Abstract
In bacteria, small RNAs (sRNAs) make important regulatory contributions to an ever increasing number of cellular processes. To expand the repertoire of known sRNAs, we sought to identify novel sRNAs in the differentiating, multicellular bacterium Streptomyces coelicolor. We describe a combined bioinformatic and experimental approach that enabled the identification and characterization of nine novel sRNAs in S. coelicolor, including a cis-encoded antisense sRNA. We examined sRNA expression throughout the S. coelicolor developmental cycle, which progresses from vegetative mycelium formation, to aerial mycelium formation and finally sporulation. We further determined the effects of growth medium composition (rich versus minimal medium) on sRNA gene expression, and compared wild-type sRNA expression profiles with those of four developmental mutants. All but two of the sRNAs exhibited some degree of medium dependence, with three sRNAs being expressed exclusively during growth on one medium type. Unlike most sRNAs characterized thus far, several sRNA genes in S. coelicolor were expressed constitutively (apart from during late sporulation), suggesting a possible housekeeping role for these transcripts. Others were expressed at specific developmental stages, and their expression profiles were altered in response to developmental mutations. Expression of one sRNA in particular was dependent upon the sporulation-specific sigma factor σWhiG.
INTRODUCTION
Biological complexity can often be correlated with regulatory complexity. It is well established that bacteria living in a highly variable environment, or having a complex life cycle, devote a large proportion of their genome to regulatory and signaling functions (1). In the case of the soil-dwelling bacterium Streptomyces coelicolor, >10% of all protein-coding genes are predicted to have a regulatory function (2). This reflects not only the variable habitat of S. coelicolor, but also its intricate life cycle that involves a series of remarkable morphological and metabolic transformations (3).
S. coelicolor is a filamentous bacterium that grows by hyphal tip extension to form a vegetative mycelium. The mycelium is then presumed to be partially cannibalized to provide nutrients for the production of secondary metabolites (including many antibiotics) and the raising of aerial hyphae, which mature to form chains of exospores. Morphogenesis and antibiotic production are stringently controlled, and are genetically coupled (sharing some regulatory elements) (4). Whilst progress has been made in elucidating the regulatory networks governing development and antibiotic production, many of the components involved in these networks have yet to be identified.
The cellular regulatory arsenal comprises not only regulatory proteins, but also non-coding RNAs, or small RNAs (sRNAs) as they are commonly referred to in bacteria because of their small size [50–500 nucleotides (nt)]. sRNAs exert their regulatory influence through interactions with mRNAs (modulating transcript stability or translation efficiency) or proteins (affecting protein activity). In Escherichia coli, sRNAs have roles in processes ranging from the control of outer membrane protein expression to the sequestration of sigma factors (5–8). sRNAs have been further implicated in sporulation in Bacillus subtilis (9), quorum sensing in Vibrio harveyi and V. cholerae (10), life cycle differentiation in Chlamydia trachomatis (11) and virulence in Shigella dysenteriae (12). Despite the obvious importance of sRNAs as regulators, their relative abundance across the bacteria has not yet been fully elucidated, and it is likely that they have an important regulatory function in Streptomyces.
Bioinformatic predictions have facilitated the identification of many sRNAs. In particular, comparative genomic searches of intergenic regions have proven productive, using criteria such as orphaned promoter sequences, orphaned terminator sequences, and conserved secondary structure in related organisms (13–17). More direct approaches to sRNA identification have involved the cloning of sRNA species (as cDNAs) from total RNA samples (18), microarray analysis of intergenic regions (9,17,19,20) and immunoprecipitation of sRNAs associated with the RNA chaperone Hfq (21,22). Hfq plays an important role in mediating the interaction of trans-encoded sRNAs with their mRNA targets, presumably stabilizing the inherently imperfect basepairing between these two RNA molecules (23).
Searching for sRNAs in S. coelicolor presents a number of challenges. For one, S. coelicolor has no obvious Hfq orthologue (24). The S. coelicolor genome is also extremely G + C rich (>70%) (2), and thus the identification of rho-independent terminators is not straightforward. The Institute for Genomic Research (TIGR—http://rice.tigr.org/tigr-scripts/CMR2/terminators_genome.spl?db=ntsc02) predicts there to be 38 rho-independent terminators within the 8.7 Mb S. coelicolor chromosome [using algorithms developed by Ermolaeva et al. (25)]; while 110 757 transcription terminators are predicted within the same genomic sequence using the TransTermHP program (26). The identification of promoter sequences is equally challenging, as S. coelicolor has 65 sigma factors, only a handful of which have been characterized, and thus there is no clear definition of what comprises a promoter in S. coelicolor. There are, however, multiple actinomycete genome sequences available, including four publically available Streptomyces genomes (2,27,28, http://www.sanger.ac.uk/Projects/S_scabies/).
Here, we describe the use of a combined bioinformatic and experimental approach to identify and characterize nine novel sRNAs in S. coelicolor, ranging in size from 34 to 288 nt. These were shown to be expressed at defined stages during the S. coelicolor life cycle, and in several instances, expression was determined to be completely dependent upon specific nutritional conditions. We examined the expression of each sRNA in mutant strains defective in antibiotic production, aerial hyphae formation and sporulation, and found five of them to have reproducibly different transcription profiles from that observed in the wild-type.
MATERIALS AND METHODS
Intergenic sequence extraction and comparative genomic analysis
Intergenic (IG) sequences were extracted from the S. coelicolor genome sequence, using annotation information from Sco.tab and DNA sequence information from Sco.dna (both data sets are available through the Sanger Institute at ftp://ftp.sanger.ac.uk/pub/S_coelicolor/whole_genome/). 833 intergenic sequences larger than 200 nt were selected from the conserved core region of the S. coelicolor chromosome (extending from SCO1413 to SCO5820—genes are annotated in increasing numerical order along the length of the linear chromosome), and were subjected to analysis using BLAST against all sequences in the database (as of August 2008) (Supplementary Table S1). Any intergenic sequences that did not exhibit similarity to other sequences with an expected value of <0.001 were considered to be S. coelicolor specific and were excluded from further analyses. Also excluded were intergenic regions where the primary similarity was to tRNAs or rRNAs. The location of sequence similarity, relative to the flanking coding sequences, was also evaluated, and regions where similarity was within 40 nt of either flanking gene were excluded, so as to remove possible riboswitches, and conserved operator sequences.
sRNAFinder
The program sRNAFinder was used to predict candidate sRNA genes throughout intergenic regions of the S. coelicolor genome (13). sRNAFinder uses a hidden Markov model to integrate primary sequence data with comparative genomics information for the purpose of predicting non-coding RNA genes. Primary sequence data includes dinucleotide frequency information and evidence of transcription termination signals. Comparative genomics information includes evidence of compensatory basepair mutations that conserve RNA secondary structure. For the comparative genomics analysis, the following genomes were employed: Burkholderia pseudomallei, Mycobacterium avium, Nocardia farcinica, Pseudomonas aeruginosa, S. avermitilis, S. coelicolor and Xanthomonas citri. Along with each sRNA gene prediction, sRNAFinder reports a measure of confidence or likelihood, for the prediction (see Supplementary Table S1).
Cloning of small RNAs
sRNA species smaller than 50 nt were cloned from RNA samples harvested from 24 h cultures grown on solid MS medium (29), as outlined in Haiser et al. (30).
Bacterial strains and culture conditions
S. coelicolor strains used are summarized in Table 1, and were grown at 30°C on either solid R2YE (rich medium—RM) or minimal medium (MM) supplemented with 0.5% mannitol (w/v) (29). E. coli strains (DH5α or TOP10F′) were grown in liquid LB medium or on solid LB agar at 37°C.
Table 1.
Streptomyces coelicolor strains
RNA isolation and Northern blot analysis
RNA was isolated as described in Hopwood et al. (31) with few modifications. Cultures were grown on the surface of cellophane discs overlaying solid agar medium. At various timepoints, cultures were harvested by scraping cells into modified Kirby's mixture [1% w/v N-lauroylsarcosine sodium salt, 6% w/v sodium 4-amino salycilate, 6% v/v phenol mixture (pH 7.9) made in 50 mM Tris (pH 8.3)]. UV spectroscopy and agarose gel electrophoresis were used to assess the quantity and quality of total RNA samples. Northern blots were prepared as described previously (30), only after hybridizing with appropriate 5′ end-labelled oligonucleotide probes (see Supplementary Table S2 for sequences), membranes were washed twice for 5 min with 2× SSC, 0.1% SDS, followed by a five minute high stringency wash with 0.2× SSC, 0.1% SDS. Northern blots for all detectable sRNAs were conducted a minimum of three times, using RNA isolated from at least two independent time courses. Comparisons between sRNA transcripts in wild-type and bld/whi mutants were done using RNA transferred to the same (single) membrane, hybridized to the same labelled oligonucleotide probe, and exposed to X-ray film for the same length of time. This ensured that wild-type and mutant RNA samples were treated equivalently at all stages, and thus any differences observed could be attributed to differing levels of sRNA transcript, and not to inconsistent probe labelling or differing exposure times.
Mapping of 5′- and 3′-ends
5′-rapid amplification of cDNA ends (RACE) was carried out using the FirstChoice® RLM-RACE kit (Ambion), following the manufacturer's instructions, but adjusted for use with bacterial RNA. Briefly, 10 μg of total RNA, extracted from S. coelicolor M600 at specific timepoints, was treated with tobacco acid pyrophosphatase (TAP) before being ligated to a 5′-RACE adapter (see Supplementary Table S2) using T4 RNA ligase. This ligated product was then used as template for reverse transcription using a primer complementary to the sRNA sequence (Supplementary Table S2), together with SuperScript™ III reverse transcriptase (Invitrogen). The resulting cDNA then served as template for 5′-end PCR amplification, using an adaptor-specific oligonucleotide and the same oligonucleotide used to prime the reverse transcription reaction (Supplementary Table S2). The resulting PCR product was then used as template for a second reaction, involving the use of a second ‘nested’ adaptor-specific oligonucleotide, and a nested sRNA-specific oligonucleotide (Supplementary Table S2). The PCR products from the second reaction were separated on an agarose gel, excised, and purified using the Qiagen® MinElute PCR purification kit. The purified products were cloned into the TOPO® vector using TOPO TA Cloning® (Invitrogen) and transformed into One Shot® DH5α-TOP10F' competent cells (Invitrogen). Colony PCR, using M13 Fwd and Rev primers (Supplementary Table S2), was conducted to identify those colonies containing cDNA inserts, which were then grown overnight. Plasmid DNA was extracted using the QIAprep® Miniprep kit, and the cloned inserts were sequenced (Supplementary Table S3).
3′-RACE was conducted using the same kit as the 5′-RACE, following the manufacturer's instructions. Prior to initiating the 3′-RACE protocol, however, 40 µg of total RNA was polyadenylated by treatment with 5 U of poly(A) polymerase (2 U/μl, Ambion) at 37°C for 1 h. The 3′-RACE adapter was then annealed to ∼50 ng poly(A)-RNA, and this RNA was reverse transcribed using M-MLV reverse transcriptase (Ambion) for 1 h at 42°C. PCR, cloning and sequencing were conducted as described for 5′-RACE (Supplementary Table S3).
In instances where it was not possible to map the 5′- or 3′-ends, northern blot analysis was used to determine the approximate transcript ends. Overlapping oligonucleotide probes were individually hybridized to northern blots, and start and end sites were mapped to within ∼10 nt.
RESULTS
Comparative genomic and computational predictions of candidate sRNA genes in S. coelicolor
Bioinformatic prediction of trans-encoded sRNAs in S. coelicolor is hindered by a lack of readily detectable promoter and transcription terminator sequences, but is greatly facilitated by the availability of multiple actinomycete genome sequences for use in comparative genomic analyses. We therefore opted to conduct a comparative genomics-based search for sRNAs in S. coelicolor, using sequence conservation in other actinomycetes as our starting point. We took advantage of the fact that actinomycetes appear to share a conserved genetic ‘core’ (2), and focussed our search on large intergenic (IG) sequences (>200 nt) within this core region. In S. coelicolor, the conserved core covers 4.9 Mb of its linear genome and is centrally positioned, housing most essential (housekeeping) genes. The left and right ‘arm’ regions on either side of the conserved core extend 1.5 and 2.3 Mb, respectively, and contain primarily non-essential, S. coelicolor-specific genes, including those for most secondary metabolic pathways (2).
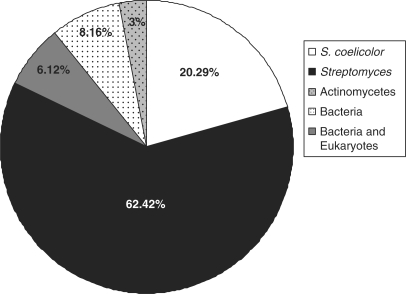
There were 833 IG regions larger than 200 nt within the conserved core region, and 169 of these regions (20.29%) were unique to S. coelicolor. The remaining 664 IG regions contained sequences that were conserved (E-value <0.001) in other organisms (Supplementary Table S1), with the majority of these (520 or 62.42% overall) having similarity to sequences in other streptomycetes. A small proportion of IG regions (6.12%) contained sequences with similarity to those in other actinomycetes, and included sequences corresponding to degenerate transposases, tRNAs and predicted riboswitches (e.g. thiamine pyrophosphate riboswitch). Beyond the actinomycetes, sequences within 68 IG regions (8.16%) were conserved in more divergent bacterial species, and again, included predominantly tRNA genes (>50% of conserved sequences), and other well characterized sequences such as the RNase P gene (32) and the FMN-sensing riboswitch (33). Finally, a small number of sequences had similarity to eukaryotic sequences (3%), including those from Homo sapiens and Oryza sativa; none of these have a characterized role or function in any organism (Figure 1).
Figure 1.
Sequence similarity of 833 core intergenic regions >200 nt in S. coelicolor. IG regions were grouped on the basis of their sequence similarity profiles into one of five different categories: S. coelicolor-specific (white), Streptomyces-specific (black), actinomycetes-specific (grey), bacteria-specific (white with dots) or widely conserved in bacteria and eukaryotes (grey with dots).
To focus our attention on those sequences most likely to encode sRNAs, we excluded regions where conservation was directly adjacent to the start of flanking coding genes (within 40 nt), as these could represent conserved promoter or operator regions. We also excluded segments containing rRNAs, tRNAs and transposase remnants. Of the remaining sequences, we concentrated on those conserved in at least three Streptomyces species. Finally, we compared the sites of sequence conservation with sites of predicted sRNA genes, as determined using the sRNAFinder program (13). Based upon these analyses and the sRNAFinder predictions, 114 IG regions were identified as containing possible sRNA genes (Supplementary Table S1).
Experimental identification of sRNAs in S. coelicolor
To experimentally validate our sRNA predictions, we selected 20 sequences for examination using northern blot analysis (Table 2). We have designated these regions ‘scr’, for Streptomyces coelicolor RNA, followed by the gene number of the adjacent annotated protein-coding gene. These sequences were conserved in syntenous regions in other Streptomyces species, were unlikely to represent operator or promoter regions given their positioning relative to adjacent open reading frames, and did not appear to encode small peptides as they lacked appropriately positioned start and stop codons. Many of these sequences were also flanked by inverted repeats that could represent rho-independent transcription terminators in Streptomyces, where poly U tails are not required for effective transcription termination (34–36). For each putative sRNA region, we designed two complementary oligonucleotide primers corresponding to 25–30 nt of the most highly conserved portion of the sequence. Using the complementary 32P-end labelled primers as probes, we examined sRNA expression in RNA samples isolated over an ∼3-day time course from S. coelicolor grown under two very different nutritional conditions: rich, glucose-containing medium (RM) and minimal medium supplemented with mannitol as carbon source (MM). This time course encompassed the full S. coelicolor life cycle (vegetative hyphae formation → aerial hyphae formation/antibiotic production → sporulation) on both media types. Upon transcript detection, northern blots were probed with individually labelled oligonucleotide primers to determine the DNA strand from which the putative sRNA was expressed. sRNA transcript ends were then delineated using 5′- and 3′-RACE experiments (see Supplementary Table S3) or, where this was not possible due to RNA secondary structure, by sequential northern blot analysis using ‘primer walking’ with overlapping oligonucleotide probes.
Table 2.
Putative sRNA-containing intergenic regions examined by northern blot analysis
| sRNA | sRNAFinder prediction score | IG length | Flanking gene orientation | Probe location in IG regiona |
|---|---|---|---|---|
| scr1906b | 0.759 | 499 | < < | 326–353 |
| scr2094 | 0.901 | 317 | > > | 79–102 |
| scr2736 | 0.887 | 643 | > < | 179–201 |
| scr3000 | 0.994 | 503 | < < | 333–357 |
| scr3045b | 0.988 | 390 | > > | 206–233 |
| scr3124 | 0.693 | 213 | > > | 124–152 |
| scr3261b | 0.975 | 698 | > > | 299–323 |
| scr3558b | 0.997 | 663 | < > | 459–482 |
| scr3974b | 0.278 | 246 | > < | 79–96 |
| scr4069 | 0.278 | 307 | > > | 161–185 |
| scr4389 | 0.852 | 333 | > > | 161–184 |
| scr4701 | 0.937 | 501 | > > | 178–200 |
| scr4784 | 0.421 | 347 | > < | 181–203 |
| scr4797 | 0.919 | 443 | < > | 233–256 |
| scr4868 | 0.719 | 243 | > < | 89–112 |
| scr4908 | 0.648 | 350 | < > | 194–218 |
| scr5028 | 0.855 | 1062 | < > | 802–826 |
| scr5334 | 0.919 | 519 | < < | 155–180 |
| scr5541 | 0.827 | 335 | < < | 225–248 |
| scr5676b | 0.862 | 400 | > < | 166–192 |
aProbe location is given relative to the start (+1) of each intergenic region (sequences are given in Supplementary Table S1).
bConfirmed sRNAs as determined by northern blot analysis and 5′-RACE mapping.
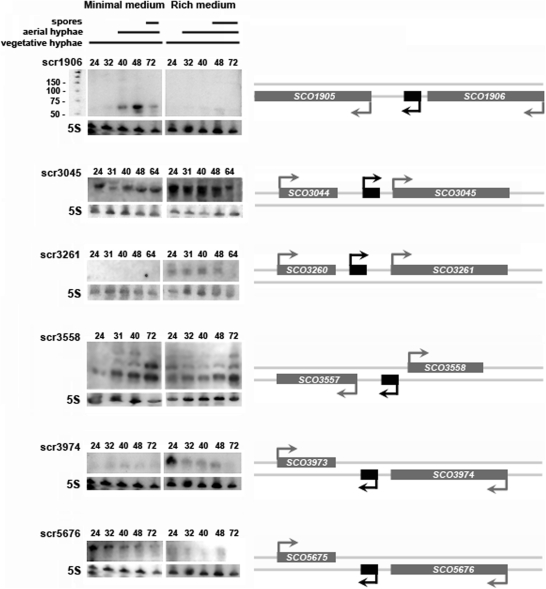
We successfully identified transcripts corresponding to sRNAs in six different intergenic regions (Table 3 and Figure 2). The candidate regions that did not yield detectable transcripts may contain sRNA genes that are either expressed at low levels, or are simply not transcribed under the experimental conditions examined here. Alternatively, they may represent conserved DNA sequences having an as yet unknown function. Of the six identified sRNA genes, two were transcribed in a medium-specific manner: scr1906 (61–63 nt) was expressed primarily on minimal medium, while scr3261 transcripts (∼185 nt) were observed only on rich medium. In contrast, scr3045 (244 and 288 nt), scr3558 (∼67, 90 and 130 nt), scr3974 (42 and 44 nt) and scr5676 (184 nt) transcripts were observed on both minimal and rich media, although higher levels of expression were observed for scr3045 and scr3974 on rich medium, and scr5676 was more highly expressed on minimal medium. The three transcripts observed for scr3558 were examined using the ‘primer walking’ method, and were found to have a shared common core sequence, but different 5′- and 3′-ends.
Table 3.
Analysed sRNAs
| sRNA | ID method | Starta | Enda | Length (nt)b | Flanking genes | Flanking gene Orientation | sRNA Orientation |
|---|---|---|---|---|---|---|---|
| scr1906 | Predicted | 2 040 854 | 204 091, 92, 93 | 61–63 | SCO1905/1906 | < < | < |
| scr2101 | Cloned | 2 258 135 | ∼2 257 986 | ∼150 | SCO2100/2101 | > > | < |
| scr3045 | Predicted | 3 334 576, 3 334 532 | 3 334 819 | 244 and 288 | SCO3044/3045 | > > | > |
| scr3261 | Predicted | ∼3 610 382 | ∼3 610 566 | ∼185 | SCO3260/3261 | > > | > |
| α3287 | Cloned | 3 636 567 | 3 636 512, 3 636 506 | 56 and 62 | SCO3287 | > | < |
| scr3558 | Predicted | ∼3 933 641, 3933612, 3 933 597 | ∼3 933 512, 3 933 523, 3 933 531 | ∼130, ∼90, ∼67 | SCO3557/3558 | < > | < |
| scr3974 | Predicted | 4 375 744, 46 | 4 375 703 | 42 and 44 | SCO3973/3974 | > < | < |
| scr4677 | Cloned | 5 108 143, 5 108 166 | ∼5 108 202, 5 108 199 | ∼70 and 34 | SCO4676/4677 | < < | > |
| scr5676 | Predicted | 6 176 413 | 6 176 230 | 184 | SCO5675/gabT | > < | < |
aBolded values were determined through 5′- or 3′-RACE mapping experiments; Bolded, italicized values were determined by cloning of the corresponding cDNA fragment.
bIn absence of 5′- or 3′-RACE data, length estimations are based on mapped transcription start site (if determined), approximate transcript size as determined by northern blot analysis, and associated terminator sequences (if appropriate).
Figure 2.
Experimental verification of sRNAs identified using a comparative genomic search of intergenic regions. Panels on the right indicate the positioning of each sRNA (shown in black) relative to adjacent protein-coding genes (shown in grey), whilst panels on the left show northern blots probed with labelled DNA oligonucleotides (together with a 25 basepair ladder shown adjacent to scr1906). For northern blot analysis, RNA was harvested from S. coelicolor grown on either minimal or rich medium at four or five different timepoints (indicated as hours post-inoculation above each sRNA blot). 5S rRNA was used as a positive control for RNA loading and RNA integrity. For each sRNA, northern blot analysis was carried out using at least three different RNA samples to ensure the reproducibility of expression profiles.
In addition to exhibiting distinct media-specific transcription patterns, each sRNA also had a unique temporal expression profile that could be correlated with particular stages of the S. coelicolor life cycle (Figure 2). Nearly all sRNAs exhibited decreased expression during later sporulation (64 or 72 h), apart from scr3558, whose expression was strongest at this time on both media types. Relatively constitutive expression was observed during vegetative growth and aerial hyphae formation for scr3045, scr3261 and scr5676. scr3974 was also expressed constitutively on minimal medium, albeit at low levels; however, on rich medium, it was expressed most highly during vegetative growth (24 h), before transcript levels decreased to nearly undetectable levels by late sporulation. scr1906 had an equally distinctive expression profile. The scr1906 transcript was observed exclusively during aerial hyphae formation and sporulation on minimal medium.
Cloning of sRNAs
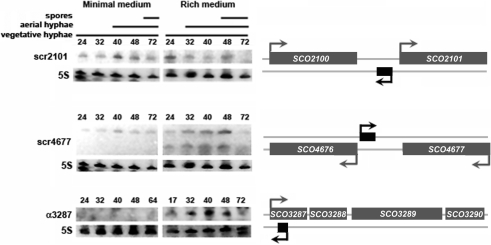
To complement our bioinformatic approach to identifying candidate sRNAs, we also conducted a small-scale cDNA cloning experiment to identify novel sRNAs. This is a technique we had used previously in our discovery of a unique population of tRNA ‘halves’ (30). In addition to identifying tRNA cleavage products (30), we detected a variety of sequences that corresponded to functional RNAs (Supplementary Table S4). These included the RNA component of the signal recognition particle (4.5S RNA); 5S, 16S and 23S rRNA degradation products; and a number of sequences that corresponded to protein coding genes. We also successfully cloned a number of sequences that appeared to represent novel sRNA candidates, based upon their position within intergenic regions or in antisense orientations relative to protein coding genes. We focussed our attention on three of these sequences (Table 3), and conducted northern blot hybridization experiments to verify gene expression, using oligonucleotide probes complementary to the cloned cDNA sequences. We detected transcripts from IG regions between SCO2100-2101 (scr2101) and SCO4676-4677 (scr4677), and antisense to the protein-coding gene SCO3287 (α3287). Hybridization to the scr2101 sequence revealed a transcript of ∼150 nt, which was constitutively present on rich and minimal media, apart from the final timepoint on rich medium where transcript abundance decreased (Figure 3), as observed for many of the previously examined sRNAs (Figure 2). This transcript was larger than the cloned cDNA fragment, suggesting that the cloned cDNA represented a degradation product of the larger transcript. For scr4677, two transcripts were detected: the larger transcript (∼70 nt) was constitutively expressed during growth on both media types (again, apart from the final timepoint on rich medium), while the smaller transcript was observed exclusively on rich medium. This smaller transcript (∼35 nt) likely corresponded to the cloned cDNA sequence (34 nt), and may be a processed version of the larger transcript, as its expression profile closely mirrored that of the larger transcript. The single SCO3287 antisense transcript was detected only during growth on rich medium, where its expression increased slightly during aerial hyphae formation and sporulation. Like scr2101, the α3287 transcript was larger than the cloned cDNA product, as determined by the mapping of both 5′- and 3′-transcript ends. Intriguingly, mapping of the 3′-end of the α3287 transcript reproducibly revealed multiple ends (Supplementary Table S3); this could explain the relatively diffuse banding pattern observed on the northern blot (Figure 3), and may suggest that termination of α3287 transcription is not a precise event.
Figure 3.
Northern blot analysis of sRNAs identified through cDNA cloning. As for Figure 2, panels on the right indicate the positioning of each sRNA (shown in black) relative to surrounding protein-coding genes (shown in grey), whilst panels on the left show northern blots probed with labelled DNA oligonucleotides. The scr4677 transcript initiates 15 nt upstream of the SCO4676 start codon, while the α3287 transcript starts 239 nt downstream of the SCO3287 translation start site, and terminates 56 or 62 nt later (184 or 178 nt downstream of the SCO3287 translation start, respectively). RNA was harvested at the timepoints indicated (in hours) above each blot, and 5S rRNA was used as a positive control for loading levels and integrity of RNA samples. Shown are representative results of northern blots that were repeated at least three times with independent RNA samples.
Expression of sRNAs in developmental mutants
Given that many sRNAs exhibited expression profiles that varied throughout the S. coelicolor developmental cycle, we wanted to determine whether the expression of any of these was affected by mutations in developmental genes. In S. coelicolor, developmental mutants are divided into two classical groups: the bld mutants and the whi mutants. bld mutants are impaired in their ability to raise aerial hyphae, and in many instances are also defective in antibiotic production, while whi mutants are blocked during the maturation of aerial hyphae and fail to form mature spores (3). We examined sRNA expression in two bld (bldA and bldB) and whi (whiG and whiB) mutant strains. bldA and bldB mutants share a defect in antibiotic production, but have distinct developmental phenotypes: the bldA mutant is a conditional mutant whose inability to raise aerial hyphae can be rescued by growth on minimal medium with mannitol, whilst bldB has a non-conditional mutant phenotype and consequently is defective in aerial hyphae formation under all growth conditions (37). In contrast, whiB and whiG disruption strains have similar mutant phenotypes in that both mutants are blocked early in the sporulation process; however, the whiB and whiG gene products are predicted to act in different pathways leading to mature spore formation (the function of WhiB has yet to be definitively determined, whilst WhiG acts as a sporulation-specific sigma factor) (38).
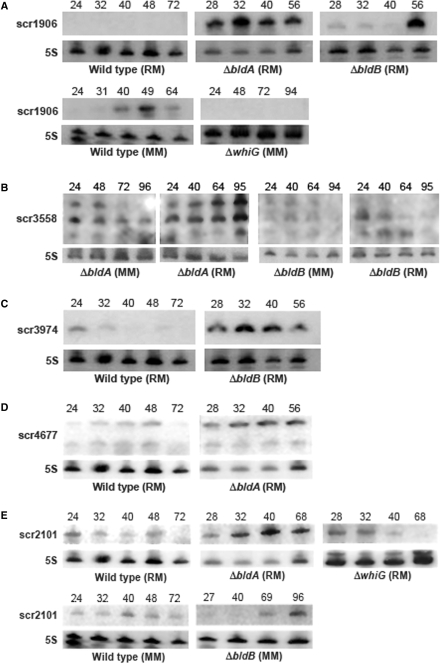
The bld and whi mutations had no effect on the transcription of four sRNAs: scr3045, scr3261, α3287 and scr5676. Expression of the remaining sRNAs was affected to varying degrees, with scr1906 in particular exhibiting a dramatically altered transcription profile. In a wild-type genetic background, scr1906 was expressed during aerial development on minimal medium; however, both this media dependence and temporally regulated expression were lost in the bldA and bldB mutant strains (Figure 4A). Both bld mutant strains exhibited wild-type expression of scr1906 on minimal medium (data not shown), while much higher expression levels were detected during growth on rich medium (Figure 4A)—a condition where little expression was typically seen in the wild-type. In contrast, there was no detectable scr1906 expression under any growth condition in a whiG mutant (Figure 4A) (wild-type expression profiles were observed in a whiB mutant, data not shown). This suggests that scr1906 expression is completely dependent upon the product of whiG (a sporulation-specific sigma factor), and that the medium-specific expression of scr1906 is contingent upon functional bld gene activity.
Figure 4.
sRNAs exhibiting altered expression profiles in bld or whi developmental mutants. (A) Top panels: comparison of scr1906 expression in bldA and bldB mutants relative to wild-type on rich medium (RM); bottom panels: scr1906 expression in wild-type versus whiG mutant on minimal medium (MM). (B) scr3558 expression in a bldA mutant (left panels), and a bldB mutant (right panels) during growth on RM and MM. (C) Expression of scr3974 in a bldB mutant (right panel) relative to wild-type strain (left panel) on RM. (D) Comparison of scr4677 expression in a wild-type strain (left panel) and a bldA mutant strain (right panel) on RM. (E) scr2101 expression in wild-type, bldA and whiG mutants during growth on RM (top panels), and expression in wild-type relative to a bldB mutant after culturing on MM (bottom panels). To ensure equivalent comparisons, wild-type and mutant RNA samples were transferred to the same membrane after separation by PAGE, and thus were subjected to identical hybridization and washing conditions, and were exposed to X-ray film for the same period of time. Time of RNA harvest (post-inoculation) is indicated on the top of each sRNA northern blot. 5S rRNA served as a control for RNA loading and RNA integrity. Northern blot analysis of wild-type and mutant RNA samples were repeated at least three times with independent RNA samples.
Like scr1906, scr3558 was expressed most highly during aerial hyphae formation and sporulation in wild-type S. coelicolor, but unlike scr1906, transcription was observed on both rich and minimal media (Figure 2). scr3558 expression was unaffected by whiB or whiG mutations (data not shown), but was affected by both bld mutations. Overall expression levels for scr3558 were maintained in a bldA mutant; however, the relative proportion of each transcript was different than had been observed in the wild-type strain (Figures 2 and 4B). In contrast, in a bldB mutant, transcription levels were significantly decreased under all growth conditions (Figure 4B). The bldB mutation had the converse effect on scr3974, where increased expression was seen during growth on rich medium, relative to wild-type (Figure 4C); a similar increase in transcript abundance was observed in both whiB and whiG mutant strains (data not shown).
The bldA mutation conferred increased expression for two of the sRNAs identified through cDNA cloning (scr4677 and scr2101). This increased expression was specific to growth on rich medium (Figure 4D and E), although for scr4677, only expression of the larger transcript was affected. The expression profile of scr2101 was also altered in bldB and whiG mutants, where it was no longer constitutively expressed, as had been observed in the wild-type strain, but instead, accumulated later in development on minimal medium in the bldB mutant, and earlier in development on rich medium in the whiG mutant (Figure 4E). These differing expression profiles may reflect altered stability of scr2101 sRNA transcripts in these mutant backgrounds. In all instances, sRNA expression in the developmental mutants was compared with wild-type expression profiles using identical experimental conditions.
DISCUSSION
sRNA detection in S. coelicolor
Our bioinformatic prediction of sRNAs in S. coelicolor involved the combined analysis of IG sequences using BLAST, to determine the extent of sequence conservation in other organisms, and the sRNAFinder program, to evaluate both primary sequence and structural conservation of putative sRNAs (13). This two-tiered approach led us to successfully identify six sRNAs that were expressed under defined conditions. We complemented our in silico sRNA predictions with a direct cloning approach to detecting sRNAs. This enabled us to identify a further three sRNAs, two of which would have been excluded from our bioinformatic search due to their close proximity to adjacent coding sequences (scr4677), and antisense orientation relative to a protein-coding gene (α3287). A previous bioinformatic screen for sRNAs in the S. coelicolor genome had predicted 32 candidate sRNA sequences, and verified expression using RT-PCR and microarray analysis for ∼24 of these (apart from 5S RNA and tmRNA, they could not detect any transcripts using northern blotting) (39). Two of the predicted 32 sRNAs overlapped with sRNAs identified here: scr3974 and scr3558.
Features of identified sRNAs in S. coelicolor
sRNA expression was examined under two distinct culture conditions—rich and minimal solid media. The expression of most sRNAs showed some degree of medium-specificity, with all but scr2101 and scr3558 being expressed more highly on one medium type than the other. The most extreme examples of this were seen for scr1906, scr3261 and α3287, which were expressed exclusively under one culture condition. Such media-specific expression is unusual but not unprecedented in S. coelicolor, with the best studied example being the ram genes, which play an important role in aerial hyphae formation (40) and are expressed exclusively on rich medium (41).
Several of the identified sRNAs appear to be expressed at relatively constitutive levels throughout development (excepting late sporulation), and not surprisingly, these were unaffected by developmental mutations. We examined the expression of all sRNAs in two different bld mutants: bldB (mutant in which the bldB gene, encoding a protein of unknown function, has been deleted), and bldA (mutant carrying a point mutation in the anticodon of the bldA-encoded leucyl tRNA). The bldB deletion had a variable effect on the expression of four different sRNAs, while the bldA mutation consistently resulted in increased expression of three different sRNAs. scr1906 expression on rich medium in the bld mutants is particularly noteworthy. The observed loss of media dependence for scr1906 expression in both bld mutants may be the consequence of defective catabolite repression—a phenotype associated with many bld mutations, including bldA and bldB (42). Previous work has also demonstrated that RNaseE activity is upregulated during S. coelicolor development, and that this upregulation depends upon a functional bldA gene product (43). The increased expression seen for scr1906, scr2101 and scr4677 (and the different transcript ratios observed for scr3558) in a bldA mutant suggests that these sRNAs, and possibly their mRNA targets, may be degraded by RNaseE, and that they may be stabilized in the presence of a bldA mutation. Interestingly, the bldA mutant effects were primarily observed during growth on rich medium; it will be interesting to determine whether RNaseE activity is in any way dependent upon growth medium composition.
The most significant effect of either whi mutation was the complete loss of scr1906 expression in a whiG mutant. whiG encodes the sporulation sigma factor σWhiG, and thus may be responsible for directing the transcription of scr1906. The promoter sequence recognized by σWhiG has been defined, as its control of two other whi genes, whiH and whiI, has been well characterized (44,45). An examination of the scr1906 promoter region did not reveal a σWhiG-like recognition sequence [T(A/C)AA (N)16 GCCGA] (45), suggesting instead that the σWhiG dependence might be an indirect effect. Both whiH and whiI encode DNA-binding transcription factors (44,45), and thus it is possible that one of these two gene products is needed for scr1906 expression. We are currently working to test this model.
The existence of scr3558 has been predicted twice previously, once in an earlier sRNA screen of the S. coelicolor genome (39), and once in a comparative genomic search for structured RNAs in bacteria (46). In the latter instance, this region was shown to be conserved within the actinobacteria (including Streptomyces, Corynebacterium, Mycobacterium, Frankia and Nocardia), and was termed the ‘6C’ RNA, due to six conserved cytosine residues found in the loop region of a conserved stem-loop structure (46). It was, however, not clear whether this ‘6C RNA’ represented a non-coding extension of an adjacent coding sequence, or whether it was an independent sRNA. Our work here confirms that distinct sRNA species are indeed encoded within this region. We detected three transcripts, each of which had a different 5′- and 3′-end (as determined by sequential northern blot analysis using overlapping primers). This could mean that each transcript initiates from its own promoter, or that one or both of the two shorter transcripts is processed from a larger transcript. The expression profile of the two shorter transcripts in particular, suggests that they are upregulated later in development, at a time that corresponds to spore formation. Not all actinobacteria in which this sequence is conserved, however, form spores, so the role of scr3558/6C RNA may instead reflect a general dormancy or metabolic slow-down response.
The location of scr4677 and α3287 suggest potential regulatory targets for both sRNAs. In the case of scr4677, transcription initiates only 15 nt upstream of the SCO4676 start codon (in a divergent orientation), suggesting that its product may interact with the 5′ untranslated region of SCO4676, possibly modulating transcript stability. SCO4676 encodes a protein of unknown function that is conserved in other Streptomyces species. For α3287, its primary target would be expected to be SCO3287, the first gene of a four gene operon, which encodes a product bearing no sequence similarity to any known protein. Preliminary investigations into SCO3287 expression profiles have, however, revealed extremely low transcript levels in wild-type strains at all times during development (Hindra and Elliot, M.A., unpublished data).
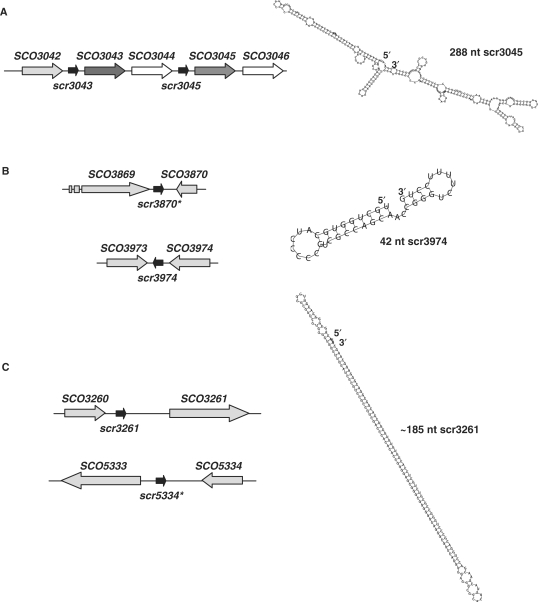
Interestingly, several sRNA sequences appear to be duplicated, or repeated, within the S. coelicolor genome (Figure 5). Based upon comparisons with S. avermitilis and S. griseus, the sequence encompassing scr3045, together with the adjacent genes SCO3045 and SCO3046, appears to have been duplicated in S. coelicolor, generating scr3043, SCO3043 and SCO3044 (Figure 5A), as these sequences are all present only once in the other Streptomyces species. We detected transcripts from both sRNA regions using 3′-end mapping (the two transcripts could not be differentiated by northern blotting); however, only the 5′-end of the scr3045 transcript was successfully mapped. As shown in Figure 5A, the transcript is predicted to be highly structured. There also appear to be two copies of scr3974 in S. coelicolor, with the second located between SCO3869 and SCO3870 (Figure 5B). These two regions were examined using differential northern blotting, which, along with 5′- and 3′-transcript end mapping, suggested that only the scr3974 region was expressed. As for scr3045, there is only a single copy of the scr3974 gene found in S. avermitilis and S. griseus, at a site adjacent to the SCO3974 orthologue. Investigation of scr3261 expression also required careful analysis, as it bore a striking resemblance to a sequence within the SCO5333–SCO5334 intergenic region (Figure 5C). The scr3261 sequence contains an enormous stem loop structure, with the stem itself being 66 nt long (Figure 5C); this feature is also conserved in the SCO5333–5334 region. Northern blots were probed with unique oligonucleotides flanking this inverted repeat region, and no transcript was ever detected with the scr5334 primers (Supplementary Table S2), while transcripts were observed when using scr3261-specific primers. As in S. coelicolor, the stem-loop structure of scr3261 is found at multiple sites in S. avermitilis and S. griseus; however, their locations are not syntenous with the scr3261 sequence, suggesting that this sequence might be associated with some type of mobile genetic element.
Figure 5.
Predicted structure and location of duplicated sRNA sequences within the S. coelicolor chromosome. (A) The sequence encompassing scr3045, SCO3045 and SCO3046 is extremely similar to that of scr3043, SCO3043 and SCO3044, suggesting a possible duplication event in S. coelicolor (other Streptomyces species have only one copy of each gene). Homologous genes are indicated in the same colours (scr3043 and scr3045: black arrows; SCO3043 and SCO3045: dark grey arrows; SCO3044 and SCO3046: white arrows). To the right is the predicted structure of scr3045. (B) The scr3974 sequence is extremely similar to that of a region found between SCO3869 and SCO3870, designated scr3870, although the flanking genes do not bear similarity to each other. To the right is the predicted structure of scr3974. (C) The scr3261 sequence bears striking similarity to a sequence located between SCO5333 and SCO5334, although, again, the flanking sequences are not similar. The scr3261 structure is distinctive due to its extensive internal complementarity. The asterisks in (B) and (C) indicate putative sRNA genes for which transcripts have never been experimentally detected.
The unusually extensive self-complementarity of the scr3261 transcript would make it a reasonable target for RNase III, which cleaves double stranded RNA. The transcript, however, appears to be stable during growth on rich medium. A possible explanation for this observation would be that scr3261 functions in sequestering a protein and that this in turn protects it from degradation; however, the biological target of scr3261 has yet to be identified.
RNA chaperones in S. coelicolor
The regulatory activity of many trans-acting sRNAs is mediated through base-pairing with their mRNA targets, and the stability of these interactions depends upon the RNA chaperone Hfq. In 2002, a thorough phylogenetic analysis revealed Hfq to be conspicuously absent from three bacterial clades, one of which included the streptomycetes and all other actinomycetes (24). Hfq is structurally, and functionally, analogous to the Sm-like proteins of eukaryotes: both proteins have important roles in RNA metabolism, and share a common fold consisting of an N-terminal helix followed by five β-strands (47,48). They do, however, have limited similarity at a sequence level. As BLAST searches rely upon sequence similarity and do not take protein structure into account, we wanted to determine whether an Hfq/Sm-equivalent protein could be identified in S. coelicolor using a structure-based approach to surveying the genome. We examined the secondary structure of all proteins smaller than 130 amino acids that were conserved in multiple Streptomyces species, using the JPRED program (49). An Hfq-like fold was not predicted for any of the proteins examined, suggesting that if S. coelicolor encodes a protein with a function analogous to that of Hfq, it is likely to have a unique sequence and structure. We predict that some proportion of the sRNAs identified here will interact with trans-encoded mRNA targets, and therefore it will be interesting to determine whether additional components are required to stabilize these interactions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health grant (R15-GM078080 to B.T.); the Grant Agency of the Czech Republic (No. 204/07/P361 to J.B.); the Canada Research Chairs programme (to M.A.E.); a Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant (No. 312495 to M.A.E.). Funding for open access charge: McMaster University.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are extremely grateful to Stephen Miller for his computational expertise and assistance, and Mark Buttner, Turlough Finan, David Capstick and Andrew Duong for helpful discussions and/or comments on the manuscript.
REFERENCES
- 1.McAdams HH, Srinivasan B, Arkin AP. The evolution of genetic regulatory systems in bacteria. Nat. Rev. Genet. 2004;5:169–178. doi: 10.1038/nrg1292. [DOI] [PubMed] [Google Scholar]
- 2.Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 3.Elliot MA, Buttner MJ, Nodwell JR. Multicellular Development in Streptomyces. In: Whitworth DE, editor. Myxobacteria: Multicellularity and Differentiation. Washington, DC: ASM Press; 2007. pp. 419–438. [Google Scholar]
- 4.Chater KF. Genetics of differentiation in Streptomyces. Annu. Rev. Microbiol. 1993;47:685–711. doi: 10.1146/annurev.mi.47.100193.003345. [DOI] [PubMed] [Google Scholar]
- 5.Wassarman KM. 6S RNA: a small RNA regulator of transcription. Curr. Opin. Microbiol. 2007;10:164–168. doi: 10.1016/j.mib.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 6.Gottesman S. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu. Rev. Microbiol. 2004;58:303–328. doi: 10.1146/annurev.micro.58.030603.123841. [DOI] [PubMed] [Google Scholar]
- 7.Guillier M, Gottesman S, Storz G. Modulating the outer membrane with small RNAs. Genes Dev. 2006;20:2338–2348. doi: 10.1101/gad.1457506. [DOI] [PubMed] [Google Scholar]
- 8.Vogel J, Papenfort K. Small non-coding RNAs and the bacterial outer membrane. Curr. Opin. Microbiol. 2006;9:605–611. doi: 10.1016/j.mib.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 9.Silvaggi JM, Perkins JB, Losick R. Genes for small, noncoding RNAs under sporulation control in Bacillus subtilis. J. Bacteriol. 2006;188:532–541. doi: 10.1128/JB.188.2.532-541.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 11.Grieshaber NA, Grieshaber SS, Fischer ER, Hackstadt T. A small RNA inhibits translation of the histone-like protein Hc1 in Chlamydia trachomatis. Mol. Microbiol. 2006;59:541–550. doi: 10.1111/j.1365-2958.2005.04949.x. [DOI] [PubMed] [Google Scholar]
- 12.Murphy ER, Payne SM. RyhB, an iron-responsive small RNA molecule, regulates Shigella dysenteriae virulence. Infect. Immun. 2007;75:3470–3477. doi: 10.1128/IAI.00112-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tjaden B. Prediction of small, noncoding RNAs in bacteria using heterogeneous data. J. Math. Biol. 2008;56:183–200. doi: 10.1007/s00285-007-0079-5. [DOI] [PubMed] [Google Scholar]
- 14.Livny J, Brencic A, Lory S, Waldor MK. Identification of 17 Pseudomonas aeruginosa sRNAs and prediction of sRNA-encoding genes in 10 diverse pathogens using the bioinformatic tool sRNAPredict2. Nucleic Acids Res. 2006;34:3484–3493. doi: 10.1093/nar/gkl453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gruber AR, Neubock R, Hofacker IL, Washietl S. The RNAz web server: prediction of thermodynamically stable and evolutionarily conserved RNA structures. Nucleic Acids Res. 2007;35:W335–W338. doi: 10.1093/nar/gkm222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rivas E, Klein RJ, Jones TA, Eddy SR. Computational identification of noncoding RNAs in E. coli by comparative genomics. Curr. Biol. 2001;11:1369–1373. doi: 10.1016/s0960-9822(01)00401-8. [DOI] [PubMed] [Google Scholar]
- 17.Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawano M, Reynolds AA, Miranda-Rios J, Storz G. Detection of 5'- and 3'-UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli. Nucleic Acids Res. 2005;33:1040–1050. doi: 10.1093/nar/gki256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu Z, Zhang A, Storz G, Gottesman S, Leppla SH. An antibody-based microarray assay for small RNA detection. Nucleic Acids Res. 2006;34:e52. doi: 10.1093/nar/gkl142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landt SG, Abeliuk E, McGrath PT, Lesley JA, McAdams HH, Shapiro L. Small non-coding RNAs in Caulobacter crescentus. Mol. Microbiol. 2008;68:600–614. doi: 10.1111/j.1365-2958.2008.06172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- 22.Christiansen JK, Nielsen JS, Ebersbach T, Valentin-Hansen P, Søgaard-Andersen L, Kallipolitis BH. Identification of small Hfq-binding RNAs in Listeria monocytogenes. RNA. 2006;12:1383–1396. doi: 10.1261/rna.49706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aiba H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr. Opin. Microbiol. 2007;10:134–139. doi: 10.1016/j.mib.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 24.Sun X, Zhulin I, Wartell RM. Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic Acids Res. 2002;30:3662–3671. doi: 10.1093/nar/gkf508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ermolaeva MD, Khalak HG, White O, Smith HO, Salzberg SL. Prediction of transcription terminators in bacterial genomes. J. Mol. Biol. 2000;301:27–33. doi: 10.1006/jmbi.2000.3836. [DOI] [PubMed] [Google Scholar]
- 26.Kingsford CL, Ayanbule K, Salzberg SL. Rapid, accurate, computational discovery of Rho-independent transcription terminators illuminates their relationship to DNA uptake. Genome Biol. 2007;8:R22. doi: 10.1186/gb-2007-8-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, Shiba T, Sakaki Y, Hattori M, Omura S. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotech. 2003;21:526–531. doi: 10.1038/nbt820. [DOI] [PubMed] [Google Scholar]
- 28.Ohnishi Y, Ishikawa J, Hara H, Suzuki H, Ikenoya M, Ikeda H, Yamashita A, Hattori M, Horinouchi S. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J. Bacteriol. 2008;190:4050–4060. doi: 10.1128/JB.00204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Norwich, UK: The John Innes Foundation; 2000. [Google Scholar]
- 30.Haiser HJ, Karginov FV, Hannon GJ, Elliot MA. Developmentally regulated cleavage of tRNAs in the bacterium Streptomyces coelicolor. Nucleic Acids Res. 2008;36:732–741. doi: 10.1093/nar/gkm1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hopwood DA, Bibb MJ, Chater KF, Kieser T, Bruton CJ, Kieser HM, Lydiate DJ, Smith CP, Ward JM, Schrempf H. Genetic Manipulation of Streptomyces – A Laboratory Manual. Norwich, UK: The John Innes Foundation; 1985. [Google Scholar]
- 32.Kazantsev AV, Pace NR. Bacterial RNase P: a new view of an ancient enzyme. Nat. Rev. Micro. 2006;4:729–740. doi: 10.1038/nrmicro1491. [DOI] [PubMed] [Google Scholar]
- 33.Winkler WC, Cohen-Chalamish S, Breaker RR. An mRNA structure that controls gene expression by binding FMN. Proc. Natl Acad. Sci. 2002;99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ingham CJ, Hunter IS, Smith MCM. Rho-independent terminators without 3′ poly-U tails from the early region of actinophage φC31. Nucleic Acids Res. 1995;23:370–376. doi: 10.1093/nar/23.3.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Unniraman S, Prakash R, Nagaraja V. Conserved economics of transcription termination in eubacteria. Nucleic Acids Res. 2002;30:675–684. doi: 10.1093/nar/30.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Unniraman S, Prakash R, Nagaraja V. Alternate paradigm for intrinsic transcription termination in eubacteria. J. Biol. Chem. 2001;276:41850–41855. doi: 10.1074/jbc.M106252200. [DOI] [PubMed] [Google Scholar]
- 37.Kelemen GH, Buttner MJ. Initiation of aerial mycelium formation in Streptomyces. Curr. Opin. Microbiol. 1998;1:656–662. doi: 10.1016/s1369-5274(98)80111-2. [DOI] [PubMed] [Google Scholar]
- 38.Chater KF. Regulation of sporulation in Streptomyces coelicolor A3(2): a checkpoint multiplex? Curr. Opin. Microbiol. 2001;4:667–673. doi: 10.1016/s1369-5274(01)00267-3. [DOI] [PubMed] [Google Scholar]
- 39.Pánek J, Bobek J, Mikulik K, Basler M, Vohradský J. Biocomputational prediction of small non-coding RNAs in Streptomyces. BMC Genomics. 2008;9:217. doi: 10.1186/1471-2164-9-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Willey JM, Willems A, Kodani S, Nodwell JR. Morphogenetic surfactants and their role in the formation of aerial hyphae in Streptomyces coelicolor. Mol. Microbiol. 2006;59:731–742. doi: 10.1111/j.1365-2958.2005.05018.x. [DOI] [PubMed] [Google Scholar]
- 41.Keijser BJF, van Wezel GP, Canters GW, Vijgenboom E. Developmental regulation of the Streptomyces lividans ram genes: involvement of RamR in regulation of the ramCSAB operon. J. Bacteriol. 2002;184:4420–4429. doi: 10.1128/JB.184.16.4420-4429.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pope MK, Green BD, Westpheling J. The bld mutants of Streptomyces coelicolor are defective in the regulation of carbon utilization, morphogenesis and cell–cell signalling. Mol. Microbiol. 1996;19:747–756. doi: 10.1046/j.1365-2958.1996.414933.x. [DOI] [PubMed] [Google Scholar]
- 43.Hagège JM, Cohen SN. A developmentally regulated Streptomyces endoribonuclease resembles ribonuclease E of Escherichia coli. Mol. Microbiol. 1997;25:1077–1090. doi: 10.1046/j.1365-2958.1997.5311904.x. [DOI] [PubMed] [Google Scholar]
- 44.Ryding NJ, Kelemen GH, Whatling CA, Flärdh K, Buttner MJ, Chater KF. A developmentally regulated gene encoding a repressor-like protein is essential for sporulation in Streptomyces coelicolor A3(2) Mol. Microbiol. 1998;29:343–357. doi: 10.1046/j.1365-2958.1998.00939.x. [DOI] [PubMed] [Google Scholar]
- 45.Aínsa JA, Parry HD, Chater KF. A response regulator-like protein that functions at an intermediate stage of sporulation in Streptomyces coelicolor A3(2) Mol. Microbiol. 1999;34:607–619. doi: 10.1046/j.1365-2958.1999.01630.x. [DOI] [PubMed] [Google Scholar]
- 46.Weinberg Z, Barrick JE, Yao Z, Roth A, Kim JN, Gore J, Wang JX, Lee ER, Block KF, Sudarsan N, et al. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res. 2007;35:4809–4819. doi: 10.1093/nar/gkm487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang A, Wassarman KM, Ortega J, Steven AC, Storz G. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell. 2002;9:11–22. doi: 10.1016/s1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]
- 48.Møller T, Franch T, Højrup P, Keene DR, Bächinger HP, Brennan RG, Valentin-Hansen P. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol. Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- 49.Cuff JA, Clamp ME, Siddiqui AS, Finlay M, Barton GJ. JPred: a consensus secondary structure prediction server. Bioinformatics. 1998;14:892–893. doi: 10.1093/bioinformatics/14.10.892. [DOI] [PubMed] [Google Scholar]
- 50.Chakraburtty R, Bibb M. The ppGpp synthetase gene (relA) of Streptomyces coelicolor A3(2) plays a conditional role in antibiotic production and morphological differentiation. J. Bacteriol. 1997;179:5854–5861. doi: 10.1128/jb.179.18.5854-5861.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chater KF, Bruton CJ, King AA, Suarez JE. The expression of Streptomyces and Escherichia coli drug-resistance determinants cloned into the Streptomyces phage phi C31. Gene. 1982;19:21–32. doi: 10.1016/0378-1119(82)90185-8. [DOI] [PubMed] [Google Scholar]
- 52.Eccleston M, Ali RA, Seyler R, Westpheling J, Nodwell J. Structural and genetic analysis of the BldB protein of Streptomyces coelicolor. J. Bacteriol. 2002;184:4270–4276. doi: 10.1128/JB.184.15.4270-4276.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Piret JM, Chater KF. Phage-mediated cloning of bldA, a region involved in Streptomyces coelicolor morphological development, and its analysis by genetic complementation. J. Bacteriol. 1985;163:965–972. doi: 10.1128/jb.163.3.965-972.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Flärdh K, Findlay KC, Chater KF. Association of early sporulation genes with suggested developmental decision points in Streptomyces coelicolor A3(2) Microbiology. 1999;145:2229–2243. doi: 10.1099/00221287-145-9-2229. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.