Abstract
BACKGROUND
Increasing rates of methicillin-resistant Staphylococcus aureus (MRSA) infections on a global scale is a major health concern. In Canada, there are 10 known epidemic types of MRSA as determined by pulsed-field gel electrophoresis (PFGE). Despite the excellent discriminatory power of PFGE, there are several disadvantages of using this technique, such as high degree of labour intensity and the inability to easily develop an MRSA typing database due to the subjective interpretation of results.
OBJECTIVES
The purpose of the present study was to determine whether spa typing, an established DNA sequence-based typing method, could be used as an alternative to PFGE for the typing of Canadian MRSA (CMRSA) epidemic isolates.
RESULTS
spa types were determined for 1488 CMRSA isolates, and the method was analyzed for its ability to identify and cluster CMRSA1-10 strains. Minimal spanning tree analysis of 1452 spa types revealed individual clonal clusters for PFGE epidemic types CMRSA1, 2, 7 and 8, but spa typing could not distinguish CMRSA5 from CMRSA9 and CMRSA10, and CMRSA3 from CMRSA4 and CMRSA6. However, specific spa types were generally associated with only one PFGE epidemic type. Based on these results, a spa typing guideline for CMRSA isolates was developed and tested using the first 300 MRSA isolates received in 2007 through the Canadian Nosocomial Infection Surveillance Program.
CONCLUSIONS
The high concordance of spa types with PFGE epidemic types using this guideline demonstrated the feasibility of spa typing as a more rapid and less technically demanding alternative typing method for MRSA in Canada.
Keywords: Canada, CNISP, Methicillin-resistant Staphylococcus aureus, Pulsed-field gel electrophoresis, spa typing
Abstract
HISTORIQUE
Le taux croissant d’infections à Staphylococcus aureus méthicillinorésistant (ou MRSA, pour Methicillin-Resistant Staphylococcus Aureus) à l’échelle mondiale est un important problème de santé publique. Au Canada, on dénombre dix types épidémiques connus de MRSA déterminés par électrophorèse en champ pulsé (ÉCP). Malgré l’excellent pouvoir discriminant de l’ÉCP, cette technique comporte plusieurs inconvénients, notamment son coefficient de main-d’œuvre et la difficulté de mettre facilement au point une base de données pour le typage du MRSA en raison de l’interprétation suggestive des résultats.
OBJECTIF
Le but de la présente étude était de déterminer si le typage du gène spa, méthode éprouvée établie selon les séquences d’ADN, pourrait être utilisé à titre de solution de rechange à l’ÉCP pour le typage des isolats épidémiques de MRSA au Canada (CMRSA).
RÉSULTATS
Les types spa ont été déterminés pour 1 488 isolats de CMRSA et la méthode a été analysée sur le plan de sa capacité à identifier et à regrouper les souches 1 à 10 de CMRSA. L’analyse de l’arbre maximal-minimal de 1 452 types spa a révélé des complexes clonaux individuels pour les types épidémiques de CMRSA 1, 2, 7 et 8, déterminés par ÉCP, mais le typage spa n’a pas permis de distinguer le CMRSA5, du CMRSA9 et du CMRSA10, ni le CMRSA3 du CMRSA4 et du CMRSA6. Par contre, les types spa spécifiques ont en général été associés à un seul type épidémique déterminé par ÉCP. Selon ces résultats, des directives pour le typage du gène spa appliqués aux isolats de CMRSA ont été mises au point et testées sur les 300 premiers isolats de MRSA reçus en 2007 par l’entremise du Programme canadien de surveillance des infections nosocomiales
CONCLUSION
La forte concordance des types spa et des types épidémiques déterminés par ÉCP à l’aide de cette directive a démontré l’applicabilité du typage spa à titre de méthode de rechange plus rapide et moins exigeante sur le plan technique pour le typage du MRSA au Canada.
Methicillin-resistant Staphylococcus aureus (MRSA) has had a dramatic impact on the health care systems of many countries worldwide. With rates of MRSA infections steadily increasing each year, it has become important to quickly distinguish strains of MRSA for the purpose of epidemiological investigation. MRSA isolates are generally characterized by pulsed-field gel electrophoresis (PFGE), which is a proven and very powerful technique for determining the degree of relatedness among isolates (1,2). The Canadian PFGE typing scheme and identification of Canadian ‘epidemic’ strains has allowed comparison with international MRSA clones (1,3).
In Canada, a 13-year national surveillance study through the Canadian Nosocomial Infection Surveillance Program (CNISP) has revealed 10 Canadian MRSA (CMRSA1-10) epidemic PFGE fingerprint clusters (1,3). In the early stages of surveillance, CMRSA1 (resembling USA600) was the most prevalent (1), but it is quickly being replaced by CMRSA2 (resembling USA100/USA800/New York), which accounted for approximately 55% of all strains in 2004 (3). CMRSA3 has virtually disappeared, being replaced by the closely related CMRSA6 (1,3). CMRSA8 is genetically similar to EMRSA15, a common European epidemic strain type (4). PFGE patterns of CMRSA7 and CMRSA10 were indistinguishable from USA400/MW2 and USA300, respectively, which are strain types linked with community-associated MRSA (CA-MRSA) outbreaks in Canada and the United States (5,6). The occurrence of CMRSA7 and CMRSA10 has increased in Canada over the past few years; however, CMRSA10 has become the more prevalent of the two (3). This has also been observed in the United States, where USA300 is more prevalent than USA400 (7).
Despite the excellent discriminatory power of PFGE, there are several disadvantages of using this technique, such as long wait times to obtain final results, a high degree of labour intensity, lack of standardization between laboratories and the inability to easily develop an MRSA typing database due to the subjective interpretation of results associated with gel-based systems (8,9). The desire to quickly and easily type MRSA led to the development of spa typing, a DNA sequencing technique that examines the polymorphic X or short-sequence repeat (SSR) region of the protein A gene (spa) (8). The SSR region consists of a variable number of short-sequence repeats, usually 24 base pairs long, and is located immediately upstream of the region encoding the C-terminal cell wall attachment sequence. Changes in the SSR region arise due to deletions, duplications and point mutations, which results in a diverse collection of ‘spa types’, where each spa type consists of a specific combination of SSRs (10). Advantages of DNA sequencing as a method for strain typing include speed, ease of use, unambiguous data interpretation and the ability to easily develop an MRSA typing database.
The present study examines the validity and feasibility of using spa typing for national surveillance by comparing PFGE and spa types for 1488 CMRSA isolates obtained from across Canada over a 13-year time period. Results from the present study were used to create a preliminary guideline for Canadian laboratories interested in using spa typing as a technically less demanding method to identify CMRSA isolates.
METHODS
Bacterial isolates
A total of 1488 CMRSA isolates were selected from a collection of over 14,000 MRSA isolates obtained through the CNISP (1) and through routine services performed at the National Microbiology Laboratory (Winnipeg, Manitoba). Isolates were chosen on the basis of unique PFGE patterns that were associated with one of the 10 CMRSA strains (3). When possible, multiple isolates of the same epidemic PFGE pattern were chosen by varying geographical distribution and/or time of isolation. The samples were collected over a 13-year time span, between 1995 and 2007, and from across Canada (245 from British Columbia, 191 from Alberta, 83 from Saskatchewan, 119 from Manitoba, 471 from Ontario, 189 from Quebec, 78 from Newfoundland, 48 from New Brunswick, 50 from Nova Scotia and 14 from Prince Edward Island).
spa sequencing
DNA was prepared using a glass bead DNA extraction method. Briefly, a 1 μL loop was used to collect colonies from an overnight culture grown on Tryptic Soy agar plates containing 5% sheep blood, and then emulsified in a microfuge tube containing 600 μL neutralization buffer (500 mL stock solution [15 mL 1 M Tris-HCl pH 8.4, 2 mL 0.5 M EDTA pH 9 and 483 mL deionized distilled water]) and 50 μL of cell disruption media (0.1 mM) (Scientific Industries Inc, USA). The samples were heated for approximately 3 min at 95°C to 100°C in a heating block, followed by disruption of cells for 3 min using a Genie vortex adaptor (Ambion, USA). The samples were then centrifuged at 3000 rpm for 1 min. Five microlitres of the supernatant was used as the template for the polymerase chain reactions (PCRs). PCR amplification of the spa repeat region was performed as previously described using primers, spa-1113f (5′-TAA AGA CGA TCC TTC GGT GAG C-3′) and spa-1514r (5′-CAG CAG TAG TGC CGT TTG CTT-3′) (10). For individual PCRs, amplicons were purified using YM-100 Microcon filtres (Millipore, USA). For high throughput, Agencourt AMPure Reagent (Agencourt Bioscience Corporation, USA) was used to purify amplicons in a 96-well format. Purified amplicons were sequenced in-house by the DNA core facility (National Microbiology Laboratory). The DNA sequences of the spa repeat region in both directions were imported as ABI files and analyzed using the spa typing program provided with BioNumerics version 4.6 (Applied Maths Inc, USA). DNA sequences were compared using the spa typing Web sites <http://tools.egenomics.com/public/login.aspx> and <http://www.spaserver.ridom.de>, the latter of which was developed by Ridom GmbH (Germany) and curated by SeqNet.org <http://www.SeqNet.org/> (10).
Cluster analysis was performed using the spa typing module of BioNumerics version 4.6. The default cost matrix was used to correct for the evolutionary distances between the repeats. Cluster analysis settings were set to 400% gap creation cost, 70% gap extension cost, 25% duplicate creation cost, 25% duplicate extension and a maximum duplication length of three repeats. The minimal spanning tree was calculated from the distance matrix using the default distance bin size of 100%. A total of 36 isolates had repeat successions (successions less than five), and were excluded from the cluster analysis as previously discussed (11).
PFGE
PFGE was performed as previously described (12). CMRSA1-10 were determined using guidelines as previously described (1,3).
RESULTS
spa typing
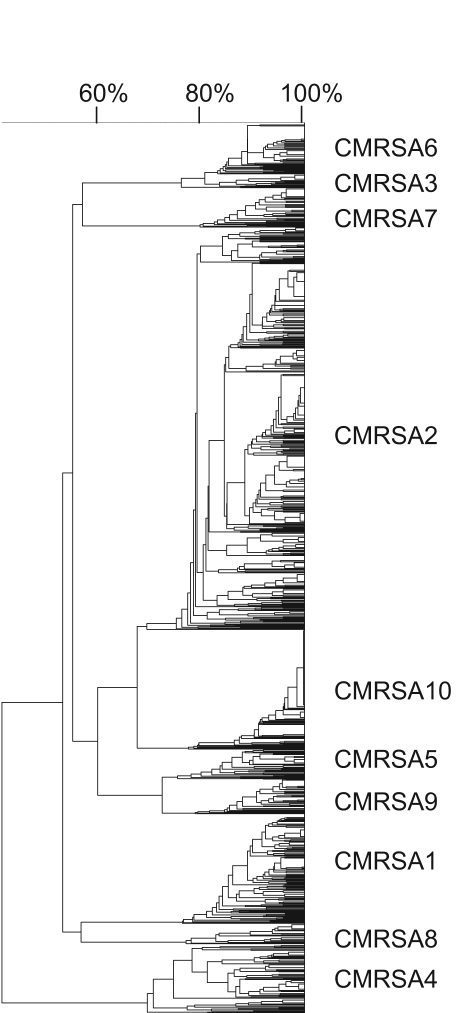
In total, 1488 CMRSA isolates representing 802 unique PFGE patterns (Figure 1) were chosen for spa typing in the present study. Multiple isolates (range two to 119) from 256 indistinguishable PFGE patterns were chosen by varying geographical distribution and/or time of isolation, when possible (refer to Table S1 provided online <www.pulsus.com>).
Figure 1.
Pulsed-field gel electrophoresis dendrogram of 1488 Canadian methicillin-resistant Staphylococcus aureus (CMRSA) isolates (types 1 to 10)
For the 1488 CMRSA isolates tested, spa typing revealed a total of 114 unique spa types averaging eight to nine repeats (Table 1). Comparison of spa types obtained from multiple isolates of 256 indistinguishable PFGE patterns revealed that 167 were associated with only one spa type. The remaining isolates from 89 indistinguishable PFGE patterns contained anywhere between two and five different spa types (Table S1).
TABLE 1.
Preliminary guideline for the assignment of Canadian epidemic types using spa typing (see also Table S1 <www.pulsus.com>). spa types indicated in bold were identified as Canadian methicillin-resistant Staphylococcus aureus (CMRSA) pulsed-field gel electrophoresis (PFGE) epidemic types from the 300 isolates used for validation from the 2007 Canadian Nosocomial Infection Surveillance Program and were subsequently added to the table (see also Table S2 <www.pulsus.com>)
| Epidemic type | Ridom spa type | Number of isolates observed | Unique PFGE types | Ridom repeat succession | Kreiswirth repeat succession |
|---|---|---|---|---|---|
| CMRSA1 (n=183) | t004 | 123 | 85 | r09r02r16r13r13r17r34r16r34 | A2AKEEMBKB |
| t015 | 2 | 2 | r08r16r02r16r34r13r17r34r16r34 | XKAKBEMBKB | |
| t026 | 3 | 2 | r08r16r34 | XKB | |
| t040 | 1 | 1 | r09r02r16r13r17r34r16r34 | A2AKEMBKB | |
| t050 | 1 | 1 | r08r16r02r16r34r34r17r34r16r34 | XKAKBBMBKB | |
| t061 | 5 | 4 | r09r02r16r13r34r17r34r16r34 | A2AKEBMBKB | |
| t065 | 8 | 7 | r09r02r16r34r13r17r34r16r34 | A2AKBEMBKB | |
| t130 | 3 | 2 | r09r34r13r17r34r16r34 | A2BEMBKB | |
| t132 | 1 | 1 | r09r34r16r34 | A2BKB | |
| t1946 | 1 | 1 | r09r34r13r13r17r34r16r34 | A2BEEMBKB | |
| t1996 | 1 | 1 | r08r16r02r16r34r34r34r16r34 | XKAKBBBKB | |
| t266 | 1 | 1 | r09r02r16r13r13r13r17r34r16r34 | A2AKEEEMBKB | |
| t2757 | 1 | 1 | r08r181r34 | X??B | |
| t2758 | 1 | 1 | r08r16r182r16r34r17r34r16r34 | XK??KBMBKB | |
| t282 | 1 | 1 | r09r02r16r34 | A2AKB | |
| t2988 | 1 | 1 | r09r08r16r13r17r34 | A2AKEMB | |
| t3031 | 1 | 1 | r09r02r16r13r13r16r34 | A2AKEEKB | |
| t330 | 2 | 2 | r09r02r16r34r34r17r34r16r34 | A2AKBBMBKB | |
| t362 | 13 | 13 | r09r34 | A2B | |
| t390 | 1 | 1 | r08r16r16r34 | XKKB | |
| t553 | 1 | 1 | r09r13r13r17r34r16r34 | A2EEMBKB | |
| t589 | 2 | 2 | r08r16r02r16r34r34r13r17r34r16r34 | TK | |
| t630 | 7 | 4 | r08r16r02r16r34r17r34r16r34 | XKAKBMBKB | |
| t779 | 1 | 1 | r08 | X | |
| CMRSA2 (n=674) | t002 | 495 | 381 | r26r23r17r34r17r20r17r12r17r16 | TJMBMDMGMK |
| t003 | 14 | 13 | r26r17r20r17r12r17r17r16 | TMDMGMMK | |
| t010 | 4 | 3 | r26r17r34r17r20r17r12r17r16 | TMBMDMGMK | |
| t014 | 4 | 4 | r26r17r20r17r12r17r17r17r16 | TMDMGMMMK | |
| t045 | 23 | 13 | r26r17r20r17r12r17r16 | TMDMGMK | |
| t062 | 7 | 4 | r26r23r17r12r17r16 | TJMGMK | |
| t067 | 11 | 9 | r26r23r17r34r17r20r17r12r17 | TJMBMDMGM | |
| t088 | 7 | 6 | r26r23r17r34r17r20r17r12r12r17r16 | TJMBMDMGGMK | |
| t105 | 3 | 3 | r26r23r17r34r17r20r17r17r16 | TJMBMDMMK | |
| t1080 | 1 | 1 | r26r23r17r34r17r20r17r34r17r16 | TJMBMDMBMK | |
| t111 | 1 | 1 | r26r23r17r16 | TJMK | |
| t1154 | 1 | 1 | r26r20r17r12r17r16 | TDMGMK | |
| t1265 | 1 | 1 | r26r23r17r34r17r20r17r12r12r12r16 | TJMBMDMGGGK | |
| t1781 | 2 | 2 | r26r16r16 | TKK | |
| t179 | 2 | 2 | r26r23r17r34r17r20r17r12r12r16 | TJMBMDMGGK | |
| t1791 | 1 | 1 | r26r23r17r34r17r20r17r20r17r12r17r16 | TJMBMDMDMGMK | |
| t2066 | 1 | 1 | r26r23r17r34r17r20r17r12r17r12r17r16 | TJMBMDMGMGMK | |
| t214 | 2 | 2 | r26r23r17r34r17r20r17r12r17r16r16 | TJMBMDMGMKK | |
| t2212 | 2 | 2 | r26r23r17r34r22r17r12r17r16 | TJMBLMGMK | |
| t2235 | 5 | 5 | r26r23 | TJ | |
| t2302 | 2 | 2 | r26r23r17r34 | TJMB | |
| t2358 | 1 | 1 | r26r23r17r34r17r20r17r12r17r20r17r12r17r16 | TJMBMDMGMDMGMK | |
| t242 | 29 | 28 | r26r23r17r13r17r20r17r12r17r16 | TJMEMDMGMK | |
| t2528 | 2 | 2 | r26r23r17r34r12r12r12r16 | TJMBGGGK | |
| t306 | 1 | 1 | r26r23r17r34r17r20r17r12r17r17r16 | TJMBMDMGMMK | |
| t311 | 21 | 19 | r26r23r17r34r20r17r12r17r16 | TJMBDMGMK | |
| CMRSA2 (n=674) | t450 | 1 | 1 | r26r23r17r34r16 | TJMBK |
| t539 | 15 | 11 | r26r23r17r34r17r12r17r16 | TJMBMGMK | |
| t548 | 6 | 6 | r26r23r17r34r17r20r17r12r16 | TJMBMDMGK | |
| t586 | 2 | 2 | r26r16 | TK | |
| t601 | 1 | 1 | r26r23r17r34r34r17r20r17r12r17r16 | TJMBBMDMGMK | |
| t640 | 1 | 1 | r26r23r17r12r16 | TJMGK | |
| t653 | 2 | 2 | r26r17r12r17r16 | TMGMK | |
| t668 | 1 | 1 | r26r23r17r34r17r20r17r16 | TJMBMDMK | |
| t688 | 1 | 1 | r26r23r17r34r17r16 | TJMBMK | |
| t854 | 1 | 1 | r26r23r17r34r17r20r17r12r16r16 | TJMBMDMGKK | |
| t954 | 1 | 1 | r26r23r17r34r17r17r16 | TJMBMMK | |
| CMRSA3 (n=17) | t030* | 1 | 1 | r15r12r16r02r24r24 | WGKAQQ |
| t037* | 15 | 13 | r15r12r16r02r25r17r24 | WGKAOMQ | |
| t138 | 1 | 1 | r08r16r02r25r17r24 | XKAOMQ | |
| CMRSA4 (n=116) | t007 | 3 | 1 | r15r12r16r16r16r16r02r25r17 | WGKKKKAOM |
| t012 | 38 | 25 | r15r12r16r02r16r02r25r17r24r24 | WGKAKAOMQQ | |
| t018 | 35 | 25 | r15r12r16r02r16r02r25r17r24r24r24 | WGKAKAOMQQQ | |
| t019 | 15 | 8 | r08r16r02r16r02r25r17r24 | XKAKAOMQ | |
| t021 | 7 | 6 | r15r12r16r02r16r02r25r17r24 | WGKAKAOMQ | |
| t037* | 1 | 1 | r15r12r16r02r25r17r24 | WGKAOMQ | |
| t122 | 1 | 1 | r08r16r02r16r02r25r17r24r24 | XKAKAOMQQ | |
| t238 | 1 | 1 | r15r21r12r16r02r16r02r25r17r24r24 | WFGKAKAOMQQ | |
| t253 | 4 | 4 | r15r12r16r02r16r02r25r17r24r24r24r24 | WGKAKAOMQQQQ | |
| t275* | 1 | 1 | r15r12r16r02r25r17r24r24 | WGKAOMQQ | |
| t318 | 3 | 2 | r15r12r16r16r02r16r02r25r17r24 | WGKKAKAOMQ | |
| t323 | 4 | 3 | r15r12r16r02r16r34r17r24r24r24 | WGKAKBMQQQ | |
| t338 | 2 | 1 | r15r21r16r02r25r17r24 | WFKAOMQ | |
| t638 | 1 | 1 | r15r12r16r02r17 | WGKAM | |
| CMRSA5 (n=50) | t008* | 2 | 2 | r11r19r12r21r17r34r24r34r22r25 | YHGFMBQBLO |
| t051 | 1 | 1 | r11r19r21r12r21r17r34r24r34r22r25 | YHFGFMBQBLO | |
| t064 | 37 | 27 | r11r19r12r05r17r34r24r34r22r25 | YHGCMBQBLO | |
| t068 | 1 | 1 | r11r19r19r12r21r17r34r24r34r22r25 | YHHGFMBQBLO | |
| t1171 | 1 | 1 | r11r19r12r17r34r24r34r22r25 | YHGMBQBLO | |
| t190 | 2 | 2 | r11r17r34r24r34r22r25 | YMBQBLO | |
| t451 | 4 | 3 | r11r12r05r17r34r24r34r22r25 | YGCMBQBLO | |
| t460 | 1 | 1 | r11r19r21r17r34r24r24r34r22r25 | YHFMBQQBLO | |
| t951 | 1 | 1 | r11r10r05r17r34r24r34r22r25 | YC2CMBQBLO | |
| CMRSA6 (n=92) | t030* | 1 | 1 | r15r12r16r02r24r24 | WGKAQQ |
| t037* | 87 | 47 | r15r12r16r02r25r17r24 | WGKAOMQ | |
| t074 | 1 | 1 | r15r21r12r16r02r25r17r24 | WFGKAOMQ | |
| t129 | 1 | 1 | r15r12r24 | WGQ | |
| t275* | 1 | 1 | r15r12r16r02r25r17r24r24 | WGKAOMQQ | |
| t388 | 1 | 1 | r15r12r16r02r25r24 | WGKAOQ | |
| CMRSA7 (n=66) | t127 | 5 | 5 | r07r23r21r16r34r33r13 | UJFKBPE |
| t1274 | 1 | 1 | r07r23r21r16r16r33r21r16r33r21r16r33r13 | UJFKKPFKPFKPE | |
| t128 | 50 | 21 | r07r23r23r21r16r34r33r13 | UJJFKBPE | |
| t1508 | 1 | 1 | r15r16r34r33r13 | WKBPE | |
| t175 | 7 | 6 | r07r23r21r16r16r33r21r16r33r13 | UJFKKPFKPE | |
| t1784 | 1 | 1 | r07r34r33r13 | UBPE | |
| t2207 | 1 | 1 | r07r33r13 | UPE | |
| CMRSA7 (n=66) | t2593 | 1 | 1 | r07r23r21r34r33r13 | UJFBPE |
| t555 | 1 | 1 | r07r23r23r23r21r16r34r33r13 | UJJJFKBPE | |
| CMRSA8 (n=34) | t005 | 1 | 1 | r26r23r13r23r31r05r17r25r17r25r16r28 | TJEJNCMOMOKR |
| t020 | 1 | 1 | r26r23r31r29r17r31r29r17r25r17r25r16r28 | TJNF2MNF2MOMOKR | |
| t022 | 8 | 7 | r26r23r13r23r31r29r17r31r29r17r25r17r25r16r28 | TJEJNF2MNF2MOMOKR | |
| t032 | 17 | 13 | r26r23r23r13r23r31r29r17r31r29r17r25r17r25r16r28 | TJJEJNF2MNF2MOMOKR | |
| t1465 | 1 | 1 | r26r23r23r13r23r31r29r17r31r29r17r24r25r17r25r16r28 | TJJEJNF2MNF2MQOMOKR | |
| t2618 | 1 | 1 | r26r23r13r23r31r05r17r25r17r25r17r25r16r28 | TJEJNCMOMOMOKR | |
| t2818 | 1 | 1 | r26r23r23r13r23r31r29r17r31r29r17r25r17r24r25r16r28 | TJJEJNF2MNF2MOMQOKR | |
| t515 | 1 | 1 | r26r23r23r13r23r31r29r17r31r29r17r25r16r16r28 | TJJEJNF2MNF2MOKKR | |
| t578 | 2 | 2 | r26r23r23r13r23r31r29r17r31r29r17r25r17r25r28 | TJJEJNF2MNF2MOMOR | |
| t891 | 1 | 1 | r26r23r13r23r31r05r17r25r17r25r28 | TJEJNCMOMOR | |
| CMRSA9 (n=58) | t008* | 57 | 35 | r11r19r12r21r17r34r24r34r22r25 | YHGFMBQBLO |
| t955 | 1 | 1 | r11r19r12r21r17r34r24r34r22r33r25 | YHGFMBQBLPO | |
| CMRSA10 (n=198) | t008* | 186 | 37 | r11r19r12r21r17r34r24r34r22r25 | YHGFMBQBLO |
| t024 | 4 | 1 | r11r12r21r17r34r24r34r22r25 | YGFMBQBLO | |
| t068 | 1 | 1 | r11r19r19r12r21r17r34r24r34r22r25 | YHHGFMBQBLO | |
| t1705 | 1 | 1 | r11r34r24r34r22r25 | YBQBLO | |
| t1767 | 1 | 1 | r11r19r12r21r17r34r24r24r34r22r25 | YHGFMBQQBLO | |
| t2067 | 1 | 1 | r11r19r12r21r66r34r24r34r22r25 | YHGFF4BQBLO | |
| t3023 | 2 | 1 | r11r19r12r197r17r34r24r34r22r25r25 | YHG??MBQBLOO | |
| t3135 | 1 | 1 | r11r19r34r24r34r24r34r22r25 | YHBQBQBLO | |
| t3154 | 1 | 1 | r11r19r12r21r17r34r24r05r25 | YHGFMBQCO | |
| t351 | 1 | 1 | r11r19r12r21r22r25 | YHGFLO | |
| t574 | 2 | 1 | r11r19r12r12r34r24r34r22r25 | YHGGBQBLO | |
| t723 | 1 | 1 | r11r19r12r34r22r25 | YHGBLO | |
| t818 | 1 | 1 | r11r19r12r21r17r34 | YHGFMB |
Refer to discussion section for assignment of spa types t008, t030, t037 and t275 to a CMRSA epidemic type
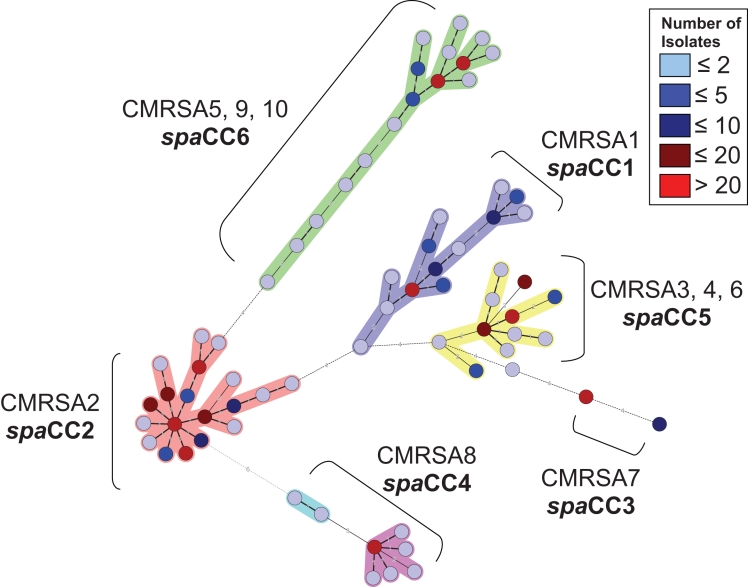
Cluster analysis, based on the degree of similarity between spa repeat successions, was performed for 1452 isolates using the spa minimal spanning tree application of BioNumerics version 4.6. This analysis revealed six spa clonal complexes (spaCC), which were arbitrarily named spaCC1-6 (Figure 2). spaCC1 contained only isolates with PFGE epidemic fingerprint types CMRSA1, spaCC2 contained only isolates with PFGE epidemic fingerprint types CMRSA2, spaCC3 contained only isolates with PFGE epidemic fingerprint types CMRSA7 and spaCC4 contained only isolates with PFGE epidemic fingerprint types CMRSA8. The remaining two clusters consisted of a mixture of PFGE types, which included isolates of PFGE epidemic fingerprint types CMRSA3, 4, and 6 in spaCC5 and CMRSA5, 9 and 10 in spaCC6 (Figure 1).
Figure 2.
Minimal spanning tree analysis for the spa types of 1452 Canadian methicillin-resistant Staphylococcus aureus (CMRSA) isolates (types 1 to 10). Clusters of related spa types were arbitrarily assigned clonal complexes (spaCC1-6) and are indicated in bold. Pulsed-field gel electrophoresis epidemic types (CMRSA1-10) associated with each spaCC is also indicated
Despite the inability to clearly differentiate all PFGE epidemic types using spa clustering (Figure 2), the majority of individual spa types within these clusters (110 of 114) were only associated with a single PFGE epidemic type, which was even noted for spaCC4 (Table 1). On the basis of these results, a table is provided to be used as a preliminary guideline for spa typing of CMRSA epidemic isolates (Table 1).
Validation of the spa guideline
The first 300 MRSA isolates received in 2007 through the CNISP were used to validate the spa typing guideline. Resolved spa types and PFGE banding patterns were independently assigned CMRSA epidemic types and were then compared (refer to Table S2 provided online <www.pulsus.com>). Of the 300 isolates, 285 were assigned Canadian epidemic types (CMRSA1-10) using spa typing and the guideline alone. Of the 15 isolates that were not assigned epidemic types, eight contained spa types that were not listed in the guideline, but based on similarities of repeat successions; Panton-Valentine leukocidin (PVL) results were assigned to their corresponding PFGE epidemic types (CMRSA10 [t068, t1767, t2067, t3135 and t3154], CMRSA2 [t688] and CMRSA7 [t2593 and t555]) (Tables 1 and S2). Six isolates did not cluster with any of the 10 Canadian epidemic spa types, which included four isolates with spa types t216, t186 and t901. The other two were spa type t437, which is associated with a ST59 pandemic CA-MRSA strain (also referred to as USA1000/WA MRSA15), and one with t044, which is associated with a ST80 pandemic CA-MRSA strain (also referred to as the ‘European CA-MRSA clone’) (Table S2). The last nonassigned epidemic isolate was spa type t008 and was PVL-negative and, therefore, required PFGE for designation.
Of the same 300 isolates, 283 were assigned Canadian epidemic types (CMRSA1-10) using PFGE. PFGE was consistent for the six non-CMRSA epidemic isolates predicted using spa typing (Table S2). However, the PFGE banding pattern for the remaining 11 isolates clustered within a PFGE epidemic type, but they were not assigned to that epidemic cluster because they differed by seven or more bands from the epidemic type strain. These 11 nonepidemic-assigned isolates clustered within the same PFGE epidemic fingerprint types that were predicted using spa typing (Table S2).
Cost and time comparison of spa and PFGE
To compare the costs and required time for either spa typing or PFGE, a number of assumptions had to be made. For PFGE, the availability of two PFGE units for MRSA was assumed and for spa typing, in-house capability of DNA sequencing was assumed. Using these assumptions, the cost and time required to process 48 MRSA isolates were compared. For spa typing, the time required for DNA extraction, PCR, amplicon cleanup, DNA sequencing and the analysis of 48 isolates was estimated to be approximately 13 h. Whereas for PFGE, the time required to prepare and digest plugs, load and run gels, stain and destain gels, and the analysis of 48 isolates was estimated to be approximately 58 h. Estimated ‘hands-on’ technical times were approximately 6 h for spa typing and 12 h for PFGE. The estimated cost for spa typing of a single isolate was approximately between $9.50 and $12.00. Nonaccess to in-house DNA sequencing would obviously add to the cost and turnaround time of spa typing, but could be worth the increased objectivity of the analysis. The estimated cost of PFGE for a single isolate was approximately between $9.50 and $10.00. The following estimates will vary depending on laboratory capabilities and availability of equipment, but were used as baselines to compare the two methods.
DISCUSSION
spa typing and PFGE comparison
CMRSA isolates (n=1488) representing 802 unique PFGE patterns (Figure 1) were chosen for spa typing in the present study. A comparison of spa types and unique PFGE patterns revealed that spa types were not confined to a specific PFGE banding pattern, and that indistinguishable PFGE patterns could contain anywhere from one to at least five different spa types. As a result, spa typing could not reliably be used as an indicator of a specific PFGE fingerprint type. However, closer examination of the repeat successions of the spa region for these isolates were similar (Table 1) and, therefore, a less discriminatory approach involving the clustering of related spa and PFGE epidemic types was examined.
Analysis of the clustering of related spa types revealed that two of the spa clonal clusters contained multiple PFGE epidemic types and, therefore, could not be used for the specific classification of PFGE epidemic types. The clustering of PFGE epidemic types CMRSA5, 9 and 10 in spaCC6 (Figure 2) was not surprising because a dendrogram generated from the PFGE fingerprint types of these strains indicated that they were closely related (Figure 1). Secondly, all three PFGE epidemic types, CMRSA5, 9 and 10, have the same multilocus sequence typing (MLST) designation (ST8) (Table 2). However, the clustering of PFGE epidemic types CMRSA4 with CMRSA3 and CMRSA6 in spaCC5 was initially surprising (Figure 2). CMRSA3 and CMRSA6 are genetically similar by both MLST (Table 2) and PFGE (Figure 1), and would therefore be expected to be closely related. However, CMRSA4 is distantly related to both CMRSA3 and CMRSA6 when comparing these epidemic types in the PFGE dendrogram (Figure 1). In addition, CMRSA4 belongs to a different MLST CC in comparison with CMRSA3 and CMRSA6 (Table 2). Previously observed ‘violations’ of MLST ST assignment by spa typing have been attributed to intergenomic recombination involving the small region of the chromosome that spa typing focuses on (13). This was previously shown for lineages belonging to MLST CC8 and CC30 (14), and likely explains the clustering of spa types obtained from CMRSA4 with CMRSA3 and CMRSA6. As a result, isolates identified as unrelated by MLST, could be seen as related by spa typing because they would both share the same repeats in the polymorphic X region of the protein A gene. It is also interesting to note that CMRSA3 and CMRSA6 do not cluster with any of the MLST CC8 and CC30 lineages, thereby suggesting that PFGE is also unable to correctly classify recombinants from multiple genetic backgrounds. These violations cannot be resolved by spa typing alone, but are observed when both spa typing and MLST analyses are used.
TABLE 2.
International comparison of spa types commonly associated with Canadian methicillin-resistant Staphylococcus aureus strains 1–10 (CMRSA1-10)
| Ridom spa type | Canadian epidemic type | MLST (CC) | Other associated MRSA clones* |
|---|---|---|---|
| t002 | CMRSA2 | ST5 (CC5) | CC5, Rhine Hesse MRSA (prototype), EMRSA-3, New York clone, Japan clone, Pediatric, USA100 ORSA II, USA800 ORSA IV, ST 5 ORSA I |
| t004 | CMRSA1 | ST45 (CC45) | CC45, Berlin MRSA (prototype), USA600 ORSA II, USA600 ORSA IV |
| t008 | CMRSA9, CMRSA10 | ST8 (CC8) | CC8, Northern German MRSA (subclone), USA300 ORSA IV (cMRSA in the United States), Archaic/Iberian, ST250 ORSA I |
| t018 | CMRSA4 | ST36 (CC30) | CC30, prototype of ST-36, EMRSA-16, USA200 ORSA II |
| t032 | CMRSA8 | ST22 (CC22) | Barnim MRSA (prototype & subclone), EMRSA-15, prototype of ST-22, CC22 |
| t037 | CMRSA3 | ST241 (CC8) | CC8/239, Vienna MRSA, Brazilian/Hungarian, ST239 ORSA III, ST240 ORSA III, EMRSA-1, -4, -7, -9, -11 |
| t064 | CMRSA5 | ST8 (CC8) | CC8, Archaic/Iberian, USA500 ORSA IV, USA500 ORSA II, ST8 ORSA I, ST8 ORSA IV, ST8 ORSA III |
| t128 | CMRSA7 | ST1 (CC1) | ST1 (related to USA400/MW2) |
Obtained from the Ridom Web site <http://www.spaserver.ridom.de> (10). CC Clonal complex; cMRSA Community-acquired MRSA; MLST Multilocus sequence typing
Despite the inability to assign epidemic types on the basis of spa clustering analysis, small genetic differences within the spa region still enabled us to differentiate a CMRSA4 isolate from a CMRSA3 or a CMRSA6 isolate, which could be the result of micro- and/or macroevolution of this small chromosomal region over time following the recombination event (15).
Assignment of Canadian epidemic types using spa typing
In the present study, and many others (13,15,16), spa typing displayed excellent concordance with PFGE fingerprint clusters. For the four spa types, t008, t030, t037 and t275, where multiple PFGE epidemic types were found, additional molecular typing would be required for classification. For instance, spa type t008 was associated with three different PFGE epidemic types of CMRSA (CMRSA5, CMRSA9 and CMRSA10). However, of these three epidemic types, only CMRSA10 is known to carry PVL. Therefore, for isolates with spa type t008, an additional PCR reaction is proposed to detect the presence or absence of PVL. A PVL-positive result for a spa type t008 isolate would likely differentiate CMRSA10 from CMRSA5 and CMRSA9 isolates and could, therefore, be reported as CMRSA10. For PVL-negative strains, a PFGE is proposed for identification of CMRSA5, CMRSA9 or a PVL-negative CMRSA10 isolate. To date, CMRSA5 and CMRSA9 strains containing PVL have not been reported, and PVL-negative CMRSA10 isolates in Canada appear to be rare and should, therefore, not result in a large number of samples requiring PFGE.
Although not examined in the present study, additional PCR-based methods for delineating different strains of the same spa type could also include the use of staphylococcal cassette chromosome mec typing (17) or the arginine catabolic mobile element (18), alone or in combination with PVL, as an indicator of CMRSA10 strains.
spa type t037 was associated with three different PFGE epidemic types of CMRSA (CMRSA3, CMRSA4 and CMR-SA6); however, the occurrence of spa type t037 in CMRSA4 was only seen once and might represent a rare recombination event. Both CMRSA3 and CMRSA6 are staphylococcal cassette chromosome mec type III (3), but MLST could be useful in differentiating spa type t037 isolates. However, MLST requires DNA sequencing of seven loci, which is an expensive and time consuming method. PFGE is, therefore, recommended to differentiate MRSA isolates of spa type t037. It should be noted that CMRSA3 has dramatically declined in Canada, and it is, therefore, expected that the majority of t037 isolates would be CMRSA6 (3). PFGE would also be recommended for spa types t030 and t275, which were both seen in two different epidemic PFGE types. To date, these spa types appear to be rare in Canada, having only been seen in four of the 1488 isolates tested, and would, therefore, not be expected to result in large numbers of samples requiring PFGE for identification.
The assignment of CMRSA epidemic types using the provided spa guideline (Table 1) was highly successful for the molecular classification of the first 300 MRSA isolates received in 2007 through the CNISP. The high discriminatory ability of spa typing has been attributed to the small size of repeats, which are more prone to duplication and deletion via slip-strand mispairing, and a high number of synonymous substitutions per synonymous site (dS=0.72) (15). These slow point mutations and fast-occurring changes in the number of repeats enables spa typing to be effectively used in both long- and short-term epidemiologic studies (15). It should be noted that the use of spa typing in an outbreak setting was not assessed in the present study, but has previously been shown to be successful in outbreak investigations (8,10) and early warning systems for the detection of outbreaks (19). It is, therefore, suggested that the combination of PFGE and spa typing would be useful for obtaining additional comparative data in such settings and will be important for the continued national surveillance and reporting of MRSA in Canada. For instance, spa typing of the first 300 CNISP MRSA isolates received in 2007 revealed that performing PFGE alone potentially misclassified 11 isolates (3.6%) as nonepidemic (Table S2). This has identified a potential limitation of the past definition of epidemic CMRSA isolates in Canada, which is the assignment of epidemic types based primarily on comparisons of PFGE banding patterns with one representative epidemic type strain. spa typing of additional nonepidemic-assigned isolates is, therefore, warranted to examine possible alternative definitions of epidemic MRSA types in Canada.
International comparison of spa types
spa types are freely comparable using international databases such as Ridom (http://www.spaserver.ridom.de) (10), which currently contains over 3300 spa types that were obtained from the submission of over 47,000 isolates from 45 different countries. Briefly, spa types of MRSA strains defined as epidemic in Canada were also prevalent around the world (Table 2). Comparison of spa types obtained from the present study to the Ridom database also revealed two spa types, t044 and t437, associated with two pandemic CA-MRSA strains (ST80 ‘European’ and ST59 clones) that have not previously been reported in Canada. The potential emergence of these two CA-MRSA clones in Canada should continue to be monitored.
SUMMARY
The high discriminatory power of PFGE provides an excellent tool for studying short-term outbreaks, and was supported in the present study and others as a useful method for long-term epidemiological surveillance studies. However, PFGE is labour-intensive and technically demanding. The dependence on strict adherence to standardization protocols between laboratories and the subjective interpretation of PFGE results has led to an inability to easily develop MRSA typing databases on a large scale. In comparison with PFGE, spa typing was capable of yielding results faster, enabled the processing of more isolates, did not require any type of subjective interpretation and was comparable in cost with PFGE. The spa data were also easily exportable, and could be uploaded rapidly into databases for ‘real-time’ surveillance of MRSA on a national or international scale. The provided guideline for the assignment of Canadian epidemic PFGE types using spa typing is preliminary and, therefore, a periodically updated table, including newly defined spa types, will be made accessible on-line <www.nml.ca> or available on request.
ACKNOWLEDGEMENTS
Funding was provided by the Public Health Agency of Canada and by the Canadian Institute of Health Research-funded Northern Antibiotic Resistance Partnership. The authors thank Lisa Louie for her critical comments on this manuscript.
Footnotes
MEMBERS OF THE CANADIAN NOSOCOMIAL INFECTION SURVEILLANCE PROGRAM: David Boyd, (National Microbiology Laboratory, Public Health Agency of Canada), Elizabeth Bryce (Vancouver General Hospital, Vancouver, British Columbia), John Conly (Foothills Medical Centre, Calgary, Alberta), Gordon Dow (The Moncton Hospital, Moncton, New Brunswick), John Embil and Joanne Embree (Health Sciences Centre, Winnipeg, Manitoba), Charles Frenette (Hôpital Charles LeMoyne, Longueil, Quebec), Michael Gardam, (University Health Network, Toronto, Ontario), Denise Gravel (Centre for Infectious Disease Prevention and Control, Public Health Agency of Canada), Elizabeth Henderson (Peter Lougheed Centre, Calgary, Alberta), James Hutchinson (Health Sciences Centre, St John’s, Newfoundland), Michael John (London Health Sciences Centre, London, Ontario), Lynn Johnston (Queen Elizabeth II Health Sciences Centre, Halifax, Nova Scotia), Pamela Kibsey (Victoria General Hospital, Victoria, British Columbia), Joanne Langley (IWK Health Science Centre, Halifax, Nova Scotia), Mark Loeb (Hamilton Health Sciences Corporation, Hamilton, Ontario), Anne Matlow (Hospital for Sick Children, Toronto, Ontario), Allison McGeer (Mount Sinai Hospital, Toronto, Ontario), Sophie Michaud (CHUS-Hôpital Fleurimont, Sherbrooke, Quebec), Mark Miller (SMBD-Jewish General Hospital, Montreal, Quebec), Dorothy Moore (Montreal Children’s Hospital, Montreal, Quebec), Michael Mulvey (National Microbiology Laboratory, Public Health Agency of Canada), Marianna Ofner-Agostini (Centre for Infectious Disease Prevention and Control, Public Health Agency of Canada), Shirley Paton (Centre for Infectious Disease Prevention and Control, Public Health Agency of Canada), Virginia Roth (The Ottawa Hospital, Ottawa, Ontario), Andrew Simor (Sunnybrook Health Sciences Centre, Toronto, Ontario), Jacob Stegenga (Centre for Infectious Disease Prevention and Control, Public Health Agency of Canada), Tammy Stuart (Canadian Field Epidemiology Program, Public Health Agency of Canada), Kathryn Suh (Children’s Hospital of Eastern Ontario, Ottawa, Ontario), Geoffrey Taylor (University of Alberta Hospital, Edmonton, Alberta), Eva Thomas (Children’s and Women’s Health Center, Vancouver, British Columbia), Nathalie Turgeon (Hôtel-Dieu de Quebec du CHUQ, Quebec), Mary Vearncombe (Sunnybrook Health Sciences Centre, Toronto, Ontario), Joseph Vayalumkal (Canadian Field Epidemiology Program, Public Health Agency of Canada), Karl Weiss (Maisonneuve-Rosemont Hospital, Montreal, Quebec), Alice Wong (Royal University Hospital, Saskatoon, Saskatchewan) and Dick Zoutman (Kingston General Hospital, Kingston, Ontario).
Supplementary Material
REFERENCES
- 1.Simor AE, Ofner-Agostini M, Bryce E, McGeer A, Paton S, Mulvey MR Canadian Hospital Epidemiology Committee and Canadian Nosocomial Infection Surveillance Program, Health Canada. Laboratory characterization of methicillin-resistant Staphylococcus aureus in Canadian hospitals: Results of 5 years of National Surveillance, 1995–1999. J Infect Dis. 2002;186:652–60. doi: 10.1086/342292. [DOI] [PubMed] [Google Scholar]
- 2.McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: Establishing a national database. J Clin Microbiol. 2003;41:5113–20. doi: 10.1128/JCM.41.11.5113-5120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christianson S, Golding GR, Campbell J, Mulvey MR The Canadian Nosocomial Infection Surveillance Program. Comparative genomics of Canadian epidemic lineages of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2007;45:1904–11. doi: 10.1128/JCM.02500-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richardson JF, Reith S. Characterisation of a strain of methicillin-resistant Staphylococcus aureus (EMRSA-15) by conventional and molecular methods. J Hosp Infect. 1993;25:45–52. doi: 10.1016/0195-6701(93)90007-m. [DOI] [PubMed] [Google Scholar]
- 5.Mulvey MR, MacDougall L, Cholin B, Horsman G, Fidyk M, Woods S Saskatchewan CA-MRSA Study Group. Community-associated methicillin-resistant Staphylococcus aureus, Canada. Emerg Infect Dis. 2005;11:844–50. doi: 10.3201/eid1106.041146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seybold U, Kourbatova EV, Johnson JG, et al. Emergence of community-associated methicillin-resistant Staphylococcus aureus USA300 genotype as a major cause of health care-associated blood stream infections. Clin Infect Dis. 2006;42:647–56. doi: 10.1086/499815. [DOI] [PubMed] [Google Scholar]
- 7.Tenover FC, McDougal LK, Goering RV, et al. Characterization of a strain of community associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. J Clin Microbiol. 2006;44:108–18. doi: 10.1128/JCM.44.1.108-118.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shopsin B, Gomez M, Montgomery SO, et al. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J Clin Microbiol. 1999;37:3556–63. doi: 10.1128/jcm.37.11.3556-3563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trindade PA, McCulloch JA, Oliveira GA, Mamizuka EM. Molecular techniques for MRSA typing: Current issues and perspectives. Braz J Infect Dis. 2003;7:32–43. doi: 10.1590/s1413-86702003000100005. [DOI] [PubMed] [Google Scholar]
- 10.Harmsen D, Claus H, Witte W, et al. Typing of methicillin-resistant Staphylococcus aureus in a university setting by using a novel software for spa repeat determination and database management. J Clin Microbiol. 2003;41:5442–8. doi: 10.1128/JCM.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mellmann A, Weniger T, Berssenbrügge C, et al. Based upon repeat pattern (BURP): An algorithm to characterize the long-term evolution of Staphylococcus aureus populations based on spa polymorphisms. BMC Microbiol. 2007;7:98. doi: 10.1186/1471-2180-7-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mulvey MR, Chui L, Ismail J, et al. Canadian Committee for the Standardization of Molecular Methods. Development of a Canadian standardized protocol for subtyping methicillin-resistant Staphylococcus aureus using pulsed-field gel electrophoresis. J Clin Microbiol. 2001;39:3481–5. doi: 10.1128/JCM.39.10.3481-3485.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cookson BD, Robinson DA, Monk AB, et al. Evaluation of molecular typing methods in characterizing a European collection of epidemic methicillin-resistant Staphylococcus aureus strains: The HARMONY collection. J Clin Microbiol. 2007;45:1830–7. doi: 10.1128/JCM.02402-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson DA, Enright MC. Evolution of Staphylococcus aureus by large chromosomal replacements. J Bacteriol. 2004;186:1060–4. doi: 10.1128/JB.186.4.1060-1064.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koreen L, Ramaswamy SV, Gravis EA, Naidich S, Musser JM, Kreiswirth BN. spa typing method for discriminating among Staphylococcus aureus isolates: Implications for use of a single marker to detect genetic micro- and macrovariation. J Clin Microbiol. 2004;42:792–9. doi: 10.1128/JCM.42.2.792-799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hallin M, Deplano A, Denis O, et al. Validation of pulsed-field gel electrophoresis and spa typing for long-term, nationwide epidemiological surveillance studies of Staphylococcus aureus infections. J Clin Microbiol. 2007;45:127–33. doi: 10.1128/JCM.01866-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oliveira DC, de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46:2155–61. doi: 10.1128/AAC.46.7.2155-2161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goering RV, McDougal LK, Fosheim GE, Bonnstetter KK, Wolter DJ, Tenover FC. Epidemiological distribution of the arginine catabolic mobile element among selected methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates. J Clin Microbiol. 2007;45:1981–4. doi: 10.1128/JCM.00273-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mellmann A, Friedrich AW, Rosenkötter N, et al. Automated DNA sequence-based early warning system for the detection of methicillin-resistant Staphylococcus aureus outbreaks. PLoS Med. 2006;3:e33. doi: 10.1371/journal.pmed.0030033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.