Abstract
Depending on the circumstances, autophagy can prevent, cause or regulate the kinetics of cell death. Consequently, it is particularly important that experimental strategies to study these responses are designed carefully. Here, I discuss some of the issues to be considered when designing such experiments.
Keywords: autophagy, cell death, apoptosis, clonogenicity
Not Dead Fred is a character in the Broadway musical Spamalot and the film Monty Python and the Holy Grail. Fred is dragged through the streets of a Black Death-ravaged village towards a cart pulled by a man crying out “Bring out your dead”. The villager dragging Fred calls out “Here's one” but before the cart driver can accept his payment, Fred points out that he's “Not dead”. The cart driver explains that if the body says he's not dead then its against regulations to take him, to which the villager replies “Oh go on, take him, he's nearly dead”. Fred pipes up, “I'm getting better.” The villager argues with Fred “No you're not, you'll be stone dead in a minute” and asks the cart driver if he can hang on. The cart driver is too busy to wait so the villager asks for help, and the cart driver uses a club to make sure that Fred is really dead. The joke works because it plays on the fact that you can only be dead or not dead, and that “nearly dead” or “getting better” are not particularly useful terms. We have a similar problem in biology – how do we tell if a cell is dead or dying? This issue is of particular importance in autophagy research because, in different circumstances, autophagy can cause death or prevent death and can also delay, but not actually prevent, death. Thus our problem is that autophagy might be the equivalent of the club that finally does Fred in, it might be a way to avoid the swing of the club and allow recovery from a bout with the Black Death or it might be equivalent to the negotiation between the villager and the cart driver that delays things but doesn't change Fred's fate. Surprisingly, despite the fact that everyone gets the joke about Fred, many experiments that are intended to discriminate between these possibilities don't really do so and instead run the risk of measuring activities that are analogous to “nearly dead” and “getting better”. So, how should we design such experiments?
Most mammalian cell viability assays measure an activity associated with living cells. For example MTT assays measure mitochondrial metabolism and similar assays measure other cellular enzymatic activities while counting cells that stain with trypan blue tests if they have intact cell membranes. Other common assays detect characteristics that are considered typical of dying cells such as the phosphatidylserine translocation to the cell surface, which occurs in apoptotic cells but is also regulated by autophagy 1 or the fragmentation of DNA that occurs in cells that are undergoing apoptosis. In some papers, the tests are more subjective and one sometimes sees experiments where cells are scored as alive or dead based just on visual observation e.g. counting percentage of rounded and flat cells. The problem is that while these kinds of experiments can give you a measure of the number of alive and dead cells at a given time point, they don't tell us anything about whether the cells that are scored as alive are “nearly dead”. For example, after treatment with a toxic agent a cell might be scored as alive now, but if it is going to lose membrane integrity an hour from now, then the conclusion that the cell is alive and well is not really valid– it's more like Fred just before the cart driver hits him with the club. Moreover, some assays that are used to measure cell viability- e.g. those that measure mitochondrial activity may be particularly problematic when one is studying autophagy because increased or decreased mitophagy (i.e. the degradation of mitochondria after their sequestration in autophagosomes) might affect the signal independently of any real effect on cell viability.
Many studies aim to determine the role of autophagy in response to a toxic evente.g. treatment with a drug that kills cells, expression of a toxic protein, exposure to a stressful stimulus such as ischemia/hypoxia or removal of nutrients or growth factors. The experiments are usually conceptually simple. Control cells and cells where autophagy has been inhibited (e.g. by siRNA knockdown of autophagy regulators such as Atg5, Atg7, Beclin1 etc.) are exposed to the stimulus, or, control cells are compared to cells where autophagy has been stimulated (e.g. with treatment by rapamycin or other agents that activate autophagy such as trehalose 2). After the cells are treated with the drug or other stimulus, differences in the efficiency of cell death as a result of autophagy manipulation are determined. Often these experiments are performed within a fairly short time (24−72 hours) after the cells were exposed to the stimulus that is being studied; and, often the stimulus is delivered at one fairly high dose that has a robust effect on its own. I suggest that more reliable data would be obtained if two straightforward elements in the design of experiments where routinely applied.
The first design element is to use a dose response if at all possible. This is important because measuring a shift in the slope of the dose response curve or a difference in the drug's EC50 (i.e. the dose required for a 50% inhibition of cell viability) provides a more sensitive way to detect both positive and negative effects of a manipulation (in our case, a more sensitive way to test what autophagy does) than using agents at single doses. The use of dose response curves also allows one to more easily detect effects when the autophagy manipulation is not fully effective- e.g. if the siRNA knockdown does not completely remove an autophagy regulator. With drugs and other toxic agents it is easy to perform dose response experiments because all one needs to do is use different concentrations of the drug. For some stimuli such as hypoxia or growth factor removal, there may be a threshold that one needs to get to before any effect is observed and it may not be meaningful to manipulate the amount of the stimulus around this threshold. In this case, a better choice would be to remove the growth factor or oxygen for increasing amounts of time and develop a dose response curve this way– for example, this approach was used by Degenhardt et.al. in their study of tumor cell ischemia and the ability of autophagy to protect from cell death due to ischemia3.
The second (and, in my view, more important) design element is that one should use long-term assays of cell survival in conjunction with short-term viability assays measuring metabolic activity, membrane integrity etc. This is important because by comparing results from the short- and long-term assays it is possible to detect if autophagy alters the kinetics of cell death or alters the actual amount of death. To do this, one measures cell viability using a short-term assay (usually a day or two after treatment with the drug) and also tests if the cells are capable of long-term survival after removal of the stimulus (i.e. after the drug is taken away, the nutrients/ growth factors are returned etc.). Experiments like this are often performed in cultured immortal and/or transformed cell lines. For such cells the most rigorous test of true survival is to ask if the cells can recover their capacity for growth after removal of the stimulus and replacement with normal growth media. The easiest assay for this kind of experiment is to plate cells at low density and follow the formation of colonies as a measure of the number of cells that were alive and capable of division. This type of experiment is easy to perform and is quantitative however, it requires that the colonies grow big enough that one can stain and count them- this may require that the cells go through several cell division cycles, therefore for cells where this may be difficult (e.g. primary cells), an alternate approach is to follow several cells over just a few days using time-lapse microscopy and ask if each individual cell is capable of division. For example, Colell et.al. used time-lapse microscopy of individual cells to show that GAPDH-induced autophagy protects against caspase-independent death even after mitochondria have released their cytochrome c. We know that the cells were truly protected by the autophagy because, after a few days, they reform functional mitochondria and regain their ability to divide 4.
For cells that do not divide such as neurons or cardiac myocytes, monitoring cell division is not possible. However, the central point is that to determine how autophagy affects the life or death decision, it is important to determine if the cell is alive over an extended period of time after removal of the death stimulus. It is possible to do this with non-dividing cells by assessing markers of continued metabolic activity after the removal of the death stimulus at various times to provide convincing data that the cells are still alive. For example, in the case of a neuron where autophagy has been inhibited or increased and the cell then subjected to an external stress, one might design an experiment to monitor the dynamic restructuring of dendritic spines in the same GFP-labeled cell over a week using fluorescence microscopy– if the cell retains the ability to grow and contract dendritic spines and the dynamic nature of these changes are maintained several days after removal of the death stimulus, then one is on firmer ground in concluding that the cell is truly recovered from the insult than if one just makes a single assessment at a particular time point. And, if manipulation of autophagy changes this effect, the resulting conclusion about the role of autophagy in this mechanism is also stronger.
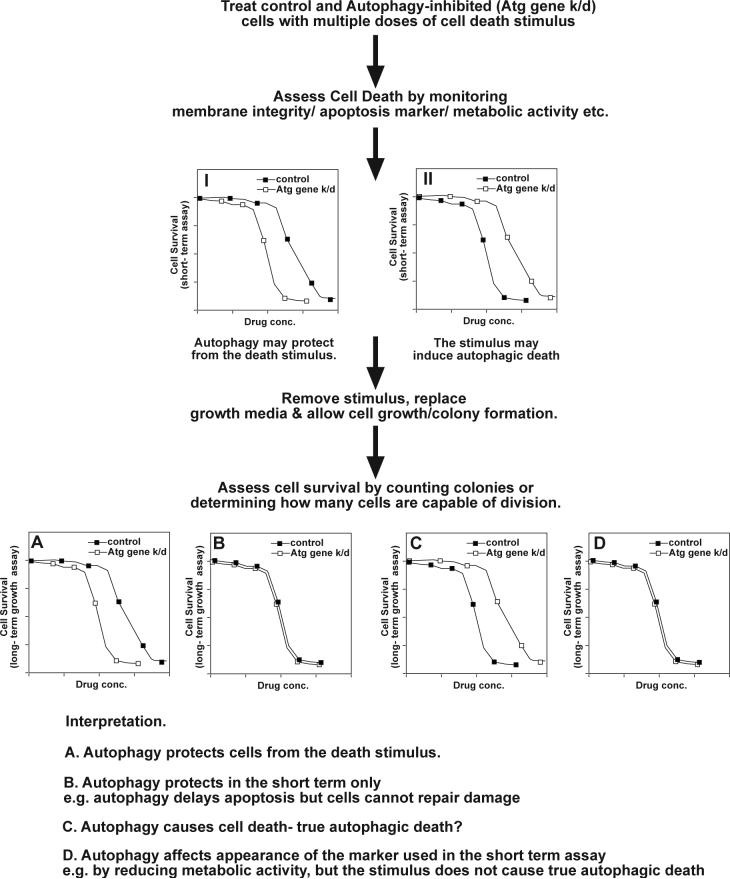
An example of this approach is shown in Fig.1. Curve I shows an experiment where on a short-term assay it appears that autophagy may protect cells (i.e. Atg gene knockdown increases cell death as shown by a leftward shift in the dose response curve). Curve II represents the opposite – i.e. on short-term assays it appears that autophagy promotes cell death as shown by protection (rightward shift in the dose response curve) when Atg genes are knocked down. However, when the stimulus is removed and cell survival is determined at a later time point (this allows cells that are terminally damaged by the stimulus that is being tested to die while allowing cells that have been protected from the stimulus to recover and regain their normal growth behavior), different results may be obtained. In the example here, the apparent protection seen in curve I may be real (curve A) or may be lost when long-term survival is assessed (curve B). This allows one to discriminate between true protection by autophagy and a situation where autophagy merely delays death. The latter situation might, for example, occur if autophagy blocks the activation of caspases and thus prevents apoptosis but does not prevent the cells from dying via a slower method. On the other hand true protection suggests either that the autophagy prevents the damage caused by the stimulus or (probably more common) that the protection provided by autophagy kept the cell alive until it had time to repair the damage. The same experimental strategy allows one to definitively demonstrate autophagic cell death (i.e. death that is caused by autophagy). This is shown in curves C and D following on from Curve II. Autophagy may appear to promote cell death when in fact it is merely altering the marker that is being measured– this would provide a curve like that seen in D. For example, this scenario can arise if one measures mitochondrial enzyme activity as a measure of cell viability – increased autophagy might cause a reduction in the signal that is measured because of mitophagy however it would not be correct to conclude that there were fewer viable cells in the sample instead those cells may be quite viable but have fewer functioning mitochondria. The same approaches can be applied to test the effects of increasing autophagy on cell survival. It is also possible that one would detect no effect in short-term assays but see autophagy-dependent protection (or death) in the long-term assays. This situation might arise when the assay used for the short-term viability test is performed too early after exposure to the stimulus that is being tested.
Figure 1.
Schematic representation of experimental design to determine the role of autophagy in drug-induced cell death. The extent of protection/potentiation caused by autophagy knockdown can provide an estimate of how much of the death is dependent upon autophagy. The curves show typical cases where inhibition of autophagy is not completely effective and the role of autophagy may not be absolute. In an extreme case where all cell death was due to autophagy, complete inhibition of autophagy might provide complete protection against the stimulus at all doses.
The recommendations outlined here seem obvious and should be applied to any study of cell death whether it involves autophagy or not. However, because of the varied roles that autophagy can play related to cell death, these design elements may be especially important when studying autophagy and cell death. Unfortunately, the rapidly expanding autophagy literature shows that many papers that address the role of autophagy in cell death do not incorporate these kinds of design elements into their study. While this certainly doesn't mean that the conclusions of those papers are all wrong, it is quite likely that at least some mistaken conclusions have made it into the literature that could have been avoided by incorporating relatively easy experiments into the study and all of us when we are designing and performing experiments or reviewing the papers that report them would, I suggest, benefit from incorporating these ideas. It is of course also important to incorporate other aspects of good experimental design into these kinds of experiments. For example, because the same proteins (e.g. Bcl-2, Bcl-xL, Atg5, FADD and possibly Beclin-1) can regulate both autophagy and apoptosis (for review see 5, 6), an effect on cell survival that is induced by knockdown of one such protein could, for example, mislead you to conclude that autophagy had an effect when in fact the response was due to altered apoptosis. Obviously, inhibiting the process using different methods can minimize this possibility. Because many of the opportunities to use autophagy-targeted therapeutics 7 ultimately rely on altering cell death, careful consideration of experimental design when studying autophagy's roles in any given death process is of critical importance. Lastly, these issues are relevant to in vitro studies and will also need to be demonstrated in an in vivo context. However, the value of designing the most rigorous experiments in cell culture is that it will provide a stronger framework for designing the best experiments to determine the role of autophagy in regulating cell death vivo.
Acknowledgements
Work in my laboratory is supported by grants from the National Cancer Institute.
References
- 1.Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, Hogan RN, Gilpin C, Levine B. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–46. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 2.Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem. 2007;282:5641–52. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- 3.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, Nelson DA, Jin S, White E. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colell A, Ricci JE, Tait S, Milasta S, Maurer U, Bouchier-Hayes L, Fitzgerald P, Guio-Carrion A, Waterhouse NJ, Li CW, Mari B, Barbry P, Newmeyer DD, Beere HM, Green DR. GAPDH and Autophagy Preserve Survival after Apoptotic Cytochrome c Release in the Absence of Caspase Activation. Cell. 2007;129:983–97. doi: 10.1016/j.cell.2007.03.045. [DOI] [PubMed] [Google Scholar]
- 5.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–52. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 6.Thorburn A. Apoptosis and autophagy: regulatory connections between two supposedly different processes. Apoptosis. 2007 doi: 10.1007/s10495-007-0154-9. In Press doi 10.1007/s10495-007-0154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ. Potential therapeutic applications of autophagy. Nat Rev Drug Discov. 2007;6:304–12. doi: 10.1038/nrd2272. [DOI] [PubMed] [Google Scholar]