Abstract
Emerging evidence suggests that components of the ubiquitin–proteasome system are involved in the regulation of gene expression. A variety of factors, including transcriptional activators, coactivators, and histones, are controlled by ubiquitylation, but the mechanisms through which this modification can function in transcription are generally unknown. Here, we report that the Saccharomyces cerevisiae protein Asr1 is a RING finger ubiquitin-ligase that binds directly to RNA polymerase II via the carboxyl-terminal domain (CTD) of the largest subunit of the enzyme. We show that interaction of Asr1 with the CTD depends on serine-5 phosphorylation within the CTD and results in ubiquitylation of at least 2 subunits of the enzyme, Rpb1 and Rpb2. Ubiquitylation by Asr1 leads to the ejection of the Rpb4/Rpb7 heterodimer from the polymerase complex and is associated with inactivation of polymerase function. Our data demonstrate that ubiquitylation can directly alter the subunit composition of a core component of the transcriptional machinery and provide a paradigm for how ubiquitin can influence gene activity.
Keywords: transcription, ubiquitin
The correct regulation of gene transcription depends on mechanisms that regulate the formation and dynamics of large multiprotein complexes during various stages of the transcription process. One of the most prominent of these mechanisms is posttranslational protein modification. Phosphorylation is frequently used to promote and stabilize the interaction of various proteins; recruitment of capping and splicing factors to elongating RNA polymerase II (pol II), for example, is signaled by phosphorylation within the carboxyl-terminal domain (CTD) of its largest subunit (1), although modifications such as methylation (2) and acetylation (3) can also influence critical protein–protein interactions. One modification that has received attention in recent years is ubiquitylation, as it has become evident that modification of transcription proteins by ubiquitin (Ub) (4, 5) plays a role in diverse aspects of gene regulation.
Ub is a 76-amino acid protein that is covalently linked to other proteins by the action of an enzymatic cascade, the last step of which is mediated via a Ub-protein ligase (or E3) that recognizes a specific element within the substrate and promotes transfer of Ub to a lysine residue(s) within that protein (6). The utility of ubiquitylation stems from its specificity and its ability to function as either a “classic” modification or as a signal for substrate destruction by the 26S proteasome. By varying the extent of protein ubiquitylation, the type of poly-Ub chains, or the sites of ubiquitylation in the substrate, Ub can act as either a reversible modifier of protein function or an irreversible mechanism for limiting protein levels.
Several recent sets of studies have revealed that ubiquitylation influences multiple steps in the transcription process. Our particular interest has centered on the connection between ubiquitylation of transcriptional activators and the regulation of gene activity, and we and others have proposed that either ubiquitylation or ubiquitylation and destruction (4, 5) are a positive signal for transcriptional activation and may be important for tight regulation of gene activity. A similar “Ub-clock” model has been proposed for transcriptional coactivators (7). In addition to proteolysis, however, there appear to be significant roles for nondestructive ubiquitylation in transcription. Monoubiquitylation of histone H2B, for example, signals methylation of histones H3 and H4 (8). Oligoubiquitylation of the Met-30 transcription factor can regulate its interaction with important transcriptional partners (9). And ubiquitylation can also control recruitment of the mRNA export machinery protein to sites of transcription (10), coordinating transcription with mRNA export. The diversity of the transcriptional processes that are thus far known to be regulated by ubiquitylation suggests that this modification may participate in many stages of transcription.
We are interested in identifying additional ways in which ubiquitylation impacts transcription and in understanding mechanisms through which this modification can function. This interest has led us to study Asr1, a Saccharomyces cerevisiae Ub-ligase that binds directly to the pol II CTD and, via ubiquitylation, inactivates the enzyme by ejecting 2 subunits from the pol II complex. These data reveal that the activity of a core component of the transcriptional machinery can be modified by nonproteolytic ubiquitylation, and demonstrate how Ub can change the composition of a large multiprotein complex.
Results
RPC Proteins.
The goal of our work was to identify and characterize proteins that directly connect the transcription and Ub systems. To this end, we surveyed the literature for examples of proteins with probable links to both systems. This analysis led us to rA9, a mammalian protein that was identified in a 2-hybrid screen for factors that bind the CTD of Rpb1, the largest subunit of pol II (11). rA9 contains a domain that binds the CTD (CTD-binding domain; CBD), as well as a series of SR repeats, commonly found in splicing factors. Interestingly, we observed that rA9 also contains a RING finger and a PHD domain, both of which are associated with Ub-ligase activity (12, 13). The combination of CTD-binding and potential E3 activities in rA9 suggested to us that it may function as a Ub-ligase within the context of transcription.
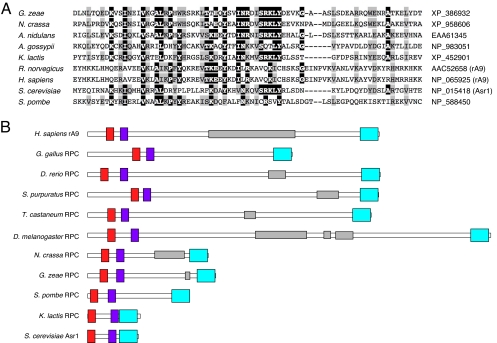
To study rA9, we asked whether a related protein is present in the yeast S. cerevisiae. Although BLAST searches failed to identify homologous proteins in yeast, we were able to identify sequences with small, but significant, homology to the CBD in a variety of organisms, including fungi (Fig. 1A). Within these proteins, the region of CBD homology is located at the carboxyl terminus of the protein, and, remarkably, the majority contain either an amino-terminal RING finger, or a RING/PHD combination [Fig. 1B and supporting information (SI) Methods]. We refer to proteins with this architecture as RC (RING/CBD) or RPC (RING/PHD/CBD) proteins. We chose to study the single RPC member in S. cerevisiae, Asr1 (14), a nonessential protein that has been implicated in the alcohol stress response.
Fig. 1.
The RPC group of proteins. (A) ClustalW alignment of CBD segments from the indicated species. (B) Architecture of RPC proteins. Proteins are depicted to scale. RING domains are red, PHD domains are purple, and the CBD is cyan. RS domains are shown in gray.
Asr1 Is a Ub-ligase That Binds the Pol II CTD.
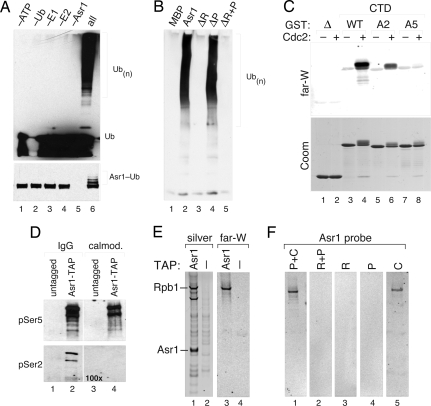
Neither rA9 nor Asr1 has been shown to possess Ub-ligase activity. To determine whether the RING and/or PHD domains within Asr1 are functional, we asked whether Asr1 is a bona fide E3. We found that recombinant Asr1 directs the formation of unanchored poly-Ub chains (Fig. 2A Upper) and is also capable of significant autoubiquitylation (Fig. 2A Lower). Notably, the ligase activity of Asr1 is disrupted by mutations in the RING finger (Fig. 2B, lane 3), but not the PHD domain (lane 4), revealing that Asr1 is indeed a RING finger Ub-ligase.
Fig. 2.
Asr1 is an E3 that binds the pol II CTD. (A) In vitro ubiquitylation assay, with recombinant MBP-Asr1, examining formation of unanchored poly-Ub chains (Upper) or Asr1 autoubiquitylation (Lower). (B) In vitro ubiquitylation analysis with selected Asr1 mutants (ΔR, RING mutant; ΔP, PHD mutant). (C) Far-Western (far-W) analysis of Asr1 binding to GST-CTD proteins. (D) Western blot of Asr1–TAP complex with phospho-specific antibodies against Ser-5 or Ser-2 of the CTD repeats. IgG is the Asr1-associated material found in the first step of the TAP purification; calmod is the more stringently purified material from the second purification step. One hundred times more material is present in the calmod samples than in the IgG samples. (E) FW analysis of Asr1 against the Asr1–TAP preparation. Rpb1 and Asr1, identified by mass spectrometry, are indicated. (F) FW analysis of Asr1 mutants against the Asr1–TAP preparation. For Asr1 mutants: P+C is residues 91–310, R+P is residues 1–198, R is residues 1–90, P is residues 91–198, C is residues 199–310 (i.e., the CBD).
To test whether Asr1 can bind the CTD, we used Far-Western (FW) analysis to probe interaction of Asr1 with recombinant GST-CTD (ref. 15 and Fig. 2C). Asr1 bound strongly to the wild-type (WT) CTD in a manner that depended on its phosphorylation (lanes 3 and 4). Analysis of mutations that disrupt the major sites of phosphorylation, at Ser-2 and Ser-5 within the CTD repeats (15), revealed that alanine substitution at Ser-2 reduced Asr1 binding (lane 6), whereas the corresponding mutation at Ser-5 blocked Asr1 interaction (lane 8). Thus, Asr1 recognizes the CTD in a manner that depends on Ser-5 phosphorylation. Consistent with this notion, recovery of Asr1 from yeast cells by tandem affinity purification (TAP) showed that it associates tightly with the Ser-5-phosphorylated form of Rpb1 (Fig. 2D, lanes 2 and 4), but not the Ser-2-phosphorylated form. Additionally, we identified Rpb1 as a major Asr1-TAP-associated protein by mass spectrometry (Fig. 2E, lanes 1 and 2) and found by FW that Asr1 directly interacts with Rpb1 (lanes 3 and 4) via the region corresponding to the CBD (Fig. 2F, lane 5). We conclude that Asr1 is a Ub-ligase that directly associates with the Ser-5-phosphorylated CTD of pol II.
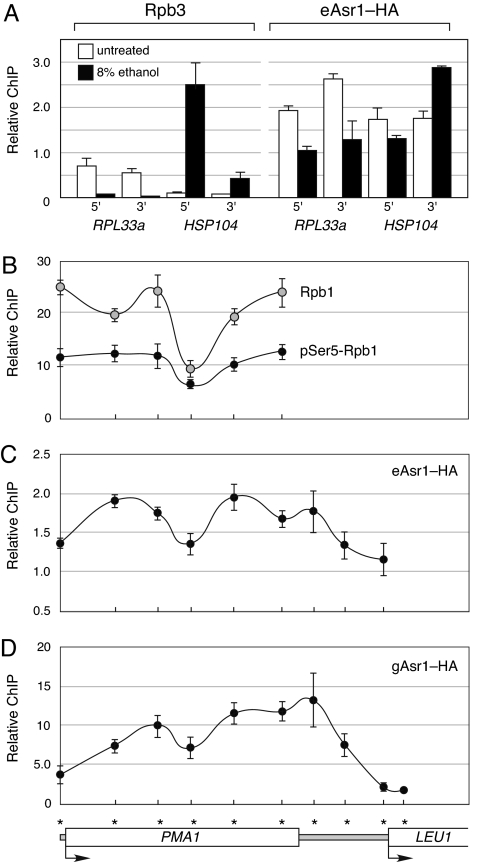
As expected from its ability to bind Rpb1, Asr1 associates with genes in a manner that correlates with their activity (Fig. 3). Using chromatin immunoprecipitation (ChIP) analysis, we found that ethanol shock, which represses the RPL33a gene and activates HSP104, resulted in the loss of Asr1 from RPL33a and its recruitment to HSP104. Scanning across the PMA1 gene (Fig. 3 B–D), we found that the pattern of distribution of both endogenous Asr1 (eAsr1-HA) and galactose-inducible Asr1 (gAsr1-HA) correlates with the distribution of both total (as measured by Rpb3) and Ser-5-phosphorylated Rpb1. These ChIP data support the notion that Asr1 interacts with Ser-5-phosphorylated Rpb1 on chromatin.
Fig. 3.
ChIP analysis of Asr1. (A) Yeast were shocked with 8% ethanol for 5 min, and levels of Rpb3 or endogenous (HA-tagged) Asr1 (eAsr1-HA) at the 5′ and 3′ ends of the RPL33a and HSP104 genes were determined by ChIP. (B) Exponentially growing cultures of yeast were subject to ChIP analysis, examining distribution of total Rpb1 or phospho-Ser-5 Rpb1 along the PMA1 gene. (C and D) As in B, except that the distribution of endogenous (C) or galactose-induced (D) Asr1-HA was measured. Diagram at the Bottom depicts the relative location of the primer sets used for analysis of ChIP DNA. Shown is the mean of 4 ChIP experiments ± S.E.M.
Asr1 Targets Pol II Subunits for Ubiquitylation.
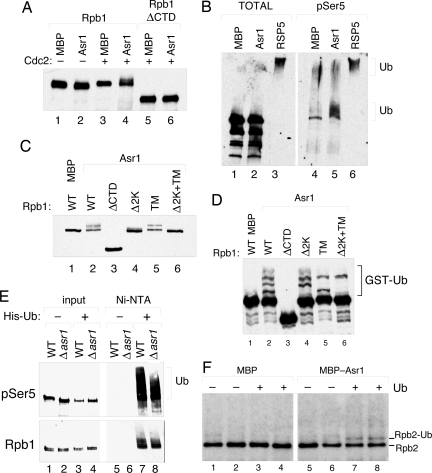
Given that Asr1 binds directly to the CTD repeat of Rpb1, we speculated that this pol II subunit might be a substrate for Asr1-dependent ubiquitylation. We had observed that recombinant Asr1 can ubiquitylate a GST-CTD fusion protein after its phosphorylation at Ser-5 (Fig. S1). We therefore asked whether Rpb1 itself is a substrate for Asr1 (Fig. 4). In vitro translated Rpb1 was ubiquitylated by recombinant Asr1 (Fig. 4A) in a manner that depends on Rpb1 phosphorylation (compare lanes 2 and 4) and the CTD (compare lanes 4 and 6). Curiously, Asr1 did not induce significant polyubiquitylation of Rpb1; we observed a small, discrete, shift in the molecular weight of Rpb1 upon ubiquitylation, consistent with the addition of only a few Ub moieties to the Rpb1 protein. This pattern of ubiquitylation was not altered by use of methylated Ub (data not shown), suggesting that Rpb1 is monoubiquitylated at several sites by Asr1.
Fig. 4.
Ubiquitylation of Rpb1 and Rpb2 by Asr1. (A) In vitro ubiquitylation of in vitro translated Rpb1 and an Rpb1 CTD deletion mutant by recombinant MBP-Asr1 (Asr1) or an MBP control. Where indicated, Rpb1 was phosphorylated by recombinant Cdc2 before the ubiquitylation reaction. (B) In vitro ubiquitylation of purified pol II complexes by recombinant MBP, MBP-Asr1, or Rsp5. Either the total pool of Rpb1 (Left) or the Ser-5-phosphorylated pool of Rpb1 (Right) was detected by WB. (C) In vitro ubiquitylation of in vitro translated Rpb1 mutants by Asr1. The Δ2K mutant is missing lysine residues 1720 and 1725 of Rpb1. In the TM mutants, residues K1452/K1458/K1487 have been changed to arginine. (D) As in C except using GST-Ub. (E) In vivo ubiquitylation of Rpb1 in congenic WT or Δasr1 yeast, assayed by expression of polyhistidine-tagged and recovery of ubiquitylated proteins by chromatography on Ni–nitrilotriacetic acid resin. (F) In vitro ubiquitylation of Rpb2. Purified pol II complexes (isolated by TAP-tagged Rpb1) were ubiquitylated by recombinant MBP or MBP-Asr1 as in B and detected by WB.
To ask whether Asr1 can ubiquitylate Rpb1 within the context of pol II, we subjected purified pol II complex to a ubiquitylation assay similar to that above. We compared Asr1 with the Rsp5 Ub-ligase, an E3 that mediates the ubiquitylation and destruction of Rpb1 in response to DNA damage (16). In these assays, Asr1 had little effect on the total pool of Rpb1 (Fig. 4B, lanes 1 and 2), whereas Rsp5 directed the formation of high-molecular-weight Rpb1–Ub conjugates (lane 3), consistent with its role in Rpb1 destruction. Analysis of the Ser-5-phosphorylated pool of Rpb1, however (lanes 4–6), revealed that almost all of the Ser-5-phosphorylated Rpb1 was shifted into a discrete cluster of Rpb1–Ub conjugates in the presence of Asr1 (lane 5). These data are consistent with those obtained for free Rpb1 and reinforce the notion that Asr1 catalyzes a limited set of ubiquitylation events on the Rpb1 protein.
We attempted to map the sites of ubiquitylation within Rpb1 by analysis of a set of Rpb1 deletion mutants (Fig. S2). Although there are 95 lysine residues within Rpb1, we found that an ≈350-aa segment of Rpb1, encompassing the CTD, could be efficiently ubiquitylated by Asr1 in vitro (Fig. S2, lane 9). This segment contains 5 lysine residues, 3 amino-terminal (K1452, K1458, and K1487) and 2 carboxyl-terminal (K1720 and K1725) to the CTD. Interestingly, disruption of either K1720/K1755 (Fig. 4C, lane 4) or K1452/K1458/K1487 (lane 5) resulted in the disappearance of specific sets of ubiquitylated Rpb1 species; this can be seen more clearly by using a GST-tagged Ub as a substrate (Fig. 4D). When all 5 lysine residues were disrupted (the Δ2K+TM mutant), the majority of Rpb1–Ub conjugates disappeared (lanes 6 in Fig. 4 C and D). There are, however, several sites that we have not been able to map, as clearly seen in the GST-Ub experiment, where a high-molecular-weight GST-Ub–Rpb1 species persists in the Δ2K+TM mutant (lane 6). From these data, we conclude that Asr1 likely ubiquitylates a set of 5 lysine residues adjacent to the CTD and several sites elsewhere in the protein.
To determine whether Asr1 contributes to the ubiquitylation of Rpb1 in cells, we performed an in vivo ubiquitylation assay, comparing the extent of Rpb1 ubiquitylation in WT and asr1-null yeast (Fig. 4E). We were surprised to find that in the absence of UV-induced DNA damage, Ser-5-phosphorylated Rpb1 was extensively ubiquitylated (lane 7). This level of ubiquitylation was not observed when we probed for total Rpb1 protein (compare Fig. 4E Upper and Lower), suggesting that it is restricted to the initiated form of pol II. Most importantly, however, deletion of ASR1 (lane 8) significantly decreased the average molecular weight of the Rpb1–Ub species. From this result, we conclude that Asr1 contributes to ubiquitylation of Ser-5-phosphorylated Rpb1 in vivo. Moreover, because Rpb1 remains extensively ubiquitylated in asr1-null yeast, we conclude that a redundant mechanism for Rpb1 ubiquitylation must exist.
Rpb1 is not the only subunit of pol II that is ubiquitylated by Asr1 in vitro. As shown in Fig. 4F, the Rpb2 subunit is also modified by Ub in the presence of recombinant Asr1. This type of ubiquitylation is similar to that observed with Rpb1, with apparently a single Ub conjugate being formed during the reaction. We have also detected such high-molecular-weight forms of Rpb2 associated with Asr1 in vivo (data not shown). This result establishes that Asr1 can ubiquitylate multiple subunits within the pol II complex.
Asr1 Interacts with Pol II and Modifies Its Subunit Composition.
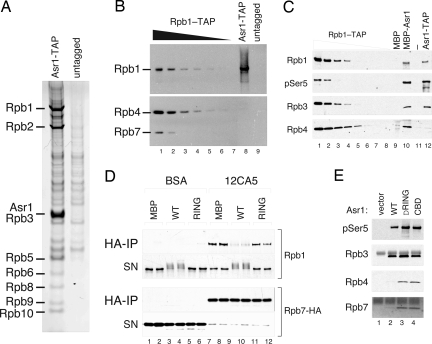
The biological function of Asr1 is unknown. Although it has been reported that deletion of ASR1 confers sensitivity to ethanol stress (14), this finding is disputed (17). In our hands, disruption of ASR1 has no pronounced phenotype (SI Methods). To begin to understand the function of Asr1, therefore, we purified TAP-tagged Asr1–TAP complexes and analyzed them by mass spectrometry (Fig. 5A; note that this is the same gel as presented in Fig. 2E). This analysis revealed that Asr1 purifies with a total of 10 of 12 core pol II subunits, Rpb1, 2, 3, 5, 6, 8, 9, 10, 11, and 12 (SI Methods); we did not reproducibly detect any other proteins in these preparations. It should be noted that Asr1 is a very low-abundance protein, most likely because of its RING-dependent rapid turnover (Fig. S3); and although a large fraction of the total Asr1 population is associated with these pol II subunits, only a small percentage of total pol II is in complex with Asr1 at steady state (data not shown).
Fig. 5.
Asr1-mediated ubiquitylation excludes Rpb4/7 from the RNA pol II complex. (A) Asr1–TAP purification. TAP purification was performed on extract from either untagged yeast (negative control) or yeast carrying TAP-tagged Asr1. The indicated proteins were identified by mass spectrometry of individual bands and by shotgun analysis in solution. (B) The Asr1 complex is devoid of Rpb4/7. Increasing amounts of Rpb1–TAP complexes were run against an Asr1–TAP preparation and probed for Rpb1, Rpb7, and Rpb7 by WB. (C) Asr1 can bind pol II that includes Rpb4. Yeast extract was run over an MBP-Asr1 column, and the indicated pol II subunits were detected by WB. Asr1–TAP and Rpb1–TAP complexes are shown for comparison. (D) The Ub-ligase activity of Asr1 excludes Rpb7 from the pol II complex. Purified pol II complexes were ubiquitylated by Asr1, and Rpb7-HA was recovered by immunoprecipitation (BSA is the negative control). Coprecipitating Rpb1 complexes in the immunoprecipitant (IP) or supernatant (SN) were detected by WB. (E) The RING finger of Asr1 is required to exclude Rpb4 and Rpb7 from the Asr1 complex. Asr1 and the indicated mutants were immunoprecipitated from yeast and probed for pol II subunits by WB.
Most notable from the mass spectrometry analysis was our inability to detect Rpb4 and Rpb7, which form a dissociable heterodimer that docks onto pol II principally via Rpb7 (18). Western blotting confirmed that Rpb4/7 were not present in the Asr1-pulldown (Fig. 5B; compare lanes 1 and 8), revealing that Asr1 associates with pol II that is specifically lacking these subunits. One possible explanation for this result would be that Asr1 only recognizes pol II in the absence of Rpb4/7. This, however, does not appear to be the case. When we passed ATP-depleted yeast extract over a column containing immobilized Asr1 (Fig. 5C), we found that it efficiently captured Rpb4 (lane 10) at levels similar to those in the control Rpb1–TAP purification. Thus, Asr1 can recognize pol II that contains Rpb4 (and thus Rpb7).
An alternative explanation would be that it is the Ub-ligase activity of Asr1 that excludes Rpb4/7 from the pol II complex. We therefore examined the fate of the Rpb7–pol II interaction during the course of Asr1-dependent pol II ubiquitylation (Fig. 5D). Purified pol II was ubiquitylated by Asr1, and the Rpb1–Rpb7 interaction was measured by determining the amount of Rpb1 that coimmunoprecipitated with Rpb7. As before, WT Asr1 (but not the RING mutant) efficiently modified Rpb1 under these conditions, resulting in the formation of a discrete set of oligoubiquitylated Rpb1 proteins (lanes 3–4 and 9–10). Recovery of Rpb7 from these reactions (via the HA tag; lanes 7–12) showed that Asr1-mediated ubiquitylation did indeed disrupt the Rpb1–Rpb7 interaction (compare lanes 7 and 8 with 9 and 10). Not only was significantly less Rpb1 recovered after the ubiquitylation reaction, but what was recovered appeared to be the unmodified form (Rpb1 IP; lanes 9 and 10); the higher-molecular-weight Rpb1–Ub conjugates present in the supernatant were not represented in the Rpb7-bound material. From this result, we conclude that ubiquitylated Rpb1 does not associate with Rpb7 and that Asr1-mediated ubiquitylation of pol II disrupts its interaction with Rpb7.
If Asr1-mediated ubiquitylation modulates the association of Rpb4/7 with the remainder of the pol II complex, we would expect that recovery of a catalytically inactive form of Asr1 would bring down all subunits of pol II, including Rpb4/7. Indeed, immunoprecipitation of the Asr1 RING mutant, or the Asr1 CBD, from yeast cells resulted in the efficient recovery of both Rpb4 and Rpb7 (Fig. 5E; compare lane 2 with lanes 3 and 4), demonstrating that it is the Ub-ligase activity of Asr1 that modulates Rpb4/7 association with pol II in vivo.
Discussion
Our attempts to identify a protein that connects the transcription and Ub systems led to our analysis of Asr1 and the finding that it is a Ub-ligase that directly recognizes the Ser-5-phosphorylated form of RNA polymerase II and ubiquitylates at least 2 subunits within the enzyme, leading to ejection of the Rpb4/7 heterodimer from the remainder of the complex. Asr1 associates with RNA polymerase within the context of chromatin and defines a group of structurally conserved proteins present in many eukaryotes. The ability of Asr1 to interact specifically with a discrete subset of pol II molecules demonstrates how a component of the Ub–proteasome system can “sense” the modification status of a basal transcription factor and act to alter its subunit composition. Asr1 is thus one of the most explicit examples of how the transcription and Ub systems can intersect. Importantly, our study of Asr1 also demonstrates how ubiquitylation can modulate the composition of a large, multiprotein, complex such as pol II.
Perhaps the most intriguing question raised from this work is the biological function of Asr1. Its biochemical properties and its effect on Rpb4/7 imply that Asr1 is directly involved in some aspect of the transcription process. In S. cerevisiae, the Rpb4/7 heterodimer is present at substoichiometric levels within pol II (19) and may shuttle between different polymerase complexes (20). Rpb4/7 are not required for the catalytic activity of RNA polymerase II but are essential for promoter-driven initiation of transcription (20). There is considerable evidence that Rpb4/7 are involved in aspects of RNA processing (21), and it has been suggested that Rpb4/7 dissociates from RNA polymerase and travels with mRNA to the cytoplasm (21). There is thus a significant body of work suggesting that Rpb4/7 association with pol II is dynamic and that Rpb4/7 may have functions outside of the context of the polymerase. One possibility is that Asr1-mediated ejection of Rpb4/7 allows these 2 subunits to exchange with other polymerases or to perform their role as an uncomplexed heterodimer.
We favor the notion, however, that Asr1 is a negative regulator of pol II. In vivo, Rpb4/7 appears to co-occupy all sites of transcription where core pol II subunits are located (22), making it unlikely that these 2 proteins are jettisoned at a specific stage of the transcription cycle. RNA pol II lacking Rpb4/7 interacts less stably with DNA than does the 12-subunit polymerase (23), and it is reasonable to assume that ejection of Rpb4/7 would cause pol II to disengage from the template. A negative role for Asr1 is further supported by our finding that the Asr1 complex is devoid of significant polymerase activity (Fig. S4), despite the presence of all 10 subunits that are sufficient for catalytic activity (20). This result implies that some other ubiquitylation event, elsewhere in pol II, leads to inactivation of polymerase function. By specifically interacting with pol II species in which the CTD is phosphorylated at Ser-5 but not Ser-2 (Fig. 2), Asr1 may be targeting a subset of incorrectly initiated or immature pol II complexes, such as those that result from abortive or cryptic transcription events, for disassembly. It also possible, based on the association of Asr1 with the 3′ end of genes (Fig. 3), that this ligase acts in part to disassociate pol II complexes from DNA as part of the transcription termination process. The finding that a second mechanism exists for ubiquitylation of Ser-5-phosphorylated Rpb1 in vivo (Fig. 4) means that it may be difficult to expose the underlying biological function of Asr1 without uncovering the second pathway.
The observation that RPC proteins (and the related RC proteins that lack the PHD domain) broadly exist throughout eukaryotes is consistent with the idea that these proteins perform an important function. Their role in the transcription process is further supported by the presence of PHD domains in the RPC proteins. PHD-containing proteins frequently interact with chromatin, and in some cases PHD domains can directly bind modified histones (24). It is also interesting to note that several of the RPC/RC proteins also contain SR domains that are frequently present in pre-mRNA splicing factors and can participate in protein–protein and protein–RNA interactions that are important for spliceosome assembly (25). Perhaps, therefore, this group of proteins can also exploit their Ub-ligase activity to drive changes in protein–protein interactions that are required for appropriate cotranscriptional pre-mRNA splicing.
Finally, our data establish that ubiquitylation can act to change directly the composition of a multiprotein complex. There are examples of ubiquitylation leading to a loss of protein–protein interactions, but these examples typically require Ub-dependent chaperones (26) to recognize the ubiquitylated substrate and remodel the interactions by ATP-dependent hydrolysis. Our ability to reconstitute Ub-dependent disruption of the Rpb1–Rpb7 interaction by using highly purified or recombinant proteins (Fig. 5D) suggests that this process does not require a Ub-directed chaperone. It is possible that ubiquitylation of 1 or more subunits in the complex acts as a “wedge” to drive Rpb4/7 from the complex, in which case no chaperone would be formally required. It will be interesting to determine whether this mechanism functions in other Ub-dependent cellular processes. It will also be interesting to determine whether other Ub-ligases that associate with the transcriptional machinery (27) similarly act to drive changes in the assembly of active transcription complexes.
Methods
Detailed procedures are described in SI Methods.
Yeast and Plasmids.
Yeast used in this work are described in Table S1. For expression of recombinant Asr1 as an MBP fusion, Asr1-coding sequences were cloned into the expression vector pMAL-c2x (NEB), and site-directed mutagenesis was used to introduce mutations within the RING domain (C29A and C50A) and/or the PHD domain (C143A and C146A) of Asr1. Galactose-inducible Asr1 expression constructs were created by cloning the relevant ASR1 sequences into pYES2 (Invitrogen). Plasmid p425-pCUP1-UbG76A was used to express polyhistidine-tagged Ub under the control of the CUP1 promoter. Plasmids for expressing GST-CTD fusion proteins in Escherichia coli were a gift from J. Corden (Johns Hopkins Medical School, Baltimore, MD) (15).
Antibodies.
HA-tagged proteins were detected by 12CA5 or 3F10 (Roche). Anti-Rpb1 antibodies 8WG16, H5, H14, y-80, and CTD4H8 were from Covance. Anti-Rpb4 and anti-Rpb7 antibodies were a gift from A. Sentenac (Commissariat à l'Énergie Atomique, Saclay, France) (28). Anti-Rpb3 and anti-Rpb4 antibodies were from NeoClone, and anti-Rpb7 antibody (yC-19) was from Santa Cruz Biotechnology. Horseradish peroxidase-conjugated anti-GST antibody was from Abcam.
Protein Purification and Production.
Asr1–TAP and Rpb1–TAP complexes were isolated according to the published method (29), and purifications were analyzed by mass spectrometry. Where indicated, Asr1, Rpb1, and the appropriate mutants were synthesized in vitro by using the TNT-coupled transcription/translation kit (Promega). Recombinant GST-CTD and MBP-Asr1 proteins were isolated from DH5α bacterial cells and purified by chromatography on glutathione–agarose, or amylose resin, respectively.
Biochemical Assays.
In vitro ubiquitylation assays were carried out as described in ref. 16. Substrates were either recombinant GST-CTD (phosphorylated by Cdc2; NEB), in vitro synthesized Rpb1-HA protein, or purified Rpb1–TAP complexes. Reactions were analyzed by SDS/PAGE/Western blotting (WB). For analysis of the effects of Asr1 on the Rpb1–Rpb7 interaction Rpb1–TAP complexes were purified from yeast expressing both Rpb1–TAP and Rpb7-HA, subject to ubiquitylation, and Rpb7-HA was recovered by nondenaturing immunoprecipitation with the 12CA5 antibody (or BSA control).
For FW analyses, phosphorylated GST-CTD proteins or purified Asr1–TAP purifications were resolved by SDS/PAGE (4–12% gradient), transferred to nitrocellulose membranes, and probed with in vitro translated, radiolabeled, Asr1 proteins (30).
In Vivo Assays.
To analyze the role of the Asr1-RING finger on the interaction of Rpb1 with Rpb4/7 in vivo, inducible forms of Asr1-HA (in the pYES2 vector, as described above) were expressed in Δasr1 yeast cells (BY4742) for 4 h by galactose induction. Cells were harvested, lysates were prepared, and HA-tagged Asr1 was recovered by immunoprecipitation. Coprecipitating Rpb1, Rpb4, and Rpb7 were visualized by SDS/PAGE and WB analysis.
ChIP analyses were performed by using a published method (31). For detection of endogenous, HA-tagged, Asr1 at the PMA1 gene, parallel ChIP assays were performed by using the 12CA5 monoclonal antibody from congenic yeast at which the ASR1 locus was either untagged (BY4742) or HA-tagged (BY4742 ASR1-HA). Specific signals from ChIP DNA from the Asr1-HA strain were normalized to the corresponding signals (per primer set) from the untagged yeast. For detection of overexpressed Asr1, cultures (BY4742 Δasr1) carrying galactose-inducible expression vectors for Asr1, or Asr1-HA, were grown overnight in raffinose, induced for 2 h by the addition of 2% galactose, and ChIP was performed as above. Total (Y-80) and pSer5 (H14) Rpb1 levels across PMA1 were quantified by ChIP with the indicated antibodies. For ChIP at the RPL33a and HSP104 genes, cultures were first treated with 8% ethanol for 5 min before cross-linking. ChIP DNA was quantified by real-time PCR. Primer sequences are available upon request.
Supplementary Material
Acknowledgments.
We thank J. Corden, B. Futcher (State University of New York, Stony Brook, NY), J. Huibregtse (University of Texas, Austin, TX), A. Sentenac, and R. Li (Stowers Institute for Medical Research, Kansas City, MO) for reagents; M. Myers for performing mass spectrometry; and G. Collins, A. Leung, and D. Simpson for comments on the manuscript. This work was supported by National Institutes of Health Grant GM067728. W.P.T. was a Leukemia and Lymphoma Society Scholar. A.D. was supported by the Philippe Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809372105/DCSupplemental.
References
- 1.Buratowski S. The CTD code. Nat Struct Biol. 2003;10:679–680. doi: 10.1038/nsb0903-679. [DOI] [PubMed] [Google Scholar]
- 2.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 3.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 4.Lipford JR, Smith GT, Chi Y, Deshaies RJ. A putative stimulatory role for activator turnover in gene expression. Nature. 2005;438:113–116. doi: 10.1038/nature04098. [DOI] [PubMed] [Google Scholar]
- 5.Salghetti SE, Caudy AA, Chenoweth JG, Tansey WP. Regulation of transcriptional activation domain function by ubiquitin. Science. 2001;293:1651–1653. doi: 10.1126/science.1062079. [DOI] [PubMed] [Google Scholar]
- 6.Varshavsky A. The ubiquitin system. Trends Biochem Sci. 1997;22:383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]
- 7.Lonard DM, O'Malley BW. SRC-3 transcription-coupled activation, degradation, and the ubiquitin clock: Is there enough coactivator to go around in cells? Sci Signal. 2008;1:pe16. doi: 10.1126/stke.113pe16. [DOI] [PubMed] [Google Scholar]
- 8.Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 9.Kaiser P, Flick K, Wittenberg C, Reed SI. Regulation of transcription by ubiquitination without proteolysis: Cdc34/SCF(Met30)-mediated inactivation of the transcription factor Met4. Cell. 2000;102:303–314. doi: 10.1016/s0092-8674(00)00036-2. [DOI] [PubMed] [Google Scholar]
- 10.Gwizdek C, et al. Ubiquitin-associated domain of Mex67 synchronizes recruitment of the mRNA export machinery with transcription. Proc Natl Acad Sci USA. 2006;103:16376–16381. doi: 10.1073/pnas.0607941103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuryev A, et al. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc Natl Acad Sci USA. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freemont PS. RING for destruction? Curr Biol. 2000;10:R84–R87. doi: 10.1016/s0960-9822(00)00287-6. [DOI] [PubMed] [Google Scholar]
- 13.Ivanov AV, et al. PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol Cell. 2007;28:823–837. doi: 10.1016/j.molcel.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Betz C, Schlenstedt G, Bailer SM. Asr1p, a novel yeast ring/PHD finger protein, signals alcohol stress to the nucleus. J Biol Chem. 2004;279:28174–28181. doi: 10.1074/jbc.M401595200. [DOI] [PubMed] [Google Scholar]
- 15.Patturajan M, et al. Growth-related changes in phosphorylation of yeast RNA polymerase II. J Biol Chem. 1998;273:4689–4694. doi: 10.1074/jbc.273.8.4689. [DOI] [PubMed] [Google Scholar]
- 16.Huibregtse JM, Yang JC, Beaudenon SL. The large subunit of RNA polymerase II is a substrate of the Rsp5 ubiquitin-protein ligase. Proc Natl Acad Sci USA. 1997;94:3656–3661. doi: 10.1073/pnas.94.8.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Izawa S, Ikeda K, Kita T, Inoue Y. Asr1, an alcohol-responsive factor of Saccharomyces cerevisiae, is dispensable for alcoholic fermentation. Appl Microbiol Biotechnol. 2006;72:560–565. doi: 10.1007/s00253-005-0294-1. [DOI] [PubMed] [Google Scholar]
- 18.Choder M. Rpb4 and Rpb7: Subunits of RNA polymerase II and beyond. Trends Biochem Sci. 2004;29:674–681. doi: 10.1016/j.tibs.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 19.Choder M, Young RA. A portion of RNA polymerase II molecules has a component essential for stress responses and stress survival. Mol Cell Biol. 1993;13:6984–6991. doi: 10.1128/mcb.13.11.6984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edwards AM, Kane CM, Young RA, Kornberg RD. Two dissociable subunits of yeast RNA polymerase II stimulate the initiation of transcription at a promoter in vitro. J Biol Chem. 1991;266:71–75. [PubMed] [Google Scholar]
- 21.Runner VM, Podolny V, Buratowski S. The Rpb4 subunit of RNA polymerase II contributes to cotranscriptional recruitment of 3′ processing factors. Mol Cell Biol. 2008;28:1883–1891. doi: 10.1128/MCB.01714-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jasiak AJ, et al. Genome-associated RNA polymerase II includes the dissociable RPB4/7 subcomplex. J Biol Chem. 2008;283:26423–26427. doi: 10.1074/jbc.M803237200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jensen GJ, Meredith G, Bushnell DA, Kornberg RD. Structure of wild-type yeast RNA polymerase II and location of Rpb4 and Rpb7. EMBO J. 1998;17:2353–2358. doi: 10.1093/emboj/17.8.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wysocka J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 25.Hertel KJ, Graveley BR. RS domains contact the pre-mRNA throughout spliceosome assembly. Trends Biochem Sci. 2005;30:115–118. doi: 10.1016/j.tibs.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 26.Rape M, Jentsch S. Productive RUPture: Activation of transcription factors by proteasomal processing. Biochim Biophys Acta. 2004;1695:209–213. doi: 10.1016/j.bbamcr.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 27.Takagi Y, et al. Ubiquitin ligase activity of TFIIH and the transcriptional response to DNA damage. Mol Cell. 2005;18:237–243. doi: 10.1016/j.molcel.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 28.Huet J, Phalente L, Buttin G, Sentenac A, Fromageot P. Probing yeast RNA polymerase A subunits with monospecific antibodies. EMBO J. 1982;1:1193–1198. doi: 10.1002/j.1460-2075.1982.tb00012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Puig O, et al. The tandem affinity purification (TAP) method: A general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 30.Decoville M, Giraud-Panis MJ, Mosrin-Huaman C, Leng M, Locker D. HMG boxes of DSP1 protein interact with the rel homology domain of transcription factors. Nucleic Acids Res. 2000;28:454–462. doi: 10.1093/nar/28.2.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuo MH, Allis CD. In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods. 1999;19:425–433. doi: 10.1006/meth.1999.0879. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.