Abstract
The transcription factor E2F-1 drives proliferation and death, but the mechanisms that differentially regulate these divergent actions are poorly understood. The hypoxia-inducible death factor Bnip3 is an E2F-1 target gene and integral component of the intrinsic mitochondrial death pathway. The mechanisms that govern Bnip3 gene activity remain cryptic. Herein we show that the transcription factor NF-κB provides a molecular switch that determines whether E2F-1 signals proliferation or death under physiological conditions. We show under basal nonapoptotic conditions that NF-κB constitutively occupies and transcriptionally silences Bnip3 gene transcription by competing with E2F-1 for Bnip3 promoter binding. Conversely, in the absence of NF-κB, or during hypoxia when NF-κB abundance is reduced, basal Bnip3 gene transcription is activated by the unrestricted binding of E2F-1 to the Bnip3 promoter. Genetic knock-down of E2F-1 or retinoblastoma gene product over-expression in cardiac and human pancreatic cancer cells deficient for NF-κB signaling abrogated basal and hypoxia-inducible Bnip3 transcription. The survival kinase PI3K/Akt inhibited Bnip3 expression levels in cells in a manner dependent upon NF-κB activation. Hence, by way of example, we show that the transcriptional inhibition of E2F-1-dependent Bnip3 expression by NF-κB highlights a survival pathway that overrides the E2F-1 tumor suppressor program. Our data may explain more fundamentally how cells, by selectively inhibiting E2F-1-dependent death gene transcription, avert apoptosis down-stream of the retinoblastoma/E2F-1 cell cycle pathway.
Keywords: apoptosis, cardiac myocytes, gene transcription, hypoxia
In the postmitotic heart, the loss of cardiac cells by an apoptotic process has been posited as an underlying cause of ventricular remodeling and contractile failure after ischemic injury (1, 2). NF-κB regulates a variety of cellular processes including inflammation, immune cell maturation, and survival (3, 4). The underlying mechanism that accounts for the diverse and varied properties of NF-κB likely reflects its ability to impinge on multiple signaling pathways within the cell (5, 6). NF-κB exists in nonstimulated cells as an inactive heterodimeric complex comprised of p50 and p65 kDa protein subunits bound to its cytoplasmic inhibitor protein IκBα (i.e., inhibitor of NF-κB). Classical NF-κB activation involves the phosphorylation-dependent degradation of IκBα mediated by the IκB kinase (IKK) signaling complex (7). Several kinases including PI3K/Akt kinase signal through NF-κB for cell survival. Despite this well established and accepted paradigm for signal-induced NF-κB activation, there is growing evidence, including data from our laboratory, that NF-κB can localize to the nucleus in nonstimulated cells (8, 9). The significance of this finding is unknown but raises the intriguing possibility that basal nuclear levels may regulate the expression of certain genes required for cell survival (9). This notion is largely substantiated by increased sensitivity of p65 NF-κB-deficient cells to death signals (5, 6, 10). In this context, much of the known biological properties of NF-κB, including cell survival, have been ascribed to its well established and proven role as a transcriptional activator (4, 11). However, a less defined but emerging property, at least in the context of cell survival, includes its role as a transcriptional repressor (12–14).
Mitochondrial perturbations have been postulated to be an underlying feature of the intrinsic cell death pathway (reviewed in ref. 15). Several lines of investigation have implicated members of the Bcl-2 gene family as regulators of the intrinsic death pathway. Notably, Bnip3 (i.e., Bcl-2 19-kDa interacting protein) was first identified in cells permissive for adenovirus infection as an E1B (i.e., early region 1B) 19-kDa interacting protein (16, 17). Since these initial observations, our laboratory established Bnip3 as a hypoxia-inducible factor crucial for provoking mitochondrial perturbations and cell death of ventricular myocytes (9, 18–20). That inappropriate Bnip3 gene expression would otherwise be lethal to cells implies that the Bnip3 promoter must be highly regulated and under tight transcriptional control. Indeed, the Bnip3 promoter is subject to strong negative repression under normoxic conditions but is highly induced in cells during hypoxia in a manner dependent on the cellular factor E2F-1 (21).
We have identified canonical DNA-binding elements for NF-κB and E2F-1 within the proximal Bnip3 promoter (14, 21). The significance of this is unproven, however, because E2F-1 and NF-κB have opposing actions on cell fate. We therefore reasoned that the regulation of Bnip3 by these factors may be a crucial determinant for basal cell survival. In this report, we provide compelling evidence that NF-κB averts cell death by a mechanism that antagonizes the transcriptional activation properties of E2F-1 at the level of the Bnip3 promoter. Our data highlight a tumor suppressor pathway that may explain more fundamentally how cells differentially regulate E2F-1 gene expression and avert apoptosis for basal cell survival.
Results
IKK-NF-κB Signaling Pathway Suppresses E2F-1-Induced Cell Death.
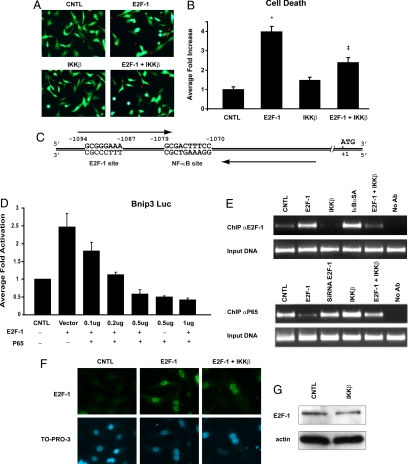
As earlier work established a survival role for NF-κB in ventricular myocytes, we tested whether IKKβ-mediated activation of NF-κB would suppress cell death induced by E2F-1. To test this possibility, cells were infected with recombinantadenoviruses encoding IKKβ or E2F-1. As shown in Fig. 1 A and B, vital staining revealed that cells expressing IKKβ were indistinguishable from virally infected control cells with respect to cell viability. In contrast, a fourfold increase (P < 0.01) in myocyte death was observed in cells expressing E2F-1. Importantly, cell death induced by E2F-1 was suppressed by IKKβ-mediated NF-κB activation, indicating that signaling pathways leading to NF-κB activation by IKKβ are functionally intact and operational for suppressing E2F-1-induced apoptosis of ventricular myocytes.
Fig. 1.
Viability assay of postnatal ventricular myocytes. (A) Representative fluorescent images of cell stained with vital dyes calcein AM and ethidium homodimer, indicating living (green) and dead (red) cells, respectively. (B) Quantitative data for A. (*Statistically different from control [CNTL], ‡Statistically different from E2F-1.) (C) Schematic diagram of the Bnip3 promoter depicting the proximity of the NF-κB and E2F-1 promoter elements. (D) Dose-response curve for inhibition of E2F-1-mediated Bnip3 promoter luciferase activity by p65 NF-κB. E2F-1 (0.5 μg DNA) was used for all conditions tested. Data are mean activation (in fold) versus control ± SE. (*Statistically different from control [CNTL].) (E) ChIP assays for E2F-1 (Upper) and P65 NF-κB (Lower) for Bnip3 promoter binding; respective DNA input for ChIP assays are shown. (F) Immunocytochemistry for E2F-1 (green) and nuclear staining by TO-PRO-3 (blue). (G) Western blot for E2F-1 protein (Upper). α-sarcomeric actin for protein loading (Lower).
Regulation of Bnip3 Gene Transcription by p65 NF-κB and E2F-1.
To begin to dissect the underlying mode by which NF-κB suppresses E2F-1-induced cell death, we focused our attention on the p65 NF-κB subunit because the biological properties conferred by NF-κB have largely been attributed to the actions of the p65. Our earlier work demonstrated the importance of the p65 and not the p50 subunit for suppressing apoptosis in ventricular myocytes (9, 22). We reasoned that p65 NF-κB likely impinged on one or more key factors required for E2F-1-induced cell death. Preliminary findings by our laboratory showed that the cellular factor E2F-1 was crucial for basal and inducible transcription of Bnip3 (21). Coincident with these findings, we observed consensus elements for E2F-1 adjacent to the canonical elements for NF-κB within the Bnip3 promoter (Fig. 1C). Given the close proximity of E2F-1 and NF-κB consensus elements, we reasoned that p65NF-κB may suppress cell death and Bnip3 transcription by interfering with the actions of E2F-1 at the level of the Bnip3 promoter.
To test this possibility, Bnip3 gene transcription was monitored in postnatal ventricular myocytes in the presence and absence of eukaryotic expression vectors encoding E2F-1 or p65 NF-κB. As shown in Fig. 1D, in contrast to vector control cells, basal Bnip3 gene transcription was markedly repressed in cells expressing the p65 NF-κB in a dose-dependent manner. Importantly, in contrast to vector alone, a 2.5-fold increase (P < 0.01) in Bnip3 gene transcription was observed in cells over-expressing E2F-1. Mutations of E2F-1 defective for DNA binding had no influence on basal Bnip3 promoter activity (21). Chromatin immunoprecipitation (ChIP) analysis verified that E2F-1 directly engaged the Bnip3 promoter coincident with the increased Bnip3 gene transcription (Fig. 1E Upper). Interestingly, a marked reduction in E2F-1 Bnip3 promoter binding and Bnip3 gene transcription was observed in cells after IKKβ-mediated NF-κB activation (Fig. 1E Upper and Fig. 2B), suggesting that p65 protein inhibits E2F-1 Bnip3 promoter binding. Importantly, no apparent change in E2F-1 protein levels or nuclear localization was observed in cells after IKKβ-mediated NF-κB activation, excluding the possibility that the reduced binding of E2F-1 to the Bnip3 promoter in NF-κB activated cells was related to alterations in E2F-1 protein abundance, expression, or nuclear targeting (Fig. 1 F and G). Furthermore, in reciprocal ChIP experiments, we tested whether over-expression of E2F-1 would inhibit p65 Bnip3 promoter binding. As shown in Fig. 2E Lower, binding of p65NF-κB to the Bnip3 promoter was reduced in cells over-expressing E2F-1 compared with vector control cells or cells expressing siRNA directed against E2F-1—a finding consistent with the increased Bnip3 promoter activity by E2F-1 (Fig. 1D).
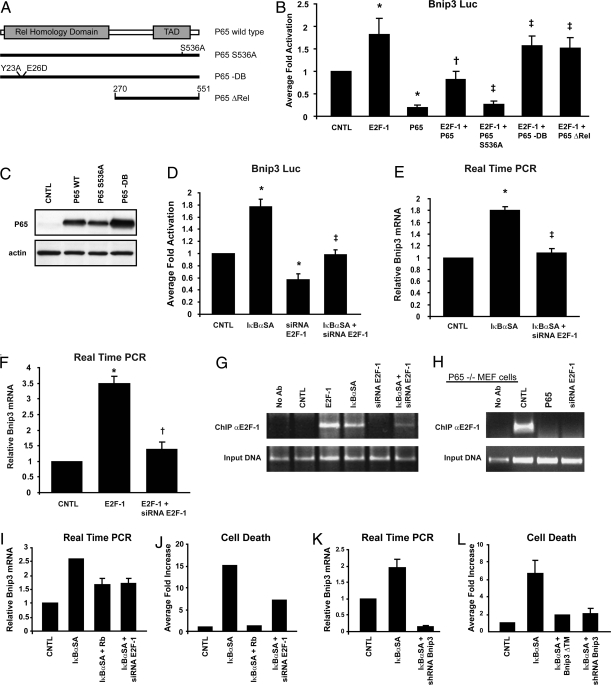
Fig. 2.
Repression of E2F-1-dependent Bnip3 gene transcription by NF-κB. (A) Schematic of WT p65 NF-κB and mutations: p65S536A is transactivation defective (11), p65-DB is DNA binding defective (30), and p65ΔRel is Rel domain deleted (12). (B) Bnip3 Promoter luciferase reporter assay (*Statistically different from control [CNTL], ‡Statistically different from E2F-1 + p65.) (C Upper) Western blot analysis for p65NF-κB proteins shown in B Lower, cytoplasmic actin to demonstrate equivalent protein loading. (D) Luciferase reporter assay for Bnip3 promoter activity. Data are presented as described in Fig. 1A. (*Statistically different from control [CNTL], ‡Statistically different from IκBαSA.) (E) Real-time quantitative PCR for endogenous Bnip3 mRNA levels. (*Statistically different from control [CNTL], ‡Statistically different from IκBαSA.) (F) qPCR for endogenous Bnip3 mRNA transcript levels. (*Statistically different from control [CNTL], †Statistically different from E2F-1.) (G Upper) ChIP assay for E2F-1 Bnip3 promoter binding in ventricular myocytes. (Lower) input DNA for the ChIP assay. (H Upper) ChIP assay depicting E2F-1 Bnip3 promoter binding in p65−/− mouse embryonic fibroblast cells (5). (Lower) input DNA for the ChIP assay. (I and J) Quantitative PCR for endogenous Bnip3 mRNA and cell viability in control ventricular myocytes and myocytes rendered defective for NF-κB activation with IκBαSA (9). (K and L) Panc-1 cells; analysis is as described for I and J. (Bnip3ΔTM, Bnip3 mutant defective for mitochondrial targeting; shRNA, short hairpin interference RNA directed against Bnip3.)
Inhibition of E2F-1-Mediated Bnip3 Gene Transcription by p65 NF-κB.
To verify that the inhibitory actions imposed by NF-κB on E2F-1-dependent Bnip3 transcription were not related to the de novo activation of NF-κB-regulated genes, we tested whether a mutation of p65 NF-κB previously shown to be defective for transactivation would influence E2F-1-mediated transcription of Bnip3 (Fig. 2A). As shown in Fig. 2B, E2F-1-dependent Bnip3 gene transcription was repressed by either the WT p65 NF-κB or mutations of p65NF-κB defective for transactivation, verifying that inhibitory actions imposed by p65 NF-κB were not likely contingent on gene de novo transcription by the p65 NF-κB. Interestingly, however, E2F-1-mediated Bnip3 transcription was unaffected by a mutation of the p65 NF-κB defective for DNA binding, indicating that DNA binding properties of p65NF-κB are required to suppress Bnip3 transcription by E2F-1. Importantly, as shown in Fig. 2C, all p65 proteins tested were expressed to comparable levels, substantiating that the observed findings on Bnip3 gene transcription were related to differences in p65 protein function and not caused by differences in efficiency of delivery or protein expression among WT p65 and mutant proteins. Because the majority of the regulatory properties conferred by p65 NF-κB have been attributed to the Rel homology domain, we tested whether this domain of the p65 NF-κB subunit was involved in the repression of Bnip3 transcription. As shown in Fig. 2B, E2F-1-directed Bnip3 transcription was relatively unaffected by the p65 Rel domain deletion mutation, indicating that the negative regulation of Bnip3 gene transcription by p65 likely involves this domain. Further, deletion or point-substitution mutations of the NF-κB/RelA elements within the Bnip3 promoter abrogated the inhibitory actions of p65 NF-κB on Bnip3 gene transcription, substantiating that repression of Bnip3 by p65 protein is mediated through the NF-κB elements (9).
E2F-1 Bnip3 Gene Transcription in Cardiac and Pancreatic Cancer Cells.
To validate the notion that NF-κB suppresses basal Bnip3 transcription by antagonizing the actions of E2F-1 at the level of the Bnip3 promoter, we tested this possibility by rendering cells defective for NF-κB activation with a nonphosphorylatable form of IκBα (IκBαSA), shown by our laboratory to inhibit NF-κB signaling in ventricular myocytes (9). As shown in Fig. 2D, basal Bnip3 gene expression was increased twofold (P < 0.01) in cells defective for NF-κB signaling—a finding concordant with our earlier work and the notion that the Bnip3 promoter is repressed by NF-κB signaling. To prove that the resultant increase in basal Bnip3 transcription in the absence of NF-κB signaling was directly related to actions of E2F-1 at the Bnip3 promoter, we next tested whether genetic knockdown of E2F-1 in cells defective for NF-κB activation would inhibit Bnip3 gene transcription. As shown in Fig. 2 D and E, the observed increase in Bnip3 gene transcription in cells defective for NF-κB activation was markedly suppressed by genetic knockdown with siRNA directed against E2F-1. Earlier work by our laboratory authenticated the specificity of siRNA-E2F-1 used for impairing E2F-1 (21). Furthermore, quantitative real-time PCR verified the Bnip3 promoter luciferase assay data, demonstrating the induction of the endogenous Bnip3 gene by E2F-1 in the absence of NF-κB signaling (Fig. 2 E and F). Moreover, ChIP analysis of the Bnip3 promoter verified that E2F-1 binding to the Bnip3 promoter was increased in cardiac cells rendered defective for NF-κB signaling with the IκBαSA (21), a finding consistent with our Bnip3 transcription data (Fig. 2D). To conclusively prove that p65 NF-κB disrupts E2F-1 Bnip3 promoter binding, we assessed by ChIP analysis the extent of E2F-1 binding to the Bnip3 promoter in cells derived from p65−/− mouse embryonic fibroblasts (5). As shown in Fig. 2H, E2F-1 bound the Bnip3 promoter under basal conditions in p65−/− cells. Importantly, repletion of p65 NF-κB subunit back into p65−/− cells prevented E2F-1 Bnip3 promoter binding (Fig. 2H). Furthermore, earlier we showed that deletion of the NF-κB DNA element base pairs (-1070/-1079) within the proximal Bnip3 promoter in increased basal Bnip3 gene transcription (9), highlighting the importance of this NF-κB site for inhibiting Bnip3 promoter activity. Together, our data substantiate that negative regulation imposed by NF-κB antagonizes E2F-1 at the level of the Bnip3 promoter. Because defects in apoptosis signaling have been postulated to be an underling feature of many cancers, we next assessed whether E2F-1-dependent Bnip3 transcription is regulated in human pancreatic ductal carcinoma cells (Panc-1), which contain elevated levels of NF-κB. Inactivation of NF-κB in ventricular myocytes (Fig. 2 I and J) and Panc-1 cells (Fig. 2 K and L) resulted in a 2.6-fold and 2.3-fold increase, respectively, in endogenous Bnip3 gene transcription and cell death. Furthermore, genetic knock-down of E2F-1 with siRNA or by overexpression of retinoblastoma gene product (Rb) as a means to inhibit E2F-1 activity suppressed Bnip3 gene transcription and cell death in cardiac defective for NF-κB signaling (Fig. 2 I and J). These findings not only verify that Bnip3 promoter is repressed by NF-κB, but importantly highlight that its induction is contingent on E2F-1 and not lineage-restricted. Importantly, the increased cell death observed in the Panc-1 cells rendered defective for NF-κB activation was contingent on Bnip3 activation, because loss of Bnip3 function by dominant-negative inhibition or genetic knock-down of Bnip3 independently suppressed cell death (Fig. 2L). Collectively, our data substantiate that the link between NF-κB and E2F-1 for the regulation of Bnip3 is functionally conserved in cardiac and pancreatic cancer cells.
E2F-1 Bnip3 Promoter Binding During Hypoxia.
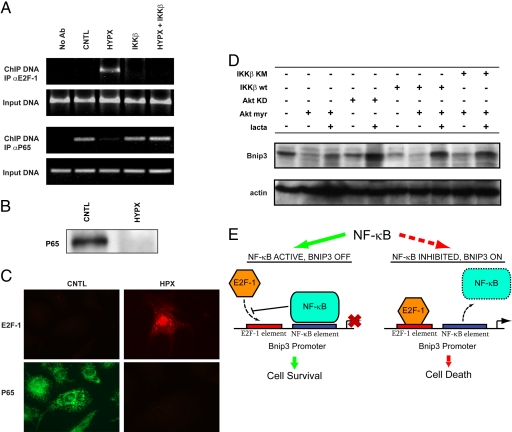
As earlier work by our laboratory established that IKKβ-mediated activation of NF-κB was sufficient to suppress hypoxia-induced mitochondrial defects and cell death of ventricular myocytes imposed by Bnip3 (9, 23), we were interested in determining whether the observed repression of Bnip3 by NF-κB during hypoxia was related to alterations in E2F-1 activity at the level of the Bnip3 promoter. For these studies, we performed ChIP assays of the Bnip3 promoter to assess E2F-1 and p65 DNA binding under normoxic and hypoxic conditions. As shown in Fig. 3A, in contrast to normoxic control cells, a marked reduction in p65NF-κB Bnip3 promoter binding was observed in cells subjected to hypoxia. Concordant with these findings was a dramatic reduction in p65 protein levels in hypoxic cells (Fig. 3 B and C), a finding consistent with our earlier work (9). The hypoxia-induced loss of p65 NF-κB Bnip3 promoter binding was accompanied by a reciprocal increase in E2F-1 nuclear staining and E2F-1 Bnip3 promoter binding (Fig. 3 A and C). These findings are in complete agreement with the de-repression of the Bnip3 promoter and induction of Bnip3 gene transcription by E2F-1 during hypoxia (9). Importantly, activation of NF-κB by IKKβ restored p65 binding to the Bnip3 promoter and repressed hypoxia-induced binding of E2F-1. This finding is in complete agreement with the ability of IKKβ mediated NF-κB activation to repress Bnip3 gene transcription and cell death in hypoxic cells (9). Collectively, our data strongly suggest that Bnip3 gene transcription is negatively regulated by NF-κB by a mechanism that antagonizes the ability of E2F-1 to engage the Bnip3 promoter.
Fig. 3.
NF-κB activation prevents hypoxia-induced E2F-1 binding to the Bnip3 promoter. (A) ChIP analysis for E2F-1 (Upper) and p65 (Lower) Bnip3 promoter binding input DNA for ChIP assay shown. (B) Western blot analysis for p65 NF-κB under normoxia (CNTL) and hypoxia (HYPX). (C) Epifluorescence microscopy for endogenous E2F-1 (red) and p65 (green) proteins in ventricular myocytes (magnification 600×). (D Upper) Western blot analysis for Bnip3 in the absence and presence of constitutively active Akt (Akt myr), kinase-dead Akt (Akt KD), WT IKKβ (IKKβ wt), kinase inactive IKKβ (IKKβ KM), and proteasome inhibitor lactacystine (1 μM, lacta). (Lower) α-sarcomeric actin for protein loading. (E) Model for regulation of basal cell survival (see text for details), NF-κB blocks basal E2F-1-dependent Bnip3 promoter binding and Bnip3 transcription (solid green arrow). Loss of NF-κB activity de-represses the Bnip3 promoter, allowing E2F-1 Bnip3 promoter binding, Bnip3 gene transcription, and cell death (dotted red arrow).
PI3K/Akt Survival Pathway Blocks Bnip3 Expression.
Because the PI3K/Akt survival kinases are known to signal through NF-κB, we next assessed whether Akt activation would influence Bnip3 expression levels in ventricular myocytes. For these experiments, cardiac cells were infected with recombinant adenoviruses encoding constitutively active or kinase inactive forms of Akt. As shown by Western blot (Fig. 3D), Bnip3 expression levels were markedly decreased in cells in the presence of the constitutively active form of Akt. However, Bnip3 expression was relatively unaffected in cells expressing the kinase inactive form Akt. Furthermore, the inhibitory actions imposed by Akt on Bnip3 expression were contingent on a functional IKKβ-NF-κB signaling pathway, because the ability of Akt to suppress Bnip3 expression was abrogated by a kinase inactive form of IKKβ. This suggests that IKKβ-mediated NF-κB signaling is down-stream of Akt-mediated repression of Bnip3.
Discussion
Bnip3 is uniquely distinguished from other Bcl-2 gene family members known to provoke cell death by at least two important features. First, Bnip3 expression is transcriptionally induced by ischemic or hypoxic stress, and second, the Bnip3 promoter contains DNA binding elements for NF-κB and E2F-1 that are absent from the other Bcl-2 death factors. These unique properties of Bnip3 highlight its importance as a key regulator of cell death during hypoxic injury and its transcriptional divergence from the other Bcl-2 death proteins. Given that de-regulated Bnip3 gene expression would otherwise provoke mitochondrial defects and cell death, it is implied that the Bnip3 promoter must be under tight transcriptional control (9). Earlier work by our laboratory demonstrated that part of the survival paradigm elicited by NF-κB involves the strong transcriptional repression of the Bnip3 promoter (9, 14). The de-repression of Bnip3 in the absence of NF-κB signaling is intriguing and implies that NF-κB may suppress or impinge on a factor that may otherwise be required for basal Bnip3 gene transcription.
E2F-1 has been shown to provoke apoptosis in a variety of cell types, including cardiac myocytes (21, 24, 25). Thus, the findings of the present study are consistent with a model in which NF-κB suppresses cell death by directly antagonizing the actions of E2F-1 on Bnip3 gene transcription. The relationship between E2F-1 and p65 NF-κB for the regulation of basal Bnip3 gene transcription is even more profound given that we have shown that, even under normoxic conditions, in the absence of HIF-1α activation, genetic knock-down of E2F-1 completely suppressed the basal increase in Bnip3 gene transcription in cardiac and Panc-1 cells rendered defective for NF-κB. These findings not only substantiate the importance of E2F-1 for activation of the Bnip3 promoter, but highlight the interplay between NF-κB and E2F-1 for regulating cell survival. Because the inability of cells to effectively mount an apoptotic response is a primary defect in most cancers, it is tempting to speculate that the inhibition of E2F-1-dependent death gene expression by NF-κB may confer a growth advantage for cells in hypoxic tumors. Concordant with this view is a recent report documenting the inhibition of a subset of E2F-1- regulated apoptotic genes by the PI3K/Akt pathway in breast and ovarian cancers (26). However, in that study, the underlying mechanism by which PI3K/Akt inhibited E2F-1-dependent death gene expression was not determined. Our data strongly suggest that NF-κB activation down-stream of the Akt signaling pathway is crucial for the inhibitory actions imposed on Bnip3 gene transcription. Indeed, the de-repression of Bnip3 transcription in cardiac myocytes and Panc-1 cells upon NF-κB inhibition, as seen here, is consistent with this notion.
We provide the following tenable models to explain our findings. First, given that E2F-1 and NF-κB elements are located adjacent to each other, the p65 NF-κB subunit may simply obscure or sterically impair the transactivation potential of E2F-1, thereby inhibiting Bnip3 gene transcription. This notion is supported by the fact that genetic knock-down of E2F-1 under basal conditions inhibited Bnip3 transcription. However, the reduction in E2F-1 bound to the Bnip3 promoter after NF-κB activation argues against this possibility. An alternative, more plausible, explanation purports that NF-κB likely displaces or prevents recruitment of E2F-1 to the Bnip3 promoter under nonapoptotic conditions. This view is supported by several salient and key observations: E2F-1 binding to the Bnip3 promoter was dramatically increased in p65−/− cells; mutations of p65 defective for DNA binding prevented the inhibitory actions of p65NF-κB on E2F-1-mediated Bnip3 gene transcription; and similarly, a nonphosphorylatable form of IκBα, which prevents nuclear targeting of p65NF-κB, increased basal E2F-1 binding to the Bnip3 promoter and Bnip3 gene transcription—a finding consistent with the reduction in p65NF-κB protein levels during hypoxia (9). Perhaps most compelling was our finding that E2F-1-Bnip3 promoter binding was abrogated upon repletion of p65 subunit into p65−/− cells. Last, the fact that IKKβ-mediated NF-κB activation suppressed basal and hypoxia-inducible E2F-1 binding to the Bnip3 promoter and Bnip3 gene transcription (9) without influencing E2F-1 protein levels or nuclear targeting suggests that NF-κB actively prevents E2F-1-Bnip3 promoter binding.
Based on our findings, we envision a model for the regulation of Bnip3 promoter activity and cell death by NF-κB and E2F-1 (Fig. 3E). In this paradigm, p65NF-κB serves as a “molecular switch” or functional check-point that regulates E2F-1-dependent death gene expression. Because inappropriate Bnip3 activation would otherwise have catastrophic consequences in postmitotic organs such as the heart, the basal repression of E2F-1-dependent Bnip3 expression by NF-κB is crucial for actively averting apoptosis. However, in tumor cells, the inability to effectively mount an apoptotic response in the face of excessive NF-κB- and E2F-1-mediated proliferative signals is viewed as a requisite for cell transformation. This is perhaps best exemplified by the counterintuitive and unexpected tumor development in mice deficient for E2F-1 (27). Thus, the cell death machinery must be precisely balanced to ensure tissue homeostasis in a cell- and context-specific manner. Although unproven, we speculate that therapeutic abrogation of NF-κB activity in E2F-1-dependent tumors would de-repress Bnip3 gene transcription and render cells more susceptible to conventional chemotherapeutic agents. This notion is in line with our earlier work and the increased Bnip3 expression and apoptosis in p65−/− cells (9, 14, 23). Thus, our data highlight a survival paradigm for NF-κB that includes the active transcriptional inhibition of E2F-1-dependent death gene expression down-stream of the Rb/E2F-1 pathway. Our data may fundamentally explain how cardiac cells discriminate between the growth- and apoptosis-inducing properties of E2F-1 for basal cell survival and why cancer cells fail to effectively undergo apoptosis when NF-κB is chronically activated.
Materials and Methods
Cell Culture and Transfection.
Postnatal cardiac myocytes from 2-d-old Sprague–Dawley rat hearts were isolated and submitted to primary culture (17, 18). Myocytes were transfected with the 2.3-kbp human Bnip3 promoter luciferase reporter (Bnip3 wt Luc) as we reported (9) (Fig. S1). Expression plasmids encoding epitope-FLAG-tagged WT p65, and derivatives defective for transactivation (p65 S529A, p65 S536A, p65 Rel deletion) or DNA binding (p65 Y23A,E26D, designated p65-DB) have been reported (12, 28). Luciferase activity was normalized to β-galactosidase activity.
Immunoprecipitation and Western Blot.
For immunodetection of p65-NF-κB and E2F-1 proteins, cardiac myocytes cell lysates (100 μg) were resolved on a 10% SDS/PAGE gel. p65 NF-κB or E2F-1 proteins were detected by using antibodies directed against p65 protein (clone C20, cat. no. sc-372) or E2F-1 protein (clone KH95, cat. no. sc-251; 1 μg/ml; Santa Cruz Biotechnology). Bound proteins were detected by enhanced chemiluminescence reagents (Amersham Pharmacia).
ChIP Assay.
ChIP assays were performed by using a ChIP assay kit (Upstate Biotechnology) as reported (29). DNA was amplified by using PCR primer pairs designed to amplify a 278-bp segment of the Bnip3 promoter spanning the E2F-1 and NF-κB elements: forward, 5′-AAAGCGGGAAATGAGAAAGC-3′; reverse, 5′-TCGAGCAGAGTCGAAAGAGTC-3′. PCR amplification products were analyzed on a 2% agarose gel as we reported (21).
Supplementary Material
Acknowledgments.
We thank the late Dr. Arnold H. Greenberg (University of Manitoba, Winnipeg, Canada) for his friendship and generous gift of reagents, Dr. James Davie (University of Manitoba, Winnipeg, Canada) for Panc-1 cells, and Drs. Dean Ballard (Vanderbilt University Medical Center, Nashville, TN), Joseph Nevins (Duke University, Durham, NC), and Gioacchino Natoli (Campus IFOM-IEO, Milan) for reagents. We also thank Dr. Albert Baldwin for critical comments on the manuscript and Steven Zamick, Lindsey MacDonald, and Ravi Jayas for expert technical assistance. This work was supported by grants from the Canadian Institute for Health Research (L.A.K.). J.S. holds a studentship from the Manitoba Health Research Council and L.A.K. is a Canada Research Chair in Molecular Cardiology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807735105/DCSupplemental.
References
- 1.Foo RS, Mani K, Kitsis RN. Death begets failure in the heart. J Clin Invest. 2005;115:565–571. doi: 10.1172/JCI24569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gottlieb RA, Kitsis RN. Seeing death in the living. Nat Med. 2001;7:1277–1278. doi: 10.1038/nm1201-1277. [DOI] [PubMed] [Google Scholar]
- 3.Baeuerle PA, Baltimore D. NF-kappa B: Ten years after. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 4.Karin M, Lin A. NF-κB at the crossroads of life and death. Nat Immunol. 2002;3:221–227. doi: 10.1038/ni0302-221. [DOI] [PubMed] [Google Scholar]
- 5.Beg AA, Sha WC, Bronson RT, Ghosh S, Baltimore D. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-κ B. Nature. 1995;376:167–170. doi: 10.1038/376167a0. [DOI] [PubMed] [Google Scholar]
- 6.Van Antwerp DJ, Martin SJ, Kafri T, Green DR, Verma IM. Suppression of TNF-α-induced apoptosis by NF-κB. Science. 1996;274:787–789. doi: 10.1126/science.274.5288.787. [DOI] [PubMed] [Google Scholar]
- 7.Karin M. The beginning of the end: IκB kinase (IKK) and NF-κB activation. J Biol Chem. 1999;274:27339–27342. doi: 10.1074/jbc.274.39.27339. [DOI] [PubMed] [Google Scholar]
- 8.Carlotti F, Dower SK, Qwarnstrom EE. Dynamic shuttling of nuclear factor κB between the nucleus and cytoplasm as a consequence of inhibitor dissociation. J Biol Chem. 2000;275:41028–41034. doi: 10.1074/jbc.M006179200. [DOI] [PubMed] [Google Scholar]
- 9.Baetz D, et al. Nuclear factor-κB-mediated cell survival involves transcriptional silencing of the mitochondrial death gene BNIP3 in ventricular myocytes. Circulation. 2005;112:3777–3785. doi: 10.1161/CIRCULATIONAHA.105.573899. [DOI] [PubMed] [Google Scholar]
- 10.Li ZW, et al. The IKKbeta subunit of IκB kinase (IKK) is essential for nuclear factor κB activation and prevention of apoptosis. J Exp Med. 1999;189:1839–1845. doi: 10.1084/jem.189.11.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Madrid LV, et al. Akt suppresses apoptosis by stimulating the transactivation potential of the RelA/p65 subunit of NF-κB. Mol Cell Biol. 2000;20:1626–1638. doi: 10.1128/mcb.20.5.1626-1638.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashburner BP, Westerheide SD, Baldwin AS., Jr The p65 (RelA) subunit of NF-κB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol Cell Biol. 2001;21:7065–7077. doi: 10.1128/MCB.21.20.7065-7077.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCullough KD, Martindale JL, Klotz LO, Aw TY, Holbrook NJ. Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Mol Cell Biol. 2001;21:1249–1259. doi: 10.1128/MCB.21.4.1249-1259.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaw J, et al. Transcriptional silencing of the death gene BNIP3 by cooperative action of NF-κB and histone deacetylase 1 in ventricular myocytes. Circ Res. 2006;99:1347–1354. doi: 10.1161/01.RES.0000251744.06138.50. [DOI] [PubMed] [Google Scholar]
- 15.Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 16.Boyd JM, et al. Adenovirus E1B 19 kDa and Bcl-2 proteins interact with a common set of cellular proteins. Cell. 1994;79:341–351. doi: 10.1016/0092-8674(94)90202-x. and erratum (1994) 79: 1120. [DOI] [PubMed] [Google Scholar]
- 17.Kirshenbaum LA, Schneider MD. Adenovirus E1A represses cardiac gene transcription and reactivates DNA synthesis in ventricular myocytes, via alternative pocket protein- and p300-binding domains. J Biol Chem. 1995;270:7791–7794. doi: 10.1074/jbc.270.14.7791. [DOI] [PubMed] [Google Scholar]
- 18.Regula KM, Ens K, Kirshenbaum LA. Inducible expression of BNIP3 provokes mitochondrial defects and hypoxia-mediated cell death of ventricular myocytes. Circ Res. 2002;91:226–231. doi: 10.1161/01.res.0000029232.42227.16. [DOI] [PubMed] [Google Scholar]
- 19.Kubasiak LA, Hernandez OM, Bishopric NH, Webster KA. Hypoxia and acidosis activate cardiac myocyte death through the Bcl-2 family protein BNIP3. Proc Natl Acad Sci USA. 2002;99:12825–12830. doi: 10.1073/pnas.202474099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bruick RK. Expression of the gene encoding the proapoptotic Nip3 protein is induced by hypoxia. Proc Natl Acad Sci USA. 2000;97:9082–9087. doi: 10.1073/pnas.97.16.9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yurkova N, et al. The cell cycle factor E2F-1 activates Bnip3 and the intrinsic death pathway in ventricular myocytes. Circ Res. 2008;102:472–479. doi: 10.1161/CIRCRESAHA.107.164731. [DOI] [PubMed] [Google Scholar]
- 22.Regula KM, Ens K, Kirshenbaum LA. IKK beta is required for Bcl-2-mediated NF-κB activation in ventricular myocytes. J Biol Chem. 2002;277:38676–38682. doi: 10.1074/jbc.M206175200. [DOI] [PubMed] [Google Scholar]
- 23.Regula KM, Baetz D, Kirshenbaum LA. Nuclear factor-κB represses hypoxia-induced mitochondrial defects and cell death of ventricular myocytes. Circulation. 2004;110:3795–3802. doi: 10.1161/01.CIR.0000150537.59754.55. [DOI] [PubMed] [Google Scholar]
- 24.Kirshenbaum LA, Abdellatif M, Chakraborty S, Schneider MD. Human E2F-1 reactivates cell-cycle progression in ventricular myocytes and represses cardiac gene transcription. Dev Biol. 1996;179:402–411. doi: 10.1006/dbio.1996.0270. [DOI] [PubMed] [Google Scholar]
- 25.Hauck L, Hansmann G, Dietz R, von Harsdorf R. Inhibition of hypoxia-induced apoptosis by modulation of retinoblastoma protein-dependent signaling in cardiomyocytes. Circ Res. 2002;91:782–789. doi: 10.1161/01.res.0000041030.98642.41. [DOI] [PubMed] [Google Scholar]
- 26.Hallstrom TC, Mori S, Nevins JR. An E2F1-dependent gene expression program that determines the balance between proliferation and cell death. Cancer Cell. 2008;13:11–22. doi: 10.1016/j.ccr.2007.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cloud JE, et al. Mutant mouse models reveal the relative roles of E2F1 and E2F3 in vivo. Mol Cell Biol. 2002;22:2663–2672. doi: 10.1128/MCB.22.8.2663-2672.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madrid LV, Mayo MW, Reuther JY, Baldwin AS., Jr Akt stimulates the transactivation potential of the RelA/p65 Subunit of NF-κB through utilization of the IκB kinase and activation of the mitogen-activated protein kinase p38. J Biol Chem. 2001;276:18934–18940. doi: 10.1074/jbc.M101103200. [DOI] [PubMed] [Google Scholar]
- 29.Nowak DE, Tian B, Brasier AR. Two-step cross-linking method for identification of NF-κB gene network by chromatin immunoprecipitation. Biotechniques. 2005;39:715–725. doi: 10.2144/000112014. [DOI] [PubMed] [Google Scholar]
- 30.Saccani S, Marazzi I, Beg AA, Natoli G. Degradation of promoter-bound p65/RelA is essential for the prompt termination of the nuclear factor κB response. J Exp Med. 2004;200:107–113. doi: 10.1084/jem.20040196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.