Abstract
COP1 is a Ring-Finger E3 ubiquitin ligase that is involved in plant development, mammalian cell survival, growth, and metabolism. Here we report that COP1, whose expression is enhanced by insulin, regulates FoxO1 protein stability. We found that in Fao hepatoma cells, ectopic expression of COP1 decreased, whereas knockdown of COP1 expression increased the level of endogenous FoxO1 protein without impacting other factors such as C/EBPα and CREB (cAMP-response element-binding protein). We further showed that COP1 binds FoxO1, enhances its ubiquitination, and promotes its degradation via the ubiquitin-proteasome pathway. To determine the biological significance of COP1-mediated FoxO1 protein degradation, we have examined the impact of COP1 on FoxO1-mediated gene expression and found that COP1 suppressed FoxO1 reporter gene as well as FoxO1 target genes such as glucose-6-phosphatase and phosphoenolpyruvate carboxykinase, two key targets for FoxO1 in the regulation of gluconeogenesis, with corresponding changes of hepatic glucose production in Fao cells. We suggest that by functioning as a FoxO1 E3 ligase, COP1 may play a role in the regulation of hepatic glucose metabolism.
COP1 is an E3 ubiquitin ligase characterized by the presence of three signature domains; that is, a RING-finger motif followed by a coiled-coil domain and seven WD40 repeats (1). COP1 is evolutionarily conserved through higher eukaryotes so that mammalian COP1 was first cloned based on its sequence homology to Arabidopsis thaliana COP1 (2, 3).
In plants COP1 targets transcription factors including LAF1, HY5, HYH, and HFR1 (4) and photoreceptors including phytochrome A and cryptochrome (1, 5) for ubiquitin-dependent proteasomal degradation. Because all of these factors are involved in photomorphogenesis, COP1 is considered a central switch in light signal transduction during plant development. In mammals, COP1 has been shown to regulate the stabilities of p53, c-Jun, and acetyl-CoA carboxylase 1 (ACC1),4 each through different mechanisms. COP1 directly interacts with and functions as an E3 ligase for the tumor suppressor p53 to induce its degradation and inhibit p53-mediated gene expression (6). Consequently, COP1 promotes cell cycle progression and cell survival (6). COP1 also inhibits c-Jun transcriptional activity. However, COP1 does not function as a c-Jun E3 ligase. Instead, COP1 recruits c-Jun to an E3 complex containing DET1, DDB1, cullin4A, and Roc (7) for c-Jun protein degradation. By interacting with the transactivation domain of c-Jun, COP1 can also inhibit its activity without altering c-Jun protein expression (3, 7). Currently, the biological significance of c-Jun regulation by COP1 is not clear. Because c-Jun is a stress responsive transcription factor, it has been speculated that COP1 may be involved in cellular stress responses (8). COP1 also functions as ACC1 E3 ligase. However, in this instance COP1 does not directly interact with ACC1. Instead, COP1 is recruited to ACC1 (9) via its interaction with TRB3, a pseudokinase and negative regulator of Akt in muscle and the liver (10, 11). ACC 1 is a biotin-containing enzyme that catalyzes the carboxylation of acetyl-CoA to form malonyl-CoA, an intermediate metabolite with a key role in the regulation of fatty acid metabolism (12). Therefore, by modulating ACC1, COP1 regulates lipid metabolism. More recently, COP1 was also found to interact with TORC2, a signal-dependent CREB co-activator. In this study it was found that COP1 regulates TORC2 protein stability and thereby regulates liver glucose metabolism (13).
COP1 activity is subject to signal-dependent regulation. In plants COP1 is localized in cytoplasm under light conditions and accumulates in the nucleus under dark conditions, but the mechanism by which light signaling regulates COP1 cellular localization remains unresolved (14). In mammalian cells, COP1 has been found to localize in different cellular compartments including the cytoplasm and nucleus (3, 4) and may be exported to cytoplasm in response to DNA damage by UV radiation (15).
FoxO proteins are a family of transcription factors within the larger Forkhead family of proteins, named for their characteristic “forkhead box” DNA binding domain. The mammalian FoxO subfamily consists of at least four members including FoxO1, FoxO3, FoxO4, and FoxO6 (16). FoxO proteins are direct targets for Akt kinase, which phosphorylates these factors at multiple sites (e.g. Thr-32, Ser-256, and Thr-319 in FoxO1) (17). Phosphorylation by Akt provides multiple mechanisms to regulate FoxO1-mediated gene expression including nuclear exclusion, FoxO1 protein degradation (for a review, see Ref. 16), and disrupting interaction between FoxO1 and its co-activator PGC1 (18). Acting as downstream targets of Akt, FoxO proteins are involved in a wide range of cellular processes including growth, survival, differentiation, and metabolism. Of the different FoxO proteins, FoxO1 is recognized as a critical regulator of liver glucose metabolism (17). G6Pase and PEPCK, two major targets of FoxO1 (19–21), are critical rate determinants of gluconeogenesis. Consistent with the view that FoxO1 plays an important role in glucose homeostasis, inactivation of FoxO1 in mouse liver either by liver-specific knock-out or by FoxO1 antisense RNA decreases the expression of G6Pase and PEPCK and improves glucose tolerance (21–23).
Because COP1 interacts with TRB3 and because TRB3 has been shown to regulate glucose metabolism (10, 11), we set out to further investigate COP1 function in hepatoma cells. During these studies we uncovered that COP1 expression is induced by insulin and that COP1 specifically modulates FoxO1 protein stability. Our data show that by down-regulation of FoxO1 COP1 regulates FoxO1 target genes such as G6Pase and PEPCK gene expression and, thereby, hepatic glucose production.
MATERIALS AND METHODS
Reagents—Human insulin, dexamethasone, forskolin, and reagents for glucose production were from Sigma-Aldrich. The proteasome inhibitor ALLN was from EMD Biosciences (Gibbstown, NJ). Commercial antibodies used in this study include anti-HA mouse monoclonal antibody (Covance Inc.), anti-β-tubulin and anti-FLAG mouse monoclonal antibodies (Sigma-Aldrich), anti-p53 and anti-FoxO1 antibodies (Cell Signaling, Danvers, MA), anti-C/EBPα and anti-C/EBPβ antibodies (Santa Cruz Biotechnology Inc.), and anti-Fox2A, anti-Akt, and anti-p110γ (Millipore). Anti-CREB, anti-PGC-1α and anti-TRB3 antibodies have been described (10). The anti-COP1 antibodies were generated in our laboratory by using His-tagged COP1 peptides (amino acids 1–240). All antibodies generated in our laboratory were affinity-purified using corresponding antigens.
Plasmids—HA-tagged FoxO1, FoxO3, TRB3, Akt expression vectors, and 3XIRS-luciferase reporter have been described (10). pGL3 SV40 promoter luciferase reporter is from Promega. The PEPCK luciferase and G6Pase reporter have been described (24, 25). G6Pase mutant luciferase reporters are the gift of Dr. R. M. O'Brien of Vanderbilt University Medical School (26). FLAG-tagged human COP1 expression plasmids were a gift from Dr. X. W. Deng of Yale University (4). The E3 ligase-defective COP1 (E-COP1) was generated with site-direct mutagenesis kit from Stratagene with forward primer 5′-TGTGGCCACAGCTTTGCCTACAAGGCTATTCATCAGAGTTTG-3′ and reverse primer 5′-CAAACTCTGATGAATAGCCTTGTAGGCAAAGCTGTGGCCACA-3′. The mutation was verified by automated sequencing. The Δ3WD-COP1 and Δ6WD-COP1 constructs were generated using XbaI and NdeI sites within the human COP1 cDNA to remove the corresponding sequence from the C terminus. To generate COP1 shRNA, forward oligonucleotide, 5′GGACCACTCAGTGAGTAGCAAAGCTTTGCTACTCACTGAGTGGTCCTTTTTG-3′, and reverse oligonucleotide, 5′-AATTCAAAAAGGACCACTCAGTGAGTAGCAAAGCTTTGCTACTCACTGAGTGGTCC-3′, were annealed and cloned into pKSU6 expression vector between ApaI and EcoRI sites as described (27). The shRNA sequence that corresponded to mouse COP1 cDNA (739–758) and is identical to human COP1 is in italics. As a control for COP1 shRNA, we used a scrambled shRNA from Addgene Co (plasmid number 1864). To generate COP1 adenoviral expression vectors, HA-tagged COP1 was cloned into pAdTrack (28). The production of adenovirus has been described (10). The COP1-expressing pMIGR1 was generated by cloning HA-tagged COP1 into BglII and XhoI sites of pMIRG1, and retrovirus was produced as previously described (29).
Cell Culture, Virus Transduction, Transient Transfection, and Luciferase Assay—Fao rat hepatoma cells were grown in RPMI 1640 supplemented with 10% (v/v) new born calf serum, 2 mm l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). Where applicable, the cells were treated with 100 nm insulin for 4 h. HEK293 cells and HepG2 cells were grown in Dulbecco's modified Eagle's medium-high glucose supplemented with 10% (v/v) fetal bovine serum, 2 mm l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). Transfections of HEK293 and HepG2 cells were carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Transient transfection and luciferase assays have been described (30). The luciferase activity was measured using FB12 luminometer (Zylux Co., Oak Ridge, TN). For adenoviral transduction (10), CsCl-purified adenovirus were incubated with cells for 4–8 h. The amount of virus used was to achieve more than 80% infection of cells. To transduce Fao cells with pMIGR1 retroviruses, the retroviruses were incubated with the cell in the presence of 6 μg/ml Polybrene (Sigma) overnight. The amount of virus used was to achieve more than 80% transduction judged by the GFP-expressing cells.
Western Blot and Co-immunoprecipitation Assay—Cells were treated as described in the figure legends, washed twice with cold phosphate-buffered saline, and extracted with o-IP buffer (20 mm Tris, pH 7.6, 250 mm NaCl, 0.5 mm EDTA, 0.5 mm dithiothreitol, 10 mm β-glycerophosphate, 1% Nonidet P-40, 10% glycerol, and protease inhibitors). For immunoprecipitation assays, total cell lysates were incubated overnight with primary antibodies (e.g. anti-FLAG antibodies) followed by a 45-min incubation with protein A/G-agarose. Immunoprecipitates bound to agarose beads were washed and subjected to SDS-PAGE and Western blotting. For Western blot analysis of cell lysates, equal amounts of protein (typically 20–30 μg) were subjected to electrophoresis on SDS-PAGE and transferred to nitrocellulose membranes (Bio-Rad) and blocked in 5% dry milk, and the membranes were incubated with each primary antibody and probed with horseradish peroxidase-conjugated secondary antibody. The protein bands were visualized using the ECL detection system (Amersham Biosciences). All controls were resolved and probed on parallel blots loaded with the same amount of proteins, and no blots were stripped for re-probing.
RNA Analysis—Total RNA was prepared from cells using RNeasy kit (Qiagen, Inc. Valencia, CA) according to the manufacturer's instructions. RT-PCR reactions were carried out using a kit (Ambion, Austin TX) according to the manufacturer's instructions. The primers used for PCR were: PEPCK, forward 5′-TGGTCTGGACTTCTTGACAAG-3′, and reverse, 5′-ACCGTCTTGCTTTCGATCCTGG-3′; G6Pase, forward, 5′-TGTCTTGGTGTCTGTGATCGCTG-3′, and reverse, 5′-AAGTGAGCCGCAAGGTAGATCC-3′; 36B4 (the acidic ribosomal phosphoprotein P0 for control (31)), forward, 5-ATGATTATCCAAAATGCTTCATTG-3, and reverse, 5-AACAGCATATCCCGAATCTCA-3; COP1, forward, 5′-AGAATACAGCCAACCTCCAG-3, and reverse, 5′-TCCACTGCATCCTGGATGAC-3′. Real time PCR were carried out using Option 2 Cycler (MJ Research) with the reagents from Applied Biosystem. The PCR cycles were: 94 °C for 45 s, 54 °C for 30 s, and 72 °C 45 s for 24 cycles.
Chromatin Immunoprecipitation Assays—Chromatin immunoprecipitation assay was performed as previously described (32). Briefly, Fao cells expressing GFP, COP1, and COP1 shRNA were treated with or without 100 nm insulin for 4 h. Then the cells were cross-linked with 1% formaldehyde, and nuclei were isolated. Next, the nuclei were lysed in lysis buffer (5 mm PIPES, pH 8.0, 100 mm KCl, 0.5% Nonidet P-40), sonicated, and immunoprecipitated with anti-FoxO1 antibodies. The immunoprecipitates of FoxO1 were analyzed with PCR. The primers designed to amplify the rat G6Pase promoter are 5′-CAGACTCTGCCCTGAGCCTCTGGCCTG-3′ and 5-CCCTGGATTCAGTCTGTAGGTCAACCTAGC-3, which amplify the region of rat G6Pase promoter from –307 to –64. The PCR cycles were 95 °C for 30 s, 70 °C for 30 s (–1 °C/cycle × 4), 72 °C for 30s, and then 95 °C for 30 s, 66 °C for 30 s, 72 °C for 30 s for 25 cycles. PCR products were visualized by electrophoresis on 2% agarose gels containing ethidium bromide.
Glucose Output Assays—Fao rat hepatoma cells infected with proper amount adenovirus (>80% infection) were treated with a combination of 0.25 μm dexamethasone and 2.5 μm forskolin or a combination of 0.25 μm dexamethasone, 2.5 μm forskolin, and 100 nm insulin for 5 h at 37°C. Cells were incubated for an additional 3 h in glucose production buffer (glucose-free Dulbecco's modified essential medium, pH 7.4, containing 20 mm sodium lactate and 2 mm sodium pyruvate without phenol red), at the end of which 0.5 ml of medium was used to measure the glucose concentration using a glucose assay kit (Sigma 510-A). Cells were collected and lysed, and the total protein concentration was measured by Bradford assay (Bio-Rad) to normalize for cell count.
RESULTS
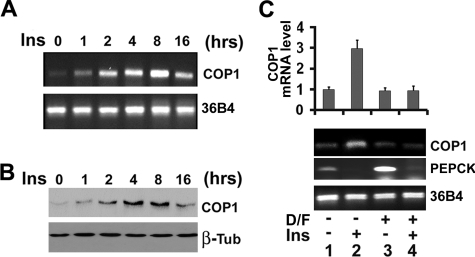
COP1 Expression Is Regulated by Insulin and cAMP in Fao Hepatoma Cells—COP1 interacts with TRB3 (9) and TRB3 has been shown to regulate glucose metabolism in the liver and muscle (10, 11). We, therefore, examined COP1 expression in Fao hepatoma cells, chosen for their fidelity to liver cells and their well preserved hormone responses (33, 34). We examined the effect of 100 nm insulin on expression of both COP1 mRNA and protein using RT-PCR and Western blotting assays. As shown in Fig. 1A, the level of COP1 mRNA is low in Fao cells under basal conditions and was incrementally induced after insulin stimulation with the maximum induction observed at 8 h after insulin treatment. The difference in the level of COP1 mRNA was not due to the variation of samples, as comparable levels of 36B4 were observed in all lanes (bottom panel). This increase in COP1 message also translated into increased levels of COP1 protein (Fig. 1B).
FIGURE 1.
Expression of COP1 in Fao cells. A, RT-PCR. B, Western blot analysis of COP1 from Fao cells treated with 100 nm insulin (Ins) after an overnight period of serum deprivation. Total RNA and whole cell lysates were prepared at the indicated times for RT-PCR analysis using primers to COP1 (top) and 36B4 (bottom). Western blots were probed with anti-COP1 (top) and anti-TRB1 antibodies (bottom). C, real time PCR analysis of mRNA from Fao cells treated with a combination of dexamethasone (0.5 μm) and forskolin (2.5 μm) (D/F) for 4 h (lanes 3 and 4) in the presence (lanes 2 and 4) or absence (lanes 1 and 3) of 100 nm insulin. The RT-PCR was repeated three times with independent RNA sample preparations. A representative result of the RT-PCR is shown. Top, average COP1 mRNA expression from three different quantitative PCRs. The level of COP1 mRNA in control cells was set as 1. The error bars represent S.D.
In the liver, insulin, which is secreted in the fed state, displays an inverse regulatory effect on target gene expression compared with glucagon (cAMP) and glucocorticoids, which mediate the effects of fasting on glucose regulation. With this in mind we examined the effect of cAMP and glucocorticoids on insulin-induced COP1 expression. To this end, Fao cells were treated with a mix of dexamethasone and forskolin (cAMP agonist) (Dex/Fsk) for 4 h in the presence and absence of insulin. As presented in Fig. 1C, 4 h of insulin treatment of Fao cells increased COP1 mRNA by 3-fold. Dex/Fsk mix alone had no impact on COP1 expression (compare lanes 1 and 3). However, it strongly suppressed insulin-induced COP1 expression (compare lanes 2 and 4). We also examined expression of PEPCK gene, a known target for the above hormones during nutritional manipulations of fasting and refeeding. In keeping with their roles in regulating PEPCK, insulin suppressed, whereas Dex/Fsk induced expression of PEPCK gene. Comparable levels of 36B4 control were observed in each lane, suggesting an inverse hormonal regulation of COP1 and PEPCK mRNA. Taken together our data demonstrate that expression of COP1 is modulated by the same hormonal conditions that regulate hepatic glucose metabolism.
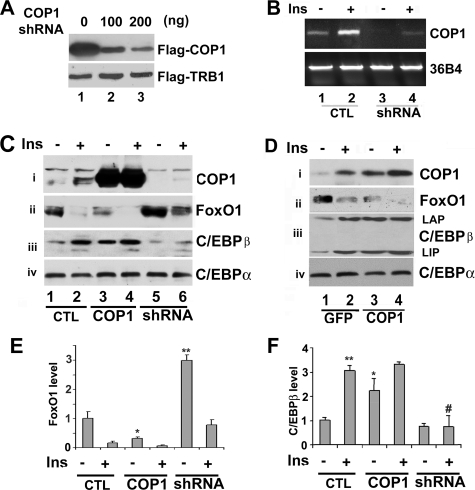
COP1 Inhibits FoxO1 Protein Expression—After showing that insulin induces COP1 expression, we sought to determine whether COP1 might regulate the factors that are important for insulin-mediated cellular events, in particular, glucose metabolism. To this end, we decided to knock down or ectopically overexpress COP1 in Fao cells. To suppress COP1 expression, we generated a COP1 shRNA expression vector. Shown in Fig. 2A are Western blots of lysates from HEK293 cells co-transfected with FLAG-tagged COP1 and FLAG-tagged TRB1 in the presence of increasing amounts of COP1 shRNA expression vector. COP1 shRNA suppressed expression of transfected FLAG-tagged COP1 in a dose-dependent manner (top panel, compare lanes 1, 2, and 3) and had no effect on FLAG-tagged TRB1 (bottom panel, used here as a control), indicating the specificity of this shRNA to COP1. We also examined the efficacy of this shRNA to suppress endogenous COP1 expression in Fao cells. Presented in Fig. 2B, when introduced into Fao cells via adenoviral transfer, this shRNA abrogated endogenous COP1 gene expression (compare lanes 1 and 3 or 2 and 4).
FIGURE 2.
A, COP1 shRNA suppresses protein expression from COP1-transfected cells. HEK293 cells were transfected with FLAG-tagged COP1 and FLAG-tagged TRB1 expression vectors (200 ng/well) along with increasing amounts of COP1 shRNA expression vector (0, 100, and 200 ng, respectively). 48 h post-transfection, total cell lysates were prepared for Western blot analysis using anti FLAG-antibodies. B, COP1 shRNA suppresses endogenous COP1 expression. Fao cells were transduced with adenovirus expressing GFP or COP1 shRNA (shRNA). 36 h post-transduction the cells were treated with (or without) insulin (Ins) for 4 h, and total RNA was prepared for RT-PCR analysis using primers for COP1 (top) or 36B4 (bottom), respectively. C, COP1 suppresses FoxO1 protein expression in Fao hepatoma cells. Fao cells were transduced with GFP (–)-, COP1 (WT)-, and COP1 shRNA (shRNA)-expressing adenoviruses. 24 h post-transduction cells were serum-deprived overnight followed by 4 h of 100 nm insulin treatment (+). Total cell lysates prepared after insulin treatment were subjected to Western blot analysis using anti-COP1 (i), anti-FoxO1 (ii), anti-C/EBPα (iii), and anti-C/EBPβ (iv), antibodies, respectively. CTL, control. D, the experiments are carried out as in C, except COP1 expressing pMIGR-1 retroviruses were used. E and F, quantification of FoxO1 and C/EBPβ expression in C and D. The levels of FoxO1 or C/EBPβ were set as 1 under control condition (no effector expression and insulin treatment). Each bar represents the means ± S.D. (n = 3). The p values were related to the control conditions. *, p < 0.02. **, p < 0.012. #, p < 0.023.
Next, we assessed the impact of COP1 overexpression or knockdown on several hepatic factors that are important for insulin-mediated glucose homeostasis. Specifically, Fao cells were transduced with either GFP, COP1, or COP1 shRNA-expressing adenoviruses. Twenty-four hours post-transduction cells were serum-starved overnight and treated with insulin (100 nm) or 4 h. First, we evaluated COP1 protein expression by Western blotting. As shown in Fig. 2C, HA-COP1 adenovirus resulted in a strong expression of COP1 over endogenous protein (panel i, compare lanes 1 and 3) in Fao cells, whereas COP1 shRNA-expressing adenovirus effectively suppressed endogenous COP1 (compare lanes 1 and 5). Consistent with our earlier finding, insulin promoted COP1 protein expression (compare lanes 1 and 2). Next, we examined the effect of COP1 knockdown and overexpression on insulin action in hepatoma cells. In keeping with previous reports of insulin-dependent FoxO1 protein degradation (35), insulin treatment decreased the level of FoxO1 protein by more than 70% (Fig. 2C, panel ii, compare lanes 1 and 2, and E). Ectopic expression of COP1 alone decreased the level of FoxO1 by more than 50% and by more than 90% with further insulin treatment (Fig. 2C, panel ii, compare lanes 1, 3, and 4, and E). Conversely, knockdown of COP1 increased the level of FoxO1 protein by about 3-fold (Fig. 2C, panel ii, compare lanes 1 and 5, and E). Insulin treatment still decreased the level of FoxO1 (Fig. 2C, panel ii, compare lanes 5 and 6, but the level of FoxO1 remained comparable with control conditions (compare lanes 1 and 6), Taken together, our data show that COP1 is likely a regulator of FoxO1.
We also examined the expression of C/EBPα and CEBPβ. No changes were observed in the expression of C/EBPα (panel vi). However, insulin induced C/EBPβ expression by more than 3-fold (panel iii, compare lanes 1 and 2, and E). COP1, whose expression increased in conjunction with insulin stimulation, also induced C/EBPβ expression (about 2.5-fold, panel iv, compare lanes 1 and 3, and F). Correspondingly, knockdown of COP1 reduced C/EBPβ expression induced by insulin (Fig. 2C, panel iv, lanes 5 and 6, and F).
Expression of other proteins with critical roles in hepatic gene expression including Fox2A, PGC1α, and CREB showed no change under any of the experimental conditions used here (supplemental Fig. 1) COP1 has been shown to down-regulate p53 protein in response to DNA damage (6). Thus, we examined the level of p53 in response to COP1 overexpression or knockdown in Fao cells. In contrast to its effects on FoxO1 and C/EBPβ, no change of p53 protein was detected regardless of COP1 expression (supplemental Fig. 1).
In the preceding experiments we noticed that COP1 expression mediated by adenovirus transfer was more than 10-fold over endogenous COP1, raising the possibility that suppression of FoxO1 protein by COP1 may be an adverse effect of strong overexpression. To address this potential problem, we expressed HA-tagged COP1 with pMIGR-1 retrovirus in Fao cells. Shown in Fig. 2D, COP1 expressed from pMIGR-1 was about 2–3-fold higher over endogenous COP1 (compare lanes 1 and 3). Under these conditions, we observed similar expression patterns of all factors examined in Fig. 2C (and supplemental Fig. 1B). In addition, in these experiments we also examined the expression of two isoforms of C/EBPβ liver-enriched activating protein (LAP) and liver-enriched transcription inhibitory protein (LIP). We observed that both isoforms of C/EBPβ were concomitantly induced by insulin (panel iii).
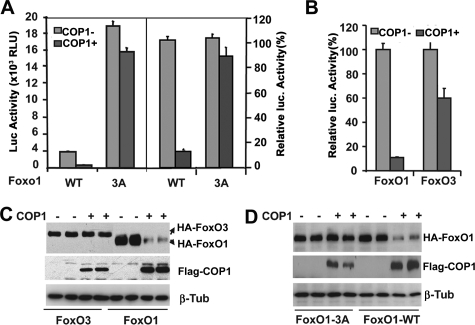
COP1 Regulates FoxO1-mediated Reporter Activity—Suppression of FoxO1 expression by COP1 prompted us to investigate whether COP1 modulates FoxO1-mediated gene expression. We, therefore, examined the impact of COP1 on FoxO1-dependent transactivation using FoxO1 effector plasmids and a luciferase reporter driven by three copies of FoxO1 consensus binding sites in the IGFBP1 gene promoter (3xIRS-luc) (36) in transient transfection and luciferase assay. As shown in Fig. 3A, ectopic expression of COP1 strongly reduced FoxO1-mediated 3xIRS-luciferase activity (∼90%) (left panel, the raw luciferase activity; right panel, the same data as in left panel but presented as relative percent activity). To determine whether inhibition of FoxO1 activity by COP1 depends on functional Akt phosphorylation sites in FoxO1, we examined the impact of COP1 on the activity of FoxO1-3A, in which all three Akt phosphorylation sites (Thr-32, Ser-256, and Thr-319) are substituted with alanine. In agreement with earlier studies that FoxO1 transcriptional activity is inhibited by phosphorylation, FoxO1-3A displayed a 5-fold higher activation compared with wild type FoxO1 (37). In marked contrast to its effect on wild-type FoxO1, COP1 only marginally inhibited FoxO1-3A mediated reporter activation (10–15% inhibition). These data demonstrated that COP1 inhibits FoxO1 activity, and this inhibition requires the functional Akt phosphorylation in FoxO1.
FIGURE 3.
COP1 specifically regulates FoxO1 activity. A, transient transfection and luciferase assay of HEK293 cells transfected with 3xIRS luciferase reporter and the wild type FoxO1 (WT) or Akt phosphorylation defective FoxO1 (3A) with or without COP1 expressing vectors. The data were normalized to β-galactosidase expressed from cotransfected pCMV-β-galactosidase plasmids. The presented representative dataset reflects results from transient transfection performed in triplicate. The bars indicate S.D. The left part of figure shows the luciferase (Luc) activity as relative light units (RLU). The right part of figure shows the luciferase activity expressed as a percentage of values obtained in the absence of COP1 expression. B, experiments similar to those seen in 3A except that here FoxO3 expression vector was used. C, Western blot analysis of total cell lysates from B to assess the expression of FoxO1 and COP1 with the indicated antibodies. D, Western blot analysis of total cell lysates prepared from 3A with the indicated antibodies. β-Tub, β-tubulin.
To determine whether COP1 also similarly regulates the other members of the FoxO family, we assessed the ability of COP1 to regulate the activity of FoxO3. In a similar transient transfection and luciferase assay, FoxO1 mediated luciferase reporter activity was profoundly inhibited by COP1 (∼90%), whereas that of FoxO3 was affected to a far lesser degree (∼35%) (Fig. 3B), implying that COP1 differentially regulates the activity of the individual FoxO family members.
COP1 Induces Proteasome-dependent Degradation of FoxO1—Multiple mechanisms account for inhibition of FoxO transcriptional activity including nuclear export, protein degradation, and disruption of co-activator interaction, all of which depend on Akt-regulated phosphorylation of FoxO (18, 37, 38). Because COP1 showed a very high capacity for inhibiting FoxO1 activity and is a known E3 ubiquitin ligase, we reasoned that COP1 might specifically induce FoxO1 protein degradation. To test this, we assessed the protein level of transfected HA-tagged FoxO protein in cell lysates prepared for luciferase assays. Using anti-HA antibodies in Western blot analysis (Fig. 3C), we found that overexpression of COP1 had no appreciable impact on the level of FoxO3 expression (compare the first and second lanes and the third and fourth lanes). In sharp contrast, COP1 reduced the level of FoxO1 protein by more than 80% (compare the fourth and fifth lanes and the sixth and eighth lanes). These differences were not due to the variation in protein loading, as judged by similar levels of β-tubulin in all lanes. The specific down-regulation of FoxO1 protein expression by COP1 is consistent with our finding that COP1 specifically suppresses FoxO1-mediated luciferase reporter activity.
We also assessed the ability of COP1 to regulate protein levels of FoxO1-3A. As shown in Fig. 3D, although COP1 induced FoxO1 protein degradation, it had no impact on the level of FoxO-3A protein (compare the first and second lanes and the third and fourth lanes). These results are consistent with the observation that COP1 did not significantly affect FoxO1-3A activity and, in agreement with the previous studies, that degradation of FoxO1 requires phosphorylation by Akt (35, 39).
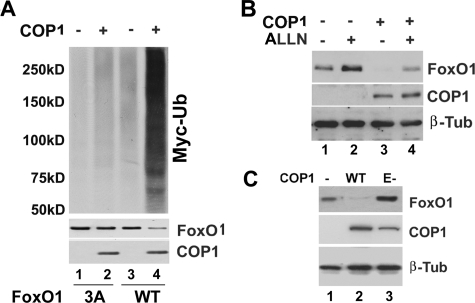
COP1 is an E3 ubiquitin ligase. We, therefore, reasoned that COP1 likely down-regulates FoxO1 protein via the ubiquitin-proteasome pathway. To test this, we examined whether COP1 promotes FoxO1 ubiquitination. Specifically, HEK293 cells were co-transfected with HA-tagged FoxO1 and Myc-tagged ubiquitin expression vectors with or without COP1 expression vectors. After 30 hours, total cell lysates were prepared for immunoprecipitation with anti-HA antibodies, and the immunoprecipitates were analyzed in Western blot with anti-Myc antibodies. Shown in Fig. 4A, Myc-Ub was detected in HA-FoxO1 immunoprecipitates in a characteristic smeary pattern (left, lanes 1 and 3), an indication of FoxO1 ubiquitination. Overexpression of COP1 strongly increased the amount of Myc-Ub associated with HA-FoxO1 immunoprecipitates (top panel, compare lanes 3 and 4) with corresponding a decrease in the level of FoxO1 protein (middle panel compare lanes 2 and 4), suggesting that COP1 promotes FoxO1 ubiquitination. We also examined the impact of COP1 on FoxO1-3A ubiquitination. In agreement with the observation that COP1 did not down-regulate FoxO1-3A protein, COP1 had minimum effect on FoxO1-3A ubiquitination (Fig. 4A, compare lanes 1 and 2, top panel). Next, we examined whether treatment with a proteasome inhibitor blocked COP1-induced FoxO1 protein degradation. To this end, we treated HEK293 cells coexpressing HA-tagged FoxO1 and FLAG-tagged COP1 with the proteasome inhibitor ALLN (40). As shown in Fig. 4B, ALLN treatment alone elevated the level of FoxO1 protein (compare lanes 1 and 2) as previously reported (35) and could reverse the down-regulation of FoxO1 by COP1 (compare lanes 3 and 4). Taken together, we conclude that COP1 may be instrumental in Akt-regulated FoxO1 protein degradation through the ubiquitin-proteasomal pathway.
FIGURE 4.
COP1 promotes FoxO1 degradation via the ubiquitin-proteasome pathway. A, COP1 increases FoxO1 ubiquitination. HEK293 cells were co-transfected with wild type (WT) FoxO1 or FoxO1-3A mutant along with Myc-tagged ubiquitin (Ub) and COP1 or control expression vector. 40 h post-transfection total cell lysates were prepared and immunoprecipitated using anti-HA-antibodies. The HA-FoxO immunoprecipitates were analyzed in Western blot analysis using anti-Myc antibodies (top). The middle and bottom panels show the respective cellular level of HA-tagged FoxO1 and FLAG-COP1 protein. β-Tub, β-tubulin. B, proteasome inhibitor ALLN reverses COP-induced degradation of FoxO1 in HEK293 cells transfected with HA-FoxO1 long with COP1 or control plasmids (pcDNA3.1). The cells were treated with either DMSO (–) or 10μm ALLN (+) for 4 h, and cell lysates were used to detect levels of COP1 and FoxO1 protein expression by Western blot analysis. C, COP1 E3 (E-) ligase activity is required for induction of FoxO1 protein degradation. The experiments were carried out as in Fig. 3C, except that an expression vector for E-3 ligase deficient COP1 (E-COP1) was also included.
COP1 has been shown to regulate protein stability either as an E3 ligase (e.g. for p53) (6) or as an anchor protein for recruiting other E3 ligases (e.g. for c-Jun) (7). We examined whether COP1-induced FoxO1 protein degradation requires COP1 E3 ligase activity by generating E3 ligase defective COP1 mutant (E-COP1) in which Cys-154 and Cys-157 within the Ring finger domain were substituted with alanines. Previous studies have demonstrated that this mutant has no E3 ligase activity (6). Presented in Fig. 4C, instead of decreasing FoxO1 protein levels, E-COP1 led to a 1.5-fold increase in FoxO1 protein (compare lanes 1 and 3), indicating that COP1 E3 ligase activity is likely required for FoxO1 protein degradation.
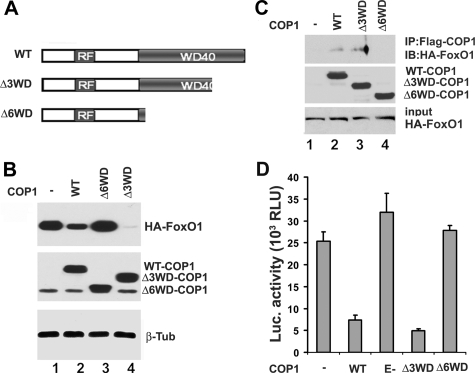
To further characterize the regulation of FoxO1 by COP1, we generated C-terminal COP1 deletion mutants for protein-protein interaction studies. The COP1 C terminus includes six repeats of a conserved WD40 domain, which is often found to mediate the association between E3 ligase and its substrates (41). The construct Δ3WD-COP1 consists of a 185-amino acid deletion at the C-terminal end consisting of three WD40 motifs. The Δ6WD-COP1 construct is deleted for 310 amino acids including 6 WD40 motifs at the C-terminal end of COP1 (Fig. 5A). After obtaining these different COP1 mutants, we first examined their abilities to induce FoxO1 protein degradation. As presented in Fig. 5B, similar to early observations (Fig. 4), COP1 decreased the level of FoxO1 (compare lanes 1 and 2). On the other hand, COP-Δ6WD had no impact on the level of FoxO1 protein (compare lanes 1 and 3), whereas COP1-Δ3WD exhibited a higher capability of down-regulating FoxO1 compared with wild type COP1 (compare lanes 2 and 4).
FIGURE 5.
Interaction between COP1 and FoxO1 is required for COP1 to inhibit FoxO1 activity. A, schematic representation of COP1 protein domains and the respective mutants. RF, ring figure; WD40, structural motifs with tryptophan-aspartic acid repeats; WT, wild type; Δ3WD, 3 WD40 motif deletion; Δ6WD, 6 WD40 motif deletion. B, the role of WD40 motif in regulation of FoxO1. HA-tagged FoxO1 was cotransfected with parental vector (pCMV-FLAGII), wild type, Δ3WD, and 63WD COP1 into HEK293 cells. 36 h host-transfection the total cell lysates were prepared for Western blot with anti-HA (top), anti-FLAG (middle), and anti-β-tubulin (β-Tub) (antibodies (bottom). Top, HA-tagged FoxO1 in total cell lysates. Middle, FLAG-tagged COP1 and its mutants in total cell lysates. Bottom, β-tubulin in total cell lysates. C, co-immunoprecipitation (IP) assay with HA-tagged FoxO1 and FLAG-tagged COP1 proteins to demonstrate that WD40 motifs of COP1 mediate the association with FoxO1. HEK293 cells were transfected with HA-tagged FoxO1 along with the expression vectors encoding wild type or Δ3WD or Δ6WD deletion mutants of COP1. 30 h post-transfection the cells were treated with 10 μm ALLN for 6 h. Anti-FLAG immunoprecipitates from the transfected cells were analyzed in Western blot (IB) using anti-HA antibodies (Top). Middle, input of FLAG-tagged COP1 and its mutants. Bottom, input of HA-tagged FoxO1. D, transient transfection and luciferase (Luc) assay examining the effect of different COP1 mutants on FoxO1 activity. RLU, relative light units. The experiments were carried out as in Fig. 3A.
The major function of WD40 motifs within E3 ligase proteins is to mediate interaction between the E3 ligase and its substrates (41). One might expect that the differential regulation of FoxO1 stability by the two COP1 mutants could be due to differential interaction between FoxO1 and the two COP1 peptides. To examine this, we employed a co-immunoprecipitation assay in HEK293 cell lysates co-expressing HA-tagged FoxO1 and the above FLAG-tagged COP1 proteins. Because COP1 promotes FoxO1 protein degradation, the HEK293 cells were pretreated with 10 μm ALLN for 6 h before being harvested for the co-immunoprecipitation assay. We observed that HA-tagged FoxO1 could be recovered from anti-FLAG immunoprecipitates of cells expressing FLAG-tagged COP1 and not from control cells (Fig. 5C, lane 1 and 2). No HA-tagged FoxO1 was detected in FLAG-tagged COP1-Δ6WD immunoprecipitates (Fig. 5C, top panel, lane 4), suggesting that the WD40 motifs of COP1 mediate association with FoxO1. In agreement with the finding that COP1-Δ3WD exhibits a higher capacity to induce FoxO1 protein degradation, a higher amount of FoxO1 was recovered from COP1-Δ3WD immunoprecipitates compared with wild type COP1 (Fig. 5C compare lanes 2 and 3, top panel). Next, we examined how these different COP1 mutants affected FoxO1-mediated luciferase reporter activity. In keeping with their abilities to regulate the level of FoxO1 protein, COP1-Δ6WD and E-COP1 did not significantly inhibit FoxO1-mediated gene expression. By contrast, COP1-Δ3WD showed a higher ability to inhibit FoxO1 activity (Fig. 5D), suggesting that inhibition of FoxO1 by COP1 is directly related to COP1-dependent FoxO1 protein degradation.
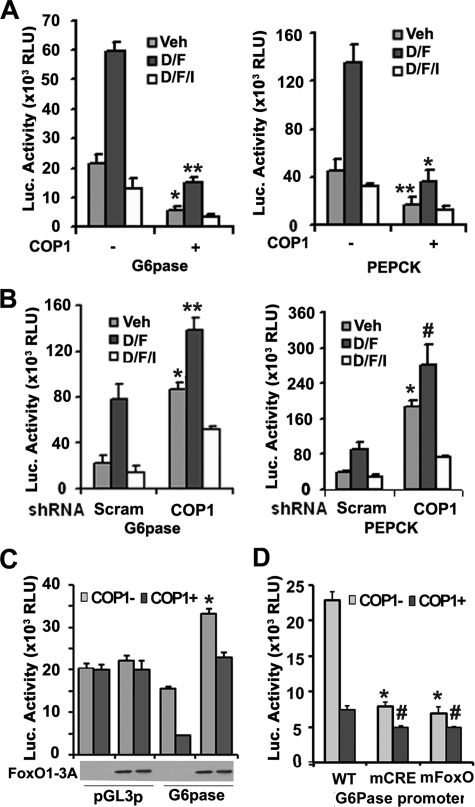
COP1 Regulates Expression of G6Pase and PEPCK Genes—As presented above, we have shown that COP1 suppresses FoxO1 activity. Next, we sought to determine the biological impact of COP1 regulated FoxO1 activity. As a critical mediator of insulin action in the liver (17), FoxO1 regulates the expression of rate-determining gluconeogenic genes G6Pase and PEPCK (for a review, see Ref. 42). Therefore, we examined whether COP1 was able to regulate the promoter activities of these two genes in transient transfection assays. As shown in Fig. 6A, both G6Pase and PEPCK promoters are activated by Dex/Fsk stimulation, which are suppressed by insulin treatment as previously reported (43, 44). Ectopic expression of COP1 suppressed basal promoter activity of both G6Pase (Fig. 6A, left panel) and PEPCK (Fig. 6A, right panel) by more than 60% and Dex/Fsk-induced promoter activity by more than 70%. Conversely, expression of COP1 shRNA increased the activity of both promoters by more than 4-fold (Fig. 6B, left panel for G6Pase and right panel for PEPCK). Interestingly, regardless of the expression of COP1, insulin suppressed the promoter activity of G6Pase and PEPCK, implying that COP1 expression likely plays an accessory role in the regulating expression of G6Pase and PEPCK genes.
FIGURE 6.
COP1 regulates PEPCK and G6Pase promoter activity. A, overexpression of COP1 represses G6Pase and PEPCK promoter activity. HepG2 cells were transfected with G6Pase or PEPCK luciferase (Luc) reporter plus empty vector (EV) or COP1 expression vector (COP1). 24 h post-transfection, the cells were serum-deprived overnight and treated with either vehicle (Veh) or dexamethasone (0.5 μm) and forskolin (2.5 μm (D/F) or dexamethasone (0.5 μm), forskolin (2.5μm), and 100 nm insulin (D/F/I) for 4 h. The experiments were carried out in triplicate. Luciferase activity was normalized to the activity of β-galactosidase used here as an internal cotransfected control. The bar shows the S.D. RLU, relative light units. B, knockdown of COP1 increases G6Pase and PEPCK promoter activity. The experiments were essentially carried out as in A except a scramble shRNA (Scram) and COP1 shRNA were used. C, the role of FoxO1 in COP1 modulated G6Pase promoter activity. HepG2 cells were transfected with G6Pase luciferase reporter with or without COP1 expression vector at the presence or absence of FoxO1-3A expression vectors. 36 h post-transfection, total cell lysates were prepared for luciferase assay as described in Fig. 3A. The experiments were carried out in triplicates and repeated twice. The expression of HA-tagged FoxO1-3A is shown below. D, effect of COP1 on activity o G6Pase promoter that harbors either a mutated CREB binding site (mCRE) or a mutated FoxO1 binding site (mFoxo). The experiments were carried out as in A. Each bar represents the means ± S.D. (n = 3). The p values were related to the control conditions. *, p < 0.01; **, p < 0.003; #, p < 0.014. WT, wild type.
To determine the specific contexts in which COP1 inhibition of target gene expression depends on FoxO1, we also used FoxO1-3A mutant in transient transfection assays. As previously reported (19), overexpression of FoxO1-3A increased G6Pase promoter activity by 60% and partially blocked inhibition of G6Pase promoter activity by COP1 (Fig. 6C). The effects of FoxO1-3A and COP1 are specific to G6Pase promoter as no significant change in luciferase activity was observed when SV40 promoter was used to drive reporter gene expression (pGL3-SV40) in the presence of either FoxO1-3A or COP1 (Fig. 6C). Partial blocking of COP1 to inhibit G6Pase promoter activity implies that COP1 might modulate other factors that are also important for G6Pase gene expression. More recently, Dentin et al. (13) showed that COP1 suppressed expression of G6Pase by down-regulating the CREB co-activator TORC2. We, therefore, examined how COP1 regulates the activity of G6Pase promoter in which either CREB or FoxO1 binding sites were mutated. Similar to previous reports (26), mutation of each site in the G6Pase promoter decreases the total activity of this promoter (Fig. 6D). Again ectopic expression of COP1 suppressed activity of wild type promoter ∼3-fold, and COP1-dependent suppression of wild-type G6Pase promoter resulted in activity levels that matched those of G6Pase-CRE (CREB response element) and G6Pase FoxO-site mutants in the absence of COP1. COP1 only modestly suppressed activity of the CRE and Foxo mutants (about 1.5-fold), suggesting that COP1 modulates G6Pase promoter activity via the FoxO1 and CREB binding sites.
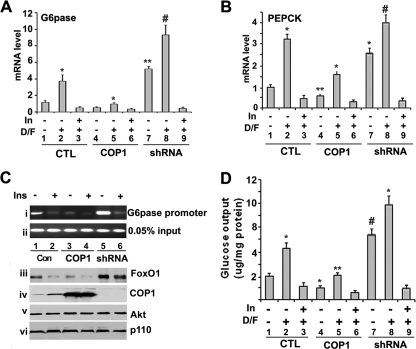
Next, we examined whether COP1 could similarly modulate expression of endogenous PEPCK and G6Pase genes. To this end, Fao cells were infected with GFP, COP1, or COP1 shRNA-expressing adenoviruses. Thirty hours post-infection, the cells were serum-deprived overnight. After either a 4-h Dex/Fsk or Dex/Fsk/insulin treatment, total RNA was prepared and subjected to conventional RT-PCR (supplemental Fig. 2) and real time RT-PCR analysis with primers specific to G6Pase and PEPCK, respectively. As shown in Fig. 7A, Fao cells express G6Pase under basal conditions, and treatment of cells with Dex/Fsk led to a 3-fold increase in G6Pase expression (compare lanes 1 and 2), and insulin treatment suppressed induction of G6Pase expression induced by Dex/Fsk (compare lanes 2 and 3). When COP1 was overexpressed, the level of G6Pase mRNA decreased by 3-fold under basal conditions (Fig. 7A, compare lanes 1 and 5) and Dex/Fsk treatment (compare lane 3 and 5). Conversely, when COP1 shRNA was introduced into Fao cells, it enhanced the expression of G6Pase by 3-fold (compare lanes 1 and 7 or 3 and 8). Consistent with the fact that insulin/Akt-dependent phosphorylation of FoxO1 is a step that precedes FoxO1 nuclear exclusion and degradation, insulin treatment suppressed G6Pase expression induced by COP1 shRNA as well as Dex/Fsk (compare lanes 7, 8, and 9). Expression of PEPCK was also similarly regulated (Fig. 7B).
FIGURE 7.
COP1 regulates the expression of endogenous PEPCK and G6Pase gene. A, real time PCR analysis of endogenous G6Pase and PEPCK genes expression. Fao cells were infected with GFP (CTL) or COP1- or COP1-shRNA (shRNA)-expressing adenoviruses. 30 h post-infection the cells were serum-starved overnight followed by 4 h of treatment with either DMSO, 0.5 μm dexamethasone, 2.5 μm forskolin (D/F), or 0.5 μm dexamethasone, 0.5 μm forskolin, 100 nm insulin (In). RNA was prepared for real time-RT-PCR to detect G6Pase gene expression. B, same as A, except that here we assessed expression of PEPCK. C, chromatin immunoprecipitation assay to show FoxO1 binding to G6Pase promoter. Fao cells in 15-cm dishes were processed as in A except for the chromatin immunoprecipitation assay. The presence of the G6Pase promoter (panel i) in the chromatin immunoprecipitate of anti-FoxO1 and in the chromatin preparation before immunoprecipitation (input, panel ii)) was then assayed using PCR as described under “Experimental Procedures.” In addition, a portion of cells were used to prepare the total cell lysates for Western blotting with anti-FoxO1 (panel iii), anti-COP1 (panel iv), anti-Akt (panel v), and anti-p110γ (panel vi) antibodies. D, glucose output assay. The same cells as in A were serum-deprived overnight, and the medium was replaced with glucose-free, phenol-free medium. After 4 h of treatment with the indicated reagents, the medium was collected and subjected to glucose assay with Sigma glucose assay reagents, and d-glucose served as a standard for the assay. The experiments were carried out in triplicate, and the amount of glucose in the medium was normalized to levels of total cellular protein. In all the experiments, each bar represents the mean ± S.D. (n = 3). The p values were related to the control conditions. *, p < 0.015; **, p < 0.005; #, p < 0.011.
To demonstrate that regulation of gene expression by COP1 is related to FoxO1, we performed chromatin immunoprecipitation assays to determine whether COP1 altered FoxO1 binding/occupancy of G6Pase promoter. Similar to previous studies (32), we observed that FoxO1 was associated by G6Pase promoter recovery by PCR from FoxO1 immunoprecipitates (Fig. 7C, lane 1). Insulin decreased the level of FoxO1 binding (panel i, lanes 1 and 2), in agreement with the notion that insulin regulates FoxO1 nuclear exclusion and its subsequently degradation. Consistent with its ability to regulate FoxO1 protein expression (Fig. 7C, panel iii), ectopic expression of COP1 decreases, whereas knockdown of COP1 increases FoxO1 binding to G6Pase promoter under basal conditions (compare panel i and iii, lanes 1, 3, and 5). However, insulin still decreases the binding activity of FoxO1 even with COP1 shRNA knockdown (compare lanes 5 and 6, in panel i and iii). Insulin induced a decrease in FoxO1 promoter occupancy even in the presence of COP1 shRNA is consistent with that notion that insulin/Akt-dependent nuclear exclusion of FoxO1 provides a fundamental mechanism to regulate FoxO1 activity. The difference in FoxO1 binding to G6Pase promoter is not due to the sample variation, as comparable amount of input DNA was observed in each sample (panel ii). In addition, we also assessed the levels of Akt (panel v) and p110γ (panel vi) of PI3K. No change of either one was observed, providing further evidence that COP1 modulates FoxO1 protein expression.
Both G6Pase and PEPCK are key enzymes in hepatic glucose production (45). Inhibition of these genes by COP1 prompted us to examine the impact of COP1 on hepatic glucose production. We carried out glucose output assays in Fao cells with COP1 overexpression or COP1 knockdown. As shown in Fig. 7D, Dex/Fsk treatment increased glucose production 2.5-fold (compare lanes 1 and 2), which are inhibited by insulin (lane 3). Consistent with COP1 suppression of G6Pase and PEPCK gene expression, overexpression of COP1 led to a 2-fold decrease in glucose output by Fao cells (compare lanes 1 and 4 or 2 and 6). On the other hand, expression of COP1 shRNA in Fao cells increased glucose production by more than 3-fold (compare lane 3 and 7 or 4 and 8). Again, insulin suppressed the glucose production induced by Dex/Fsk and COP shRNA (compare lanes 7, 8, and 9) Taken together, our data demonstrate that COP1 regulates FoxO1 protein levels by targeting it for proteasomal degradation. Such a regulatory role for COP1 impacts the expression of both G6Pase and PEPCK genes and, accordingly, regulates glucose production in Fao cells without impairing insulin signaling.
DISCUSSION
COP1 is an evolutionarily conserved E3 ubiquitin ligase. In plants, COP1 plays an essential role in photomorphogenesis (1), and in mammals COP1 regulates cell survival (6) and lipid metabolism (9). In this report, we found that COP1 expression is induced by insulin, and COP1 induces FoxO1 protein degradation. Because COP1 specifically regulates FoxO1 but not FoxO3A, another member of FoxO protein family, we characterized the regulation of FoxO1 by COP1. Our data show that COP1 induces FoxO1 protein degradation via the ubiquitin-proteasome pathway and, thus, functions as a FoxO1 E3 ligase. We found that COP1 directly interacts with FoxO1 and enhances FoxO1 protein ubiquitination (Figs. 3 and 4). The down-regulation of FoxO1 by COP1 is also consistent with the finding that overexpression of COP1 decreased FoxO1 expression, and more significantly, knockdown of COP1 expression in Fao cells increased the level of FoxO1 protein (Fig. 2C). Interestingly, COP1 only modulates the stability of FoxO1 but not that of FoxO3 (Fig. 3), demonstrating that that COP1 functions as a specific FoxO1 E3 ligase. COP1 contains several tandem WD motifs in its C terminus, some of which interact with FoxO1 (Fig. 5). Removal of last 3 WD-40 motifs increased the affinity of COP1 interaction with FoxO1, implying the presence of a negative regulator that can probably modulate the interaction between COP1 and FoxO1. In this regard it will be of great interest to identify this negative regulator(s) in future studies.
It is well documented that insulin induces FoxO1 phosphorylation which in turn promotes nuclear exclusion and degradation of FoxO1 protein (16). In fact, studies by Fukamizu and co-workers (35) showed that FoxO1 nuclear exclusion is necessary for insulin-mediated degradation of FoxO1. However, Tindall and co-workers (38) showed that SKP2, a nuclear F-box protein, promotes FoxO1 protein ubiquitination and degradation. Furthermore, Accili and co-workers (46) reported that oxidative stress promotes FoxO1 protein nuclear inclusion and ubiquitination. COP1 is localized in the nucleus as well as the cytoplasm (Refs. 3 and 4) and data not shown). Thus, it is tempting to speculate that COP1 may promote FoxO1 protein ubiquitination in either the nucleus or the cytoplasm, an issue that remains to be explored.
It is also noteworthy that, although less profound, COP1 still exhibits significant inhibition of FoxO3 activity. The mechanism by which COP1 inhibits FoxO3-activated reporter gene expression remains undetermined. One possibility is that COP1 may enhance insulin signaling, an issue that is under investigation. Should this be true, it may explain why COP1 induces FoxO1 protein degradation even under basal conditions (Fig. 2D).
Members of the FoxO family are involved a wide range of biological processes including cell survival, growth, and metabolism (22). Interestingly, although it may participate in multiple biological processes, FoxO1 is extensively studied in glucose metabolism through its regulation of metabolic genes in the liver (17). To explore the biological significance of COP1-mediated FoxO1 protein degradation, we investigated the potential effect of COP1 on G6Pase and PEPCK, two crucial hepatic genes that are positively regulated by FoxO1. Our data indicated that consistent with the negative regulation of FoxO1 protein levels, increasing COP1 expression inhibits, whereas decrease in COP1 expression promotes, the expression of PEPCK and G6Pase genes (Fig. 7). Because both genes are key rate determinants of hepatic gluconeogenesis and induce glucose production, overexpression of COP1 inhibits, whereas knockdown of COP1 enhances, hepatic glucose production in Fao cells. These data show that by regulating FoxO1 protein stability, COP1 likely exerts important effects on hepatic glucose metabolism.
Regulation of G6Pase and PEPCK by metabolic hormones such as glucagon and insulin is complex and involves numerous factors including FoxA2, CREB, C/EBP, glucocorticoid receptor, and FoxO1 (47, 48). Our data indicate that COP1 does not regulate the stability of C/EBPα, CREB, Fox2, A and PGC1α, ruling out a potential role of these proteins in COP1-modulated G6Pase or PEPCK gene expression.
In Fao cells, COP1 expression, similar to insulin, induces the expression of LAP and LIP, two isoforms of C/EBPβ (Fig. 2D). Previously, Duong et al. (49) observed that insulin induced expression of LIP in H4IIE rat hepatoma. It was concluded that insulin suppressed PEPCK gene expression in part through induction of LIP expression. Thus, the question raised is whether induction of C/EBPβ by COP1 also contributes to inhibition of G6Pase and PEPCK by COP1. COP1 has been shown to interact with TRB3 in adipocytes (9). Studies of TRB3 in preadipocytes reveal that TRB3 interacts with C/EBPβ and impairs C/EBPβ DNA binding activity (50). Therefore, it is possible that COP1 modulates C/EBPβ activity via TRB3. Interestingly, TRB3 only interacts with the LAP but not LIP (50). We speculate that in such a scenario, although insulin and COP1 expression induce both isoforms of CEBPβ, the potential regulation of LAP by COP1 via TRB3 would result in LIP becoming a more potent dominant inhibitory form of C/EBPβ and contributing to inhibition of gluconeogenic genes by COP1. In this regard, induction of both isoforms of C/EBPβ and potential inhibition of LAP may constitute an additional pathway by which COP1 regulates expression of G6Pase. Further studies are required to define such a pathway.
In any event, given the established role of FoxO1 in the regulation of G6Pase and PEPCK expression and induction of FoxO1 protein degradation by COP1, we believe it is reasonable to conclude that by regulating FoxO1 protein stability, COP1 plays a role in the regulation G6Pase and PEPCK gene expression via FoxO1.
Previously, Montminy and co-workers (13) reported that COP1 induced TORC2 degradation and suppressed PEPCK and G6Pase expression in mouse liver. In agreement with these studies, we have found that mutating the CRE site in the G6Pase promoter partially impaired the ability of COP1 to suppress G6Pase promoter activity (Fig. 6D). Thus, COP1 suppresses expression of G6Pase and PEPCK genes by down-regulation of FoxO1 and TORC2. Collectively, this observation not only provides further evidence that COP1 is an important regulator of glucose metabolism but also might explain why FoxO1-3A only partially rescued G6Pase promoter activity suppressed by COP1 (Fig. 6C).
It may be noted that althoughCOP1 suppresses the overall expression of G6Pase and PEPCK genes, the abilities of these genes to respond to glucocorticoids and cAMP still remain (Figs. 6 and 7). This is consistent with the studies of liver-specific FoxO1 knock-out mice in which reduction of FoxO1 activity decreases the overall expression of G6Pase and PEPCK genes, whereas the responsiveness of these genes to fasting and cAMP remains (23) and in agreement with the observation that overexpression of constitutively active FoxO1-3A increases the basal level of G6Pase expression with the minimal effects on the responsiveness of G6Pase genes to cAMP (51). Furthermore, induction of FoxO1 protein degradation by COP1 required functional Akt phosphorylation sites in FoxO1 (Fig. 3). All these data indicate that regulation of FoxO1 by COP1 is an event that occurs post-Akt activation. Regardless, regulation of G6Pase and PEPCK gene expression by COP1 adds a new player in the regulation of G6Pase and PEPCK gene expression. Currently, how insulin modulates COP1 expression remains unexplored and will be the focus of future studies.
Supplementary Material
Acknowledgments
We thank Dr. XW Deng for providing mammalian COP1 expression plasmids and Dr. Richard M. O'Brien for mutant G6Pase luciferase reporter plasmids.
This work was supported, in whole or in part, by National Institutes of Health Grant DK08147 (to U. S. J.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
Footnotes
The abbreviations used are: ACC1, acetyl-CoA carboxylase 1; COP1, constitutive photomorphogenic 1; FoxO1, Forkhead Box O1; PEPCK, phosphoenolpyruvate carboxykinase; G6Pase, glucose-6-phosphatase; ACC1, acetyl-CoA carboxylase 1; WD motif, tryptophan-aspartic acid motif; RT, reverse transcriptase; ALLN, N-acetyl-l-leucinyl-l-leucinyl-l-norleucinal; LAP, liver-activating protein; LIP, liver-inhibitory protein; CREB, cAMP-response element (CRE)-binding protein; HA, hemagglutinin; shRNA, short hairpin RNA; Dex, dexamethasone; Fsk, forskolin; GST, green fluorescent protein; PGC1, PPAR gamma coactivator 1.
References
- 1.Yi, C., and Deng, X. W. (2005) Trends Cell Biol. 15 618–625 [DOI] [PubMed] [Google Scholar]
- 2.Wang, H., Kang, D., Deng, X. W., and Wei, N. (1999) Curr. Biol. 9 711–714 [DOI] [PubMed] [Google Scholar]
- 3.Bianchi, E., Denti, S., Catena, R., Rossetti, G., Polo, S., Gasparian, S., Putignano, S., Rogge, L., and Pardi, R. (2003) J. Biol. Chem. 278 19682–1969012615916 [Google Scholar]
- 4.Yi, C., Wang, H., Wei, N., and Deng, X. W. (2002) BMC Cell Biol. 3 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Subramanian, C., Kim, B. H., Lyssenko, N. N., Xu, X., Johnson, C. H., and von Arnim, A. G. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 6798–6802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dornan, D., Wertz, I., Shimizu, H., Arnott, D., Frantz, G. D., Dowd, P., O'Rourke, K., Koeppen, H., and Dixit, V. M. (2004) Nature 429 86–92 [DOI] [PubMed] [Google Scholar]
- 7.Wertz, I. E., O'Rourke, K. M., Zhang, Z., Dornan, D., Arnott, D., Deshaies, R. J., and Dixit, V. M. (2004) Science 303 1371–1374 [DOI] [PubMed] [Google Scholar]
- 8.Yi, C., Li, S., Chen, X., Wiemer, E. A., Wang, J., Wei, N., and Deng, X. W. (2005) Cancer Res. 65 5835–5840 [DOI] [PubMed] [Google Scholar]
- 9.Qi, L., Heredia, J. E., Altarejos, J. Y., Screaton, R., Goebel, N., Niessen, S., Macleod, I. X., Liew, C. W., Kulkarni, R. N., Bain, J., Newgard, C., Nelson, M., Evans, R. M., Yates, J., and Montminy, M. (2006) Science 312 1763–1766 [DOI] [PubMed] [Google Scholar]
- 10.Du, K., Herzig, S., Kulkarni, R. N., and Montminy, M. (2003) Science 300 1574–1577 [DOI] [PubMed] [Google Scholar]
- 11.Koh, H. J., Arnolds, D. E., Fujii, N., Tran, T. T., Rogers, M. J., Jessen, N., Li, Y., Liew, C. W., Ho, R. C., Hirshman, M. F., Kulkarni, R. N., Kahn, C. R., and Goodyear, L. J. (2006) Mol. Cell. Biol. 26 8217–8227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tong, L., and Harwood, H. J., Jr. (2006) J. Cell. Biochem. 99 1476–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dentin, R., Liu, Y., Koo, S. H., Hedrick, S., Vargas, T., Heredia, J., Yates, J., III, and Montminy, M. (2007) Nature 449 366–369 [DOI] [PubMed] [Google Scholar]
- 14.Yamamoto, N., and Deng, X. W. (1999) Genes Cells 4 489–500 [DOI] [PubMed] [Google Scholar]
- 15.Dornan, D., Shimizu, H., Mah, A., Dudhela, T., Eby, M., O'Rourke, K., Seshagiri, S., and Dixit, V. M. (2006) Science 313 1122–1126 [DOI] [PubMed] [Google Scholar]
- 16.Greer, E. L., and Brunet, A. (2005) Oncogene 24 7410–7425 [DOI] [PubMed] [Google Scholar]
- 17.Barthel, A., Schmoll, D., and Unterman, T. G. (2005) Trends Endocrinol. Metab. 16 183–189 [DOI] [PubMed] [Google Scholar]
- 18.Puigserver, P., Rhee, J., Donovan, J., Walkey, C. J., Yoon, J. C., Oriente, F., Kitamura, Y., Altomonte, J., Dong, H., Accili, D., and Spiegelman, B. M. (2003) Nature 423 550–555 [DOI] [PubMed] [Google Scholar]
- 19.Hall, R. K., Yamasaki, T., Kucera, T., Waltner-Law, M., O'Brien, R., and Granner, D. K. (2000) J. Biol. Chem. 275 30169–30175 [DOI] [PubMed] [Google Scholar]
- 20.Nakae, J., Kitamura, T., Silver, D. L., and Accili, D. (2001) J. Clin. Investig. 108 1359–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samuel, V. T., Choi, C. S., Phillips, T. G., Romanelli, A. J., Geisler, J. G., Bhanot, S., McKay, R., Monia, B., Shutter, J. R., Lindberg, R. A., Shulman, G. I., and Veniant, M. M. (2006) Diabetes 55 2042–2050 [DOI] [PubMed] [Google Scholar]
- 22.Accili, D., and Arden, K. C. (2004) Cell 117 421–426 [DOI] [PubMed] [Google Scholar]
- 23.Matsumoto, M., Pocai, A., Rossetti, L., Depinho, R. A., and Accili, D. (2007) Cell Metab. 6 208–216 [DOI] [PubMed] [Google Scholar]
- 24.Herzig, S., Long, F., Jhala, U. S., Hedrick, S., Quinn, R., Bauer, A., Rudolph, D., Schutz, G., Yoon, C., Puigserver, P., Spiegelman, B., and Montminy, M. (2001) Nature 413 179–183 [DOI] [PubMed] [Google Scholar]
- 25.Cho, C. Y., Koo, S. H., Wang, Y., Callaway, S., Hedrick, S., Mak, P. A., Orth, A. P., Peters, E. C., Saez, E., Montminy, M., Schultz, P. G., and Chanda, S. K. (2006) Cell Metab. 3 367–378 [DOI] [PubMed] [Google Scholar]
- 26.Vander Kooi, B. T., Onuma, H., Oeser, J. K., Svitek, C. A., Allen, S. R., Vander Kooi, C. W., Chazin, W. J., and O'Brien, R. M. (2005) Mol. Endocrinol. 19 3001–3022 [DOI] [PubMed] [Google Scholar]
- 27.Kato, S., and Du, K. (2007) Biochem. Biophys. Res. Commun. 353 933–938 [DOI] [PubMed] [Google Scholar]
- 28.He, T. C., Zhou, S., da Costa, L. T., Yu, J., Kinzler, K. W., and Vogelstein, B. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pear, W. S., Miller, J. P., Xu, L., Pui, J. C., Soffer, B., Quackenbush, R. C., Pendergast, A. M., Bronson, R., Aster, J. C., Scott, M. L., and Baltimore, D. (1998) Blood 92 3780–3792 [PubMed] [Google Scholar]
- 30.Du, K., Asahara, H., Jhala, U. S., Wagner, B. L., and Montminy, M. (2000) Mol. Cell. Biol. 20 4320–4327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laborda, J. (1991) Nucleic Acids Res. 19 3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vander Kooi, B. T., Streeper, R. S., Svitek, C. A., Oeser, J. K., Powell, D. R., and O'Brien, R. M. (2003) J. Biol. Chem. 278 11782–11793 [DOI] [PubMed] [Google Scholar]
- 33.Kahn, C. R., Bertolotti, R., Ninio, M., and Weiss, M. C. (1981) Nature 290 717–720 [DOI] [PubMed] [Google Scholar]
- 34.Fehlmann, M., Crettaz, M., and Kahn, C. R. (1983) Biochem. J. 214 845–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsuzaki, H., Daitoku, H., Hatta, M., Tanaka, K., and Fukamizu, A. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 11285–11290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang, E. D., Nunez, G., Barr, F. G., and Guan, K. L. (1999) J. Biol. Chem. 274 16741–16746 [DOI] [PubMed] [Google Scholar]
- 37.Biggs, W. H., III, Meisenhelder, J., Hunter, T., Cavenee, W. K., and Arden, K. C. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 7421–7426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang, H., Regan, K. M., Wang, F., Wang, D., Smith, D. I., van Deursen, J. M., and Tindall, D. J. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 1649–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Plas, D. R., and Thompson, C. B. (2003) J. Biol. Chem. 278 12361–12366 [DOI] [PubMed] [Google Scholar]
- 40.Vinitsky, A., Michaud, C., Powers, J. C., and Orlowski, M. (1992) Biochemistry 31 9421–9428 [DOI] [PubMed] [Google Scholar]
- 41.Pickart, C. M. (2001) Annu. Rev. Biochem. 70 503–533 [DOI] [PubMed] [Google Scholar]
- 42.Gross, D. N., van den Heuvel, A. P., and Birnbaum, M. J. (2008) Oncogene 27 2320–2336 [DOI] [PubMed] [Google Scholar]
- 43.Sasaki, K., Cripe, T. P., Koch, S. R., Andreone, T. L., Petersen, D. D., Beale, E. G., and Granner, D. K. (1984) J. Biol. Chem. 259 15242–15251 [PubMed] [Google Scholar]
- 44.Streeper, R. S., Svitek, C. A., Chapman, S., Greenbaum, L. E., Taub, R., and O'Brien, R. M. (1997) J. Biol. Chem. 272 11698–11701 [DOI] [PubMed] [Google Scholar]
- 45.Postic, C., Dentin, R., and Girard, J. (2004) Diabetes Metab. 30 398–408 [DOI] [PubMed] [Google Scholar]
- 46.Kitamura, Y. I., Kitamura, T., Kruse, J. P., Raum, J. C., Stein, R., Gu, W., and Accili, D. (2005) Cell Metab. 2 153–163 [DOI] [PubMed] [Google Scholar]
- 47.Hanson, R. W., and Reshef, L. (1997) Annu. Rev. Biochem. 66 581–611 [DOI] [PubMed] [Google Scholar]
- 48.O'Brien, R. M., Streeper, R. S., Ayala, J. E., Stadelmaier, B. T., and Hornbuckle, L. A. (2001) Biochem. Soc. Trans. 29 552–558 [DOI] [PubMed] [Google Scholar]
- 49.Duong, D. T., Waltner-Law, M. E., Sears, R., Sealy, L., and Granner, D. K. (2002) J. Biol. Chem. 277 32234–32242 [DOI] [PubMed] [Google Scholar]
- 50.Bezy, O., Vernochet, C., Gesta, S., Farmer, S. R., and Kahn, C. R. (2007) Mol. Cell. Biol. 27 6818–6831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakae, J., Cao, Y., Daitoku, H., Fukamizu, A., Ogawa, W., Yano, Y., and Hayashi, Y. (2006) J. Clin. Investig. 116 2473–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.