Abstract
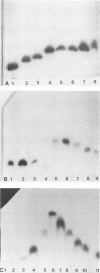
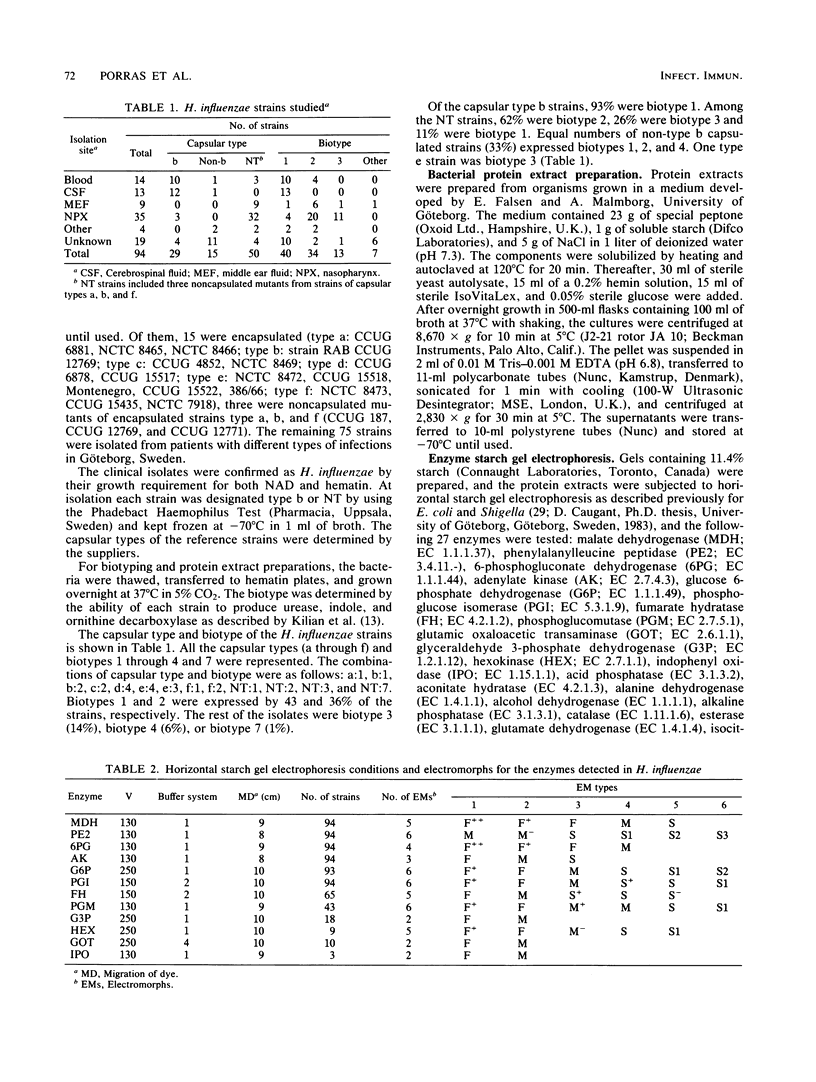
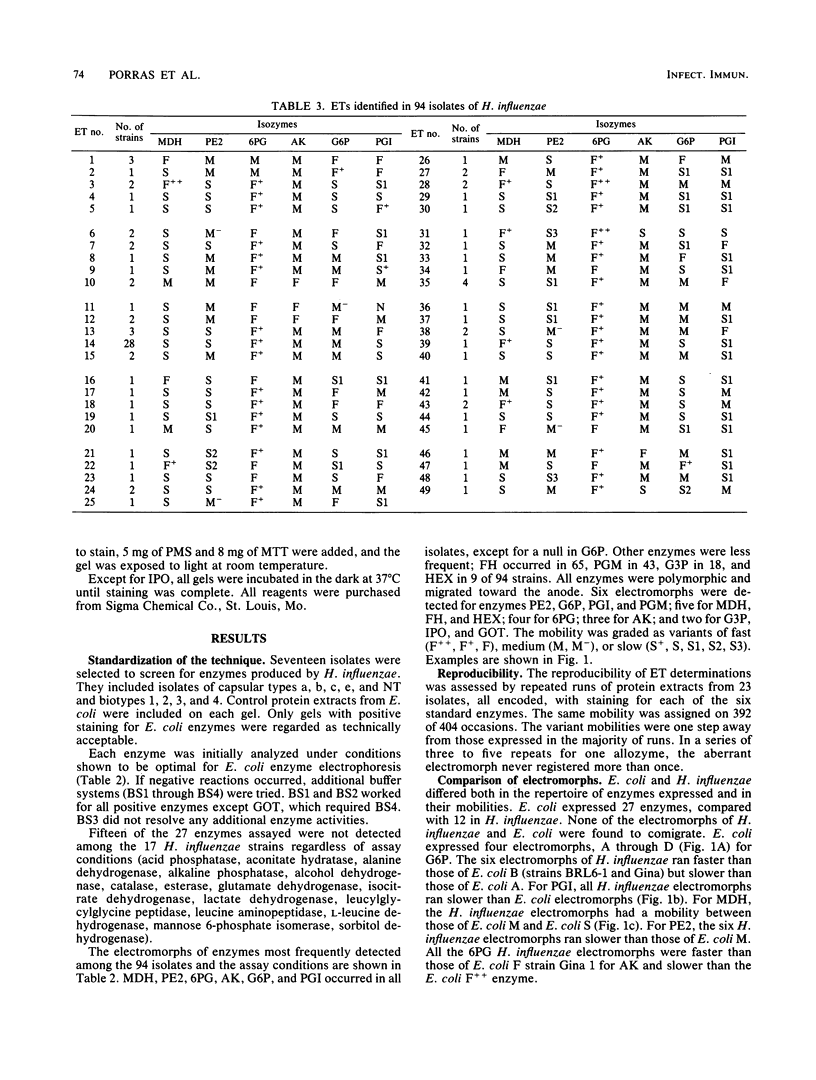
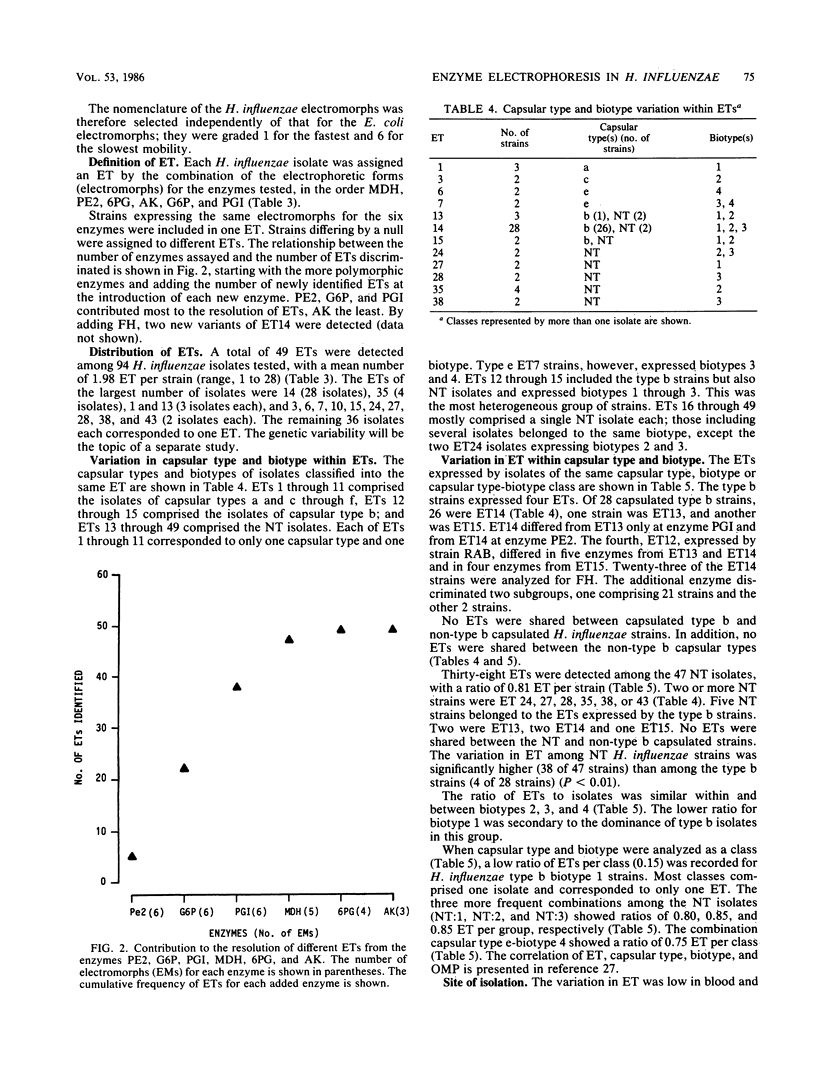
Multilocus enzyme electrophoresis was adapted to the study of Haemophilus influenzae. Protein extracts from sonicated whole bacteria were subjected to starch gel electrophoresis. After staining with substrates, the position of each isoenzyme (electromorph) was registered. Each isolate was assigned an electrophoretic type (ET) by the combination of electromorphs for the enzymes stained. Twenty-seven enzymes were tested; 12 were expressed in H. influenzae. Six enzymes were selected for subsequent study: malate dehydrogenase (MDH), phenylalanylleucine peptidase (PE2), 6-phosphogluconate dehydrogenase (6PG), adenylate kinase (AK), glucose 6-phosphate dehydrogenase (G6P), and phosphoglucose isomerase (PGI). They were polymorphic and occurred in all isolates. Six electromorphs were found for PE2, G6P, and PGI, five for MDH, four for 6PG, and three for AK. PE2, G6P, and PGI contributed most of the ET resolution (48 of 49 ETs). Multilocus enzyme electrophoresis showed several advantages over previous typing techniques. An ET could be assigned to both typable and nontypable (NT) isolates. The technique was powerful in resolving differences among isolates. The 94 isolates comprised 49 ETs, five biotypes, and six capsular types and NT isolates. Strains known to be related expressed the same ET, e.g., RAB b+ and b-, ET12; Ma a+ and a-, ET1. ET variability among type b isolates was low; 26 of 28 clinical isolates expressed ET14; 2 of 28 expressed ET13 and ET15, differing from ET14 by one electromorph each. In contrast, the 47 NT isolates comprised 38 different ETs. No ETs were shared between non-type b capsulated strains and type b or NT strains. Interestingly, five NT isolates expressed the same ET as type b strains. (iv) Strains of the same capsular type but different biotypes expressed different ETs. ET determinations will thus be useful in studying the epidemiology and evolution of H. influenzae.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Achtman M., Mercer A., Kusecek B., Pohl A., Heuzenroeder M., Aaronson W., Sutton A., Silver R. P. Six widespread bacterial clones among Escherichia coli K1 isolates. Infect Immun. 1983 Jan;39(1):315–335. doi: 10.1128/iai.39.1.315-335.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J., Low K. B. Linkage map of Escherichia coli K-12, edition 6. Microbiol Rev. 1980 Mar;44(1):1–56. doi: 10.1128/mr.44.1.1-56.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barenkamp S. J., Munson R. S., Jr, Granoff D. M. Outer membrane protein and biotype analysis of pathogenic nontypable Haemophilus influenzae. Infect Immun. 1982 May;36(2):535–540. doi: 10.1128/iai.36.2.535-540.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barenkamp S. J., Munson R. S., Jr, Granoff D. M. Subtyping isolates of Haemophilus influenzae type b by outer-membrane protein profiles. J Infect Dis. 1981 May;143(5):668–676. doi: 10.1093/infdis/143.5.668. [DOI] [PubMed] [Google Scholar]
- Brenner D. J., Fanning G. R., Skerman F. J., Falkow S. Polynucleotide sequence divergence among strains of Escherichia coli and closely related organisms. J Bacteriol. 1972 Mar;109(3):953–965. doi: 10.1128/jb.109.3.953-965.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caugant D. A., Levin B. R., Orskov I., Orskov F., Svanborg Eden C., Selander R. K. Genetic diversity in relation to serotype in Escherichia coli. Infect Immun. 1985 Aug;49(2):407–413. doi: 10.1128/iai.49.2.407-413.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caugant D. A., Levin B. R., Selander R. K. Genetic diversity and temporal variation in the E. coli population of a human host. Genetics. 1981 Jul;98(3):467–490. doi: 10.1093/genetics/98.3.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris H. Enzyme polymorphisms in man. Proc R Soc Lond B Biol Sci. 1966 Mar 22;164(995):298–310. doi: 10.1098/rspb.1966.0032. [DOI] [PubMed] [Google Scholar]
- Hoiseth S. K., Connelly C. J., Moxon E. R. Genetics of spontaneous, high-frequency loss of b capsule expression in Haemophilus influenzae. Infect Immun. 1985 Aug;49(2):389–395. doi: 10.1128/iai.49.2.389-395.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubby J. L., Lewontin R. C. A molecular approach to the study of genic heterozygosity in natural populations. I. The number of alleles at different loci in Drosophila pseudoobscura. Genetics. 1966 Aug;54(2):577–594. doi: 10.1093/genetics/54.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inzana T. J. Electrophoretic heterogeneity and interstrain variation of the lipopolysaccharide of Haemophilus influenzae. J Infect Dis. 1983 Sep;148(3):492–499. doi: 10.1093/infdis/148.3.492. [DOI] [PubMed] [Google Scholar]
- Kilian M., Sørensen I., Frederiksen W. Biochemical characteristics of 130 recent isolates from Haemophilus influenzae meningitis. J Clin Microbiol. 1979 Mar;9(3):409–412. doi: 10.1128/jcm.9.3.409-412.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin R. C., Hubby J. L. A molecular approach to the study of genic heterozygosity in natural populations. II. Amount of variation and degree of heterozygosity in natural populations of Drosophila pseudoobscura. Genetics. 1966 Aug;54(2):595–609. doi: 10.1093/genetics/54.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb M. R., Smith D. H. Outer membrane protein composition in disease isolates of Haemophilus influenzae: pathogenic and epidemiological implications. Infect Immun. 1980 Dec;30(3):709–717. doi: 10.1128/iai.30.3.709-717.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S. S., Teter M. J., Gilligan P. H. Biotype of Haemophilus influenzae: correlation with virulence and ampicillin resistance. J Infect Dis. 1983 May;147(5):800–806. doi: 10.1093/infdis/147.5.800. [DOI] [PubMed] [Google Scholar]
- Losonsky G. A., Santosham M., Sehgal V. M., Zwahlen A., Moxon E. R. Haemophilus influenzae disease in the White Mountain Apaches: molecular epidemiology of a high risk population. Pediatr Infect Dis. 1984 Nov-Dec;3(6):539–547. doi: 10.1097/00006454-198411000-00012. [DOI] [PubMed] [Google Scholar]
- Marshall D. R., Brown A. H. The charge-state model of protein polymorphism in natural populations. J Mol Evol. 1975 Nov 4;6(3):149–163. doi: 10.1007/BF01732353. [DOI] [PubMed] [Google Scholar]
- Milkman R. Electrophoretic variation in Escherichia coli from natural sources. Science. 1973 Dec 7;182(4116):1024–1026. doi: 10.1126/science.182.4116.1024. [DOI] [PubMed] [Google Scholar]
- Moxon E. R., Deich R. A., Connelly C. Cloning of chromosomal DNA from Haemophilus influenzae. Its use for studying the expression of type b capsule and virulence. J Clin Invest. 1984 Feb;73(2):298–306. doi: 10.1172/JCI111214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy T. F., Dudas K. C., Mylotte J. M., Apicella M. A. A subtyping system for nontypable Haemophilus influenzae based on outer-membrane proteins. J Infect Dis. 1983 May;147(5):838–846. doi: 10.1093/infdis/147.5.838. [DOI] [PubMed] [Google Scholar]
- Oberhofer T. R., Back A. E. Biotypes of Haemophilus encountered in clinical laboratories. J Clin Microbiol. 1979 Aug;10(2):168–174. doi: 10.1128/jcm.10.2.168-174.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H., Whittam T. S., Caugant D. A., Selander R. K. Enzyme polymorphism and genetic population structure in Escherichia coli and Shigella. J Gen Microbiol. 1983 Sep;129(9):2715–2726. doi: 10.1099/00221287-129-9-2715. [DOI] [PubMed] [Google Scholar]
- Porras O., Caugant D. A., Gray B., Lagergård T., Levin B. R., Svanborg-Edén C. Difference in structure between type b and nontypable Haemophilus influenzae populations. Infect Immun. 1986 Jul;53(1):79–89. doi: 10.1128/iai.53.1.79-89.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porras O., Svanborg-Edén C., Lagergård T., Hanson L. A. Method for testing adherence of Haemophilus influenzae to human buccal epithelial cells. Eur J Clin Microbiol. 1985 Jun;4(3):310–315. doi: 10.1007/BF02013659. [DOI] [PubMed] [Google Scholar]
- Selander R. K., Levin B. R. Genetic diversity and structure in Escherichia coli populations. Science. 1980 Oct 31;210(4469):545–547. doi: 10.1126/science.6999623. [DOI] [PubMed] [Google Scholar]
- Wilson A. C., Carlson S. S., White T. J. Biochemical evolution. Annu Rev Biochem. 1977;46:573–639. doi: 10.1146/annurev.bi.46.070177.003041. [DOI] [PubMed] [Google Scholar]