Abstract
Recently, two-photon imaging has allowed intravital tracking of lymphocyte migration and cellular interactions during germinal center (GC) reactions. The implications of two-photon measurements obtained by several investigators are currently the subject of controversy. With the help of two mathematical approaches, we reanalyze these data. It is shown that the measured lymphocyte migration frequency between the dark and the light zone is quantitatively explained by persistent random walk of lymphocytes. The cell motility data imply a fast intermixture of cells within the whole GC in approximately 3 h, and this does not allow for maintenance of dark and light zones. The model predicts that chemotaxis is active in GCs to maintain GC zoning and demonstrates that chemotaxis is consistent with two-photon lymphocyte motility data. However, the model also predicts that the chemokine sensitivity is quickly down-regulated. On the basis of these findings, we formulate a novel GC lymphocyte migration model and propose its verification by new two-photon experiments that combine the measurement of B cell migration with that of specific chemokine receptor expression levels. In addition, we discuss some statistical limitations for the interpretation of two-photon cell motility measurements in general.
Affinity maturation of B cells occurs within the microenvironment of germinal centers (GCs), and this localized immune response gives rise to long-lived antibody-secreting plasma and memory cells. In the course of the GC reaction, a specific spatial cell organization is observed with two main compartments: the light zone and the dark zone (1). In the dark zone, B cells proliferate and undergo somatic hypermutation of their immunoglobulin genes (2–4). In the light zone, follicular DCs (FDCs) retain antigen in the form of immune complexes. B cells in the light zone engage these immune complexes held on FDCs and compete for survival signals provided by both FDCs (5) and T helper cells (6), which are required for their differentiation into plasma and memory cells (7–8).
Intravital two-photon microscopy allows the visualization of fluorescently labeled cells as they move through living tissue. This minimally invasive imaging technique generates time-resolved data of cell shape, motility, and contact (9–11). Recently, data obtained with intravital microscopy have been published by three groups detailing the dynamic features of lymphocytes in GCs during the process of affinity maturation (12–14). These results have the potential to unravel the functional implications of the peculiar migratory behavior and cellular interactions of GC B cells, as well as the specific spatial organization of the GC into two zones. Both migration and zoning are connected problems and subject to long-standing speculation and conflicting conclusions (15–19).
All three groups (12–14) agree on the interpretation that B cell motility follows random walk migration with a directional persistence time of ∼1 min. This means that during the persistence time, a cell migrates in one direction before changing randomly to a new migration direction. Furthermore, all three groups agree that during measurements of 1 h, 5–10% of the observed B cells will have migrated from one zone to the other. As two-photon data are largely descriptive, the general conclusions drawn from these data for a GC migration model of B cells, however, are quite different (18, 19). Three migration models have been proposed, the widely accepted cyclic reentry model (20–22), the intra-zonal circulation model (14, 18), and the one-way migration model (17).
The cyclic reentry model assumes that a functional dependence exists between the light and the dark zone. According to this view, B cells proliferate and mutate in the dark zone and then follow a gradient provided by the chemokine CXCL13 to the light zone, where they compete for FDC- and T cell–derived survival signals. The successful B cell clones emerging from this selection process may either differentiate into output cells (plasma and memory cells) or migrate back to the dark zone attracted by the chemokine CXCL12 to reproliferate (23). Despite the measured motility data suggesting persistent random walk, the results of two experimental groups are considered to be in accordance with this chemokine-driven migration model (12, 13).
The intra-zonal circulation model (14, 18) views the light and the dark zones as functionally independent zones. Each B cell primarily circulates only within one of the two zones. The exchange of cells between the two zones occurs rarely and is considered to be of minor importance.
The third model, referred to as the one-way migration model (17), suggests that cells perform a persistent random walk and that reentry of selected B cells from the light zone to the dark zone is neither necessary for reproliferation nor are these cells actively moving toward the dark zone. As a consequence, selected B cells also reproliferate in the light zone and migration between the zones occurs only by chance as a result of random migration.
We use a mathematical approach to clarify the interpretation of intravital data. Although mathematical analyses of two-photon data have proven to be conclusive (24–26), suitable spatially resolved mathematical analyses of these data are limited. In this article, statistical and functional modeling approaches are used to interpret the experimental results from the three aforementioned studies (12–14). In addition, predictions of the B cell behavior on a time scale not yet accessible by two-photon imaging are made.
The statistical model assumes a minimum number of parameters, relies exclusively on the two-photon data, and allows statistical analysis of B cell trans-zone migration to be performed. The results of applying this model demonstrate that the experimental frequency of trans-zone migration can be explained by the assumption of persistent random walk. A very high number of trans-zone migration events must be collected and analyzed to deduce statistically reliable statements about GC migration models, even more than the currently available datasets (12–14). In addition, it is shown that a threshold for the migration distance of cells improves analysis of trans-zone migration events.
The functional model relies on mathematical techniques previously applied to the GC reaction (17, 21, 22, 27–32). It captures the migration and interaction of individual GC cells and permits analysis of affinity maturation during the GC reaction. In addition, the spatiotemporal GC organization, contact times between cells, and other dynamic quantities can be assessed. Application of this approach reveals that the GC zones, although not strictly required for affinity maturation, are only observed if B cells are sensitive to the soluble factors CXCL12 and CXCL13 (23). Furthermore, this model predicts that a down-regulation in sensitivity occurs for these chemokines. Collectively, our simulations reconcile an interesting controversy, demonstrating that two-photon motility data that support random walk migration turn out to be compatible with transient sensitivity to chemokines.
RESULTS
Statistical model analysis
This approach involves a three-dimensional reconstruction of B cell tracks that relies on the speed and turning angle distributions obtained by two-photon imaging. Statistical analysis of B cell tracks allows one to test whether the observed B cell motility and inter-zonal migration frequency can be reconciled with the hypothesis of a random walk with directional persistence time. Details of the statistical model and the fitting procedure, which is similar to the approach used in a recent study (26), are described in the Materials and methods and the Supplemental materials and methods (available at http://www.jem.org/cgi/content/full/jem.20081160/DC1).
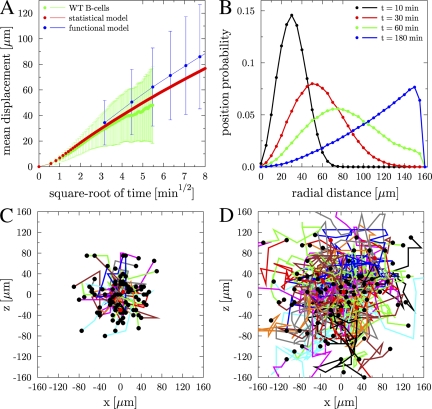
From the simulation of 105 cell tracks of homogeneously distributed B cells within a spherical GC of radius R = 160 μm, we compute the motility coefficient M = <|ri(t)|>2/6t from the time-dependent mean displacement <|ri(t)|>, where ri(t) denotes the position of the ith cell as a function of time (t) and the average is taken over all cells). The resulting time dependence of the mean displacement is shown in Fig. 1 A (red line) and compared with experimental data for B cells in WT mice (green line). When this is performed for B cells from WT versus CXCL13 knockout (KO) mice, we obtain motility coefficients of M = 21.4 μm2/min and M = 8.5 μm2/min for WT and CXCL13 KO mice, respectively. These values are in agreement with the experimentally determined motility coefficients of M = 21.4 ± 4.2 μm2/min and M = 8.5 ± 3.5 μm2/min, respectively, reported by Allen et al. (12). Importantly, this agreement is found for persistence times of Δtp = 1.24 min and Δtp = 1.05 min, respectively. We conclude from the consistent description of the motility data (Fig. 1 A and Figs. S1–S3, available at http://www.jem.org/cgi/content/full/jem.20081160/DC1, for WT mice) that the B cell motility in GCs can be interpreted as random walk migration with a directional persistence time. The Δtp for B cells in WT versus CXCL13 KO mice is 20% longer and reflects the impact of chemotactic signaling on persistence time.
Figure 1.
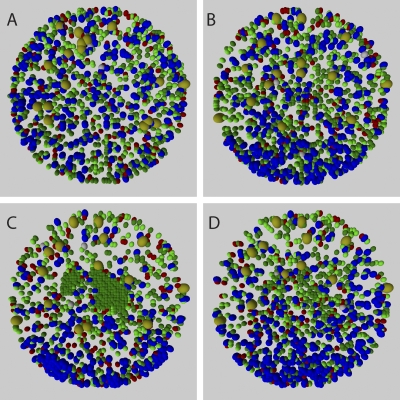
GC B cell migration data as obtained from the mathematical modeling. (A) Mean displacement of B cells in WT mice as a function of the square root of time. Simulation results of the statistical model (red) and the functional model (blue) are compared with experimental results (green) by Allen et al. (12). The linear relation after a time-lag, which is induced by the persistence time, is a qualitative indication for random walk migration. (B) Position probability distribution P(r,t) depending on the radial distance as obtained from the statistical model for GC B cells in WT mice at four different times. At t = 60 min, the distribution P(r,t) is affected by the finite GC volume, whereas at t = 180 min, the quadratic dependence of P(r,t) reflects a homogeneous distribution of B cells in the GC volume. (C) GC B cell tracks as obtained from the functional model at day five after onset of proliferation (transient model with weak chemotaxis). The three-dimensional cell tracks have been projected on the x–z plane with the cell starting positions set relative to the center of the plane. The 90 cells have been randomly chosen and tracked over a period of half an hour. (D) The same as in C, but with a tracking period of 3 h.
Experimental frequency of trans-zone migration is compatible with random walk
The experimental two-photon data consistently report that 5–10% of B cells migrate between the GC zones per hour (12–14), which is generally considered to be a small value. However, a rough estimate reveals that this value corresponds to 40% of the maximal possible frequency: for a homogeneous distribution of B cells in zones of equal size, the probability to randomly choose a B cell from one of the two zones is 50%. With a probability of ∼50%, the chosen B cell migrates toward the other zone and will reach it after a sufficiently long time. Thus, the maximal frequency of this event is estimated to be 25%. Relative to this value, the observed trans-zone migration frequency of ∼10% per hour corresponds to 40% of the maximal possible frequency.
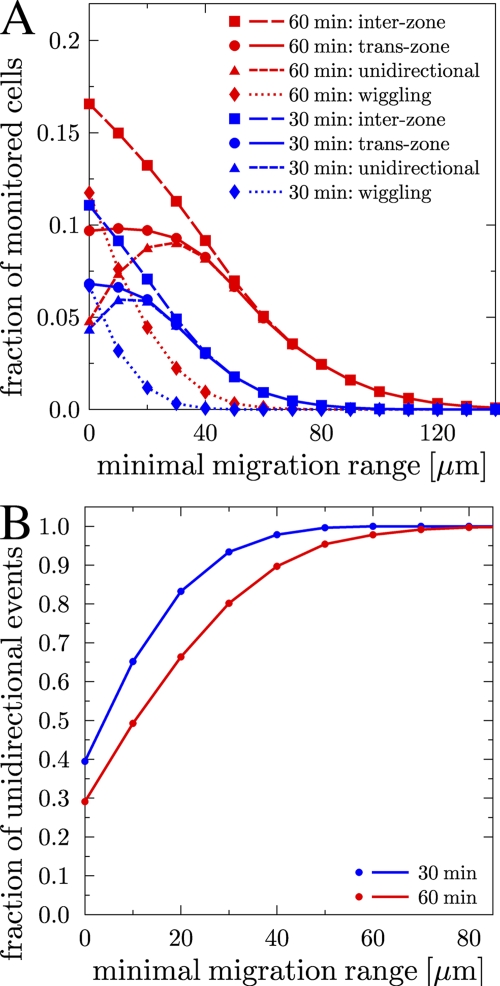
Using the statistical model, we investigate whether the experimentally determined trans-zone migration frequency of 5–10% is consistent with the assumption of random walk migration. To address this question, we have to distinguish between the inter-zone migration, i.e., the frequency of B cells crossing the boundary between zones (zone boundary) and ending up in either of the two zones, and the more restricted trans-zone migration, i.e., the frequency of B cells crossing the zone boundary and ending up in the opposite zone. Inter-zone migration comprises cells that cross the zone boundary unidirectionally, i.e., that are traversing the zone boundary only once, and cells that are wiggling at the zone boundary. The unidirectional migration paths are relatively straight across the zone boundary, whereas wiggling is characterized by the indecisive tossing of cells at the zone boundary as a consequence of the random walk migration. The number of wiggling migration events at the zone boundary is defined as the difference of inter-zone and unidirectional migration events.
We compute fractions for all types of zonal migration, i.e., the number of cells traversing the zone boundary divided by the total B cell population per hour. Here, it is assumed that the zone boundary divides the GC into two equal parts. We start again from a homogeneous distribution of 105 WT B cells in the spherical GC with radius R = 160 μm. The fraction of wiggling cells (Fig. 2 A, diamonds) and of unidirectional migration events (Fig. 2 A, triangles) depend strongly on the minimal migration range (rmin). The minimal migration range is defined as the thickness in micrometers of the zone boundary that has to be traversed by a cell to be counted as a zonal transition. The contribution of wiggling cells is an exponentially decreasing function of rmin (Fig. 2 A, diamonds, and Fig. S4, available at http://www.jem.org/cgi/content/full/jem.20081160/DC1), such that for sufficiently large rmin all zonal transitions correspond to cells that cross the boundary unidirectionally (Fig. 2 A, triangles, and Fig. S4). For a minimal migration range of 40 μm, the model predicts trans-zone migration frequencies of 8% per hour (Fig. 2 A, circles), which is in agreement with the experimental data.
Figure 2.
B cell migration between the dark zone and the light zone in the statistical model. (A) Fraction of cells that perform migration of type inter-zone, trans-zone, unidirectional, and wiggling as a function of the minimal migration range rmin across the zone boundary during simulation times of half an hour (blue) and 1 h (red). The fraction of cells that perform inter-zone migration consists of the cell fractions that perform unidirectional migration and that are wiggling at the zone boundary. (B) Fraction of unidirectional migration events across the zone boundary relative to all inter-zone migration events. B cells that traverse the zone boundary by migrating a minimal range of 25 μm (40 μm) within 30 min (60 min) perform with high probability of ∼90% unidirectional migration events.
Trans-zone migration is dominated by cells that change between zones unidirectionally for a sufficiently large zone boundary thickness rmin > 25 μm after a simulation time of half an hour (Fig. 2 B, blue line). These results agree with the observation that monitored cell tracks of unidirectional migration across the boundary during half-hour measurements follow relatively straight paths that are of a typical length of >20 μm (12). Furthermore, in one-hour measurements, the corresponding zone boundary thickness is rmin > 40 μm (Fig. 2 B, red line). Therefore, meaningful experimental analyses that distinguish wiggling cells from unidirectionally moving cells require the definition of a minimal migration range across the zone boundary.
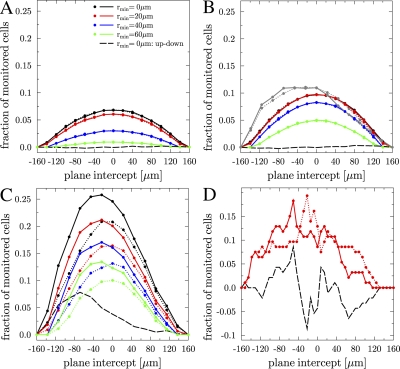
Next, we address the question whether B cell trans-zone migration exhibits a directional preference between zones. As a directional preference may depend on the position of the zone boundary in the GC, we introduce an intercepting plane that is oriented parallel to the zone boundary and can be moved through the GC volume. We measure trans-zone migration through this intercepting plane in silico. Results are shown for different rmin after measurements of 30 min (Fig. 3 A) and 1 h (Fig. 3 B, all but gray lines). The frequency of trans-zone migration is maximal for the plane intercept located at 0 μm, i.e., where the GC consists of two equal parts, as there the area of the intercepting plane is maximal. The directional preference of trans-zone migration is measured as the difference of B cells moving up (Fig. 3, solid lines) and down (Fig. 3, dotted lines) through the intercepting plane. No statistically relevant difference is observed in the statistical model (Fig. 3, A and B, dashed line).
Figure 3.
B cell migration between the dark zone and the light zone. (A) Fraction of cells performing trans-zone migration after half an hour simulation time within the statistical model as a function of the location of the intercepting plane along the z direction and for various minimal migration ranges rmin. A very small difference is observed between up-moving (solid lines) and down-moving (dotted lines) B cells within the statistical model. The black dashed line corresponds to the fraction of up-moving minus down-moving B cells for rmin = 0 μm. (B) As before, with the same color coding but for 1 h of simulation time. In addition, for rmin = 0 μm, the corresponding result from the functional model (transient weak chemotaxis model) at day five after onset of proliferation is shown for comparison (gray lines). The position of the maximum coincides with the position of the boundary between the dark and light zone in the functional model (Fig. 5 B). (C) The same color coding as before, but for a simulation time of 10 h using the transient weak chemotaxis model. After 10 h, a clear preference of up-moving (solid lines) over down-moving (dotted lines) cells is observed. (D) Results from the functional model (transient weak chemotaxis model) for 50 centrocytes and 50 centroblasts tracked for 1 h at day five after onset of proliferation. The observed curves are dominated by statistical fluctuations.
Intermixture of B cells in GCs is quick
Having validated the statistical model, we can now go beyond the time scale that limits two-photon experiments and perform simulations of B cell migration on a time scale of several hours. Computed cell tracks allow the calculation of the position distribution P(r,t) of the probability to find a B cell after a time interval (t) at a radial distance (r) from its initial position at the center of the GC (Fig. 1 B). We use P(r,t) to compute the migration distance <|ri(t)|> for WT B cells in a GC with radius R = 160 μm. During the time intervals of 10, 30, 60, and 180 min, the cells migrate distances of 31, 57, 82, and 113 μm, respectively. These results deviate from a Gaussian random walk in free space, which leads to the migration distances (<|ri(t)|> = [6Mt]1/2) of 36, 62, 88, and 152 μm for the same time intervals, respectively. For shorter time intervals, the deviation is related to the cell's persistence time, whereas during longer intervals the boundary of the GC volume adds to the difference.
B cells can reach the GC boundary already after 1 h (Fig. 1 B, green line). Most B cells travel this distance within 3 h, which is reflected by the quadratic dependence of the position probability on the radial distance (Fig. 1 B, blue line). This means that in the absence of additional factors, the GC quickly evolves into an intermixed cell system within 3 h, which is even shorter than the estimate of 18 h by Schwickert et al. (13).
Functional model analysis
The quick intermixture, as observed in the statistical model, requires mechanisms to maintain the observed zonal GC morphology for several days of the GC reaction. This is investigated with a functional mathematical model. The essential feature of this model is that it captures the whole GC reaction comprising the population kinetics, cellular interactions, and affinity maturation. This level of description allows one to distinguish between centroblasts, proliferating B cells that undergo somatic hypermutation, and centrocytes, nondividing B cells with activated apoptosis competing for survival signals.
For a functional analysis, the GC reaction has to be represented either by a set of differential equations for each possible clone (33) or by discrete event simulators (17, 30–32, 34). Here, a stochastic discrete event simulator in three spatial dimensions is used, which is based on the extension of a previously published agent-based model (32). The included mechanisms follow, to a large extent, the classical model of the GC reaction (1, 15). A detailed description of the model's assumptions is provided in the Supplemental materials and methods.
The simulated GC reactions are validated by experimental data. For example, the resulting population kinetics are typical of secondary immunizations (Fig. S5 A, available at http://www.jem.org/cgi/content/full/jem.20081160/DC1). Furthermore, affinity maturation takes place (Fig. S5B), and a dominance of high affinity clones is found at day eight after onset of proliferation (35, 36). The number of accumulated mutations that impact on the antibody-affinity is five to eight in high affinity clones (37, 38). Finally, the ratio of output cells at day twelve to output cells at day six measured earlier (29, 39) is reproduced. Note that all simulations discussed from this point forward exhibit the aforementioned dynamic properties unless stated otherwise (Fig. S5).
Short centrocyte–FDC contacts are dominant in vivo and in silico
Next, the duration of contacts between centrocytes and FDCs is analyzed. The functional model assumes, similar to biological observations (40), that centrocytes bind antigen on FDCs to obtain survival signals and that the binding process is affinity dependent. Furthermore, depending on the affinity of the encoded antibody, the B cells either contact FDCs in a static condition of 30 min or in a transient manner that lasts until the B cell moves on, i.e., continues to migrate. No parameters other than those related to cell motility and antibody affinity enter this description of cell–cell contact.
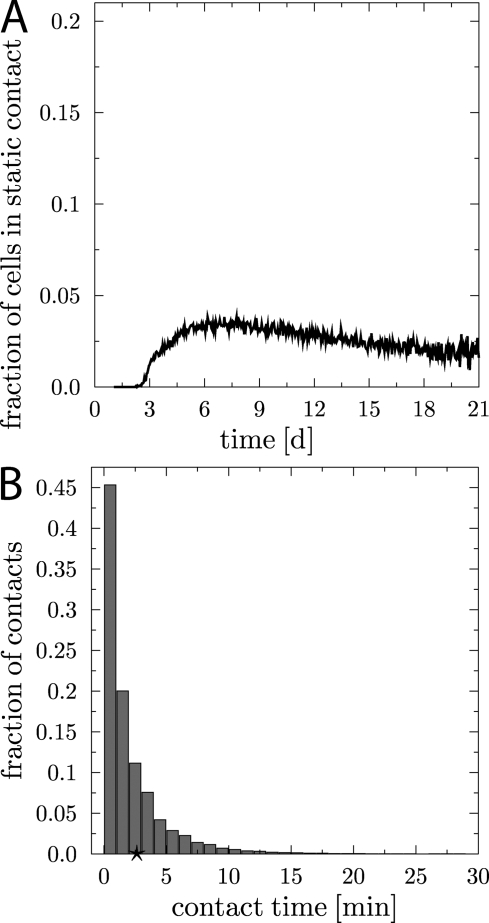
Contacts between centrocytes and FDCs observed in vivo have rarely been found to be longer than 5 min (13), although the finding that apoptosis is switched off after 2 h of signaling (41) would have suggested longer contact times. We have therefore investigated how long centrocyte–FDC contacts would appear under the imposed condition that static functional contacts would last for 30 min. As can be seen in Fig. 4, only 2–4% of the cells are in a static contact in the simulations. The histogram (Fig. 4 B) depicts the duration of all (static and transient) centrocyte–FDC contacts in the simulations. In agreement with the two-photon measurements (13), the simulation suggests that the majority of B cells in the functional model exhibit only short contacts with FDCs, i.e., ≤ 5 min, and that static contacts are rare events. Indeed, around day five of the reaction, affinity maturation is not fully accomplished and most cells are deleted because of disadvantageous mutations. Thus, it cannot be concluded from the experimental contact data that centrocytes integrate signals from short contacts with FDCs (13). Instead, even though signal integration cannot be ruled out, the simulations lead us to propose that it is the rare static contacts that are sufficient to drive affinity maturation.
Figure 4.
Analysis of centrocyte–FDC contacts obtained in the functional model. Despite the model assuming static contacts of 30 min for signaling to high-affinity clones, the vast majority of contacts monitored during a tracking period of 1 h is of dynamic nature and last only ∼3 min. Only a few cells exhibit contact times of >10 min. This is reflected in the fraction of centrocytes in static contact to FDCs, which is in the range of only a few percent (A) and in the histogram of centrocyte–FDC contact times (B). The star indicates the mean contact time of 2–3 min.
Experimental data supporting B cell random walk migration are compatible with chemotaxis
The experimental two-photon data (12–14) suggest that B cells perform a random walk with directional persistence time in the absence of chemotaxis. Thus, we investigate using the functional model under which circumstances the measured two-photon imaging data can be reconciled with chemotaxis. Based upon the results of the statistical model a directional persistence time of 1.24 min is used in the simulations, performed with or without chemotaxis. When the simulations are performed with chemotaxis, centrocytes are assumed to respond to the chemokine CXCL13 and centroblasts to CXCL12. Furthermore, it is assumed that CXCL13 is FDC-derived, whereas CXCL12 is secreted by stromal cells at the border of the follicle with the T zone. The spatial distribution of chemokines is shown in Fig. S6 (available at http://www.jem.org/cgi/content/full/jem.20081160/DC1).
The simulations in the absence of chemokines reproduce the experimentally observed motility coefficient. We refer to this scenario as the random model. When chemotaxis is permanently switched on, interestingly, this does not lead to a larger motility coefficient. On the contrary, an attraction occurs to the center of the chemotactic sources leading to unphysiological cell accumulation (unpublished data) and concomitant reduction of the motility coefficient (Fig. S5, C and D). This cell accumulation is not the result of an artificially induced space restriction on the lattice, as cell motility is maintained in all simulations by allowing cells to exchange lattice positions even if no free lattice site is available (Supplemental materials and methods, section Cell motility and chemotaxis).
As an intermediate solution, the transient model is introduced, that is, the sensitivity of newly differentiated centrocytes for CXCL13 is assumed to be down-regulated after 6 h, or after successful encounter with antigen on FDC. Similarly, the sensitivity of centroblasts for CXCL12 is down-regulated after 6 h or upon differentiation to centrocytes. The transient model is still compatible with the measured mean displacement curve (Fig. 1 A and Fig. S5 C). These results suggest that B cells undergo temporarily directed migration, implying that transient chemotaxis might well be hidden in the experimental motility data. Ignoring the complex functional environment of the GC, a similar result was found for Brownian particles in weak external fields (42). Temporarily directed migration is further supported by the observation that trans-zone migration paths are relatively straight as compared with B cell migration within the zones (12).
GC zoning requires B cell chemotaxis with desensitization
Next, the morphological implications of chemotaxis are addressed using the functional model. As in the statistical model, the experimentally observed high motility of B cells leads to a quick intermixture of cells between the GC zones. For example, this is shown in Fig. 1 (C and D) when applying the transient model. After half an hour, the distribution of cell tracks (Fig. 1 C) is once again similar to experimental results (19). In the absence of chemotaxis, the quick intermixture implies that the maintenance of dark and light zones is not possible (Fig. 5 A).
Figure 5.
GC morphology for different B cell migration models. 25-μm-thick slices through the center of the three-dimensional GC showing its morphology at day five after onset of proliferation. Results are obtained from the functional model simulation for different B cell migration models. Color code: FDC (large, dark yellow); proliferating B cells (large, blue); centrocytes (small, green); selected centrocytes (small, light green); T cells (red); output cells (gray). (A) Without chemotaxis, the fast intermixture of cells leads to the dissolution of any zonal structure. (B) With weak chemotaxis, two zones develop with a substantial number of proliferating B cells in the light zone. (C) In the case of moderate chemotaxis, where cell polarity is determined to equal parts by randomness and chemotactic signals, cells artificially accumulate in their respective zones. (D) Moderate chemotaxis together with chemokine concentration-dependent desensitization yields clear zonal separations as for weak chemotaxis (B).
To maintain a dark zone, newly differentiated centrocytes could be actively driven out of the dark zone into the light zone by chemotaxis. Therefore, we analyze the GC morphology using different chemotaxis models (Supplemental materials and methods, section Cell motility and chemotaxis). As permanent chemotaxis leads to artificial cell accumulations and unphysiological motility coefficients (Fig. S5 D), only the results using the transient model are reported here.
Applying this model, cells repolarize both randomly as well as in the direction of chemokine gradients. The extent of random versus chemokine contribution defines the three levels of chemotactic strength. In the case of weak chemotaxis, cell repolarization occurs with a weight of one tenth because of chemokine gradients. These simulations demonstrate that the dark zone is maintained for a physiological period of time during the GC reaction (Fig. 5 B). Increasing the strength of chemotaxis to a moderate level, where both contributions to repolarization are of equal weight, already induces an unphysiological cell accumulation (Fig. 5 C). This effect of cell accumulation is even more pronounced for strong chemotaxis (unpublished data), where the contribution of the chemokine gradient is further increased 10-fold.
Unphysiological cell accumulation may be avoided by introducing an additional mechanism, i.e, desensitization. The desensitization model describes a scenario in which the cell sensitivity also depends on the absolute chemokine concentration and is shut-off when a critical concentration is encountered (Supplemental materials and methods, section Cell motility and chemotaxis). Indeed, cell accumulation is dissolved for moderate chemotaxis in the desensitization model (Fig. 5 D).
Trans-zone migration is subject to statistical fluctuations and sensitive to tracking durations
An important issue of the recent series of intravital GC experiments refers to trans-zone migration of B cells. Our analysis within the statistical model revealed that undirected persistent cell migration leads to the amount of trans-zone migration per hour that is found in intravital experiments (12–14). How does this result, then, depend on the functional environment and on the presence of chemotaxis? In the functional model, the frequency of trans-zone migration events is evaluated by monitoring 6,000 cell tracks (Fig. 3, B [gray line] and C) and, within 1 h of tracking, the frequency of trans-zone migration is consistent with experimental data. Compared with the statistical model, the maximum is shifted to the actual boundary between dark and light zone (Fig. 3 B, gray line) which is around the plane intercept at −50 μm (dark zone at even more negative values). This reflects a different density of cells in both zones. After 10 h of tracking, the amount of observed trans-zone migration is increased to a maximal value of 25% (Fig. 3 C, black solid line), which is expected on the basis of the simple estimate given above. In contrast to shorter tracking times, more B cells move from the dark to the light zone than from the light to the dark zone (Fig. 3 C, black dashed line). It is important to realize that tracking >1,000 cells is required to obtain statistically reliable results. Tracking 100 cells, as is typically done in today's tracking experiments, leads to trans-zone migration frequencies that are dominated by statistical fluctuations (Fig. 3 D and Discussion).
DISCUSSION
There is agreement in all three two-photon microscopy studies (12–14) that the B cell motility in GCs can be interpreted as a random walk with persistence time. This interpretation can be reconciled with both the statistical and the functional model. The simulations show that chemotaxis and desensitization mechanisms have a prominent impact on the GC organization and guarantee the existence of distinguishable zones without getting into conflict with the two-photon motility data.
Experimental results also consistently suggest that the frequency of B cells that migrate between the GC zones is as small as a few percent per 1 h of cell tracking. In silico, both the statistical and functional models yield a frequency of trans-zone migration events that is in agreement with the experimental data. We have shown that the seemingly limited trans-zone migration events imply a fast intermixture of all B cells in the whole GC within 3 h.
Trans-zone migration data have to be interpreted very carefully. If no minimal migration range for cells crossing between the two zones is imposed, the observed migration events may be dominated by cells wiggling along the boundary without any directional preference for one of the zones (Fig. 2). Our calculations provide an experimental guideline for the required minimal migration range (Fig. 2 B).
All three experiments suggest a slight preference for trans-zone migration from the dark zone to the light zone, even though they are statistically not very firm. Although the imaging duration and the number of tracked cells in current two-photon measurements is limited for technical reasons, mathematical models do not suffer from these restrictions. Tracking several thousand B cells within the functional model, a significant preference is only observed for long tracking times of 10 h, which is in the order of the centrocyte lifetime (Fig. 3, black dashed lines). In this range, selection of centrocytes acts as a sink of cells on the side of the intercepting plane containing the FDC network and affects the frequency of trans-zone migration events. Thus, the model is in agreement with the measured small preference of trans-zone migration events but predicts that for much longer tracking times a more substantial directional preference is expected.
Two-photon experiments are to some extent limited by the cell track analysis, which today cannot be done fully automatically such that the number of tracked cells is typically not more than a few hundreds. Because of the small frequency of trans-zone migration events, monitoring several thousand cells is necessary to obtain a reliable certainty of the numerical results. For 1,000 monitored cells, the statistical fluctuations are in the order of 10%, which increases to 100% for 100 cells. Next, rather irregular trans-zone migration patterns are observed (Fig. 3 D). These are in parts comparable to experimental results of trans-zone migration (14). Therefore, we think that the conclusion that trans-zone migration is suppressed at the interface between dark zone and light zone of the GC (14) has to be reconsidered.
The simulations show that the measured quick random migration of all cells throughout the GC makes a durable dark zone impossible. This implies that the specific morphology of the GC requires an additional mechanism that conducts freshly differentiated centrocytes from the dark zone to the light zone and recycled B cells back to the dark zone. Natural candidates for such a random walk with a drift are the chemokines CXCL12 and CXCL13 (23).
The model predicts that chemotactic sensitivity of B cells has to be down-regulated again after a characteristic duration, or in response to signals provided by FDCs or T cells, or after encounter of overcritical chemokine concentrations in some regions of the GC. Desensitization is known to be related to chemokine receptor internalization (43). CXCR5 up-regulation after contact with dendritic cells was observed for T cells (44,45) and CCR7 up-regulation after contact with antigen was observed for follicular B cells (46). B cells also regulate CXCR4-receptor expression in dependence of SDF-1α (47). A concentration-dependent response of T cells to SDF-1 was observed (48). A bell-shaped response curve was also shown for B cells (49), and for CXCL13 in particular (50). Thus, experimental evidence exists for the regulation of B cell sensitivity to chemokines.
If B cells were not desensitized by at least one of the aforementioned mechanisms, chemotaxis would get into conflict with experimentally observed motility data (Fig. S5 D) and chemotaxis would induce artificial cell accumulation in the FDC network and at the T zone boundary (Fig. 5 C). It turns out that transient chemotactic sensitivity of B cells is consistent with all motility data and induces a realistic GC morphology (Fig. 5, B and D). Then, a dominance of proliferating B cells in the dark zone and a dominance of centrocytes in the light zone is observed. However, the simulations exhibit a nonnegligible amount of B cell proliferation in the light zone, as is also observed in in vivo experiments (12, 51, 52).
It is striking that the population kinetics (Fig. S5 A) and affinity maturation (Fig. S5 B) are widely independent of the chosen chemotaxis model and, by this, of the morphological organization of the GC. Thus, GCs without dark zones might well exist (17). In fact, CXCR5-deficient mice exhibit disorganised follicles but still show signs of affinity maturation (53). From this vantage point, the function of the dark zone may be less related to affinity maturation than to GC onset by providing space with reduced selection pressure (30). It would be very interesting to investigate experimentally how affinity maturation compares with and without antagonists of B cell chemokine receptors in normal mice.
In conclusion, we come to the result that the intra-zonal circulation model (14, 18) is unlikely and that the data support a compromise between the one-way (17) and the cyclic reentry model (20–22); the intra-zonal circulation model is incompatible with the trans-zone migration data that are fully explained by the assumption of persistent random walk migration. We conclude from the functional model simulations that an intra-zonal circulation model would induce far less trans-zone migration than that found in all three experiments during 1 h. The one-way migration model, which relies on pure random walk migration, would give rise to an unphysiological GC morphology caused by the high motility of B cells and by this the fast intermixture of all cells. Finally, the cyclic reentry model assumes a strong and chemokine-driven recirculation of recycled B cells to the dark zone. If CXCL12 sensitivity was sufficiently strong to redirect almost all recycled B cells back to the dark zone, the motility coefficient and the morphology would contradict experimental data. The motility data and the GC morphology impose a maximum strength of chemotaxis in respect to a randomly chosen repolarization.
Our simulations predict a novel migration model for B cells in GC reactions that reconciles all experimental and theoretical data. Most of the cells in the dark zone and light zone actively perform persistent random walk migration. Centrocytes acquire additional weak sensitivity to CXCL13 after or during differentiation from centroblasts and orient toward the FDC network. In the FDC network, the centrocytes lose CXCL13 sensitivity. This might be induced by contact with FDC, by overcritical CXCL13 concentrations and CXCR5 internalization, or simply by down-regulation of CXCR5 after a characteristic time. Similarly, a return to the dark zone of positively selected centrocytes is facilitated by a weak transient chemotactic sensitivity for CXCL12. However, random walk migration remains the dominant pathway of repolarization and reproliferation of positively selected centrocytes frequently occurs in the light zone (12, 51, 52).
Intravital experiments that visualize B cell migration in combination with chemokine receptor levels might one day unravel whether transient chemotaxis underlies the apparently pure random walk. Alternatively, it might be possible in two-photon experiments to track B cells that were just positively selected. A suitable marker may be a reporter indicating B cell stimulation, like fluorescent protein expressed after immunoglobulin class switching (7, 54). Freshly selected B cells would be expected to exhibit a stronger sensitivity to chemokine signals. In addition, tracking of these B cells might allow the observation of further cell divisions, which would be the first direct proof of recycling in the GC.
MATERIALS AND METHODS
Statistical model.
This model of B cell motility is exclusively based on two-photon imaging data, i.e., the B cell speed and turning angle distributions as obtained from two-photon microscopy experiments by Allen et al. (12). The data are derived from the recorded cell tracks and are plotted in Fig. S1. Cellular interactions are effectively accounted for via these distributions.
We reconstruct B cell tracks by computing the position vector ri(t + Δt) of the ith cell at time t + Δt from its position ri(t) and its velocity vi(t) at time t: ri(t + Δt) = ri(t) + Δt vi(t). At each time step Δt, we decide whether or not to update the velocity vi(t) = vi(t) ei(t) in terms of the cell speed vi(t) and the unit vector ei(t) of the cell polarity. The change in the cell's polarity vector at subsequent times is related to the turning angle α(t) = arccos[ei(t) • ei(t − Δt)].
The persistent random walk hypothesis is incorporated by choosing the speed and turning angle independently. This is realized using a Monte Carlo acceptance-rejection method to generate random speeds and random turning angles according to the underlying distributions (Fig. S1). Furthermore, in three spatial dimensions, ei(t) is not uniquely determined by the turning angle, but may refer to any point on a circle with radius rα = |sin(α)|. The point on this circle is chosen at random from a uniform distribution.
The turning angle and the speed are permanently changing after characteristic time steps Δtp and Δtv, respectively. Because it is an experimental fact that changes in the cell speed occur on the time scale of seconds, whereas changes in the cell polarity occur on the time scale of minutes, we set Δt = Δtv = 20 s corresponding to the time interval between two consecutive speed measurements (12). Thus, a new speed value is randomly drawn from the corresponding distribution (Fig. S1 A) at each time step Δt = 20 s, whereas the parameter Δtp is fitted to reproduce the measured curve of the mean displacement versus the square root of time (Fig. 1 A). We draw turning angles at random persistence time steps δtp = Δtp (0.5 + ρ), where ρ is a uniformly distributed random variable between 0 and 1, such that <δtp> = Δtp on average.
It is important to note that even though the directional persistence time Δtp is not known a priori, we can nevertheless draw turning angles from the experimental distribution that has been evaluated with a time resolution of 20 s. As shown in the supplemental materials (Fig. S2), the turning angle distribution in Fig. S1 B is extremely robust and represents a reasonable approximation for different values of Δtp ranging from 20 s up to 160 s. Determining Δtp from the condition that the experimentally observed mean displacement curve is recovered for B cells in WT mice and in CXCL13 KO mice, yields the reasonable values Δtp = 1.24 min and Δtp = 1.05 min, respectively. Typical time sequences of the turning angle and the speed for a B cell in WT mice are presented in Fig. S3 during 30 min of cell tracking.
Functional model.
We developed a hybrid agent-based model for simulations of the GC reaction in three spatial dimensions that relies on previous mathematical models for the GC reaction (17, 29, 30, 32). The model consists of the following three coupled levels: (a) The first level contains the main lattice corresponding to the physical space in which cells can migrate and interact. Each node of the lattice can carry up to one biological cell. The cells migrate and interact according to the assumptions explained in the Supplemental materials and methods. (b) The second level refers to the shape space (55), which is represented by a four-dimensional lattice encoding the antibody type of each B cell. Somatic hypermutation is represented by switching the antibody type to a neighboring point in the shape space. Without loss of generality, the optimal clone for a given antigen is in the center of the shape space. The Hamming distance of a clone from the optimal clone is mapped to a quantity associated with the antibody-antigen affinity (29, 56). This affinity determines the binding probability of a cell object to an FDC. (c) The third level deals with solving a system of reaction-diffusion equations for the solubles. An Alternating-Direction-Implicit method is applied to solve the partial differential equations on a lattice. The solubles considered in the following are the chemokines CXCL12 and CXCL13. It is assumed that the source for CXCL12 are stromal cells at the border of the follicle to the T zone, and the FDCs in the light zone for CXCL13.
A detailed description is given in the Supplemental materials and methods. The parameters of the model are listed in Table S1 (available at http://www.jem.org/cgi/content/full/jem.20081160/DC1).
Online supplemental material.
The applied mathematical framework is explained in the Supplemental materials and methods. Detailed information about the statistical model and its validity is provided and presented in Figs. S1–S3. Fig. S4 shows B cell migration between the dark zone and the light zone as a function of the number of zonal transitions within the statistical model. Additional information on the GC reaction in terms of the population kinetics, affinity maturation, and B cell mean displacement is presented in Figure S5. The corresponding GC chemokine distributions of CXCL12 and CXCL13 are shown in Figure S6. The functional model and its relation to physiology are described and all model parameters are summarized in Table S1. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20081160/DC1.
Supplementary Material
Acknowledgments
We thank C.D.C. Allen, T. Okada, H. Lucy Tang, and J.G. Cyster for providing in vivo cell track data from their two-photon imaging experiments.
This research is financially supported by the European Union within the NEST project MAMOCELL. Frankfurt Institute of Advanced Study is supported by the ALTANA AG.
The authors have no conflicting financial interests.
Abbreviations used: FDC, follicular DC; GC, germinal center.
References
- 1.Nossal, G. 1992. The molecular and cellular basis of affinity maturation in the antibody response. Cell. 68:1–2. [DOI] [PubMed] [Google Scholar]
- 2.Berek, C., and C. Milstein. 1987. Mutation drift and repertoire shift in the maturation of the immune response. Immunol. Rev. 96:23–41. [DOI] [PubMed] [Google Scholar]
- 3.Toellner, K.M., W. Jenkinson, D. Taylor, M. Khan, D. Sze, D. Sansom, C. Vinuesa, and I. MacLennan. 2002. Low-level hypermutation in T cell-independent germinal centers compared with high mutation rates associated with T cell-dependent germinal centers. J. Exp. Med. 195:383–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunn-Walters, D.K., A. Belelovsky, H. Edelman, M. Banerjee, and R. Mehr. 2002. The dynamics of germinal centre selection as measured by graph-theoretical analysis of mutational lineage trees. Dev. Immunol. 9:233–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kosco-Vilbois, M.H. 2003. Are follicular dendritic cells really good for nothing? Nat. Rev. Immunol. 3:764–769. [DOI] [PubMed] [Google Scholar]
- 6.Walker, L.S.K., A. Gulbranson-Judge, S. Flynn, T. Brocker, and P.J.L. Lane. 2000. Costimulation and selection for T-cell help for germinal centres: the role of CD28 and OX40. Immunol. Today. 21:333–337. [DOI] [PubMed] [Google Scholar]
- 7.Toellner, K.M., S. Luther, D. Sze, R. Choy, D. Taylor, I. MacLennan, and H. Acha-Orbea. 1998. T helper 1 (Th1) and Th2 characteristics start to develop during T cell priming and are associated with an immediate ability to induce immunoglobulin class switching. J. Exp. Med. 187:1193–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Vinuesa, C.G., M.C. Cook, J. Ball, M. Drew, Y. Sunners, M. Cascalho, M. Wabl, G.G.B. Klaus, and C.M. MacLennan. 2000. Germinal centers without T cells. J. Exp. Med. 191:485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cahalan, M.D., I. Parker, S. Wei, and M. Miller. 2002. Two-photon tissue imaging: seeing the immune system in a fresh light. Nat. Rev. Immunol. 2:872–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sumen, C., T. Mempel, I. Mazo, and U. von Andrian. 2004. Intravital microscopy: visualizing immunity in context. Immunity. 21:315– 329. [DOI] [PubMed] [Google Scholar]
- 11.Gunzer, M., C. Weishaupt, A. Hillmer, Y. Basoglu, P. Friedl, K.E. Dittmar, W. Kolanus, G. Varga, and S. Grabbe. 2004. A spectrum of biophysical interaction modes between T cells and different antigen-presenting cells during priming in 3-D collagen and in vivo. Blood. 104:2801–2809. [DOI] [PubMed] [Google Scholar]
- 12.Allen, C.D., T. Okada, H.L. Tang, and J. Cyster. 2007. Imaging of germinal center selection events during affinity maturation. Science. 315:528–531. [DOI] [PubMed] [Google Scholar]
- 13.Schwickert, T.A., R. Lindquist, G. Shakhar, G. Livshits, D. Skokos, M. Kosco-Vilbois, M. Dustin, and M. Nussenzweig. 2007. In vivo germinal center imaging reveals a dynamic open structure. Nature. 446:83–87. [DOI] [PubMed] [Google Scholar]
- 14.Hauser, A.E., T. Junt, T. Mempel, M. Sneddon, S. Kleinstein, S. Henrickson, U. von Andrian, M. Shlomchik, and A. Haberman. 2007. Definition of germinal-center B cell migration in vivo reveals predominant intrazonal circulation patterns. Immunity. 26:655–667. [DOI] [PubMed] [Google Scholar]
- 15.MacLennan, I.C.M. 1994. Germinal centers. Annu. Rev. Immunol. 12:117–139. [DOI] [PubMed] [Google Scholar]
- 16.Manser, T. 2004. Textbook germinal centers? J. Immunol. 172:3369–3375. [DOI] [PubMed] [Google Scholar]
- 17.Meyer-Hermann, M.E., and P. Maini. 2005. Back to one-way germinal centers. J. Immunol. 174:2489–2493. [DOI] [PubMed] [Google Scholar]
- 18.Hauser, A.E., M. Shlomchik, and A. Haberman. 2007. In vivo imaging studies shed light on germinal-centre development. Nat. Rev. Immunol. 7:499–504. [DOI] [PubMed] [Google Scholar]
- 19.Allen, C.D., T. Okada, and J. Cyster. 2007. Germinal-center organization and cellular dynamics. Immunity. 27:190–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacLennan, I.C., G. Johnson, Y. Liu, and J. Gordon. 1991. The heterogeneity of follicular reactions. Res. Immunol. 142:253–257. [DOI] [PubMed] [Google Scholar]
- 21.Kepler, T.B., and A. Perelson. 1993. Cyclic re-entry of germinal center B cells and the efficiency of affinity maturation. Immunol. Today. 14:412–415. [DOI] [PubMed] [Google Scholar]
- 22.Oprea, M., and A. Perelson. 1997. Somatic mutation leads to efficient affinity maturation when centrocytes recycle back to centroblasts. J. Immunol. 158:5155–5162. [PubMed] [Google Scholar]
- 23.Allen, C.D., K.M. Ansel, C. Low, R. Lesley, H. Tamamura, N. Fujii, and J.G. Cyster. 2004. Germinal center dark and light zone organization is mediated by CXCR4 and CXCR5. Nat. Immunol. 5:943–952. [DOI] [PubMed] [Google Scholar]
- 24.Meyer-Hermann, M.E., and P. Maini. 2005. Interpreting two-photon imaging data of lymphocyte motility. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 71:061912. [DOI] [PubMed] [Google Scholar]
- 25.Beltman, J.B., A.F. Mar'ee, J.N. Lynch, M.J. Miller, and R.J. de Boer. 2007. Lymph node topology dictates T cell migration behavior. J. Exp. Med. 204:771–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beauchemin, C., N.M. Dixit, and A.S. Perelson. 2007. Characterizing T cell movement within lymph nodes in the absence of antigen. J. Immunol. 178:5505–5512. [DOI] [PubMed] [Google Scholar]
- 27.Radmacher, M.D., G. Kelsoe, and T.B. Kepler. 1998. Predicted and inferred waiting-times for key mutations in the germinal center reaction - evidence for stochasticity in selection. Immunol. Cell Biol. 76:373–381. [DOI] [PubMed] [Google Scholar]
- 28.Kesmir, C., and R.J. de Boer. 1999. A Mathematical Model on Germinal Center Kinetics and Termination. J. Immunol. 163:2463–2469. [PubMed] [Google Scholar]
- 29.Meyer-Hermann, M., A. Deutsch, and M. Or-Guil. 2001. Recycling probability and dynamical properties of germinal center reactions. J. Theor. Biol. 210:265–285. [DOI] [PubMed] [Google Scholar]
- 30.Meyer-Hermann, M. 2002. A mathematical model for the germinal center morphology and affinity maturation. J. Theor. Biol. 216:273–300. [DOI] [PubMed] [Google Scholar]
- 31.Kesmir, C., and R.J. De Boer. 2003. A spatial model of germinal center reactions: cellular adhesion based sorting of B cells results in efficient affinity maturation. J. Theor. Biol. 222:9–22. [DOI] [PubMed] [Google Scholar]
- 32.Meyer-Hermann, M.E., P.K. Maini, and D. Iber. 2006. An analysis of B cell selection mechanisms in germinal centers. Math. Med. Biol. 23:255–277. [DOI] [PubMed] [Google Scholar]
- 33.Meyer-Hermann, M. 2007. A concerted action of B cell selection mechanisms. Adv. Complex Syst. 10:557–580. [Google Scholar]
- 34.Figge, M.T. 2005. Stochastic discrete event simulation of germinal center reactions. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 71:051907. [DOI] [PubMed] [Google Scholar]
- 35.Jacob, J., J. Przylepa, C. Miller, and G. Kelsoe. 1993. In situ studies of the primary response to (4-hydroxy-3-nitrophenyl)acetyl. III. The kinetics of V region mutation and selection in germinal center B cells. J. Exp. Med. 178:1293–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith, K.G., A. Light, G.J. Nossal, and D.M. Tarlinton. 1997. The extent of affinity maturation differs between the memory and antibody-forming cell compartments in the primary immune response. EMBO J. 16:2996–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pascual, V., Y.-J. Liu, A. Magalski, O. De Bouteiller, J. Banchereau, and J.D. Capra. 1994. Analysis of somatic mutation in five B cell subsets of human tonsil. J. Exp. Med. 180:329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.England, P., R. Nageotte, M. Renard, A.-L. Page, and H. Bedouelle. 1999. Functional characterization of the somatic hypermutation process leading to antibody D1.3, a high affinity antibody directed against lysozyme. J. Immunol. 162:2129–2136. [PubMed] [Google Scholar]
- 39.Han, S., K. Hathcock, B. Zheng, T. Kepler, R. Hades, and G. Kelsoe. 1995. Cellular interaction in germinal centers. J. Immunol. 155:556–567. [PubMed] [Google Scholar]
- 40.Batista, F.D., and M. Neuberger. 1998. Affinity dependence of the B cell response to antigen: a threshold, a ceiling and the importance of off-rate. Immunity. 8:751–759. [DOI] [PubMed] [Google Scholar]
- 41.Lindhout, E., A. Lakeman, and C. de Groot. 1995. Follicular dendritic cells inhibit apoptosis in human B lymphocytes by rapid and irreversible blockade of preexisting endonuclease. J. Exp. Med. 181:1985–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bouchaud, J.-P., and A. Georges. 1990. Anomalous diffusion in disordered media: Statistical mechanisms, models and physical applications. Phys. Rep. 195:127–293. [Google Scholar]
- 43.Moser, B., M. Wolf, A. Walz, and P. Loetscher. 2004. Chemokines: multiple levels of leukocyte migration control. Trends Immunol. 25:75–84. [DOI] [PubMed] [Google Scholar]
- 44.Flynn, S., K.-M. Toellner, C. Raykundalia, M. Goodall, and P. Lane. 1998. CD4 T cell cytokine differentiation: the B cell activation molecule, OX40 ligand, instructs CD4 T cells to express interleukin 4 and upregulates expression of the chemokine receptor, Blr-1. J. Exp. Med. 188:297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ansel, K.M., L.J. McHeyzer-Williams, V.N. Ngo, M.G. McHeyzer-Williams, and J.G. Cyster. 1999. In vivo-activated CD4 T cells upregulate CXC chemokine receptor 5 and reprogram their response to lymphoid chemokines. J. Exp. Med. 190:1123–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reif, K., E.H. Ekland, L. Ohl, H. Nakano, M. Lipp, R. Förster, and J.G. Cyster. 2002. Balanced responsiveness to chemoattractants from adjacent zones determines B-cell position. Nature. 416:94–99. [DOI] [PubMed] [Google Scholar]
- 47.Vicente-Manzanares, M., M.C. Montoya, M. Mellado, J.M.R. Frade, M.A. del Pozo, M. Nieto, M.O. de Landazuri, C. Martinez-A., and F. Sanchez-Madrid. 1998. The chemokine SDF-1α triggers a chemotactic response and induces cell polarization in human B lymphocytes. Eur. J. Immunol. 28:2197–2207. [DOI] [PubMed] [Google Scholar]
- 48.Poznansky, M.C., I.T. Olszak, R. Foxall, R.H. Evans, A.D. Luster, and D.T. Scadden. 2000. Active movement of T cells away from a chemokine. Nat. Med. 6:543–548. [DOI] [PubMed] [Google Scholar]
- 49.Kim, C.H., L.M. Pelus, J.R. White, E. Applebaum, K. Johanson, and H.E. Broxmeyer. 1998. CK beta-11/macrophage inflammatory protein-3 beta/EBI1-ligand chemokine is an efficacious chemoattractant for T and B cells. J. Immunol. 160:2418–2424. [PubMed] [Google Scholar]
- 50.Gunn, M.D., V.N. Ngo, K.M. Ansel, E.H. Ekland, J.G. Cyster, and L.T. Williams. 1998. A B-cell-homing chemokine made in lymphoid follicles activates Burkitt's lymphoma receptor-1. Nature. 391:799–803. [DOI] [PubMed] [Google Scholar]
- 51.Gerdes, J., U. Schwab, H. Lemke, and H. Stein. 1983. Production of a mouse monoclonal antibody reactive with a human nuclear antigen associated with cell proliferation. Int. J. Cancer. 31:13–20. [DOI] [PubMed] [Google Scholar]
- 52.Kosco-Vilbois, M.H., H. Zentgraf, J. Gerdesc, and J.-Y. Bonnefoy. 1997. To ‘B’ or not to ‘B’ a germinal center? Immunol. Today. 18:225–230. [DOI] [PubMed] [Google Scholar]
- 53.Voigt, I., S.A. Camacho, B.A. de Boer, M. Lipp, R. Förster, and C. Berek. 2000. CXCR5-deficient mice develop functional germinal centers in the splenic T cell zone. Eur. J. Immunol. 30:560–567. [DOI] [PubMed] [Google Scholar]
- 54.Casola, S., G. Cattoretti, N. Uyttersprot, S.B. Koralov, J. Seagal, Z. Hao, A. Waisman, A. Egert, D. Ghitza, and K. Rajewsky. 2006. Tracking germinal center B cells expressing germ-line immunoglobulin gamma1 transcripts by conditional gene targeting. Proc. Natl. Acad. Sci. USA. 103:7396–7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perelson, A.S., and G.F. Oster. 1979. Theoretical Studies of Clonal Selection: Minimal Antibody Repertoire Size and Reliability of Self-Non-self Discrimination. J. Theor. Biol. 81:645–670. [DOI] [PubMed] [Google Scholar]
- 56.Meyer-Hermann, M., and T. Beyer. 2004. The type of seeder cells determines the efficiency of germinal center reactions. Bull. Math. Biol. 66:125–141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.