Abstract
Accurate positioning of the mitotic spindle is important for the genetic material to be distributed evenly in dividing cells, but little is known about the mechanisms that regulate this process. Here we report that two microtubule-associated proteins important for spindle positioning interact with several proteins in the sumoylation pathway. By two-hybrid analysis, Kar9p and Bim1p interact with the yeast SUMO Smt3p, the E2 enzyme Ubc9p, an E3 Nfi1p, as well as Wss1p, a weak suppressor of a temperature-sensitive smt3 allele. The physical interaction between Kar9p and Ubc9p was confirmed by in vitro binding assays. A single-amino-acid substitution in Kar9p, L304P disrupted its two-hybrid interaction with proteins in the sumoylation pathway, but retained its interactions with the spindle positioning proteins Bim1p, Stu2p, Bik1p, and Myo2p. The kar9-L304P mutant showed defects in positioning the mitotic spindle, with the spindle located more distally than normal. Whereas wild-type Kar9p-3GFP normally localizes to only the bud-directed spindle pole body (SPB), Kar9p-L304P-3GFP was mislocalized to both SPBs. Using a reconstitution assay, Kar9p was sumoylated in vitro. We propose a model in which sumoylation regulates spindle positioning by restricting Kar9p to one SPB. These findings raise the possibility that sumoylation could regulate other microtubule-dependent processes.
IN eukaryotic cell division, the mitotic spindle must be accurately positioned relative to the plane of cytokinesis for each cell to inherit a complete set of genetic information. In the budding yeast Saccharomyces cerevisiae, spindle positioning is dependent upon the interaction of cytoplasmic microtubules with the cortical actin cytoskeleton (Sullivan and Huffaker 1992; Carminati and Stearns 1997; Beach et al. 2000; Yin et al. 2000; Hwang et al. 2003). In this yeast, cytoplasmic microtubules are anchored to the spindle at the spindle pole body (SPB), a plaque-like structure embedded in the nuclear envelope equivalent to the centrosome of higher organisms (Byers and Goetsch 1975; Byers 1981).
During mitosis, only one of the two SPBs is transferred across the neck into the bud, while the other is retained in the mother cell (Pereira et al. 2001). The asymmetric nature of the spindle is defined in part by Kar9p, which localizes to the SPB that is transferred to the bud (Liakopoulos et al. 2003; Maekawa et al. 2003; Moore and Miller 2007). Kar9p is first localized at the SPB and is then transported by the kinesin Kip2p along the cytoplasmic microtubule to the plus end (Liakopoulos et al. 2003; Maekawa et al. 2003), where it links the cytoplasmic microtubule to the actin cytoskeleton by binding the type V myosin Myo2p (Beach et al. 2000; Yin et al. 2000; Hwang et al. 2003). The Kar9p linker interacts with the cytoplasmic microtubule through the microtubule binding protein Bim1p, the yeast representative of the highly conserved EB1 family of microtubule plus-end tracking proteins (Schwartz et al. 1997; Muhua et al. 1998; Lee et al. 2000; Miller et al. 2000). When the Myo2p-Kar9p-Bim1p linkage is formed, the myosin motor can steer the attached microtubules into the bud, aligning the spindle along the mother-bud axis and positioning it near the mother-bud neck (Beach et al. 2000; Yin et al. 2000; Hwang et al. 2003).
In addition to the role of Bim1p in the Kar9p-microtubule attachment complex, Bim1p also influences cytoplasmic microtubule dynamics. Bim1p binds to both growing and shrinking microtubules and promotes their dynamic instability, especially during G1 (Tirnauer et al. 1999; Wolyniak et al. 2006). Bim1p interacts with two other microtubule plus-end binding proteins, the XMAP215 homolog Stu2p and the CLIP-170 homolog Bik1p. These promote microtubule dynamics not only in G1 but also in preanaphase (Wolyniak et al. 2006). It is not known what regulates this aspect of Bim1p and Stu2p function (Tirnauer et al. 1999; Wolyniak et al. 2006). In addition to spindle positioning, Bim1p also functions on intranuclear microtubules (Schwartz et al. 1997; Tanaka et al. 2005; Wolyniak et al. 2006).
To date, phosphorylation is the only regulatory mechanism shown to control the asymmetric localization of Kar9p. Liakopoulos et al. (2003) identified two Cdc28p-dependent phosphorylation sites on Kar9p, serines 197 and 496 (Liakopoulos et al. 2003). Mutating either serine to alanine results in the association of Kar9p with both poles (Liakopoulos et al. 2003; Moore et al. 2006; Moore and Miller 2007). Serine 197 is likely to be targeted for phosphorylation by the cyclin Clb4p, whereas serine 496 is phosphorylated in conjunction with Clb5p (Moore et al. 2006; Moore and Miller 2007). Phosphorylation of serine 197 is critical for the essential function of Kar9p revealed by the absence of dynein, whereas phosphorylation of serine 496 is not (Moore and Miller 2007). Further, Kar9p interacts with Bik1p, the yeast homolog of the CLIP-170 microtubule binding protein, which acts to restrict Kar9p to one SPB by promoting phosphorylation at serine 496 (Moore et al. 2006).
In this report, we investigate whether sumoylation also plays a role in regulating the localization of Kar9p to one SPB. Conserved from yeast to human, SUMO/SMT3 (small ubiquitin-like modifier) encodes the single SUMO species in S. cerevisiae and is essential. Smt3p/SUMO is covalently attached to target proteins through an isopeptide bond with the ɛ-amino group of lysine residues. Sumoylation regulates a wide variety of cellular processes, including sister chromatid cohesion (Biggins et al. 2001; Stead et al. 2003), septin ring formation (Johnson and Blobel 1999; Takahashi et al. 1999; Johnson and Gupta 2001; Martin and Konopka 2004), DNA repair (Johnson 2004; Ulrich 2004; Aragón 2005; Bartek and Lukas 2006), and transcriptional regulation (Smolen et al. 2004; Gill 2005; Savare et al. 2005; Girard and Goossens 2006; Gupta and Bei 2006). Unlike polyubiquitination, which targets proteins for degradation, classically sumoylation exerts its regulatory effect on target proteins by altering protein–protein interactions, protein localization, and by antagonizing ubiquitin conjugation resulting in protein stabilization (Kamitani et al. 1997; Desterro et al. 1998; Dohmen 2004; Gill 2004; Johnson 2004). Recently, cross-talk between sumoylation and ubiquitination processes has been observed in which sumoylation acts as a target for SUMO-directed ubiquitin ligases (Prudden et al. 2007; Sun et al. 2007; Uzunova et al. 2007; Xie et al. 2007). However, it is currently unknown whether sumoylation directly regulates microtubule-based processes, such as spindle positioning.
The enzymes within the sumoylation pathway are analogous to those required for ubiquitination. The covalent attachment of SUMO/Smt3p in yeast requires a number of highly conserved enzymes for the processing, activation (E1's), conjugation (E2's), and ligation (E3's) of SUMO to target proteins. Like ubiquitin, Smt3p is synthesized as a precursor protein. Processing removes the last three amino acids of SUMO/Smt3p to expose a new carboxy-terminal glycine (Gly98) (Johnson and Blobel 1997; Kamitani et al. 1997; Li and Hochstrasser 1999). Because this glycine is used in the formation of the isopepetide bond with the target lysine, mutation of this residue to alanine precludes the conjugation of SUMO to its substrate. Mature Smt3p is activated by a heterodimer of Uba2p and Aos1p, proteins with sequence similarity to ubiquitin-activating enzymes, or E1's (Johnson et al. 1997; Dohmen 2004). Activated Smt3p forms a transient thioester bond with a single E2-like conjugating enzyme, Ubc9p (Johnson and Blobel 1997; Sampson et al. 2001; Bencsath et al. 2002; Gill 2004). Although SUMO can be conjugated to target proteins in the absence of an E3 ligase in vitro (Okuma et al. 1999), E3-like ligases can enhance the efficiency of conjugation and specificity for SUMO targets in vivo (Hochstrasser 2001; Johnson 2004; Ihara et al. 2005; Liu et al. 2006). In yeast, four E3 enzymes have been identified, Siz1p, Siz2p/Nfi1p, Mms21p, and Zip3p (Johnson and Gupta 2001; Takahashi et al. 2001a,b, 2003; Zhao and Blobel 2005; Cheng et al. 2006).
Classically, Smt3p is conjugated to a lysine residue within a “standard” consensus site, ΨKxE/D, where Ψ is a large hydrophobic amino acid and x is any amino acid (Melchior 2000; Rodriguez et al. 2001; Johnson 2004). However, many examples of conjugation at nonconsensus sites have also been identified (Comerford et al. 2003; Chung et al. 2004; Johnson 2004; Pichler et al. 2005). Recently, secondary structural elements have been found to play an important part in SUMO conjugation (Pichler et al. 2005). Further, sumoylation at multiple lysines has also been observed (Rui et al. 2002; Comerford et al. 2003).
In this study we evaluate the hypothesis that sumoylation is a novel mechanism for the regulation of the microtubule-dependent process of spindle positioning in yeast. We show that Kar9p and Bim1p interact with SUMO/Smt3p and several enzymes within the yeast sumoylation machinery. A mutation in KAR9, kar9-L304P, disrupts the interaction between KAR9 and the sumoylation pathway enzymes. This mutant exhibits a spindle-positioning defect, most likely arising from the mislocalization of Kar9p to both SPBs and subsequent misorientation of cytoplasmic microtubules. This work implicates sumoylation as a novel mechanism for the regulation of spindle positioning in yeast.
MATERIALS AND METHODS
Yeast strains and growth conditions:
S. cerevisiae strains and plasmids used in this study are listed in Table 1. Primers and oligonucleotides used for strain constructions can be found in supplemental Table 1S. Cells were grown in yeast–peptone–dextrose (YPD) or synthetic complete (SC) media as previously described (Moore et al. 2006).
TABLE 1.
Strains and plasmids used in this study
| Genotype/comments | Source | |
|---|---|---|
| Yeast strains | ||
| yRM425/MS4304 | MATadhc1Δ∷URA3 his3Δ200 ura3-52 trp1Δ1 leu2-3 leu2-112 | M. D. Rose |
| yRM435/MS4308 | MATakip2Δ∷URA3 trp1Δ1 ade2-101 his3Δ200 ura3-52 leu2-3 leu2-112 | M. D. Rose |
| yRM373/MS4262 | MATα dhc1Δ∷URA3 leu2-3 leu2-112 ura3-52 trp1Δ1 | M. D. Rose |
| yRM1340/MS4308 | MATα kar9Δ∷LEU2 leu2-3 leu2-112 ura3-52 ade2-101 his3Δ200 | M. D. Rose |
| yRM1756/PJ69-4α | MATα trp1-901 leu2-3 leu2-112 ura3-52 his3Δ200 gal4Δ gal80Δ LYS2∷GAL1-HIS3 GAL2-ADE2 met3∷GAL7-lacZ | James et al. (1996) |
| yRM1757/PJ69-4A | MATatrp1-901 leu2-3 leu2-112 ura3-52 his3Δ200 gal4Δ gal80Δ LYS2∷GAL1-HIS3 GAL2-ADE2 met3∷GAL7-lacZ | James et al. (1996) |
| yRM2147/MS1556 | MATaleu2-3 leu2-112 ura3-52 ade2-101 his3Δ200 | M. D. Rose |
| yRM2057 | MATabim1Δ∷KAN trp1-901 leu2-3 leu2-112 ura3-52 his3Δ200 gal4Δ gal80Δ LYS2∷GAL1-HIS3 GAL2-ADE2 met3∷GAL7-lacZ | Miller et al. (2000) |
| yRM2258 | MATabik1Δ∷TRP trp1-901 leu2-3 leu2-112 ura3-52 his3Δ200 gal4Δ gal80Δ LYS2∷GAL1-HIS3 GAL2-ADE2 met3∷GAL7-lacZ | Moore et al. (2006) |
| yRM3399 | MATα kar9Δ∷KAN lys2Δ his3Δ leu2Δ ura3Δ | Open Biosystems |
| yRM4366 | MATaKAR9-tap∷URA3 bar1Δ∷LEU2 ura3-52 leu2-3 leu2-112 his3Δ200 trp1Δ1 | Moore et al. (2006) |
| yRM4769 | MATasiz1Δ∷KANRtrp1-901 leu2-3 leu2-112 ura3-52 his3Δ200 gal4Δ gal80Δ LYS2∷GAL1-HIS3 GAL2-ADE2 met3∷GAL7-lacZ | This study |
| yRM4755 | MATα siz2Δ∷KANRtrp1-901 leu2-3 leu2-112 ura3-52 his3Δ200 gal4Δ gal80Δ LYS2∷GAL1-HIS3 GAL2-ADE2 met3∷GAL7-lacZ | This study |
| yRM5084 | MATaCFP-TUB1∷URA3 ura3-52 leu2-3 leu2-112 trp1Δ1 his3Δ200 ade2-101 | Moore et al. (2006) |
| yRM5494 | MATaleu2-3 leu2-112 ura3-52 ade2-101 his3Δ200 [pGAL-KAR9-V5] | This study |
| yRM5500 | MATaleu2-3 leu2-112 ura3-52 ade2-101 his3Δ200 [pGAL-KAR9] | This study |
| yRM5970 | MATα CFP-TUB1∷URA3 KAR9-3GFP∷TRP1 ura3-52 leu2-3 leu2-112 trp1Δ1 his3Δ200 ade2-101 | This study |
| yRM5973 | MATaCFP-TUB1∷URA3 kar9-L304P-3GFP∷TRP1 ura3-52 leu2-3 leu2-112 trp1Δ1 his3Δ200 ade2-101 | This study |
| yRM6172 | MATakar9Δ∷KANRtrp1-901 leu2-3 leu2-112 ura3-52 his3Δ200 gal4Δ gal80Δ LYS2∷GAL1-HIS3 GAL2-ADE2 met3∷GAL7-lacZ | This study |
| yRM6398 | MATα jnm1Δ∷LEU2 ura3-52 leu2-3 leu2-112 trp1Δ1 ade2-101 | This study |
| yRM6491/PM1176 | MATaSMT3∷HIS3 ura3-52 leu2-3 leu2-112 ade2-101 his3Δ200 | This study/P. Meluh |
| yRM6492/PM1177 | MATasmt3-12∷HIS3 ura3-52 leu2-3 leu2-112 ade2-101 his3Δ200 | This study/P. Meluh |
| yRM6493/PM1178 | MATasmt3-11∷HIS3 ura3-52 leu2-3 leu2-112 ade2-101 his3Δ200 | This study/P. Meluh |
| yRM6727 | MATα jnm1Δ∷LEU2 leu2-3 leu2-112 ura3-52 ade2-101 his3Δ200 | This study |
| yRM6739 | MATα Kar9p-L304P-3GFP-TRP1 trp1Δ1 ura3-52 leu2-3 leu2-112 | This study |
| yRM6740 | MATaKar9p-L304P-3GFP-TRP1 trp1Δ1 ade2-101 leu2-3 leu2-112 | This study |
| yRM6764 | MATabim1Δ∷KANRtrp1Δ1 his3Δ200 ura3-52 leu2-3 leu2-112 | This study |
| yRM6836 | MATa pCUP-GST-KAR9-URA3-LEU2-d leu2-3 leu2-112 his3Δ200 ura3-52 pep4∷HIS3 | E. Phyzicky/E. Grayhack |
| yRM6903 | MATα kar9-A196E S197E-3GFP∷TRP1 SPC110-DsRed∷KanR leu2-3 leu2-112 ura3-52 trp1Δ1 [pAFS92 GFP-TUB1∷URA3] | Moore and Miller (2007) |
| yRM7140 | MATasiz1Δ∷KAN leu2-3 leu2-112 ura3-52 trp1Δ1 ade2-101 his3Δ200 | This study |
| yRM7166 | MATacst9Δ∷KAN leu2-3 leu2-112 ura3-52 trp1Δ1 ade2-101 his3Δ200 | This study |
| yRM7248 | MATanfi1Δ∷KAN leu2-3 leu2-112 ura3-52 trp1Δ1 ade2-101 his3Δ200 | This study |
| yRM7258 | MATα KAR9-3GFP∷TRP1 GFP-TUB1∷URA3 leu2-3 leu2-112 ura3-52 trp1Δ1 ade2-101 his3Δ200 | This study |
| yRM7261 | MATα kar9-L304P-3GFP∷TRP1 GFP-TUB1∷URA3 leu2-3 leu2-112 ura3-52 trp1Δ1 [pAFS92 GFP-TUB1∷URA3] | This study |
| yRM7271 | MATanfi1Δ∷KAN GFP-TUB1∷URA3 leu2-3 leu2-112 ura3-52 trp1Δ1 ade2-101 his3Δ200 | This study |
| yRM7309 | MATaGFP-TUB1∷URA3 leu2-3 leu2-112 ura3-52 trp1Δ1 ade2-101 his3Δ200 | This study |
| yRM7311 | MATacst9Δ∷KAN GFP-TUB1∷URA3 leu2-3 leu2-112 ura3-52 trp1Δ1 his3Δ200 | This study |
| yRM7361 | MATα KAR9-3GFP∷TRP1 kar9Δ∷LEU2 leu2-3 leu2-112 ura3-52 ade2-101 his3Δ200 | This study |
| yRM7363 | MATaKAR9-3GFP∷TRP1 dhc1Δ∷LEU2 leu2-3 leu2-112 ura3-52 ade2-101 his3Δ200 | This study |
| yRM7364 | MATasiz1Δ∷KAN GFP-TUB1∷URA3 leu2-3 leu2-112 ura3-52 trp1Δ1 ade2-101 his3Δ200 | This study |
| yRM7575 | MATa pCUP-GST-kar9-L304P-URA3-LEU2-d leu2-3 leu2-112 his3Δ200 ura3-52 pep4∷HIS3 | This study |
| yRM7604 | MATα smt3-11∷HIS3 KAR9-3GFP∷TRP1 CFP-TUB1∷URA3 leu2-3 leu2-112 ura3-52 trp1Δ1 | This study |
| yRM7635 | MATα smt3-12∷HIS3 KAR9-3GFP∷TRP1 GFP-TUB1∷URA3 leu2-3 leu2-112 ura3-52 trp1Δ1 his3Δ200 | This study |
| yRM7712 | MATα SMT3∷HIS3 KAR9-3GFP∷TRP1 GFP-TUB1∷URA3 leu2-3 leu2-112 ura3-52 trp1Δ1 his3Δ200 | This study |
| EAS0456/DFS188 | MATaura3-52 leu2-112 lys his3∷HindIII arg8∷hisG rho− | E. Sia |
| Plasmids | ||
| pRM443 | KIP2 URA3 CEN AmpR | Miller et al. (1998) |
| pRM493/pDB65/B3102 | BIK1 LEU2 CEN AmpR | G. Fink |
| pRM773/pET30c | Vector for tagging proteins with his6KANR | Novagen |
| pRM1151/pMR3758 | GAD-C1 LEU2 2μ AmpR | James et al. (1996) |
| pRM1153/pMR3760 | GAD-C3 LEU2 2μ AmpR | James et al. (1996) |
| pRM1154/pMR3761 | GBDU URA3 2μ AmpR | James et al. (1996) |
| pRM1493/pMR4150 | GBDU-KAR9 URA3 2μ AmpR | Miller et al. (2000) |
| pRM1765 | pGAL-BIM1-V5-his6 URA3 AmpR | Invitrogen |
| pRM1893/pMR4755 | GAD-BIM1 LEU2 2μ AmpR | Miller et al. (2000) |
| pRM1895/pMR4764 | GAD-kar91-393aaLEU2 2μ AmpR | Miller et al. (2000) |
| pRM1916/pMR4769 | GAD-STU2649-888aaLEU2 2μ AmpR | Miller et al. (2000) |
| pRM2138/EBL473 | BIM1 CEN URA3 AmpR | E. Elion |
| pRM2345 | GBDU empty URA3 2μ with BglII restriction site in the polylinker replaced with SacI AmpR | Moore et al. (2006) |
| pRM2473/pHY312 | GAD-myo2 tail LEU2 2μ AmpR | Yin et al. (2000) |
| pRM2627 | GAD-BIK1 LEU2 2μ AmpR | Moore et al. (2006) |
| pRM3145 | GBDU-BIM1 URA3 2μ AmpR | This study |
| pRM3369 | Empty GST vector AmpR | This study |
| pRM3366 | GST-KAR9390–644aa AmpR | Moore et al. (2006) |
| pRM3367 | GST-KAR9470-580aa AmpR | Moore et al. (2006) |
| pRM3368 | GST-KAR9533-580aa AmpR | Moore et al. (2006) |
| pRM3448/pAFS125C | pCFP-TUB1 URA3 AmpR | Straight et al. (1997) |
| pRM3595 | GBDU-KIP2 URA3 2μ AmpR | This study |
| pRM3634 | 3XGFP TRP1 integration plasmid with SalI–XmaI–SacI sites in the polylinker AmpR | Moore et al. (2006) |
| pRM3662 | KAR9117-644aa-3XGFP TRP1 AmpR integration plasmid | Moore et al. (2006) |
| pRM4125 | GAL1-KAR9 URA3 2μ AmpR | This study |
| pRM4319 | GAL1-KAR9-V5 URA3 2μ AmpR | This study |
| pRM4380 | GAD424 LEU2 2μ AmpR | E. Sia |
| pRM4382/pLAJ20 | GAD-SMT3-GG LEU2 2μ AmpR | This study |
| pRM4383/pLAJ21 | GAD-SMT3-GA LEU2 2μ AmpR | This study |
| pRM4419/AFS92 | pGFP-TUB1∷URA3 AmpR | A. Straight |
| pRM4472 | GBDU-kar9-K301R URA3 2μ AmpR | This study |
| pRM4495 | GAD-UBC9 LEU2 2μ AmpR | This study |
| pRM4496 | GAD-NFI1 LEU2 2μ AmpR | This study |
| pRM4544 | GBDU-kar9-K76R URA3 2μ AmpR | This study |
| pRM4594 | GAD-UFD1 LEU2 2μ AmpR | This study |
| pRM4595 | GAD-NIS1 LEU2 2μ AmpR | This study |
| pRM4596 | GAD-RIS1 LEU2 2μ AmpR | This study |
| pRM4597 | GAD-WSS1 LEU2 2μ AmpR | This study |
| pRM4698 | GBDU-kar9-K594R URA3 2μ AmpR | This study |
| pRM4699 | GBDU-bim1-K25R K26R URA3 2μ AmpR | This study |
| pRM4700 | GBDU-bim1-K110R URA3 2μ AmpR | This study |
| pRM4701 | GBDU-bim1-K266R K267R URA3 2μ AmpR | This study |
| pRM4920/pLAJ19 | GAD-SMT3 LEU2 2μ AmpR | This study |
| pRM5169 | His6-UBC9 AmpR | Johnson and Blobel (1997) |
| pRM5415 | GBDU-kar9-K307R URA3 2μ AmpR | This study |
| pRM5421 | GBDU-kar9-L304P URA3 2μ AmpR | This study |
| pRM5424 | GBDU-kar9-K529R URA3 2μ AmpR | This study |
| pRM5617 | GBDU-kar9-S197E URA3 2μ AmpR | Moore and Miller (2007) |
| pRM5619 | GBDU-kar9-S496E URA3 2μ AmpR | Moore and Miller (2007) |
| pRM5777 | GBDU-kar9-S496A URA3 2μ AmpR | Moore and Miller (2007) |
| pRM5785 | kar9117-644aa-L304P-3GFP TRP1 AmpR integration plasmid | This study |
| pRM5829 | GAD-URM1 LEU2 2μ AmpR | This study |
| pRM5880 | GAD-UBI4 LEU2 2μ AmpR | This study |
| pRM6028/pMR3548 | His6-RVS161 KANR | Brizzio et al. (1998) |
| pRM6113 | GBDU-kar9-S197A S496A URA3 2μ AmpR | Moore and Miller (2007) |
| pRM6050 | GBDU-kar9-S197A URA3 2μ AmpR | Moore and Miller 2007 |
| pRM6255 | GBDU-kar9-A196E S197E URA3 2μ AmpR | Moore and Miller (2007) |
| pRM6294 | GBDU-kar9-S197A S496E URA3 2μ AmpR | Moore and Miller (2007) |
| pRM6295 | GBDU-kar9-A196E S197E S496A URA3 2μ AmpR | Moore and Miller (2007) |
| pRM6510 | GBDU-kar9-K301R K307R URA3 2μ AmpR | This study |
| pRM6575 | GBDU-kar9-K120R URA3 2μ AmpR | This study |
| pRM6576 | GBDU-kar9-K224R K227R URA3 2μ AmpR | This study |
| pRM6707 | GBDU-kar9-M293P URA3 2μ AmpR | This study |
| pRM6713 | his6-S-tag-Smt3p-gg KanR | This study |
| pRM6760 | GST-AOS1/UBA2 AmpR | Bencsath et al. (2002) |
| pRM7831 | GBDU-kar9-K76R K120R K301R K307R URA3 2μ AmpR | This study |
Two-hybrid assay:
The two-hybrid system of James et al. (1996) was used. Full-length KAR9 fused to the binding domain (BD) (pRM1493/pMR4150) was generated as described (Miller et al. 2000).
To create a reporter strain containing both plasmids, DNA binding domain (DBD) and activation domain (AD) plasmids were transformed into the MATa PJ69-4A (yRM1757) and MATα PJ69-4α (yRM1756) reporter strains, respectively, and mated on YPD overnight at 30° (James et al. 1996). Diploids were selected on SC media lacking uracil and leucine. Interactions were assayed by transferring cells with a multi-prong transfer device to SC plates lacking uracil and leucine (−ura −leu) or histidine (−his). Growth was scored after incubation at 30° for 2–3 days.
UBI4-AD:
To generate a ubiquitin fusion with the GAL4-AD, UBI4 was synthesized by PCR with terminal BamHI and PstI restriction sites using primers 436 and 437 and genomic DNA from the wild-type strain yRM2147 as template. This product was cloned into the BamHI and PstI restriction sites of pGAD-C1/pRM1151 and verified by sequencing, generating pRM5880.
URM1-AD:
A URM1 fusion to the GAL4-AD was generated by synthesizing URM1 with terminal EcoRI and BamHI restriction sites using primers 429 and 430 and genomic DNA from yRM2147 as template. This was cloned into the EcoRI and BamHI sites of pGAD-C1/pRM1151 and verified by sequencing, generating pRM5829.
BIM1-BD:
BIM1 was synthesized with terminal SalI and SacI sites using primers 70 and 71 and pRM2138 as a template. This was cloned into the SalI and SacI sites of the DBD vector, pRM2345, to generate pRM3145.
KIP2-BD:
KIP2 was synthesized by PCR with terminal BamHI and EcoRI sites using primers 183 and 184 and pRM443 as a template. This product was cloned into the terminal BamHI and EcoRI sites of pGBDU/pRM1154 and confirmed by sequencing to generate pRM3595.
BIK1-AD:
BIK1 was synthesized by PCR with terminal BamHI and EcoRI sites using primers 59 and 60 and pRM493 as a template. The PCR product was cloned into pGAD/pRM1153 and confirmed by sequencing to generate pRM2627.
AD-SMT3, AD-SMT3-GG, and AD-SMT3-GA:
Three plasmids were constructed to express the full-length, truncated, and mutated forms of Smt3p fused to the GAL4 activation domain. Full-length SMT3 was amplified with primers 5′-CGGCCGATACGGATCCCGA TGTCGGACTCAGAAGTCAATC-3′ and 5′-CGCCGAAGGCTGCAGCCTAATACGTAGCA CCACCAATC-3′ using EAS0456/DFS188 genomic DNA as a template. The product was fused to the GAL4 activation domain to create pLAJ19/pRM4920. The mature truncated form of SMT3 was amplified with primers 5′-CGGCCGATACGGATCCCGATGTCGGACTCAGAAGTCAATC-3′ and 5′-GACATGT CGCTGCAGCACCACCAATCTGTTCTCTGTG-3′ using EAS0456/DFS188 genomic DNA as a template. This resulted in pLAJ20/pRM4382. The mature truncated form in which the C-terminal glycine changed to alanine was amplified with this mutation using primers 5′-CGGCCGATACGGATCCCGATG TCGGACTCAGAAGTCAATC-3′and 5′-GACA TGTCGCTGCAGCAGCACCAATCTGTTC TCTGTG-3′ to generate pLAJ21/pRM4383. In each case, the insert was cloned into pGAD424 at BamHI and PstI sites.
AD-UBC9, AD-NFI1, AD-WSS1, AD-NIS1, AD-RIS1, and AD-UFD1:
The UBC9, NFI1, WSS1, NIS1, RIS1, and UFD1 genes fused with the GAL4 activation domain were isolated from a S. cerevisiae genomic library in a two-hybrid screen for proteins that interact with a fragment of a mitochondrial protein (L. Pogorzala and E. Sia, unpublished results). Sequencing was used to determine the identity and point at which the insert was fused in frame to the activation domain of GAL4. pGAD-UBC9 (pRM4495) contains the full-length UBC9 gene without its intron fused in frame with the GAL4 activation domain. In the pGAD-NFI1 (pRM4496) plasmid, the fusion begins with codon 339 of NFI1, truncating the amino half of the protein. In the pGAD-WSS1 (pRM4597) plasmid, the fusion begins with codon 235 of WSS1, expressing only the carboxy-terminal domain. In the pGAD-NIS1 (pRM4595) plasmid, the fusion begins with codon 40 of NIS1. pGAD-RIS1 (pRM4596) contains the full-length RIS1. pGAD-UFD1 (pRM4594) contains the GAL4 activation domain fused with full-length UFD1.
Two-hybrid disruptions:
To generate a two-hybrid reporter strain disrupted for KAR9, we amplified by PCR a genomic region containing KAR9∷KAN from the Open Biosystems/ATCC collection (yRM3399) using primers 453 and 454. The PCR product contains 500-bp upstream and 500-bp downstream sequence of KAR9 with the coding region of KAR9 replaced by KAN. The product was then transformed into PJ69-4A (yRM1757) (James et al. 1996). Integrants were selected on YPD plates containing 200 mg/liter geneticin (Invitrogen, Carlsbad, CA). The disruption was confirmed by PCR and phenotype analysis by scoring defects in nuclear positioning as described (Miller and Rose 1998). This process generated yRM6172.
Two-hybrid reporter strains disrupted for BIM1 (yRM2057) were generated as described in Miller et al. (2000). Two-hybrid reporter strains disrupted for BIK1 (yRM2258) were generated as described in Moore et al. (2006).
Site-directed mutagenesis of potential SUMO sites in KAR9:
Point mutations were introduced into either KAR9 (pRM1493) or BIM1 (pRM3145) on the two-hybrid DBD-plasmid to generate lysine-to-arginine mutations by site-directed mutagenesis using the QuikChange kit (Stratagene, La Jolla, CA). The oligonucleotides used in the mutagenesis are listed in supplemental Table 1S. The mutant L304P was generated using primers 396 and 397 and pRM1493 as the template. The constructs were sequenced to confirm the absence of additional mutations. Two-hybrid analysis was carried out in the haploid two-hybrid reporter strain, yRM1757, unless noted otherwise.
Affinity chromatography:
Isolation of bacterial extracts containing His6-Ubc9 and His6-Rvs161:
Bacterial strains containing his6-Ubc9p (pRM 5169, a gift from Erica Johnson), empty Kan vector (pRM 773) and his6-Rvs161 (pRM6028/pMR3548) (Brizzio et al. 1998) were diluted 1:100 and grown for 2 hrs in LB amp at 37°. Cells were induced with 1 mm IPTG for 3 hr and harvested by centrifugation. His6-Ubc9 and His6-empty extracts were prepared by resuspending the bacterial cells in B150 + 1% Sigma protease inhibitors for his6-tagged proteins (Sigma, St. Louis) and 1 mm PMSF, lysed by sonication at 0° and clarified at 20,000 g for 20 min. The supernatant containing the Ubc9 protein was diluted with 1/5 volume of glycerol and flash frozen in liquid nitrogen. His6-Rvs161 extract was prepared by resuspending cells in binding buffer (5 mm imidazole, 500 mm NaCl, 20 mm Tris-HCl, pH 7.9) with lysozyme at 30° for 15 min, followed by addition of 1% Sigma protease inhibitors for his6-tagged proteins (Sigma) and 1 mm PMSF, and lysed by sonication at 0°. The pellet was obtained by centrifugation at 20,000 g for 20 min and was resuspended in 8 m urea/binding buffer.
Isolation of yeast extracts:
Strains containing pGAL-KAR9-V5 (yRM 5494) and pGAL-KAR9-no tag (yRM 5500) were grown to mid-exponential phase in synthetic –ura liquid media containing 2% sucrose, a noninducing sugar. KAR9 expression was induced for 3 hr with 2% galactose. Cells were washed 1× in water, resuspended in 15% glycerol, pelleted, excess fluid removed, and flash frozen in liquid nitrogen. Cells were thawed at 0°, resuspended in B150 buffer containing 1% Sigma protease inhibitors for his-tagged proteins (Sigma) and 1 mm PMSF. Cells were lysed by vortexing with glass beads, and extracts were clarified by centrifugation at 16,000 g for 25 min at 4°.
Pull-down assays:
For the binding assay, 10 mg of total bacterial extract in 1000 μl B150 or 8 m urea/binding buffer were added to 200 μl Talon CellThru cobalt beads (Novagen Madison, WI) that had been prewashed several times with B150 buffer (His6Ubc9 and His6-empty) or 8 m urea/binding buffer (His6-Rvs161). Extracts were incubated with the beads on a rotisserie mixer for 4 hr at 4°. The beads were then loaded onto 0.7 ml microspin columns and washed extensively with B150 or 8.0 m urea in binding buffer. For the His6-Rvs161, beads were washed 2 times in 8 m urea/binding buffer, once in 4 m urea/binding buffer, once in 2 m urea/binding buffer, once in 0 m urea/binding buffer, and at least 5 times in B150 with the last two washes containing 2 mgs/ml BSA to block nonspecific binding. For the his6-Ubc9, and his6-empty samples, the beads were washed at least 10 times in B150 with the last two washes containing 2 mgs/ml BSA. Four milligrams of yeast extract in 0.5 ml B150 plus protease inhibitors were loaded onto each of the columns. The extract and beads were rotated at 4° for 2 hr to allow binding. The columns were washed 8 times with B150 buffer, followed by two washes with B150 containing 5 mm imidazole. Samples were eluted with two washes (150 μl) of 150 mm EDTA in B150 buffer.
Gel samples were prepared for SDS–PAGE gels and Western blotting. The V5 epitope was detected using mouse anti-V5 (1:5000) (Novagen) for 4 hr. The his6-epitope was detected using mouse anti-His6 primary antibody (1:2000) (Novagen). Actin was detected using chicken anti-actin (1:20,000), with all solutions prepared in TBS.
GST-Kar9p binding assays:
The expression of Kar9p truncations fused to GST and GST alone was induced in bacteria by the addition of 1 mm IPTG at 37° for 2 hr. Cells were harvested and sonicated in 1% Triton X-100/1× PBS supplemented with bacterial protease inhibitor (Sigma) and 1 mm PMSF. The extract was clarified at 13,000 rpm for 20 min, and the pellet was discarded. Whole cell extract (2 mg) was applied to the equilibrated glutathione conjugated beads (Amersham Bioscience, Piscataway, NJ) and incubated at 4° for 30 min. The beads were washed five times with 1× PBS. His6-Ubc9p extract was prepared as described above. Whole cell extract (1 mg) containing his6-Ubc9p was added to the GST bound beads, incubated at 4° for 30 min and then washed five times with 1× PBS. Bound proteins were eluted with 100 μl of 10 mm reduced glutathione. Samples were prepared for analysis by 12% SDS–PAGE. For detection by Western blot analysis, anti-GST (1:8000) (Sigma) and anti-his6 (1:2000) (Novagen) were used to detect GST fusions and his6-Ubc9p, respectively.
In vitro sumoylation assay:
To purify Kar9p for the in vitro sumoylation assay, Kar9p was tagged with N-terminal glutathione-S-transferase (GST) under a copper inducible promoter and expressed in yeast (yRM6836) (Grayhack and Phizicky 2001; Phizicky et al. 2002). In this Kar9p fusion, an aspartic acid-to-valine mutation was present at amino acid position 559. Expression was induced for 2 hr with 0.5 mm CuSO4 and GST-Kar9p was purified on GST-conjugated beads (Amersham Bioscience) in B150 buffer supplemented with yeast protease inhibitor cocktail and 1 mm PMSF.
Sumoylation proteins used in this assay include Smt3p-gg (pRM6713), Ubc9p (pRM5169), and Aos1p/Uba2p (pRM6760). Smt3p-gg and Ubc9p were tagged with six histidine residues at the N terminus and purified from bacteria using nickel affinity column chromatography. GST-Aos1p and Uba2p were purified as described (Bencsath et al. 2002). Briefly, the two proteins coexpressed from a bicistronic vector were copurified from bacteria using glutathione affinity chromatography. To elute the protein from the beads, the fusion protein was cleaved with thrombin overnight on ice. The protein was then dialyzed into sumoylation assay buffer (50 mm Tris, pH 7.6, 5 mm MgCl2). In the assay, 5 μg of GST-Kar9p was incubated with 5 μg his6-Smt3p-gg, 2 μg His6-Ubc9p, 2 μg Aos1p/Uba2p, 4 mm ATP, and 7 μl of an ATP regeneration system (3.5 units/ml creatine kinase (Sigma), 10 mm creatine phosphate (Sigma), and 0.6 units/ml inorganic pyrophosphatase (Sigma), and sumoylation assay buffer (50 mm Tris, pH 7.6, 5 mm MgCl2). The mixture was incubated at 30° for 2 hr. To stop the reaction, the sample buffer containing 5% β-mercaptoethanol was added and the samples were boiled for 5 min. Reaction products were subjected to 6% SDS–PAGE and visualized by Western blot analysis. The presence of GST-Kar9p was detected using anti-GST (Sigma).
Fluorescence microscopy:
To study the effect of the L304P mutation on Kar9p localization, genomic KAR9 was tagged with 3XGFP at the C terminus. A fragment of KAR9 (349–1932 bp), which includes the L304P mutation, was amplified using primer 69 and 194 and pRM5421 as the template. The PCR fragment was cloned into pRM3634 at the SalI–SacI site to generate pRM5785. To integrate the 3XGFP-tagged form into the genome, the plasmid was linearized at the ClaI site in KAR9 and transformed into yRM5084, which contains cyan fluorescent protein (CFP)-tagged TUB1 to label the microtubules to create yRM5973. Wild-type KAR9-3XGFP (yRM5970) was generated as described in Moore et al. 2006.
Microscopy was carried out on a motorized Zeiss Axioplan 2 microscope equipped with a 100× Plan-Neofluor lens (1.3NA) (Carl Zeiss, Thornwood, NY), a cooled charged coupled device camera (ORCA-ER, Hamamatsu, Hamamatsu City, Japan) using Openlab 3.5.2 software (Improvision, Lexington, MA) as previously described (Moore et al. 2006).
High-density culture Kar9-tap:
A strain expressing KAR9 with a C-terminal TAP tag at the genomic KAR9 locus yRM4366 and a wild-type strain yRM2147/MS1556 were grown to increasing densities overnight in YPD. The cell density was determined by cell counts on a hemacytometer.
Protein levels in the Kar9p-L304P mutant:
Strains containing either wild-type Kar9p-3XGFP (yRM5970) or Kar9p-L304P-3XGFP (yRM5973) were cultured to midexponential phase in YPD. To prepare whole cell extracts, the cells were collected and resuspended in B150 buffer (50 mm Tris pH 7.4, 150 mm NaCl, 0.2% Triton X-100) containing 1% protease inhibitor (Sigma), and 1 mm PMSF. The cells were lysed by vortexing with glass beads. Centrifugation at 13,000 rpm for 30 min was used to remove cell debris. The Bradford protein assay (BioRad, Hercules, CA) was used to calculate protein concentration using BSA as a standard. Protein samples were prepared in 3× Laemmli sample buffer for SDS–PAGE and Western blot analysis. The blots were blocked in PBS containing 5% milk. To detect Kar9p-3XGFP, 1:100 anti-GFP (Clontech, Mountain View, CA) diluted in PBS containing 5% milk was applied to the blot for 3 hr at room temperature (RT). Anti-rabbit with an HRP conjugate (Santa Cruz Biotechnology, Santa Cruz, CA) was used at 1:2000 dilution for 1 hr at room temperature. As a loading control to detect actin, 1:20,000 anti-actin in TBS containing 5% milk was used for 4 hr at RT, followed by a 1 hr incubation at RT with 1;10,000 anti-chicken (Aves Labs, Tigard, OR). Alternatively, anti-phosphoglycerate kinase (Invitrogen-Molecular Probes, Eugene, OR) was also used as a loading control at 2 μg/ml in PBS with 5% milk.
To prepare cell extracts by a modification of the Ohashi method (Ohashi et al. 1982), cells were washed one time in water and resuspended in 17% trichloroacetic acid (TCA). Glass beads were added and vortexed for 1 min at RT and placed on ice for 1 min. This cycle was repeated four times. Cell extract was transferred to a new microfuge tube. The glass beads were washed with 1 ml 5% TCA and added to the cell extract and centrifuged at 13,000 rpm for 20 min. The pellet was then resusupended in 3× Laemmli sample buffer and 1 m Tris pH 9.0 was added to adjust pH. β-mercaptoethanol was added and the sample was boiled for 10 min. For Western blotting, the sample was run on 7% SDS–PAGE.
Sumoylation levels in SUMO mutants:
Cells were broken open by a modification of the Ohashi method (see above) and prepared for Western blotting with anti-Smt3p at 1:10,000 dilution for 3 hr at RT. Anti-rabbit HRP conjugated secondary antibody (1:2000 dilution, Santa Cruz Biotechnology) was used for 1 hr at RT. The same blot was cut to probe for actin using 1:10,000 anti-actin in TBS for 3 hr at RT. The blot was incubated with anti-chicken-HRP (1:10,000, Aves Labs) for 1 hr at RT.
RESULTS
Kar9p and Bim1p interact with Smt3p by two-hybrid analysis:
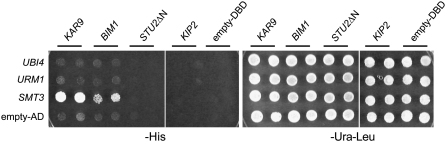
Ubiquitin and ubiquitin-like proteins post-translationally modify lysine residues on target proteins (Goehring et al. 2003; Bossis and Melchior 2006; Kerscher et al. 2006). Because two forms of Kar9p are known (Miller et al. 2000), we investigated whether ubiquitin or the ubiquitin-related proteins, Ubi4p, Urm1p, or Smt3p/SUMO, might interact with Kar9p using a two-hybrid approach. As shown in Figure 1, KAR9 interacted with SMT3, but not with ubiquitin UBI4 or the ubiquitin-related modifier URM1. We then tested three other genes that are involved in spindle positioning and known to interact with KAR9. The MAP encoded by BIM1 also interacted with SMT3, although less strongly than KAR9 (Figure 1). In contrast, two other genes that are involved in spindle positioning, the kinesin KIP2 and the carboxy-terminal end of the MAP encoded by STU2, did not interact with any of the ubiquitin-like proteins tested (Figure 1).
Figure 1.—
KAR9 and BIM1 interact with SUMO (SMT3) by two-hybrid analysis, but not with ubiquitin (UBI4) and another ubiquitin-related protein (URM1). Diploid two-hybrid reporter strains were generated by crossing yRM1757/PJ69-4A containing KAR9-BD (pRM1493), BIM1-BD (pRM3145), KIP2-BD (pRM3595), STU2-BD (pRM1916), or empty BD (pRM1154) with yRM1756/PJ69-4α containing AD-UBI4 (pRM5880), AD-URM1 (pRM5829), AD-SMT3 (pRM4920), or empty AD (pRM1151). Diploids were selected on SD −ura −leu media and tested for interaction by growth on SD −his media at 30° for 2–3 days. Two independent diploid colonies were tested. These results indicate that although UBI4, URM1, and SMT3 encode related molecules, only SMT3 interacts with KAR9 and BIM1.
Kar9p and Bim1p interact with Smt3p-GG but not Smt3p-GA:
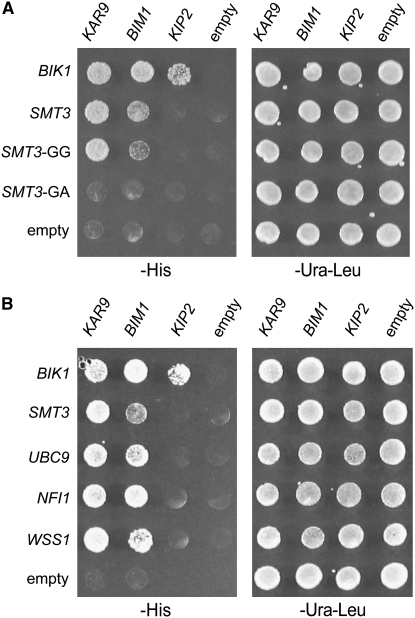
Ulp1p cleaves off the three terminal amino acids from Smt3p to expose glycine 98 as the terminal amino acid residue (Johnson et al. 1997). This exposed residue is essential for the conjugation of SUMO to target proteins and its absence is predicted to abrogate conjugation of Smt3p to its targets (Johnson et al. 1997). We therefore deleted the last three amino acids of the pro-form of SMT3 and mutated glycine 98 to alanine, exposing it as the carboxy-terminal residue in the two-hybrid fusion protein. In contrast to the full-length SMT3 and the preprocessed SMT3-GG control, the SMT3-GA mutation did not interact with either KAR9 or BIM1 (Figure 2A). Western blot analysis was used to show that the SMT3-GA construct was indeed expressed (data not shown). This suggests that these interactions may represent a conjugation event between the AD-SMT3 and the KAR9-BD and/or BIM1-BD. The interactions of BIK1 with KAR9, BIM1, and KIP2 serve as positive controls (Carvalho et al. 2004; Moore et al. 2006; Wolyniak et al. 2006).
Figure 2.—
KAR9 and BIM1 interact with multiple proteins in the sumoylation pathway by two-hybrid analysis. Two-hybrid reporter strains (yRM1757/PJ69-4A) containing KAR9-BD (pRM1493), BIM1-BD (pRM3145), KIP2-BD (pRM3595), or empty BD (pRM1154) were mated to reporter strains (yRM1756/PJ69-4α) containing AD-BIK1 (pRM2627), AD-SMT3 (pRM4920), AD-SMT3-GG (pRM4382), AD-SMT3-GA (pRM4383), AD-UBC9 (pRM4495), AD-NFI1 (pRM4496), AD-WSS1 (pRM4597), or empty AD (pRM4380) plasmids. Diploids were selected on media lacking uracil and leucine (−ura −leu) and assayed for interactions on media lacking histidine (−his). These were scored after incubation at 30° for 2–3 days. (A) The yeast two-hybrid interaction is specific to forms of SUMO that are competent for conjugation. AD-SMT3 is the full-length construct. AD-SMT3-GG truncates the last three amino acids of SMT3, leaving glycine 98 as the last amino acid. In the AD-SMT3-GA construct, glycine 98 was mutated to an alanine residue, creating a conjugation-incompetent form of Smt3p. The kinesin Kip2p, which transports Kar9p to the plus end, is included as an additional negative control, indicating that not all proteins involved in spindle positioning interact with SUMO/Smt3p. Bik1p has been shown to interact with Kip2p previously (Carvalho et al. 2004) and therefore the BIK1-KIP2 interaction demonstrates that the KIP2-BD construct is functional. (B) KAR9 and BIM1 interact with other proteins in the sumoylation pathway. Ubc9p is an E2 enzyme and Nfi1p is an E3. Wss1p was identified as a weak suppressor of a temperature-sensitive allele of SMT3 (Biggins et al. 2001).
Kar9p and Bim1p interact with other enzymes in the sumoylation pathway:
A number of enzymes function in the sumoylation pathway to assist in the conjugation of Smt3p/SUMO to target proteins. If the interaction of KAR9 and BIM1 with Smt3p represents bona fide interactions, then we reasoned that Kar9p and Bim1p might interact with other proteins that function in the sumoylation pathway. We therefore tested whether the E2 Ubc9p and the E3 protein Nfi1p might also interact with Kar9p or Bim1p. As shown in Figure 2B, KAR9 and BIM1 interacted with both UBC9 and NFI1. KAR9 and BIM1 also interacted with WSS1 (Figure 2B). WSS1 was previously identified as a high-copy suppressor of the smt3-331 temperature-sensitive allele and as a Smt3p/SUMO interacting protein in a two-hybrid screen, although its role is unclear (Biggins et al. 2001; Hannich et al. 2005). We also tested the E3 encoded by SIZ1 for a two-hybrid interaction with KAR9 and BIM1, but none was detected (data not shown). The kinesin encoded by KIP2 did not interact with UBC9, NFI1, or WSS1. These data support the idea that these two-hybrid interactions represent a biological connection between the processes of spindle positioning and sumoylation.
Several other proteins have been shown to interact with Smt3p/SUMO in yeast by two-hybrid analysis, including Nis1p, Ris1p, and Ufd1p (Hannich et al. 2005). Nis1p localizes to the bud neck and interacts with the septins, which are the most abundant sumoylated proteins in the yeast cell (Iwase and Toh-e 2001). Ris1p, which has been previously implicated in silencing, contains a RING motif, suggesting that it may have E3 ligase activity (Zhang and Buchman 1997; Hannich et al. 2005). Ufd1p is involved in the recognition of polyubiquitinated proteins and functions in delivering these proteins to the proteasome for disposal (Hitchcock et al. 2001; Ye et al. 2001; Bays and Hampton 2002; Braun et al. 2002). We tested whether these proteins might also interact with either Kar9p or Bim1p. All three interacted with KAR9 and BIM1 (data not shown). Although the molecular basis for the connection between Nis1p, Ris1p, and Ufd1p and the process of sumoylation requires further investigation, these data are consistent with a link between ubiquitin-like proteins and Kar9p and Bim1p.
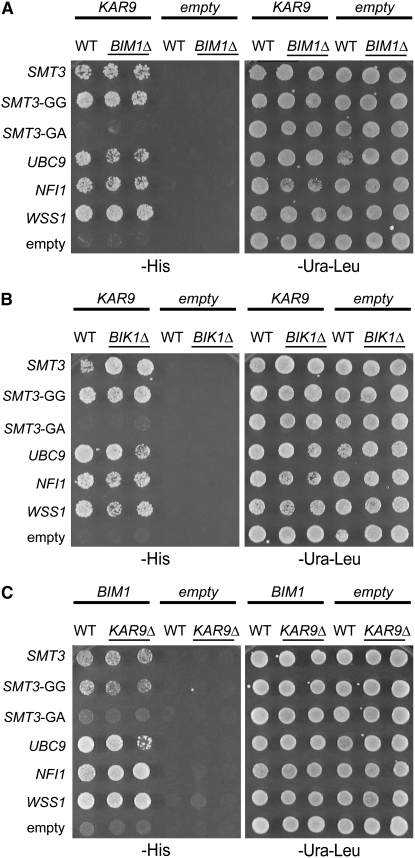
The interaction between Smt3p and Kar9p does not occur through a tertiary complex with Bim1p or Bik1p:
The two-hybrid assay does not differentiate between the direct interaction of two proteins and the interaction of two proteins resulting from a third bridging protein. Kar9p physically interacts with the microtubule-associated proteins, Bim1p and Bik1p (Miller et al. 2000; Moore et al. 2006). Therefore, we first tested whether the KAR9-SMT3 interaction required either of these proteins by deleting BIM1 or BIK1 from the two-hybrid reporter strain. As observed in the wild-type strain, KAR9 retained its interaction with SMT3 and SMT3-GG in both the bim1Δ and bik1Δ two-hybrid reporter strains (Figure 3, A and B). Furthermore, KAR9 also maintained its interactions with UBC9, NFI1, and WSS1 in both these reporter strains (Figure 3, A and B). These results suggest that the interactions between Kar9p and the sumoylation proteins do not require either Bim1p or Bik1p.
Figure 3.—
Testing the bridging model. Either wild-type (yRM1757) or two-hybrid reporter strains deleted for BIM1 (yRM2057), or disrupted for BIK1 (yRM2258), or deleted for KAR9 (yRM6172) were transformed with the following constructs, KAR9-BD, BIM1-BD, empty BD, AD-SMT3, AD-SMT3-GG, AD-SMT3-GA, AD-UBC9, AD-NFI1, AD-WSS1, or empty AD, as indicated. For these assays, the two-hybrid reporter strains were haploid. The interactions were assayed on media lacking histidine (−his) after 2–3 days at 30°. (A) Kar9p maintained its interactions with sumoylation proteins in the absence of Bim1p. Two independent colonies containing KAR9-BD or the empty DBD were tested in the reporter strains deleted for BIM1 or BIK1. One colony containing KAR9-BD or the empty BD was tested in the wild-type reporter strain. (B) Kar9p retained its interactions with sumoylation proteins in reporter strains deleted for BIK1. Transformations, plasmids, and strains were as described in A. (C) Bim1p maintained its two-hybrid interactions with sumoylation proteins in the absence of Kar9p.
We next tested the reciprocal possibility that the interaction between Bim1p and the sumoylation proteins might require KAR9. In the two-hybrid reporter deleted for KAR9, the interaction between BIM1 and SMT3 was maintained. BIM1 also retained its interaction with UBC9, NFI1, and WSS1 in the absence of Kar9p (Figure 3C). These results suggest that Kar9p is not required for Bim1p to interact with sumoylation-pathway proteins.
Because E3 enzymes facilitate the conjugation of SUMO to substrates, we also tested whether reporter strains lacking the E3's SIZ1 or NFI1 would affect the interaction of either BIM1 or KAR9 with the sumoylation machinery. Indeed, we found this to be the case. For BIM1, the deletion of either SIZ1 or NFI1 disrupted interactions with both SMT3-GG and UBC9 (data not shown). For KAR9, the two E3's had different effects. Deletion of SIZ1 had little or no effect on either SMT3-GG or UBC9. In contrast, the deletion of NFI1 disrupted KAR9's interaction with UBC9, but had little or no effect on its interaction with SMT3-GG (data not shown). The differing requirements for the E3 enzymes in these two-hybrid interactions indicate that the sumoylation system may utilize distinct mechanisms to regulate different microtubule-associated proteins.
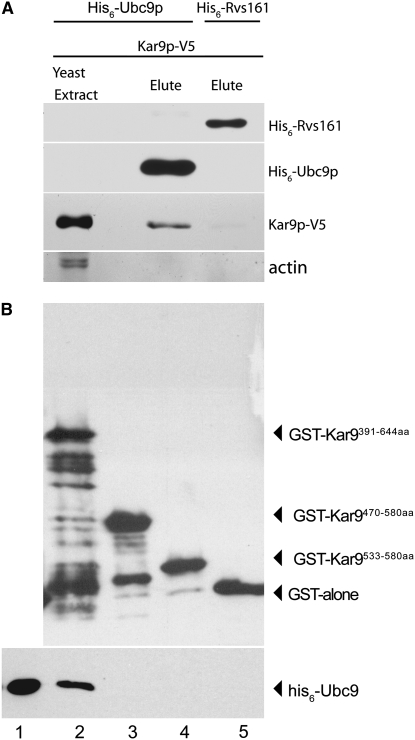
His6-Ubc9p binds the basic domain of Kar9p:
To confirm these two-hybrid interactions by other methods, we sought to determine whether Kar9p and Ubc9p could physically interact. We focused on Ubc9p first because it is the only E2 SUMO-conjugating enzyme in yeast and is essential for SUMO conjugation (Johnson and Blobel 1997; Schwarz et al. 1998; Sampson et al. 2001; Bencsath et al. 2002). Ubc9p tagged with six histidine residues was expressed in bacteria and purified on a Talon affinity column (Johnson and Blobel 1997). The amphiphysin-like protein Rvs161p tagged with his6 served as a negative control. KAR9 was tagged with the viral V5 epitope and expressed under the control of the GAL1-inducible promoter. Yeast extracts containing overexpressed Kar9p-V5 were applied to both columns (see materials and methods). Bound proteins were eluted from the columns using EDTA. Using Western blot analysis, Kar9p-V5 was detected in the eluant of the his6-Ubc9p column, whereas little or none was detected in the his6-Rvs161p column (Figure 4). These data suggest that Kar9p and Ubc9p interact physically.
Figure 4.—
Kar9p binds Ubc9p. (A) His6-Ubc9p can pull Kar9p out of a yeast extract. Kar9p tagged with V5 was expressed in yeast using the GAL1 inducible promoter (pRM4319). His6-tagged Ubc9p (pRM5169) and Rvs161p (pRM6028/pMR3548) were expressed in bacteria and purified by Talon affinity column chromatography. Yeast extract containing Kar9p-V5 was then added to the column. Bound protein was eluted by the addition of EDTA and prepared for SDS–PAGE and Western blotting. The presence of his6-Ubc9p or his6-Rvs161 and Kar9p-V5 was detected using anti-his6 and anti-V5 antibodies, respectively. Kar9p-V5 coeluted with his6-Ubc9p (lane 2) but not with his6-Rvs161p (lane 3). (B) Ubc9p binds to the basic domain of Kar9p. GST alone (pRM3369) and three truncations of the basic domain of Kar9p fused to glutathione-S-transferase [390–644 aa (pRM3366), 470–580 aa (pRM3367), and 533–580 aa (pRM3368)] were harvested from bacteria and bound to glutathione agarose beads (see materials and methods). One milligram of bacterial extract containing his6-Ubc9 was applied to the beads complexed with either the GST-Kar9p truncations (lanes 2–4) or GST alone (lane 5). Bound protein was eluted with the addition of reduced glutathione (lanes 2–4) and samples were prepared for Western blotting. Lane 1 represents 1/200 of the his6-Ubc9 that was loaded onto the GST-Kar9p beads.
We next investigated whether Ubc9p could bind to Kar9p directly. Because full-length Kar9p is not stable in bacteria, a series of three truncations of Kar9p containing various lengths of its basic domain were fused to GST, expressed in bacteria, and purified on glutathione beads (see materials and methods). His6-Ubc9p contained in bacterial extracts was tested for its association with the immobilized Kar9p protein. As shown in Figure 4B, his6-Ubc9p was present in the eluant from columns containing the fusions to the carboxy-terminal third of Kar9p (391–644 aa), but not in the shorter Kar9p truncations or GST alone (Figure 4B). We conclude that Ubc9p interacts with the basic domain of Kar9p and that no other yeast protein is required for this interaction.
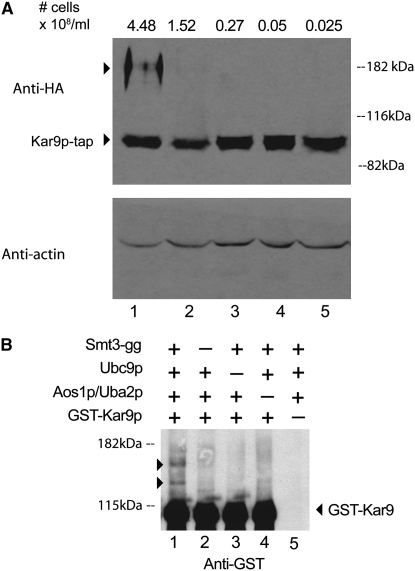
Higher molecular weight forms of Kar9p are observed in cultures grown to high density:
Sumoylation of some proteins is upregulated in response to cell stress (Zhou et al. 2004; Hannich et al. 2005). To explore the possibility that Kar9p could be modified when grown to high density, cultures of Kar9p-tap were grown to varying densities and analyzed by Western blot analysis. In cultures grown to 4.48 × 108 cells/ml or higher, we observed two slower migrating bands that reacted specifically with anti-HA, in addition to the Kar9p band migrating at the predicted molecular weight (Figure 5A). These bands were present in much lower intensities or not at all in cultures in midexponential phase (lanes 2–5). Although we have not yet identified the molecular basis for this shift, it is tempting to speculate that these slower migrating bands represent sumoylated forms of Kar9p. However, these bands migrate slower than what would be predicted by the addition of single or double SUMO moieties.
Figure 5.—
Higher molecular weight forms of Kar9p. (A) Kar9p in densely grown cultures. A yeast strain containing Kar9p-tap was grown to the indicated cell densities in YPD. The cell number was determined using a hemacytometer. Whole cell extracts were analyzed by Western blotting. The tap epitope used in the Kar9p-tap fusion contains his6, HA, and protein A epitopes (Moore et al. 2006). Anti-HA was used to detect the HA epitope. Anti-actin was used as a loading control (bottom). (B) Smt3p can be conjugated to Kar9p in vitro. GST-Kar9p was purified from yeast (yRM6836). The processed form of Smt3p, his6-Smt3p-gg (pRM6713), and Ubc9p (pRM5169) were purified from bacteria by nickel affinity chromatography. The two components of the SUMO E1 complex, Aos1p and Uba2p (pRM6760), were copurified from bacteria using glutathione affinity chromatography. The assay was performed by combining the indicated components of the SUMO pathway with GST-Kar9p at 30° for 2 hr (see materials and methods). The same amount of ATP and an ATP regeneration system were added to each reaction. The reactions were prepared for SDS–PAGE and Western blot analysis using anti-GST. GST-Kar9p can be conjugated by Smt3p-gg in vitro (lane 1) judging by the presence of the shifted bands (arrowheads). These bands were consistently absent in reactions lacking Smt3p-gg (lane 2), Ubc9p (lane 3), Aos1p/Uba2p (lane 4), and Kar9p substrate (lane 5).
An in vitro sumoylation assay results in higher molecular weight Kar9p bands:
To further investigate the relationship between sumoylation and Kar9p, we asked whether Smt3p could be transferred to Kar9p in vitro. We employed an in vitro sumoylation assay modeled after that of Bencsath et al. (2002) and Bhaskar et al. (2002), purifying the sumoylation components from bacteria (see materials and methods). Smt3p was used in the processed form, Smt3p-gg. Using GST-Kar9p purified from yeast (see materials and methods) (Grayhack and Phizicky 2001; Phizicky et al. 2002), two shifted forms of Kar9p were generated that were dependent upon the presence of all the sumoylation enzymes in the reaction (Figure 5B, arrowheads). This indicates that Kar9p can be sumoylated in vitro on at least two residues. This finding helps explain why mutagenesis of single lysine residues in Kar9p did not disrupt the two-hybrid interaction (see below). In a parallel experiment, no shifted bands specific to Bim1p-V5 were observed (data not shown).
A leucine 304-to-proline mutation in Kar9p disrupts its two-hybrid interaction with Smt3p and proteins involved in sumoylation:
Although other attachment sites are known, Smt3p/SUMO conjugation to target proteins is often observed at lysines within the standard consensus sequence, ΨKxE/D. KAR9 contains one of these sites, at K301, and two partial consensus sites containing KxD, at K76 and K120. To determine whether any of these sites were responsible for the interaction of Smt3p with Kar9p, we mutated these lysines to arginine within the KAR9-BD two-hybrid construct by site-directed mutagenesis. As shown in Table 2, these single mutations as well as the triple mutation did not diminish the two-hybrid interactions between KAR9 and SMT3. Mutagenesis of lysines within less well-conserved sites at K224R, K307R, K529R, and K594R also failed to disrupt the KAR9-SMT3 two-hybrid interaction (Table 2).
TABLE 2.
Mutagenesis of potential sites for Smt3p interaction
| Potential SUMO sites | Flanking sequence | Interaction with AD-SMT3 |
|---|---|---|
| KAR9 | ||
| K76 | NKKRL GKDD ILLFM | + |
| K120 | LEDEFS KDD QDSDK | + |
| K224, K227 | TETTE PKVPK FSPA | + |
| K281, K282 | QTILQ KKYE LIMKD | NDa |
| K301 | SEFRE LKVE LIDKR | + |
| K307 | SEFRE LKVE LIDKR | + |
| K301, K307 | SEFRE LKVE LIDKR | + |
| K76, K120, K301, K307 | NKKRL GKDD ILLFM | + |
| LEDEFS KDD QDSDK | ||
| SEFRE LKVE LIDKR | ||
| L304P | KVELIDKRWN | − |
| K529 | SRGEN EKSP DSFIT | + |
| K594 | ENTPI AKVF QTPPT | + |
| M293P | IMKDYRF M NSEFRE | − |
| BIM1 | ||
| K25, K26 | NLNYKKIEECGTG | + |
| K110 | HWIRHKDESVYDP | + |
| K266, K267 | LRFVKKVESILYA | + |
The lysine residues (underlined) were mutated by site-directed mutagenesis in the KAR9-BD plasmid (pRM1493). In addition to the “standard” site identified by inspection (K301), additional potential SUMO attachment sites were identified by the SUMOplot Prediction software program (Abgent, http://www.abgent.com/tools/sumoplot). Leucine 304 lies immediately adjacent to a “standard” SUMO consensus site at K301. For the kar9 mutants, the following plasmids contain the indicated mutations: K76R (pRM4544), K120R (pRM6575), K224R K227R (pRM6576), K301R (pRM4472), K307R (pRM5415), K301RK307R (pRM6510), K76R K120R K301R K307R (pRM7831), L304P (pRM5421), K529R (pRM5424), K594R (pRM4698), and M293P (pRM6707). For the bim1 mutants, the plasmids are as follows: K25RK26R (pRM4699), K110R (pRM4700), and K266RK267R (pRM4701). Interaction with AD-SMT3 was tested as described in Figure 2, A and B. The boldface font represents residues of interest, such as potential sumoylation sites.
We were unable to obtain the K281R K282R mutant, as either a single or double mutation. This is most likely due to the highly AT-rich region surrounding these residues.
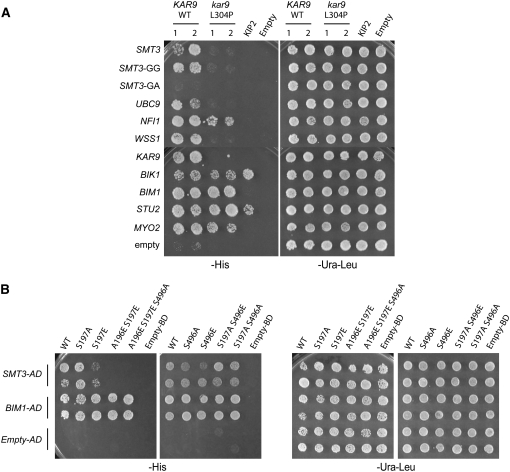
In the course of these mutagenesis efforts, we discovered a serendipitous mutation resulting in the conversion of leucine 304 to proline, kar9-L304P. Two-hybrid analysis of this mutant revealed that its two-hybrid interaction with Smt3p was lost (Table 2, Figure 6A). We then analyzed its interactions with other proteins of the sumoylation pathway and found that its interactions with UBC9 and WSS1 were also abolished. Its interaction with the E3 factor Nfi1p was diminished in comparison to wild type (WT). Thus, the interactions of kar9-L304P with the entire panel of sumoylation proteins were disrupted.
Figure 6.—
(A) The kar9-L304P mutation disrupts interactions with sumoylation proteins, but not with microtubule-associated proteins or Myo2p. AD-SMT3, AD-SMT3-GG, AD-SMT3-GA, AD-UBC9, AD-NFI1, AD-WSS1, AD-KAR9 (aa 1–393) (pRM1895), AD-BIK1 (pRM2627), AD-BIM1 (pRM1893), AD-STU2 (aa 649–888) (pRM1916), and AD-MYO2 (aa 1083–1574) (pRM2473) or empty AD was transformed into the two-hybrid reporter strain (yRM1756/PJ69-4α) and selected on media lacking leucine. KAR9-BD or kar9-L304P-BD was transformed into the two-hybrid reporter strain with the opposite mating type (yRM1757/PJ69-4A) and selected on media minus uracil. After mating, diploids were selected on media lacking uracil and leucine (−ura −leu). Two colonies containing either wild-type KAR9 or kar9-L304P were analyzed. Growth was scored on plates lacking histidine (−his) at 30° after 2–3 days. (B) Phosphorylation of Kar9p at serine 197 inhibits the interaction between KAR9 and SMT3. Phospho-mimetic and phospho-inhibitory mutants of Kar9p were generated in which serine 197 and/or serine 496 were mutated to alanine (A) or glutamic acid (E) by site-directed mutagenesis (Moore and Miller 2007). GAL4-DNA binding domain fusions of these mutants were tested against the activation domain fusions of SMT3 and BIM1 in a two-hybrid reporter strain lacking BIM1 (yRM2057) by scoring growth on media lacking histidine (−his) after incubation at 30° for 2–3 days. The following strains containing BD fusions were used: kar9-S197A (pRM6050), kar9-S197E (pRM5617), kar9-A196E S197E (pRM6255), kar9-A196E S197E S496A (pRM6295), kar9-S496A (pRM5777), kar9-S496E (pRM5619), kar9-S197A S496E (pRM6294), and kar9-S197A S496A (pRM6113). The A196E S197E mutant disrupts the interaction between KAR9 and SMT3, but not with BIM1.
Since sumoylation is known to affect protein–protein interactions, we next asked whether the kar9-L304P mutant maintained the two-hybrid interactions with previously identified interactors of Kar9p that function in spindle positioning. To perform these experiments, Bik1p, Bim1p, the amino terminal half of Kar9p (1–393 aa), the amino-terminal half of Stu2p, and the carboxy-terminal tail domain of Myo2p were fused to the GAL4 activation domain. The kar9-L304P mutant retained its interaction with all the spindle positioning proteins, except for Kar9p itself (Figure 6A). Because the L304P mutant maintains interactions with many of its known binding partners, these findings suggest that the L304P mutant is likely to be at least partially functional.
Because the L304P mutation resides in a region of Kar9p predicted with moderate certainty to form a coiled-coil structure at amino acids 263–335 (Miller and Rose 1998), we analyzed the effect this mutation would have on the predicted secondary structure of Kar9p. Using the COILs program (Lupas et al. 1991) with an analysis window of 28, this mutation is predicted to abolish the coiled-coil motif. In an attempt to separate the predicted structure of Kar9p from the binding of Smt3p, we engineered in a separate mutation that is also predicted to disrupt the coiled-coil structure of Kar9p, M293P, converting the methionine residue at position 293 to a proline. However, this mutation also resulted in the disruption of the two-hybrid interaction between Kar9p and the SUMO pathway proteins (Table 2). These findings suggest the possibility that the coiled-coil structure is required for the interaction with Smt3p.
In some instances, sumoylation occurs on multiple lysine residues (Johnson and Blobel 1999; Hofmann et al. 2000; Steffan et al. 2004). Because the kar9-L304P mutation is flanked by a canonical sumoylation site containing a lysine residue at position 301 and another lysine at position 307, we considered the possibility that either lysine 301 or 307, or both, might be targets for SUMO conjugation. To test this we generated a K301R K307R double mutant. However, this mutant maintained its interaction with Smt3p (Table 2), Ubc9p, Nfi1p, Wss1p, and with the spindle positioning proteins (data not shown), suggesting that the kar9-L304P mutant does not exert its effect by blocking SUMO conjugation at these nearby lysines. This is also consistent with multiple sumoylation sites.
The lack of interaction of L304P in the two-hybrid assay for Smt3p and Ubc9 suggested that it should display a diminished shift in the in vitro sumoylation assay. To investigate this, GST-Kar9p-L304P was purified side by side with wild-type Kar9p. To our surprise, L304P did shift, producing two higher molecular weight bands (data not shown). However, in contrast to wild type, the intensity of the top band in L304P is diminished with a concomitant increase in the lower band. This result could be explained if Kar9p has at least three sumoylation sites that are used in pairs and if only one of the three sites is impaired in L304P.
In Bim1p, lysine-to-arginine mutations were also engineered at three locations containing the partial consensus site for sumoylation, KxE, at residues K25 K26, K110, and K266 K267. These single residue mutations did not disrupt the interaction of Bim1p with Smt3p (Table 2). Because sumoylation at multiple sites and at unconventional-consensus sites is not uncommon, our failure to disrupt the interaction by mutation of these selected lysines does not argue strongly for or against a model involving conjugation.
The phosphorylation status of Kar9p affects its interaction with Smt3p:
Kar9p is phosphorylated at serines 197 and 496 by the cyclin-dependent kinase Cdc28p (Liakopoulos et al. 2003; Maekawa and Schiebel 2004; Moore et al. 2006; Moore and Miller 2007). Phosphorylation can also regulate sumoylation, either positively or negatively (Müller et al. 1998; Hietakangas et al. 2003; Yang et al. 2003; Hietakangas et al. 2006). We therefore tested whether phospho-inhibited or phospho-mimetic forms of Kar9p would affect its two-hybrid interaction with Smt3p. The phospho-inhibited serine-to-alanine mutations at positions 197 and 496, either singly or in combination, had little or no effect on the interaction (Figure 6B). A phospho-mimetic mutation at position 496 in which serine was replaced by a glutamic acid residue also had no obvious defect. However, the phospho-mimetic mutation S197E significantly reduced the interaction. Phosphorylation introduces two negative charges, whereas only one negative charge is present on glutamic acid. To simulate this charge difference, a second glutamic acid was introduced at position 196, creating A196E S197E (Moore and Miller 2007). This mutation eliminated the interaction with Smt3p. As reported previously, the phosphorylation status of Kar9p did not affect its interaction with Bim1p (Moore and Miller 2007). These findings suggest that phosphorylation at serine 197 may act to antagonize the sumoylation of Kar9p.
To study the phosphorylation status of L304P in more detail, we examined the phospho-banding pattern of Kar9p in B150 buffer as described previously (Moore et al. 2006; Moore and Miller 2007) and by preparation of samples in 17% TCA (Ohashi method, see materials and methods). For both methods, L304P was phosphorylated, but the levels of the top band were diminished, with an increase in the middle band. This suggests that L304P has reduced levels of phosphorylation at just one site (data not shown). Side-by-side comparisons of the phospho-banding patterns showed that the L304P banding pattern was distinct from that of both S197A and S496A alone (data not shown) (Moore and Miller 2007). This is consistent with our previous finding that a third phosphorylation site exists on Kar9p (Moore and Miller 2007). Determining the identity of this altered site will require additional investigation. Finally, we tested the kar9-L304P mutant for interaction with Clb5p, which is implicated in Kar9p phosphorylation at serine 496 (Moore et al. 2006; Moore and Miller 2007). This interaction was maintained, but at a reduced level in comparison to wild-type Kar9p (data not shown). Whether the reduced Clb5p interaction is responsible for the reduced L304P phosphorylation will be determined by future work.
The kar9-L304P mutant displays a defect in spindle positioning:
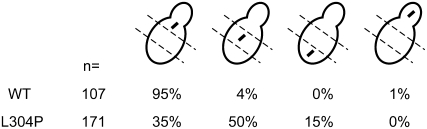
To assess the effects of the L304P mutation on Kar9p function, we first analyzed the position of the mitotic spindle marked with CFP-Tub1p. In wild type, 95% of cells have the spindle positioned near the bud neck (n = 107), in agreement with previous findings (Figure 7) (Liakopoulos et al. 2003; Moore et al. 2006; Moore and Miller 2007). In contrast, 65% of kar9-L304P cells have mispositioned spindles, with 50% of the mutant cells displaying spindles positioned in the center of the mother cell and 15% of the spindles in the distal third of the mother cell. Thus, the L304P mutation results in the short bipolar spindle being positioned farther away from the mother-bud neck than in wild type.
Figure 7.—
kar9-L304P displays a defect in positioning the mitotic spindle. The kar9-L304P mutant (yRM5973) and wild type (yRM5970) were scored for the position of the mitotic spindle marked with CFP-Tub1p (pRM3448). Only cells containing short bipolar spindles were counted. The data represent the percentages of cells in the following categories: cells with the spindle at the bud neck (column 1), in the middle area of the mother cell (column 2), in the bottom area of the mother cell (column 3), and in the bud (column 4).
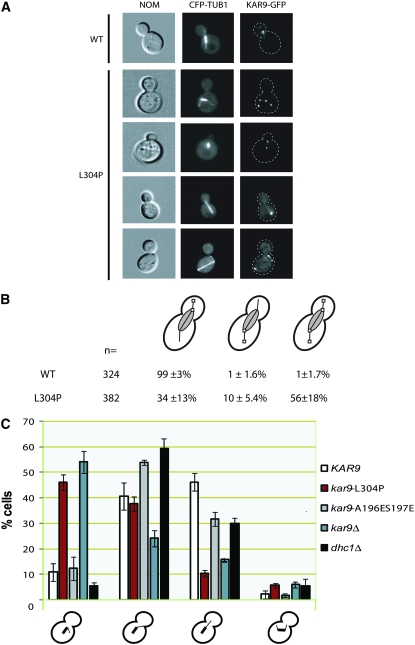
kar9-L304P-3GFP is mislocalized to both spindle poles:
Because sumoylation can alter the localization of proteins, we investigated the localization of the kar9-L304P mutant by tagging its carboxy terminus with three copies of the GFP at the endogenous KAR9 locus, as previously described (Moore et al. 2006; Moore and Miller 2007). Consistent with previous reports, wild-type Kar9p localized primarily to the bud-directed spindle pole body and/or the microtubules emanating from this pole (99%) (Figure 8) (Liakopoulos et al. 2003; Moore et al. 2006; Moore and Miller 2007). In contrast, kar9-L304P localized to the correct pole in only 34% of cells (Figure 8B). Instead, 56% of the L304P cells had Kar9p on both poles and/or both sets of microtubules. These data are consistent with the hypothesis that an interaction with Smt3p is involved in restricting Kar9p to one SPB.
Figure 8.—
kar9-L304P-3GFP localizes to both sets of cytoplasmic microtubules. (A) Examples of Kar9p-3GFP (yRM5970) and kar9-L304P-3GFP (yRM5973) localization in cells expressing CFP-Tub1p labeled microtubules. (B) Quantification of Kar9p localization. Cells with short bipolar spindles and anaphase cells were scored. The percentages shown are the mean of each category ± the standard deviation. These values were calculated from five independent counts of n > 50. Kar9p-3GFP is depicted as white squares. (C) The kar9-L304P mutant displays a cytoplasmic microtubule-orientation defect. Microtubules were marked with GFP-Tub1p. The orientation of cytoplasmic microtubules was scored in preanaphase cells of the following strains: WT (yRM7258), kar9-L304P (yRM7261), kar9-A196ES197E (yRM6903), kar9Δ (yRM7361), and dhc1Δ (yRM7263). Cells with no cytoplasmic microtubule oriented toward the bud or neck but instead seen in the mother cell were scored as “misoriented” (first bar grouping). If no microtubule entered the bud but at least one ended at the bud neck, it was categorized as “to the neck” (second bar grouping). Cells with at least one microtubule entering the bud were scored as “into the bud” (third bar grouping). Cells with both microtubules associated with daughter and mother SPB orient toward the daughter cell were categorized as “both mother and daughter microtubule going toward the bud” (fourth bar grouping). Error bars represent the standard error of the mean. For each strain, the data were collected with at least five replicates containing at least 50 cells each (>280 total, with the exception of the dhc1Δ strain in which 130 cells were scored).
kar9-L304P displays a microtubule orientation defect:
Because KAR9 is important for cytoplasmic microtubule orientation, we next assayed whether microtubules labeled with GFP were oriented properly into the bud. As shown in Figure 8C, kar9-L304P displayed a significant increase in the number of cells with misoriented microtubules (46%) in comparison to wild type (11%). Concomitantly, L304P showed a decrease in the number of cells with microtubules going into the bud (10.5%) compared to wild type (46%). Kar9p is also important for the linkage of microtubules to the bud neck (Liakopoulos et al 2003; Moore and Miller 2007). However, 37% of kar9-L304P cells had microtubules interacting with the neck, similar to that seen in wild type (40%). These findings imply that while kar9-L304P can link microtubules to the bud neck, its function in orienting microtubules into the bud is disrupted. Consistent with previous reports (Moore and Miller 2007), microtubule orientation in kar9-A196ES197E was similar to wild type (Figure 8C).
kar9-L304P is not viable in the absence of dynein:
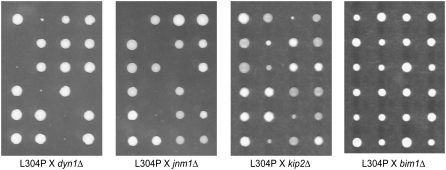
Spindle positioning is carried out by two partially redundant pathways, the Kar9p pathway and the dynein pathway (Miller and Rose 1998). Prior to anaphase, the Kar9p pathway positions the nucleus at the bud neck and orients the mitotic spindle along the mother-bud axis (Miller and Rose 1998; Kusch et al. 2002). The dynein pathway is responsible for providing the pulling force that moves the nucleus into the mother-bud neck at or just after the initiation of anaphase (Kahana et al. 1995; Yeh et al. 1995; Adames and Cooper 2000; Lee et al. 2005). Neither pathway by itself is essential for cell viability (Muhua et al. 1994; Miller and Rose 1998). However, disruption of both pathways simultaneously is lethal (Miller et al. 1998; Miller and Rose 1998). Therefore, to investigate whether the kar9-L304P mutation disrupted the essential function of Kar9p that is revealed in the absence of dynein, we crossed it to mutants in the dynein pathway. Of the 23 segregants predicted to be kar9-L304P dyn1Δ double mutants on the basis of the distribution of markers in sister spores, all produced colonies that displayed a severe growth defect or were dead (Figure 9, Table 3). A similar synthetic phenotype was observed when kar9-L304P was crossed to a deletion of JNM1, which encodes a component of the yeast dynactin complex (Figure 9, Table 3). When kar9-L304P was combined with a deletion of KIP2, which encodes a kinesin, a slightly less severe synthetic phenotype was observed; the kar9-L304P kip2Δ double mutant appeared synthetically sick. These results suggest that the kar9-L304P mutation disrupted a critical aspect of Kar9p function.
Figure 9.—
kar9-L304P displays a severe growth defect in combination with mutations in the dynein pathway. Examples of dissection plates are shown of crosses between kar9-L304P-3GFP and strains deleted for components of the dynein pathway, the dynein heavy chain DHC1, the dynamitin component of the dynactin complex JNM1, and the kinesin KIP2. kar9-L304P crossed to a strain deleted for BIM1, which functions in the KAR9 pathway, is shown for comparison. Colonies were grown on YPD at 30° for 2 days. Strains are as listed in Table 3. Tetrads are aligned horizontally.
TABLE 3.
Kar9p-L304P displays a synthetic lethal phenotype in combination with dynein mutants
| Cross | Viability | Tetrads (PD:TT:NPD) | Double mutants viable/predicted |
|---|---|---|---|
| kar9-L304P × dyn1Δ | Not viable | 25 (6:15:4) | 0/23 |
| kar9-L304P × jnm1Δ | Not viable | 24 (5:15:4) | 0/23 |
| kar9-L304P × kip2Δ | Synthetic sick | 22 (4:17:1) | 0/19 |
| kar9-L304P × bim1Δ | Viable | 31 (6:23:2) | 27/27 |
Double mutants were generated by meiotic crosses and scored for growth after 2–3 days. Progeny exhibiting no growth or forming microcolonies ∼10-fold smaller than single mutant or wild-type cells (see Figure 9) were scored as “not viable.” The kar9-L304P kip2Δ double mutants were scored as “synthetic sick” as these colonies were smaller than those produced by single mutants but did not exhibit phenotypes as severe as those observed for other double mutants. The following parental strains were used in these crosses: kar9-L304P (yRM6739 or yRM6740), dyn1Δ (yRM425), jnm1Δ (yRM6398), kip2Δ (yRM435), and bim1Δ (yRM6764).
Because sumoylation can affect the stabilization of some proteins (Steffan et al. 2004), we investigated whether the steady-state levels of kar9-L304P were changed in comparison to wild type. For this, whole cell extracts were prepared for Western blot analysis by bead beading either in B150 buffer or trichloroacetic acid in water (Ohashi method). For samples prepared by the Ohashi method, this analysis showed that the levels of full-length Kar9p were equivalent in kar9-L304P and wild-type strains in comparison to the Pgk1 loading control, albeit, more breakdown products were seen for L304P (data not shown). However for B150 buffer, the apparent levels of L304P were more than fivefold higher than wild-type Kar9p when compared to the equal loading control (data not shown). Similar results were seen with both Kar9p-L304P-tap and Kar9p-L304P-3GFP. Because the samples were prepared side by side in either B150 buffer or TCA, we conclude that the L304P mutation does not significantly affect steady-state levels of Kar9p protein. Instead, the L304P mutation significantly increases the solubility of Kar9p in B150 buffer. This is consistent with L304P having an altered protein conformation or protein–protein interactions.
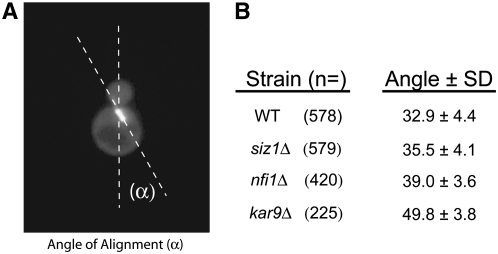
The mitotic spindle in E3 mutants displays a modest increase in its angle of alignment:
Our data suggests the hypothesis that sumoylation may regulate spindle positioning. If true, then one might expect that mutants deficient in sumoylation would display defects in spindle positioning. To investigate this question, we measured the angle at which short bipolar spindles visualized with GFP-Tub1p were aligned along the mother-bud axis (Figure 10) in strains deleted for KAR9, or two genes encoding the E3 sumoylation enzymes SIZ1 and SIZ2/NFI1. As expected, the average angle of spindle alignment in kar9Δ cells (49.8° ± SD 3.8, n = 225) was greater than that in wild type (32.0° ± SD 4.4, n = 578). In contrast, the angle of alignment for siz1Δ strains (35.5° ± SD 4.1, n = 579) was only slightly increased in comparison to wild type. This small difference was nevertheless statistically significant using an unpaired t-test (P < 0.0001). nfi1Δ mutants also displayed a small but significant increase in the average angle of alignment (39° ± SD 3.6, n = 420, P < 0.0001) compared to wild type. No obvious difference in the angle of alignment was observed in strains deleted for WSS1 (data not shown). These measurements were conducted in the two-hybrid reporter strain deleted for the indicated mutants. To confirm these findings, we remade the SIZ1 and NFI1 deletions in our standard laboratory strain and repeated the analysis. In this analysis the angles were binned into three categories, aligned (0°–45°), partially aligned (45.1°–70°), and not aligned (70.1°–90°). Again, we found that nfi1Δ displayed a moderate spindle positioning defect, with 27% of the cells containing spindles that were not aligned (70.1°–90°) compared to 14% in wild type (see supplemental Figure S1). These results suggest that sumoylation exerts a modest effect on spindle positioning.
Figure 10.—
nfi1Δ strains display a moderate increase in the angle at which the spindle aligns along the mother-bud axis. (A) The angle of alignment (α). (B) Preanaphase cells displaying short bipolar spindles labeled with GFP-Tub1p (pRM4419) were analyzed in strains deleted for SIZ1 (yRM4769), NFI1 (yRM4755), kar9Δ (yRM6172), and wild type (yRM1756 or yRM1757). Each strain was counted on at least 4 separate days.
Sumoylation mutants do not display an obvious synthetic phenotype in combination with kar9Δ or dyn1Δ mutants:
Spindle positioning requires two partially redundant pathways, the Kar9p pathway and the dynein pathway (Muhua et al. 1994; Miller and Rose 1998). To determine whether the moderate spindle positioning defect seen for nfi1Δ would be affected by the absence of either of these mutants, we crossed deletions of the E3 enzymes encoded by CST9, SIZ1, and NFI1 to deletions in either KAR9 or DYN1. In all cases, the colony size of the germinated double-mutant spores displayed no growth defect that was obviously different than either single mutant (Table 4). Similarly, we crossed smt3-11 and smt3-12 to deletions in either KAR9 or JNM1, which encodes a component of the dynactin complex. These smt3 alleles are temperature sensitive, precluding analysis at 37°, but show only mild perturbations at the permissive temperature. Therefore, we analyzed the double mutants at 23°. However, no obvious growth defect was seen in the colony size of these double mutants.
TABLE 4.
Sumoylation mutants do not display synthetic lethality with either KAR9 or dynein pathway mutants
| Cross | Viability | Tetrad (PD:TT:NPD) | Double mutant (viable/predicted) |
|---|---|---|---|
| cst9Δ × jnm1Δ | Viable | 25 (3:16:6) | 27/28 |
| siz1Δ × jnm1Δ | Viable | 24 (3:19:2) | 22/23 |
| cst9Δ × dhc1Δ | Viable | 27 (3:20:4) | 28/28 |
| siz1Δ × dhc1Δ | Viable | 22 (4:14:4) | 22/22 |
| nfi1Δ × dhc1Δ | Viable | 27 (2:21:4) | 28/29 |
| SMT3 × jnm1Δ | Viable | 27 (8:15:4) | 20/23 |
| smt3-11 × jnm1Δ | Viable | 28 (6:16:6) | 28/28 |
| smt3-12 × jnm1Δ | Viable | 16 (2:12:2) | 16/16 |
| cst9Δ × kar9Δ | Viable | 31 (7:18:6) | 28/30 |
| siz1Δ × kar9Δ | Viable | 28 (2:21:5) | 30/31 |
| nfi1Δ × kar9Δ | Viable | 30 (9:19:2) | 23/23 |
| SMT3 × kar9Δ | Viable | 33 (5:25:3) | 30/31 |
| smt3-11 × kar9Δ | Viable | 19 (1:14:4) | 20/22 |
| smt3-12 × kar9Δ | Viable | 20 (3:11:6) | 23/23 |
Double mutants between sumoylation mutants and KAR9 or dynein pathway mutants were generated by meiotic crosses and scored for growth after 2–3 days. The tetrads were incubated at 30° except SMT3, smt3-11, and smt3-12 crosses, which were grown at 23°. The double mutant, single mutant, and wild type exhibited no difference in growth at 30° or 23°. Thus the double mutants were scored as viable. The following parental strains were used in the crosses: cst9Δ (yRM7166), siz1Δ (yRM7140), nfi1Δ/siz2Δ (yRM7248), SMT3 (yRM6491), smt3-11 (yRM6493), smt3-12 (yRM6492), jnm1Δ (yRM6727), dhc1Δ (yRM373), and kar9Δ (yRM1340).
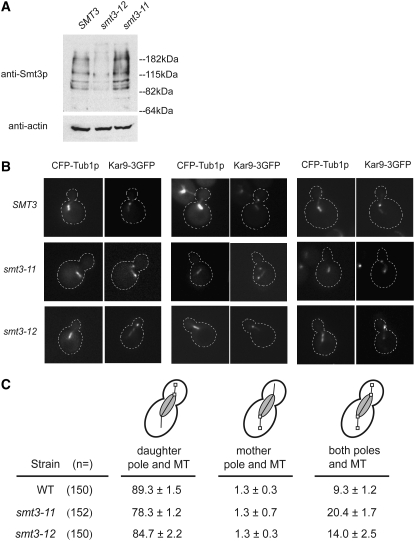
Asymmetric localization of Kar9p is perturbed in smt3-11 mutants:
The localization of Kar9p is usually restricted to the bud-directed spindle pole body and the associated microtubules (Liakopoulos et al. 2003; Moore and Miller 2007). To investigate the role of sumoylation on this asymmetric localization, we studied the localization of Kar9p in two smt3 mutants, smt3-11 and smt3-12, which have opposing effects on the overall sumoylation levels in the cell. The smt3-12 mutant displays a decreased level of sumoylation in comparison to wild type, whereas the smt3-11 mutant displays an increased level of sumoylation (Figure 11A).
Figure 11.—
Kar9p-GFP localization is altered in smt3-11 mutants. (A) Global sumoylation levels are increased in smt3-11 and decreased in smt3-12 mutants. Whole cell extracts were prepared by the Ohashi method from SMT3 (yRM6491), smt3-11 (yRM6493), or smt3-12 strains (yRM 6492) grown at room temperature in YPD. Western blots were probed with rabbit anti-Smt3p. (B) Kar9p shows an increase in the symmetric localization in smt3-11. Kar9p-3GFP (pRM3662) at the chromosomal locus was analyzed in a wild-type SMT3 background (yRM7712), in smt3-11 (yRM7604), and smt3-12 (yRM7635) mutants, each containing CFP-marked microtubules. Cells were grown in SC media at room temperature. Three examples of Kar9p-3GFP localization are shown for each strain. (C) Quantification of Kar9p-3GFP localization in the same strains shown in B. Cells with short bipolar spindles were scored for Kar9p-3GFP localization and classified as having Kar9p on the daughter-bound spindle pole body and its associated microtubules, or on the mother spindle pole body and associated microtubules, or on both spindle pole bodies. Percentages were calculated from averages of three independent experiments conducted on different days, each with an N of at least 50. These averages represent the mean percentages of each category ± the standard error of the mean.
To study Kar9p localization, Kar9p-3GFP and CFP-Tub1p were introduced into these strains by genetic crosses. Kar9p was scored in preanaphase cells with short bipolar spindles as localized either on the daughter-bound spindle pole body and its associated microtubules or on the mother pole and its microtubules, or localized on both poles. In wild-type cells, 89% of cells Kar9p localized to the daughter pole with about 9% of cells displaying Kar9p on both poles, consistent with previous observations (Figure 11, B and C) (Moore and Miller 2007). In contrast, 20% of smt3-11 cells displayed Kar9p on both poles (Figure 11, B and C). This is a statistically significant difference from wild type using a Fischer's exact test (P < 0.01). In smt3-12, 14% of cells showed Kar9p on both poles. While this frequency is slightly increased, it is not statistically different from the wild-type value (P = 0.280) (Figure 11C). Thus, the two SUMO mutants had different effects on Kar9p localization.
DISCUSSION
Sumoylation regulates many vital processes in the cell. The two-hybrid and physical interaction data presented here establish for the first time a connection between sumoylation and two microtubule-associated proteins important for spindle positioning in S. cerevisiae. Two-hybrid analysis suggests that Kar9p and Bim1p interact not only with Smt3p, but also with several enzymes in the sumoylation pathway. A Kar9p mutant that is deficient in its interactions with sumoylation pathway proteins but not with many of its other binding partners displays a spindle-positioning defect. Kar9p can be sumoylated at two sites in vitro. Furthermore, deletion of the E3 sumoylation enzyme Nfi1p/Siz2p results in a small increase in the angle at which the mitotic spindle aligns along the mother-bud axis. We also find that Kar9p is mislocalized in smt3-11 mutants. We propose a model in which sumoylation regulates the microtubule-based process of spindle positioning in yeast.
How do Kar9p and Bim1p interact with SUMO?
Three models have been proposed for how proteins can interact with SUMO (Hannich et al. 2005). In the first model, SUMO is conjugated to the target protein. In the second model, SUMO interacts directly with the target protein through noncovalent molecular recognition events. In the third model, the protein binds to a third (sumoylated) protein that acts as a bridge between the protein and SUMO. While these models are not necessarily mutually exclusive, we favor a model in which Kar9p is directly conjugated to SUMO on the basis of three lines of evidence. Glycine 98 of Smt3p is critical for its conjugation to target proteins (Johnson and Blobel 1997; Kamitani et al. 1997; Li and Hochstrasser 1999). In a previous study, deletion of glycines 97 and 98 resulted in the loss of interaction between two-hybrid partners which was interpreted as the inability of SUMO to conjugate to its partners (Hannich et al. 2005). In our study, mutation of glycine 98 to alanine in the SMT3-GA construct should similarly abrogate the ability of Smt3p to conjugate to substrates. Indeed, Kar9p and Bim1p did not interact with this form of Smt3p by two-hybrid analysis. Second, three independent methods, two-hybrid analysis, physical binding studies, and direct binding studies with bacterially expressed proteins, have been used to demonstrate that Kar9p interacts with the E2 enzyme necessary for sumoylation, Ubc9p (Figures 2B and 4, A and B), supporting the hypothesis that Kar9p is likely to be sumoylated. Third, the ability of Kar9p to be conjugated with Smt3p in vitro suggests that SUMO may be covalently attached in vivo (Figure 5B). However despite several attempts, we have not yet been able to demonstrate the conjugation of Smt3p to Kar9p in vivo.
In an attempt to verify that the two-hybrid interaction was dependent on conjugation, we added a high-copy plasmid containing either the deconjugating enzyme ULP1 or ULP2 to the two-hybrid reporter strain (Hannich et al. 2005). However, no disruption was seen in the Kar9p-Smt3p interaction (data not shown). While this is consistent with a noncovalent interaction between Kar9p and Smt3p, we cannot however rule out the possibility that steric interference from the DBD and AD fusions prevented deconjugation by Ulp1p and Ulp2p.
Kar9p interacts physically with Bim1p and Bik1p (Lee et al. 2000; Miller et al. 2000; Moore et al. 2006). To test the bridging model, we deleted either KAR9 or BIM1 from the two-hybrid reporter strain (Figure 3, A and C). For both strains, the deletion did not prevent the interaction between the remaining protein and the sumoylation proteins. This analysis suggests that neither Kar9p nor Bim1p acts as a bridging protein for the other. Further, deletion of BIK1 did not prevent the interaction of KAR9 with the sumoylation proteins, suggesting that Bik1p also does not act as a bridging protein. Lastly, the absence of an interaction between the sumoylation proteins and Stu2p and Kip2p suggests that these proteins also do not bridge the two-hybrid interactions between Smt3p and Kar9p. However, the possibility still remains that the interaction between Smt3p and Kar9p or Bim1p could be indirect, resulting from a bridging interaction with an untested protein.
A Kar9p mutant defective in interaction with sumoylation proteins shows a spindle positioning defect and a defect in Kar9p-Kar9p interactions:
The kar9-L304P mutant lacks interaction with Smt3p and several other sumoylation proteins. This mutant also displays a dramatic increase in the frequency of spindle mislocalization away from the bud neck. Consistent with this, Kar9p-L304P localizes to both SPBs and the associated microtubules. This is in contrast to the wild-type protein, which is restricted to the bud-directed SPB and microtubules. The fact that Kar9p-L304P interacts with Bim1p, Bik1p, Stu2p, and Myo2p suggests that it is at least a partially functioning allele. Nevertheless, this mutation disrupts the essential function of the KAR9 pathway seen in the absence of dynein, consistent with the hypothesis that sumoylation plays an important role in spindle positioning.
Kar9p self associates, but little is known about whether this self-association is important for spindle positioning or the mechanisms that may regulate it (Miller et al. 2000). Analysis by the COILS program (Lupas et al. 1991) predicts with moderate certainty that Kar9p contains two segments of coiled-coil structure in the center of the molecule, with the first coiled-coil predicted more strongly than the second (Miller and Rose 1998). Both the L304P and M293P mutations lie within this structure and eliminate the prediction for the first coiled coil, while leaving the prediction for the second predicted coiled coil unchanged. Because both the M293P and the L304P mutations disrupt the interactions with Kar9p itself and with Smt3p, we cannot distinguish between the possibility that sumoylation promotes Kar9p's self-interaction and the possibility that the Kar9p-Kar9p interaction or this putative secondary structure is a prerequisite for sumoylation. To differentiate between these two possibilities, new mutations will be required that separate the Kar9p self-interaction and its interaction with Smt3p. However, we cannot dismiss the possibility that the kar9-L304P mutation disrupts a critical aspect of Kar9p function unrelated to sumoylation. In this scenario, the loss of interaction with Smt3p would be coincidental.
Phosphorylation of Kar9p and its interaction with Smt3p:
Phosphorylation can influence the sumoylation of proteins either positively or negatively (Daniel et al. 2007; Li et al. 2007; Tremblay et al. 2008; Vanhatupa et al. 2008). Kar9p has been shown previously to be phosphorylated at serines 197 and 496 which regulates its asymmetric localization to the daughter-bound spindle pole body and function in microtubule orientation (Liakopoulos et al. 2003; Moore et al. 2006; Moore and Miller 2007). Our finding that the phospho-mimetic mutant kar9-A196E S197E does not interact with Smt3p by two-hybrid suggests that phosphorylation at this site is likely to antagonize sumoylation of Kar9p.
Conversely, sumoylation can also influence phosphorylation. Consistent with this, we found that while Kar9p-L304P was phosphorylated, the levels of the slowest migrating phosphorylation species were greatly diminished (data not shown). We also investigated the phosphorylation status of Kar9p in the temperature-sensitive smt3-11 mutant. Here, Kar9p appeared to be maximally phosphorylated, but with significantly lower levels of the nonphosphorylated species present (data not shown). Taken together, these data are consistent with crosstalk between the sumoylation pathway and Kar9p phosphorylation, but additional work is required to elucidate the molecular details of this control.
Two kar9 mutants in this study A196ES197E and L304P displayed reduced interactions with Smt3p by two-hybrid analysis. However, these two mutants have different effects on microtubule orientation. Consistent with previous reports, we found here that kar9-A196E S197E displayed microtubule orientation profiles similar to the wild type (Figure 8C) (Moore and Miller 2007). In contrast, kar9-L304P has a significant microtubule orientation defect. L304P is also synthetically lethal with dyn1Δ (this study), whereas kar9-A196E S197E is not (Moore and Miller 2007). Thus, the kar9-A196E S197E allele is highly functional, making it unlikely that its loss of interaction with Smt3p is due to its being a hypomorphic allele. The phospho-inhibited version of this residue, S197A, however, does display several similarities with L304P, including its synthetic lethality with dynein, its defect in cytoplasmic microtubule orientation, and its defect in its asymmetric localization (Moore and Miller 2007). Together these data suggest that serine 197 is important for Kar9p's interaction with Smt3p.
Is Kar9p sumoylated in vivo?
Two-hybrid analysis can suggest that two proteins interact, but is not informative about the particular cell cycle or the life-cycle stage during which that interaction occurs. The two-hybrid and physical interactions reported here could represent interactions that occur during only short phases of the cell cycle, such as S phase, or for only a small population of Kar9p molecules. This could explain why we have not yet been able to identify Smt3p conjugated to either Kar9p or Bim1p by biochemical approaches in vivo. Indeed, it is clear from the literature that many proteins are sumoylated at specific points in the cell cycle and/or at low levels (Johnson and Blobel 1999; Bachant et al. 2002; Stead et al. 2003; Dieckhoff et al. 2004). It is also possible that maximal Kar9p sumoylation might occur during a different life-cycle stage such as during stationary phase, mating, or meiosis. For example, cells lacking the SUMO protease Smt4p show a severe defect in sporulation (Li and Hochstrasser 2000; Melchior 2000). While we have observed high molecular weight bands of Kar9p in densely grown culture but not cultures in log phase (Figure 5), future work will be required to elucidate whether these bands are in fact the result of sumoylation.
In evaluating our hypothesis that sumoylation is a novel mechanism for regulating spindle positioning in yeast, two pieces of evidence indicate that it may not play a major role under standard laboratory growth conditions. First, mutations in the sumoylation pathway did not display an obvious synthetic growth defect when combined with mutations in either the Kar9p or dynein pathways (Table 4). Second, the spindle-orientation defects seen in deletions of the E3 enzyme, Nfi1p, while statistically significant, were nevertheless modest (Figure 10 and supplemental Figure S1). Since sumoylation levels have been observed to increase during cell stresses such as ethanol and hydrogen peroxide treatment and heat stress (Hong et al. 2001; Hannich et al. 2005; Zhao and Blobel 2005), it is possible that the modest defects seen in these mutants might be exacerbated under stressful conditions. Whether Kar9p's interaction with Smt3p increases in response to cell stress is also an area that merits further investigation.
The sumoylation of target proteins is reversible, owing to the action of deconjugation enzymes Ulp1p and Ulp2p in the cell (Li and Hochstrasser 1999; Schwienhorst et al. 2000). As a dynamic process, the potential exists for sumoylation to act as a rapid “on/off” switch to regulate Kar9p function, and in turn, to regulate the microtubule-dependent processes requiring Kar9p. The finding by the Hochstrasser laboratory that strains deleted for ULP2/SMT4 and strains containing mutations in the E2 Ubc9p are sensitive to the microtubule destabilizing drug, benomyl is consistent with our hypothesis (Li and Hochstrasser 2000; Hannich et al. 2005). This phenotype has been attributed to defects in the mitotic spindle, but could also be explained by defects associated with cytoplasmic microtubules.
While this article was in the final stages of review, we became aware of a work in press from the Barral and Liakopoulos laboratories describing the in vivo isolation of Kar9p-SUMO conjugates (Leisner et al. 2008). In their work, they identify four lysine residues important for Kar9p sumoylation. Kar9p lacking these four sites accumulates on the mother-directed SPB. Thus, both our work and theirs are in agreement that sumoylation is important for the asymmetric localization of Kar9p and spindle positioning.
Recently, several microtubule-associated proteins have been identified as substrates of sumoylation. Tau, a microtubule-binding protein associated with Alzheimer's disease, is sumoylated but it is not yet known whether this regulates its binding to microtubules (Dorval and Fraser 2006). The kinetochore protein Ndc80p binds microtubules and the Dam1 complex. The sumoylation of Ndc80p appears to be regulated differently under microtubule depolymerizing conditions than other sumoylated kinetochore proteins, such as Ndc10p, Cep3p, and Bir1p (Montpetit et al. 2006). In mammals, sumoylated Ran-GAP1 is targeted to the mitotic spindle and kinetochores, and is required there for accurate microtubule attachment to kinetochores (Joseph et al. 2002, 2004). Together, these studies highlight that a great deal remains to be understood about how microtubule-associated proteins in general are regulated by sumoylation. In yeast, Kar9p has been postulated to be the paralogue of the adenomatous polyposis coli (APC) protein, a tumor-suppressor protein and regulator of Wnt signaling (Berrueta et al. 1998; Mimori-Kiyosue et al. 2000; Bienz 2001; Nathke 2004; Hanson and Miller 2005; Kita et al. 2006). EB1, the mammalian homolog of Bim1p, also binds APC (Morrison et al. 1998). APC forms a complex with axin, a multi-functional protein, which is sumoylated (Nishida et al. 2001; Rui et al. 2002). While our findings suggest that two additional microtubule-associated proteins in yeast interact with SUMO, it will be exciting for future research to determine whether sumoylation is a general paradigm for the regulation of other microtubule-based processes in other systems.
Acknowledgments
We thank Bruce Goode, Mark Hochstrasser, Erica Johnson, and Mark Rose for providing yeast strains, plasmids, and reagents. We thank Henri Jasper for his critical reading of the manuscript. This work was supported by a grant to R.K.M. from the National Science Foundation (MCB-0414768).
References
- Adames, N. R., and J. A. Cooper, 2000. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J. Cell Biol. 149 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragón, L., 2005. Sumoylation: a new wrestler in the DNA repair ring. Proc. Natl. Acad. Sci. USA 102 4661–4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachant, J., A. Alcasabas, Y. Blat, N. Kleckner and S. J. Elledge, 2002. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol. Cell. 9 1169–1182. [DOI] [PubMed] [Google Scholar]
- Bartek, J., and J. Lukas, 2006. The stress of finding NEMO. Science. 311 1110–1111. [DOI] [PubMed] [Google Scholar]
- Bays, N. W., and R. Y. Hampton, 2002. Cdc48-Ufd1-Npl4: stuck in the middle with Ub. Curr. Biol. 12 R366–R371. [DOI] [PubMed] [Google Scholar]
- Beach, D. L., J. Thibodeaux, P. Maddox, E. Yeh and K. Bloom, 2000. The role of the proteins Kar9 and Myo2 in orienting the mitotic spindle of budding yeast. Curr. Biol. 10 1497–1506. [DOI] [PubMed] [Google Scholar]
- Bencsath, K. P., M. S. Podgorski, V. R. Pagala, C. A. Slaughter and B. A. Schulman, 2002. Identification of a multifunctional binding site on Ubc9p required for Smt3p conjugation. J. Biol. Chem. 277 47938–47945. [DOI] [PubMed] [Google Scholar]
- Berrueta, L., S. K. Kraeft, J. S. Tirnauer, S. C. Schuyler, L. B. Chen et al., 1998. The adenomatous polyposis coli-binding protein EB1 is associated with cytoplasmic and spindle microtubules. Proc. Natl. Acad. Sci. USA 95 10596–10601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar, V., M. Smith and A. J. Courey, 2002. Conjugation of Smt3 to dorsal may potentiate the Drosophila immune response. Mol. Cell. Biol. 22 492–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienz, M., 2001. Spindles cotton on to junctions, APC and EB1. Nat. Cell Biol. 3 E67–E68. [DOI] [PubMed] [Google Scholar]
- Biggins, S., N. Bhalla, A. Chang, D. L. Smith and A. W. Murray, 2001. Genes involved in sister chromatid separation and segregation in the budding yeast Saccharomyces cerevisiae. Genetics 159 453–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossis, G., and F. Melchior, 2006. SUMO: regulating the regulator. Cell Div. 1 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, S., K. Matuschewski, M. Rape, S. Thoms and S. Jentsch, 2002. Role of the ubiquitin-selective CDC48 (UFD/NPL4) chaperone (segregase) in ERAD of OLE1 and other substrates. EMBO J. 21 615–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brizzio, V., A. E. Gammie and M. D. Rose, 1998. Rvs161p interacts with Fus2p to promote cell fusion in Saccharomyces cerevisiae. J. Cell Biol. 141 567–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers, B., 1981. Cytology of the yeast life cycle, pp. 59–96 in The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance, edited by J. N. Strathern, W. W. Jones and J. R. Broach. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Byers, B., and L. Goetsch, 1975. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J. Bacteriol. 124 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carminati, J. L., and T. Stearns, 1997. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J. Cell Biol. 138 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho, P., M. L. Gupta, Jr., M. A. Hoyt and D. Pellman, 2004. Cell cycle control of kinesin-mediated transport of Bik1(CLIP170) regulates microtubule stability and dynein activation. Dev. Cell 6 815–829. [DOI] [PubMed] [Google Scholar]
- Cheng, C.H., Y.-H. Lo, S.-S. Liang, S.-C. Ti, F.-M. Lin et al., 2006. SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev. 20 2067–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, T. L., H.-H. Hsiao, Y.-Y. Yeh, H.-L. Shia, Y.-L. Chen et al., 2004. In vitro modification of human centromere protein CENP-C fragments by small-ubiquitin like modifier (SUMO) protein: definitive identification of modification sites by tandem mass spectrometry analysis of the isopeptides. J. Biol. Chem. 279 39653–39662. [DOI] [PubMed] [Google Scholar]
- Comerford, K. M., M. O. Leonard, J. Karhausen, R. Carey, S. P. Colgan et al., 2003. Small ubiquitin related modifier-1 modification mediated resolution of CREB-dependent responses to hypoxia. Proc. Natl. Acad. Sci. USA 100 986–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel, A. R., E. J. Faivre and C. A. Lange, 2007. Phosphorylation-dependent antagonism of sumoylation derepresses progesterone receptor action in breast cancer cells. Mol. Endocrinol. 21 2890–2906. [DOI] [PubMed] [Google Scholar]
- Desterro, J. M., M. S. Rodriguez and R. T. Hay, 1998. SUMO-1 modification of IκBα inhibits NF-κB activation. Mol. Cell 2 233–239. [DOI] [PubMed] [Google Scholar]
- Dieckhoff, P., M. Bolte, Y. Sancak, G. H. Braus and S. Irniger, 2004. Smt3/SUMO and Ubc9 are required for efficient APC/C-mediated proteolysis in budding yeast. Mol. Microbiol. 51 1375–1387. [DOI] [PubMed] [Google Scholar]
- Dorval, V., and P. E. Fraser, 2006. Small ubiquitin-like modifier (sumo) modification of native unfolded proteins Tau and α-synuclein. J. Biol. Chem. 281 9919–9924. [DOI] [PubMed] [Google Scholar]
- Dohmen, R. J., 2004. Review: SUMO protein modification. Biochim. Biophys. Acta 1695 113–131. [DOI] [PubMed] [Google Scholar]
- Gill, G., 2004. SUMO and ubiquitin in the nucleus: Different functions, similar mechanisms? Genes Dev. 18 2046–2059. [DOI] [PubMed] [Google Scholar]
- Gill, G., 2005. Something about SUMO inhibits transcription. Curr. Opin. Genet. Dev. 15 536–541. [DOI] [PubMed] [Google Scholar]
- Girard, M., and M. Goossens, 2006. Sumoylation of the SOX10 transcription factor regulates its transcriptional activity. FEBS Lett. 580 1635–1641. [DOI] [PubMed] [Google Scholar]
- Goehring, A. S., D. M. Rivers and G. F. Sprague Jr., 2003. Attachment of the ubiquitin-related protein Urm1p to the antioxidant protein Ahp1p. Eukaryot. Cell 2 930–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayhack, E. J., and E. M. Phizicky, 2001. Genomic analysis of biochemical function. Curr. Opin. Chem. Biol. 5 34–39. [DOI] [PubMed] [Google Scholar]
- Gupta, V., and M. Bei, 2006. Modification of Msx1 by SUMO-1. Biochem. Biophys. Res. Comm. 345 74–77. [DOI] [PubMed] [Google Scholar]
- Hannich, J. T., A. Lewis, M. B. Kroetz, S. J. Li, H. Heide et al., 2005. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J. Biol. Chem. 280 4102–4110. [DOI] [PubMed] [Google Scholar]
- Hanson, C. A., and J. R. Miller, 2005. Non-traditional roles for the adenomatous polyposis coli (APC) tumor suppressor protein. Gene 361 1–12. [DOI] [PubMed] [Google Scholar]
- Hietakangas, V., J. K. Ahlskog, A. M. Jakobsson, M. Hellesuo, N. M. Sahlberg et al., 2003. Phosphorylation of serine 303 is a prerequisite for the stress-inducible SUMO modification of heat shock factor 1. Mol. Cell. Biol. 23 2953–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietakangas, V., J. Anckar, H. A. Blomster, M. Fujimoto, J. J. Palvimo et al., 2006. PDSM, a motif for phosphoylation-dependent SUMO modification. Proc. Natl. Acad. Sci. USA 103 45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock, A. L., H. Krebber, S. Frietze, A. Lin, M. Latterich et al., 2001. The conserved Npl4 protein complex mediates proteasome-dependent membrane-bound transcription factor activation. Mol. Biol. Cell 12 3226–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser, M., 2001. SP-RING for SUMO: new functions bloom for a ubiquitin-like protein. Cell 107 5–8. [DOI] [PubMed] [Google Scholar]
- Hofmann, H., S. Flöss and T. Stamminger, 2000. Covalent modification of the transactivator protein IE2-p86 of human cytomegalovirus by conjugation to the ubiquitin-homologous proteins SUMO-1 and hSMT3b. J. Virol. 74 2510–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, Y., R. Rogers, M. J. Matunis, C. N. Mayhew, M. L. Goodson et al., 2001. Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J. Biol. Chem. 276 40263–40267 (erratum: J. Biol. Chem. 277: 26708). [DOI] [PubMed] [Google Scholar]
- Hwang, E., J. Kusch, Y. Barral and T. C. Huffaker, 2003. Spindle orientation in Saccharomyces cerevisiae depends on the transport of microtubule ends along polarized actin cables. J. Cell Biol. 161 483–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara, M., H. Yamamoto and A. Kikuchi, 2005. SUMO-1 modification of PIASy, an E3 ligase, is necessary for PIASy-dependent activation of Tcf-4. Mol. Cell. Biol. 25 3506–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase, M., and A. Toh-e, 2001. Nis1 encoded by YNL078W: a new neck protein of Saccharomyces cerevisiae. Genes Genet. Syst. 76 335–343. [DOI] [PubMed] [Google Scholar]
- James, P., J. Halladay and E. A. Craig, 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, E. S., 2004. Protein modification by SUMO. Ann. Rev. Biochem. 73 355–382. [DOI] [PubMed] [Google Scholar]
- Johnson, E. S., and G. Blobel, 1997. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J. Biol. Chem. 272 26799–26802. [DOI] [PubMed] [Google Scholar]
- Johnson, E. S., and G. Blobel, 1999. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J. Cell Biol. 147 981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, E. S., and A. A. Gupta, 2001. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106 735–744. [DOI] [PubMed] [Google Scholar]
- Johnson, E. S., I. Schwienhorst, R. J. Dohmen and G. Blobel, 1997. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 16 5509–5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph, J., S. H. Tan, T. S. Karpova, J. G. McNally and M. Dasso, 2002. SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J. Cell Biol. 156 595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph, J., S. T. Liu, S. A. Jablonski, T. J. Yen and M. Dasso, 2004. The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr. Biol. 14 611–617. [DOI] [PubMed] [Google Scholar]
- Kahana, J. A., B. J. Schnapp and P. A. Silver, 1995. Kinetics of spindle pole body separation in budding yeast. Proc. Natl. Acad. Sci. USA 92 9707–9711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani, T., H. P. Nguyen and E. T. Yeh, 1997. Preferential modification of nuclear proteins by a novel ubiquitin-like molecule. J. Biol. Chem. 272 14001–14004. [DOI] [PubMed] [Google Scholar]
- Kerscher, O., R. Felberbaum and M. Hochstrasser, 2006. Modification of proteins by ubiquitin and ubiquitin-like proteins. Ann. Rev. Cell Dev. Biol. 22 159–180. [DOI] [PubMed] [Google Scholar]
- Kita, K., T. Wittmann, I. S. Nathke and C. M. Waterman-Storer, 2006. Adenomatous polyposis coli on microtubule plus ends in cell extensions can promote microtubule net growth with or without EB1. Mol. Biol. Cell 17 2331–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch, J., A. Meyer, M. P. Snyder and Y. Barral, 2002. Microtubule capture by the cleavage apparatus is required for proper spindle positioning in yeast. Genes Dev. 16 1627–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, L., J. S. Tirnauer, J. Li, S. C. Schuyler, J. Y. Liu et al., 2000. Positioning of the mitotic spindle by a cortical-microtubule capture mechanism. Science 287 2260–2262. [DOI] [PubMed] [Google Scholar]
- Lee, W. L., M. A. Kaiser and J. A. Cooper, 2005. The offloading model for dynein function: differential function of motor subunits. J. Cell Biol. 168 201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leisner, C., D. Kammerer, A. Denoth, M. Britschi, Y. Barral et al., 2008. Regulation of mitotic spindle asymmetry by SUMO and the spindle assembly checkpoint in yeast. Curr. Biol. 18 1249–1255. [DOI] [PubMed] [Google Scholar]
- Li, S.-J., and M. Hochstrasser, 1999. A new protease required for cell-cycle progression in yeast. Nature 398 246–251. [DOI] [PubMed] [Google Scholar]
- Li, S.-J., and M. Hochstrasser, 2000. The yeast ULP2(SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol. Cell. Biol. 20 2367–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Y. K. Lee, J. C. Jeng, Y. Yen, D. C. Schultz et al., 2007. Role for KAP1 serine 824 phosphorylation and sumoylation/desymoylation switch in regulating KAP1-mediated transcriptional repression. J. Biol. Chem. 282 36177–36189. [DOI] [PubMed] [Google Scholar]
- Liakopoulos, D., J. Kusch, S. Grava, J. Vogel and Y. Barral, 2003. Asymmetric loading of Kar9 onto spindle poles and microtubules ensures proper spindle alignment. Cell 112 561–574. [DOI] [PubMed] [Google Scholar]
- Liu, S. T., W. H. Wang, Y. R. Hong, J. Y. Chuang, P. J. Lu et al., 2006. Sumoylation of Rta of Epstein-Barr virus is preferentially enhanced by PIASxbeta. Virus Res. 19 163–190. [DOI] [PubMed] [Google Scholar]
- Lupas, A., M. Van Dyke and J. Stock, 1991. Predicting coiled coils from protein sequences. Science 252 1162–1164. [DOI] [PubMed] [Google Scholar]
- Maekawa, H., T. Usui, M. Knop and E. Schiebel, 2003. Yeast Cdk1 translocates to the plus end of cytoplasmic microtubules to regulate bud cortex interactions. EMBO J. 22 438–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa, H., and E. Schiebel, 2004. Cdk1-Clb4 controls the interaction of astral microtubule plus ends with subdomains of the daughter cell cortex. Genes Dev. 18 1709–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, S. W., and J. B. Konopka, 2004. SUMO modification of septin-interacting proteins in Candida albicans. J. Biol. Chem. 279 40861–40867. [DOI] [PubMed] [Google Scholar]
- Melchior, F., 2000. SUMO-nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. 16 591–626. [DOI] [PubMed] [Google Scholar]
- Miller, R. K., and M. D. Rose, 1998. Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J. Cell Biol. 140 377–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, R. K., K. K. Heller, L. Frisen, D. L. Wallack, D. Loayza et al., 1998. The kinesin-related proteins, Kip2p and Kip3p, function differently in nuclear migration in yeast. Mol. Biol. Cell 9 2051–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, R. K., S.-C. Cheng and M. D. Rose, 2000. Bim1p/Yeb1p mediates the Kar9p-dependent cortical attachment of cytoplasmic microtubules. Mol. Biol. Cell 11 2949–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue, Y., N. Shiina and S. Tsukita, 2000. Adenomatous polyposis coli (APC) protein moves along microtubules and concentrates at their growing ends in epithelial cells. J. Cell Biol. 148 505–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montpetit, B., T. R. Hazbun, S. Fields and P. Hieter, 2006. Sumoylation of the budding yeast kinetochore protein Ndc10 is required for Ndc10 spindle localization and regulation of anaphase spindle elongation. J. Cell Biol. 174 653–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, J. K., and R. K. Miller, 2007. The CDK, Cdc28p, regulates multiple aspects of Kar9p function in yeast. Mol. Biol. Cell 18 1187–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, J. K., S. D'Silva and R. K. Miller, 2006. The CLIP-170 homologue Bik1p promotes the phosphorylation and asymmetric localization of Kar9p. Mol. Biol. Cell 17 178–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison, E. E., B. N. Wardleworth, J. M. Askham, A. F. Markham and D. M. Meredith, 1998. EB1, a protein which interacts with the APC tumour suppressor, is associated with the microtubule cytoskeleton throughout the cell cycle. Oncogene 17 3471–3477. [DOI] [PubMed] [Google Scholar]
- Müller, S., M. J. Matunis and A. Dejean, 1998. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 17 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhua, L., T. S. Karpova and J. A. Cooper, 1994. A yeast actin-related protein homologous to that in vertebrate dynactin complex is important for spindle orientation and nuclear migration. Cell 78 669–679. [DOI] [PubMed] [Google Scholar]
- Muhua, L., N. R. Adames, M. D. Murphy, C. R. Shields and J. A. Cooper, 1998. A cytokinesis checkpoint requiring the yeast homologue of an APC- binding protein. Nature 393 487–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathke, I. S., 2004. The adenomatous polyposis coli protein: the Achilles heel of the gut epithelium. Ann. Rev. Cell Dev. Biol. 20 337–366. [DOI] [PubMed] [Google Scholar]
- Nishida, T., F. Kaneko, M. Kitagawa and H. Yasuda, 2001. Characterization of a novel mammalian SUMO-1/Smt3-specific isopeptidase, a homologue of rat axam, which is an axin-binding protein promoting beta-catenin degradation. J. Biol. Chem. 276 39060–39066. [DOI] [PubMed] [Google Scholar]
- Ohashi, A., J. Gibson, I. Gregor and G. Schatz, 1982. Import of proteins into mitochondria. The precursor of cytochrome C1 is processed in two steps, one of them heme-dependent. J. Biol. Chem. 257 13042–13047. [PubMed] [Google Scholar]
- Okuma, T., R. Honda, G. Ichikawa, N. Tsumagari and H. Yasuda, 1999. In vitro SUMO-1 modification requires two enzymatic steps, E1 and E2. Biochem. Biophys. Res. Comm. 254 693–698. [DOI] [PubMed] [Google Scholar]
- Pereira, G., T. U. Tanaka, K. Nasmyth and E. Schiebel, 2001. Modes of spindle pole body inheritance and segregation of the Bfa1p-Bub2p checkpoint protein complex. EMBO J. 20 6359–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky, E. M., M. R. Martzen, S. M. McCraith, S. L. Spinelli, F. Xing et al., 2002. Biochemical genomics approach to map activities to genes. Methods Enzymol. 350 546–559. [DOI] [PubMed] [Google Scholar]
- Pichler, A., P. Knipscheer, E. Oberhofer, W. J. van Dijk, R. Korner et al., 2005. SUMO modification of the ubiquitin-conjugating enzyme E2–25K. Nat. Struct. Mol. Biol. 12 264–269. [DOI] [PubMed] [Google Scholar]
- Prudden, J., S. Pebernard, G. Raffa, D. A. Slavin, J. J. Perry et al., 2007. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 26 4089–4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, M. S., C. Dargemont and R. T. Hay, 2001. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 276 12654–12659. [DOI] [PubMed] [Google Scholar]
- Rui, H. L., E. Fan, H. M. Zhou, Z. Xu, Y. Zhang et al., 2002. SUMO-1 modification of the C-terminal KVEKVD of Axin is required for JNK activation but has no effect on Wnt signaling. J. Biol. Chem. 277 42981–42986. [DOI] [PubMed] [Google Scholar]
- Sampson, D. A., M. Wang and M. J. Matunis, 2001. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J. Biol. Chem. 276 21664–21669. [DOI] [PubMed] [Google Scholar]
- Savare, J., N. Bonneaud and F. Girard, 2005. SUMO represses transcriptional activity of the Drosophila SoxNeuro and human Sox3 central nervous system-specific transcription factors. Mol. Biol. Cell 16 2660–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, K., K. Richards and D. Botstein, 1997. BIM1 encodes a microtubule-binding protein in yeast. Mol. Biol. Cell 8 2677–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, S. E., K. Matuschewski, D. Liakopoulos, M. Scheffner and S. Jentsch, 1998. The ubiquitin-like proteins SMT3 and SUMO-1 are conjugated by the UBC9 E2 enzyme. Proc. Natl. Acad. Sci. USA 95 560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwienhorst, I., E. S. Johnson and R. J. Dohmen, 2000. SUMO conjugation and deconjugation. Mol. Gen. Genet. 263 771–786. [DOI] [PubMed] [Google Scholar]
- Smolen, G. A., M. T. Vassileva, J. Wells, M. J. Matunis and D. A. Haber, 2004. SUMO-1 modification of the Wilms' tumor suppressor WT1. Cancer Res. 64 7846–7851. [DOI] [PubMed] [Google Scholar]
- Stead, K., C. Aguilar, T. Hartman, M. Drexel, P. Meluh et al., 2003. Pds5p regulates the maintenance of sister chromatid cohesion and is sumoylated to promote the dissolution of cohesion. J. Cell Biol. 163 729–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan, J. S., N. Agrawal, J. Pallos, E. Rockabrand, L. C. Trotman et al., 2004. SUMO modification of Huntingtin and Huntington's disease pathology. Science 304 100–103. [DOI] [PubMed] [Google Scholar]
- Straight, A. F., W. F. Marshall, J. W. Sedat and A. W. Murray, 1997. Mitosis in living budding yeast: anaphase A but no metaphase plate. Science 277 574–578. [DOI] [PubMed] [Google Scholar]
- Sullivan, D. S., and T. C. Huffaker, 1992. Astral microtubules are not required for anaphase B in Saccharomyces cerevisiae. J. Cell Biol. 119 379–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, H., J. D. Leverson and T. Hunter, 2007. Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 26 4102–4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, Y., M. Iwase, M. Konishi, M. Tanaka, A. Toh-E et al., 1999. Smt3, a SUMO-1 homolog, is conjugated to Cdc3, a component of septin rings at the mother-bud neck in budding yeast. Biochem. Biophys. Res. Comm. 259 582–587. [DOI] [PubMed] [Google Scholar]
- Takahashi, Y., T. Kahyo, A. Toh-E, H. Yasuda and Y. Kikuchi, 2001. a Yeast Ull1/Siz1 is a novel SUMO1/Smt3 ligase for septin components and functions as an adaptor between conjugating enzyme and substrates. J. Biol. Chem. 276 48973–48977. [DOI] [PubMed] [Google Scholar]
- Takahashi, Y., A. Toh-e and Y. Kikuchi, 2001. b A novel factor required for the SUMO1/Smt3 conjugation of yeast septins. Gene 275 223–231. [DOI] [PubMed] [Google Scholar]
- Takahashi, Y., A. Toh-E and Y. Kikuchi, 2003. Comparative analysis of yeast PIAS-type SUMO ligases in vivo and in vitro. J. Biochem. 133 415–422. [DOI] [PubMed] [Google Scholar]
- Tanaka, K., N. Mukae, H. Dewar, M. van Breugel, E. K. James et al., 2005. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature 434 987–994. [DOI] [PubMed] [Google Scholar]
- Tirnauer, J. S., E. O'Toole, L. Berrueta, B. E. Bierer and D. Pellman, 1999. Yeast Bim1p promotes the G1-specific dynamics of microtubules. J. Cell Biol. 145 993–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay, A. M., B. J. Wilson, X. J. Yang and V. Giguère, 2008. Phosphorylation-dependent sumoylation regulates estrogen-related receptor-alpha and gamma transcriptional activity through a synergy control motif. Mol. Endocrinol. 22 570–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich, H. D., 2004. How to activate a damage-tolerant polymerase: consequences of PCNA modifications by ubiquitin and SUMO. Cell Cycle 3 15–18. [PubMed] [Google Scholar]
- Uzunova, K., K. Göttsche, M. Miteva, S. R. Weisshaar, C. Glanemann et al., 2007. Ubiquitin-dependent proteolytic control of SUMO conjugates. J. Biol. Chem. 282 34167–34175. [DOI] [PubMed] [Google Scholar]
- Vanhatupa, S., D. Ungureanu, M. Paakkunainen and O. Silvennoinen, 2008. MAPK-induced ser727 phosphorylation promotes SUMOylation of STAT1. Biochem. J. 409 179–185. [DOI] [PubMed] [Google Scholar]
- Wolyniak, M. J., K. Blake-Hodek, K. Kosco, E. Hwang, L. You et al., 2006. The regulation of microtubule dynamics in S. cerevisiae by three interacting plus-end tracking proteins. Mol. Biol. Cell 7 2789–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Y., O. Kerscher, M. B. Kroetz, J. F. McConchie, P. Sung et al., 2007. The yeast Hex3-Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J. Biol. Chem. 282 34176–34184. [DOI] [PubMed] [Google Scholar]
- Yang, S.-H., E. Jaffray, R. T. Hay and A. D. Sharrocks, 2003. Dynamic interplay of the SUMO and ERK pathways in regulating Elk-1 transcriptional activity. Mol. Cell 12 63–74. [DOI] [PubMed] [Google Scholar]
- Ye, Y., H. H. Meyer and T. A. Rapoport, 2001. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414 652–656. [DOI] [PubMed] [Google Scholar]
- Yeh, E., R. V. Skibbens, J. W. Cheng, E. D. Salmon and K. Bloom, 1995. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 130 687–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, H., D. Pruyne, T. C. Huffaker and A. Bretscher, 2000. Myosin V orientates the mitotic spindle in yeast. Nature 406 1013–1015. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., and A. R. Buchman, 1997. Identification of a member of a DNA-dependent ATPase family that causes interference with silencing. Mol. Cell. Biol. 17 5461–5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., and G. Blobel, 2005. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl. Acad. Sci. USA 102 4777–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, W., J. J. Ryan and H. Zhou, 2004. Global analyses of sumoylated proteins in Saccharomyces cerevisiae: induction of protein sumoylation by cellular stresses. J. Biol. Chem. 279 32262–32268. [DOI] [PMC free article] [PubMed] [Google Scholar]