Abstract
A library of drugs that are in clinical trials or use was screened for inhibitors of hypoxia-inducible factor 1 (HIF-1). Twenty drugs inhibited HIF-1-dependent gene transcription by >88% at a concentration of 0.4 μM. Eleven of these drugs were cardiac glycosides, including digoxin, ouabain, and proscillaridin A, which inhibited HIF-1α protein synthesis and expression of HIF-1 target genes in cancer cells. Digoxin administration increased latency and decreased growth of tumor xenografts, whereas treatment of established tumors resulted in growth arrest within one week. Enforced expression of HIF-1α by transfection was not inhibited by digoxin, and xenografts derived from these cells were resistant to the anti-tumor effects of digoxin, demonstrating that HIF-1 is a critical target of digoxin for cancer therapy.
Keywords: cancer therapy, hypoxia, tumor xenograft
Many human cancers contain regions of hypoxia because of rapid cell proliferation and the presence of intratumoral blood vessels that are structurally and functionally abnormal, resulting in both spatial and temporal heterogeneity of blood flow (1, 2). The presence of intratumoral hypoxia is significantly associated with increased risk of treatment failure, invasion, metastasis, and patient mortality (3). A principal mechanism by which cancer cells adapt to the hypoxic microenvironment is through the activity of hypoxia-inducible factor 1 (HIF-1). HIF-1 is a transcription factor that regulates the expression of hundreds of genes in response to hypoxia, including VEGF, which encodes vascular endothelial growth factor, a key regulator of angiogenesis; GLUT1, which encodes glucose transporter 1; and HK1 and HK2, which encode hexokinase, the first enzyme of the glycolytic pathway (4, 5). Expression of these proteins serves either to increase O2 delivery (VEGF) or to allow metabolic adaptation to reduced O2 availability (GLUT1, HK1, HK2). HIF-1 also controls the expression of genes involved in tumor cell immortalization, stem cell maintenance/de-differentiation, genetic instability, autocrine growth, invasion/metastasis, and treatment failure (6, 7).
HIF-1 is a heterodimeric protein that is composed of O2-regulated HIF-1α and constitutively expressed HIF-1β subunits (8, 9). Under normoxic conditions, HIF-1α is hydroxylated on proline residue 564 and/or 402 by proline hydroxylase domain protein 2 (PHD2), which is required for the binding of the von Hippel-Lindau protein (VHL), the recognition subunit of an E3 ubiquitin-protein ligase that targets HIF-1α for proteasomal degradation (10). Under hypoxic conditions, PHD2 activity is inhibited, then HIF-1α accumulates, dimerizes with HIF-1β, and activates the transcription of target genes. VHL loss-of-function results in constitutive expression of HIF-1α in the majority of clear cell renal carcinomas (11). HIF-2α is also regulated by PHD2 and VHL, dimerizes with HIF-1β, and transactivates an overlapping but distinct group of target genes (12).
Whereas VHL loss-of-function leading to dysregulated HIF-1α expression is restricted to renal cell carcinoma and cerebellar hemangioblastoma, many common human cancers have mutations in phosphatidylinositol 3-kinase or upstream signaling pathways (13). These genetic alterations activate the mammalian target of rapamycin (mTOR), a serine-threonine protein kinase that activates p70 S6 kinase, which phosphorylates ribosomal protein S6 (RPS6) and induces increased translation of HIF-1α mRNA into protein (14). Derivatives of rapamycin that inhibit mTOR are in clinical trials as anti-cancer agents and HIF-1α overexpression has been shown to sensitize kidney cancers to these drugs (15).
The combination of increased synthesis and decreased degradation leads to increased HIF-1α protein levels in many cancers (16, 17). HIF-1α overexpression in tumor biopsies is associated with increased patient mortality in human cancers of the bladder, brain, breast, cervix, endometrium, oropharynx, lung, skin, and stomach, which reflects the large battery of HIF-1 target genes encoding proteins that play important roles in many key aspects of cancer biology (6, 7, 12). Based on these findings, there is considerable interest in identifying compounds that inhibit HIF-1 activity and testing their ability to inhibit tumor growth (6, 18). In the present study, we surveyed drugs that are already in clinical use to identify novel inhibitors of HIF-1.
Results
Cell-Based Screen for Inhibitors of HIF-1 Transcriptional Activity.
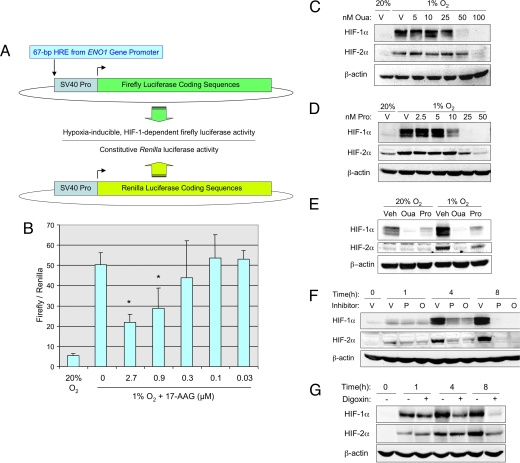
To screen for inhibitors of HIF-1 by using a cell-based assay, we engineered a reporter cell line Hep3B-c1, which contains reporter genes for hypoxia-inducible expression of firefly luciferase and constitutive expression of Renilla luciferase. Human Hep3B hepatoblastoma cells were stably transfected with plasmid p2.1, in which expression of firefly luciferase coding sequences is driven by a 68-bp hypoxia response element (HRE) from the human ENO1 gene inserted upstream of a basal SV40 promoter (Fig. 1A). The HRE contains essential binding sites for HIF-1, which mediates increased transcription in cells that are either exposed to hypoxia or co-transfected with an HIF-1α expression vector (19). In addition to p2.1, Hep3B-c1 cells were stably co-transfected with pSVRenilla, in which expression of Renilla luciferase coding sequences is driven by the SV40 promoter alone (Fig. 1A). By determining the ratio of firefly/Renilla luciferase activity under nonhypoxic and hypoxic conditions, we could screen for compounds that specifically inhibited hypoxia-induced firefly luciferase activity driven by HIF-1.
Fig. 1.
Inhibition of HIF-1α and HIF-2α by cardiac glycosides. (A) Hep3B cells were stably co-transfected with: p2.1, a plasmid containing firefly luciferase coding sequences downstream of a hypoxia response element (HRE) and the SV40 early region promoter (Top); and pSV-Renilla, a plasmid containing Renilla luciferase coding sequences downstream of the SV40 early region promoter. The ratio of firefly/Renilla luciferase activity in cells exposed to nonhypoxic (20% O2) or hypoxic (1% O2) culture conditions was determined. (B) The effect of the known HIF-1 inhibitor 17-AAG on the ratio of firefly/Renilla luciferase activity in hypoxic cells was determined; mean ± SD (n = 3) are shown. *, P < 0.05 compared to untreated (Student's t test). (C and D) Hep3B cells were exposed to vehicle (V) or the indicated concentration (nM) of ouabain (Oua) or proscillaridin A (Pro) for 24 h under hypoxic (1% O2) or nonhypoxic (20% O2) conditions and cell lysates were subjected to immunoblot assays for HIF-1α, HIF-2α, and β-actin. (E) PC3 cells were exposed to vehicle (V), 50 nM ouabain (Oua), or 50 nM proscillaridin A (Pro) for 24 h. (F) Hep3B cells were exposed to vehicle (V), 50 nM proscillaridin A (P), or 100 nM ouabain (O) under hypoxic conditions for the indicated time and immunoblot assays were performed. (G) Hep3B cells were exposed to vehicle (−) or 100 nM digoxin (+) under hypoxic conditions for the indicated time and immunoblot assays were performed.
When Hep3B-c1 cells were cultured under hypoxic conditions (1% O2), the ratio of firefly/Renilla luciferase activity was ten-fold higher than when the cells were cultured under nonhypoxic conditions (20% O2) for 24 h (Fig. 1B). To validate the use of these cells in screening for inhibitors of HIF-1, Hep3B-c1 cells were exposed to hypoxia in the presence of increasing amounts of the HSP90 inhibitor 17-allylaminogeldanamycin (17-AAG). Inhibition of HSP90 binding to HIF-1α by 17-AAG and other geldanamycin derivatives leads to increased binding of RACK1, which promotes the ubiquitination and proteasomal degradation of HIF-1α protein (20). HIF-1 transcriptional activity, as measured by the ratio of firefly/Renilla luciferase activity, decreased in a dose-dependent manner in cells treated with 17-AAG (Fig. 1B).
We next screened the Hopkins Drug Library, a collection of 3,120 drugs that have been approved by the Food and Drug Administration or have entered phase II clinical trials (21). Three hundred thirty-six (336) drugs were found to result in at least 50% reduction of firefly/Renilla luciferase activity in hypoxic Hep3B-c1 cells at a concentration of 10 μM [supporting information (SI) Fig. S1]. The top 220 hits were re-screened at a concentration of 2 μM and 153 of these were re-screened at a concentration of 0.4 μM. Thirty-two hits were identified, of which the top 20 resulted in >88% inhibition of firefly/Renilla luciferase activity. Among these 20 drugs were two known inhibitors of HIF-1, rapamycin (14) and rotenone (22). Remarkably, 11 of these 20 drugs (digoxin, ouabain, proscillaridin A, digitoxin, acetyldigitoxin, convallatoxin, peruvoside, strophanthin K, nerifolin, cymarin, and periplocymarin) were cardiac glycosides.
Cardiac Glycosides Inhibit HIF-1α and HIF-2α Protein Expression.
A dose-response study revealed that exposure of Hep3B cells to ouabain (Fig. 1C) or proscillaridin A (Fig. 1D) at concentrations ≥50 nM for 24 h inhibited hypoxia-induced expression of HIF-1α and HIF-2α protein. In human PC3 prostate cancer cells, which express detectable levels of HIF-1α under nonhypoxic as well as hypoxic conditions, 50 nM ouabain or proscillaridin A inhibited HIF-1α expression at both at 20% and 1% O2 (Fig. 1E). To investigate the kinetics of inhibition, Hep3B cells were concurrently exposed to 50 nM proscillaridin A or 100 nM ouabain and to 1% O2. HIF-1α and HIF-2α were strongly induced by exposure to hypoxia for 4–8 h in the presence of vehicle whereas, in the presence of proscillaridin A or ouabain, expression was reduced in a time-dependent manner with complete inhibition achieved after 8 h (Fig. 1F). Similar results were obtained in Hep3B cells treated with 100 nM digoxin (Fig. 1G). Drug washout experiments indicated that HIF-1 transcriptional activity gradually returned to normal levels by 24 h after removal of digoxin from the culture media (Fig. S2).
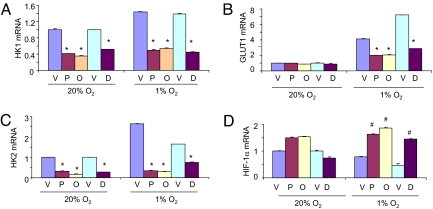
To investigate the effect of drug treatment on the expression of mRNAs encoded by HIF-1 target genes, Hep3B cells were cultured in the presence of vehicle, proscillaridin A, ouabain, or digoxin for 24 h at 20% or 1% O2 and total RNA was isolated. Analysis by quantitative real-time reverse transcription-PCR (RT-PCR) revealed significantly decreased levels of mRNA encoding HK1, HK2, and GLUT1 in cardiac glycoside-treated cells cultured at 1% O2, as well as decreased HK1 and HK2 mRNA levels in drug-treated cells cultured at 20% O2 (Fig. 2 A–C). In contrast, drug treatment did not inhibit HIF-1α mRNA levels at 20% O2 and significantly increased HIF-1α mRNA levels at 1% O2 (Fig. 2D). These results indicate that digoxin inhibits the expression of HIF-1 target genes by a mechanism that does not involve inhibition of HIF-1α mRNA expression.
Fig. 2.
Cardiac glycosides inhibit hypoxia-induced expression of HIF-1 target genes. In two separate experiments, Hep3B cells were exposed to 20% or 1% O2 for 24 h in the presence of: vehicle (V), 50 nM proscillaridin A (P), or 100 nM ouabain (O); and vehicle (V) or 100 nM digoxin (D). Total RNA was extracted from the cells and analyzed by quantitative real-time RT-PCR for mRNAs encoding hexokinase 1 (HK1; A), glucose transporter 1 (GLUT1; B), hexokinase 2 (HK2; C), and hypoxia-inducible factor 1α (HIF-1α; D). For each mRNA in each experiment, expression was normalized to the levels in vehicle-treated cells at 20% O2. The bars show mean ± SD (n = 4 each). [*, significant decrease compared to vehicle (P < 0.05, Student's t test); #, significant increase compared to vehicle (P < 0.05, Student's t test).]
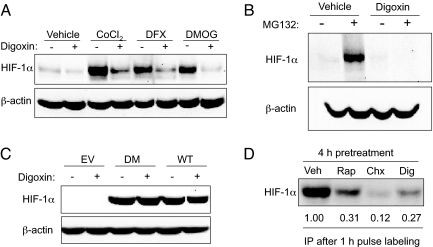
In addition to hypoxia, HIF-1 can also be induced by exposing cells to cobalt chloride, desferrioxamine, or dimethyloxalylglycine, each of which inhibits the prolyl hydroxylases that target HIF-1α for VHL-dependent ubiquitination and proteasomal degradation (23). HIF-1α induction by each of these compounds was blocked by treatment with digoxin (Fig. 3A). Treatment with digoxin (Fig. 3B), proscillaridin A or ouabain (Fig. S3A) blocked HIF-1α expression even in the presence of the proteasome inhibitor MG132. Taken together these results indicate that cardiac glycosides inhibit HIF-1α expression independently of the PHD-VHL-proteasome pathway.
Fig. 3.
Cardiac glycosides inhibit HIF-1α protein synthesis. (A) Hep3B cells were exposed to vehicle or the HIF-1α inducer cobalt chloride (CoCl2), desferrioxamine (DFX), or dimethyloxalylglycine (DMOG) in the presence of 100 nM digoxin (+) or vehicle (−) for 24 h and whole cell lysates were subjected to immunoblot assays. (B) Hep3B cells were treated with vehicle or digoxin in the absence (−) or presence (+) of 5 μM MG132. (C) 293 cells were transfected with empty vector (EV) or expression vector encoding FLAG epitope-tagged HIF-1α that was wild-type or double mutant (DM) because of Pro → Ala substitutions at residues 402 and 564 of the protein. The cells were cultured in the presence of 100 nM digoxin (+) or vehicle (−) for 24 h, and cell lysates were subjected to immunoblot assays by using anti-FLAG antibody. (D) Hep3B cells were pretreated for 4 h with vehicle (Veh), 25 nM rapamycin (Rap), 20 μg/ml cycloheximide (Chx), or 100 nM digoxin (Dig), [35S]methionine/cysteine was added for 1 h followed by cell lysis, HIF-1α immunoprecipitation (IP), SDS/PAGE, and autoradiography. Signal intensity was quantified by densitometry.
Although endogenous HIF-1α was potently inhibited by digoxin, proscillaridin A, or ouabain, these drugs had no effect on HIF-1α expressed from a transiently transfected plasmid vector, regardless of whether the protein was wild type HIF-1α or the double-mutant HIF-1αDM, which contains Pro-to-Ala missense mutations at the residues that are subjected to hydroxylation (Fig. 3C and Fig. S3B). A major difference between endogenous and exogenous HIF-1α is that the latter is translated from mRNA that lacks 5′ and 3′ untranslated sequences, which regulate mRNA translation and stability. Because the cardiac glycosides did not reduce HIF-1α mRNA levels (Fig. 2D), this result implied that cardiac glycosides inhibit the translation of HIF-1α mRNA into protein. To test this hypothesis, we pulse-labeled Hep3B cells with [35S]-methionine/cysteine in the presence of: cycloheximide, a global protein synthesis inhibitor; rapamycin, which is known to selectively inhibit the translation of HIF-1α mRNA (14); or digoxin. HIF-1α was then immunoprecipitated from cell lysates and analyzed by PAGE and autoradiography. At the concentration used, digoxin effectively inhibited the synthesis of HIF-1α protein, similar to the effect of cycloheximide or rapamycin (Fig. 3D). Digoxin reduced de novo HIF-1α synthesis by 73%, whereas overall protein synthesis was inhibited by only 19% in digoxin-treated cells (Fig. S3C). In contrast, cycloheximide had similar inhibitory effects on overall (85%) and HIF-1α (88%) protein synthesis.
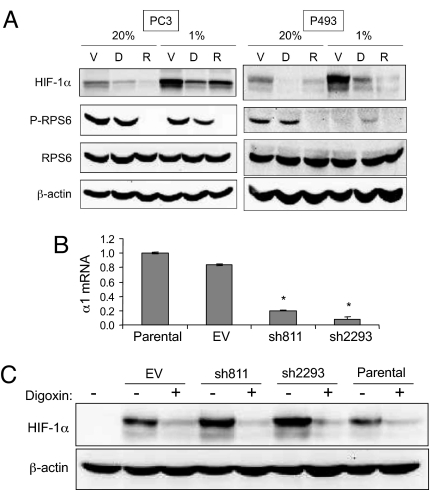
HIF-1α translation is regulated by mTOR (14), which activates S6 kinase, which in turn phosphorylates ribosomal protein S6 and the levels of phospho-RPS6 can be used as a read-out of mTOR activity (24). In PC3 cells, RPS6 was phosphorylated under both nonhypoxic and hypoxic conditions in the absence, but not in the presence, of rapamycin (Fig. 4A Left). In P493 lymphoma cells, RPS6 phosphorylation was observed only in nonhypoxic cells and was rapamycin-sensitive (Fig. 4A Right). Similar results were observed in 293T cells (data not shown). The inhibitory effect of hypoxia on RPS6 phosphorylation is consistent with published reports (24). In contrast, treatment of these three cell lines with digoxin at concentrations that potently inhibited HIF-1α protein expression, had no inhibitory effect on RPS6 phosphorylation (Fig. 4A). These results indicate that, unlike rapamycin, digoxin inhibits HIF-1α translation by a mechanism that is independent of mTOR.
Fig. 4.
Inhibition of HIF-1 by digoxin is independent of mTOR and Na+/K+ ATPase α1 subunit. (A) PC3 (Left) and P493 (Right) cells were cultured at 20% or 1% O2 for 24 h in the presence of vehicle (V), 100 nM digoxin (D), or 25 nM rapamycin (R) and whole cell lysates were subjected to immunoblot assays with antibodies specific for HIF-1α, phosphorylated ribosomal protein S6 (P-RPS6), total RPS6, or β-actin. (B) RNA was isolated from untreated 293T cells (Parental) or cells transfected with empty vector (EV) or expression vectors encoding two different short hairpin RNAs against the Na+/K+ ATPase α1 subunit (sh811, sh2293). The levels of α1 subunit mRNA (mean ± SD) were quantified by real-time RT-PCR. *, P < 0.05 vs. Parental or EV (Student's t test). (C) 293T cells were transiently transfected with EV or vector encoding sh811 or sh2293 and treated with vehicle (−) or 100 nM digoxin (+) at 1% O2 for 24 h. Whole cell lysates were subjected to immunoblot assays for HIF-1α and β-actin.
In patients with congestive heart failure, the therapeutic target of digoxin and other cardiac glycosides is the α1 subunit of the Na+/K+ ATPase (25). Transfection of 293T cells with vectors encoding two short hairpin RNAs, which target different sequences within the α1 mRNA, effectively reduced the levels of α1 mRNA (Fig. 4B). However, knockdown of the Na+/K+ ATPase α1 subunit had no effect on the levels of HIF-1α or HIF-2α protein, either in the presence or absence of digoxin (Fig. 4C) or ouabain (Fig. S4).
The cardiac glycosides digoxin, ouabain, and proscillaridin A have been reported to inhibit topoisomerases I and II in MCF7 cells (26). Camptothecin analogues that inhibit topoisomerase I have been shown to inhibit HIF-1α expression (27), whereas the compound NSC-644221 has been reported to inhibit HIF-1α expression in a topoisomerase II-dependent manner (28). However, transfection of 293T cells with short hairpin RNA that reduced the levels of mRNA for topoisomerase I and/or topoisomerase II had no effect on HIF-1α protein levels in the presence or absence of digoxin (Fig. S5).
Effects of Digoxin on Cell Growth and Survival.
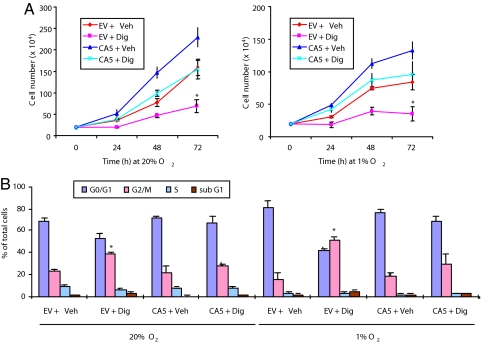
To analyze the consequences of cardiac glycoside treatment of cancer cells ex vivo and in vivo, we focused on digoxin because of its extensive clinical use for the treatment of congestive heart failure. We analyzed PC3 prostate cancer cells that were stably transfected with an expression vector encoding a constitutively active form of HIF-1α (PC3-CA5) or an empty vector (PC3-EV). The expression of endogenous HIF-1α was inhibited by digoxin treatment of cultured PC3-EV and PC3-CA5 cells, whereas HIF-1αCA5 expression was unaffected (Fig. S6A). These results are in agreement with those obtained after transient transfection of 293 cells with expression vector encoding wild type HIF-1α or HIF-1αDM (Fig. 3C), indicating that it is the forced expression of mRNA containing HIF-1α coding sequences in the absence of the untranslated sequences that confers resistance to digoxin. The growth of PC3-EV and PC3-CA5 cells was significantly inhibited by digoxin regardless of O2 concentration (Fig. 5A). The growth of digoxin-treated PC3-CA5 cells was similar to that of vehicle-treated PC3-EV cells. These results suggest that digoxin has both HIF-1α-dependent and HIF-1α-independent effects on cell growth. Analysis of propidium iodide-stained cells by flow cytometry revealed a significant increase in the percentage of PC3-EV cells at the G2-M phase of the cell cycle after 72-h digoxin treatment, as compared to vehicle-treated PC3-EV cells or digoxin-treated PC3-CA5 cells (Fig. 5B). An increased number of subG1 digoxin-treated PC3-EV cells was also observed. Annexin V staining revealed increased apoptosis in PC3-EV, but not PC3-CA5, cells after digoxin treatment (Fig. S6B). These results indicate that digoxin induced both cell cycle arrest and apoptosis that were at least in part dependent on its inhibition of HIF-1α expression.
Fig. 5.
Effect of digoxin on PC3 cell growth. (A) PC3-EV and PC3-CA5 cells were cultured at 20% O2 (Left) or 1% O2 (Right) in the presence of vehicle (Veh) or digoxin (Dig) for 0–72 h and the number of viable cells was determined by trypan blue counting. (B) PC3 subclones were cultured for 72 h in the presence of vehicle or digoxin, stained with propidium iodide, and subjected to flow cytometry to determine the percentage of cells in each cell cycle stage. [*, P < 0.01 vs. all other conditions by 3-way ANOVA (multiple comparisons performed with Holm-Sidak method).]
Anti-Cancer Effects of Digoxin In Vivo.
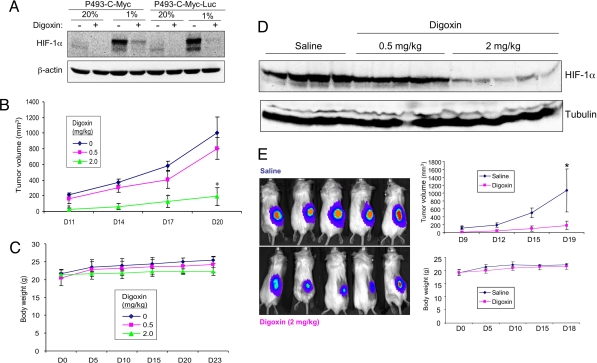
P493-Myc cells, which are transformed human B-lymphocytes, form tumors in SCID mice in a Myc-dependent manner, as treatment of the cells with doxycycline represses Myc expression and blocks tumor growth (29). We chose these cells for in vivo studies because we previously demonstrated that inhibition of HIF-1α expression by RNA interference dramatically inhibited the growth of P493-Myc tumor xenografts in severe combined immunodeficiency (SCID) mice (29). Digoxin treatment blocked hypoxia-induced HIF-1α expression in cultured P493-Myc cells (Fig. 6A). Daily intraperitoneal (i.p.) injections of mice with 0.5 or 2 mg/kg of digoxin significantly inhibited the growth of P493-Myc tumor xenografts in a dose-dependent manner (Fig. 6B), with only modest effects on body weight (Fig. 6C). The anti-tumor effect of digoxin paralleled the dose-dependent inhibition of HIF-1α protein expression in the tumors (Fig. 6D). Subcutaneous (s.c.) injection of P493-Myc-Luc cells that were engineered to express a luciferase expression vector allowed the effects of drug treatment to be analyzed at an early time point (day 8) and replicated the effects on tumor volume at later time points (Fig. 6E).
Fig. 6.
Effect of digoxin on HIF-1α expression and tumor xenograft growth. (A) Parental P493-Myc cells and a subclone expressing firefly luciferase (P493-Myc-Luc) were cultured at 20% or 1% O2 for 24 h in the presence of vehicle (−) or 100 nM digoxin (+). (B–D) 2.5 × 107 P493-Myc cells were implanted in s.c. tissue on the flanks of SCID mice (n = 4–5 in each group), which were treated with daily i.p. injections of 0, 0.5, or 2 mg/kg of digoxin in saline, starting 3 days before tumor cell implantation. Tumor volume was determined every 3 days based on caliper measurements; means ± SEM are shown. *, P < 0.05 (Student's t test) (B). Body weight was determined every 5 days; means ± SEM are shown (C). Tumors were harvested on day 23, and tissue lysates were subjected to immunoblot assays (D; each lane represents an independent tumor). (E) P493-Myc-Luc cells were implanted in s.c. tissue on the flanks of SCID mice (n = 5 each), which were treated with daily i.p. injections of saline or digoxin (2 mg/kg) starting 3 days before tumor cell implantation. Luciferase activity was measured on day 8 after tumor cell implantation (Left). Tumor volume (Top Right) and body weight (Bottom Right) measurements were performed on the indicated days; mean ± SEM are shown. [*, P < 0.05 (Student's t test).]
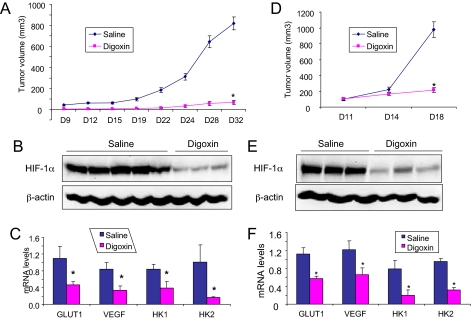
Significant inhibition of tumor growth (Fig. 7A), with no significant effect on body weight (Fig. S7A), was also observed in SCID mice harboring s.c. xenografts of PC3 cells that were treated with 2 mg/kg of digoxin by daily i.p. injection. In this experiment, tumors were palpable in all five vehicle-treated mice by day 9, whereas tumors in the five digoxin-treated mice were first palpable on day 9, 15, 15, 24, and 28, respectively. Tumors from digoxin-treated mice expressed significantly reduced levels of HIF-1α protein (Fig. 7B) and mRNA for GLUT1, HK1, HK2, and VEGF (Fig. 7C).
Fig. 7.
Pretreatment of PC3 and delayed treatment of P493-Myc xenografts with digoxin. (A–C) PC3 cells were implanted in s.c. tissue on the flanks of SCID mice, which were treated with daily i.p. injections of saline or 2 mg/kg of digoxin (n = 5 each) starting 3 days before tumor cell implantation. Tumor volume was determined every 2–4 days based on caliper measurements; means ± SEM are shown (A). Tumors were harvested on day 35 and split in half. Tissue protein lysates were subjected to immunoblot assays (B; each lane represents an independent tumor). Total RNA was extracted and relative levels of GLUT1, VEGF, HK1, and HK2 mRNA were determined by quantitative real-time RT-PCR; means ± SD are shown (C). (D–F) P493-Myc cells were implanted in s.c. tissue on the flanks of SCID mice (n = 3 for each group) and tumors were allowed to grow to a volume of ≈100 mm3 before initiation of treatment on day 11 with saline or digoxin (2 mg/kg). Tumor volume was measured on days 11, 14, and 18 (D). Tumors were harvested on day 18 and subjected to protein (E) and mRNA (F) analyses. [*, P < 0.05 (Student's t test).]
When treatment with digoxin was delayed until day 11 after s.c. implantation of P493-Myc cells, at which time mean tumor volume had reached 100 mm3, tumor growth arrested within 7 days (Fig. 7D) without significant effects on body weight (Fig. S7B). Treatment for 7 days also resulted in a significant reduction in the intratumoral levels of HIF-1α protein (Fig. 7E) as well as GLUT1, HK1, HK2, and VEGF mRNA (Fig. 7F). Similar results were obtained when the treatment of PC3 tumor-bearing mice was delayed until day 20, at which time mean tumor volume had reached 200 mm3 (Fig. S8).
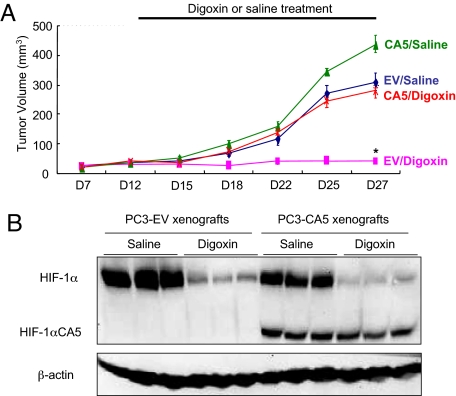
These results demonstrate that digoxin effectively blocks HIF-1α expression in P493 and PC3 cells in vitro and in vivo and that this inhibition is associated with a significant reduction in tumor xenograft growth. To demonstrate that the anti-tumor effects of digoxin were dependent on its inhibition of HIF-1α expression, we injected SCID mice with the PC3-EV and PC3-CA5 subclones that were previously analyzed for cell growth and survival (Fig. 5). When tumor-bearing mice were treated with digoxin, the growth of xenografts derived from PC3-EV cells was significantly inhibited, whereas the growth of digoxin-treated PC3-CA5 xenografts was similar to that of untreated PC3-EV xenografts (Fig. 8A). Immunoblot analysis of tumor lysates revealed that expression of endogenous HIF-1α in PC3-EV and PC3-CA5 xenografts was inhibited by digoxin treatment, whereas expression of HIF-1αCA5 in PC3-CA5 xenografts was unaffected (Fig. 8B). These results demonstrate that inhibition of HIF-1α expression is the critical mechanism by which digoxin inhibits the growth of PC3 tumor xenografts.
Fig. 8.
PC3 cells with enforced expression of HIF-1α are resistant to digoxin. (A) PC3-EV and PC3-CA5 cells were implanted in s.c. tissue of SCID mice, which were treated with daily i.p. injections of saline or 2 mg/kg of digoxin, starting on day 12 after implantation. Tumor volumes were determined every 2–4 days (means ± SD shown; n = 5 for each group). [*, P < 0.05 vs. all other conditions (Student's t test).] (B) Tumor lysates were prepared on day 27 for immunoblot assays.
Discussion
Our screen of 3,120 drugs revealed that cardiac glycosides are potent inhibitors of HIF-1α synthesis. Cardiac glycosides are a class of drugs derived from plants of the genera Digitalis, Strophanthus, and others, which have been prescribed for centuries to treat congestive heart failure and arrhythmias. In these conditions, cardiac glycosides bind to the Na+/K+ ATPase and inhibit its activity (30). Endogenous ouabain and proscillaridin A-like compounds have been detected in mammalian tissues and body fluids. Studies performed over the last decade have suggested that cardiac glycosides may have activity as anti-cancer agents (30, 31). Long-term follow-up of 175 breast cancer patients who were taking digitalis preparations revealed a 6% death rate compared to 34% in those who were not on digitalis treatment (32). A study of over 9,000 patients taking digitoxin revealed an association between high plasma levels and decreased risk of leukemia/lymphoma and kidney/urinary tract cancers (33). Digitoxin inhibited the development of skin papillomas in mice treated with dimethylbenzanthracene and tetradecanoylphorbol ester (34). Digoxin inhibited the growth of neuroblastoma tumor xenografts in mice and angiogenesis in chick chorioallantoic membrane assays (35). UNBS1450, a hemisynthetic compound with increased affinity for the Na/K ATPase, inhibited the growth of A549 nonsmall cell lung cancer orthotopic xenografts (36).
Although inhibition of HIF-1α synthesis by cardiac glycosides is clearly a class effect, the mechanism of action of these drugs remains unclear. In our studies, knockdown of mRNA encoding the Na+/K+ ATPase α1 subunit in 293 cells by >80% did not affect HIF-1α expression (in the absence of cardiac glycosides) or the ability of cardiac glycosides to inhibit HIF-1α expression. The levels of mRNA encoding the α2 subunit in 293 cells were very low (CT = 36 cycles compared to 22 cycles for α1). However, in the absence of a complete knockout of both Na+/K+ ATPase α1 and α2 subunit expression, this mechanism of action cannot be excluded.
Both digoxin and rapamycin share in common the fact that they have modest effects on global protein synthesis but very potently inhibit HIF-1α mRNA translation. Rapamycin blocks HIF-1α protein synthesis by inhibiting mTOR, but our studies demonstrated that cardiac glycosides did not affect mTOR activity. Exposure of cancer cells to hypoxia induces the formation of stress granules containing mRNAs that are not translated until the cells are reoxygenated (37). If cardiac glycosides were to promote the sequestration of HIF-1α mRNA in stress granules, it might explain both the loss of HIF-1α protein expression and the increase in HIF-1α mRNA levels in drug-treated hypoxic cells. Further analysis of the mechanism of action of cardiac glycosides may lead to novel insights into the regulation of HIF-1α protein synthesis. Finally, whereas inhibition of HIF-1 activity is required for the anti-cancer effect of digoxin in PC3 prostate cancer cells, cardiac glycosides have been reported to affect multiple signaling pathways, suggesting that the anti-cancer effects of digoxin may be multifactorial and context dependent.
In the present study, we have demonstrated that digoxin (i) prolongs tumor latency and inhibits tumor xenograft growth in mice when treatment is initiated before the s.c. implantation of P493-Myc, P493-Myc-Luc, PC3, and Hep3B (data not shown) cells; (ii) arrests tumor growth when treatment is initiated after the establishment of PC3 and P493-Myc tumor xenografts; (iii) inhibits the expression of HIF-1α protein and the HIF-1 target genes VEGF, GLUT1, HK1, and HK2; and (iv) does not effectively inhibit the growth of xenografts derived from transfected cells with enforced HIF-1α expression. Further studies are required to determine the range of tumor cell types in which digoxin or other cardiac glycosides effectively inhibit intratumoral expression of HIF-1α and tumor growth. Therapeutic plasma concentrations of digoxin in cardiac patients (≈10–30 nM) are somewhat lower than the concentration required for maximal inhibition of HIF-1α expression in hypoxic cancer cells after 24 h of drug treatment. However, effective inhibition at lower concentrations might be observed after more prolonged exposure of cells to the drug. The ability to both inhibit HIF-1α protein synthesis with digoxin or rapamycin (14) and to promote HIF-1α protein degradation with HSP90 inhibitors such as 17-AAG (20) or with an antioxidant such as N-acetyl cysteine or ascorbate (29) suggests that combination therapy may provide synergistic effects leading to increased efficacy and decreased resistance.
Clinical trials of digoxin in nonsmall cell lung cancer (in combination with Erlotinib) and ErbB2-positive breast cancer (with Lapatinib) are currently recruiting patients (ClinicalTrials.gov identifiers NCT00281021 and NCT00650910, respectively). A dominant tenet of contemporary chemotherapy is that treatments are targeted to patients based on specific molecular characteristics of their cancer. For example, HIF-1α over-expression identifies patients with kidney cancer that are sensitive to mTOR inhibitors (15). Increased HIF-1α expression is associated with increased patient mortality in both ErbB2-positive breast cancer (38) and nonsmall cell lung cancer (39). Knowledge of the relevant therapeutic target of digoxin in human cancer will allow this drug to be used in a more rational manner. Specifically, our findings suggest that immunohistochemical analysis of HIF-1α in tumor biopsy sections (16, 17) may identify a subset of breast and lung cancer patients in current and future trials who are most likely to respond favorably to the inclusion of digoxin in combination chemotherapy regimens. Furthermore, our dose-response (Fig. 6) and cell transfection (Fig. 8) studies demonstrated that efficient inhibition of HIF-1α expression is required for anti-tumor efficacy of digoxin. It remains to be determined whether intratumoral HIF-1α expression is effectively inhibited at the digoxin doses being administered in ongoing clinical trials.
Materials and Methods
Tissue Culture Cells and Reagents.
Human Hep3B, Hep3B-c1, 293, and 293T cells were cultured in Dulbecco's modified essential medium (DMEM). PC3, PC3-EV, PC3-CA5 (29) P493-Myc, and P493-Myc-Luc cells were cultured in RPMI-1640. The media were supplemented with 10% FBS and 1% penicillin-streptomycin.
Drug Library Screening.
Hep3B cells were stably transfected with reporter plasmids p2.1 and pSV-Renilla (14, 19). 1 × 104 Hep3B-c1 cells were seeded in each well of duplicate 96-well plates and cultured in DMEM with 10% FBS. The following day, the cells were pretreated with chemical compounds from the Hopkins Drug Library (21) or vehicle (dimethyl sulfoxide) for 2 h. The plate was then placed in a modular incubator chamber, which was flushed with a gas mixture consisting of 1% O2, 5% CO2, and balance N2. The sealed chamber was incubated at 37°C for 24 h. The ratio of firefly/Renilla luciferase activity was determined by using the Dual Luciferase Assay System (Promega) and a Victor3 Microplate Reader (PerkinElmer). For subsequent validation studies in cultured cells, digoxin, ouabain, and proscillaridin A were purchased from Sigma–Aldrich.
Immunoblot Analysis.
Proteins extracted from cells with RIPA buffer were fractionated by 10% SDS/PAGE. Antibodies against HIF-1α (16), HIF-2α (Novus Biologicals), FLAG (Sigma-Aldrich), phospho-RPS6 and total RPS6 (Cell Signaling Technology) were used for immunoblot assays. Blots were stripped and re-probed with a polyclonal β-actin antibody (Santa Cruz Biotechnology) to confirm equal protein loading.
Quantitative Real-time RT-PCR.
Total RNA was isolated by using TRIzol reagent (Invitrogen) followed by DNase (Ambion) treatment according to the manufacturers' instructions. One microgram of total RNA was used for first-strand cDNA synthesis by using iScript cDNA Synthesis system (BioRad Laboratories). cDNA samples were diluted 1:10 and real-time PCR was performed by using iQ SYBR Green Supermix and the iCycler Real-time PCR Detection System (Bio-Rad). Primers for GLUT1, HK1, HK2, VEGF, Na+/K+ ATPase α1 subunit, topoisomerase I, and topoisomerase II mRNA (Table S1) were designed by using Beacon Designer software (BioRad) and annealing temperature was optimized by gradient PCR. The fold change in expression of each target mRNA relative to 18S rRNA was calculated based on the threshold cycle (Ct) as 2−Δ(ΔCt), where ΔCt = Cttarget − Ct18S and Δ(ΔCt) = ΔCttreatment − ΔCtcontrol.
Transient Transfection Assays.
293 or 293T cells were transfected with Fugene-6 (Roche Applied Science). Expression vectors encoding FLAG-tagged HIF-1α and HIF-1α-DM were previously described (20). The shRNA targeting sequences are shown in Table S2. shRNA oligonucleotides were annealed and ligated into BglII/HindIII-digested pSUPER-retro-neo-GFP (OligoEngine).
Pulse-Labeling.
Cells were pretreated with vehicle, rapamycin, cycloheximide, or digoxin for 4 h and labeled with [35S]-methionine/cysteine (200 μCi/ml; Amersham Pharmacia Biotech) for 1 h. Whole cell lysates were prepared by using 10 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 0.1% SDS, 1% deoxycholate, and 1 mM PMSF. HIF-1α was immunoprecipitated with monoclonal antibody H1α67 (16), and subjected to SDS/PAGE followed by autoradiography.
Apoptosis Assays.
Apoptosis was measured by flow cytometry by using the Annexin V-PE Apoptosis Detection Kit (BD Bioscience) according to the manufacturer's instructions.
Xenograft Assays.
All animal studies were performed according to protocols approved by the Johns Hopkins University Animal Care and Use Committee and were in accordance with the NIH Guide for the Care and Use of Laboratory Animals. Digoxin and saline for injection were obtained from the pharmacy of The Johns Hopkins Hospital. Subcutaneous implantation of P493-Myc, P493-Myc-Luc, PC3, PC3-EV, and PC3-CA5 cells into athymic SCID mice (National Cancer Institute) and Xenogen imaging were performed as previously described (29). Tumors were measured in three dimensions (a, b, c) and volume (V) was calculated: V = abc × 0.52. Tumors were harvested 4 h after the last dose of digoxin. One-third of each tumor was processed for RNA isolation and a tissue protein lysate was prepared from the remaining two-thirds.
Supplementary Material
Acknowledgments.
We thank Karen Padgett (Novus Biologicals, Littleton, CO) for providing anti-HIF-2α antibodies and David Kass and Philip Cole (Johns Hopkins University) for helpful discussions. This work was supported by the Flight Attendant Medical Research Institute and The Johns Hopkins Institute for Cell Engineering.
Footnotes
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected in 2008.
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809763105/DCSupplemental.
References
- 1.Fukumura D, Jain RK. Tumor microenvironment abnormalities: Causes, consequences, and strategies to normalize. J Cell Biochem. 2007;101:937–949. doi: 10.1002/jcb.21187. [DOI] [PubMed] [Google Scholar]
- 2.Dewhirst MW, Cao Y, Moeller B. Cycling hypoxia and free radicals regulate angiogenesis and radiotherapy response. Nat Rev Cancer. 2008;8:425–437. doi: 10.1038/nrc2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vaupel P, Mayer A. Hypoxia in cancer: Significance and impact on clinical outcome. Cancer Metastasis Rev. 2007;26:225–239. doi: 10.1007/s10555-007-9055-1. [DOI] [PubMed] [Google Scholar]
- 4.Manalo DJ, et al. Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood. 2005;105:659–669. doi: 10.1182/blood-2004-07-2958. [DOI] [PubMed] [Google Scholar]
- 5.Elvidge GP, et al. Concordant regulation of gene expression by hypoxia and 2-oxoglutarate-dependent dioxygenase inhibition: The role of HIF-1α, HIF-2α, and other pathways. J Biol Chem. 2006;281:15215–15226. doi: 10.1074/jbc.M511408200. [DOI] [PubMed] [Google Scholar]
- 6.Semenza GL. Evaluation of HIF-1 inhibitors as anticancer agents. Drug Discov Today. 2007;12:853–859. doi: 10.1016/j.drudis.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 7.Rankin EB, Giaccia AJ. The role of hypoxia-inducible factors in tumorigenesis. Cell Death Differ. 2008;15:678–685. doi: 10.1038/cdd.2008.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang GL, Semenza GL. Purification and characterization of hypoxia-inducible factor 1. J Biol Chem. 1995;270:1230–1237. doi: 10.1074/jbc.270.3.1230. [DOI] [PubMed] [Google Scholar]
- 10.Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: The central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 11.Maxwell PH, et al. The tumor suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 12.Garcia JA. HIFing the brakes: Therapeutic opportunities for treatment of human malignancies. Sci STKE. 2006;2006(337):pe25. doi: 10.1126/stke.3372006pe25. [DOI] [PubMed] [Google Scholar]
- 13.Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441:424–430. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- 14.Laughner E, Taghavi P, Chiles K, Mahon PC, Semenza GL. HER2 (neu) signaling increases the rate of hypoxia-inducible factor 1α (HIF-1α) synthesis: Novel mechanism for HIF-1-mediated vascular endothelial growth factor expression. Mol Cell Biol. 2001;21:3995–4004. doi: 10.1128/MCB.21.12.3995-4004.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas GV, et al. Hypoxia-inducible factor determines sensitivity to inhibitors of mTOR in kidney cancer. Nat Med. 2006;12:122–127. doi: 10.1038/nm1337. [DOI] [PubMed] [Google Scholar]
- 16.Zhong H, et al. Overexpression of hypoxia-inducible factor 1α in common human cancers and their metastases. Cancer Res. 1999;59:5830–5835. [PubMed] [Google Scholar]
- 17.Talks KL, et al. The expression and distribution of the hypoxia-inducible factors HIF-1α and HIF-2α in normal human tissues, cancers, and tumor-associated macrophages. Am J Pathol. 2000;157:411–421. doi: 10.1016/s0002-9440(10)64554-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melillo G. Targeting hypoxia cell signaling for cancer therapy. Cancer Metastasis Rev. 2007;26:341–352. doi: 10.1007/s10555-007-9059-x. [DOI] [PubMed] [Google Scholar]
- 19.Semenza GL, et al. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J Biol Chem. 1996;271:32529–32537. doi: 10.1074/jbc.271.51.32529. [DOI] [PubMed] [Google Scholar]
- 20.Liu YV, et al. RACK1 competes with HSP90 for binding to HIF-1α and is required for O2-independent and HSP90 inhibitor-induced degradation of HIF-1α. Mol Cell. 2007;25:207–217. doi: 10.1016/j.molcel.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chong CR, et al. Inhibition of angiogenesis by the antifungal drug itraconazole. ACS Chem Biol. 2007;2:263–270. doi: 10.1021/cb600362d. [DOI] [PubMed] [Google Scholar]
- 22.Agani FH, Pichiule P, Chavez JC, LaManna JC. Inhibitors of mitochondrial complex I attenuate the accumulation of hypoxia-inducible factor-1 during hypoxia in Hep3B cells. Comp Biochem Physiol A. 2002;132:107–109. doi: 10.1016/s1095-6433(01)00535-9. [DOI] [PubMed] [Google Scholar]
- 23.Epstein AC, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 24.Arsham AM, Howell JJ, Simon MC. A novel hypoxia-inducible factor-independent hypoxic response regulating mammalian target of rapamycin and its targets. J Biol Chem. 2003;278:29655–29660. doi: 10.1074/jbc.M212770200. [DOI] [PubMed] [Google Scholar]
- 25.Al-Ghoul M, Valdes R., Jr Mammalian cardenolides in cancer prevention and therapeutics. Ther Drug Monit. 2008;30:234–238. doi: 10.1097/FTD.0b013e31816b90ff. [DOI] [PubMed] [Google Scholar]
- 26.Bielawski K, Winnicka K, Bielawska A. Inhibition of DNA topoisomerases I and II, and growth inhibition of breast cancer MCF-7 cells by ouabain, digoxin and proscillaridin A. Biol Pharm Bull. 2006;29:1493–1497. doi: 10.1248/bpb.29.1493. [DOI] [PubMed] [Google Scholar]
- 27.Rapisarda A, et al. Identification of small molecule inhibitors of hypoxia-inducible factor 1 transcriptional activation pathway. Cancer Res. 2002;62:4316–4324. [PubMed] [Google Scholar]
- 28.Creighton-Gutteridge M, et al. Cell type-specific, topoisomerase II-dependent inhibition of hypoxia-inducible factor-1α protein accumulation by NSC 644221. Clin Cancer Res. 2007;13:1010–1018. doi: 10.1158/1078-0432.CCR-06-2301. [DOI] [PubMed] [Google Scholar]
- 29.Gao P, et al. HIF-dependent antitumorigenic effect of antioxidants in vivo. Cancer Cell. 2007;12:230–238. doi: 10.1016/j.ccr.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mijatovic T, et al. Cardiotonic steroids on the road to anti-cancer therapy. Biochim Biophys Acta. 2007;1776:32–57. doi: 10.1016/j.bbcan.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 31.Newman RA, Yang P, Pawlus AD, Block KI. Cardiac glycosides as novel cancer therapeutic agents. Mol Interv. 2008;8:36–49. doi: 10.1124/mi.8.1.8. [DOI] [PubMed] [Google Scholar]
- 32.Stenkvist B. Is digitalis a therapy for breast carcinoma? Oncol Rep. 1999;6:493–496. [PubMed] [Google Scholar]
- 33.Haux J, Klepp O, Spigset O, Tretli S. Digitoxin medication and cancer; Case control and internal dose-response studies. BMC Cancer. 2001;1:11. doi: 10.1186/1471-2407-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inada A, et al. Anti-tumor promoting activities of natural products. II. Inhibitory effects of digitoxin on two-stage carcinogenesis of mouse skin tumors and mouse pulmonary tumors. Biol Pharm Bull. 1993;16:930–931. doi: 10.1248/bpb.16.930. [DOI] [PubMed] [Google Scholar]
- 35.Svensson A, Azarbayjani F, Bäckman U, Matsumoto T, Christofferson R. Digoxin inhibits neuroblastoma tumor growth in mice. Anticancer Res. 2005;25:207–212. [PubMed] [Google Scholar]
- 36.Mijatovic T, et al. The cardenolide UNBS1450 is able to deactivate nuclear factor κB-mediated cytoprotective effects in human non small cell lung cancer cells. Mol Cancer Ther. 2006;5:391–399. doi: 10.1158/1535-7163.MCT-05-0367. [DOI] [PubMed] [Google Scholar]
- 37.Moeller BJ, Cao Y, Li CY, Dewhirst MW. Radiation activates HIF-1 to regulate vascular radiosensitivity in tumors: Role of reoxygenation, free radicals, and stress granules. Cancer Cell. 2004;5:429–441. doi: 10.1016/s1535-6108(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 38.Giatromanolaki A, et al. c-erbB-2 related aggressiveness in breast cancer is hypoxia inducible factor-1α dependent. Clin Cancer Res. 2004;10:7972–7977. doi: 10.1158/1078-0432.CCR-04-1068. [DOI] [PubMed] [Google Scholar]
- 39.Swinson DE, et al. Hypoxia-inducible factor-1α in non small cell lung cancer: Relation to growth factor, protease and apoptosis pathways. Int J Cancer. 2004;111:43–50. doi: 10.1002/ijc.20052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.