Abstract
Telomeres shield the natural ends of chromosomes from nucleolytic attack, recognition as double-strand breaks, and inappropriate processing by DNA repair machinery. The trimeric Stn1/Ten1/Cdc13 complex is critical for chromosome end protection in Saccharomyces cerevisiae, while vertebrate telomeres are protected by shelterin, a complex of six proteins that does not include STN1 or TEN1. Recent studies demonstrate that Stn1 and Ten1 orthologs in Schizosaccharomyces pombe contribute to telomere integrity in a complex that is distinct from the shelterin components, Pot1 and Tpp1. Thus, chromosome-end protection may be mediated by distinct subcomplexes of telomere proteins. Here we report the identification of a STN1 gene in Arabidopsis that is essential for chromosome-end protection. AtSTN1 encodes an 18-kDa protein bearing a single oligonucleotide/oligosaccharide binding fold with significant sequence similarity to the yeast Stn1 proteins. Plants null for AtSTN1 display an immediate onset of growth and developmental defects and reduced fertility. These outward phenotypes are accompanied by catastrophic loss of telomeric and subtelomeric DNA, high levels of end-to-end chromosome fusions, increased G-overhang signals, and elevated telomere recombination. Thus, AtSTN1 is a crucial component of the protective telomere cap in Arabidopsis, and likely in other multicellular eukaryotes.
Keywords: telomere, anaphase bridges, oligonucleotide/oligosaccharide binding fold, G-overhang
Telomeres distinguish the natural ends of chromosomes from double-strand breaks by virtue of their unusual architecture and protein composition. Vertebrate telomeres are bound by a core complex of six proteins, termed “shelterin,” which regulates the length of the telomeric DNA tract, suppresses the activation of a DNA damage response at the terminus, and protects the ends from inappropriate recombination, nuclease attack, and end-to-end fusion (1, 2). Shelterin is composed of two double-strand telomere binding proteins, TRF1 and TRF2, a single-strand telomere binding protein, POT1, and three bridging proteins TIN2, RAP1, and TPP1 (1, 2). TRF2 and the oligonucleotide/oligosaccharide binding fold (OB-fold) containing protein POT1 are critical for chromosome end protection (3–6). Studies in Schizosaccharomyces pombe confirm the presence of several shelterin homologs, including Taz1 (a TRF1/TRF2 homolog), Rap1, Pot1, and Tpz1 (a TPP1 homolog) (7–11).
In contrast, budding yeast telomeres are protected by a trimeric complex of three OB-fold proteins, Stn1/Ten1/Cdc13 (12–14). Recent studies demonstrate that Stn1 and Ten1 orthologs in S. pombe also contribute to telomere capping (15). Notably, SpStn1 and SpTen1 interact with each other, but thus far evidence is lacking for a physical interaction between these proteins and SpPot1 (15). Furthermore, Tpz1, but not Stn1/Ten1, was identified by mass spectrometry of Pot1-associated proteins in S. pombe (11), indicating that in S. pombe chromosome ends are protected by two distinct telomere protein subcomplexes.
Ten1 has so far only been discerned in fungi (15, 16). However, several candidate orthologs of the SpStn1 protein can be found in the genomes of higher eukaryotes, including human beings, by position-specific iterative BLAST (PSI-BLAST) (15, 17). Here we use a genetic approach to demonstrate that the STN1 gene in the flowering plant Arabidopsis is essential for chromosome-end protection. In striking contrast to plants lacking telomerase, which display a progressive but gradual loss of telomeric DNA that ultimately leads to end-to-end chromosome fusions and worsening growth and developmental defects beginning in the sixth generation (G6) (18), telomeres are immediately and catastrophically compromised in Arabidopsis mutants null for STN1. Telomeric and subtelomeric DNA are extensively eroded and mutants exhibit increased G-overhang signals, elevated telomere recombination, and massive telomere fusion, resulting in severe growth defects and sterility. These findings not only indicate that AtSTN1 is required for telomere capping in Arabidopsis, but further suggest that additional key components of the telomere complex remain to be elucidated in metazoa.
Results
Identification of AtSTN1.
To search for a STN1 protein in the plant kingdom, PSI-BLAST was employed using the protein sequence of SpStn1 as the query. In the second iteration, a previously uncharacterized protein, NP_563781, from Arabidopsis thaliana was uncovered with an E-value of 2e−06, well above the program threshold (0.005). The corresponding single-copy gene, At1g07130, was designated AtSTN1. A combination of EST database searches and 3′ RACE was used to verify the boundaries of the AtSTN1 coding region. AtSTN1 lacks introns and is predicted to encode a small protein of 160 aa that can assume a single OB-fold (Fig. 1A). AtSTN1 mRNA is expressed in all plant tissues examined [supporting information (SI) Fig. S1], unlike the mRNA for TERT, the catalytic subunit of telomerase, which accumulates only in highly proliferative organs (19).
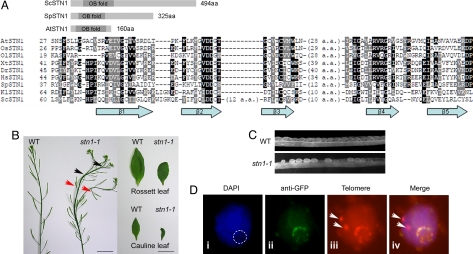
Fig. 1.
Identification of AtSTN1 and severe morphological defects in STN1 deficient plants. (A) (Top) Diagram showing the OB-fold domain structure of STN1 homologs from S. cerevisiae (Sc), S. pombe (Sp) and A. thaliana (At). (Bottom) Alignment of putative STN1 orthologs from plants and other organisms generated by Macvector and Boxshade software. The secondary structure was predicted by PSIPRED (20). At, A. thaliana; Dr, D. rerio; Hs, H. sapiens; Kl, K. lactis; Ol, O. lucimarinus; Os, O. sativa; Sc, S. cerevisiae; Sp, S. pombe; Xt, X. tropicalis (see SI). (B) Morphological defects in stn1–1 mutants. Stems (Left), rosette leaves (Top right), and cauline leaves (Bottom right) are shown for WT plants and stn1–1 mutants. Fused stems (black arrows) and altered phyllotaxy (red arrows) are indicated. (Scale bars, 1 cm.) (C) Aborted seed development in stn1–1 mutants. Siliques from WT plants and stn1–1 mutants were visualized by microscopy. (D) STN1 colocalizes with telomeres. Isolated nuclei from STN1-YFP transformants were stained with DAPI (i) [position of the nucleolus (absence of DAPI staining) indicated by the dashed line]; STN1-YFP was detected with an anti-GFP antibody (ii); and the telomeres were labeled by FISH with a telomere probe (iii) (see Materials and Methods for details). Panels (i) to (iii) were superimposed to produce panel (iv). Arrows in (iii) and (iv) indicate internal stretches of telomere signals as described in (21).
Database searches revealed potential STN1 homologs from other sequenced plant genomes, including rice and single-celled green algae (see Fig. 1A). As expected (15, 17), putative STN1 homologs were also uncovered in a wide variety of other eukaryotes, including fishes, amphibians, birds, rodents, and primates (see Fig. 1A and data not shown). In contrast to STN1 orthologs from yeast, the plant STN1 proteins lack a C-terminal extension (see Fig. 1A).
Protein sequence alignment indicated that AtSTN1 displays limited sequence similarity to SpStn1 (see Fig. 1A), but this similarity is statistically significant within the predicted OB-fold domain. Positions 7–143 of AtSTN1 align to positions 16–136 of SpStn1 with 23% identity and 40% similarity. Secondary-structure prediction by PSIPRED (20) indicated that residues within four of the five essential beta strands of the OB-fold (β1, β2, β3, and β4) in AtSTN1 share significant similarity to that of functionally verified STN1 protein from yeasts, as well as the putative STN1 proteins from other higher eukaryotes (see Fig. 1A). The sequence conservation of β5 is reduced in the Stn1 sequences from higher eukaryotes relative to their counterparts in yeasts. PFAM analysis confirmed that both AtSTN1 and SpStn1 proteins contain a “tRNA_anti” OB-fold nucleic acid-binding domain, arguing that the OB-fold domain of the two proteins belongs to the same family. Results of PFAM analysis can be retrieved for AtSTN1 (http://pfam.sanger.ac.uk/protein?entry=Q9LMK5) and for SpStn1 (http://pfam.sanger.ac.uk/protein?entry=Q0E7J7).
Severe Morphological Defects in Arabidopsis stn1 Mutants.
We examined the in vivo function of AtSTN1 by studying two T-DNA insertion lines, designated stn1–1 and stn1–2, which were obtained from the Arabidopsis Biological Resource Center. RT-PCR analysis of homozygous mutants confirmed that full-length AtSTN1 mRNA was disrupted in both lines (Fig. S2). Both mutant lines displayed a fasciated phenotype with severe morphological abnormalities in G1, although the severity of the defects varied somewhat in different individuals. In nearly all mutants, apical dominance was completely abolished, leading to multiple inflorescence bolts that were often fused (Fig. 1B, black arrows). In addition, floral phyllotaxy was perturbed and siliques developed at irregular positions on the inflorescence bolt (see Fig. 1B, red arrows). Similar to what has been observed in late generation (G8–G9) tert mutants (18), leaf size was substantially reduced in stn1 mutants, likely reflecting defects in cell proliferation (see Fig. 1B, Right). stn1 mutants produced numerous undeveloped ovules (Fig. 1C) and the germination rate declined dramatically through successive generations. Only 17% (n = 144) of the seeds from G1 mutants germinated to produce G2 plants. G2 progeny (G3) arrested early in vegetative development without producing a germline (data not shown). Many of these phenotypes are reminiscent of late generation tert mutants (18).
AtSTN1 Colocalizes with Arabidopsis Telomeres in Vivo.
To monitor the subcellular localization of AtSTN1, we generated a stn1–1 line expressing a C-terminal YFP tagged version of AtSTN1 under the control of the CaMV 35S promoter. The transgene fully complemented the telomere defects in stn1–1 mutants (see below). In root-tip meristems, distinct spots of YFP signal formed a ring around the periphery of the nucleolus (data not shown). The arrangement of Arabidopsis telomeres at the nucleolar periphery has previously been noted in meiotic interphase (21). FISH with a telomere probe also produced signals at the nucleolar periphery in somatic cells from roots and immature pistils (e.g., Fig. 1D, iii). Immunolocalization using an anti-GFP antibody (Fig. 1D, ii) combined with telomere FISH on the same nuclei produced colocalizing signals (Fig. 1D, iv). This localization was specific to terminal telomeric DNA sequences as the STN1-YFP signal did not overlap with internal stretches of telomeric DNA sequence on chromosome 1 (21) (Fig. 1D, iv, arrows). We conclude that AtSTN1 colocalizes with telomeres in Arabidopsis.
Extensive Telomere Erosion in Plants Lacking AtSTN1.
In S. pombe, the absence of Stn1 leads to an immediate and profound loss of terminal DNA sequences (15). To determine if AtSTN1 protects chromosome ends in Arabidopsis, terminal restriction fragment (TRF) analysis was performed to examine bulk telomere length. In both stn1–1 and stn1–2 mutants, telomere tracts appeared as a broad, heterogeneous smear (Fig. 2A). Although the average length of bulk telomeres was only slightly shorter than in WT siblings (2.4 kb versus 2.7 kb, respectively), the shortest telomere tracts in stn1–1 mutants were significantly shorter than in WT, trailing down to ≈600 bp (1.4 kb shorter than the shortest WT telomeres) (see Fig. 2A). In contrast, telomeres in tert mutants decline much more gradually, reaching 600 bp in G6 or G7 (18).
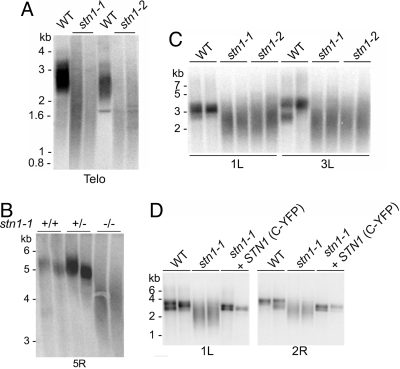
Fig. 2.
Extensive telomere erosion in stn1 mutants. (A) TRF analysis of WT, stn1–1, and stn1–2 mutants. For each genotype, data from two individual sibling plants are shown. The blot was hybridized with a radio-labeled G-rich telomeric probe. Molecular weight makers are indicated. (B) Subtelomeric TRF analysis of WT, heterozygous, and homozygous stn1–1 mutants. The blot was hybridized with a probe specific for the right arm of chromosome 5 (5R). (C) PETRA analysis of WT, stn1–1, and stn1–2 mutants. The blot was hybridized with a telomeric probe. Telomere length on the left arm of chromosomes 1 and 3 (1L and 3L) was measured. (D) PETRA analysis of stn1 mutants expressing a C-terminal YFP tagged WT STN1 transgene. Telomere length was examined on the chromosome arms indicated.
Next, we monitored telomere length dynamics on individual chromosome arms using subtelomeric TRF and primer extension telomere repeat amplification (PETRA). For subtelomeric TRF, we used a probe corresponding to the right arm of chromosome 5 (5R) (Fig. 2B). For PETRA, the left arms of chromosomes 1 and 3 (1L and 3L) were assessed (Fig. 2C). Consistent with conventional TRF analysis, both assays revealed dramatic telomere erosion in plants lacking AtSTN1. Moreover, individual telomere tracts in stn1 mutants spanned a broader size range than those in WT (see Fig. 2 B and C). By contrast, telomere tracts on homologous chromosomes in tert mutants are even more homogenous in size than in WT, typically forming a single sharp band that spans 100 to 200 bp on an agarose gel (22). We confirmed that the telomere defect in stn1–1 mutants was because of the T-DNA insertion in the AtSTN1 gene by complementation. Bulk telomere analysis (data not shown) and PETRA demonstrated that the profile of telomere tracts in stn1–1 plants expressing an AtSTN1 transgene was restored to WT (Fig. 2D).
Finally, we asked whether telomerase activity was diminished in stn1 mutants using a real-time telomere repeat amplification protocol (23, 24). In vitro telomerase activity levels in stn1–1 mutants were approximately the same as in WT plants (Fig. S3). Thus, the loss of telomeric DNA observed in stn1 mutants is not because of telomerase enzyme deficiency, but we cannot rule out the possibility that telomerase access to the telomere is impeded in the absence of AtSTN1.
AtSTN1 is Required to Prevent Telomere Fusions.
Extensive loss of telomeric DNA can trigger end-to-end chromosome fusions. To determine whether telomeres in stn1 mutants engage in end-joining reactions, we monitored the frequency of anaphase bridges in the pistils of these plants. As expected, no bridged chromosomes were observed in WT plants (Fig. 3A and Table S1). However, up to 29% of the anaphases in stn1–1 mutants showed evidence of fused chromosomes (Fig. 3 B–D, see Table S1). This degree of genome instability is not observed in tert mutants until G8 or G9 (18). The immediate and catastrophic onset of genome instability in stn1 mutants reinforces the conclusion that AtSTN1 plays a critical role in chromosome end protection in Arabidopsis.
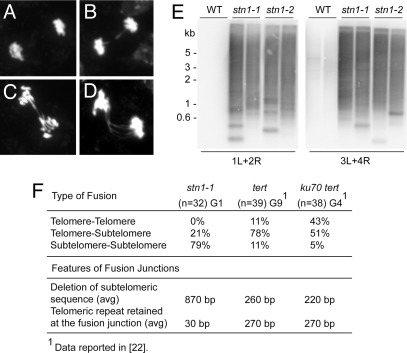
Fig. 3.
STN1 is required to prevent telomere fusions. (A–D) Cytology of mitotic chromosomes in WT (A) and stn1–1 mutants (B–D) is shown. DAPI-stained chromosome spreads were prepared from pistils. Examples of stn1–1 anaphases with one (B), two (C) or four (D) bridges are shown. (E) Telomere fusion PCR products obtained from WT, stn1–1, and stn1–2 mutants were hybridized using a telomeric probe. Primer pairs used to amplify specific subtelomeric regions are indicated. (F) Summary of DNA sequence analysis of cloned telomere fusion junctions in stn1–1 (G1) mutants. Data for tert (G9) and tert ku70 (G4) were taken from a previous study (22).
To further characterize the architecture of chromosome fusion junctions in stn1 mutants, we used telomere fusion PCR using primers directed at unique subtelomeric sequences on different chromosome arms (22). Abundant telomere fusion PCR products were generated with G1 stn1–1 DNA, which appeared as an intense, heterogeneous smear (Fig. 3E). This observation is consistent with our previous studies, showing that telomere fusion is initiated when telomeres shorten below 1 kb (22). Sequence analysis of cloned PCR products showed that the majority (79%) of end-joining events in stn1–1 mutants involved subtelomere-to-subtelomere fusion (Fig. 3F). In contrast, chromosome fusion junctions primarily reflect telomere-to-subtelomere joining in late generation tert mutants (78%), and telomere-to-telomere (43%) or telomere-to-subtelomere (51%) fusions in ku70 tert mutants (see Fig. 3F) (22). Notably, the average deletion of subtelomeric DNA was fourfold greater in stn1–1 mutants (≈870 bp) (see Fig. 3F) than in tert (G9, ≈260 bp) or ku70 tert mutants (G4, ≈220 bp) (22). Because bulk telomere length is much shorter in tert (G9) and in ku70 tert (G4) mutants where an equivalent level of genome instability is observed, our G1 stn1–1 results indicate that at least a subset of telomeres in these mutants suffer extensive nucleolytic attack before being recruited into end-to-end chromosome fusions.
AtSTN1 Is Required to Maintain Proper Telomere Architecture and to Block Formation of Extra-Chromosomal Telomeric Circles.
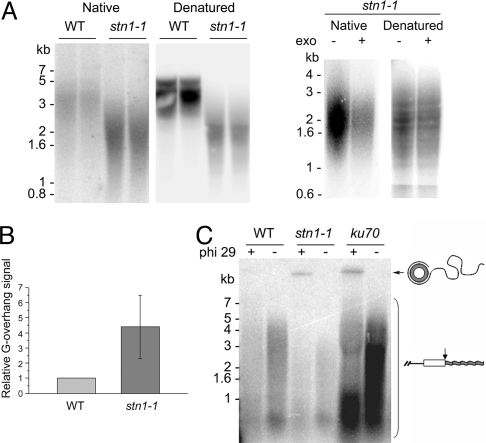
Mutations in Stn1, Ten1, or Cdc13 in Saccharomyces cerevisiae (16, 25, 26) and Stn1 in Kluyveromyces lactis (27) lead to gross elongation of the G-overhang. These data are interpreted to mean that the Stn1 complex protects the telomeric C-strand from degradation. In-gel hybridization was used to determine if AtSTN1 contributes to the maintenance of telomere end structure in Arabidopsis. Relative to WT, the G-overhang signal was increased by approximately fourfold in stn1–1 mutants [Fig. 4A (Left) and B]. As expected, exonuclease treatment indicated that the hybridization signal detected in the native gel correlated with terminal G-overhangs (Fig. 4A, Right). Thus, AtSTN1 is required to maintain the proper architecture of the chromosome terminus.
Fig. 4.
Loss of STN1 leads to increased G-overhang signals and increased telomeric circle formation. (A) In-gel hybridization analysis of DNA isolated from WT and stn1–1 mutants using a C-strand telomeric probe under native and denaturing conditions (Left). The hybridization signal in the native gel was strongly reduced by exonuclease treatment, demonstrating that the signal was dependent on G-overhangs (Right). (B) Quantification of the G-overhang signal. The relative G-overhang signal was determined from five independent experiments as described in Materials and Methods. Data are represented as mean ± SEM. (C) Telomeric circle amplification (TCA) was performed with WT, stn1–1, and ku70 mutant DNA in the presence or absence of phi 29 polymerase. DNA from ku70 mutants served as a positive control. The hybridization signal for linear telomere tracts is indicated by the bracket.
The frequency of telomere recombination is dramatically increased in K. lactis stn1 mutants (27). To determine whether this is also true in plants lacking AtSTN1, we looked for evidence of telomere rapid deletion (TRD). TRD results in large, stochastic deletions of telomere tracts and is thought to occur when the t-loop on the chromosome terminus undergoes branch migration, giving rise to a Holliday junction intermediate that is subsequently resolved to produce a truncated telomere and an extrachromosomal telomeric circle (28). We monitored TRD using telomeric circle amplification (TCA), which detects the telomeric circle by-products of TRD (29). In this procedure, phi29 polymerase is used to amplify telomeric DNA circles into extremely long ssDNA, which is distinguished from endogenous linear telomere fragments based on its slower migration on a denaturing agarose gel. As expected, telomeric circles were enriched in our ku70-mutant control reaction (29) (Fig. 4C). A similar high molecular-weight product was generated in stn1–1 mutants, but not in the WT control. We conclude that STN1 suppresses telomere recombination in Arabidopsis.
Taken together, our data indicate that AtSTN1 is an essential component of the protective telomere cap in Arabidopsis that prevents nucleolytic attack, end-to-end chromosome fusions, and telomere recombination.
Discussion
Although Barbara McClintock described the protective “capping” function of maize telomeres nearly 70 years ago (30), we still know relatively little about why natural chromosome ends are recalcitrant to nuclease attack and end-joining reactions, especially in higher eukaryotes. In part, our understanding has been stymied by the rapid evolution of the telomere protein complex. Here we provide evidence that STN1 is conserved in metazoa and plays an essential role in chromosome-end protection.
AtSTN1 was identified in the second iteration of PSI-BLAST as a protein bearing sequence similarity to the OB-fold domain of S. pombe Stn1. Subsequent analysis revealed putative STN1 orthologs in a variety of plants and vertebrates [present study; (15, 17)]. Structure-based alignment shows significant sequence similarity within four of the five essential beta strands of the core of the OB-fold. While the overall similarity among the Stn1 orthologs is not high, minimal sequence similarity among telomere proteins from different taxa is not without precedent. For example, Pot1 from S. pombe shows only 19% identity and 40% similarity to the TEBP alpha subunit in ciliates, and yet the two proteins are functional and structural homologs (10, 31).
One distinction between the STN1 proteins from plants and yeasts is the absence of a C-terminal extension in the former. Recent studies indicate that the N and C terminus of ScStn1 encode independent and separable functions at the telomere (32, 33). The N-terminal OB-fold of ScStn1 is required for cell viability and mutation of this domain leads to an increase of single-strand DNA at the chromosome terminus (33), arguing that the N-terminal OB-fold is essential for chromosome-end protection. In contrast, the C-terminal domain of ScStn1 is required for telomere-length control and plays no detectable role in telomere capping (33). Like Arabidopsis stn1 mutants, a null mutation in the S. pombe Stn1 leads to severe telomere deprotection phenotype, suggesting the major role of Stn1 in S. pombe and Arabidopsis may be in chromosome-end protection. Notably, S. pombe Stn1 protein is significantly truncated relative to S. cerevisiae Stn1 (325 aa versus 494 aa), consistent with rapid evolution of the C-terminal domain. We hypothesize that the C-terminal domain of STN1 is not crucial for its telomere-capping function in plants and hence was lost in the 1.5 billion years since plants and yeasts shared a common ancestor.
The strongest evidence that AtSTN1 is a functional homolog of the yeast Stn1 proteins is based on genetic data. Plants lacking STN1 display phenotypes that strongly parallel S. pombe stn1 null mutants (15). In both cases, stn1 mutants exhibit an immediate and profound telomere deprotection phenotype. In Arabidopsis mutants, both telomeric and subtelomeric tracts are subjected to extensive nuclease attack. Telomeric C-strands are particularly vulnerable to digestion, creating extended G-overhangs. As a likely consequence, stn1 mutants exhibit increased intrachromosomal telomere recombination as evidenced by an accumulation of telomere circles. TRD may further fuel the erosion of terminal DNA sequences in this setting. The degraded telomeres engage in end-joining reactions, triggering genome-wide instability and the cell-proliferation arrest typical of plants experiencing severe telomere dysfunction (18). We conclude that STN1 is a crucial component of the telomere complex in Arabidopsis essential for chromosome-end protection.
Shelterin homologs have not been clearly defined in plants. Arabidopsis harbors at least six myb-related proteins that bind double-strand telomeric DNA in vitro in a manner similar to vertebrate TRF1 and TRF2 (34), as well as three putative POT1 paralogs. Although the functions of AtPOT1b and AtPOT1c are still under investigation (35) (A.D.L. Nelson and D.E.S., unpublished work), AtPOT1a is a physical component of the telomerase RNP that is required for telomerase action in vivo (36). Strikingly, homologs for RAP1, TPP1, and TIN2 cannot be discerned in the Arabidopsis genome with the current search algorithms, underscoring the conclusion that telomere proteins are evolving at a rapid pace.
Besides STN1, the only other plant protein directly implicated in chromosome end protection is from rice. Like mammalian TRF2, rice telomere binding-protein 1 (RTBP1) bears a myb-like DNA binding domain (37). However, in contrast to TRF2-depleted mammalian telomeres, which activate a strong DNA damage response and massive end-to-end chromosome fusions (3, 38, 39), plants lacking RTBP1 display very gradual telomere lengthening over successive plant generations and only in G2 do telomere fusions become evident (37). This mild phenotype may reflect functional redundancy of myb-bearing telomere proteins in plants (40). STN1, by contrast, is a single-copy gene in all of the sequenced plant genomes we surveyed.
The conserved function of Stn1 in yeasts (13, 15, 25) and STN1 in flowering plants (present study), and the existence of putative homologs in vertebrates (15, 17) argues that this family of proteins contributes to chromosome-end protection in a broad range of eukaryotes. Notably, STN1 was not identified as a component of the shelterin complex (41–43) in mammals. It is conceivable that STN1 interacts only transiently with telomeres (e.g., during a specific period of the cell cycle). Alternatively, STN1 may be part of an end-protection complex distinct from shelterin. In support of this hypothesis, a TPP1 homolog, Tpz1, but not Stn1/Ten1, was recently identified by mass spectrometry of Pot1-associated proteins in S. pombe (11). Interestingly, SpPot1 does not interact with Stn1/Ten1 in a yeast two-hybrid assay (15), implying that S. pombe telomeres are composed of two distinct capping complexes, one bearing Pot1 and Tpz1 (from shelterin) and a second containing Stn1 and Ten1. Given that mammalian shelterin contains orthologs only from the former complex, POT1/TPP1, and that STN1 is a key component of the telomere cap in plants, the data suggest that higher eukaryotic telomeres are protected by a network of telomere protein subcomplexes, the full constituency of which is yet to be elucidated.
Materials and Methods
Plant Materials and Plasmids.
The stn1 mutants were obtained from the Arabidopsis Biological Resource Center. The T-DNA insertion lines, stn1–1 (CS023504) and stn1–2 (CS846727), were genotyped by PCR using primers 5′-ATGGATCGATCCCTCCAAAG-3′ and 5′- TTGAATACGAACACGATAACAAC-3′. Plants were grown according to the conditions described (36). Siliques from WT and stn1–1 mutants were dissected ≈10 days after fertilization and photographed using a Zeiss Axiocam digital camera coupled to a Zeiss microscope. A transgenic construct of STN1 was prepared by inserting a C-terminal YFP tag using an Ala (Gly)5 Ala linker sequence. Tagged STN1 was cloned into a Gateway entry vector pENTR (Invitrogen) and then subcloned into a binary vector pB7WG2 (Invitrogen) according to manufacturer instructions. The resultant binary vector was used to transform plants as described (36).
RT-PCR.
Total RNA was extracted from plant tissues using an RNA purification kit (Fisher Scientifics). Reverse transcription was performed using SuperScript III reverse transcriptase (Invitrogen) per manufacturer's instructions. PCR of STN1 cDNA was performed using the above primers, with the following program: 95 °C 3 min; 25 cycles of 94 °C 20 sec, 55 °C 30 sec, 72 °C 1 min 30 sec; 72 °C 7 min. 3′ RACE was used to map the 3′ end of the STN1 transcript using RLM-RACE kit (Ambion) according to manufacturer's instructions.
Cytology, Immunofluorescence, and FISH.
To monitor anaphase bridge formation, cells were prepared from pistils, stained with DAPI Vectashield (Vector Laboratories), and then analyzed with an epifluorescence microscope (Zeiss) as described (18). Anaphase bridges were scored as a percentage of total anaphase cells. For combined immunolocalization and FISH, second generation transformants (T2) expressing a C-terminal YFP-tagged version of STN1 were grown to seedlings (≈7-days old) and fixed with 4% formaldehyde for 30 min on ice. Root nuclei from the seedlings were extracted and dried onto polylysine-coated slides, and immunolocalization was performed as described (44). A rabbit anti-GFP antibody (Abcam) was used as the primary antibody and a FITC-conjugated donkey anti-rabbit antibody (Jackson ImmunoResearch) was used as the secondary antibody. After immunolocalization, the nuclei were postfixed with 4% formaldehyde and 0.1% glutaraldehyde for 30 min before FISH. Nuclei were washed with 1 × PBS, passed through an ethanol series (70%, 80%, 90%, 100%) at −20 °C and then dried. Digoxygenin-dUTP-labeled telomere probe was prepared as described (21). FISH was performed as described (45). Detection of digoxygenin-labeled probes was with a rhodamine conjugated anti-digoxygenin antibody (Roche). Nuclei were counterstained with DAPI Vectashield and analyzed with an epifluorescence microscope (Zeiss).
TRF, PETRA, and Telomere Fusion PCR.
DNA from individual whole plants was extracted as described (46). TRF analysis was performed using 50 μg of DNA digested with Tru1I (Fermentas) and hybridized with a [32P] 5′ end-labeled (T3AG3)4 oligonucleotide probe (19). The average length of bulk telomeres was determined by Telometric 1.2 (47); the range of telomere length was obtained using ImageQuant software. Subtelomeric TRF analysis was performed using 100 μg of DNA digested with SpeI and PvuII (New England Biolabs) and hybridized with a 5R probe (48). Telomere fusion PCR and PETRA were performed as described (22).
In-Gel Hybridization and Telomeric Circle Amplification.
In-gel hybridization was performed as described (49). The relative amount of single-strand G-overhang was calculated by quantifying the hybridization signal obtained from the native gel and then normalizing this value with the loading control of either interstitial telomere signal from the denaturing blot or ethidium bromide staining of the agarose gel. The single-strand G-overhang signal obtained from WT DNA was set to one and each sample was normalized to this value. Exonuclease treatment was performed by incubating DNA samples with T4 DNA polymerase (New England Biolabs) before in-gel hybridization at 12 °C for 30 min. TCA was performed as described (29).
Supplementary Material
Acknowledgments.
We thank Eugene Shakirov for the initial identification of the AtSTN1 gene in the Arabidopsis database and Anand Venkatraman for invaluable help with the bioinformatics analysis. We are grateful to Barbara Zellinger and Karel Riha for detailed advice on TCA and to Susan Armstrong for the digoxygenin-dUTP-labeled telomere probe for FISH. We also thank Tom McKnight for thoughtful comments on the manuscript and Jeff Kapler and members of the Shippen laboratory for helpful advice. This work was supported by National Institutes of Health Grant GM065383 and National Science Foundation Grant MCB0615928 (to D.E.S.) and Ruth L. Kirschstein National Research Service Award GM800052 (to J.C.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807867105/DCSupplemental.
References
- 1.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 2.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–314. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 3.van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 4.Yang Q, Zheng YL, Harris CC. POT1 and TRF2 cooperate to maintain telomeric integrity. Mol Cell Biol. 2005;25:1070–1080. doi: 10.1128/MCB.25.3.1070-1080.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hockemeyer D, Daniels JP, Takai H, de Lange T. Recent expansion of the telomeric complex in rodents: two distinct POT1 proteins protect mouse telomeres. Cell. 2006;126:63–77. doi: 10.1016/j.cell.2006.04.044. [DOI] [PubMed] [Google Scholar]
- 6.Wu L, et al. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell. 2006;126:49–62. doi: 10.1016/j.cell.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 7.Ferreira MG, Cooper JP. The fission yeast Taz1 protein protects chromosomes from Ku-dependent end-to-end fusions. Mol Cell. 2001;7:55–63. doi: 10.1016/s1097-2765(01)00154-x. [DOI] [PubMed] [Google Scholar]
- 8.Kanoh J, Ishikawa F. spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr Biol. 2001;11:1624–1630. doi: 10.1016/s0960-9822(01)00503-6. [DOI] [PubMed] [Google Scholar]
- 9.Chikashige Y, Hiraoka Y. Telomere binding of the Rap1 protein is required for meiosis in fission yeast. Curr Biol. 2001;11:1618–1623. doi: 10.1016/s0960-9822(01)00457-2. [DOI] [PubMed] [Google Scholar]
- 10.Baumann P, Cech TR. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- 11.Miyoshi T, Kanoh J, Saito M, Ishikawa F. Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science. 2008;320:1341–1344. doi: 10.1126/science.1154819. [DOI] [PubMed] [Google Scholar]
- 12.Lustig AJ. Cdc13 subcomplexes regulate multiple telomere functions. Nat Struct Mol Biol. 2001;8:297–299. doi: 10.1038/86157. [DOI] [PubMed] [Google Scholar]
- 13.Pennock E, Buckley K, Lundblad V. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell. 2001;104:387–396. doi: 10.1016/s0092-8674(01)00226-4. [DOI] [PubMed] [Google Scholar]
- 14.Lundblad V. In: Telomeres. de Lange T, Lundblad V, Blackburn EH, editors. Cold Spring Harbor Laboratory Press; 2006. pp. 345–386. [Google Scholar]
- 15.Martin V, Du LL, Rozenzhak S, Russell P. Protection of telomeres by a conserved Stn1-Ten1 complex. Proc Natl Acad Sci USA. 2007;104:14038–14043. doi: 10.1073/pnas.0705497104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grandin N, Damon C, Charbonneau M. Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J. 2001;20:1173–1183. doi: 10.1093/emboj/20.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V. RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol. 2007;14:208–214. doi: 10.1038/nsmb1205. [DOI] [PubMed] [Google Scholar]
- 18.Riha K, McKnight TD, Griffing LR, Shippen DE. Living with genome instability: plant responses to telomere dysfunction. Science. 2001;291:1797–1800. doi: 10.1126/science.1057110. [DOI] [PubMed] [Google Scholar]
- 19.Fitzgerald MS, et al. Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc Natl Acad Sci USA. 1999;96:14813–14818. doi: 10.1073/pnas.96.26.14813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 21.Armstrong SJ, Franklin FCH, Jones GH. Nucleolus-associated telomere clustering and pairing precede meiotic chromosome synapsis in Arabidopsis thaliana. J Cell Sci. 2001;114:4207–4217. doi: 10.1242/jcs.114.23.4207. [DOI] [PubMed] [Google Scholar]
- 22.Heacock M, Spangler E, Riha K, Puizina J, Shippen DE. Molecular analysis of telomere fusions in Arabidopsis: multiple pathways for chromosome end-joining. EMBO J. 2004;23:2304–2313. doi: 10.1038/sj.emboj.7600236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herbert BS, Hochreiter AE, Wright WE, Shay JW. Nonradioactive detection of telomerase activity using the telomeric repeat amplification protocol. Nat Protoc. 2006;1:1583–1590. doi: 10.1038/nprot.2006.239. [DOI] [PubMed] [Google Scholar]
- 24.Kannan K, Nelson ADL, Shippen DE. Dyskerin is a component of the Arabidopsis telomerase RNP required for telomere maintenance. Mol Cell Biol. 2008;28:2332–2341. doi: 10.1128/MCB.01490-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grandin N, Reed SI, Charbonneau M. Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev. 1997;11:512–527. doi: 10.1101/gad.11.4.512. [DOI] [PubMed] [Google Scholar]
- 26.Nugent CI, Hughes TR, Lue NF, Lundblad V. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- 27.Iyer S, Chadha AD, McEachern MJ. A mutation in the STN1 gene triggers an alternative lengthening of telomere-like runaway recombinational telomere elongation and rapid deletion in yeast. Mol Cell Biol. 2005;25:8064–8073. doi: 10.1128/MCB.25.18.8064-8073.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lustig AJ. Clues to catastrophic telomere loss in mammals from yeast telomere rapid deletion. Nat Rev Genet. 2003;4:916–923. doi: 10.1038/nrg1207. [DOI] [PubMed] [Google Scholar]
- 29.Zellinger B, Akimcheva S, Puizina J, Schirato M, Riha K. Ku suppresses formation of telomeric circles and alternative telomere lengthening in Arabidopsis. Mol Cell. 2007;27:163–169. doi: 10.1016/j.molcel.2007.05.025. [DOI] [PubMed] [Google Scholar]
- 30.McClintock B. The behavior in successive nuclear divisions of a chromosome broken at meiosis. Proc Natl Acad Sci USA. 1939;25:405–416. doi: 10.1073/pnas.25.8.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lei M, Podell ER, Baumann P, Cech TR. DNA self-recognition in the structure of Pot1 bound to telomeric single-stranded DNA. Nature. 2003;426:198–203. doi: 10.1038/nature02092. [DOI] [PubMed] [Google Scholar]
- 32.Petreaca RC, Chiu HC, Nugent CI. The role of Stn1p in Saccharomyces cerevisiae telomere capping can be separated from its interaction with Cdc13p. Genetics. 2007;177:1459–1474. doi: 10.1534/genetics.107.078840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puglisi A, Bianchi A, Lemmens L, Damay P, Shore D. Distinct roles for yeast Stn1 in telomere capping and telomerase inhibition. EMBO J. 2008;27:2328–2339. doi: 10.1038/emboj.2008.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karamysheva ZN, Surovtseva YV, Vespa L, Shakirov EV, Shippen DE. A C-terminal myb extension domain defines a novel family of double-strand telomeric DNA-binding proteins in Arabidopsis. J Biol Chem. 2004;279:47799–47807. doi: 10.1074/jbc.M407938200. [DOI] [PubMed] [Google Scholar]
- 35.Shakirov EV, Surovtseva YV, Osbun N, Shippen DE. The Arabidopsis Pot1 and Pot2 proteins function in telomere length homeostasis and chromosome end protection. Mol Cell Biol. 2005;25:7725–7733. doi: 10.1128/MCB.25.17.7725-7733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Surovtseva YV, et al. Arabidopsis POT1 associates with the telomerase RNP and is required for telomere maintenance. EMBO J. 2007;26:3653–3661. doi: 10.1038/sj.emboj.7601792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hong JP, et al. Suppression of RICE TELOMERE BINDING PROTEIN1 results in severe and gradual developmental defects accompanied by genome instability in rice. Plant Cell. 2007;19:1770–1781. doi: 10.1105/tpc.107.051953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karlseder J, Broccoli D, Dai Y, Hardy S, de Lange T. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science. 1999;283:1321–1325. doi: 10.1126/science.283.5406.1321. [DOI] [PubMed] [Google Scholar]
- 39.Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 40.Shippen DE. In: Telomeres. de Lange T, Lundblad V, Blackburn EH, editors. Cold Spring Harbor Laboratory Press; 2006. pp. 525–550. [Google Scholar]
- 41.Liu D, et al. PTOP interacts with POT1 and regulates its localization to telomeres. Nat Cell Biol. 2004;6:673–680. doi: 10.1038/ncb1142. [DOI] [PubMed] [Google Scholar]
- 42.O'Connor MS, Safari A, Liu D, Qin J, Songyang Z. The human Rap1 protein complex and modulation of telomere length. J Biol Chem. 2004;279:28585–28591. doi: 10.1074/jbc.M312913200. [DOI] [PubMed] [Google Scholar]
- 43.Ye JZ-S, et al. TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J Biol Chem. 2004;279:47264–47271. doi: 10.1074/jbc.M409047200. [DOI] [PubMed] [Google Scholar]
- 44.Onodera Y, et al. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 45.Kato A, Lamb JC, Birchler JA. Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc Natl Acad Sci USA. 2004;101:13554–13559. doi: 10.1073/pnas.0403659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cocciolone SM, Cone KC. Pl-Bh, an anthocyanin regulatory gene of maize that leads to variegated pigmentation. Genetics. 1993;135:575–588. doi: 10.1093/genetics/135.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grant JD, et al. Telometric: a tool providing simplified, reproducible measurements of telomeric DNA from constant field agarose gels. BioTechniques. 2001;31:1314–1318. doi: 10.2144/01316bc02. [DOI] [PubMed] [Google Scholar]
- 48.Shakirov EV, Shippen DE. Length regulation and dynamics of individual telomere tracts in wild-type Arabidopsis. Plant Cell. 2004;16:1959–1967. doi: 10.1105/tpc.104.023093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heacock ML, Idol RA, Friesner JD, Britt AB, Shippen DE. Telomere dynamics and fusion of critically shortened telomeres in plants lacking DNA ligase IV. Nucleic Acids Res. 2007;35:6490–6500. doi: 10.1093/nar/gkm472. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.