Summary
Gene expression requires proper messenger (m) RNA export and translation. However, the functional links between these consecutive steps have not been fully defined. Gle1 is an essential, conserved mRNA export factor whose export function is dependent on the small molecule inositol hexakisphosphate (IP6). Here we show that both Gle1 and IP6 are required for efficient translation termination in Saccharomyces cerevisiae, and Gle1 interacts with termination factors. In addition, Gle1 has a conserved physical association with the initiation factor eIF3, and gle1 mutants display genetic interactions with the eIF3 mutant nip1-1. Strikingly, gle1 mutants have defects in initiation, whereas strains lacking IP6 do not. We propose that Gle1 functions together with IP6 and the DEAD-box protein Dbp5 to regulate termination. However, Gle1 also independently mediates initiation. Thus, Gle1 is uniquely positioned to coordinate the mRNA export and translation mechanisms. These results directly impact models for perturbation of Gle1 function in pathophysiology.
Introduction
A series of molecular events are required during the lifespan of eukaryotic messenger (m) RNA, including transcription, processing, export, translation, and degradation. Completion of these steps underlies all gene expression, and specific quality control mechanisms are predicted to control each (Moore, 2005). To date, functional connections have been documented between transcription, nuclear processing, and export (Hagiwara and Nojima, 2007). For example, the recruitment of mRNA export factors is coupled to proper transcription and splicing (Kohler and Hurt, 2007). Links between mRNA export and translation also exist (Culjkovic et al., 2006; Gross et al., 2007; Jin et al., 2003; Rosenwald et al., 1995); however, the molecular mechanisms are less well defined. Such linkages presumably allow efficient cellular regulation of gene expression at multiple levels simultaneously.
Eukaryotic translation is a well-conserved mechanism with four major phases: initiation, elongation, termination, and recycling (for review, see Kapp and Lorsch, 2004). Initiation, which involves the assembly of translation-competent ribosomes on mRNA, depends on eukaryotic initiation factors (eIFs) that stimulate ribosome loading. One of these, eIF3, has 6 to 13 subunits (in S. cerevisiae and humans, respectively) and functions by stimulating complex formation on the 40S subunit (forming the 43S preinitiation complex), recruiting the mRNA, and facilitating start site scanning (Hinnebusch, 2006). Interestingly, eIF3 also regulates dissociation and recycling of the ribosome after termination (Pisarev et al., 2007).
Proper termination of translation is equally critical for gene expression. Termination is primarily regulated by release factors (eRFs) 1 and 3 (Sup45 and Sup35, respectively, in yeast) (Kapp and Lorsch, 2004). The stop codon is recognized by eRF1/Sup45, which stimulates ribosomal peptidyl transferase activity to release the completed polypeptide. This activity is enhanced by eRF3, although eRF3 also binds poly(A) binding protein (PABP/Pab1) and might have a role in ribosome recycling (Hoshino et al., 1999).
Before translation, however, an mRNA must exit the nucleus via the nuclear pore complex (NPC). Initial mRNA export steps require recognition of an export-competent mRNA-protein complex (mRNP) by the transport factor Mex67 (TAP/NXF1 in vertebrates) (Gruter et al., 1998; Katahira et al., 1999; Segref et al., 1997). Following Mex67-dependent targeting to the NPC, several other factors are necessary for proper export. Gle1 is an essential mRNA export factor in both yeast and human cells (Murphy and Wente, 1996; Watkins et al., 1998). Gle1 associates with the NPC through nucleoporin-42 (Nup42) in yeast and hCG1 and Nup155 in human cells (Kendirgi et al., 2005; Murphy and Wente, 1996; Rayala et al., 2004). In conjunction with the small molecule inositol hexakisphosphate (IP6), Gle1 stimulates the RNA-dependent ATPase activity of Dbp5 (Alcazar-Roman et al., 2006; Weirich et al., 2006). Dbp5 is an essential member of the DEAD-box helicase family, with a cytoplasmic NPC binding site at Nup159 in yeast that is juxtaposed to the Gle1-Nup42 complex (Hodge et al., 1999; Schmitt et al., 1999; Snay-Hodge et al., 1998). Gle1/IP6-activated Dbp5 is converted to a Dbp5-ADP state that triggers mRNP remodeling, displacing proteins to facilitate export directionality (Tran et al., 2007).
Prior studies have suggested coupling between the export and translation mechanisms. The vertebrate TAP/NXF1 promotes the translation of RNA harboring a viral constitutive transport element (CTE) (Jin et al., 2003). Evidence also suggests that the cap-binding protein eIF4E regulates export of a subset of cell cycle-related mRNAs (Culjkovic et al., 2006; Rosenwald et al., 1995). Intriguingly, a recent report has revealed a novel function of Dbp5 in translation termination (Gross et al., 2007). Thus, we hypothesized that Gle1 might also have a role in translation via Dbp5 activation. Although Gle1 is enriched at the nuclear rim, Gle1 is also found in the cytoplasm (Del Priore et al., 1996; Watkins et al., 1998), supporting the existence of a Gle1 pool for the translation machinery.
Fully delineating Gle1 cellular functions is especially important given the causal links reported between mutations in human GLE1 and a severe form of human motor neuron degeneration (Nousiainen et al., 2008). Here we show that Gle1 has genetic and physical interactions with translation factors and plays roles in both termination and initiation. Surprisingly, IP6 production is only required for translation termination. This suggests that Gle1 plays two independent roles in translation, one in termination via IP6-dependent activation of Dbp5, and one in initiation that is IP6 and Dbp5-independent. This work reveals an extensive molecular linkage between mRNA export and translation and provides a mechanism for co-regulation of critical gene expression steps.
Results
Gle1 plays a role in translation
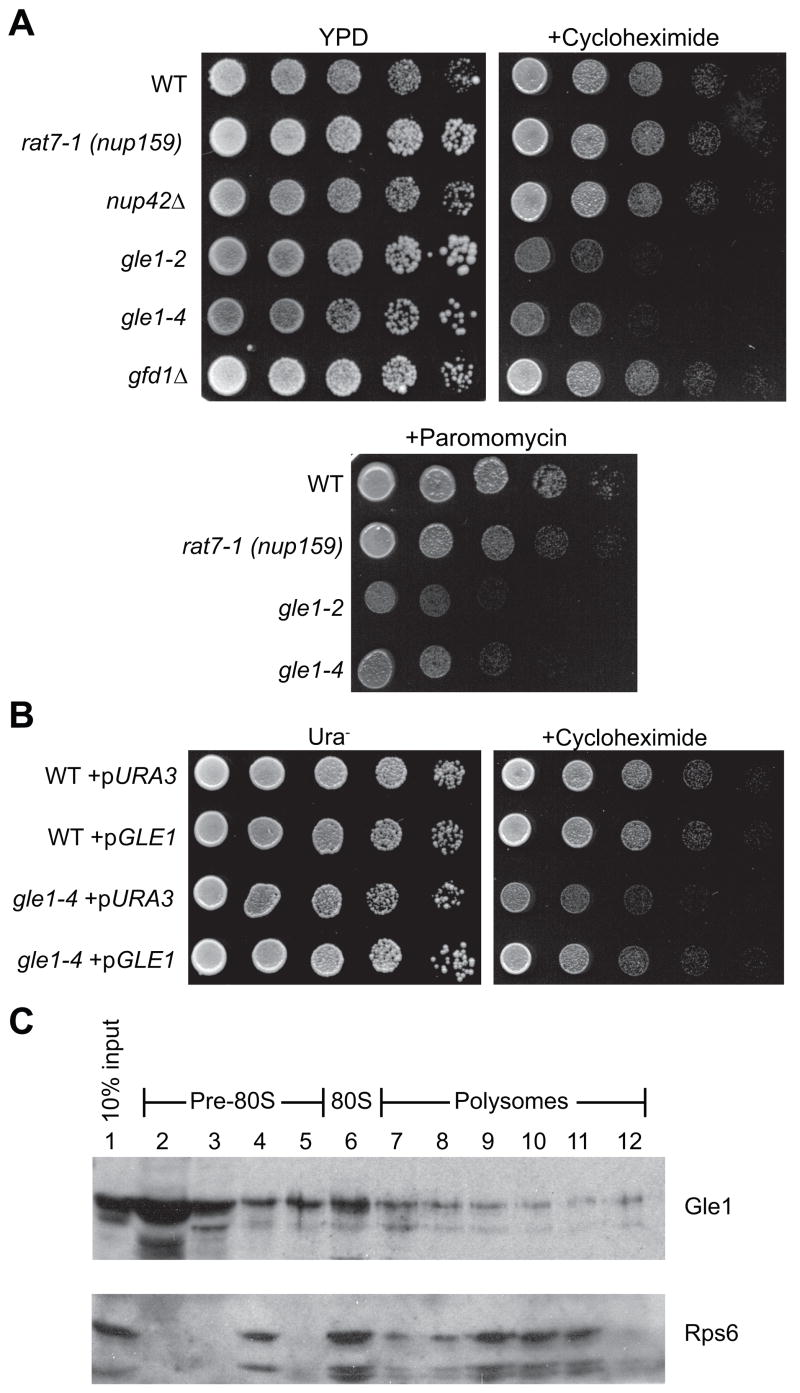
To investigate whether Gle1 functions in translation, we employed the S. cerevisiae model system and analyzed a panel of mutant strains for growth defects in the presence of the translation inhibitors cycloheximide and paromomycin (Figure 1A). These inhibitors have been previously used to identify mutants with defects in different aspects of translation (Greenberg et al., 1998; Gross et al., 2007; Wakem and Sherman, 1990) and allow a rapid assessment of general translation perturbations. Two temperature-sensitive gle1 mutant strains (gle1-2 and gle1-4) grew similarly to a wild-type control strain at 23°C on rich media. However, at 23°C, both had significantly reduced growth in the presence of cycloheximide and paromomycin (Figure 1A). As controls, strains harboring a wild type GLE1-expressing plasmid were rescued for the cycloheximide sensitive growth defect (Figure 1B). Importantly, strains harboring mutations in other genes required for mRNA export through NPCs (rat7-1 (nup159), nup42Δ) were not inhibited for growth under these conditions (Figure 1A). Of note, Nup159 and Nup42 are the respective NPC docking sites for Dbp5 and Gle1 (Hodge et al., 1999; Murphy and Wente, 1996; Schmitt et al., 1999), and the rat7-1 (nup159) mutant has a severe mRNA export defect (Gorsch et al., 1995). This indicated that the hypersensitivity of the gle1 mutants to translation inhibition is not attributed to general defects in mRNA export.
Figure 1. A role for Gle1 in translation.
(A) Cultures of gle1-2, gle1-4, rat7-1 (nup159), nup42Δ, gfd1Δ, and wild-type (WT) strains were spotted in five-fold serial dilutions on YPD alone or YPD containing 0.1 μg/ml cycloheximide or 0.4 mg/ml paromomycin, then incubated 2 days at 23°C.
(B) Cultures of gle1-4 and wild-type strains transformed with empty URA3/CEN plasmid (pURA3) or a GLE1/URA3/CEN (pGLE1) plasmid were spotted as in (A) on Ura− selective media with or without cycloheximide.
(C) Lysates of wild-type strains grown at 23°C were subjected to sucrose density fractionation and immunoblotted with α-Gle1 and α-Rps6 (ribosomal protein control). Ribosome distribution was determined by OD254.
If Gle1 is involved in translation, we speculated that it might be associated with ribosomes. Sucrose density fractionation was performed with lysates from wild-type strains and samples were analyzed by immunoblotting for Gle1. The majority of Gle1 was found in lightest (non-ribosome) fractions; however, a significant portion of Gle1 was in the heavier ribosomal fractions, particularly the 80S fraction (Figure 1C, lane 6). This indicated that Gle1 might functionally associate with ribosomal complexes.
Gle1 interacts with the termination factor Sup45
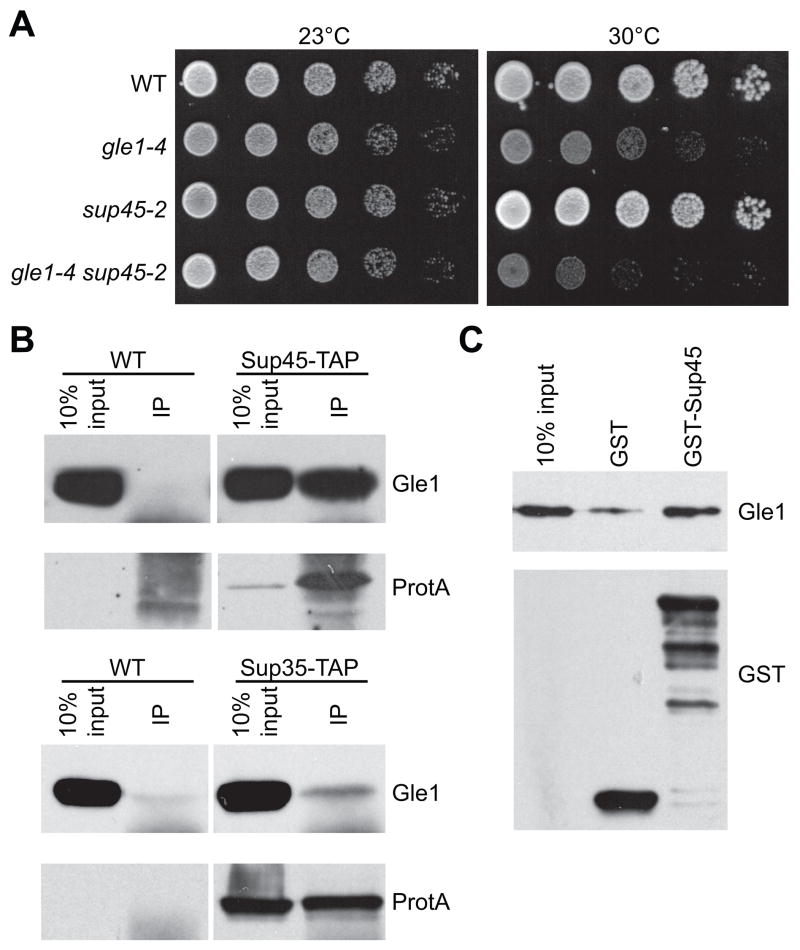
We next used genetic and biochemical assays to test whether Gle1 plays a role in translation termination. First, gle1 sup45 double mutant strains were tested for synthetic fitness defects at elevated temperatures (Figure 2A). At 30°C, a semi-permissive growth temperature for the gle1-4 strain, growth of the gle1-4 sup45-2 double mutant was inhibited compared to either single mutant. Second, co-immunoprecipitation experiments were conducted to test for Gle1 physical association with termination factors. Using strains with Sup45 and Sup35 epitope-tagged by a tandem affinity purification (TAP) sequence, complexes were isolated from cell lysates and immunoblotted (Figure 2B, top panel). Substantial levels of endogenous Gle1 were co-immunoprecipitated with Sup45-TAP. Minimal Gle1 was detected in isolations from control lysates lacking a TAP-tagged protein. The Gle1:Sup45-TAP interaction was not altered by RNase treatment, indicating that the association is not mediated through RNA (data not shown). Although substantially lower, Gle1 also co-immunoprecipitated with Sup35-TAP (Figure 2B, bottom panel).
Figure 2. Gle1 interacts with termination factors.
(A) Cultures of gle1-4, sup45-2, gle1-4 sup45-2, and wild-type control (WT) strains were serially diluted, spotted on YPD, and incubated 2–3 days at 23°or 30°C.
(B) Immunoprecipitations were performed with lysates from SUP35-TAP, SUP45-TAP, and wild-type (WT) yeast strains. Lysates (10% of input) and total immunoprecipitates (IP) were immunoblotted with α-mouse IgG (to detect TAP-tagged proteins (ProtA) or α-Gle1.
(C) Soluble binding assays were performed with GST-Sup45 or GST from bacterial lysates and recombinant purified Gle1. Gle1 input (10%) and bound samples were immunoblotted with α-GST or α-Gle1.
To investigate whether Gle1 and Sup45 directly bind in vitro, soluble binding assays were performed with bacterially expressed Gle1 and GST-tagged Sup45. Compared to a control assay with GST alone, significantly higher levels of purified Gle1 bound to GST-Sup45 (Figure 2C). Further, Gle1 still bound to GST-Sup45 after RNase treatment (data not shown). Dbp5 also associates with Sup45 (Gross et al., 2007). Therefore, we concluded that Gle1 and Dbp5 are both physically connected to the translational termination complex.
Gle1 is required for efficient translation termination
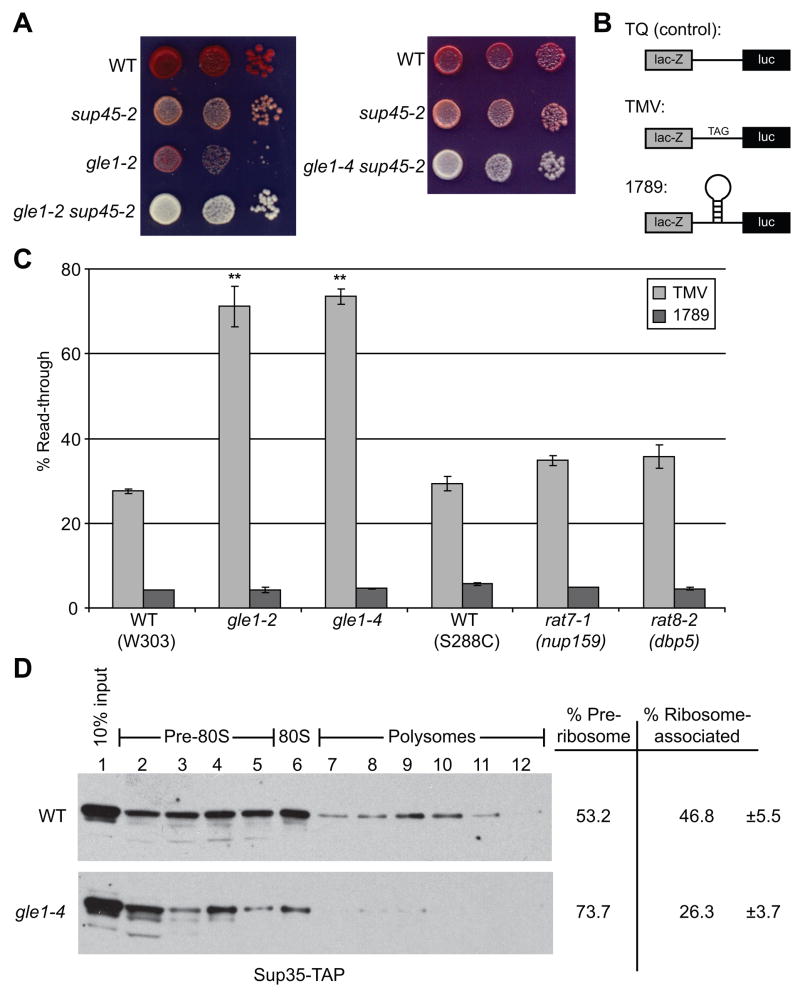
To further examine the functional requirement for Gle1 in termination, two independent assays for termination efficiency were conducted. First, we used strains with an ade2-1 allele harboring an ochre nonsense mutation that are red in color due to lack of functional Ade2 enzyme. Nonsense codon read-through, as a result of termination defects, produces functional Ade2 protein and reduces red pigmentation (Cox, 1965). Colonies of a wild-type control strain were red in color, whereas the temperature-sensitive sup45-2 mutant was pink (Figure 3A). In stark contrast, gle1-2 sup45-2 and gle1-4 sup45-2 double mutant colonies were white. This indicated a more severe termination defect in the double mutants than in the respective single mutant strains. Of note, this effect was observed at the permissive temperature for gle1 mutants (23°C).
Figure 3. Gle1 is required for efficient translation termination.
(A) Cultures of gle1-2, sup45-2, gle1-2 sup45, gle1-4 sup45-2 and wild-type controls (WT) were serially diluted and spotted on 0.25 YPD. Plates were incubated 4 days at 23°C and colony color assessed.
(B) The tandem β-galactosidase/luciferase reporter constructs are shown: TQ (control) lacks a stop codon in the linker, TMV has a stop codon inserted in frame into the linker region, and 1789 has a stem-loop from HIV-1 inserted into the linker.
(C) gle1-2, gle1-4, rat8-2 (dbp5), rat7-1 (nup159), and wild-type control (WT) strains were transformed with the tandem reporter constructs and luciferase and β-galactosidase assays were performed. Activities were normalized to cell number, and ratios of luciferase to β-galactosidase activity determined. Read-through activity is expressed as the percentage of the TMV or 1789 reporter compared to TQ control for each strain, n = 3–5 independent experiments. Error bars = 1 SEM above and below the mean. ** p<0.01
(D) Lysates of SUP35-TAP and gle1-4 SUP35-TAP strains grown at 23°C were subjected to sucrose density fractionation and immunoblotted with α-mouse IgG to detect Sup35-TAP. Ribosome distribution was determined by OD254, and blots were analyzed by densitometry to assign ribosome association percentages (n = 4). Error = 1 SEM above and below the mean. p<0.05
We also performed an assay that utilizes a tandem β-galactosidase/luciferase reporter separated by a short linker sequence (Stahl et al., 1995). Three related plasmid-based reporters are shown in Figure 3B: one with a stop codon inserted in frame into the linker region (TMV), a control lacking the stop codon (TQ), and a control in which a stem-loop from HIV-1 has been inserted in the linker (1789). These plasmids were transformed into control and gle1 mutant strains, and the read-through efficiency was determined at 23°C. Comparing ratios of luciferase to β-galactosidase activity between the TQ control and the other two plasmids yielded the relative level of abnormal termination read-through. Roughly consistent with previous results (Stahl et al., 1995), wild-type strains showed ~27% read-through with the TMV construct (Figure 3C). Strikingly, read-through in both gle1-2 and gle1-4 strains was dramatically increased to 71% and 73%, respectively. The effects were specific to normal termination, as read-through efficiency of the stem loop did not change (Figure 3C). Similar results were observed using other stop codon sequences (data not shown). A rat7-1 (nup159) strain showed minimal change (34%), indicating that a general perturbation of mRNA export did not impact translation termination. Another mRNA export mutant strain, nab2-ΔN, also had no appreciable defect in read-through efficiency (Supplemental Figure 1A). The defect was also notably less (36%) in a rat8-2 (dbp5) mutant compared to gle1 mutants at 23°C, and others previously reported a modest increase (from ~16 to 25%) in read-through for the rat8-2 (dbp5) mutant after a brief pulse at the restrictive temperature (Gross et al., 2007). We concluded that the differences between the rat8-2 (dbp5) and gle1 mutants reflect mutant specificity and highlight the robustness of the gle1 mutant defect. Interestingly, GLE1 overexpression partially rescued the read-through defect of a sup35-21 termination mutant (Supplemental Figure 1B). Taken together, Gle1 is required for efficient translation termination.
We next analyzed the direct functional consequences on termination. Association of Sup35 (eRF3) with the termination complex is a critical step in termination, and others have shown that Dbp5 is required for Sup35 incorporation into termination complexes (Gross et al., 2007). Sucrose density fractionation was performed with lysates from control and gle1-4 strains containing TAP-tagged Sup35 (Figure 3D). In wild-type samples, significant levels of Sup35-TAP co-fractionated with ribosomes (80S and polysomes, determined by OD254). Densitometric analysis revealed that approximately 47% of Sup35-TAP was ribosome-associated in wild type lysates. However, the levels of ribosome-associated Sup35-TAP significantly decreased in lysates from gle1-4 mutant cells, particularly in the later polysome fractions (compare lanes 7–12 in wild-type vs. gle1-4), and the majority of the Sup35-TAP shifted to the low density fraction (lane 2). Quantification showed that the level of ribosome-associated Sup35-TAP in gle1-4 samples (26%) dropped to nearly half that in wild-type. In the gle1-4 mutant, Sup45, Dbp5, and Gle1 did not show significant decreases in ribosome association (Supplemental Figure 1C). We concluded that Gle1 promotes ribosome association of Sup35 in termination.
Gle1 interacts with the initiation factor eIF3
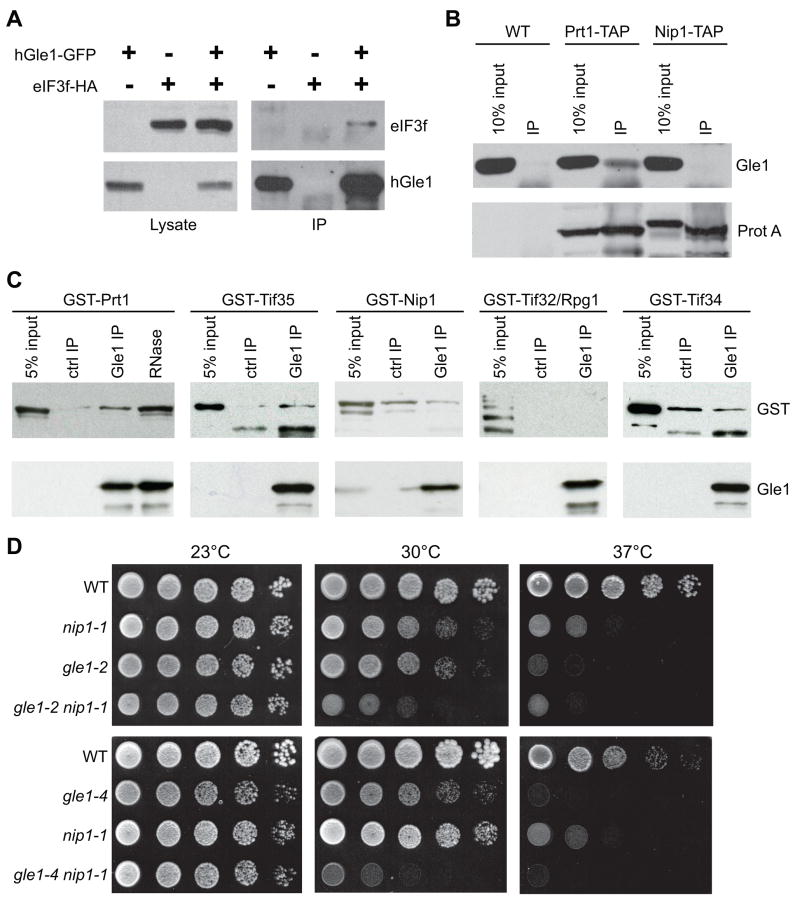
Although consistent with the reported Dbp5 interactions, the association of Gle1 with the translation termination complex was intriguing in light of our prior discovery of a yeast two-hybrid interaction between human Gle1 (hGle1) and the f subunit (p47/S5/ε) of the translation initiation factor eIF3 (Rayala et al., 2004). To further examine this interaction in human cells, plasmids expressing GFP-tagged hGle1 and HA-tagged eIF3f were transfected into HeLa cells. Immunoprecipitations with anti-GFP antibodies were performed from total cell lysates (Figure 4A). Immunoblot analysis showed that eIF3f-HA was specifically co-isolated with hGle1-GFP. This result confirmed that hGle1 interacts with the eIF3 subunit.
Figure 4. Gle1 has conserved interactions with eIF3 subunits.
(A) Plasmids expressing hGle1-GFP and eIF3f-HA were cotransfected into HeLa cells, and immunoprecipitations were performed using α-GFP antibodies. Lysates (5%) and immunoprecipitates were immunoblotted using α-GFP or α-HA.
(B) Immunoprecipitations were performed with lysates from PRT1-TAP, NIP1-TAP, and wild-type (WT) yeast strains. Lysates (10% of input) and total immunoprecipitates (IP) were immunoblotted with α-mouse IgG (for TAP-tagged proteins (ProtA)) or α-Gle1.
(C) Recombinant purified Gle1 was incubated with bacterial lysates for GST-Prt1, -Nip1, -Rpg1/Tif32, -Tif34, or -Tif35, and bound proteins were isolated by immunoprecipitation with α-Gle1. In controls, lysates were incubated without Gle1. RNase A treatment was conducted with the Gle1/GST-Prt1 sample. Total bound and input (5%) were immunoblotted with α-GST or α-Gle1.
(D) Cultures of the respective gle1 and nip1 single and double mutant strains with wild-type controls (WT) were serially diluted and spotted on YPD. Plates were incubated 3 days at 23°C, 30°C or 37°C.
As the human eIF3f subunit is not conserved in S. cerevisiae (Hinnebusch, 2006), we performed immunoprecipitations from yeast strains expressing TAP-tagged versions of different yeast eIF3 subunits. Endogenous Gle1 was co-immunoprecipitated with Prt1-TAP (eIF3b) (Figure 4B). We did not consistently detect significant co-isolation of Gle1 with the other eIF3 subunits tested (Nip1/eIF3c, Rpg1/eIF3a, and Tif34/eIF3i), though this might reflect the stringency of the experimental conditions (Figure 4B and data not shown). Of note, we did not detect Dbp5 in either the Prt1-TAP or Nip1-TAP complexes (data not shown). We concluded that Gle1 physically associates with eIF3 subunits, and that these interactions are conserved from yeast to humans.
To test for direct physical interactions between Gle1 and eIF3, we performed in vitro binding experiments. Lysates from bacterial cells expressing GST-tagged fusions for each of the five core subunits of yeast eIF3 (Prt1, Nip1, Tif32/Rpg1, Tif34, and Tif35) were incubated with recombinant purified Gle1. Immunoprecipitations were performed with anti-Gle1 antibodies and the GST-tagged eIF3 subunits were detected by immunoblotting. As shown in Figure 4C, GST-Prt1 was co-isolated with Gle1, and the association was maintained after RNase treatment. GST-Tif35, which is part of a sub-complex with Prt1 (Phan et al., 2001), also bound Gle1. The other three subunits did not specifically bind Gle1. Thus, Gle1 physically associates with eIF3, with two potential binding surfaces in Prt1 and Tif35.
To test for functional links between Gle1 and eIF3, we generated gle1 nip1 double mutant strains and growth was assayed at a range of different temperatures. At 23°C, no differences were observed when comparing growth between single and double mutants (Figure 4D). However, synthetic fitness defects were detected at 30°C, wherein both the gle1-2 nip1-1 and the gle1-4 nip1-1 strains had inhibited growth compared to the single mutants. In contrast, no synthetic growth defects were observed in rat8-2 (dbp5) nip1-1 or rat7-1 (nup159) nip1-1 double mutants (Supplemental Figure 2A and data not shown). Further indicating interaction specificity, no synthetic growth defects were observed in gle1 prt1-1 or gle1 rpg1-1 double mutants (data not shown). Thus, both physical and genetic interactions were found between Gle1 and eIF3, and these functions appear to be independent of Dbp5.
Gle1 functions in translation initiation
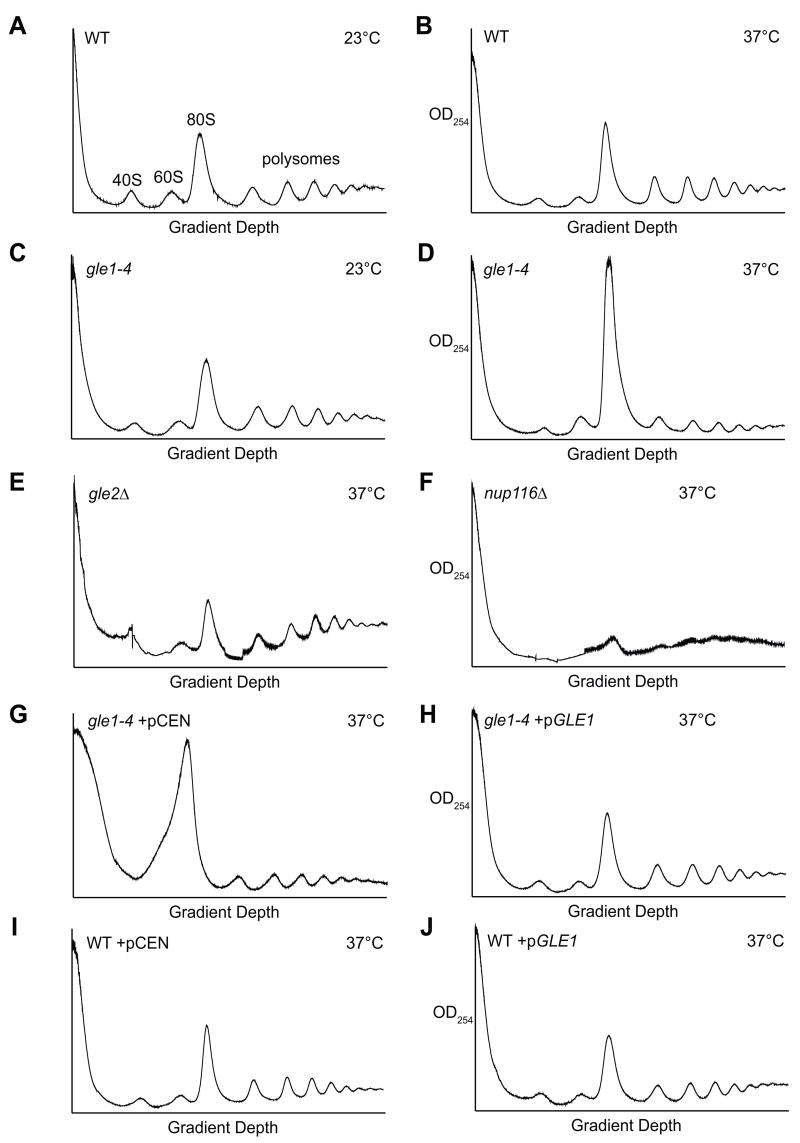
To address the hypothesis that Gle1 is required for initiation, we first analyzed the monosome to polysome content of different mutant strains using sucrose density sedimentation. At the permissive temperature (23°C), the polysome profile from the gle1-4 mutant cells was similar to that from wild type cells (Figure 5A, C). However, after a temperature shift to 37°C, gle1-4 cells displayed a dramatically elevated monosome peak and reduced polysomes (Figure 5D). This profile indicated a defect in translation initiation, as it was similar to those of strains defective for initiation factors, including nip1 (Greenberg et al., 1998; Nielsen et al., 2004). Intriguingly, the polysome profile of rat8-2 (dbp5) at 37°C is also suggestive of an initiation defect (Supplemental Figure 2B; Gross et al., 2007). As controls, we confirmed that the gle1-4 defect was rescued by plasmid-based GLE1 expression (Figure 5G, H). Importantly, as shown in Figure 5E, a strain lacking GLE2 (gle2Δ), encoding another mRNA export factor (Murphy et al., 1996), had no significant perturbation in the monosome:polysome ratio at 37°C. In addition, a nup116Δ strain displayed a different profile with reduced monosomes and polysomes (Figure 5F). This is likely due to diminished ribosome subunit export and a total block in nucleocytoplasmic trafficking at 37°C in the nup116Δ cells (Stage-Zimmermann et al., 2000; Wente and Blobel, 1993). As the gle1-4 ribosome profile is distinct from both the gle2Δ and nup116Δ profiles, we concluded that the gle1-4 defect was not due to a block in mRNA export. The altered profile of the gle1-4 mutant cells supports a role for Gle1 in translation initiation.
Figure 5. Strains harboring gle1 mutant alleles have polysome profile defects.
Polysome profiles were generated by subjecting cell lysates to 7–47% sucrose density centrifugation and OD254 analysis of the fractionated gradient. The monosome (80S) and polysome peaks are labeled in (A). Cultures of wild-type cells were grown at 23°C (A) or shifted to 37°C for 75 min prior to harvest (B). Cultures of gle1-4 were grown at 23°C (C) or shifted to 37°C for 75 min prior to harvest (D). The mRNA export mutants gle2Δ (E) and nup116Δ (F) were shifted to 37°C for 75 min prior to harvest. Note that both monosome and polysome peaks are reduced in the nup116Δ strain. The gle1-4 and wild-type control (WT) strains harboring either a URA3/CEN or GLE1/URA3/CEN plasmid were shifted to 37°C for 60 min prior to harvest (G–J).
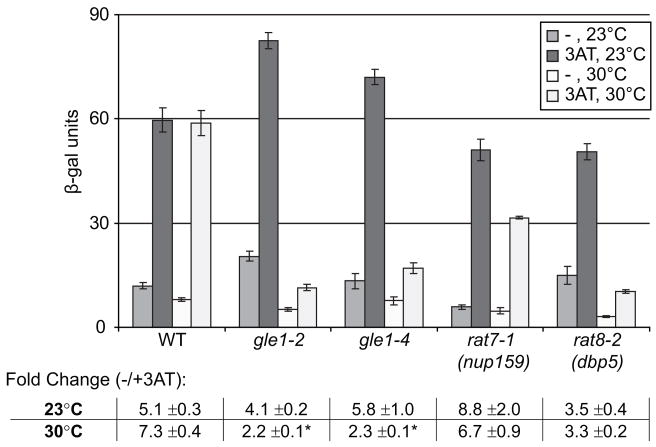
Based on the genetic and physical links between Gle1 and eIF3, we tested whether gle1 mutants have defects in GCN4 regulation. Expression of GCN4, an activator of biosynthetic pathways, is subject to translational control (for review, see Hinnebusch, 2005). Translation of the GCN4 mRNA is normally de-repressed by amino acid starvation via alteration of the eIF2α phosphorylation state. However, this regulation is impaired by defects in initiation factors such as eIF3 (e.g. Nielsen et al., 2004), and initiation mutant strains often show either constitutive repression or de-repression of GCN4 expression (Gcn− or Gcd− phenotypes, respectively). To examine GCN4 expression, we utilized a reporter in which the non-coding 5′ region of GCN4 is linked to lacZ. Incubation of a wild-type control strain with 3-aminotriazole (3AT), an inhibitor of the histidine synthesis pathway, activated β-galactosidase activity to a similar extent at 23°C or 30°C (Figure 6). In contrast, both the gle1-2 and gle1-4 strains had high activity in 3AT-treated samples at 23°C with a sharp reduction of activation at a semi-permissive temperature of 30°C. This indicated that the gle1 mutants have a Gcn− phenotype, and the dramatic reduction in activation is consistent with a role for Gle1 in translation initiation.
Figure 6. GCN4 activation defects are detected in gle1 mutants.
Wild-type (WT), gle1-2, gle1-4, rat7-1 (nup159), and rat8-2 (dbp5) strains were transformed with a GCN4-lacZ reporter plasmid. Cultures were grown in Ura− His−selective media at either 23°C or 30°C for 2h. 3-aminotriazole (3AT) was added as indicated. Following growth overnight at 23°C or 30°C, assays for β-galactosidase activity were performed. Activity is expressed as β-gal units (1 unit hydrolyzes 1 μmol of ONPG substrate per min per cell). Error bars = 1 SEM above and below the mean. Table summarizes fold activation (+3AT divided by −3AT) for each strain at each temp. * p<0.05 for the fold activation at 23°C vs. 30°C. (n = 3–5 independent experiments).
On the other hand, in rat7-1 (nup159) strains, activation was still observed at 30°C with a small decrease compared to 23°C. However, the basal level of activity in the rat7-1 (nup159) strains was also decreased, such that the fold-increase induced by 3AT changed only slightly (Figure 6). Activation by 3AT in the rat7-1 (nup159) mutant was also maintained up to 34°C (data not shown). Overall, the results with the rat7-1 (nup159) strain were fully consistent with a defect in export of the reporter mRNA. A cold-sensitive mRNA export mutant strain, nab2-ΔN (Marfatia et al., 2003), also did not display any temperature-dependent changes in GCN4 activation (Supplemental Figure 2C). Further, a sup45-2 termination mutant showed a moderate Gcd− phenotype with significant activity in the absence of 3AT (Supplemental Figure 2D), suggesting that the gle1 mutant phenotype is not due to termination defects. Interestingly, the rat8-2 (dbp5) mutant strain showed a substantial reduction in activity at a semi-permisssive temperature (Figure 6). This could indicate a Gcn− defect; however, similar to the rat7-1 (nup159) mutant, little change in the fold-increase was observed in the rat8-2 (dbp5) mutant. These results were consistent with the lack of genetic and biochemical connections to translation initiation function for Dbp5. We conclude that if Dbp5 plays a role in initiation, this role is distinct from Gle1.
IP6 production regulates termination but not initiation
n nuclear export, IP6 binds Gle1 to facilitate co-activation of Dbp5 (Alcazar-Roman et al., 2006; Weirich et al., 2006). IP6 is generated by a phospholipase C-dependent pathway and the coordinated activities of two inositol kinases (York et al., 1999). Ipk2 is a dual function kinase that converts inositol 1,4,5 trisphosphate to inositol 1,3,4,5,6 pentakisphosphate (IP5), whereas a 2-specific kinase, Ipk1, mediates production of IP6 from IP5 (York et al., 1999). In addition, a set of IP6 kinases (Vip1 and Kcs1) are responsible for the generation of distinct higher order pyrophosphorylated inositols (Mulugu et al., 2007; Saiardi et al., 1999). Roles for these soluble inositides in translation have not been previously reported.
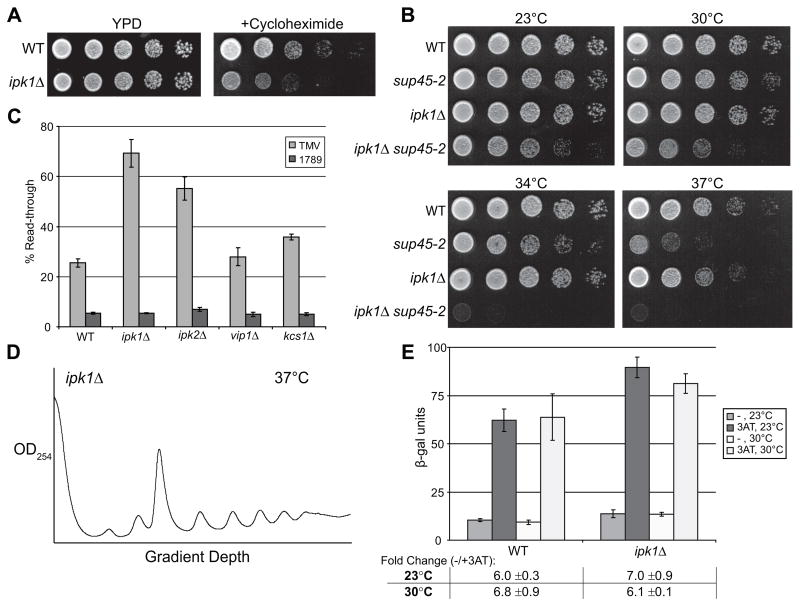
We found that the ipk1Δstrain was hypersensitive to the translation inhibitor cycloheximide (Figure 7A). In addition, ipk1Δ sup45-2 double mutant strains were generated and displayed synthetic growth defects at all temperatures with lethality at 34°C (Figure 7B). The ipk1Δ sup45-2 strains also showed increased termination read-through based on the assay for elevated Ade2 production in ade2-1 strains (data not shown). Furthermore, like gle1 mutant strains, ipk1Δ strains had highly elevated read-through activity (69%) in the tandem reporter assay (Figure 7C). Importantly, the ipk2Δ strain also had increased termination read-through. In contrast, normal or only slightly elevated read-through activity was observed in vip1Δ and kcs1Δ strains, respectively. These data strongly implicate IP6 production by Ipk1 in translation termination.
Figure 7. IP6 regulates termination but not initiation.
(A) Cultures of ipk1Δ and wild-type (WT) strains were spotted in five-fold serial dilutions on YPD alone or on YPD containing 0.1 μg/ml cycloheximide. Plates were incubated 2 days at 23°C.
(B) Cultures of ipk1Δ, sup45-2, ipk1Δ sup45-2 and wild-type (WT) strains were serially diluted, spotted on YPD, and incubated 2 days at 23°C, 30°C, 34°C, or 37°C.
(C) Read-through assays were performed with ipk1Δ, ipk2Δ, vip1Δ, kcs1Δ, and wild-type control (WT) strains using the β-galactosidase/luciferase tandem reporters. Read-through efficiency was determined as in Figure 3C, n = 4–6 independent experiments.
(D) Polysome profiles for ipk1Δ were generated as in Figure 5. Cells were shifted to 37°C for 75 min prior to harvest.
(E) GCN4 activation assays were performed with ipk1Δ and wild-type control (WT) strains using the GCN4-lacZ reporter as in Figure 6.
Because IP6 directly binds Gle1 (Alcazar-Roman et al., 2006; Weirich et al., 2006), we hypothesized that IP6 might also play a role in initiation. However, polysome profiles of lysates from an ipk1Δ strain revealed no major defects at either 23°C or after a shift to 37°C (Figure 7D and data not shown). In contrast to gle1 mutants, the ipk1Δ strain also displayed no perturbation in activation of the GCN4-lacZ reporter at any temperature tested (Figure 7E). Finally, no synthetic genetic interaction was observed with ipk1Δ nip1-1 double mutants (data not shown). Therefore, Ipk1 is not required for initiation, and the function of Gle1 in initiation is not dependent on IP6 production.
Discussion
Here we report that Gle1, a bona fide mRNA export factor, also has a role in translation. Strikingly, Gle1 is required for both translation initiation and termination. For termination, we document genetic and physical interactions between Gle1 and the termination factor Sup45/eRF1 (Figure 2), and functional relevance from a perturbation in Sup35/eRF3 recruitment in gle1 mutants (Figure 3D). We further show that Ipk1, and specifically IP6 production, is also linked to translation termination. These requirements for Gle1 and IP6 parallel those recently reported for Dbp5 in translation termination (Gross et al., 2007). Thus, we predict that the Gle1/IP6 role in termination is via activation of Dbp5. However, we also find that Gle1 influences translation initiation. There are genetic and physical interactions between Gle1 and subunits of the initiation factor eIF3 (Figure 4), as well as translation initiation defects in gle1 mutants (Figures 5 & 6). In sharp contrast, IP6 is not involved in initiation, and any Dbp5 initiation role is distinct from Gle1. Interestingly, the gle1 mutant alleles used here are temperature-sensitive with regard to the initiation defects, whereas the termination deficiencies are manifested at the permissive temperature. This lends further credence to the proposal that Gle1 exhibits distinctly different functions in initiation and termination. We conclude that the roles for Gle1 in translation termination and initiation are separable, and the Gle1 initiation function is independent of Dbp5 and IP6.
We further hypothesize that the roles for Gle1 and IP6 in translation are independent of their function in mRNA export. Specifically, the translation defects in the gle1 and ipk1 mutants are not due to indirect effects of mRNA export defects. There are four distinct lines of evidence that support this conclusion. First, other mRNA export mutant strains such as rat7-1 (nup159) do not have the same phenotypes (Figures 1, 3, & 6; Gross et al., 2007). The gle1-2, gle1-4, rat7-1 (nup159), and rat8-2 (dbp5) strains all have minor mRNA export defects at 23°C with more severe defects at 37°C (Gorsch et al., 1995; Murphy and Wente, 1996; Snay-Hodge et al., 1998). Further, these mutants are all in the same pathway for mRNA export (Alcazar-Roman et al., 2006; Hodge et al., 1999). Since they share similar defects in mRNA export, the different translation phenotypes are very likely the result of defects in a distinct translation function. Our data demonstrating termination but not initiation defects in ipk1Δ strains also supports the specificity of the gle1 mutant phenotypes. Second, at least in some assays, the gle1 and ipk1 mutant phenotypes are not the expected result for an mRNA export-restricted mutant. For instance, in the read-through assay (Figures 3C & 7C), a perturbation in only mRNA export should either decrease activity or have no effect (because the levels of cytoplasmic lac-Z and luciferase reporter would be reduced equally with decreased export). Instead, in gle1 and ipk1 mutants, we observe a dramatic increase in read-through. Third, if a defect in remodeling the mRNP during export is responsible for the perturbations in translation, then the phenotypes of the gle1, ipk1, and dbp5 mutants should be the same. Yet they are distinct, highlighted by the lack of initiation defects in the ipk1Δ mutant. Finally, Gle1 is fully implicated in translation by its specific physical interactions with translation factors both in vivo and in vitro (Figures 2B, C, & 4A, B, C). Gle1 interaction with both initiation and termination factors provides a direct connection to both translation steps.
Previous studies have shown that both human and yeast Gle1 are partially localized to the cytoplasm (Del Priore et al., 1996; Watkins et al., 1998), and hGle1 shuttles between the nucleus and cytoplasm (Kendirgi et al., 2003). This localization is consistent with additional Gle1 functions at sites other than the NPC. Furthermore, hGle1 is present in two isoforms, A and B, that are nearly identical except at the extreme carboxy terminus (Kendirgi et al., 2003). Intriguingly, whereas hGle1B localizes to the nuclear rim, hGle1A does not, and its primary function could be in the cytoplasm. However, as alternative yeast Gle1 isoforms have not been reported, the export and translation functions in S. cerevisiae are presumably performed by the same Gle1 protein.
We propose that Gle1 is a critical factor bridging multiple steps to couple mRNA export and translation. First, as the mRNP passes through the nuclear pore, the RNA-binding protein composition is remodeled in a mechanism that requires Gle1/IP6 activation of the Dbp5 ATPase (Alcazar-Roman et al., 2006; Tran et al., 2007; Weirich et al., 2006). It is possible that Gle1 binds the mRNP during this NPC translocation step and remains associated throughout translation. However, given the predicted low cellular abundance of yeast Gle1 (Ghaemmaghami et al., 2003), transient and/or regulated interactions are more likely. Following export, mRNA translation is initiated by assembly of the eIFs and the 40S subunit. The physical association between Gle1 and eIF3 subunits (Figure 4) suggests that eIF3 could recruit Gle1 to initiation complexes. The precise biochemical role for Gle1 in translation initiation is unknown. Our results suggest that Dbp5 and IP6 are not required for this Gle1-dependent initiation step. The Gcn-phenotype of gle1 mutant strains (Figure 6) might indicate a role for Gle1 in scanning and recognition of the translation start site, although GCN4 regulation is complex and dependent on many processes (Hinnebusch, 2005). Determining the mechanism for the action of Gle1 in initiation is a future goal.
The termination phase of translation begins when the ribosome encounters a stop codon. Krebber and coworkers propose that Dbp5 functions in termination by forming a complex with eRF1/Sup45 on the mRNA, and then remodeling the mRNA-protein complex to allow eRF3/Sup35 recruitment and proper positioning of the termination complex (Gross et al., 2007). As such, the Dbp5 remodeling function in termination is potentially mechanistically similar to its activity in mRNA export (Alcazar-Roman et al., 2006; Tran et al., 2007). This remodeling mechanism is fully consistent with our results showing Gle1 and IP6 functioning in termination, with both having interactions and/or mutant defects that are highly similar to those of Dbp5 (Figures 1, 2, 3 & 7). We propose that Gle1 and IP6 co-activate Dbp5 to trigger remodeling of the translationally-engaged mRNP for proper termination. One effect of this remodeling might be to stabilize the association of eRF3/Sup35, which dissociates in dbp5 and gle1 mutant strains (Figure 3C and Gross et al., 2007). Finally, although Dbp5 might dissociate from the mRNP in the later stages of termination (Gross et al., 2007), an exciting speculation is that Gle1 remains bound and contributes to ribosome dissociation and recycling via eIF3 recruitment. Others have recently reported that eIF3 stimulates dissociation of ribosomes and recycling in an in vitro termination assay (Pisarev et al., 2007). Gle1 would then be inherently available for further rounds of translation initiation by virtue of continued association with eIF3. However, it is not known if the pool of eIF3 in recycling is the same as that in initiation.
Importantly, a recent study has reported that inherited mutations of hGLE1 result in LCCS1, a fetal motor neuron disease with accompanying pleiotropic effects (Nousiainen et al., 2008). The identification of Gle1 translation roles opens up the possibility that the pathophysiology is due to defects in translation rather than, or in addition to, defects in mRNA export. This finding underscores the importance of elucidating how Gle1 activity is regulated in both export and translation. With the Gle1, IP6 and Dbp5 roles in both mRNA export and translation, there are a small but growing number of connections between these two steps in the mRNA life cycle. As an mRNA is exported, there are potentially multiple quality control and signaling steps for properly distinguishing between targeting for translation versus shunting to cytoplasmic degradation or storage granules. Moreover, during polarized trafficking of mRNPs in the cytoplasm, translation is inhibited between nuclear export and transport to the subcellular locale (Sossin and DesGroseillers, 2006). This is especially critical for neuronal development and function (Sossin and DesGroseillers, 2006). Gle1 is positioned to co-regulate export and translation and allow coordination between these different decision points. This may result in a strategy for synchronizing these consecutive processes, allowing for increased efficiency in the regulation of gene expression in specific gene subsets or in specific subcellular locales.
Experimental Procedures
Yeast Strains and Plasmids
Yeast strains and plasmids used are listed in Supplemental Table 1 and 2. Yeast growth assays were performed by serial dilution as previously described (Tran et al., 2007). For the ade2-1 suppression assay, 25-fold dilutions were plated on 0.25YPD (0.25% yeast extract, 1% peptone, 2% glucose).
Immunoprecipitations
For TAP-tagged yeast strains, mid-log phase cultures were lysed in lysis buffer (20mM Tris-HCl pH8.0, 5mM MgCl2, 150mM NaCl, 2% Triton X-100, protease inhibitors) by bead beater (Biospec). Soluble fractions were inclubated with IgG sepharose beads for ~3 h at 4°C after pre-blocking with 1%BSA. Bound proteins were subsequently washed with wash buffer (50mM Tris-HCl pH8.0, 150mM NaCl, 0.1% Triton X-100), eluted with SDS sample buffer, resolved by SDS-PAGE and immunoblotted. Rabbit anti-mouse IgG (Promega) was used to detect TAP-tagged proteins. Yeast Gle1 was detected with affinity-purified guinea pig polyclonal antibodies raised against recombinant MBP-Gle1 (Cocalico Biologicals). For hGle1 immunoprecipitations, HeLa cells were transfected with plasmids expressing hGle1-GFP (pSW1482) and eIF3f-HA using Lipofectamine (Invitrogen) and harvested after approximately 40 hours. Samples were lysed with NETN (170mM NaCl, 1mM EDTA, 20mM Tris-HCl pH8.0, 0.5% NP-40, protease inhibitors). Total protein concentration was determined by Bradford assay, and 1mg of total protein lysate was incubated with anti-GFP antibodies and Protein A-sepharose for 5h at 4°C. Samples were washed 3 times with NETN, resuspended with SDS sample buffer, separated by SDS-PAGE and immunoblotted using anti-GFP and anti-HA antibodies. Secondary antibodies were HRP-conjugated (Jackson) and blots were developed via SuperSignal West Pico ECL (Pierce).
In vitro binding reactions
Recombinant proteins were expressed in bacteria and purified as in (Tran et al., 2007). For Sup45 binding assays, lysates expressing GST-Sup45 or GST alone were incubated with glutathione resin in Buffer B (20mM HEPES pH7.5, 150mM NaCl, 20mM HEPES, 20% glycerol) followed by addition of 0.5 μg purified Gle1 and incubation for 2h at 23°C. Bound proteins were washed and eluted with SDS sample buffer, and 25–33% of the total was immunoblotted. For eIF3 binding assays, bacterial lysates expressing GST fusions of eIF3 subunits were incubated with 1.5 μg purified Gle1 in 300 μl binding buffer at 4°C for 2 hrs. Antibodies to Gle1 and Protein A beads were added, and samples were incubated an additional 1.5 hrs. When applicable, 100 μg/ml RNase A (Qiagen) was added during binding.
Termination read-through assays
Assays were performed essentially as described (Stahl et al., 1995). Yeast strains transformed with pACTQ, pACTMV, or pAC1789 plasmids were grown in liquid Leu−media. Duplicate samples were diluted into YPD and incubated overnight at 23°C until the OD600 ~1.0. Cells were pelleted and resuspended in luciferase lysis buffer (25mM Tris-Phosphate, 2mM EGTA, 10% glycerol, 0.5% Triton X-100). Following lysis, samples were centrifuged and supernatants used for luciferase and β-galactosidase assays. For luciferase assays, samples were diluted in assay buffer (15mM MgSO4, 15mM K2PO4, 4mM EGTA, 1mM DTT, 1mM ATP), and 1mM luciferin was injected before reading in a Lumimark Plus microplate reader (BioRad). Liquid β-galactosidase assays were performed using o-nitrophenyl β-D-galactopyranoside as the substrate according to Clontech protocols. Activity equals 1 μmol of ONPG hydrolyzed per min per cell. Luciferase assay values were first normalized to the OD600 of the cells, then divided by the β-gal activity of each sample. Duplicates were averaged before determining read-through efficiency. The resulting luc: β-gal ratio from TQ samples was set at 100% for each strain. Percent read-through was determined by comparing the luc: β-gal ratios from TMV and 1789 samples to the TQ ratio for each strain. All quantitative data represent the mean value of 3 to 6 independent experiments. Error bars represent standard error of means (SEM) above and below the mean. Statistical significance and p values were calculated by the student’s t-test.
Polysome profiles
Polysome analysis was performed essentially as described (Windgassen et al., 2004). Strains were grown in YPD to approximately 0.5 OD600 and then incubated on ice for 15 min in the presence of 100 μg/ml cycloheximide. Following lysis, supernatants (10 OD600 units) were loaded onto 11 ml sucrose gradients (7 to 47%) in low salt buffer and centrifuged for 2 h at 38,000 rpm in a TH-641 rotor. Fractions from the gradients were removed with continuous monitoring of absorbance at 254nm. If applicable, 1ml fractions were collected and precipitated with 25% TCA prior to resolution by SDS-PAGE and immunoblotting. Samples were separated on SDS-PAGE and immunoblotted. Band intensity was quantified by densitometry (NIH ImageJ).
GCN4 activation assays
Adapted from the published protocol (Moehle and Hinnebusch, 1991), yeast strains harboring the GCN4-lacZ expressing plasmids were grown to stationary phase in Ura− media at 23°C. Cultures were diluted into 4 samples in Ura−His− media, with 2 samples each incubated at 23°C and 30°C for 2 h. 10mM 3-aminotriazole was added to one sample of each strain at each temperature, and all samples were incubated at the respective temperature overnight. Cells were centrifuged and resuspended in Z buffer (60mM Na2HPO4, 40mM NaH2PO4, 10mM KCl, 1mM MgSO4). Liquid β-galactosidase assays were carried out as above.
Supplementary Material
Acknowledgments
We thank Drs. A. J. Link and V. R. Gerbasi for advice and equipment use; Drs. A. G. Hinnebusch, T. von der Haar, G. Stahl, M. A. Nelson, A. Corbett and C. N. Cole for plasmids and yeast strains; L. J. Terry, E. B. Shows, S. R. Carmody, and A. R. Alcazar-Roman for critical reading of the manuscript, and other Wente lab members for discussions. This work was supported by NIH grant R01-GM51219 (S. R. W.) and NIH Kirchstein NRSA 1F32-GM082065 and 1T32- CA009582 (T. A. B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alcazar-Roman AR, Tran EJ, Guo S, Wente SR. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat Cell Biol. 2006;8:711–716. doi: 10.1038/ncb1427. [DOI] [PubMed] [Google Scholar]
- Cox BS. Psi, a cytoplasmic suppressor of super-suppressor in yeast. Heredity. 1965;20:505–521. [Google Scholar]
- Culjkovic B, Topisirovic I, Skrabanek L, Ruiz-Gutierrez M, Borden KL. eIF4E is a central node of an RNA regulon that governs cellular proliferation. J Cell Biol. 2006;175:415–426. doi: 10.1083/jcb.200607020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Priore V, Snay CA, Bahr A, Cole CN. The product of the Saccharomyces cerevisiae RSS1 gene, identified as a high-copy suppressor of the rat7-1 temperature-sensitive allele of the RAT7/NUP159 nucleoporin, is required for efficient mRNA export. Mol Biol Cell. 1996;7:1601–1621. doi: 10.1091/mbc.7.10.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Gorsch LC, Dockendorff TC, Cole CN. A conditional allele of the novel repeat-containing yeast nucleoporin RAT7/NUP159 causes both rapid cessation of mRNA export and reversible clustering of nuclear pore complexes. J Cell Biol. 1995;129:939–955. doi: 10.1083/jcb.129.4.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg JR, Phan L, Gu Z, deSilva A, Apolito C, Sherman F, Hinnebusch AG, Goldfarb DS. Nip1p associates with 40 S ribosomes and the Prt1p subunit of eukaryotic initiation factor 3 and is required for efficient translation initiation. J Biol Chem. 1998;273:23485–23494. doi: 10.1074/jbc.273.36.23485. [DOI] [PubMed] [Google Scholar]
- Gross T, Siepmann A, Sturm D, Windgassen M, Scarcelli JJ, Seedorf M, Cole CN, Krebber H. The DEAD-box RNA helicase Dbp5 functions in translation termination. Science. 2007;315:646–649. doi: 10.1126/science.1134641. [DOI] [PubMed] [Google Scholar]
- Gruter P, Tabernero C, von Kobbe C, Schmitt C, Saavedra C, Bachi A, Wilm M, Felber BK, Izaurralde E. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- Hagiwara M, Nojima T. Cross-talks between transcription and post-transcriptional events within a ‘mRNA factory’. J Biochem. 2007;142:11–15. doi: 10.1093/jb/mvm123. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem Sci. 2006;31:553–562. doi: 10.1016/j.tibs.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Hodge CA, Colot HV, Stafford P, Cole CN. Rat8p/Dbp5p is a shuttling transport factor that interacts with Rat7p/Nup159p and Gle1p and suppresses the mRNA export defect of xpo1-1 cells. Embo J. 1999;18:5778–5788. doi: 10.1093/emboj/18.20.5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino S, Imai M, Kobayashi T, Uchida N, Katada T. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-Poly(A) tail of mRNA. Direct association of erf3/GSPT with polyadenylate-binding protein. J Biol Chem. 1999;274:16677–16680. doi: 10.1074/jbc.274.24.16677. [DOI] [PubMed] [Google Scholar]
- Jin L, Guzik BW, Bor YC, Rekosh D, Hammarskjold ML. Tap and NXT promote translation of unspliced mRNA. Genes Dev. 2003;17:3075–3086. doi: 10.1101/gad.1155703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapp LD, Lorsch JR. The molecular mechanics of eukaryotic translation. Annu Rev Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- Katahira J, Strasser K, Podtelejnikov A, Mann M, Jung JU, Hurt E. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. Embo J. 1999;18:2593–2609. doi: 10.1093/emboj/18.9.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendirgi F, Barry DM, Griffis ER, Powers MA, Wente SR. An essential role for hGle1 nucleocytoplasmic shuttling in mRNA export. J Cell Biol. 2003;160:1029–1040. doi: 10.1083/jcb.200211081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendirgi F, Rexer DJ, Alcazar-Roman AR, Onishko HM, Wente SR. Interaction between the shuttling mRNA export factor Gle1 and the nucleoporin hCG1: a conserved mechanism in the export of Hsp70 mRNA. Mol Biol Cell. 2005;16:4304–4315. doi: 10.1091/mbc.E04-11-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol. 2007;8:761–773. doi: 10.1038/nrm2255. [DOI] [PubMed] [Google Scholar]
- Marfatia KA, Crafton EB, Green DM, Corbett AH. Domain analysis of the Saccharomyces cerevisiae heterogeneous nuclear ribonucleoprotein, Nab2p. Dissecting the requirements for Nab2p-facilitated poly(A) RNA export. J Biol Chem. 2003;278:6731–6740. doi: 10.1074/jbc.M207571200. [DOI] [PubMed] [Google Scholar]
- Moehle CM, Hinnebusch AG. Association of RAP1 binding sites with stringent control of ribosomal protein gene transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:2723–2735. doi: 10.1128/mcb.11.5.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- Mulugu S, Bai W, Fridy PC, Bastidas RJ, Otto JC, Dollins DE, Haystead TA, Ribeiro AA, York JD. A conserved family of enzymes that phosphorylate inositol hexakisphosphate. Science. 2007;316:106–109. doi: 10.1126/science.1139099. [DOI] [PubMed] [Google Scholar]
- Murphy R, Watkins JL, Wente SR. GLE2, a Saccharomyces cerevisiae homologue of the Schizosaccharomyces pombe export factor RAE1, is required for nuclear pore complex structure and function. Mol Biol Cell. 1996;7:1921–1937. doi: 10.1091/mbc.7.12.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy R, Wente SR. An RNA-export mediator with an essential nuclear export signal. Nature. 1996;383:357–360. doi: 10.1038/383357a0. [DOI] [PubMed] [Google Scholar]
- Nielsen KH, Szamecz B, Valasek L, Jivotovskaya A, Shin BS, Hinnebusch AG. Functions of eIF3 downstream of 48S assembly impact AUG recognition and GCN4 translational control. Embo J. 2004;23:1166–1177. doi: 10.1038/sj.emboj.7600116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nousiainen HO, Kestila M, Pakkasjarvi N, Honkala H, Kuure S, Tallila J, Vuopala K, Ignatius J, Herva R, Peltonen L. Mutations in mRNA export mediator GLE1 result in a fetal motoneuron disease. Nat Genet. 2008;40:155–157. doi: 10.1038/ng.2007.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan L, Schoenfeld LW, Valasek L, Nielsen KH, Hinnebusch AG. A subcomplex of three eIF3 subunits binds eIF1 and eIF5 and stimulates ribosome binding of mRNA and tRNA(i)Met. Embo J. 2001;20:2954–2965. doi: 10.1093/emboj/20.11.2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Hellen CU, Pestova TV. Recycling of eukaryotic posttermination ribosomal complexes. Cell. 2007;131:286–299. doi: 10.1016/j.cell.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayala HJ, Kendirgi F, Barry DM, Majerus PW, Wente SR. The mRNA export factor human Gle1 interacts with the nuclear pore complex protein Nup155. Mol Cell Proteomics. 2004;3:145–155. doi: 10.1074/mcp.M300106-MCP200. [DOI] [PubMed] [Google Scholar]
- Rosenwald IB, Kaspar R, Rousseau D, Gehrke L, Leboulch P, Chen JJ, Schmidt EV, Sonenberg N, London IM. Eukaryotic translation initiation factor 4E regulates expression of cyclin D1 at transcriptional and post-transcriptional levels. J Biol Chem. 1995;270:21176–21180. doi: 10.1074/jbc.270.36.21176. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Erdjument-Bromage H, Snowman AM, Tempst P, Snyder SH. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr Biol. 1999;9:1323–1326. doi: 10.1016/s0960-9822(00)80055-x. [DOI] [PubMed] [Google Scholar]
- Schmitt C, von Kobbe C, Bachi A, Pante N, Rodrigues JP, Boscheron C, Rigaut G, Wilm M, Seraphin B, Carmo-Fonseca M, Izaurralde E. Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. Embo J. 1999;18:4332–4347. doi: 10.1093/emboj/18.15.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref A, Sharma K, Doye V, Hellwig A, Huber J, Luhrmann R, Hurt E. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. Embo J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snay-Hodge CA, Colot HV, Goldstein AL, Cole CN. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. Embo J. 1998;17:2663–2676. doi: 10.1093/emboj/17.9.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sossin WS, DesGroseillers L. Intracellular trafficking of RNA in neurons. Traffic. 2006;7:1581–1589. doi: 10.1111/j.1600-0854.2006.00500.x. [DOI] [PubMed] [Google Scholar]
- Stage-Zimmermann T, Schmidt U, Silver PA. Factors affecting nuclear export of the 60S ribosomal subunit in vivo. Mol Biol Cell. 2000;11:3777–3789. doi: 10.1091/mbc.11.11.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl G, Bidou L, Rousset JP, Cassan M. Versatile vectors to study recoding: conservation of rules between yeast and mammalian cells. Nucleic Acids Res. 1995;23:1557–1560. doi: 10.1093/nar/23.9.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran EJ, Zhou Y, Corbett AH, Wente SR. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol Cell. 2007;28:850–859. doi: 10.1016/j.molcel.2007.09.019. [DOI] [PubMed] [Google Scholar]
- Wakem LP, Sherman F. Isolation and characterization of omnipotent suppressors in the yeast Saccharomyces cerevisiae. Genetics. 1990;124:515–522. doi: 10.1093/genetics/124.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins JL, Murphy R, Emtage JL, Wente SR. The human homologue of Saccharomyces cerevisiae Gle1p is required for poly(A)+ RNA export. Proc Natl Acad Sci U S A. 1998;95:6779–6784. doi: 10.1073/pnas.95.12.6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirich CS, Erzberger JP, Flick JS, Berger JM, Thorner J, Weis K. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat Cell Biol. 2006;8:668–676. doi: 10.1038/ncb1424. [DOI] [PubMed] [Google Scholar]
- Wente SR, Blobel G. A temperature-sensitive NUP116 null mutant forms a nuclear envelope seal over the yeast nuclear pore complex thereby blocking nucleocytoplasmic traffic. J Cell Biol. 1993;123:275–284. doi: 10.1083/jcb.123.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windgassen M, Sturm D, Cajigas IJ, Gonzalez CI, Seedorf M, Bastians H, Krebber H. Yeast shuttling SR proteins Npl3p, Gbp2p, and Hrb1p are part of the translating mRNPs, and Npl3p can function as a translational repressor. Mol Cell Biol. 2004;24:10479–10491. doi: 10.1128/MCB.24.23.10479-10491.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York JD, Odom AR, Murphy R, Ives EB, Wente SR. A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science. 1999;285:96–100. doi: 10.1126/science.285.5424.96. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.