SUMMARY
The mechanism of translation in eubacteria and organelles is thought to be similar. In eubacteria, the three initiation factors IF1, IF2, and IF3 are vital. Although the homologs of IF2 and IF3 are found in mammalian mitochondria, an IF1 homolog has never been detected. Here, we show that bovine mitochondrial IF2 (IF2mt) complements E. coli containing a deletion of the IF2 gene (E. coli ΔinfB). We find that IF1 is no longer essential in an IF2mt-supported E. coli ΔinfB strain. Furthermore, biochemical and molecular modeling data show that a conserved insertion of 37 amino acids in the IF2mt substitutes for the function of IF1. Deletion of this insertion from IF2mt supports E. coli for the essential function of IF2. However, in this background, IF1 remains essential. These observations provide strong evidence that a single factor (IF2mt) in mammalian mitochondria performs the functions of two eubacterial factors, IF1 and IF2.
INTRODUCTION
In eubacteria, the process of translation involves three initiation factors, IF1, IF2, and IF3 (reviewed by Gualerzi and Pon, 1990). Interestingly, despite the overall divergent evolution of initiation factors in the three kingdoms of life, the eubacterial IF1 and IF2 have functional and structural homologs in eukaryotes as well as in Archaea (Choi et al., 1998; Lee et al., 1999). IF1 and IF2 are, therefore, referred to as “universal translation initiation factors” (Kyrpides and Woese, 1998; Roll-Mecak et al., 2001). In eubacteria, both IF1 (Cummings and Hershey, 1994) and IF2 (Laalami et al., 1991) are essential for cell viability and, in concert with IF3, serve to enhance the kinetics and fidelity of initiation complex formation (reviewed by Gualerzi and Pon, 1990; Boelens and Gualerzi, 2002; Laursen et al., 2005).
IF1, a protein of 71 amino acids in E. coli, is the smallest of the three eubacterial initiation factors. It is encoded by an essential gene (infA). IF1 is known to stabilize the binding of the initiator tRNA to the small (30S) ribosomal subunit in the presence of IF2. It also destabilizes this binding in the presence of IF3. In the presence of IF3, IF2, and initiator tRNA on the 30S subunit, IF1 reduces the rate of the large (50S) ribosomal subunit docking (Antoun et al., 2006a). These activities have been shown to enhance the accuracy of initiator tRNA selection on the ribosome (Antoun et al., 2006b) and may account for much of the importance of IF1 in eubacteria. It has been suggested that the binding for IF1 overlaps with the A site of the 30S subunit and that this factor might function to block the A site, making it unavailable for tRNA binding during initiation. This notion has been corroborated by the solution of crystal structure of the 30S subunit in complex with IF1 (Carter et al., 2001).
IF2 is the central player in prokaryotic translational initiation that promotes the binding of fMet-tRNAfMet to the P site of the 30S ribosomal subunit (Gualerzi and Pon, 1990). It is a ribosome-dependent GTPase, and in its GTP-bound form, it facilitates the joining of the two ribosomal subunits. GTP hydrolysis triggers the release of IF2 from the 70S initiation complex (Antoun et al., 2003). Recent cryo-EM reconstructions of 70S initiation complexes containing E. coli or Thermus thermophilus IF2 (Allen et al., 2005; Myasnikov et al., 2005) indicate that there are contacts between IF1 and IF2 on the ribosome.
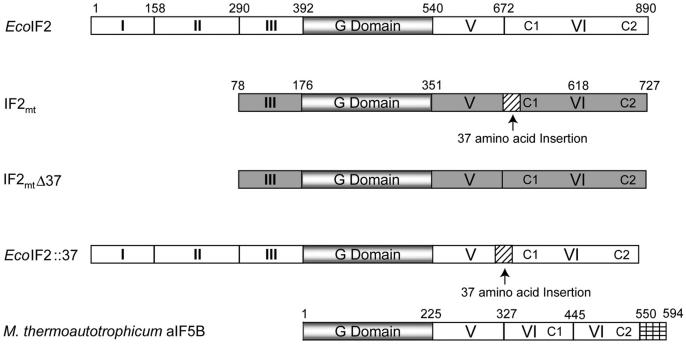
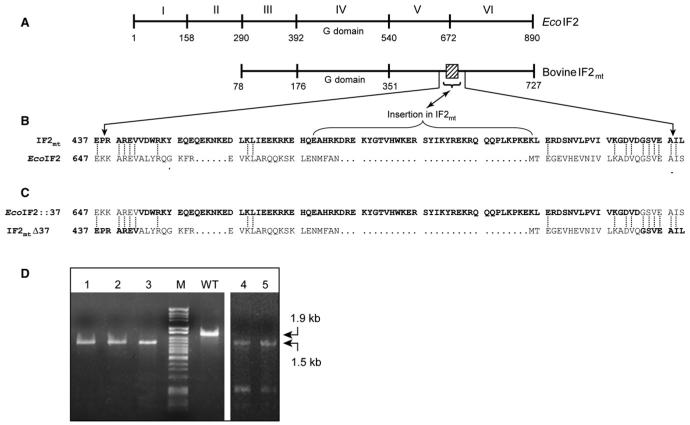
Mitochondria are believed to have arisen from the ancestors of present-day α-proteobacteria (reviewed by Gray et al., 1999), and the mitochondrial protein synthetic machinery is generally believed to resemble the eubacterial one (reviewed by Pel and Grivell, 1994). A factor equivalent to eubacterial IF2 has been characterized from mitochondria of mammals (IF2mt)(Liao and Spremulli, 1990; Ma and Spremulli, 1995) and yeast (Vambutas et al., 1991; Garofalo et al., 2003; Tibbetts et al., 2003). Using the six domain nomenclature for IF2 (Laursen et al., 2005), the alignment of IF2mt with E. coli IF2 indicates that it consists of domains III-VI (Figure 1). A peculiar feature of IF2mt is the presence of a universal translation initiation factors conserved 37 amino acid insertion between domains V and VI (Figure 1) (Spencer and Spremulli, 2005). The crystal structures of aIF5B, the archaeal IF2 homolog, in the GTP- and GDP-bound states have been determined and indicate that this factor forms a chalice-like structure encompassing domains analogous to domains IV-VI (Roll-Mecak et al., 2000). The comparison of a structural model of IF2mt with the crystal structure of aIF5B shows that the conserved insertion in IF2mt forms an α helix that protrudes out of the main body of the molecule. Mutations in this region decrease the affinity of IF2mt for mitochondrial ribosomes (Spencer and Spremulli, 2005).
Figure 1. Domain Structure of E. coli IF2, EcoIF2; Bovine Mitochondrial IF2, IF2mt; Methanobacterium thermoautotrophicum IF5B, aIF5B; and the Chimeric Constructs.
The domains are numbered according to Laursen et al. (2005). Domains of EcoIF2 are shown in white, while those of IF2mt are gray. The conserved G domain is shaded, and the insertion found in IF2mt is stripped with diagonal black lines. The short C-terminal extension observed in aIF5B is indicated by cross bars. Note that for the sake of comparison, the domains of aIF5B (Roll-Mecak et al., 2000) have also been numbered according to Laursen et al. (2005).
True to their universal nature, homologs of IF1 have been found in the organellar translational system of chloroplasts (Koc and Spremulli, 2002). However, an intriguing situation exists in mitochondria, where no IF1 homolog has been detected (Koc and Spremulli, 2002; Towpik, 2005). This observation indicates that there is a fundamental difference between translation initiation in mitochondria and in eubacteria. Interestingly, mammalian mitochondrial ribosomes have a protein to RNA ratio opposite to that found in eubacterial ribosomes (Sharma et al., 2003; reviewed by Spremulli et al., 2004). Thus, translation in mitochondria seems to fall into its own genre with similarities to as well as differences from both the eukaryotic and eubacterial systems.
The investigation of translation in mammalian mitochondria has remained cumbersome due to technical demands posed by in vitro reconstitution of the mitochondrial translational machinery (Dekker et al., 1993; Spremulli et al., 2004). In an attempt to overcome this problem, we have initiated development of a system wherein the mitochondrial initiation factor(s) can be studied in E. coli. Here we report that, in E. coli, the bovine IF2mt substitutes for the essential biological roles of both IF1 and IF2. Our observations provide an unprecedented example of a single translation initiation factor (IF2mt) performing the functions of the two essential initiation factors in E. coli.
RESULTS
Bovine IF2mt Supports the Growth of E. coli ΔIF2
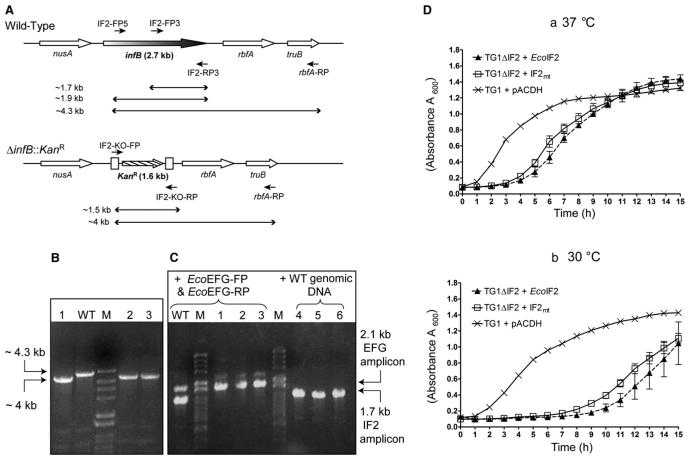
To investigate whether IF2mt can function in E. coli, we first generated a derivative of E. coli TG1 wherein infB, the gene encoding IF2, was replaced with a gene coding for kanamycin resistance (ΔinfB::KanR, Figure 2A) in the presence of a plasmid-borne copy of the EcoIF2 open reading frame cloned downstream of lacP (pACDH-EcoIF2) as support. The gene replacement relied on recombination between short regions of homology at the ends of a linear DNA fragment harboring an antibiotic resistance (KanR) marker and the flanking sequences of the chromosomal infB gene in E. coli. This linear DNA was introduced into cells by electroporation and the recombinants (knockouts) wherein the infB gene was replaced by KanR were selected on kanamycin. To enhance the efficiency of such a genetic exchange, the λ Red recombinase proteins were also expressed in the host from a plasmid, pKD46 (AmpR, temperature-sensitive origin of replication; Datsenko and Wanner, 2000).
Figure 2. Generation of E. coli ΔIF2 Strain.
(A) The organization of the nusA-infB genes. Lines with a single arrowhead indicate the location of the primers used, and lines with double arrowheads represent the expected length of amplicons with different primer combinations on wild-type or ΔinfB::KanR templates. The boxes that flank the KanR marker at the ΔinfB::KanR locus indicate the FRT sites used to excise KanR marker from the locus (see the Supplemental Data).
(B) Verification of replacement of infB with KanR was done by PCR with IF2-KO-FP, IF2-FP5, and rbfA-RP. The IF2-FP5 would anneal only to the wild-type template, while the IF2-KO-FP would anneal only to the ΔinfB::KanR template. Lanes include the following: WT, wild-type; M, the DNA size markers (λ HindII + HindIII); and numbers, three transductants (ΔIF2 + pACDH-IF2mt).
(C) PCR with primers IF2-FP3 and IF2-RP3 to check for ectopic insertion of IF2 gene in the genome of transductants (ΔIF2 + pACDH-IF2mt). As indicated by the bracket on the lanes, for one set of PCR reactions, primers EFG-FP and EFG-RP were used to amplify an unlinked gene fusA (codes for EFG) as a control. For the second set, wild-type DNA was added to the DNA from transductants. The template used in lanes 1-3 is the same as in (B); and the templates in lanes 4-6 are the same as in 1-3 except that they were spiked with the wild-type genomic DNA.
(D) Shown are growth curves of E. coli ΔIF2 strains complemented with EcoIF2 or IF2mt. (Da) E. coli and E. coli ΔIF2 strains supported by different IF2 constructs were grown at 37°C, and (Db) the same strains at 30°C. Samples of all knockout strains were taken in triplicates. The lines join the mean, and the vertical bars denote the standard deviation.
Subsequent to generation of the desired E. coli TG1 ΔinfB::KanR strain (for more details, see the Supplemental Data available online), we used the classical method of P1 phage-mediated transduction for delivering the infB::KanR locus from this strain (the donor strain) into E. coli TG1 harboring either vector alone or the constructs expressing various initiation factors (recipient strains) to effect genetic exchange of the wild-type infB locus. As expected of an essential gene, transductants (KanR) were obtained when the recipient strain harbored a plasmid borne copy of EcoIF2, but not when it harbored an empty vector. Interestingly, transductants were also obtained when the recipient strain harbored pACDH-IF2mt, which encodes bovine mitochondrial IF2. The replacement of infB by infB::KanR (Figure 2A) in these transductants was verified by diagnostic PCR. As shown in Figure 2B (lanes 1-3), the genomic DNA from the transductants yields an amplicon of 4 kb, the size expected in the event of replacement of infB by KanR. Further, to rule out the presence of infB being ectopically inserted in the transductant genome, PCR was carried out using primers to amplify a 1.7 kb internal fragment of infB. As shown in Figure 2C, no amplification of the 1.7 kb infB fragment (lanes 1-3) was observed from the transductant genomic DNA. However, PCR amplification of the expected fragment (1.7 kb) was seen when chromosomal DNA from the wild-type strain was used (lane WT). Primers for a physically unlinked gene, fusA (encoding EFG), were also included in these reactions as a control for quality of DNA and PCR conditions. Amplification of fusA occurred from both the wild-type and the transductant genomic DNAs (lanes WT, 1-3). Further, in a PCR with infB internal primers, the addition of wild-type genomic DNA allowed amplification of the infB fragment in the presence of transductant DNA (lanes 4-6), precluding the possibility of inhibitors of infB amplification in the transductant genomic DNA. Taken together, these analyses confirmed the absence of infB gene in the transductants.
The transductants produced in this experiment were, thus, knockouts of E. coli IF2 surviving solely on IF2mt. The generation of these transductants clearly demonstrates that IF2mt can complement E. coli TG1 ΔinfB::KanR. For further analyses, the KanR marker from this strain was excised (see the Supplemental Data) to generate E. coli TG1 ΔinfB (also referred to as E. coli ΔIF2) strain. As shown in Figure 2D, comparisons of the growth of various E. coli knockout strains supported by eubacterial or mitochondrial IF2s showed that, at 37°C as well as 30°C, pACDH-IF2mt is able to support the growth of E. coli ΔIF2 till saturation. The growth of this strain is similar to that of E. coli ΔIF2 supported by pACDH-EcoIF2. It is clear that E. coli ΔIF2 strains supported by either pACDH-EcoIF2 or pACDH-IF2mt experience a longer lag phase compared to the parent strain, E. coli TG1 + pACDH. However, it is also important to note that the mitochondrial factor clearly substitutes for the bacterial IF2 in vivo.
Bovine IF2mt Can Support the Growth of E. coli ΔIF1ΔIF2
The absence of IF1 in mitochondria raises a possibility that IF2mt might not require an IF1-like factor and, consequently, in the presence of IF2mt, IF1 may no longer be essential in E. coli. In order to examine this possibility, we used the method of linear DNA transformation (Datsenko and Wanner, 2000) to generate a knockout of IF1 in E. coli DY330 (this strain harbors the λ Red recombinase genes on its chromosome, Lee et al. [2001])by replacing infA, the gene for IF1, with a ChlR gene (Figure 3A). Survival of this strain (E. coli DY330 ΔinfA::ChlR + pTrc-MtuIF1; also referred to as E. coli DY330ΔIF1) is supported by the presence of pTrc-MtuIF1, which expresses IF1 from another eubacterial species, Mycobacterium tuberculosis. This strain was used as a donor to raise P1 phage lysate, and the transductions were performed to deliver infA::ChlR locus from it to E. coli TG1 recipients to generate knockouts of infA. Expectedly, we obtained transductants (ΔinfA::ChlR, also referred to as ΔIF1) in cases when the recipient harbored either pTrc-EcoIF1 or pTrc-MtuIF1, but not when it harbored pACDH-EcoIF2 or pACDH-IF2mt. At first glance, this observation suggests that the presence of IF2mt does not complement for the function of IF1. However, these strains are wild-type for IF2 (infB), and competition may occur between EcoIF2 and IF2mt for binding to ribosomes. Thus, transduction experiments were also done with E. coli ΔIF2 complemented by either pACDH-EcoIF2 or pACDH-IF2mt as recipients. Again, not unexpectedly, no transductants were obtained when the recipient was supported by pACDH-EcoIF2. Interestingly, however, transductants were obtained when the recipient E. coli ΔIF2 strain was supported by pACDH-IF2mt. This observation indicates that E. coli can survive in the absence of IF1 when IF2mt is present, suggesting that the mitochondrial factor plays the role of both IF2 and IF1 in initiation.
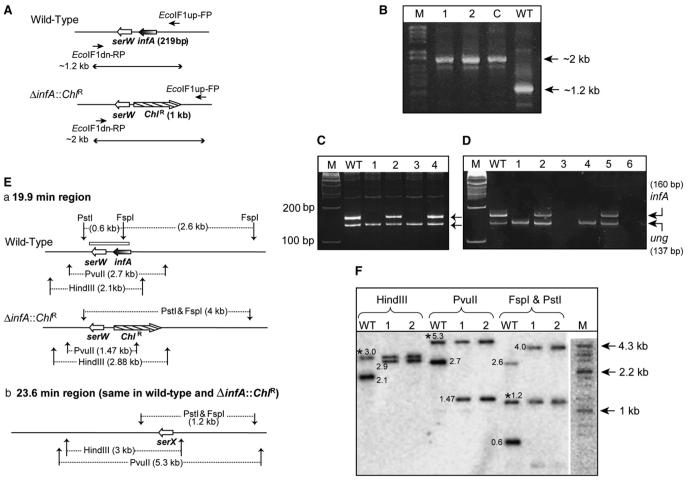
Figure 3. Generation and Analysis of E. coli ΔIF1 Strains.
(A) The organization of the infA locus. Lines with a single arrowhead indicate the location of primers (EcoIF1up-FP and EcoIF1 dn-RP) used, and lines with double arrowheads represent the expected length of amplicons with wild-type or ΔinfA::ChlR templates.
(B) Products of PCR with EcoIF1up-FP and EcoIF1 dn-RP; in lane WT, the template was from wild-type; in lane C from E. coli DY330 ΔIF1 + pTrc-MtuIF1; and in 1 and 2 from transductants obtained using E. coli TG1 ΔIF2 recipients harboring pACDH-IF2mt.
(C) PCR amplification of an internal fragment of infA open reading frame using IF1-RT-FP and IF1-RT-RP, and ung with EcoungSeq-FP and Ecoung-RT-RP. Lanes include the following: WT, wild-type; 1 and 3, ΔIF1ΔIF2; 2 and 4, ΔIF1ΔIF2 strains spiked with WT DNA. M denotes a 100 bp marker.
(D) RT-PCR to amplify an internal fragment of infA mRNA using IF1-RT-FP and IF1-RT-RP; and ung mRNA using EcoungSeq-FP and Ecoung-RT-RP. Lanes include the following: WT, wild-type; 1 and 4, ΔIF1ΔIF2 strains; 2 and 5, ΔIF1ΔIF2 spiked with RNA preparations from WT. Lanes 3 and 6 are reactions containing RNA from ΔIF1ΔIF2 strains without the reverse transcriptase. M denotes a 100 bp marker.
(E) (Ea) The 19.9 min region showing the infA locus and expected fragment lengths from digestion with various restriction enzymes. The box above infA/serW locus indicates the region corresponding to the probe used for Southern analysis. (Eb) The 23.6 min region of the E. coli genome containing serX, which remains unchanged in both strains but which was detected due to its identity to serW.
(F) Electronic autoradiography of the Southern blot using BioImager FLA5100 (Fuji Film, Japan). WT denotes the lane with wild-type genomic DNA, and 1 and 2 indicate lanes with genomic DNA of ΔinfA::ChlR ΔinfB (ΔIF1ΔIF2) transductants. M denotes the DNA size markers (λ HindII + HindIII). Bands common to the wild-type and the transductants due to hybridization to serX are indicated by an asterisk. Numbers next to the bands indicate their respective sizes in kilobases.
To confirm that the infA gene had indeed been replaced by the ChlR gene (Figure 3A), the transductants were analyzed by PCR using primers flanking infA. This analysis resulted in a 2 kb amplicon (Figure 3B, lanes 1 and 2) in comparison to a 1.2 kb amplicon from the wild-type strain (lane WT) indicating the replacement of infA with the larger ChlR gene. Thus, these transductants were knockouts of both infA and infB supported by IF2mt (E. coli TG1 ΔinfA::ChlR ΔinfB + pACDH-IF2mt; also referred to as E. coli ΔIF1ΔIF2 + pACDH-IF2mt).
To rule out the ectopic insertion of the small (219 bp) infA ORF in the genome of the ΔIF1ΔIF2 strain, PCR with internal primers to amplify a 160 bp region of the infA ORF was also carried out. As shown in Figure 3C, the DNA templates from the wild-type strain (lane WT) resulted in the amplification of the expected 160 bp fragment. Amplification of this fragment was not seen from the genomic DNA of the knockout strains (lanes 1 and 3; for the ΔIF1ΔIF2 strains) but observed upon spiking of the knockout strain DNA with the wild-type strain DNA (lanes 2 and 4). As an additional control for the DNA quality, primers for amplification of a 137 bp fragment of ung (encoding uracil DNA glycosylase; Varshney et al. [1988]) were included in all the reactions, and the expected size amplicon was visible in all the lanes, confirming that the absence of infA in lanes 1 and 3 is a genuine property of the knockout DNA. Furthermore, reverse transcriptase-PCR using DNA-free RNA from all the strains was done to show the absence of the infA mRNA (Figure 3D). While the expected 160 bp fragment (infA) is seen with the RNA prepared from the wild-type strain (lane WT), it is absent in the reactions carried out with the RNA from ΔIF1ΔIF2 strains (lane 1 and 4). Expectedly, spiking of the RNA from ΔIF1ΔIF2 with the RNA from the wild-type strain led to amplification of the 160 bp infA fragment (lanes 2 and 5). As controls, while the band corresponding to ung was seen in all five lanes (WT, 1, 2, 4, and 5), no amplifications were seen in the reactions carried out in the absence of the reverse transcriptase (lanes 3 and 6).
To unequivocally establish the absence of infA in these transductants, a Southern analysis was carried out. As shown in Figure 3E, the probe used in this analysis also included a region homologous to serW, a tRNASer gene downstream of infA. Three enzymes were used for the analysis: FspI, which has a site in infA but not in ChlR; PvuII with a site in ChlR but not in infA; and HindIII, which cuts neither of the genes. As observed in Figure 3F, the wild-type DNA shows bands of 2.1 kb (HindIII) and 2.7 kb (PvuII). Double digestion of the wild-type DNA with FspI and PstI gives bands at 0.6 and 2.6 kb. When the genomic DNA from the transductants was used, a band of 2.9 kb was detected in the HindIII digest; a new band at 1.47 kb was observed in the PvuII digest, and the FspI/PstI double digest gave a band of just about 4 kb. This new digestion pattern with the transductant genomic DNA was the pattern expected from a replacement of infA by ChlR (Figure 3Ea). Interestingly, there is another copy of tRNASer gene, serX, at the 23.6 min region (Figure 3Eb), which accounts for the extra bands of common sizes (∼3.0, 5.3, and 1.2 kb, marked by an asterisk) in both the transductant and wild-type lanes, in all three digests (Figure 3F). This band serves as an internal control for the Southern analysis. Taken together, the PCR and the Southern analyses confirm the absence of infA gene in the transductants, demonstrating that infA is dispensable in an E. coli ΔIF2 + pACDH-IF2mt strain. This observation again emphasizes that IF2mt can replace both IF1 and IF2 in E. coli.
IF2mt Supports Efficient Log Phase Growth of E. coli ΔIF1ΔIF2
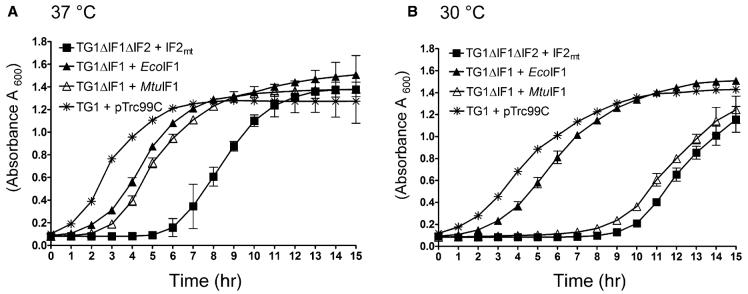
To assess how effectively IF2mt replaces E. coli IF1 and IF2 in the double knockout (E. coli ΔIF1ΔIF2 + pACDH-IF2mt), growth of the strains was monitored at both 37°C and 30°C. This analysis (Figure 4) indicated that, following a lag period, the growth of E. coli ΔIF1ΔIF2 supported by pACDH-IF2mt was quite comparable to that of the starting strain. The long lag phase seems to be a general property of the ΔIF2 strain, as the ΔIF2 strain supported by EcoIF2 also exhibits a similar response (Figure 2D). The ΔIF1 strain complemented with plasmids carrying either E. coli IF1 or M. tuberculosis IF1 also grew at a comparable rate. Interestingly, the ΔIF1 strain expressing EcoIF1 does not experience a distinct increase in lag phase at 30°C (Figure 4B), unlike the ΔIF1 strain supported by MtuIF1 or the ΔIF1ΔIF2 strain supported by IF2mt.
Figure 4. Analysis of the Growth of Various Strains.
(A) E. coli, E. coli ΔIF1, and E. coli ΔIF1ΔIF2 strains supported by different IF1 or IF2 constructs, grown at 37°C. (B) is the same as (A) except that the growth temperature was 30°C. Samples of all knockout strains were taken in triplicates. The lines join the mean, and the vertical bars denote the standard deviation.
IF2mt Lacking the Conserved 37 Amino Acid Insertion Supports E. coli ΔIF2, but Not E. coli ΔIF1ΔIF2
An alignment of the relevant portion of IF2mt with EcoIF2 (Figure 5B) shows that, though the 37 amino acid insertion is absent from EcoIF2, the portions flanking the insertion are conserved between IF2mt and EcoIF2. To investigate the possibility that the insertion in IF2mt mimics the role of IF1, we generated chimeras wherein the 37 amino acid stretch from IF2mt was swapped with the corresponding region from EcoIF2. The chimeras were made such that the swapped portion began and ended in the conserved sequences flanking the 37 amino acid insertion (Figure 5C). Such a design to “ecolinize” IF2mt (IF2mtΔ37) was adopted to minimize structural disturbances in the chimeric proteins. Similarly, the EcoIF2::37 was designed such that the 37 amino acid insertion (from IF2mt) was within the context of the same conserved flanks.
Figure 5. Chimeras of EcoIF2 and IF2mt.
(A) Schematic representation of the domain organization of EcoIF2 and bovine IF2mt. The numbers indicate the amino acid positions. IF2mt lacks the counterparts of EcoIF2 domains I and II. The 37 amino acid insertion conserved in mitochondrial IF2s is indicated by a box.
(B) Alignment of a portion of the sequences of EcoIF2 and bovine IF2mt (shown in bold letters). The identical residues are joined by a dotted line. The 37 amino acid insertion is marked by a bracket and is flanked by regions of high conservation, which were chosen for a swap between the two IF2s.
(C) The sequences of the resulting chimeras are shown.
(D) PCR analysis of ΔinfB::KanR transductants harboring either pACDH-IF2mtΔ37 (lanes 1-3) or pACDH-EcoIF2::37 (lanes 4 and 5) using IF2-FP5, IF2-RP3, IF2-KO-FP, and IF2-KO-RP (Figure 2A) to confirm replacement of infB with KanR. WT stands for wild-type, and M is DNA size marker (λ HindII + HindIII).
In order to assess the ability of the chimeras to replace EcoIF2 and permit cell growth, P1 phage-mediated transductions were carried out to transfer the ΔinfB::KanR locus from E. coli ΔinfB::KanR + pACDH-EcoIF2 (donor) to the recipient strains of E. coli TG1 harboring either pACDH-IF2mtΔ37 or pACDH-EcoIF2::37. Transductants were obtained in both the cases, and the replacement of infB with KanR was confirmed in transductants harboring either pACDH-IF2mtΔ37 (Figures 2A and 5D, lanes 1-3) or pACDH-EcoIF2::37 (Figure 5D, lanes 4 and 5) by PCR. This result indicates that both the chimeras can function as IF2 in vivo.
We then performed a second set of transductions using P1 phage raised on E. coli DY330ΔIF1 to check if infA can be replaced with ChlR in E. coli ΔIF2 harboring either pACDH-IF2mtΔ37 or pACDH-EcoIF2::37. In both cases, transductants were not obtained, suggesting that neither of the chimeras is able to support a double knockout of IF1 and IF2.
In Vitro Analysis of IF2mt Activity on E. coli 70S Ribosomes
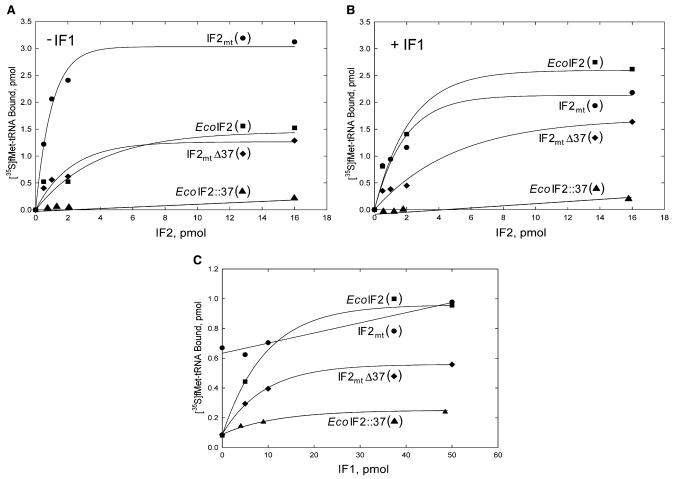
The activities of EcoIF2, IF2mt, and the chimeric IF2s (IF2mtΔ37 and EcoIF2::37) were examined by assaying the activities of these factors in promoting the binding of fMet-tRNAfMet to E. coli 70S ribosomes in the presence and absence of E. coli IF1. As indicated in Figure 6A, EcoIF2 and IF2mtΔ37 show modest activity in the absence of IF1. In contrast, IF2mt is quite active when assayed in the absence of IF1. In the presence of IF1 (Figure 6B), the activity of EcoIF2 is quite comparable to that of IF2mt. The activity of IF2mtΔ37 appears to be slightly improved by the presence of IF1.
Figure 6. The Abilities of IF2 Variants to Facilitate Binding of [35S]fMet-tRNAfMet to E. coli 70S Ribosomes.
(A and B) (A) The ability of different IF2 variants to facilitate the binding of [35S]fMet-tRNAfMet to E. coli 70S ribosomes was measured in absence of E. coli IF1 or (B) in the presence of E. coli IF1 (0.5 μM).
(C) The effect of a varying dosage of IF1 on the activities of various IF2s. Reaction mixtures contained constant amounts of the various IF2s (0.02 μM for IF2mt, EcoIF2, or IF2mtΔ37; and 0.06 μM for EcoIF2::37) and the indicated amounts of IF1.
To assess the effect of IF1 on the activities of the IF2 variants more directly, constant amounts of the various IF2s were assayed in the absence of IF1 and in the presence of increasing concentrations of IF1. As is evident from data in Figure 6C, IF2mt is quite active in the absence of IF1, and the activity of this factor on E. coli ribosomes is not stimulated significantly upon the addition of IF1. In contrast, E. coli IF2 has much lower activity than IF2mt in the absence of IF1 but is stimulated nearly 10-fold by the presence of IF1. Interestingly, IF2mtΔ37 shows significantly lower activity than IF2mt and is stimulated about 5-fold by the addition of IF1. This observation suggests that the insertion in IF2mt permits this factor to function effectively in the absence of IF1 and that its deletion results in an IF2 derivative (IF2mtΔ37) whose activity is more dependent on the addition of IF1. Such a dependency agrees with the idea that IF2mtΔ37 can no longer support an infA deletion, a property also evident in the transduction experiments (see previous section). Addition of the insertion region to EcoIF2 (EcoIF2::37) leads to a factor with very low activity (Figure 6). It shows little stimulation by IF1, but the very anemic activity of this construct makes a clear interpretation of this particular result difficult. It remains to be seen if further improvements in the design of such a construct, e.g., wherein the 37 amino acid insertion may be placed in an appropriate structural context, may allow evolution of a gain-of-function chimera of EcoIF2.
DISCUSSION
The translation initiation factors IF1 and IF2 (or their homologs) are conserved across all the three kingdoms of life (Kyrpides and Woese, 1998; Harris et al., 2003). Mitochondria from animals have homologs of IF2 and IF3 (Koc and Spremulli, 2002). However, these organelles seem to be an exception in that neither biochemical nor bioinformatics approaches have been able to detect a mitochondrial homolog of IF1. In this report, we have used a system of E. coli knockouts of the infA and infB genes to show that IF2mt can play the role of both IF1 and IF2 in E. coli.
The bovine IF2mt has significant homology to EcoIF2 in the G domain (67%, Ma et al. [1995]) but lacks the two N-terminal domains found in EcoIF2 (Figure 1). The fact that IF2mt still supports the E. coli ΔIF2 is consistent with earlier observations that the first three domains of EcoIF2 are dispensable for the growt of E. coli (Laalami et al., 1991). IF2mt was also able to complement an E. coli ΔIF1ΔIF2 strain, suggesting that IF2mt possesses both IF1- and IF2-like activities. Examination of the growth of various knockout strains indicates that deletion of infA or infB or both causes the strain to spend a longer time in lag phase compared to the parent strain. Such changes in growth of these strains are expected as levels of the initiation factors are precisely regulated in the cell (Hershey, 1987), and this regulation would be lost in the knockout strains, since these genes are neither under their natural promoters nor in their genomic context. Mutations in IF1 (Croitoru et al., 2004) and IF2 (Laursen et al., 2003) are known to cause cold sensitivity, and all three initiation factors participate in the cold shock response (Giuliodori et al., 2004). This explains the additional slowdown experienced by the knockout strains at 30°C compared to the control strain (Figures 2D and 4). Discounting the long lag phases in the growth of strains supported by IF2mt as a background property of an infB or infA deletion, the efficient log phase growth exhibited by IF2mt-supported strains (ΔIF2 as well as ΔIF1ΔIF2) suggests that translation initiation by IF2mt is efficient enough to meet the requirement of an actively dividing cell, both with and without IF1.
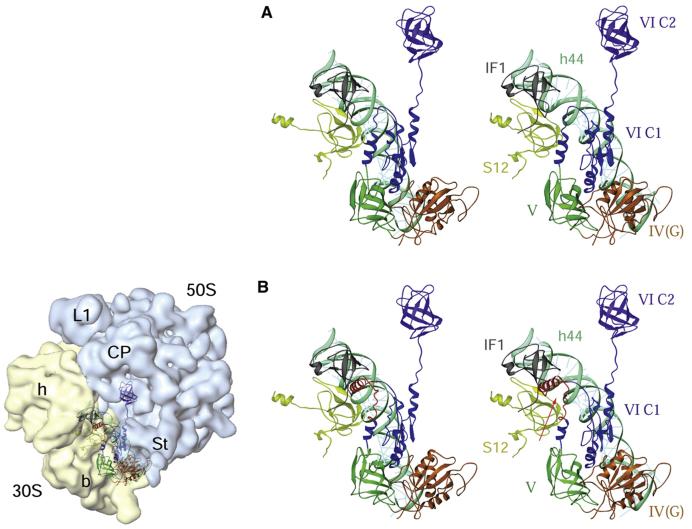
Our in vivo and in vitro data with IF2mt and EcoIF2 favor the assignment of an IF1-like function to the conserved 37 amino acid insertion in IF2mt. Also, as shown by the homology modeling of IF2mt (Figure 7; for details, see the Supplemental Data), this insertion forms a helix that projects out from the main body of IF2mt and could be responsible for its IF1-like function. Further, the model of the binding of IF2mt in the E. coli 70S initiation complex suggests that the 37 amino acid insertion reaches into the edge of the space otherwise occupied by IF1 on the ribosome. The helix formed by the conserved insertion could move relative to the main body of IF2mt. Movements in this insertion could be induced by the extensive conformational changes known to occur in IF2 upon GTP binding/hydrolysis (Roll-Mecak et al., 2000; Myasnikov et al., 2005). It has been suggested that IF1 may function by binding to and stabilizing helix 44 of the 16S rRNA during subunit association/dissociation (Carter et al., 2001). IF2 is also known to contact helix 44 (Myasnikov et al., 2005). Thus, the insertion in IF2mt could, in principle, achieve an IF1-like effect by virtue of its reach into the IF1 binding site and its proximity to helix 44. The insertion, therefore, acts like an IF1 integrated into the body of IF2mt, facilitating the binding of IF2mt onto the ribosome (Spencer and Spremulli, 2005) and stimulating initiation complex formation. In such an arrangement, both the IF1 and the IF2 equivalent are bound to leave the 70S initiation complex together, allowing us to propose that, in eubacteria, IF1 and IF2 might also dissociate concomitantly from the 70S ribosomes in vivo.
Figure 7. Interactions of Bacterial and Mammalian Mitochondrial IF2s with the E. coli 70S Ribosome.
(A) Stereo view presentation of the relative positions of the bacterial IF1 and IF2 on the E. coli 70S ribosome. The position of IF2 (identified by domains IV-VI, each depicted in different colors) was derived by docking the homology model of the factor into the cryo-EM map of an initiation complex (Allen et al., 2005), whereas the position of IF1 (gray) was obtained by docking the X-ray crystallographic structure of a 30S subunit•IF1 complex (Carter et al., 2001) into the cryo-EM map. Two 30S subunit components, protein S12 (greenish yellow) and 16S rRNA helix 44 (cyan), are also shown.
(B) A homology model of bovine IF2mt, aligned to the matching position of IF2 in (A), shows that the 37 amino acid insertion domain (red, indicated by an arrow) in the IF2mt overlaps partially with the position of IF1 in the bacterial ribosome, precluding the simultaneous binding of the two factors to the ribosome. The panel on the left of (B) depicts the orientation of the 70S ribosome, with 30S (semitransparent yellow) and 50S (semitransparent blue) ribosomal subunits and their prominent structural features identified: h and b mark the head and body, respectively, of the 30 subunit, while L1, CP, and St mark the protein L1, central protuberance, and protein L7/L12 stalk regions, respectively, of the 50S subunit (for details, refer to the Supplemental Data). Domains of IF2 are numbered according to Spencer and Spremulli (2005).
An IF1-like activity fused into an IF2 molecule is not an entirely unexpected scenario. In fact, it agrees quite well with the reports of the interaction between IF1 and IF2. Radiolabeled IF1 or IF2 have been shown to become crosslinked to each other in the presence of 30S subunits (Boileau et al., 1983), forming a structure that may mimic an EFG-like molecule (Brock et al., 1998). Direct physical interaction between the eukaryotic homologs of IF1 and IF2 (eIF1A and eIF5B) has also been reported using coimmnoprecipitation, yeast two-hybrid assay, in vitro studies, and NMR (Choi et al., 2000; Marintchev et al., 2003; Olsen et al., 2003; reviewed by Laursen et al., 2005; Londei, 2005). The IF2 density in the recent cryo-EM structure of the E. coli 70S initiation complex was observed to contact four N-terminal amino acids of IF1 (Allen and Frank, 2007). Functionally, IF1 and IF2 display a synergistic effect on initiation complex formation (Antoun et al., 2006a, 2006b) and peptidyl-tRNA drop-off (Karimi et al., 1998). IF1 and IF2, thus, form two parts of an evolutionarily conserved unit that, in the case of IF2mt, have been fused into a single protein. Interestingly, such a scenario suggests that the essential nature of IF1 is most likely for the functions that it carries out with IF2 rather than the ones it carries out with IF3 (such as ribosomal subunit dissociation).
The fusion of IF1 and IF2 activities in IF2mt would provide certain advantages to the host cell in terms of reducing the energy costs for importing two separate factors from the cytoplasm into the mitochondria and may prove to be more efficient, as the chances of a single factor encountering a small ribosomal subunit and initiating translation are much higher than those of the collision of two factors with the small ribosomal subunit. This would also allow the mitochondrion to carry out protein synthesis with a lower concentration of IF2mt compared to the levels of IF1/IF2 in other systems. A similar system in cytoplasmic translation would not be desirable, as it would be accompanied with the penalty of decreased possibilities for development of intricate regulatory mechanisms associated with independent initiation factors. Apparently, enhanced opportunities of translational regulation are one reason for higher organisms to have evolved a larger set of initiation factors.
The 37 amino acid insertion doesn't have homology to any other protein in the National Center for Biotechnology Information (NCBI) database. The presence of the amino acid insertion in IF2mt is conserved from yeast to humans (Spencer and Spremulli, 2005), although it is somewhat shorter in the lower eukaryotes. The conservation of the insertion in mitochondrial IF2s is an indication that it was present before the ancestors of eukaryotes diverged into various extant branches. The mitochondrial ribosome has a much higher rate of evolution than cytoplasmic ribosomes, and mitochondrial ribosomal proteins and rRNAs evolve at rates 13 and 23 times higher, respectively, than their cytoplasmic counterparts (Pietromonaco et al., 1986; reviewed by O'Brien, 2002). During the course of its “fast” evolution, the mitochondrial ribosome has lost several of the RNA stem structures present in eubacterial ribosomes. To fill in the gaps left by loss of the RNA moiety, the mitochondrion has adopted a dual strategy of recruiting pre-existing nonribosomal proteins onto the ribosome and lengthening some of the proteins already present in the ribosome (O'Brien, 2002; Sharma et al., 2003). Such large-scale remodeling of the mitochondrial ribosome would have necessitated changes in the mitochondrial translation factor(s) as well. IF2mt might have acquired an “extra” sequence corresponding to the 37 amino acid insertion from the pool of then-available primitive proteins to maintain functionality on the mitochondrial ribosome. Once in place, the insertion would have been retained due to the advantages it offered.
Finally, our studies illustrate the usefulness of comparing the mitochondrial translational machinery with the well-characterized eubacterial one in order to gain insight into the former. The strains developed in this study can be manipulated further to address the unsolved problems of mitochondrial translation, which demand urgent attention of researchers in wake of the increasing reports of their involvement in human disease (reviewed by Jacobs, 2003; Jacobs and Turnbull, 2005).
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Growth Conditions
The bacterial strains and plasmids used in this study are listed in Table S1. All strains were grown in LB broth or agar media (Miller, 1972). When required, ampicillin (Amp, 100 μg ml-1), kanamycin (Kan, 25 μg ml-1), chloramphenicol (Chl, 30 μg ml-1), or tetracycline (Tet, 7.5 μg ml-1, unless mentioned otherwise) were added to the medium. P1 phage-mediated transduction was used to transfer genetic material from the donor strains to the desired recipient strains (Miller, 1972). For comparative growth analysis, three separate colonies of each strain were inoculated in LB with isopropyl-β-D-thiogalactoside (IPTG, 0.5 mM) and the appropriate antibiotic, according to the selectable marker in the genome or the resident plasmid. Saturated cultures were diluted 1000-fold in LB, and 200 μl aliquots containing IPTG (0.5 mM) were shaken in 400 μl capacity wells of a microtiter plate using a Bioscreen C growth reader machine (OY Growth, Finland). The experiments were performed at 30°C and 37°C simultaneously, using two Bioscreen C machines. The absorbance was recorded at 600 nm, and the data were plotted using Prism software (GraphPad).
Cloning the IF1 Gene and Generation of IF2 Variants
See the Supplemental Data.
Generation and Analysis of Gene Knockouts in E. coli TG1
Gene knockouts were generated in E. coli TG1 as described by Datsenko and Wanner (2000). This method uses a one-step homologous recombination approach to create the deletions. To generate ΔIF1, the infA gene was replaced by ChlR (ΔinfA::ChlR), and to generate ΔIF2, the infB gene was replaced with KanR (ΔinfB::KanR). The marker cassette in ΔinfB::KanR was removed subsequently. The E. coli ΔIF2 strain thus generated was sensitive to Kan and Amp and was used to generate E. coli ΔIF1ΔIF2. To generate the latter strain, P1 phage (Miller, 1972) was used to transduce the ΔinfA::ChlR locus from E. coli ΔinfA::ChlR into an E. coli ΔIF2 recipient strain that also harbored pACDH-IF2mt. Thus, the resulting E. coli ΔIF1ΔIF2 strain is resistant to Chl and Tet (5 μg ml-1) but sensitive to Kan. Details of the knockout creation and PCR analyses are given in the Supplemental Data.
Southern analysis of E. coli ΔIF1ΔIF2 was carried out using standard procedures (Sambrook et al., 1989). The genomic DNA (1 μg) of various strains was digested overnight with the indicated restriction enzymes, transferred onto a nylon membrane by vacuum blotting, and probed with a 32P-labeled fragment generated by PCR on E. coli HB101 genomic DNA using primers, EcoIF1up-FP and EcoIF1 dn-RP. The washed blot was analyzed by phosphor imaging.
Protein Purification and In Vitro Assays
EcoIF2 and IF2mt were purified as described (Spencer and Spremulli, 2005; Ma and Spremulli, 1996). IF2mtΔ37 was subcloned into pET24b, while EcoIF2::37 remained in pET21c. These plasmids were introduced into E. coli BL21 RIL cells, and the proteins were purified as described (Grasso et al., 2007). The details of in vitro assays for IF2 activity are described in the Supplemental Data and by Spencer and Spremulli (2005).
Homology Modeling of IF2mt
See the Supplemental Data.
ACKNOWLEDGMENTS
We thank our laboratory colleagues for their suggestions on the manuscript. This work was supported by grants from the Department of Biotechnology, Department of Science and Technology, Council of Scientific and Industrial Research, and Indian Council of Medical Research, New Delhi (to U.V.) and National Institutes of Health (NIH) grants GM32734 (to L.S.) and GM61576 (to R.K.A.). R.G. and G.D. were supported by senior research fellowships of the Council of Scientific and Industrial Research, New Delhi.
Footnotes
Supplemental Data Supplemental Data include Supplemental Results, Supplemental Experimental Procedures, two tables, and Supplemental References and can be found with this article online at http://www.molecule.org/cgi/content/full/29/2/180/DC1/.
Supplementary Material
REFERENCES
- Allen GS, Frank J. Structural insights on the translation initiation complex: ghosts of a universal initiation complex. Mol. Microbiol. 2007;63:941–950. doi: 10.1111/j.1365-2958.2006.05574.x. [DOI] [PubMed] [Google Scholar]
- Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J. The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell. 2005;121:703–712. doi: 10.1016/j.cell.2005.03.023. [DOI] [PubMed] [Google Scholar]
- Antoun A, Pavlov MY, Andersson K, Tenson T, Ehrenberg M. The roles of initiation factor 2 and guanosine triphosphate in initiation of protein synthesis. EMBO J. 2003;22:5593–5601. doi: 10.1093/emboj/cdg525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoun A, Pavlov MY, Lovmar M, Ehrenberg M. How initiation factors tune the rate of initiation of protein synthesis in bacteria. EMBO J. 2006a;25:2539–2550. doi: 10.1038/sj.emboj.7601140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoun A, Pavlov MY, Lovmar M, Ehrenberg M. How initiation factors maximize the accuracy of tRNA selection in initiation of bacterial protein synthesis. Mol. Cell. 2006b;23:183–193. doi: 10.1016/j.molcel.2006.05.030. [DOI] [PubMed] [Google Scholar]
- Boelens R, Gualerzi CO. Structure and function of bacterial initiation factors. Curr. Protein Pept. Sci. 2002;3:107–119. doi: 10.2174/1389203023380765. [DOI] [PubMed] [Google Scholar]
- Boileau G, Butler P, Hershey JW, Traut RR. Direct cross-links between initiation factors 1, 2, and 3 and ribosomal proteins promoted by 2-iminothiolane. Biochemistry. 1983;22:3162–3170. doi: 10.1021/bi00282a020. [DOI] [PubMed] [Google Scholar]
- Brock S, Szkaradkiewicz K, Sprinzl M. Initiation factors of protein biosynthesis in bacteria and their structural relationship to elongation and termination factors. Mol. Microbiol. 1998;29:409–417. doi: 10.1046/j.1365-2958.1998.00893.x. [DOI] [PubMed] [Google Scholar]
- Carter AP, Clemons WM, Jr., Brodersen DE, Morgan-Warren RJ, Hartsch T, Wimberly BT, Ramakrishnan V. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science. 2001;291:498–501. doi: 10.1126/science.1057766. [DOI] [PubMed] [Google Scholar]
- Choi SK, Lee JH, Zoll WL, Merrick WC, Dever TE. Promotion of met-tRNAiMet binding to ribosomes by yIF2, a bacterial IF2 homolog in yeast. Science. 1998;280:1757–1760. doi: 10.1126/science.280.5370.1757. [DOI] [PubMed] [Google Scholar]
- Choi SK, Olsen DS, Roll-Mecak A, Martung A, Remo KL, Burley SK, Hinnebusch AG, Dever TE. Physical and functional interaction between the eukaryotic orthologs of prokaryotic translation initiation factors IF1 and IF2. Mol. Cell. Biol. 2000;20:7183–7191. doi: 10.1128/mcb.20.19.7183-7191.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croitoru V, Bucheli-Witschel M, Hagg P, Abdulkarim F, Isaksson LA. Generation and characterization of functional mutants in the translation initiation factor IF1 of Escherichia coli. Eur. J. Biochem. 2004;271:534–544. doi: 10.1046/j.1432-1033.2003.03954.x. [DOI] [PubMed] [Google Scholar]
- Cummings HS, Hershey JW. Translation initiation factor IF1 is essential for cell viability in Escherichia coli. J. Bacteriol. 1994;176:198–205. doi: 10.1128/jb.176.1.198-205.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker PJ, Papadopoulou B, Grivell LA. In-vitro translation of mitochondrial mRNAs by yeast mitochondrial ribosomes is hampered by the lack of start-codon recognition. Curr. Genet. 1993;23:22–27. doi: 10.1007/BF00336745. [DOI] [PubMed] [Google Scholar]
- Garofalo C, Trinko R, Kramer G, Appling DR, Hardesty B. Purification and characterization of yeast mitochondrial initiation factor 2. Arch. Biochem. Biophys. 2003;413:243–252. doi: 10.1016/s0003-9861(03)00119-x. [DOI] [PubMed] [Google Scholar]
- Giuliodori AM, Brandi A, Gualerzi CO, Pon CL. Preferential translation of cold-shock mRNAs during cold adaptation. RNA. 2004;10:265–276. doi: 10.1261/rna.5164904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso DG, Christian BE, Spencer AC, Spremulli LL. Over-expression and purification of mammalian mitochondrial translational initiation factor 2 and initiation factor 3. Methods Enzymol. 2007;430:59–78. doi: 10.1016/S0076-6879(07)30004-9. [DOI] [PubMed] [Google Scholar]
- Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- Gualerzi CO, Pon CL. Initiation of mRNA translation in prokaryotes. Biochemistry. 1990;29:5881–5889. doi: 10.1021/bi00477a001. [DOI] [PubMed] [Google Scholar]
- Harris JK, Kelley ST, Spiegelman GB, Pace NR. The genetic core of the universal ancestor. Genome Res. 2003;13:407–412. doi: 10.1101/gr.652803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey JW. Protein synthesis. In: Neidhart FC, editor. Escherichia coli and Salmonella typhimurium. Cellular and Molecular Biology. American Society for Microbiology; Washington, DC: 1987. pp. 613–647. [Google Scholar]
- Jacobs HT. Disorders of mitochondrial protein synthesis. Hum. Mol. Genet. 2003;12(Spec No 2):R293–R301. doi: 10.1093/hmg/ddg285. [DOI] [PubMed] [Google Scholar]
- Jacobs HT, Turnbull DM. Nuclear genes and mitochondrial translation: a new class of genetic disease. Trends Genet. 2005;21:312–314. doi: 10.1016/j.tig.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Karimi R, Pavlov MY, Heurgue-Hamard V, Buckingham RH, Ehrenberg M. Initiation factors IF1 and IF2 synergistically remove peptidyltRNAs with short polypeptides from the P-site of translating Escherichia coli ribosomes. J. Mol. Biol. 1998;281:241–252. doi: 10.1006/jmbi.1998.1953. [DOI] [PubMed] [Google Scholar]
- Koc EC, Spremulli LL. Identification of mammalian mitochondrial translational initiation factor 3 and examination of its role in initiation complex formation with natural mRNAs. J. Biol. Chem. 2002;277:35541–35549. doi: 10.1074/jbc.M202498200. [DOI] [PubMed] [Google Scholar]
- Kyrpides NC, Woese CR. Universally conserved translation initiation factors. Proc. Natl. Acad. Sci. USA. 1998;95:224–228. doi: 10.1073/pnas.95.1.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laalami S, Putzer H, Plumbridge JA, Grunberg-Manago M. A severely truncated form of translational initiation factor 2 supports growth of Escherichia coli. J. Mol. Biol. 1991;220:335–349. doi: 10.1016/0022-2836(91)90017-z. [DOI] [PubMed] [Google Scholar]
- Laursen BS, Siwanowicz I, Larigauderie G, Hedegaard J, Ito K, Nakamura Y, Kenney JM, Mortensen KK, Sperling-Petersen HU. Characterization of mutations in the GTP-binding domain of IF2 resulting in cold-sensitive growth of Escherichia coli. J. Mol. Biol. 2003;326:543–551. doi: 10.1016/s0022-2836(02)01367-0. [DOI] [PubMed] [Google Scholar]
- Laursen BS, Sorensen HP, Mortensen KK, Sperling-Petersen HU. Initiation of protein synthesis in bacteria. Microbiol. Mol. Biol. Rev. 2005;69:101–123. doi: 10.1128/MMBR.69.1.101-123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Choi SK, Roll-Mecak A, Burley SK, Dever TE. Universal conservation in translation initiation revealed by human and archaeal homologs of bacterial translation initiation factor IF2. Proc. Natl. Acad. Sci. USA. 1999;96:4342–4347. doi: 10.1073/pnas.96.8.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- Liao HX, Spremulli LL. Identification and initial characterization of translational initiation factor 2 from bovine mitochondria. J. Biol. Chem. 1990;265:13618–13622. [PubMed] [Google Scholar]
- Londei P. Evolution of translational initiation: new insights from the archaea. FEMS Microbiol. Rev. 2005;29:185–200. doi: 10.1016/j.femsre.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Ma L, Spremulli LL. Cloning and sequence analysis of the human mitochondrial translational initiation factor 2 cDNA. J. Biol. Chem. 1995;270:1859–1865. doi: 10.1074/jbc.270.4.1859. [DOI] [PubMed] [Google Scholar]
- Ma J, Spremulli LL. Expression, purification, and mechanistic studies of bovine mitochondrial translational initiation factor 2. J. Biol. Chem. 1996;271:5805–5811. doi: 10.1074/jbc.271.10.5805. [DOI] [PubMed] [Google Scholar]
- Ma J, Farwell MA, Burkhart WA, Spremulli LL. Cloning and sequence analysis of the cDNA for bovine mitochondrial translational initiation factor 2. Biochim. Biophys. Acta. 1995;1261:321–324. doi: 10.1016/0167-4781(95)00041-e. [DOI] [PubMed] [Google Scholar]
- Marintchev A, Kolupaeva VG, Pestova TV, Wagner G. Mapping the binding interface between human eukaryotic initiation factors 1A and 5B: a new interaction between old partners. Proc. Natl. Acad. Sci. USA. 2003;100:1535–1540. doi: 10.1073/pnas.0437845100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1972. [Google Scholar]
- Myasnikov AG, Marzi S, Simonetti A, Giuliodori AM, Gualerzi CO, Yusupova G, Yusupov M, Klaholz BP. Conformational transition of initiation factor 2 from the GTP- to GDP-bound state visualized on the ribosome. Nat. Struct. Mol. Biol. 2005;12:1145–1149. doi: 10.1038/nsmb1012. [DOI] [PubMed] [Google Scholar]
- O'Brien TW. Evolution of a protein-rich mitochondrial ribosome: implications for human genetic disease. Gene. 2002;286:73–79. doi: 10.1016/s0378-1119(01)00808-3. [DOI] [PubMed] [Google Scholar]
- Olsen DS, Savner EM, Mathew A, Zhang F, Krishnamoorthy T, Phan L, Hinnebusch AG. Domains of eIF1A that mediate binding to eIF2, eIF3 and eIF5B and promote ternary complex recruitment in vivo. EMBO J. 2003;22:193–204. doi: 10.1093/emboj/cdg030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pel HJ, Grivell LA. Protein synthesis in mitochondria. Mol. Biol. Rep. 1994;19:183–194. doi: 10.1007/BF00986960. [DOI] [PubMed] [Google Scholar]
- Pietromonaco SF, Hessler RA, O'Brien TW. Evolution of proteins in mammalian cytoplasmic and mitochondrial ribosomes. J. Mol. Evol. 1986;24:110–117. doi: 10.1007/BF02099958. [DOI] [PubMed] [Google Scholar]
- Roll-Mecak A, Cao C, Dever TE, Burley SK. X-ray structures of the universal translation initiation factor IF2/eIF5B: conformational changes on GDP and GTP binding. Cell. 2000;103:781–792. doi: 10.1016/s0092-8674(00)00181-1. [DOI] [PubMed] [Google Scholar]
- Roll-Mecak A, Shin BS, Dever TE, Burley SK. Engaging the ribosome: universal IFs of translation. Trends Biochem. Sci. 2001;26:705–709. doi: 10.1016/s0968-0004(01)02024-2. [DOI] [PubMed] [Google Scholar]
- Sambrook JF, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Second Edition Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Sharma MR, Koc EC, Datta PP, Booth TM, Spremulli LL, Agrawal RK. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell. 2003;115:97–108. doi: 10.1016/s0092-8674(03)00762-1. [DOI] [PubMed] [Google Scholar]
- Spencer AC, Spremulli LL. The interaction of mitochondrial translational initiation factor 2 with the small ribosomal subunit. Biochim. Biophys. Acta. 2005;1750:69–81. doi: 10.1016/j.bbapap.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Spremulli LL, Coursey A, Navratil T, Hunter SE. Initiation and elongation factors in mammalian mitochondrial protein biosynthesis. Prog. Nucleic Acid Res. Mol. Biol. 2004;77:211–261. doi: 10.1016/S0079-6603(04)77006-3. [DOI] [PubMed] [Google Scholar]
- Tibbetts AS, Oesterlin L, Chan SY, Kramer G, Hardesty B, Appling DR. Mammalian mitochondrial initiation factor 2 supports yeast mitochondrial translation without formylated initiator tRNA. J. Biol. Chem. 2003;278:31774–31780. doi: 10.1074/jbc.M304962200. [DOI] [PubMed] [Google Scholar]
- Towpik J. Regulation of mitochondrial translation in yeast. Cell. Mol. Biol. Lett. 2005;10:571–594. [PubMed] [Google Scholar]
- Vambutas A, Ackerman SH, Tzagoloff A. Mitochondrial translational-initiation and elongation factors in Saccharomyces cerevisiae. Eur. J. Biochem. 1991;201:643–652. doi: 10.1111/j.1432-1033.1991.tb16325.x. [DOI] [PubMed] [Google Scholar]
- Varshney U, Hutcheon T, van de Sande JH. Sequence analysis, expression, and conservation of Escherichia coli uracil DNA glycosylase and its gene (ung) J. Biol. Chem. 1988;263:7776–7784. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.