Abstract
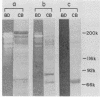
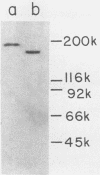
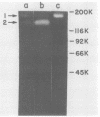
The presence of proteases in culture supernatant fluids and on the cell surface of Streptococcus sobrinus and the aggregation of multicomponent enzyme complexes make the isolation and characterization of cell surface proteins difficult. We report a simple purification procedure for dextranase and the cloning of the dextranase structural gene. S. sobrinus culture supernatant fluids were precipitated with 70% ammonium sulfate, and the precipitate was dialyzed against sodium acetate buffer and loaded onto a hemoglobin-Sepharose 4B column connected to a blue dextran-agarose column at 4 degrees C. After being washed with low concentrations of salt, the dextranase and the dextran-binding proteins were eluted with 5 M KI and further purified by gel filtration. Two dextranases (molecular weights, 175,000 and 160,000) were purified and partially characterized. The structural gene for the dextranase of S. sobrinus 6715 strain UAB66, serotype g, was cloned into the cosmid vector, pHC79. Clones were selected for expression of dextranase activity by detection of zones of enzyme-mediated hydrolysis of a blue dextran substrate incorporated into minimal medium agar plates. Release of dextranase was achieved by induction of thermoinducible, excision-defective Escherichia coli K-12 lysogens containing recombinant cosmid molecules of S. sobrinus DNA. Recombinant cosmid molecules were repackaged simultaneously into infectious lambdoid particles. Recombinant clones expressing dextranase activity which varied in size from the high-molecular-weight protein produced by S. sobrinus (i.e., 175,000) to lower-molecular-weight forms expressed by S. sobrinus have been identified and partially characterized.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barrett J. F., Curtiss R., 3rd Renaturation of dextranase activity from culture supernatant fluids of Streptococcus sobrinus after sodium dodecylsulfate polyacrylamide gel electrophoresis. Anal Biochem. 1986 Nov 1;158(2):365–370. doi: 10.1016/0003-2697(86)90562-2. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen W. H. Effects of dextranase on cariogenic and non-cariogenic dextrans. Br Dent J. 1968 Apr 16;124(8):347–349. [PubMed] [Google Scholar]
- Brown T. L., Yet M. G., Wold F. Substrate-containing gel electrophoresis: sensitive detection of amylolytic, nucleolytic, and proteolytic enzymes. Anal Biochem. 1982 May 1;122(1):164–172. doi: 10.1016/0003-2697(82)90266-4. [DOI] [PubMed] [Google Scholar]
- CURTIS S. R., 3rd CHROMOSOMAL ABERRATIONS ASSOCIATED WITH MUTATIONS TO BACTERIOPHAGE RESISTANCE IN ESCHERICHIA COLI. J Bacteriol. 1965 Jan;89:28–40. doi: 10.1128/jb.89.1.28-40.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua G. K., Bushuk W. Purification of wheat proteases by affinity chromatography on hemoglobin-Sepharose column. Biochem Biophys Res Commun. 1969 Oct 22;37(3):545–550. doi: 10.1016/0006-291x(69)90950-4. [DOI] [PubMed] [Google Scholar]
- Clark-Curtiss J. E., Jacobs W. R., Docherty M. A., Ritchie L. R., Curtiss R., 3rd Molecular analysis of DNA and construction of genomic libraries of Mycobacterium leprae. J Bacteriol. 1985 Mar;161(3):1093–1102. doi: 10.1128/jb.161.3.1093-1102.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L., Carbon J. Biochemical construction and selection of hybrid plasmids containing specific segments of the Escherichia coli genome. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4361–4365. doi: 10.1073/pnas.72.11.4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J., Hohn B. Cosmids: a type of plasmid gene-cloning vector that is packageable in vitro in bacteriophage lambda heads. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4242–4246. doi: 10.1073/pnas.75.9.4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowman R. A., Perrella M. M., Fitzgerald R. J. Caseinolytic and glyoprotein hydrolase activity of Streptococcus mutans. J Dent Res. 1976 May-Jun;55(3):391–399. doi: 10.1177/00220345760550031701. [DOI] [PubMed] [Google Scholar]
- Curtiss R., 3rd Genetic analysis of Streptococcus mutans virulence. Curr Top Microbiol Immunol. 1985;118:253–277. doi: 10.1007/978-3-642-70586-1_14. [DOI] [PubMed] [Google Scholar]
- Dewar M. D., Walker G. J. Metabolism of the polysaccharides of human dental plaque. I. Dextranase activity of streptococci, and the extracellular polysaccharides synthesized from sucrose. Caries Res. 1975;9(1):21–35. doi: 10.1159/000260139. [DOI] [PubMed] [Google Scholar]
- Ellis D. W., Miller C. H. Extracellular dextran hydrolase from Streptococcus mutans strain 6715. J Dent Res. 1977 Jan;56(1):57–69. doi: 10.1177/00220345770560011301. [DOI] [PubMed] [Google Scholar]
- Felgenhauer B., Trautner K. A comparative study of extracellular glucanhydrolase and glucosyltransferase enzyme activities of five different serotypes of oral Streptococcus mutans. Arch Oral Biol. 1982;27(6):455–461. doi: 10.1016/0003-9969(82)90084-x. [DOI] [PubMed] [Google Scholar]
- Fitzgerald R. J., Spinell D. M., Stoudt T. H. Enzymatic removal of artificial plaques. Arch Oral Biol. 1968 Jan;13(1):125–128. doi: 10.1016/0003-9969(68)90042-3. [DOI] [PubMed] [Google Scholar]
- Freedman M. L., Guggenheim B. Dextran-induced aggregation in a mutant of Streptococcus sobrinus 6715-13. Infect Immun. 1983 Jul;41(1):264–274. doi: 10.1128/iai.41.1.264-274.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germaine G. R., Harlander S. K., Leung W. L., Schachtele C. F. Streptococcus mutans dextransucrase: functioning of primer dextran and endogenous dextranase in water-soluble and water-insoluble glucan synthesis. Infect Immun. 1977 May;16(2):637–648. doi: 10.1128/iai.16.2.637-648.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons R. J., Houte J. V. Bacterial adherence in oral microbial ecology. Annu Rev Microbiol. 1975;29:19–44. doi: 10.1146/annurev.mi.29.100175.000315. [DOI] [PubMed] [Google Scholar]
- Gibbons R. J., van Houte J. On the formation of dental plaques. J Periodontol. 1973 Jun;44(6):347–360. doi: 10.1902/jop.1973.44.6.347. [DOI] [PubMed] [Google Scholar]
- Grahame D. A., Mayer R. M. The origin and composition of multiple forms of dextransucrase from Streptococcus sanguis. Biochim Biophys Acta. 1984 Apr 27;786(1-2):42–48. doi: 10.1016/0167-4838(84)90151-1. [DOI] [PubMed] [Google Scholar]
- Guggenheim B., Burckhardt J. J. Isolation and properties of a dextranase from streptococcus mutans OMZ 176. Helv Odontol Acta. 1974 Oct;18(2):101–113. [PubMed] [Google Scholar]
- Guggenheim B. Enzymatic hydrolysis and structure of water-insoluble glucan produced by glucosyltransferases from a strain of streptococcus mutans. Helv Odontol Acta. 1970 Nov;14(Suppl):89+–89+. [PubMed] [Google Scholar]
- Hamada S., Mizuno J., Murayama Y., Ooshima Y., Masuda N. Effect of dextranase on the extracellular polysaccharide synthesis of Streptococcus mutans; chemical and scanning electron microscopy studies. Infect Immun. 1975 Dec;12(6):1415–1425. doi: 10.1128/iai.12.6.1415-1425.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada S., Slade H. D. Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol Rev. 1980 Jun;44(2):331–384. doi: 10.1128/mr.44.2.331-384.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn B., Collins J. A small cosmid for efficient cloning of large DNA fragments. Gene. 1980 Nov;11(3-4):291–298. doi: 10.1016/0378-1119(80)90069-4. [DOI] [PubMed] [Google Scholar]
- Holt R. G., Abiko Y., Saito S., Smorawinska M., Hansen J. B., Curtiss R., 3rd Streptococcus mutans genes that code for extracellular proteins in Escherichia coli K-12. Infect Immun. 1982 Oct;38(1):147–156. doi: 10.1128/iai.38.1.147-156.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs W. R., Barrett J. F., Clark-Curtiss J. E., Curtiss R., 3rd In vivo repackaging of recombinant cosmid molecules for analyses of Salmonella typhimurium, Streptococcus mutans, and mycobacterial genomic libraries. Infect Immun. 1986 Apr;52(1):101–109. doi: 10.1128/iai.52.1.101-109.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura T., Shockman G. D. Purification and some properties of the endogenous, autolytic N-acetylmuramoylhydrolase of Streptococcus faecium, a bacterial glycoenzyme. J Biol Chem. 1983 Aug 10;258(15):9514–9521. [PubMed] [Google Scholar]
- Koga T., Inoue M. Effects of dextranases on cell adherence, glucan-film formation and glucan synthesis by Streptococcus mutans glucosyltransferase. Arch Oral Biol. 1979;24(3):191–198. doi: 10.1016/0003-9969(79)90139-0. [DOI] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Larrimore S., Murchison H., Shiota T., Michalek S. M., Curtiss R., 3rd In vitro and in vivo complementation of Streptococcus mutans mutants defective in adherence. Infect Immun. 1983 Nov;42(2):558–566. doi: 10.1128/iai.42.2.558-566.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodish H. F. Secondary structure of bacteriophage f2 ribonucleic acid and the initiation of in vitro protein biosynthesis. J Mol Biol. 1970 Jun 28;50(3):689–702. doi: 10.1016/0022-2836(70)90093-8. [DOI] [PubMed] [Google Scholar]
- McCabe M. M., Hamelik R. M. Multiple forms of dextran-binding proteins from Streptococcus mutans. Adv Exp Med Biol. 1978;107:749–759. doi: 10.1007/978-1-4684-3369-2_84. [DOI] [PubMed] [Google Scholar]
- McCabe M. M., Hamelik R. M., Smith E. E. Purification of dextran-binding protein from cariogenic Streptococcus mutans. Biochem Biophys Res Commun. 1977 Sep 9;78(1):273–278. doi: 10.1016/0006-291x(77)91250-5. [DOI] [PubMed] [Google Scholar]
- McGhee J. R., Michalek S. M. Immunobiology of dental caries: microbial aspects and local immunity. Annu Rev Microbiol. 1981;35:595–638. doi: 10.1146/annurev.mi.35.100181.003115. [DOI] [PubMed] [Google Scholar]
- Mullins J. I., Casey J. W., Nicolson M. O., Burck K. B., Davidson N. Sequence arrangement and biological activity of cloned feline leukemia virus proviruses from a virus-productive human cell line. J Virol. 1981 May;38(2):688–703. doi: 10.1128/jvi.38.2.688-703.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison H., Larrimore S., Curtiss R., 3rd Isolation and characterization of Streptococcus mutans mutants defective in adherence and aggregation. Infect Immun. 1981 Dec;34(3):1044–1055. doi: 10.1128/iai.34.3.1044-1055.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison H., Larrimore S., Hull S., Curtiss R., 3rd Isolation and characterization of Streptococcus mutans mutants with altered cellular morphology or chain length. Infect Immun. 1982 Oct;38(1):282–291. doi: 10.1128/iai.38.1.282-291.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T., Munoz L., Doi R. H. A procedure to remove protease activities from Bacillus subtilis sporulating cells and their crude extracts. Anal Biochem. 1977 Mar;78(1):165–170. doi: 10.1016/0003-2697(77)90020-3. [DOI] [PubMed] [Google Scholar]
- Reimerdes E. H., Klostermeyer H. Determination of proteolytic activities on casein substrates. Methods Enzymol. 1976;45:26–28. doi: 10.1016/s0076-6879(76)45005-x. [DOI] [PubMed] [Google Scholar]
- Schachtele C. F., Staat R. H., Harlander S. K. Dextranases from oral bacteria: inhibition of water-insoluble glucan production and adherence to smooth surfaces by Streptococcus mutans. Infect Immun. 1975 Aug;12(2):309–317. doi: 10.1128/iai.12.2.309-317.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg N., Tiemeier D., Enquist L. In vitro packaging of a lambda Dam vector containing EcoRI DNA fragments of Escherichia coli and phage P1. Gene. 1977 May;1(3-4):255–280. doi: 10.1016/0378-1119(77)90049-x. [DOI] [PubMed] [Google Scholar]
- Takahashi T., Tang J. Cathepsin D from porcine and bovine spleen. Methods Enzymol. 1981;80(Pt 100):565–581. doi: 10.1016/s0076-6879(81)80045-6. [DOI] [PubMed] [Google Scholar]
- Terleckyj B., Willett N. P., Shockman G. D. Growth of several cariogenic strains of oral streptococci in a chemically defined medium. Infect Immun. 1975 Apr;11(4):649–655. doi: 10.1128/iai.11.4.649-655.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G. J., Pulkownik A., Morrey-Jones J. G. Metabolism of the polysaccharides of human dental plaque: release of dextranase in batch cultures of Streptococcus mutans. J Gen Microbiol. 1981 Nov;127(1):201–208. doi: 10.1099/00221287-127-1-201. [DOI] [PubMed] [Google Scholar]
- Walker G. J. Some properties of a dextranglucosidase isolated from oral streptococci and its use in studies on dextran synthesis. J Dent Res. 1972 Mar-Apr;51(2):409–414. doi: 10.1177/00220345720510022901. [DOI] [PubMed] [Google Scholar]
- Wenham D. G., Davies R. M., Cole J. A. Insoluble glucan synthesis by mutansucrase as a determinant of the cariogenicity of Streptococcus mutans. J Gen Microbiol. 1981 Dec;127(2):407–415. doi: 10.1099/00221287-127-2-407. [DOI] [PubMed] [Google Scholar]