Abstract
Quantitative trait loci (QTL) studies have indicated growth hormone receptor (GHR) as a candidate gene affecting cattle milk yield and composition. In order to characterize genetic variation at GHR in cattle, we studied European and East African breeds with different histories of selection, and Bos grunniens, Ovis aries, Sus scrofa, Bison bison and Rangifer tarandus as references. We sequenced most of the cytoplasmic domain (900 bp of exon 10), 89 bp of exon 8, including the putative causative mutation for the QTL effect, and 390 bp of intron 8 for comparison. In the cytoplasmic domain, seven synonymous and seven non-synonymous single nucleotide polymorphisms (SNP) were identified in cattle. Three non-synonymous SNPs were found in sheep and one synonymous SNP in yak, while other studied species were monomorphic. Three major haplotypes were observed, one unique to African breeds, one unique to European breeds and one shared. Bison and yak haplotypes are derivatives of the European haplotype lineage. Most of the exon 10 non-synonymous cattle SNPs appear at phylogenetically highly conserved sites. The polymorphisms in exon 10 cluster around a ruminant-specific tyrosine residue, suggesting that this site may act as an additional signalling domain of GHR in ruminants. Alternative explanations for the persistent polymorphism include balancing selection, hitch-hiking, pleiotropic or sexually antagonistic fitness effects or relaxed functional constraints.
Keywords: growth hormone receptor, cattle, polymorphism, genetic variation
Abbreviations: QTL, quantitative trait locus, GHR, growth hormone receptor, GH, growth hormone, SNP, single nucleotide polymorphism
1. Introduction
Growth hormone receptor (GHR) is a member of the vertebrate somatotropic axis, which regulates postnatal growth and metabolism. GHR belongs to the large cytokine receptor superfamily. It consists of three domains: an extracellular ligand-binding domain, a transmembrane domain and a cytoplasmic domain. Growth hormone (GH) binds to the extracellular domains of two GH receptor molecules, causing receptor dimerization, and thereby initiates signalling cascades through the cytoplasmic domain (Frank 2001). Signal transduction begins with phosphorylation of tyrosines in the receptor and other cellular proteins. Many of the physiological functions of GH are exerted by the regulation of the transcription of other genes, including insulin-like growth factor-1, transcription factors and metabolic enzymes (Herrington & Carter-Su 2001). In cattle, GH affects directly or indirectly many aspects of growth, metabolism and lactation (Etherton & Bauman 1998).
Quantitative trait loci (QTL) mapping studies in different cattle breeds have identified effects on milk composition and yield on bovine chromosome 20 around the position of GHR (Khatkar et al. 2004), thus rendering GHR as a probable candidate for the QTL effect. The amino acid polymorphism F279Y in the transmembrane domain of GHR or a very tightly linked polymorphism has been indicated to be one cause for the lactation-related QTL effect (Blott et al. 2003; Viitala et al. 2006). Several other bovine GHR sequence polymorphisms are also known (Falaki et al. 1996; Ge et al. 2000; Blott et al. 2003; Ge et al. 2003; Sherman et al. 2008). The aim of this study was to analyse the genetic variation at the bovine cytoplasmic domain involved in signal transduction, by sequence analysis of exon 10 that codes for 90% of the intracellular part of the receptor. The putative quantitative trait nucleotide (QTN) in the exon 8, F279Y, was included in order to analyse its association with exon 10 polymorphism. Data from the GHR gene intron 8 were used as a reference to exon sequence to compare level of polymorphisms between coding and non-coding sequences.
A more general target of this study is an elucidation of the anatomy of a QTL. A common view is that quantitative traits are under selection depleting genetic variation. However, in commercial dairy populations, QTL of large effects have been shown to remain segregating (reviewed, e.g. by Khatkar et al. 2004). Furthermore, a continued response to artificial selection is usually seen, although fixation would be expected (Brotherstone & Goddard 2005). Mutation–selection balance was initially proposed as the ubiquitous explanation for the maintenance of genetic variation (Lande 1975). Although experimental data support the model of selection–mutation balance as the plausible mechanism for maintenance of quantitative genetic variation in natural populations (Zhang et al. 2004), the maintenance of genetic variance under either natural or artificial selection is not fully understood.
2. Material and methods
(a) Sampling and DNA extraction
In this study, 11 European and 3 East African cattle breeds (with 30–40 chromosomes per breed) were included. We sampled Bos taurus taurus breeds intensively selected for high milk production (Finnish Ayrshire, Holstein-Friesian, Danish Red and Danish Jersey) or beef production (Aberdeen Angus, Charolais and Hereford), less intensively selected old native breeds (Western Finncattle, Northern Finncattle, Eastern Finncattle and Jutland Cattle) and three breeds with Bos taurus indicus background from Ethiopia (Barka, morphological zebu-type; Raya, morphological sanga-type; and Fogera, zebu–sanga intermediate). As reference, we studied 2 yaks (Bos grunniens), 14 sheep (Ovis aries) representing Romanov, Finnsheep, Ålandsheep, Wrzosowska breed from Poland, Dagestan local population, Andi breed, Carpathian mountain sheep and Komi local population, 15 pigs (Sus scrofa) representing Large White, Landrace, Hampshire and Duroc breeds, 15 bison (Bison bison) from one breeding stock and 9 reindeer (Rangifer tarandus) from Helsinki Zoo and Oulu University. Individuals from the same breed were sampled to be as unrelated as possible. DNA was extracted from blood or semen as described in Miller et al. (1988).
(b) Sequence data
The GHR gene exon 10 was amplified in two fragments using the following primers: GHRex10F1 5′-GGTGTGATGTTGGGGTTAGC and GHRex10R1 5′-AGGTACCATCGCACATGTCA (starting from the preceding intron, 539 bp), and GHRex10F2 5′-ATGATGACTCTGGGCGTACC and GHRex10R2 5′-ATTGAGTACGAGGCCCTGTG (689 bp of exon 10). Exon 8 (partial, 89 bp) and part of the intron 8 (390 bp) was amplified with the primers GHRex8F 5′-TCCATGGTTCTTAATTATTATC and GHRex8R 5′-TTCACCACTTAAATTTTTGCTCA. In polymerase chain reaction (PCR), 50 ng of total DNA was used in 30 μl volume of standard DYNAZyme II (FinnZyme, Finland) PCR mix. The PCR products were purified using ExoSAP-IT enzyme (Amersham Biosciences, United Kingdom). Direct sequencing reactions were performed with DYEnamic ET Terminator Kit (Amersham Biosciences, UK) using the same primers as for amplification. The sequencing products were purified with AutoSeq96 Dye Terminator Clean-up Kit (Amersham Biosciences, UK), and separated on MegaBACE 500 (Amersham Biosciences, UK). Each fragment was sequenced at least once on both strands. Data were base-called with Cimarron 3.12 base-caller in MegaBACE Sequence Analyzer and analysed with Sequencher v. 4.0.5 (Gene Codes Corporation, USA). For six individuals with low haplotype predictions in exon 10, the amplified fragments were cloned with pGEM-T Easy Vector System kit (Promega, USA) following the manufacturer's instructions and re-sequenced.
The GenBank database contains GHR exon 10 sequences from approximately 150 mammals. All these sequences were retrieved (BLAST algorithm), aligned, and those that included indels (most rodents) were discarded from further analyses. The following species were used as background information when analysing evolutionary inference of single nucleotide polymorphisms (SNPs; conservation/non-conservation status) observed in our Artiodactyl dataset: B. t. taurus (NM_176608, AF044258, AY748827); B. t. indicus (X70041); sheep (M82912); pig (NM_214254, X54429); possum (AF467545); elephant (AF332012); horse (AF392878); giant panda (AF395535, AF367203); dog (AF133835); black bear (AF392879); Homo sapiens (NM_000163, M28466); squirrel monkey (AF339061); Rhesus monkey (U84589); olive baboon (AF150751); Senegal galago (AF540628); flying lemur (AF540627); Malayan flying lemur (AF540625); Philippine flying lemur (AF332019); large flying fox (AF392893); rabbit (AF015252); hare (AF332016); mountain beaver (AF332030); squirrel (AF332032); springhare (AF332025); fruit bat (AF392895); American beaver (AF332026); tenrec (AF392886); hedgehog (AF392882); pocket gopher (AF332027); plains pocket gopher (AF332028); hairy-tailed mole (AF392883); southern tamandua (AF332011); water shrew (AF392881); dwarf hamster (AF540640); and Caribbean manatee (AF392891).
(c) Data analysis
Multiple sequence alignment was performed with ClustalX (Thompson et al. 1997), inspected manually and corrected using Genedoc (http://www.psc.edu/biomed/genedoc; Nicholas & Nicholas 1997). To test possible effects of selection, Tajima's D for the breeds and for the non-synonymous and synonymous sites (Tajima 1989) and Fu & Li's D* (Fu & Li 1993) were calculated with the statistical analysis package DnaSP v. 4.10.4 (Rozas et al. 2003). Confidence intervals for the Tajima's D values were generated with 10 000 independent coalescent simulations. McDonald–Kreitman (McDonald & Kreitman 1991) and HKA tests (Hudson et al. 1987) were done with DnaSP.
Sliding window plot for the estimates of nucleotide diversity based on the average pairwise sequence difference (Pi; Nei 1987) was calculated with DnaSP along the exon 10 using the background sequences (34 species, sequences from GenBank and one BOS haplotype (DQ062694)). Parameter value was calculated for 10 bp windows placed at 5 bp intervals along the 900 bp sequence.
McDonald–Kreitman test finds the deviations from the prediction that if both synonymous and non-synonymous mutations are neutral, then the ratio of synonymous to non-synonymous polymorphism within a species will be similar to the ratio of synonymous to non-synonymous divergence between species. The test could not be performed with the closest species (Bos taurus versus Bison or B. taurus versus B. grunniens) due to the lack of fixed non-synonymous substitutions between species. Therefore, sheep was used as the out-group.
The Hudson–Kreitman–Aguadé (HKA) test compares levels of diversity between loci. Under neutrality, the levels of polymorphism within a species and divergence between species should be proportional to the neutral mutation rate. We used GHR partial intron 8 sequence as the neutral reference sequence. Bison was used as the closely related reference species. Owing to availability of sequences from both loci from same individuals, the dataset used for HKA was slightly smaller than that of other analyses.
The haplotypes were statistically inferred from SNP genotype data in unrelated individuals by Phase v. 2.0.2 (Stephens & Donnelly 2003). This Bayesian haplotype reconstruction method treats the unknown haplotypes as random quantities and combines prior information with the likelihood to calculate the posterior distribution, i.e. the conditional distribution of haplotypes given the observed data. The haplotypes themselves are estimated from this posterior distribution. The parameters controlling the iteration scheme were optimized as recommended (Stephens et al. 2001) by examining several independent runs using varying seed numbers. The number 100 000 was chosen for iterations, 5 as the thinning interval and 1000 as a general burn-in period. Various data partitions (breeds analysed separately versus all material pooled) were tested.
Linkage disequilibrium (LD; Weir 1979) between F279Y and exon 10 haplotypes was calculated with Genetix v. 4.05 (http://www.genetix.univmontp2.fr/genetix/genetix.htm). LD between F279Y, intron 9 and exon 10 SNPs was calculated with DnaSP applying Fisher's exact test with Bonferroni correction.
Phylogenetic reconstruction of haplotypes was not successful using the standard methods relying on bifurcation algorithms (e.g. ML or Bayesian) due to insufficient phylogenetic signal. In order to allow reticulations, which include possible recombination events, we derived a median joining network using Network v. 4.2.0.1 program (Fluxus Technology Ltd.).
3. Results
(a) Exon 10
Within the 900 bp sequence of exon 10, 14 bovine SNPs and 3 ovine SNPs were identified. One yak individual was heterozygous at one synonymous nucleotide position. All other species (pig, bison and reindeer) were monomorphic. Of the 14 bovine SNPs, 5 were present in both European and African cattle. Three of these shared SNPs were third position synonymous transitions, namely Nt1482 (T-C) and the previously reported Nt1095 (C-T) and Nt1635 (T-C) (Blott et al. 2003). Two of the shared SNPs were previously reported non-synonymous SNPs, N528T (Blott et al. 2003) and S555G (Ge et al. 2000). African samples had four private synonymous SNPs, Nt1134 (T-C), Nt1428 (A-T), Nt1458 (A-G) and Nt1575 (C-T) and two private non-synonymous ones, S439N and P519S. European samples lacked private synonymous SNPs, but had three private non-synonymous ones, N523D, A536T reported in Ge et al. (2000) and A541S. In summary, African cattle segregated for seven and four synonymous and non-synonymous SNPs, respectively, and European cattle segregated for three and five synonymous and non-synonymous SNPs, respectively. Four synonymous SNPs (Nt1134, Nt1482, Nt1575 and Nt1635), but none of the non-synonymous SNPs were located at CpG nucleotide sites.
Ovine samples had three non-synonymous SNPs: N523D (present also in Charolais cattle), P431S and H580N. Bison and yak sequences were identical with the predominant European bovine sequence, despite one synonymous difference in both species, Nt1536 (G-A) in bison and Nt1273 (T-C) in one yak individual.
In order to shed light on the nature of the amino acid polymorphisms in exon 10, we used background information from other mammalian species (figure 1). At most of the polymorphic amino acid sites, one or both of the two amino acids segregating in ruminants are not present in non-ruminant species. Only at P431S and N523D both segregating amino acids appear at least in some other Eutherian species. At A541S, only possum (Marsupialia), but no Eutherian species, has a serine. At non-polymorphic sites, ruminants have unique amino acids at positions 534 (V) and 539 (Y), which are conserved to another amino acid state in all other species (A and C, respectively).
Figure 1.
Protein alignment of the variable part of the GHR cytoplasmic domain encoded by exon 10. The ruminant sequences from this study are aligned with other mammalian sequences available in GenBank. Polymorphic amino acid sites in cattle are marked indicating the variable amino acids. The ruminant-specific tyrosine at amino acid position 539 is highlighted in bold.
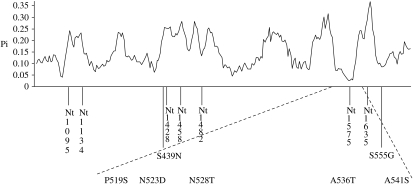
The sliding window plot of nucleotide divergence across the phylogenetic profile derived from 34 mammalian species (sequences from GenBank) along the exon 10 sequence had a modular appearance (figure 2), showing areas of high and low nucleotide divergence. In addition to occurring at conserved amino acid sites, most of the cattle missense SNPs located in a low nucleotide divergence module.
Figure 2.
Sliding window plot of nucleotide divergence across the exon 10 encoded GHR cytoplasmic domain segment among 34 different mammalian species. Locations of all synonymous (Nt) and non-synonymous polymorphisms detected in this study in cattle are shown. The segment within a low divergence area containing five amino acid substitutions between amino acid positions 519–541 is presented in a larger scale within dashed lines.
The 14 cattle SNPs were inferred to segregate as 24 different haplotypes (table 1 in the electronic supplementary material). Most individuals always received the haplotype pair suggestion with probabilities greater than 0.98, irrespective of the data partition. Among the African breeds, this was true for all but one individual that was discarded. When European breeds were analysed separately, some individuals received haplotype pair suggestions with probabilities less than 0.9. When breeds were pooled, most of the ambiguous suggestions disappeared, and the haplotype pair receiving probability greater than 0.98 always turned out to be the one that was previously (breeds separately) suggested with the highest probability. The remaining ambiguous cases were six individuals receiving alternative haplotype pairs with probabilities 0.65 and 0.35. These individuals were studied further by cloning and sequencing. The results confirmed the haplotypes with 0.65 probabilities to be the correct ones. From sheep data, the three SNPs produced haplotype pairs for all individuals with probability 1.00. The GHR haplotype sequences have been deposited in GenBank with the accession numbers DQ062692–DQ062724 in the order: BOS1–BOS25, OVIS1–OVIS4, BOGR1 and BOGR2, bison and reindeer. Theoretically, in the absence of recombination and if each mutation had occurred only once, the maximum number of different haplotypes would be 15. The higher number of observed haplotypes indicates either recombination and/or recurrent mutations within the area.
Twenty haplotypes were detected in European cattle and seven in East African breeds, three of which are shared between these two continental groups (BOS2, BOS6 and BOS10). Three major haplotypes BOS1, BOS2 and BOS3 were observed (table 1). The most common haplotypes in African breeds were BOS1 (frequency 0.38), which was not found in European cattle, and BOS2 (0.35). BOS2 was also frequent in European breeds (although absent from Holstein-Friesian and Danish Red breeds). Haplotype BOS3 was absent in African breeds, but it was the predominant haplotype among European breeds (with the exception of Jersey). Haplotypes BOS4 and BOS5 were quite widespread in the European breed pool (both present in five European breeds) and BOS6 and BOS8 in the African breeds (present in all African breeds). BOS10 had a similar frequency in Europe (0.02) and Africa (0.04). The other cattle haplotypes are rare, with frequencies of 0.1 or less within either continent.
Table 1.
Bovine haplotype frequencies, Tajima's D (D) and Fu & Li's D* (D*) values calculated from the haplotype frequencies, and QTN F279Y frequencies per breed, European breeds grouped and African breeds grouped. (Breed type is indicated by superscript: 1beef cattle, 2dairy cattle, 3local native breed, 4East African breed. The frequencies are shown as number of chromosomes. Significance is indicated by †0.10>p>0.05, *p<0.05)
| BOS haplotypes | F279Y | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | D | D* | F | Y | |
| Angus1 | 5 | 17 | 4 | 0.75 | 1.16 | 19 | 3 | |||||||||||||||||||||
| Charolais1 | 10 | 12 | 4 | 2 | 1 | 1 | 1.39 | 1.14 | 25 | 1 | ||||||||||||||||||
| Hereford1 | 12 | 7 | 3 | 1.33 | 1.18 | 10 | 12 | |||||||||||||||||||||
| Jersey2 | 29 | 5 | 2 | 0.52 | 0.93 | 37 | 1 | |||||||||||||||||||||
| Jutland3 | 1 | 21 | 9 | 2 | 2 | 1 | 0.5 | −0.69 | 27 | 3 | ||||||||||||||||||
| Red Danish2 | 16 | 9 | 1 | 0.98 | 0.09 | 40 | ||||||||||||||||||||||
| Friesian2 | 25 | 6 | 1 | 2 | 1 | 2 | 1 | 0.14 | 0.2 | 37 | 3 | |||||||||||||||||
| Ayrshire2 | 6 | 23 | 4 | 2 | 2 | 1 | 0.62 | 1.12 | 28 | 4 | ||||||||||||||||||
| W. Finncattle3 | 7 | 14 | 14 | 3 | 2 | 2.10† | 1.03 | 29 | 1 | |||||||||||||||||||
| E. Finncattle3 | 8 | 15 | 1 | 2 | 1 | 1 | 1.4 | 1.07 | 31 | 1 | ||||||||||||||||||
| N. Finncattle3 | 14 | 16 | 2 | 2 | 2 | 1 | 1 | 1.57 | 1.12 | 40 | ||||||||||||||||||
| Barka4 | 16 | 8 | 5 | 2 | 1 | 1.48 | 0.31 | 32 | ||||||||||||||||||||
| Raya4 | 11 | 16 | 3 | 2 | 2 | 2.15* | 1.32† | 6 | ||||||||||||||||||||
| Fogera4 | 9 | 9 | 4 | 2 | 1 | 2 | 1 | 1.27 | 0.35 | 22 | ||||||||||||||||||
| Europa | 1.14 | 1.16 | 323 | 28 | ||||||||||||||||||||||||
| Africa | 2.13* | 1.38† | 60 | |||||||||||||||||||||||||
A median-joining network presenting relationships and frequencies among the haplotypes including bovine, bison and yak is presented in figure 3. The SNPs Nt1134, Nt1482, Nt1575 and Nt1635 are located at highly mutable CpG sites and have most likely been targets of back and forth mutation. These sites were excluded from the network by setting the weighting parameter for these SNPs to 0 in order to reduce reticulation. By doing this, nine haplotypes were excluded from the network. The number of initially inferred haplotypes compared with the number of SNPs suggests that in addition to recurrent mutations, recombination has also increased the number of haplotypes. This would also cause reticulation in the network. Most of the haplotypes differ from each other only by one or two nucleotides.
Figure 3.
Median joining network of BOS, bison and yak haplotypes excluding nine BOS haplotypes (6,10,11,12,14,16,18,20 and 24) that include SNPs at the highly mutable CpG sites Nt1134, Nt1482, Nt1575 and Nt1635. The haplotypes found only in African cattle are underlined. The haplotype BOS2 is shared between European and African cattle. The variable nucleotide or amino acid sites are marked to illustrate differences between the haplotypes. The haplotype frequencies are given by node area.
To test whether observed nucleotide polymorphisms are neutral, we computed the Tajima's D values and Fu & Li's D* (without an out-group) for each breed and continental group (table 1). Tajima's D and Fu & Li's D* values departed statistically significantly from equilibrium neutral expectations when analysing African breeds as one group. Other groups (breeds separately or European breeds as a group) showed a trend towards elevated positive Tajima's D values, of which the D value for the African breed Raya (sanga type) was statistically significant. Coalescent simulations assuming either no recombination or free recombination confirmed the results.
Computing Tajima's D for segregating sites revealed that all non-synonymous sites in African breeds had positive Tajima's D value but the sites N523D and A541S only found in European breeds had slightly negative Tajima's D values. N528T and S555G have positive D value both in African and European breeds (table 2 in the electronic supplementary material). These Tajima's D values were not statistically significant.
We used the McDonald–Kreitman test to analyse possible excess of non-synonymous (functional) polymorphism versus synonymous (assumed neutral) polymorphism. For closest species comparisons, there were not enough fixed polymorphisms to perform the test for departure from neutrality. When using O. aries as the out-group, (Ovis versus cattle, Ovis versus European cattle, Ovis versus African cattle) the Fisher t-test or G-tests were not statistically significant, the closest to significance being Ovis versus European cattle (Fisher, p=0.12 and G-test, p=0.07), with an NI of 4.08.
(b) Exon 8–Intron 8
Within the 98 bp sequence of exon 8, we identified the previously reported F279Y (Blott et al. 2003) polymorphism in European cattle and, in addition, a synonymous nucleotide change at Nt873 in African cattle. Yak and bison showed no polymorphism, and no PCR product was obtained from sheep, pig or reindeer. Frequencies for the amino acid polymorphism F279Y are given in table 1. The Y allele that has been reported to be less favourable for milk production (Blott et al. 2003) was absent in African breeds, Danish Red and Northern Finncattle, but fairly frequent in the beef breeds. LD test did not indicate significant LD between F279Y and any of the exon 10 haplotypes or SNPs except S555G. In African samples, none of the intron 8 SNPs showed statistically significant LD with the exon 10 SNPs. In European samples, SNP Nt1635 showed significant LD with two intron 8 SNPs (data not shown). Among non-synonymous SNPs in exon 10, N528T, A536T and S555G were in LD in European samples. In African samples, the non-synonymous SNPs S439N, P519S, N528T and S555G were in LD, reflecting the major haplotypes present.
The rare Y allele was linked with divergent haplotypes, BOS2 and BOS3 or BOS3-related haplotypes (BOS5, BOS6 and BOS12). The patchy occurrence of the same polymorphism in divergent haplotype lineages suggests recurrent mutation or intragenic recombination. The physical distance between exon 8 and exon 10 is 17.5 kB.
In the 390 bp sequence of intron 8, five SNPs were identified in cattle. One of the SNPs was shared between European and African samples, two SNPs were present only in European samples and two SNPs were present only in African samples. No SNPs were found in yak samples and three SNPs were detected only in bison samples. Other species (sheep, pig and reindeer) failed to amplify in PCR.
Altogether nine haplotypes were detected in intron 8 data, six in European cattle and three in African cattle (DQ168854–DQ168862). Three bison haplotypes were detected (DQ168864–DQ168866).
HKA test results (not shown) did not show significant evidence of divergent evolutionary rates between intron 8 and exon 10 sequences for any comparisons (bison versus cattle, bison versus European cattle or bison versus African cattle, or European versus African cattle). Population subdivision may affect the power of the HKA test and hamper its use to detect deviation from the standard neutral model (Ingvarsson 2004).
4. Discussion
We have shown that cattle display interesting molecular variation in the cytoplasmic domain of GHR. The three most frequent haplotypes, BOS1, BOS2 and BOS3, are also the three most divergent haplotypes. BOS2 is common in all types of cattle breeds and geographical regions. BOS1 and BOS3 differ by seven nucleotide sites, four of which result in amino acid differences. The network of haplotypes indicates the existence of diverged African and European cattle haplogroups. Previous mtDNA studies have revealed clearly distinguishable mtDNA haplotypic profiles for the zebu and taurine lineages of domestic cattle (Bradley et al. 1996). The GHR haplotype data show a similar pattern even though we do not know the exact amount of zebu background of our African samples.
The most common European haplotype, BOS3, differs from bison and yak sequences by one synonymous nucleotide substitution, and thus the differences within European, within African and between European/African cattle haplotype sequences are much larger than between different species. This could be an indication of ancient roots of haplotype lineages.
On the basis of mtDNA D-loop sequence data, bison and yak have shown a clear genetic divergence from the domesticated cattle (Loftus et al. 1994; Ward et al. 1999). Our GHR sequence data indicate a low level of divergence between cattle, bison and yak. This may reflect the incorporation of taurine genes into the gene pools of bison and yaks. Bison were close to extinction at the end of the nineteenth century and crossing of the last animals with taurine cattle was practised (Ward et al. 1999). The estimated time since divergence between Bison and Bos is 0.46–1.23 Myr based on molecular data (Ritz et al. 2000).
Cattle were domesticated from two subspecies approximately 10 000 years ago in the region of Pakistan (B. t. indicus) and Middle East (B. taurus). According to autosomal microsatellite data, these subspecies have diverged approximately 0.5–0.8 Myr ago (MacHugh et al. 1997). The original African cattle are considered to have been taurine, and zebu cattle were introduced to Africa approximately 2000–4000 years ago. Zebu-taurine cross-breeding has influenced African breeds, and at present a mixture of morphologies with zebu and intermediate sanga forms exist, taurine populations predominating only in West Africa. The East African populations in our study represent the zebu- and sanga type. Therefore, their private GHR BOS1 haplotype may be of Bos indicus origin, explaining the large divergence between BOS1 and other haplotypes, and underlines the substantial divergence between the taurine and indicine genomes. The origin of the BOS2 haplotype, frequent in all types of breeds with different geographical background, remains an interesting question.
Trans-species polymorphism is compelling evidence for underlying balancing selection (e.g. Garrigan & Hedrick 2003). We detected one shared non-synonymous SNP (N523D) between Bos and Ovis, which is most likely a recurrent mutation. Increasing the number of analysed individuals in the related species could possibly identify trans-species polymorphisms and thus provide a test scheme for inferring whether GHR has a signature of balancing selection.
A popular way to argue in favour of the operation of selection is to use Tajima's D-test, based on the frequency distribution of polymorphisms (Tajima 1989). Simonsen et al. (1995) argue that Tajima's D-test has more power to detect selection than Fu & Li's (1993) tests. Our analysis showed that both tests provide similar results. Selection skews the distribution relative to neutral equilibrium model. Under neutral expectation, the mean Tajima's D statistic is expected to be zero. Under positive selection, the frequency spectrum is skewed towards an excess of rare polymorphisms and results in negative Tajima's D values. In contrast to positive selection, balancing selection retains genetic variants, so that there is an excess of intermediate frequency variants, and therefore positive Tajima's D values. The interpretation of our results is not, however, clear. The neutral equilibrium model also includes assumptions of random mating, as well as large and constant long-term population size. Violations of these assumptions are captured more readily as significant Tajima's D values rather than the effects of selection (Simonsen et al. 1995; Garrigan & Hedrick 2003; Wright & Gaut 2005). Consequently, statistical tests of selection may not allow argumentation in favour of selection in livestock populations, as they do not fulfil the requirements of demography-free ideal populations and some breeds might have faced population bottlenecks.
Our cattle data are from artificially selected breeds, and might thus be biased towards recent phenomena (after domestication, 10 000 years). In theory, genetic variation would have been expected to decline due to intensive artificial selection and small effective population sizes. On the other hand, the known effects of GHR on different traits such as growth or lactation (or fertility or immune response) may provide a basis for a pleiotropic explanation for persistent polymorphism at the GHR locus. One explanation for maintenance of quantitative genetic variation in domestic animals is that some alleles that are deleterious with respect to fitness are maintained by artificial selection, and thus the opposing forces counteract each other (Hill 2000). Hill–Robertson interference effects (hitch-hiking) may influence haplotype frequencies (McVean & Charlesworth 2000). Sexually antagonistic fitness effects could also be responsible for maintaining polymorphism (Turelli & Barton 2004). A final alternative is that in ruminants this domain has evolved under relaxed functional constraints, and has thus been free to capture amino acid altering mutations. Sliding window plot of GenBank sequences along the exon 10 shows that polymorphisms causing amino acid changes are located in the most conserved areas. Functional analysis of different receptor isoforms would be necessary to solve the relevance of the polymorphisms.
We did not find polymorphism in the cytoplasmic domain of GHR in pig, reindeer or bison. This might be due to smaller sample sizes. However, the probability of observing no sequence polymorphism in pig from which we sampled 14 chromosomes from one breed (Yorkshire) and 16 chromosomes from three other breeds (Landrace, Hampshire and Duroc) is low. High level of amino acid variation in the intracellular domain of the pig prolactin receptor was reported from a similar sample of 12 individuals from 6 breeds (Tomas et al. 2006). The differences in the amount of variation in the intracellular domain of these related genes in the pig and between pig and cattle suggest different evolutionary forces for these genes within and between taxa.
Thus, we have some indication for regarding the high polymorphism of GHR in cattle as a ruminant-specific characteristic. The high level of polymorphism is not the sole enigmatic characteristic that differentiates GHR sequence evolution in ruminants from other animals. The polymorphic amino acid sites (P519S, N523D, N528T, A536T, A541S and S555G) cluster around a ruminant-specific tyrosine residue at position 539. GH binding to GHR initiates JAK2 transphosphorylation and activation, resulting in the phosphorylation of five conserved tyrosines within the cytoplasmic domain of GHR. These distal phosphotyrosines of GHR, recruit STAT5 and other proteins, and are supposed to be critical in intracellular signalling (e.g. regulating IGF-I gene expression; Frank 2001). The five distal GHR tyrosine sites (469, 516, 548, 577 and 609; numbering based on human sequence) are completely conserved among all mammals, fishes and birds analysed (Jiao et al. 2006).
At position 539 there is a tyrosine in ruminants, but a cysteine is found in all other animals. This ruminant-specific tyrosine site, around which most of the cattle-specific polymorphisms cluster, may be an additional target for phosphorylation and thus allow additional protein interactions. The coexistence of two ruminant-specific and potentially functionally connected features could indicate that they are not independent. Our tentative hypothesis is that in ruminants this domain might serve as a more flexible substrate for protein interactions mediated further to signalling pathways.
Previous investigations give some indication of possible effects of GHR polymorphism in cattle. Markers in the promoter region of the GHR gene have showed association with growth (Hale et al. 2000) or serum IGF-1 concentrations (Ge et al. 2003), while the exon 10 polymorphisms A536T and S555G have not shown correlation with growth traits (Ge et al. 2003; Di Stasio et al. 2005). In our analysis of exon 10 GHR SNPs in the Finnish Ayrshire dairy cattle (Viitala et al. 2006), the polymorphisms N528T, A541S and S555G showed no significant association with milk yield or composition.
The amino acid polymorphism F279Y in the transmembrane domain of GHR has been suggested as the putative cause for the lactation-related QTL effect—or to be a tightly linked polymorphism (Blott et al. 2003). The F279Y effect is probably not mediated by the JAK2–STAT5 pathway (Zhou & Jiang 2006), but the transmembrane domain variation may be involved in receptor activation by modelling the subunit rotation within the dimer (Brown et al. 2005). The GHR F279Y allele F has a favourable effect on milk protein and fat percentage in several dairy breeds (Viitala et al. 2006). Breeds selected for meat production exhibit the highest frequencies for the allele Y, associated with increased milk volume. The linkage of the rare Y allele with two divergent exon 10 haplotype clades suggests recurrent mutation or recombination between haplotypes.
GH and GHR genes have proved to be of special interest in molecular evolutionary studies. In mammals the sequence of GH is generally strongly conserved, but in two lineages, primates and artiodactyls, the rate of GH evolution is markedly accelerated. These two episodes of rapid evolution correspond to that seen in the fastest evolving proteins. These bursts are generally regarded to result from positive selection (Forsyth & Wallis 2002). The evolutionary pattern at GHR is different for the extracellular and the intracellular domains. In the extracellular domain, the relatively slow basal evolutionary rate shows increase during the evolution of artiodactyls, primates and rodents. The intracellular signalling domain (target of this study) shows a relatively high but constant evolutionary rate, with acceleration only in rodents. Thus, in primates and artiodactyls, the episodes of rapid change in GH are matched by corresponding episodes in the extracellular (GH-binding) domain of GHR, which would accord with their being driven by adaptive changes (Liu et al. 2001; Forsyth & Wallis 2002). Although, unlike the GH, the intracellular domain of GHR does not seem to be driven by positive selection in ruminants, in cattle it harbours interesting polymorphism. The reason for the persistence of this variation in cattle remains to be elucidated, but it raises the intriguing hypothesis of being associated with cattle being malleable for artificial selection for growth and lactation traits.
Acknowledgments
The authors thank the Nordic Gene Bank for Farm Animals for the permission to use the Nordic breed samples, Jalostuspalvelu Artificial Insemination Cooperative for the pig samples, Finnish Animal Breeding Association for the bison samples, and Nurbi Marzanov for help in collecting the sheep samples. Prof. Elja Arjas and Pekka Pamilo are acknowledged for comments on previous versions of the manuscript and Miika Tapio for help in some analyses. The work is part of the Biodiversity Research Programme MOSSE 2003–2006, funded by the Ministry of Agriculture and Forestry in Finland (grant no. 3937/64/2002) and by the Department of Education in Finland (T.I.-T., Finnish Graduate School in Population Genetics). The sequences have been deposited in the GenBank database with the following accession numbers: DQ062692–DQ062724 and DQ168854–DQ168866.
Footnotes
The first two authors contributed in equal part to this work.
Supplementary Material
Table 1. GHR exon 10 haplotype sequences in the analysed cattle breeds; Table 2. Tajima's D value per synonymous or nonsynonymous site in GHR exon 10 in cattle
References
- Blott S, et al. Molecular dissection of a quantitative trait locus: a phenylalanine-to-tyrosine substitution in the transmembrane domain of the bovine growth hormone receptor is associated with a major effect on milk yield and composition. Genetics. 2003;163:253–266. doi: 10.1093/genetics/163.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley D.G, MacHugh D.E, Cunningham P, Loftus R.T. Mitochondrial diversity and the origins of African and European cattle. Proc. Natl Acad. Sci. USA. 1996;93:5131–5135. doi: 10.1073/pnas.93.10.5131. doi:10.1073/pnas.93.10.5131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brotherstone S, Goddard M. Artificial selection and maintenance of genetic variance in the global dairy cow population. Phil. Trans. R. Soc. B. 2005;360:1479–1488. doi: 10.1098/rstb.2005.1668. doi:10.1098/rstb.2005.1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R.J, et al. Model for growth hormone receptor activation based on subunit rotation within a receptor dimer. Nat. Struct. Mol. Biol. 2005;12:814–821. doi: 10.1038/nsmb977. doi:10.1038/nsmb977 [DOI] [PubMed] [Google Scholar]
- Di Stasio L, Destefanis G, Brugiapaglia A, Albera A, Rolando A. Polymorphism of the GHR gene in cattle and relationships with meat production and quality. Anim. Genet. 2005;36:138–140. doi: 10.1111/j.1365-2052.2005.01244.x. doi:10.1111/j.1365-2052.2005.01244.x [DOI] [PubMed] [Google Scholar]
- Etherton T.D, Bauman D.E. Biology of somatotropin in growth and lactation of domestic animals. Physiol. Rev. 1998;78:745–761. doi: 10.1152/physrev.1998.78.3.745. [DOI] [PubMed] [Google Scholar]
- Falaki M, Gengler N, Sneyers M, Prandi A, Massart S, Formigoni A, Burny A, Portetelle D, Renaville R. Relationships of polymorphisms for growth hormone and growth hormone receptor genes with milk production traits for Italian Holstein–Friesian bulls. J. Dairy Sci. 1996;79:1446–1453. doi: 10.3168/jds.S0022-0302(96)76503-7. [DOI] [PubMed] [Google Scholar]
- Forsyth I.A, Wallis M. Growth hormone and prolactin—molecular and functional evolution. J. Mamm. Gland Biol. Neoplasia. 2002;7:291–312. doi: 10.1023/a:1022804817104. doi:10.1023/A:1022804817104 [DOI] [PubMed] [Google Scholar]
- Frank S.J. Growth hormone signalling and its regulation: preventing too much of a good thing. Growth Horm. IGF Res. 2001;11:201–212. doi: 10.1054/ghir.2001.0237. doi:10.1054/ghir.2001.0237 [DOI] [PubMed] [Google Scholar]
- Fu Y.X, Li W.H. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigan D, Hedrick P.W. Perspective: detecting adaptive molecular polymorphism: lessons from the MHC. Evol. Int. J. Org. Evol. 2003;57:1707–1722. doi: 10.1111/j.0014-3820.2003.tb00580.x. [DOI] [PubMed] [Google Scholar]
- Ge W, Davis M.E, Hines H.C, Irvin K.M. Rapid communication: single nucleotide polymorphisms detected in exon 10 of the bovine growth hormone receptor gene. J. Anim. Sci. 2000;78:2229–2230. doi: 10.2527/2000.7882229x. [DOI] [PubMed] [Google Scholar]
- Ge W, Davis M.E, Hines H.C, Irvin K.M, Simmen R.C. Association of single nucleotide polymorphisms in the growth hormone and growth hormone receptor genes with blood serum insulin-like growth factor I concentration and growth traits in Angus cattle. J. Anim. Sci. 2003;81:641–648. doi: 10.2527/2003.813641x. [DOI] [PubMed] [Google Scholar]
- Hale C.S, Herring W.O, Shibuya H, Lucy M.C, Lubahn D.B, Keisler D.H, Johnson G.S. Decreased growth in angus steers with a short TG-microsatellite allele in the P1 promoter of the growth hormone receptor gene. J. Anim. Sci. 2000;78:2099–2104. doi: 10.2527/2000.7882099x. [DOI] [PubMed] [Google Scholar]
- Herrington J, Carter-Su C. Signaling pathways activated by the growth hormone receptor. Trends Endocrinol. Metab. 2001;12:252–257. doi: 10.1016/s1043-2760(01)00423-4. doi:10.1016/S1043-2760(01)00423-4 [DOI] [PubMed] [Google Scholar]
- Hill W.G. Maintenance of quantitative genetic variation in animal breeding programmes. Livestock Prod. Sci. 2000;63:99–109. doi:10.1016/S0301-6226(99)00115-3 [Google Scholar]
- Hudson R.R, Kreitman M, Aguade M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvarsson P.K. Population subdivision and the Hudson–Kreitman–Aguade test: testing for deviations from the neutral model in organelle genomes. Genet. Res. 2004;83:31–39. doi: 10.1017/s0016672303006529. doi:10.1017/S0016672303006529 [DOI] [PubMed] [Google Scholar]
- Jiao B, Huang X, Chan C.B, Zhang L, Wang D, Cheng C.H. The co-existence of two growth hormone receptors in teleost fish and their differential signal transduction, tissue distribution and hormonal regulation of expression in seabream. J. Mol. Endocrinol. 2006;36:23–40. doi: 10.1677/jme.1.01945. doi:10.1677/jme.1.01945 [DOI] [PubMed] [Google Scholar]
- Khatkar M.S, Thomson P.C, Tammen I, Raadsma H.W. Quantitative trait loci mapping in dairy cattle: review and meta-analysis. Genet. Sel. Evol. 2004;36:163–190. doi: 10.1186/1297-9686-36-2-163. doi:10.1051/gse:2003057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R. The maintenance of genetic variability by mutation in a polygenic character with linked loci. Genet. Res. 1975;26:221–235. doi: 10.1017/s0016672300016037. [DOI] [PubMed] [Google Scholar]
- Liu J.-C, Makova K.D, Adkins R.M, Gibson S, Li W.-H. Episodic evolution of growth hormone in primates and emergence of the species specificity of human growth hormone receptor. Mol. Biol. Evol. 2001;18:945–953. doi: 10.1093/oxfordjournals.molbev.a003895. [DOI] [PubMed] [Google Scholar]
- Loftus R.T, MacHugh D.E, Ngere L.O, Balain D.S, Badi A.M, Bradley D.G, Cunningham E.P. Mitochondrial genetic variation in European, African and Indian cattle populations. Anim. Genet. 1994;25:265–271. doi: 10.1111/j.1365-2052.1994.tb00203.x. [DOI] [PubMed] [Google Scholar]
- MacHugh D.E, Shriver M.D, Loftus R.T, Cunningham P, Bradley D.G. Microsatellite DNA variation and the evolution, domestication and phylogeography of taurine and zebu cattle (Bos taurus and Bos indicus) Genetics. 1997;146:1071–1086. doi: 10.1093/genetics/146.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald J.H, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. doi:10.1038/351652a0 [DOI] [PubMed] [Google Scholar]
- McVean G.A, Charlesworth B. The effects of Hill–Robertson interference between weakly selected mutations on patterns of molecular evolution and variation. Genetics. 2000;155:929–944. doi: 10.1093/genetics/155.2.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S.A, Dykes D.D, Polesky H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. doi:10.1093/nar/16.3.1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Columbia University Press; New York, NY: 1987. Molecular evolutionary genetics. [Google Scholar]
- Nicholas, K. B. & Nicholas Jr, H. B. 1997 Genedoc: a tool for editing and annotating multiple sequence alignments. Distributed by the author.
- Ritz L.R, Glowatzki-Mullis M.L, MacHugh D.E, Gaillard C. Phylogenetic analysis of the tribe Bovini using microsatellites. Anim. Genet. 2000;31:178–185. doi: 10.1046/j.1365-2052.2000.00621.x. doi:10.1046/j.1365-2052.2000.00621.x [DOI] [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio J.C, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. doi:10.1093/bioinformatics/btg359 [DOI] [PubMed] [Google Scholar]
- Sherman E.L, Nkrumah J.D, Murdoch B.M, Li C, Wang Z, Fu A, Moore S.S. Polymorphisms and haplotypes in the bovine neuropeptide Y, growth hormone receptor, ghrelin, insulin-like growth factor 2, and uncoupling proteins 2 and 3 genes and their associations with measures of growth, performance, feed efficiency, and carcass merit in beef cattle. J. Anim. Sci. 2008;86:1–16. doi: 10.2527/jas.2006-799. doi:10.2527/jas.2006-799 [DOI] [PubMed] [Google Scholar]
- Simonsen K.L, Churchill G.A, Aquadro C.F. Properties of statistical tests of neutrality for DNA polymorphism data. Genetics. 1995;141:413–429. doi: 10.1093/genetics/141.1.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 2003;73:1162–1169. doi: 10.1086/379378. doi:10.1086/379378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith N.J, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001;68:978–989. doi: 10.1086/319501. doi:10.1086/319501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D, Gibson T.J, Plewniak F, Jeanmougin F, Higgins D.G. The Clustal_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. doi:10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomas A, Casellas J, Ramirez O, Munoz G, Noguera J.L, Sanchez A. High amino acid variation in the intracellular domain of the pig prolactin receptor (PRLR) and its relation to ovulation rate and piglet survival traits. J. Anim. Sci. 2006;84:1991–1998. doi: 10.2527/jas.2005-664. doi:10.2527/jas.2005-664 [DOI] [PubMed] [Google Scholar]
- Turelli M, Barton N.H. Polygenic variation maintained by balancing selection: pleiotropy, sex-dependent allelic effects and G × E interactions. Genetics. 2004;166:1053–1079. doi: 10.1534/genetics.166.2.1053. doi:10.1534/genetics.166.2.1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viitala S, Szyda J, Blott S, Schulman N, Lidauer M, Maki-Tanila A, Georges M, Vilkki J. The role of the bovine growth hormone receptor and prolactin receptor genes in milk, fat and protein production in Finnish Ayrshire dairy cattle. Genetics. 2006;173:2151–2164. doi: 10.1534/genetics.105.046730. doi:10.1534/genetics.105.046730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward T.J, Bielawski J.P, Davis S.K, Templeton J.W, Derr J.N. Identification of domestic cattle hybrids in wild cattle and bison species: a general approach using mtDNA markers and the parametric bootstrap. Anim. Conserv. 1999;2:51–57. doi:10.1111/j.1469-1795.1999.tb00048.x [Google Scholar]
- Weir B.S. Inferences about linkage disequilibrium. Biometrics. 1979;35:235–254. doi:10.2307/2529947 [PubMed] [Google Scholar]
- Wright S.I, Gaut B.S. Molecular population genetics and the search for adaptive evolution in plants. Mol. Biol. Evol. 2005;22:506–519. doi: 10.1093/molbev/msi035. doi:10.1093/molbev/msi035 [DOI] [PubMed] [Google Scholar]
- Zhang X.S, Wang J, Hill W.G. Redistribution of gene frequency and changes of genetic variation following a bottleneck in population size. Genetics. 2004;167:1475–1492. doi: 10.1534/genetics.103.025874. doi:10.1534/genetics.103.025874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Jiang H. Short communication: a milk trait-associated polymorphism in the bovine growth hormone receptor gene does not affect receptor signaling. J. Dairy Sci. 2006;89:1761–1764. doi: 10.3168/jds.S0022-0302(06)72244-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 1. GHR exon 10 haplotype sequences in the analysed cattle breeds; Table 2. Tajima's D value per synonymous or nonsynonymous site in GHR exon 10 in cattle