Abstract
Misfolding and aggregation of Cu, Zn superoxide dismutase (SOD1) is implicated in neuronal death in amyotrophic lateral sclerosis. Each SOD1 monomer binds to 1 copper and 1 zinc ion and maintains its disulfide bond (Cys-57–Cys-146) in the reducing cytoplasm of cell. Mounting experimental evidence suggests that metal loss and/or disulfide reduction are important for initiating misfolding and aggregation of SOD1. To uncover the role of metals and the disulfide bond in the SOD1 folding, we systemically study the folding thermodynamics and structural dynamics of SOD1 monomer and dimer with and without metal binding and under disulfide-intact or disulfide-reduced environments in computational simulations. We use all-atom discrete molecular dynamics for sampling. Our simulation results provide dynamical evidence to the stabilizing role of metal ions in both dimer and monomer SOD1. The disulfide bond anchors a loop (Glu-49 to Asn-53) that contributes to the dimer interface. The reduction of the disulfide bond in SOD1 with metal ions depleted results in a flexible Glu-49–Asn-53 loop, which, in turn, disrupts dimer formation. Interestingly, the disulfide bond reduction does not affect the thermostability of monomer SOD1 as significantly as the metal ions do. We further study the structural dynamics of metal-free SOD1 monomers, the precursor for aggregation, in simulations and find inhomogeneous local unfolding of β-strands. The strands protected by the metal-binding and electrostatic loops are found to unfold first after metal loss, leading to a partially unfolded β-sheet prone to aggregation. Our simulation study sheds light on the critical role of metals and disulfide bond in SOD1 folding and aggregation.
Keywords: amyotrophic lateral sclerosis, discreet molecular dynamics, protein aggregation, protein stability
The Cu, Zn superoxide dismutase (SOD1) has been implicated in the fatal neurodegenerative disease amyotrophic lateral sclerosis (ALS). A large set of mutations in this ubiquitous cytosolic enzyme has been identified in familial ALS diseases (1, 2). Although the mechanism by which mutant SOD1 causes neuronal death remains elusive (3), several lines of evidence suggest that ALS is a protein aggregation disease (4–6). The SOD1 protein forms a stable homodimer in solution where each SOD1 monomer binds to 1 copper and 1 zinc ion and has an intramonomer disulfide bond (Cys-57–Cys-146). Characterization of SOD1-associated aggregation is often complicated by these modifications. Thermodynamic experiments show that SOD1 monomer remains folded upon impairment of the dimer interface (7) and loss of the zinc and copper ions (8, 9). Recently, several findings have implicated monomeric SOD1 with metal ions depleted (apoSOD1) as a precursor for aggregation (10–14). Understanding the role of metal binding and the disulfide bond (15, 16) on SOD1 dimer stability and monomer folding is thus essential to uncovering the early stages of the SOD1 aggregation.
Biochemical and biophysical studies suggested that WT Cu, Zn SOD1 dimer is exceptionally stable because of the coordination of metal ions (17). However, X-ray crystallographic studies indicate that SOD1 with metal ions bound (holoSOD1) dimer and apoSOD1 dimer have similar structures with minor rearrangement of amino acids near the metal-binding sites (9, 18). The apoSOD1 with the disulfide bond reduced has much lower dimer stability than the WT (12, 19) and forms marginally stable monomer (20). The crystal structure of disulfide-reduced apoSOD1 monomer has a displaced loop near Cys-57 that interacts with the other monomer in the WT dimer (18). Hornberg et al. (18) proposed that the low dimerization propensity of disulfide-reduced apoSOD1 is of entropic origin due to increased loop flexibility in the monomeric state. Therefore, the role of metal binding and the disulfide bond on SOD1 stability is mainly dynamic. It is important to study the dynamics to understand the role of metal ions and the disulfide bond in SOD1 folding.
Examination of the NMR-derived structural ensemble of the apoSOD1 monomer (21) indicates that loss of metal ions results into a flexible metal-binding loop and partially unstructured β-sheet that anchors the metal ions. The denaturant-induced unfolding of holoSOD1 monomer (with a mutation to disrupt the dimer interface) monitored by NMR (22) also indicates progressive unfolding in the metal-binding loop and β-sheet. Mutagenesis analysis in folding kinetics (23) suggested inhomogeneous unfolding propensity in apoSOD1 monomer. Here, we delineate the dynamical effects of metal binding and the disulfide state on SOD1 folding by using computational studies.
Biophysical properties of SOD1 have been explored in a number of computational studies, including mechanistic studies of enzyme activity (24–26), interdomain communications (27, 28), and near-native structural dynamics (28, 29). Many of these studies use traditional all-atom molecular dynamics approaches to sample protein conformations (24, 28, 29). However, the time scales accessed by traditional computational approaches are severely limited (≈10 ns) and are much smaller than those necessary for uncovering larger-scale conformational dynamics of proteins (submicrosecond to millisecond time scales). To bridge the gap in the time scale associated with SOD1 conformational dynamics, we use all-atom discrete molecular dynamics (DMD) simulations (30). DMD has been shown to have a higher sampling efficiency than traditional molecular dynamics and has been used to study protein folding thermodynamics and protein aggregation (31, 32). All-atom DMD features a transferable force field and has been successfully used to fold several small proteins ab initio (<60 aa; ref. 30).
To uncover the role of metal binding and disulfide bonding, we systematically studied the folding thermodynamics of the SOD1 monomer and dimer with and without metal coordination and with and without the disulfide bond. To enhance the sampling efficiency, we used replica exchange (33) coupled to DMD simulations. Our simulation results provide a dynamic mechanism for the stabilizing effect of metal binding and disulfide bond formation. The dynamics analysis of the monomer apoSOD1 species suggests a scenario for its potential aggregation mechanism.
Results
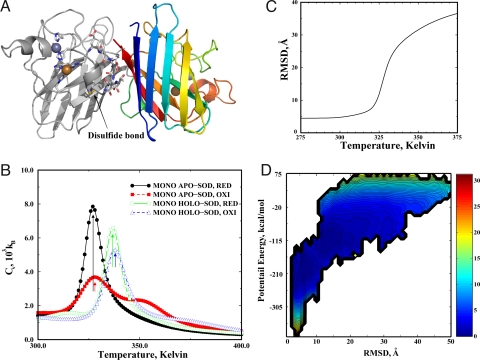
We use replica exchange (see Methods) to enhance the sampling efficiency of DMD simulations. To understand the role of metal ions and the disulfide bond in SOD1 folding and dimer formation, we performed a set of 8 replica exchange simulations of a monomer and a dimer (×2), with and without metal ions (×2), and with and without the disulfide bond (mimicking oxidative and reducing environments, respectively) (×2). We added distance constraints between the 8 metal-binding atoms and the corresponding metal ions (Fig. 1A) to mimic the strong coordination interactions. We modeled the disulfide bond formation similar to that of a hydrogen bond by considering both the distance and orientation dependence (see Methods).
Fig. 1.
The folding thermodynamics of SOD monomers. (A) The structure of native SOD dimer. One monomer is colored from blue (N-terminal) to red (C-terminal), and the other is colored in gray. The residues bound to the metal ions are shown in stick representation. A disulfide bond anchors the loop (Glu-49–Asn-53, in stick representation) that contributes to the dimer interface. (B) The specific heat plot of 4 types of SOD monomers. The arrows indicate the peaks of the corresponding specific heat. (C) The mean rmsd as a function of temperature for the disulfide-reduced apoSOD1 monomer features a sigmoidal transition. (D) The contour plot of PMF as the function of potential energy and rmsd at TF ≈325 K. The free energy difference between 2 consecutive contours is 0.6 kcal/mol. The area in the contour plot is colored according to the corresponding free energy value. The free energy unit is kcal/mol.
For each replica exchange simulation, we started from the native state (Protein Data Bank ID code 2v0a). We allocated 12 replicas for SOD1 monomer simulations and 16 replicas for SOD1 dimers. In total, we performed a total of 1.2 μs simulations for SOD1 monomers and 1.6 μs simulations for SOD1 dimers (see Methods). Because of the vast conformational space of SOD1, we did not expect DMD simulations to reach folding/unfolding equilibrium as can be done for small proteins (30). However, starting from the native state, we expected that our replica exchange DMD simulations would be able to thoroughly sample conformations in the folded state.
Metal Binding Has a Stronger Stabilizing Effect in the SOD1 Monomer Than the Disulfide Bond.
We used the weighted histogram analysis method (WHAM; SI Text) to compute the specific heat of SOD1 folding based on the replica exchange simulations (34). To reduce the artifact of structural relaxation from the starting crystallography structure in the analysis, we used the last three-quarters of the trajectory to perform various calculations (see Methods). In the low-temperature replica simulations, where the protein stays folded, the rmsds for the native state rapidly reach equilibrium (<105 time units; Fig. S1). Therefore, the initial structural relaxation did not affect our analysis.
In simulations, we did not observe equilibrium of unfolding and refolding transitions of SOD1. However, we did observe partial unfolding and refolding events in simulations (Fig. S1). Therefore, although we did not expect the computed thermodynamics to accurately quantify the free energy landscape of SOD1 folding, we do believe that by using the unfolding transition temperature TU (corresponding to the highest peak in the specific heat; indicated by arrows in Fig. 1B) as a qualitative measure of a protein's thermostability, we were able to differentiate the effects of metal binding and disulfide bond formation on SOD1 thermostability. We found that holoSOD1 monomers have a higher TU (>10 K; Fig. 1B) than apoSOD1 monomers. The disulfide bond formation has a much weaker stabilizing effect on the monomer folding with a small increase in the TU (≈1 K; Fig. 1B). Therefore, metal binding has a stronger stabilizing effect than the disulfide bond. This result is consistent with experiments where a larger melting temperature change was observed for metal loss than for disulfide reduction (20, 35). However, the magnitudes of unfolding/melting temperature changes in simulations were smaller than those determined in experiments, possibly because of insufficient sampling and inaccuracy in the force field.
The Disulfide-Reduced ApoSOD1 Monomer Folds in an Apparent 2-State Manner.
Mounting experimental evidence points to apoSOD1 as a necessary precursor to aggregation (10–13). In agreement with experimental studies, our thermodynamic analysis also suggests that apoSOD1 has a lower thermodynamic stability and is more prone to misfolding, and thus aggregation than holoSOD1. ApoSOD1 in oxidative conditions features an intermediate because of the nonnative disulfide bond formation in the unfolded state (Cys-6–Cys-111; Fig. S2). For example, in the specific heat profiles for SOD1 monomer under oxidative conditions in Fig. 1B, the shoulders at high temperatures (>TU) correspond to the formation of nonnative disulfide bonds. Removal of free cysteines in SOD1 has been shown to reduce the irreversible unfolding of SOD1 (36). However, such a decrease of irreversible unfolding in experiments can also result from the reduced interdomain disulfide formations. Further experiments are necessary to characterize the folding intermediates caused by the formation of the intradomain disulfide bond.
We evaluated the folding thermodynamics of apoSOD1 without the disulfide bond. Starting from the native state, we expected the replica exchange simulations to sample any thermodynamically stable intermediate states upon unfolding. Using WHAM, we computed the mean rmsd as the function of temperature (Fig. 1C). Because the 2 long loops (Phe-50–Gly-82; Glu-121–Ser-142) are very flexible, we excluded them from the rmsd calculation. The function was typical for a 2-folding protein with a transition from folded (small rmsd) to unfolded (large rmsd). To examine the folding dynamics more closely, we computed potential mean force (PMF) as the function of potential energy and rmsd at the melting temperature (≈325 K). The 2D-PMF at melting temperature had only 2 distinct basins (Fig. 1D), suggesting that apoSOD1 is a 2-state folding protein in the reducing environment as observed in experiment (23).
The Conformational Dynamics of SOD1 Monomers.
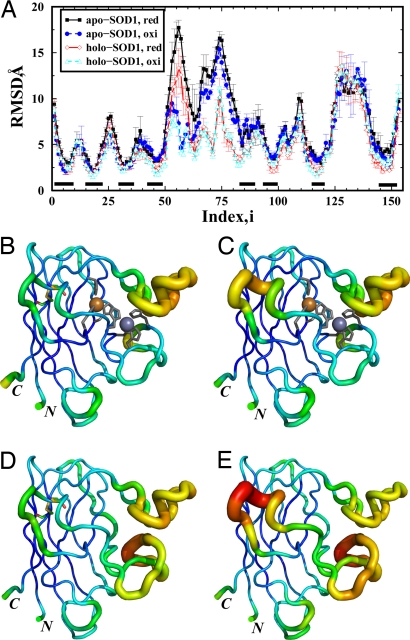
We computed the average rmsd for each residue in the folded state ensemble to study conformational dynamics on a per-residue basis. For each monomeric SOD1 system, we identified the folded conformations from all replicas. A conformation was defined as folded if the rmsd is <6 Å, which corresponds to the threshold for unfolding in Fig. 1C. The average rmsd per residue was calculated based on the identified ensemble. To test whether the conformational space of the folded SOD1 was efficiently sampled in DMD simulations, we divided the last three-quarters of each simulation trajectory into halves (0.5 × 106 to 1.25 × 106 time units and 1.25 × 106 to 2.0 × 106 time units) and constructed 2 subsets of the folded state ensemble accordingly. We calculated for each residue the average rmsd and rms fluctuations (RMSFs) from the average structure, for each subensemble separately. We found that the rmsd (error bars in Fig. 2A denote the difference of rmsd per residue between 2 subensembles) and RMSF (Fig. S3) for each residue in these subensembles were consistent with each other, suggesting a sufficient sampling of the folded state in DMD simulations.
Fig. 2.
The structural dynamics of SOD1 monomers. (A) The rmsd per residue in the folded state for SOD1 monomers. The error bars correspond to differences of the rmsd per residue between different portions of simulation trajectories. The black bars above the x axis indicate the β-strands. (B) Tube representation of holoSOD1 monomer with the disulfide bond intact: the backbone is colored according to the rmsd of the corresponding residue (blue corresponds to low rmsd value and red corresponds to large rmsd value). The backbone diameter is also proportional to the corresponding rmsd. The metal ions are shown as spheres, and the metal-coordinating residues are in stick representation. (C–E) The tube representations are also shown for holoSOD1 with disulfide reduced (C), apoSOD1 with disulfide bond (D), and apoSOD1 with disulfide reduced (E). The disulfide-bonds (Cys-57–Cys-147) are shown in stick representation in B and D. The 2 termini (N and C) are indicated in the structure.
In Fig. 2A, the regions with low rmsd correspond to the β-strands. The 2 regions with the largest rmsd are the 2 long loops, Phe-50–Gly-82 and Glu-121–Ser-142. For both apo- and holo-SOD1 monomers, the disulfide bond (Cys-57–Cys-146) mainly stabilizes the subloop (Phe-50–Ala-60). The rmsd of other residues are almost identical whether or not the disulfide bond is present in either apo- or holo-SOD1. As expected, the metal ions significantly stabilize the loop Phe-50–Gly-82, where the metal ions bind. Interestingly, compared with the rmsd profiles of the corresponding apoSOD1, the metal binding also stabilizes β-strands 4, 5, and 7. These strands also bind metal ions and form the β-sheet underneath the metal-binding loop. To illustrate the conformational dynamics, we represent the rmsd of the corresponding residue in the structure by varying the width of the backbone trace proportionally to the rmsd values (tube representation) (Fig. 2 B–E). Additionally, the backbone is colored according to the rmsd of the corresponding residue (blue corresponds to low rmsd values and red corresponds to large rmsd values). The backbone diameter is proportional to the corresponding rmsd. The tube representations of different SOD1 monomers in Fig. 2 B–E directly demonstrate the dynamical role of metal ions and the disulfide bond in reducing the conformational flexibility.
Local Unfolding of ApoSOD1 Monomers.
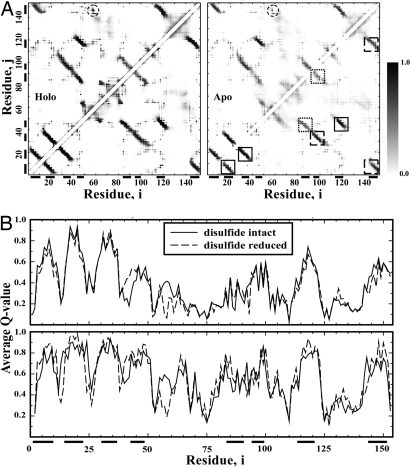
SOD1 in solution is dynamic and undergoes local unfolding (23, 37). These local unfolding events are believed to facilitate intermolecular protein–protein interactions that cause the aggregation of SOD1 (23, 37). To estimate the local unfolding propensities in apoSOD1, we computed the contact frequency and analyzed the average amount of native interactions for each residue. The contact frequency of apoSOD1 in the folded state was averaged over the conformation ensemble constructed above. A contact was defined between 2 residues having at least 1 pair of heavy atoms within a 5-Å cutoff distance. The cutoff distance was chosen to be intermediate between the distance with the lowest van der Waals (VDW) attraction energy and the interaction range. With different cutoff values tried (4.5, 5, and 5.5 Å), the results were consistent (data not shown).
We found that apoSOD1 monomers with and without the disulfide bond have a similar contact frequency map (Fig. 3A). The principal differences are in the proximity of the disulfide bond (Cys-57–Cys-146). For holoSOD1 monomers, the interstrand interactions are well maintained with high frequencies. However, for apoSOD1 monomers, interstrand interactions are weakened inhomogeneously. The interstrand interactions between strands 1 and 2, strands 2 and 3, and strands 4 and 7 have high contact frequencies; interactions between strands 3 and 6, strands 1 and 8, and strands 7 and 8 have intermediate contact frequencies; and interactions between strands 4 and 5 and between 5 and 6 have weak contact frequencies. The low contact frequencies between strands are the results of local unfolding.
Fig. 3.
The local unfolding propensity of the SOD1 monomers. (A) The contact frequency map is shown for both holoSOD1 (Left) and apoSOD1 (Right) monomers. The contact between residues i and j is colored in grayscale according to the corresponding contact frequency. The gray color bar denotes color scale. The upper triangle corresponds to SOD1 monomers with the disulfide bond. The region corresponds to the disulfide bond is circled. The lower triangle corresponds to SOD1 monomers with disulfide bond reduced. For apoSOD1, regions inside solid-line squares denote strong interstrand interactions; dotted squares refer to weak interstrand interactions; and long dashed squares correspond to intermediate interstrand interactions. The black bars along the axis indicate the corresponding β-strands. (B) The average Q value for each residue was obtained by averaging frequencies over native contacts.
In Fig. 3B, we compute the average Q value for each residue (38). The Q value quantifies the extent of native-like interactions: a high Q value corresponds to the native-like environment, whereas a low Q value suggests the loss of native-like environments caused by local unfolding (38). The Q-value plot indicates that strands 1, 2, 3, and 7 are the most stable secondary structures, strands 4, 5, 6, and 8 experience local unfolding with lower Q values, and loops are highly flexible with very low Q values. Such a local unfolding propensity along the sequence is better illustrated by mapping the Q value to the native structure (Fig. S4). The residues with average Q values >0.6 are clustered in strands 1, 2, 3, and 7. The identified local unfolding regions are consistent with experimental measurements (23).
Metal Binding and Disulfide Bond Formation Stabilizes the Dimer Interface.
For the dimer simulations, we focused on the interdomain interactions. For each of the 4 sets of replica exchange simulations of dimers, we performed WHAM analysis to compute the average number of interdomain contacts and number of native interdomain contacts (Fig. 4). We found that as temperature increases, the number of native interdomain contacts decreases. The total number of interdomain contacts, which includes the nonnative interdomain contacts, is equal to the number of native interdomain contacts at low temperatures. Interestingly, the total number of interdomain contacts displays a different behavior as temperature increases: it initially decreases, and then increases as the number of native interdomain contacts approaches 0. The number of nonnative interdomain contacts reaches maximum at temperatures corresponding to the unfolding transition temperature of the corresponding monomer. Therefore, the additional nonnative interdomain interactions are formed between unfolded chains in the small simulation box, which is promoted by the high density of SOD1 in simulation. We used a small simulation box (200 × 200 × 200 Å3), corresponding to a SOD1 monomer density of 0.42 mM. These nonnative interdomain contacts are mainly transient and nonspecific interactions. Therefore, observations at low temperatures and calculations based on ensemble averages were not affected by these transient and nonspecific interactions.
Fig. 4.
The interdomain interactions of SOD dimers. The number of interdomain contacts for apoSOD dimers without metal binding (Left) and dimers with metal binding (Right) is shown.
At 300 K, disulfide-reduced apoSOD1 dimer has 35 interdomain contacts, whereas other types of SOD1 dimers (disulfide-intact apoSOD1 dimer and holoSOD1 dimers with and without the disulfide bond) have ≈50 interdomain contacts. Without the disulfide bond, the apoSOD1 loses 80% of the total native interdomain contacts at a lower temperature (≈310 K) than the disulfide-intact apoSOD1 dimer (≈322 K; Fig. 4 Left). The reduction of the disulfide bond has a weaker effect on the loss of native interdomain contacts for holoSOD1 dimers than for apoSOD1 dimers: the holoSOD1 dimers with and without disulfides lose their interdomain contacts at a similar temperature, ≈335 K (Fig. 4 Right). Therefore, our simulations suggest that the disulfide bond is important for the dimerization of apoSOD1 and that metal-binding can rescue dimer formation for SOD1 even without the formation of the native disulfide bond. These observations are in agreement with recent experimental study on the role of disulfide bond formation and metal binding on SOD1 folding (18–20).
The Flexibility of the Disulfide Bond-Anchored Loop Is Responsible for the Weakened Disulfide-Reduced Dimer Interface of ApoSOD1.
To understand the molecular mechanism of the reduced dimer stability of the disulfide-reduced apoSOD1 dimer, we computed the frequency of interdomain contacts. The interdomain contact frequencies were averaged over conformations from all replicas where 2 monomers have at least 1 pair of interdomain residues in contact. The contact between 2 residues was defined by all heavy atoms and with a cutoff distance range of 5 Å. The ensemble included nonnative dimers sampled in simulations. In Fig. S5A, we see that native interdomain contacts formed by the disulfide bond-anchored loop (Glu-49–Asn-53; part of the long loop between strands 4 and 5) are significantly weakened. Additionally, without the disulfide bond anchoring the Glu-49–Asn-53 loop, it is highly flexible (Fig. 2A and Fig. S5B). The native interdomain interactions for the loop Glu-49–Asn-53 are not strong enough to hold the loop in the absence of the disulfide bond or metal binding, and thus result in a weakened dimer interface.
Discussion
Experimental evidence suggests that the SOD1 monomer is the precursor for aggregation (10–14, 39). Understanding the conformational dynamics and local unfolding in the SOD1 monomer may shed light on the consequent misfolding and aggregation. The analysis of SOD1 monomer simulations illustrates distinct patterns of structural dynamics for different regions of SOD1 and reveals the structural and dynamic role of metal ions and the disulfide bond. Especially, a mutant apoSOD1 monomer has been studied in NMR. The structural dispersions among the NMR ensemble exemplifies dynamics of apoSOD1 in solution (21). The conformational dynamics of apoSOD1 we observe in the folded ensemble show a striking correlation to the experimental measurement from the NMR ensemble (figure 2a in ref. 21). For example, both computational and experimental profiles show that the central region near residue Phe-64 of the metal-binding loop (Phe-50–Gly-82) has low structural flexibility compared with the rest of the loop, because it forms the core of SOD1. Also, both computational and experimental profiles indicate that strand 5 has elevated structural dynamics compared with other strands. Therefore, our all-atom DMD method was able to accurately capture the conformational dynamics of SOD1 in the folded state.
Our simulation results suggest that the binding of metal ions increases the thermostability of both SOD1 monomer and dimer. This computational finding is consistent with 2 experimental observations: loss of metal ions is obligatory for SOD1 to aggregate (40), and the holoSOD1 dimer dissociates at a higher concentration of denaturant than the apoSOD1 dimer (19). In the native state of SOD1, the copper and zinc ions are coordinated by 6 histidines (nos. 46, 48, 63, 71, 81, and 120) and 1 aspartic acid (no. 83). These residues are positioned on strands 4 and 7, and the metal-binding loop Phe-50–Gly-82 (Fig. 1A). The structural dynamics in simulations of SOD1 monomers suggest that metal binding stabilizes both the metal-binding loop and strands 4, 5, and 7, resulting in reduced structural fluctuations (Fig. 2A). Strand 7 and part of loop IV (Glu-49–Asn-53) contribute to the native dimer interface (Fig. S5A). Therefore, our molecular dynamics simulations suggest that binding of metal ions stabilizes the SOD1 dimer by reducing the structural flexibility of the elements forming the dimer interface. For SOD1 monomers, metal binding stabilizes the protein by anchoring strands 4 and 7, thus stabilizing the β-barrel in the folded state.
We found that the native disulfide bond (Cys-57–Cys-146) does not have a significant effect on the thermostability of the SOD1 monomer. However, the disulfide bond has a significant effect on the dimer association of the apoSOD1: the disulfide-reduced apoSOD1 dimer dissociates at a lower temperature than other types of SOD1 dimers. This effect is consistent with experimental studies where apoSOD1 without the native disulfide bond is mainly monomeric (18–20). The computational structural dynamics of SOD1 indicates that the disulfide bridge stabilizes part of the metal-binding loop in SOD1 monomers, which contributes significantly to the native dimer interface (Fig. S5A).
In dimer simulations, we observed that the apoSOD1 dimers dissociated before the melting of individual monomers (Figs. 1 and 4), and the holoSOD1 dimers dissociated at a similar temperature as the melting temperature of the corresponding monomers. This observation suggests that denaturation of apoSOD1 dimer features stable folded monomer, supporting a thermodynamic experimental study of a mutant apoSOD1 dimer in which the folded monomer was identified (41). For holoSOD1 dimers, our study also suggests that the dimer dissociation dimer is coupled with the denaturation of monomers, assuming that bound metal ions are not lost.
We also studied the propensity of local unfolding for SOD1 monomers. Our study suggests that the β-sheet formed by strands 1–3 is most stable, whereas strands 4, 5, 6, and 8 experience frequent local unfolding. This picture of local unfolding agrees with the experimental observation of SOD1 folding (23), where hot spots for local unfolding in SOD1 were identified. The local unfolding of strands 5, 6, and 8 exposes the edge of β-sheets and the hydrophobic core, which is prone to aggregation (42). For example, in the dimer simulations, apoSOD1 without disulfide bond forms a large amount of nonnative interdomain contacts at low temperatures (Fig. 4). In the interdomain contact frequency map (Fig. S5B), we observe significant amount of nonnative interdomain contacts with high frequencies formed between strand 1 of 1 chain and strands 1, 7, and 8 of the other chain. These nonnative interdomain interactions are consistent with the local unfolding propensity of the SOD1 monomer. However, we expect other possible modes for nonnative interdomain association besides the observed interdomain interactions because our simulations were not long enough to thoroughly sample all available conformational space of SOD1 in aggregation-associated time scales [seconds to hours (40)]. Other possible dimerization mechanisms include edge extension along exposed edges of strands 3 and 4 that are exposed after local unfolding of strands 5 and 6. Because of limited sampling and the fact that strands 1, 7, and 8 are in proximity in the native dimer, we were only able to observe the above nonnative association.
Conclusion
Our computational study of SOD1 folding thermodynamics demonstrates the dynamical role of metal ions and the disulfide bond on SOD1 folding. The binding of metal ions has profound stabilizing effects on both dimer and monomer SOD1. The coordination of metal ions increases dimer stability by reducing the structural fluctuations of the native dimer interface. The metal ions stabilize SOD1 monomers by holding strands 4 and 7 together, which experience large fluctuations in apoSOD1. The disulfide bond formation has a lesser stabilizing effect in SOD1 monomers than metal binding. However, the disulfide bond plays an important role in stabilizing the dimer interface by anchoring the loop in the dimer interface. Our study supports the hypothesis that apoSOD1 monomer is more prone for aggregation (17, 40). The simulations are consistent with a possible scenario for apoSOD1 aggregation, where metal loss induces structural fluctuations and inhomogeneous local unfolding of β-strands, and subsequently leads to exposure of the protected edge of β-sheets and hydrophobic core, promoting aggregation.
Methods
All-Atom DMD.
DMD is a special type of molecular dynamics that approximates pairwise interactions with discontinuous functions (SI Text). The all-atom DMD method uses a united atom protein model, where heavy atoms and polar hydrogen atoms are explicitly modeled (30). We included van der Waals, solvation, and environment-dependent hydrogen bond interactions. We adopted the Lazaridis-Karplus solvation model and used the fully-solvated conformation as the reference state. Because of the strong screening effect of solvent, distant charges have weak polar interactions. For salt bridges, we expected the hydrogen bonds to partially account for their polar interactions. Similar neutralization of charged residues was also used in the implicit solvent model of the effective energy function of CHARMM19 (43). We modeled the metal binding by assigning distance constraints between each metal atom and the corresponding metal-coordinating atoms. The distance and orientation dependence of disulfide bond formation was modeled by using a reaction algorithm. The details of model metal binding and disulfide bond formation are described in SI Text and Fig. S6.
Replica Exchange DMD and WHAM Analysis.
In replica exchange computing, multiple simulations or replicas of the same system are performed in parallel at different temperatures. Periodically, replicas with neighboring temperature values exchange their temperatures in a Metropolis-based stochastic manner (SI Text). For monomer simulations, we allocated 12 replicas with a set of temperatures: 270, 282, 294, 306, 318, 330, 342, 354, 366, 378, 390, and 402 K. For dimer simulations, we allocated 16 replicas with the following temperatures: 270, 279, 288, 297, 306, 315, 324, 333, 342, 351, 360, 369, 378, 387, 390, and 402 K. For each replica, we performed 2 million time units [1 time unit corresponds to 50 fs (30)] of DMD simulation, which corresponds to ≈100 ns. The exchange took place every 1,000 time units. We used WHAM analysis to compute the folding thermodynamics from replica exchange DMD simulation trajectories (SI Text). Because our simulations started from X-ray crystallography conformations, we excluded the trajectories from the first 5 × 105 time units and used those of the last 1.5 × 106 time units for WHAM analysis. We used the trajectories from all replicas to compute the histograms.
Supplementary Material
Acknowledgments.
We thank Kyle Wilcox for critical reading of the manuscript. This work is supported in part by American Heart Association Grant 0665361U and National Institutes of Health Grant R01GM080742.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803266105/DCSupplemental.
References
- 1.Rosen DR, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 2.McCord JM, Fridovich I. The utility of superoxide dismutase in studying free radical reactions. I. Radicals generated by the interaction of sulfite, dimethyl sulfoxide, and oxygen. J Biol Chem. 1969;244:6056–6063. [PubMed] [Google Scholar]
- 3.Brown RH., Jr SOD1 aggregates in ALS: Cause, correlate, or consequence? Nat Med. 1998;4:1362–1364. doi: 10.1038/3945. [DOI] [PubMed] [Google Scholar]
- 4.Okado-Matsumoto A, Fridovich I. Amyotrophic lateral sclerosis: A proposed mechanism. Proc Natl Acad Sci USA. 2002;99:9010–9014. doi: 10.1073/pnas.132260399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruijn LI, et al. Aggregation and motor neuron toxicity of an ALS-linked SOD1 mutant independent from wild-type SOD1. Science. 1998;281:1851–1854. doi: 10.1126/science.281.5384.1851. [DOI] [PubMed] [Google Scholar]
- 6.Johnston JA, Dalton MJ, Gurney ME, Kopito RR. Formation of high molecular weight complexes of mutant Cu, Zn-superoxide dismutase in a mouse model for familial amyotrophic lateral sclerosis. Proc Natl Acad Sci USA. 2000;97:12571–12576. doi: 10.1073/pnas.220417997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banci L, et al. Solution structure of reduced monomeric Q133M2 copper, zinc superoxide dismutase (SOD). Why is SOD a dimeric enzyme? Biochemistry. 1998;37:11780–11791. doi: 10.1021/bi9803473. [DOI] [PubMed] [Google Scholar]
- 8.Stroppolo ME, Malvezzi-Campeggi F, Mei G, Rosato N, Desideri A. Role of the tertiary and quaternary structures in the stability of dimeric copper, zinc superoxide dismutases. Arch Biochem Biophys. 2000;377:215–218. doi: 10.1006/abbi.2000.1780. [DOI] [PubMed] [Google Scholar]
- 9.Strange RW, et al. The structure of holo and metal-deficient wild-type human Cu, Zn superoxide dismutase and its relevance to familial amyotrophic lateral sclerosis. J Mol Biol. 2003;328:877–891. doi: 10.1016/s0022-2836(03)00355-3. [DOI] [PubMed] [Google Scholar]
- 10.Rakhit R, et al. Monomeric Cu, Zn-superoxide dismutase is a common misfolding intermediate in the oxidation models of sporadic and familial amyotrophic lateral sclerosis. J Biol Chem. 2004;279:15499–15504. doi: 10.1074/jbc.M313295200. [DOI] [PubMed] [Google Scholar]
- 11.Hough MA, et al. Dimer destabilization in superoxide dismutase may result in disease-causing properties: Structures of motor neuron disease mutants. Proc Natl Acad Sci USA. 2004;101:5976–5981. doi: 10.1073/pnas.0305143101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arnesano F, et al. The unusually stable quaternary structure of human Cu, Zn-superoxide dismutase 1 is controlled by both metal occupancy and disulfide status. J Biol Chem. 2004;279:47998–48003. doi: 10.1074/jbc.M406021200. [DOI] [PubMed] [Google Scholar]
- 13.Lynch SM, Boswell SA, Colon W. Kinetic stability of Cu/Zn superoxide dismutase is dependent on its metal ligands: Implications for ALS. Biochemistry. 2004;43:16525–16531. doi: 10.1021/bi048831v. [DOI] [PubMed] [Google Scholar]
- 14.Rakhit R, et al. An immunological epitope selective for pathological monomer-misfolded SOD1 in ALS. Nat Med. 2007;13:754–759. doi: 10.1038/nm1559. [DOI] [PubMed] [Google Scholar]
- 15.Chen Y, Dokholyan NV. A single disulfide bond differentiates aggregation pathways of β2-microglobulin. J Mol Biol. 2005;354:473–482. doi: 10.1016/j.jmb.2005.09.075. [DOI] [PubMed] [Google Scholar]
- 16.Cho SS, Levy Y, Onuchic JN, Wolynes PG. Overcoming residual frustration in domain-swapping: The roles of disulfide bonds in dimerization and aggregation. Phys Biol. 2005;2:S44–S55. doi: 10.1088/1478-3975/2/2/S05. [DOI] [PubMed] [Google Scholar]
- 17.Shaw BF, Valentine JS. How do ALS-associated mutations in superoxide dismutase 1 promote aggregation of the protein? Trends Biochem Sci. 2007;32:78–85. doi: 10.1016/j.tibs.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 18.Hornberg A, Logan DT, Marklund SL, Oliveberg M. The coupling between disulphide status, metallation, and dimer interface strength in Cu/Zn superoxide dismutase. J Mol Biol. 2007;365:333–342. doi: 10.1016/j.jmb.2006.09.048. [DOI] [PubMed] [Google Scholar]
- 19.Doucette PA, et al. Dissociation of human copper-zinc superoxide dismutase dimers using chaotrope and reductant. Insights into the molecular basis for dimer stability. J Biol Chem. 2004;279:54558–54566. doi: 10.1074/jbc.M409744200. [DOI] [PubMed] [Google Scholar]
- 20.Lindberg MJ, Normark J, Holmgren A, Oliveberg M. Folding of human superoxide dismutase: Disulfide reduction prevents dimerization and produces marginally stable monomers. Proc Natl Acad Sci USA. 2004;101:15893–15898. doi: 10.1073/pnas.0403979101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banci L, Bertini I, Cramaro F, Del CR, Viezzoli MS. Solution structure of Apo Cu, Zn superoxide dismutase: Role of metal ions in protein folding. Biochemistry. 2003;42:9543–9553. doi: 10.1021/bi034324m. [DOI] [PubMed] [Google Scholar]
- 22.Assfalg M, Banci L, Bertini I, Turano P, Vasos PR. Superoxide dismutase folding/unfolding pathway: Role of the metal ions in modulating structural and dynamical features. J Mol Biol. 2003;330:145–158. doi: 10.1016/s0022-2836(03)00533-3. [DOI] [PubMed] [Google Scholar]
- 23.Nordlund A, Oliveberg M. Folding of Cu/Zn superoxide dismutase suggests structural hotspots for gain of neurotoxic function in ALS: Parallels to precursors in amyloid disease. Proc Natl Acad Sci USA. 2006;103:10218–10223. doi: 10.1073/pnas.0601696103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen J, Subramaniam S, Wong CF, McCammon JA. Superoxide dismutase: Fluctuations in the structure and solvation of the active site channel studied by molecular dynamics simulation. Biopolymers. 1989;28:2085–2096. doi: 10.1002/bip.360281205. [DOI] [PubMed] [Google Scholar]
- 25.Wade RC, Gabdoulline RR, Ludemann SK, Lounnas V. Electrostatic steering and ionic tethering in enzyme-ligand binding: Insights from simulations. Proc Natl Acad Sci USA. 1998;95:5942–5949. doi: 10.1073/pnas.95.11.5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Falconi M, et al. Dynamics-function correlation in Cu, Zn superoxide dismutase: A spectroscopic and molecular dynamics simulation study. Biophys J. 2001;80:2556–2567. doi: 10.1016/S0006-3495(01)76227-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chillemi G, et al. The essential dynamics of Cu, Zn superoxide dismutase: Suggestion of intersubunit communication. Biophys J. 1997;73:1007–1018. doi: 10.1016/S0006-3495(97)78134-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khare SD, Dokholyan NV. Common dynamical signatures of familial amyotrophic lateral sclerosis-associated structurally diverse Cu, Zn superoxide dismutase mutants. Proc Natl Acad Sci USA. 2006;103:3147–3152. doi: 10.1073/pnas.0511266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strange RW, Yong CW, Smith W, Hasnain SS. Molecular dynamics using atomic-resolution structure reveal structural fluctuations that may lead to polymerization of human Cu-Zn superoxide dismutase. Proc Natl Acad Sci USA. 2007;104:10040–10044. doi: 10.1073/pnas.0703857104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ding F, Taso D, Nie H, Dokholyan NV. Ab initio folding of proteins using all-atom discrete molecular dynamics. Structure (London) 2008;16:1010–1018. doi: 10.1016/j.str.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dokholyan NV. Studies of folding and misfolding using simplified models. Curr Opin Struct Biol. 2006;16:79–85. doi: 10.1016/j.sbi.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 32.Khare SD, Ding F, Dokholyan NV. Folding of Cu, Zn superoxide dismutase and familial amyotrophic lateral sclerosis. J Mol Biol. 2003;334:515–525. doi: 10.1016/j.jmb.2003.09.069. [DOI] [PubMed] [Google Scholar]
- 33.Okamoto Y. Generalized-ensemble algorithms: Enhanced sampling techniques for Monte Carlo and molecular dynamics simulations. J Mol Graphics Model. 2004;22:425–439. doi: 10.1016/j.jmgm.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 34.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. The weighted histogram analysis method for free-energy calculations on biomolecules. 1. The method. J Comput Chem. 1992;13:1011–1021. [Google Scholar]
- 35.Furukawa Y, O'Halloran TV. Amyotrophic lateral sclerosis mutations have the greatest destabilizing effect on the apo- and reduced form of SOD1, leading to unfolding and oxidative aggregation. J Biol Chem. 2005;280:17266–17274. doi: 10.1074/jbc.M500482200. [DOI] [PubMed] [Google Scholar]
- 36.Lepock JR, Frey HE, Hallewell RA. Contribution of conformational stability and reversibility of unfolding to the increased thermostability of human and bovine superoxide dismutase mutated at free cysteines. J Biol Chem. 1990;265:21612–21618. [PubMed] [Google Scholar]
- 37.Shaw BF, et al. Local unfolding in a destabilized, pathogenic variant of superoxide dismutase 1 observed with H/D exchange and mass spectrometry. J Biol Chem. 2006;281:18167–18176. doi: 10.1074/jbc.M600623200. [DOI] [PubMed] [Google Scholar]
- 38.Clementi C, Nymeyer H, Onuchic JN. Topological and energetic factors: What determines the structural details of the transition state ensemble and “en-route” intermediates for protein folding? An investigation for small globular proteins. J Mol Biol. 2000;298:937–953. doi: 10.1006/jmbi.2000.3693. [DOI] [PubMed] [Google Scholar]
- 39.Rakhit R, Chakrabartty A. Structure, folding, and misfolding of Cu, Zn superoxide dismutase in amyotrophic lateral sclerosis. Biochim Biophys Acta. 2006;1762:1025–1037. doi: 10.1016/j.bbadis.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 40.Khare SD, Caplow M, Dokholyan NV. The rate and equilibrium constants for a multistep reaction sequence for the aggregation of superoxide dismutase in amyotrophic lateral sclerosis. Proc Natl Acad Sci USA. 2004;101:15094–15099. doi: 10.1073/pnas.0406650101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Svensson AK, Bilsel O, Kondrashkina E, Zitzewitz JA, Matthews CR. Mapping the folding free energy surface for metal-free human Cu, Zn superoxide dismutase. J Mol Biol. 2006;364:1084–1102. doi: 10.1016/j.jmb.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 42.Ding F, Dokholyan NV, Buldyrev SV, Stanley HE, Shakhnovich EI. Molecular dynamics simulation of the SH3 domain aggregation suggests a generic amyloidogenesis mechanism. J Mol Biol. 2002;324:851–857. doi: 10.1016/s0022-2836(02)01112-9. [DOI] [PubMed] [Google Scholar]
- 43.Lazaridis T, Karplus M. Effective energy function for proteins in solution. Proteins. 1999;35:133–152. doi: 10.1002/(sici)1097-0134(19990501)35:2<133::aid-prot1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.