOI] [PubMed] [Google Scholar]
- Bence N.F., Sampat R.M., Kopito R.R. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- Bennett E.J., Shaler T.A., Woodman B., Ryu K.Y., Zaitseva T.S., Becker C.H., Bates G.P., Schulman H., Kopito R.R. Global changes to the ubiquitin system in Huntington’s disease. Nature. 2007;448:704–708. doi: 10.1038/nature06022. [DOI] [PubMed] [Google Scholar]
- Bernales S., Papa F.R., Walter P. Intracellular signaling by the unfolded protein response. Annu. Rev. Cell Dev. Biol. 2006;22:487–508. doi: 10.1146/annurev.cellbio.21.122303.120200. [DOI] [PubMed] [Google Scholar]
- Boeddrich A., Gaumer S., Haacke A., Tzvetkov N., Albrecht M., Evert B.O., Muller E.C., Lurz R., Breuer P., Schugardt N., et al. An arginine/lysine-rich motif is crucial for VCP/p97-mediated modulation of ataxin-3 fibrillogenesis. EMBO J. 2006;25:1547–1558. doi: 10.1038/sj.emboj.7601043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun S., Matuschewski K., Rape M., Thoms S., Jentsch S. Role of the ubiquitin-selective CDC48(UFD1/NPL4)chaperone (segregase) in ERAD of OLE1 and other substrates. EMBO J. 2002;21:615–621. doi: 10.1093/emboj/21.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael J., Chatellier J., Woolfson A., Milstein C., Fersht A.R., Rubinsztein D.C. Bacterial and yeast chaperones reduce both aggregate formation and cell death in mammalian cell models of Huntington’s disease. Proc. Natl. Acad. Sci. 2000;97:9701–9705. doi: 10.1073/pnas.170280697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantuma N.P., Lindsten K., Glas R., Jellne M., Masucci M.G. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat. Biotechnol. 2000;18:538–543. doi: 10.1038/75406. [DOI] [PubMed] [Google Scholar]
- DiFiglia M., Sapp E., Chase K.O., Davies S.W., Bates G.P., Vonsattel J.P., Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- Duennwald M.L., Jagadish S., Giorgini F., Muchowski P.J., Lindquist S. A network of protein interactions determines polyglutamine toxicity. Proc. Natl. Acad. Sci. 2006a;103:11051–11056. doi: 10.1073/pnas.0604548103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duennwald M.L., Jagadish S., Muchowski P.J., Lindquist S. Flanking sequences profoundly alter polyglutamine toxicity in yeast. Proc. Natl. Acad. Sci. 2006b;103:11045–11050. doi: 10.1073/pnas.0604547103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S., Huh W.K., Bower K., Howson R.W., Belle A., Dephoure N., O’Shea E.K., Weissman J.S. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Gidalevitz T., Ben-Zvi A., Ho K.H., Brignull H.R., Morimoto R.I. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 2006;311:1471–1474. doi: 10.1126/science.1124514. [DOI] [PubMed] [Google Scholar]
- Gunawardena S., Goldstein L.S. Polyglutamine diseases and transport problems: Deadly traffic jams on neuronal highways. Arch. Neurol. 2005;62:46–51. doi: 10.1001/archneur.62.1.46. [DOI] [PubMed] [Google Scholar]
- Hampton R.Y. ER-associated degradation in protein quality control and cellular regulation. Curr. Opin. Cell Biol. 2002;14:476–482. doi: 10.1016/s0955-0674(02)00358-7. [DOI] [PubMed] [Google Scholar]
- Hay D.G., Sathasivam K., Tobaben S., Stahl B., Marber M., Mestril R., Mahal A., Smith D.L., Woodman B., Bates G.P. Progressive decrease in chaperone protein levels in a mouse model of Huntington’s disease and induction of stress proteins as a therapeutic approach. Hum. Mol. Genet. 2004;13:1389–1405. doi: 10.1093/hmg/ddh144. [DOI] [PubMed] [Google Scholar]
- Haynes C.M., Caldwell S., Cooper A.A. An HRD/DER-independent ER quality control mechanism involves Rsp5p-dependent ubiquitination and ER-Golgi transport. J. Cell Biol. 2002;158:91–101. doi: 10.1083/jcb.200201053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg C.I., Staniszewski K.E., Mensah K.N., Matouschek A., Morimoto R.I. Inefficient degradation of truncated polyglutamine proteins by the proteasome. EMBO J. 2004;23:4307–4318. doi: 10.1038/sj.emboj.7600426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E.S., Ma P.C., Ota I.M., Varshavsky A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 1995;270:17442–17456. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- Kirk N., Piper P.W. The determinants of heat-shock element-directed lacZ expression in Saccharomyces cerevisiae. Yeast. 1991;7:539–546. doi: 10.1002/yea.320070602. [DOI] [PubMed] [Google Scholar]
- Kouroku Y., Fujita E., Jimbo A., Kikuchi T., Yamagata T., Momoi M.Y., Kominami E., Kuida K., Sakamaki K., Yonehara S., et al. Polyglutamine aggregates stimulate ER stress signals and caspase-12 activation. Hum. Mol. Genet. 2002;11:1505–1515. doi: 10.1093/hmg/11.13.1505. [DOI] [PubMed] [Google Scholar]
- Krobitsch S., Lindquist S. Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc. Natl. Acad. Sci. 2000;97:1589–1594. doi: 10.1073/pnas.97.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laney J.D., Mobley E.F., Hochstrasser M. The short-lived Matα2 transcriptional repressor is protected from degradation in vivo by interactions with its corepressors Tup1 and Ssn6. Mol. Cell. Biol. 2006;26:371–380. doi: 10.1128/MCB.26.1.371-380.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S. The heat-shock response. Annu. Rev. Biochem. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- Meriin A.B., Zhang X., He X., Newnam G.P., Chernoff Y.O., Sherman M.Y. Huntington toxicity in yeast model depends on polyglutamine aggregation mediated by a prion-like protein Rnq1. J. Cell Biol. 2002;157:997–1004. doi: 10.1083/jcb.200112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer H.H., Wang Y., Warren G. Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1–Npl4. EMBO J. 2002;21:5645–5652. doi: 10.1093/emboj/cdf579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchowski P.J., Wacker J.L. Modulation of neurodegeneration by molecular chaperones. Natl. Rev. 2005;6:11–22. doi: 10.1038/nrn1587. [DOI] [PubMed] [Google Scholar]
- Muchowski P.J., Schaffar G., Sittler A., Wanker E.E., Hayer-Hartl M.K., Hartl F.U. Hsp70 and hsp40 chaperones can inhibit self-assembly of polyglutamine proteins into amyloid-like fibrils. Proc. Natl. Acad. Sci. 2000;97:7841–7846. doi: 10.1073/pnas.140202897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitoh H., Matsuzawa A., Tobiume K., Saegusa K., Takeda K., Inoue K., Hori S., Kakizuka A., Ichijo H. ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes & Dev. 2002;16:1345–1355. doi: 10.1101/gad.992302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H.T., Zoghbi H.Y. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- Outeiro T.F., Lindquist S. Yeast cells provide insight into α-synuclein biology and pathobiology. Science. 2003;302:1772–1775. doi: 10.1126/science.1090439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa F.R., Zhang C., Shokat K., Walter P. Bypassing a kinase activity with an ATP-competitive drug. Science. 2003;302:1533–1537. doi: 10.1126/science.1090031. [DOI] [PubMed] [Google Scholar]
- Patil C., Walter P. Intracellular signaling from the endoplasmic reticulum to the nucleus: The unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 2001;13:349–355. doi: 10.1016/s0955-0674(00)00219-2. [DOI] [PubMed] [Google Scholar]
- Raasi S., Wolf D.H. Ubiquitin receptors and ERAD: A network of pathways to the proteasome. Semin. Cell Dev. Biol. 2007;18:780–791. doi: 10.1016/j.semcdb.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Riley B.E., Orr H.T. Polyglutamine neurodegenerative diseases and regulation of transcription: Assembling the puzzle. Genes & Dev. 2006;20:2183–2192. doi: 10.1101/gad.1436506. [DOI] [PubMed] [Google Scholar]
- Schaffar G., Breuer P., Boteva R., Behrends C., Tzvetkov N., Strippel N., Sakahira H., Siegers K., Hayer-Hartl M., Hartl F.U. Cellular toxicity of polyglutamine expansion proteins: Mechanism of transcription factor deactivation. Mol. Cell. 2004;15:95–105. doi: 10.1016/j.molcel.2004.06.029. [DOI] [PubMed] [Google Scholar]
- Schroder M., Kaufman R.J. The mammalian unfolded protein response. Annu. Rev. Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- Tagawa K., Marubuchi S., Qi M.L., Enokido Y., Tamura T., Inagaki R., Murata M., Kanazawa I., Wanker E.E., Okazawa H. The induction levels of heat shock protein 70 differentiate the vulnerabilities to mutant huntingtin among neuronal subtypes. J. Neurosci. 2007;27:868–880. doi: 10.1523/JNEUROSCI.4522-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M., Yu Z., Dadgar N., Varambally S., Yu J., Chinnaiyan A.M., Lieberman A.P. The unfolded protein response modulates toxicity of the expanded glutamine androgen receptor. J. Biol. Chem. 2005;280:21264–21271. doi: 10.1074/jbc.M500144200. [DOI] [PubMed] [Google Scholar]
- Tiwari S., Weissman A.M. Endoplasmic reticulum (ER)-associated degradation of T cell receptor subunits. Involvement of ER-associated ubiquitin-conjugating enzymes (E2s) J. Biol. Chem. 2001;276:16193–16200. doi: 10.1074/jbc.M007640200. [DOI] [PubMed] [Google Scholar]
- Travers K.J., Patil C.K., Wodicka L., Lockhart D.J., Weissman J.S., Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Trettel F., Rigamonti D., Hilditch-Maguire P., Wheeler V.C., Sharp A.H., Persichetti F., Cattaneo E., MacDonald M.E. Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum. Mol. Genet. 2000;9:2799–2809. doi: 10.1093/hmg/9.19.2799. [DOI] [PubMed] [Google Scholar]
- Vacher C., Garcia-Oroz L., Rubinsztein D.C. Overexpression of yeast hsp104 reduces polyglutamine aggregation and prolongs survival of a transgenic mouse model of Huntington’s disease. Hum. Mol. Genet. 2005;14:3425–3433. doi: 10.1093/hmg/ddi372. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. The N-end rule. Cell. 1992;69:725–735. doi: 10.1016/0092-8674(92)90285-k. [DOI] [PubMed] [Google Scholar]
- Venkatraman P., Wetzel R., Tanaka M., Nukina N., Goldberg A.L. Eukaryotic proteasomes cannot digest polyglutamine sequences and release them during degradation of polyglutamine-containing proteins. Mol. Cell. 2004;14:95–104. doi: 10.1016/s1097-2765(04)00151-0. [DOI] [PubMed] [Google Scholar]
- Wang Q., Li L., Ye Y. Regulation of retrotranslocation by p97-associated deubiquitinating enzyme ataxin-3. J. Cell Biol. 2006;174:963–971. doi: 10.1083/jcb.200605100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrick J.M., Chan H.Y., Gray-Board G.L., Chai Y., Paulson H.L., Bonini N.M. Suppression of polyglutamine-mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat. Genet. 1999;23:425–428. doi: 10.1038/70532. [DOI] [PubMed] [Google Scholar]
- Wyttenbach A., Carmichael J., Swartz J., Furlong R.A., Narain Y., Rankin J., Rubinsztein D.C. Effects of heat shock, heat shock protein 40 (HDJ-2), and proteasome inhibition on protein aggregation in cellular models of Huntington’s disease. Proc. Natl. Acad. Sci. 2000;97:2898–2903. doi: 10.1073/pnas.97.6.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyttenbach A., Sauvageot O., Carmichael J., Diaz-Latoud C., Arrigo A.P., Rubinsztein D.C. Heat shock protein 27 prevents cellular polyglutamine toxicity and suppresses the increase of reactive oxygen species caused by huntingtin. Hum. Mol. Genet. 2002;11:1137–1151. doi: 10.1093/hmg/11.9.1137. [DOI] [PubMed] [Google Scholar]
- Ye Y., Meyer H.H., Rapoport T.A. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- Zhong X., Pittman R.N. Ataxin-3 binds VCP/p97 and regulates retrotranslocation of ERAD substrates. Hum. Mol. Genet. 2006;15:2409–2420. doi: 10.1093/hmg/ddl164. [DOI] [PubMed] [Google Scholar]