Abstract
It is well documented that dopamine can increase or decrease the activity of the Na+,K+-ATPase (NKA, sodium pump) in an organ-specific fashion. This regulation can occur, at least partially, via receptor-mediated second messenger activation and can promote NKA insertion or removal from the plasma membrane. Using co-immunoprecipitation and mass spectrometry, we now show that, in both brain and HEK293T cells, D1 and D2 dopamine receptors (DARs) can exist in a complex with the sodium pump. To determine the impact of NKA on DAR function, biological assays were conducted with NKA and DARs co-expressed in HEK293T cells. In this system, expression of NKA dramatically decreased D1 and D2 DAR densities with a concomitant functional decrease in DAR-mediated regulation of cAMP levels. Interestingly, pharmacological inhibition of endogenous or overexpressed NKA enhanced DAR function without altering receptor number or localization. Similarly, DAR function was also augmented by small interfering RNA reduction of the endogenous NKA. These data suggest that, under basal conditions, NKA negatively regulates DAR function via protein-protein interactions. In reciprocal fashion, expression of DARs decreases endogenous NKA function in the absence of dopamine, implicating DAR proteins as regulators of NKA activity. Notably, dopamine stimulation or pertussis toxin inhibition of D2 receptor signaling did not alter NKA activity, indicating that the D2-mediated decrease in NKA function is dependent upon protein-protein interactions rather than signaling molecules. This evidence for reciprocal regulation between DARs and NKA provides a novel control mechanism for both DAR signaling and cellular ion balance.
Dopamine receptors (DARs)4 are seven transmembrane-spanning G-protein-coupled receptors that mediate a diverse array of dopaminergic processes throughout the body. DARs are subdivided into two families based on their pharmacological and genetic profiles. The D1 family of receptors, containing the D1 and D5 subtypes, couple to the heterotrimeric G-protein Gs and positively regulate adenylyl cyclase activity. The D2 family of DARs consists of the D2,D3, and D4 receptor subtypes. These receptors couple to inhibitory Gi/o proteins and reduce adenylyl cyclase activity. Within the central nervous system, dopamine and its receptors regulate locomotion, addiction, cognition, and learning and memory formation (1–5); non-neuronal DARs modulate blood pressure and digestive function (6–8). Dysregulation of dopaminergic pathways and receptors has been implicated in several diseases, including schizophrenia, Parkinson disease, and hypertension. Despite the breadth of dopaminergic functions and disorders, the molecular control and coordination of dopamine signaling are still being elucidated.
It is now appreciated that DARs and other G-protein-coupled receptors can couple to a variety of proteins to form a large protein complex termed the “signalplex” (9–11). Identification of the numerous protein partners in the signalplex is critical to understanding the molecular mechanisms responsible for tissue-specific dopamine functions and various dopaminergic disorders. Several dopamine receptor-interacting proteins (DRIPs) have recently been discovered (10, 12). These identified DRIPs regulate a variety of functions, including cell signaling and receptor trafficking and retention. Using co-immunoprecipitation coupled with mass spectroscopy analysis, we have now identified the α1 subunit of the Na+,K+-ATPase (sodium pump, NKA) as a DRIP in both D1 and D2 DAR signalplexes.
The NKA is a ubiquitous membrane protein that actively hydrolyzes ATP to maintain the Na+/K+ gradient across the plasma membrane. The NKA consists of a large, catalytic α subunit and a smaller β subunit. The α1 subunit is expressed in virtually all tissues, including neurons and glia in the central nervous system (13). Neuronally, the NKA is responsible for maintenance of resting membrane potential, including restoration of membrane potential following depolarization (14).
Several investigators have demonstrated dopaminergic control of the NKA in primary striatal, lung, and kidney cells (15–20). However, in all of these studies, dopamine and other neurotransmitter receptor agonists appeared to regulate the NKA via signal transduction pathways; second messenger kinases and downstream signaling caused insertion or removal of the NKA at the plasma membrane (15, 17, 20–23). Our current data now provide the first evidence of a direct interaction between these two proteins. Furthermore, we show that the DARs and NKA are able to functionally regulate one another via protein-protein interactions in the absence of ligands or downstream signaling events. Our results indicate that, in addition to traditional second messenger-mediated communication, the DARs and NKA can associate in a complex to provide a more rapid and immediate response to external stimuli or changing the cellular environment.
EXPERIMENTAL PROCEDURES
Materials—HEK293-tsa201 (HEK293T) (24) cells were a gift from Dr. Vanitha Ramakrishnan. [3H]SCH23390 (85.0 Ci/mmol), [3H]methylspiperone (79.5 Ci/mmol), [3H]sulpiride (85.0 Ci/mmol), [3H]cAMP (26.4 Ci/mmol), and 86Rb+ (7.85–21.58 mCi/mg) were obtained from PerkinElmer Life Sciences. Bovine corpus striatum was obtained from Rockland Immunochemicals (Gilbertsville, PA). Cell culture reagents, NuPAGE gels, and gel buffers and reagents were purchased from Invitrogen. All other drugs and buffer components were purchased from Sigma, except where indicated.
Cell Culture and Transfection—HEK293T cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 1 mm sodium pyruvate, 50 μg/ml streptomycin, 50 units/ml penicillin, and 10 μg/ml gentamycin. Cells were grown at 37 °C in 5% CO2 and 90% humidity. HEK293T cells were transfected using a calcium phosphate precipitation kit (Clontech) according to the manufacturer's instructions.
Several expression constructs were used in this study. An expression construct containing only the FLAG peptide (FLAG-tag), pCMV-Tag2B, was obtained from Stratagene (La Jolla, CA). The D2L DAR expression construct containing an amino-terminal FLAG-tag epitope in pSR-D2L was created by our laboratory as described previously (25). An amino-terminal FLAG epitope-tagged construct for the rat D1 DAR (26) was created and named pSFD1, as reported previously by our laboratory (27); additionally, a FLAG-tagged D1 receptor truncated at amino acid 347 and missing most of the carboxyl terminus (designated FLAG-D1-T0) was created by our laboratory as described previously (28). A red fluorescent protein (RFP)-tagged rat D2L receptor was created by our laboratory using the pSR-D2L construct and pRFP (Evrogen, Moscow, Russia). Human NKAα1 in pCMV was obtained from OriGene (Rockville, MD), and rat enhanced green fluorescent protein (eGFP)-tagged NKAα1 was a generous gift from Dr. Alejandro Bertorello (19). Plasmid small interfering RNA (siRNA) directed against the NKAα1 subunit and scrambled siRNA were synthesized by Retrogen (San Diego, CA) in pSilencer 3.1-H1 vector.
Immunoprecipitation and Gel Electrophoresis—For HEK293T cells, cells were removed from culture flasks in calcium-free Earle's balanced salt solution (EBSS) and collected by centrifugation (200 × g). Pelleted cells were then resuspended in 1 ml of solubilization buffer (50 mm HEPES, 1 mm EDTA, 10% glycerol, 1% Triton X-100, 150 mm NaCl, 50 mm NaF, 40 mm sodium pyrophosphate) with Complete-Mini protease inhibitor mixture tablets (Roche Applied Science) and incubated on ice for 1 h. The lysate was centrifuged at 30,000 × g for 30 min to remove insoluble cell debris and then pre-cleared via incubation with protein G-agarose (Invitrogen) for 3 h. The agarose was removed by centrifugation (7,000 × g, 5 min) and discarded. Anti-FLAG M2-agarose (Sigma) was added and incubated with the lysate overnight at 4 °C. The beads were collected via centrifugation and washed three times via resuspension and re-pelleting in solubilization buffer (7,000 × g, 5 min). The agarose was then washed twice in 1× TE buffer, pH 7.4. Proteins were eluted from the beads using NuPAGE LDS sample buffer (Invitrogen) at 37 °C. Agarose was removed via centrifugation, and proteins were separated on 4–12% BisTris NuPAGE gel (Invitrogen) according to the manufacturer's instructions. For bovine corpus striatum, diced striatum was Polytron-homogenized for ∼1 min in 10 mm Tris-HCl + 1% CHAPS, pH 7.5, with Complete-Mini protease inhibitor mixture tablets and then incubated on ice for 1 h. Solubilized striatum was centrifuged at 30,000 × g for 10 min to remove insoluble cell debris; the resulting supernatant was centrifuged for an additional 30 min (30,000 × g). The supernatant was then pre-cleared via incubation with protein G-agarose for 3 h at 4 °C; agarose was removed by centrifugation (7,000 × g, 5 min) and discarded. Protein concentrations of the sample supernatants were determined using the BCA protein assay (Pierce). Cell lysate (500 μg), anti-D2S/L antibody (4 μg), and Antibody Capture Affinity ligand were added according to the manufacturer's instructions to washed Catch and Release kit beads (Pierce) and incubated overnight at 4 °C. Beads were washed three times with Catch and Release kit buffer, 3× with Tris-HCl/CHAPS, and an additional three times with Catch and Release kit buffer. Proteins were eluted from beads according to the manufacturer's instructions, heated at 37 °C for 1 h, and separated on 4–12% BisTris NuPAGE gel.
Western Blotting and Gel Staining—Proteins separated by denaturing SDS-PAGE were transferred onto polyvinylidene difluoride membranes. Membranes were blocked in Superblock (Pierce) prior to incubation with the primary antibody. Primary antibodies used in this study include the following: rat monoclonal anti-D1 dopamine receptor (clone 1-1-F11 S.E6, catalogue number D187, Sigma), rabbit polyclonal anti-D2S/L (catalogue number AB5084P, Chemicon Millipore, Billerica, MA), and mouse monoclonal anti-NKAα1 (clone C464.6, catalogue number 05-369, Upstate Millipore, Billerica, MA). After washing with Tris-buffered saline containing 0.1% Tween, membranes were incubated with HRP-conjugated secondary antibodies (Jackson ImmunoResearch, West Grove, PA; anti-rat for the D1, anti-rabbit for D2, and anti-mouse for NKAα1). Proteins were visualized via the SuperSignal West Pico chemiluminescent substrate (Pierce) according to the manufacturer's instructions, and images were recorded on film or computer using the UVP Bioimaging Systems EpiChemi3 Darkroom (UVP, Upland, CA). In some experiments, blots were stripped of all antibody using Restore Western blot stripping buffer (Pierce) according to the manufacturer's instructions. Removal of all primary antibody by the buffer was checked via incubation with HRP-secondary followed by ECL and film exposure. In experiments where total protein was examined, gels were stained using colloidal Coomassie SimplyBlue Safe Stain (Invitrogen).
Mass Spectrometry (MS)—The identification of PAGE-resolved proteins by mass spectroscopy was carried out by Prot-Tech, Inc. (Norristown, PA). In brief, each protein gel band was digested “in-gel” with modified sequencing grade trypsin (Promega, Madison, WI), and the resulting peptide mixture was subjected to tandem mass spectrometry for peptide sequencing. A Finnigan ion trap mass spectrometer LCQ coupled with a high performance liquid chromatography system running a 75 μm inner diameter C18 column was used. Data were acquired in a data-dependent mode. MS/MS spectra were used to search the most recent nonredundant protein data bases including the Protein Information Resources database and GenBank™ using ProtQuest software suite from ProtTech. The output of the data base search was manually analyzed and validated to verify proper protein identification.
Confocal Microscopy—HEK293T cells were plated in 100-mm tissue culture dishes (1 × 106 cells) and transfected as described above with 1 μg of rat D2L-RFP or 0.5 μg of rat NKAα1-eGFP, either separately or co-transfected with both constructs for dual-labeling experiments. Twenty four hours post-transfection, cells were re-seeded onto poly-d-lysine-coated 35-mm glass-bottom culture dishes and maintained for an additional 48 h. Confocal microscopy experiments were performed on a Zeiss laser scanning confocal microscope (LSM 5 Pascal). Images were collected using single line excitation (488 nm for NKAα1-eGFP or 543 nm for D2L-RFP).
Membrane Radioligand Binding Assays—HEK293T cells were plated in 150-mm tissue culture dishes (5 × 106 cells) and transfected as described above. Forty eight hours later, the cells were removed mechanically using EBSS. For experiments involving ouabain treatment, a final concentration of 5 mm ouabain was added to cells, and dishes were incubated at 37 °C for 15 min prior to removal. Intact cells were pelleted and then lysed with 5 mm Tris-HCl and 5 mm MgCl2, pH 7.4. Homogenates were centrifuged at 20,000 × g for 30 min. The membranes were resuspended with 50 mm Tris-HCl, pH 7.4. Membrane preparations were incubated for 90 min at room temperature with various concentrations of [3H]SCH23390 (D1 binding) or [3H]methylspiperone (D2 binding) in a reaction volume of 1 ml. Nonspecific binding was determined in the presence of 4 μm (+)-butaclamol. Bound ligand was separated from unbound by filtration through polyethyleneimine-soaked GF/C filters using a Brandel cell harvester. GF/C filter disks were then analyzed via liquid scintillation spectroscopy at a counting efficiency of 60%. Protein concentration was determined using a Bradford protein assay (Bio-Rad).
cAMP Accumulation Assays—Transfected HEK293T cells were seeded into 24-well poly-d-lysine-coated plates at 200,000 cells/well and cultured for 1 day prior to the experiment. For D1 accumulation assays, cells were washed three times with 400 μl of EBSS per well. Various concentrations of dopamine dissolved in stimulation buffer (Dulbecco's modified Eagle's medium, 20 μm Ro-20-1724, 0.2 mm sodium metabisulfite) were added to each well in a volume of 400 μl for 10 min at 37 °C. For D2 inhibition assays, cells were washed three times with 400 μl of EBSS per well. Various concentrations of dopamine containing 3 μm forskolin and 10 μm propranolol in stimulation buffer were added to each well in a volume of 400 μl for 10 min at 37 °C. For experiments involving ouabain treatment, stimulation buffer (for either D1 or D2L experiments) was prepared with or without 5 mm ouabain. Cells were incubated at 37 °C for 5 min in the ouabain or control stimulation buffer prior to dopamine addition. Dopamine solutions were prepared in appropriate stimulation buffer for the experiment. The reactions were terminated by adding 200 μl of 3% perchloric acid and incubated on ice for 30 min. 80 μl of 15% KHCO3 was then added to neutralize the acid. The plates remained on ice for an additional 20 min and were centrifuged at 1,300 × g for 20 min. 50 μl of the supernatant from each well was transferred to a 1.2-ml reaction tube containing 250 μl of purified cAMP-dependent protein kinase (PKA) in Tris/EDTA buffer and 50 μl of [3H]cAMP. The reaction was incubated for 90 min at 4 °C. After the incubation, 250 μl of 1% charcoal-dextran mixture was added to each tube and vortexed gently. Tubes were then incubated at 4 °C for 10 min followed by centrifugation (1,300 × g) for 20 min. Radioactivity in the supernatant was quantified by liquid scintillation spectroscopy at a counting efficiency of 60%. cAMP concentrations were determined using a standard assay curve of cAMP values from 0.1 to 100 pmol.
Intact Cell Binding Assays—Transfected HEK293T cells were seeded into poly(D)-lysine-coated 24-well plates at 200,000 cells/well. Twenty four hours later, cells were washed once with EBSS, and binding was performed by incubation for 15 min at 37 °C in 500 μl of EBSS containing increasing concentrations of [3H]sulpiride with or without 5 mm ouabain. After incubation, the binding buffer was aspirated, and the plates were washed three times with EBSS. The cells were dissolved in 500 μl of 1 mm EDTA containing 1% Triton X-100, transferred to vials, and quantified for radioactivity by liquid scintillation spectroscopy at a counting efficiency of 60%. Specific binding was defined as the difference between total and nonspecific binding, with the latter measured in the presence of 5 μm (+)-butaclamol.
Rubidium-86 Ion Uptake—HEK293T cells were plated in 150-mm tissue culture dish (5 × 106 cells) and transfected as described above. Forty eight hours later the cells were removed mechanically using EBSS. Intact cells were pelleted and then resuspended in 1.5 ml of assay buffer (5 mm HEPES-Tris, 140 mm NaCl, 5 mm KCl, 1 mm MgCl2, 1 mm CaCl2, 5 mm glucose; 0.1 mm furosemide and 0.01 mm monensin were added just prior to experimentation) with or without 5 mm ouabain. Cells were incubated at 37 °C for 10 min in assay buffer alone, then 2.5 μCi of 86Rb+ in assay buffer was added, and samples were incubated for an additional 15 min at 37 °C. Uptake was terminated by addition of 500 μl of ice-cold 10 mm HEPES, 100 mm MgCl2 followed by centrifugation at 3,000 × g. Pellets were washed a total of three times with HEPES/MgCl2, and cells were then lysed with 500 μl of NaOH and neutralized with 50 μl of concentrated HCl. Radioactivity was analyzed via liquid scintillation spectroscopy at a counting efficiency of 60%. Protein concentration was determined using a Bradford protein assay (Bio-Rad).
RESULTS
Identification of the NKA as an Interacting Protein with Both D1 and D2 DARs—As part of ongoing efforts in DRIP discovery, co-immunoprecipitation assays of epitope-tagged rat D2L receptors were coupled with MS analysis to identify D2 DAR-interacting proteins. For these experiments, HEK293T cells (which do not endogenously express the D2 DAR) were transiently transfected with constructs expressing either a FLAG peptide (FLAG tag) alone or a FLAG-D2L receptor. Proteins from solubilized cells were immunoprecipitated with anti-FLAG antibody-conjugated agarose, and the precipitated proteins were analyzed by SDS-PAGE. Staining with colloidal Coomassie Blue dye revealed multiple proteins that were precipitated from both transfection conditions (Fig. 1). All visible protein bands from the FLAG-D2L lane (Fig. 1, right lane) were excised, along with corresponding control bands (FLAG tag, Fig. 1, left lane) for comparison and selection purposes. The excised bands were digested with trypsin, and the eluted proteins were subjected to liquid chromatography-MS/MS tandem mass spectroscopy as described under “Experimental Procedures”; all identified proteins from this experiment are listed in Table 1. Proteins that were identified in both sets of immunoprecipitates were considered nonspecific interactors; no identified proteins were unique to the FLAG tag control immunoprecipitates. The NKA was identified in these experiments as a specific interacting protein with the FLAG-tagged rat D2L receptor and was chosen for further study. Similar liquid chromatography-MS/MS experiments that were conducted with the FLAG-D1 receptor revealed the NKA as a putative specific interacting protein with this receptor as well (29). Multiple nonoverlapping peptide fragments from both the D2L protein (Fig. 1, Band 13) and the NKA protein (Fig. 1, Band 3) were identified from the FLAG-D2L receptor bands; the sequence of these peptides is reported in Table 2. These proteins were absent from the corresponding control FLAG tag lanes (Fig. 1). The same NKA peptides, and three additional peptide fragments from the NKA, were identified from FLAG-D1 receptor immunoprecipitated samples. These peptides, as well as identified D1 receptor peptide sequences, are listed in Table 2.
FIGURE 1.
Identification of D2 receptor-interacting proteins from HEK293T cells by co-immunoprecipitation and mass spectrometry. The SDS-polyacrylamide gel was stained with colloidal Coomassie Blue for protein detection. Bands in the FLAG-D2L lane were derived from HEK293T cells transfected with 30 μg of FLAG-tagged rat D2L receptor and immunoprecipitated with antibody against the FLAG epitope. Bands in the FLAG Tag lane were immunoprecipitated from HEK293T cells expressing only the FLAG epitope and serve as a control. Bands were excised from both experimental and control lanes and subjected to MS-based sequencing; identified proteins are reported in Table 1. Band number indicates the position of bands that were determined by MS to contain NKAα1 subunit or rat D2L receptor peptides as reported in Table 2.
TABLE 1.
Proteins identified by mass spectroscopy that co-immunoprecipitated with the D2 dopamine receptor as shown in Fig. 1 Accession numbers for identified proteins are reported in parentheses.
| Specific rat D2L interactors | Nonspecific interactors |
|---|---|
| Cellular apoptosis susceptibility protein (AAC35008) | 26 S proteasome regulatory subunit 2 (BAG10963) |
| Ca2+-ATPase (EAW97905) | Proteasome 26 S subunit 13 (NP_787128) |
| PUM2 protein (AAH24218) | IgG light chain (ABV55856) |
| Monocarboxylic acid transporter 1 (CAI21873) | |
| Ubiquitin B (EAX04505) | |
| MCM10 protein (AAH04876) | |
| Dopamine D2 receptor (NP_036679) | |
| HSPC121 protein (AAF29085) | |
| Zinc finger protein 491 (NP_689569) | |
| SLC44A2 protein (AAH10617) | |
| Na+,K+-ATPase α1 subunit (ABW03450) | |
| Calumenin (AAB97725) | |
| Kinesin family member 11 (BAD92201) |
TABLE 2.
Peptides and corresponding parent proteins found in co-immunoprecipitation mass spectroscopy experiments
| HEK293T transfection | Band no.a | Trypsin-digested peptide | Parent protein |
|---|---|---|---|
| FLAG-D2 | 3 | VDNSSLTGESEPQTR, LNIPVSQVNPR | Na+,K+-ATPase |
| 13 | QNWSRPFNGSEGK, YTAVAMPMLYNTR | D2 DAR | |
| FLAG-D1 | VDNSSLTGESEPQTR, LSLDELHR | Na+,K+-ATPase | |
| LNIPVSQVNPR, SPDFTNENPLETR | |||
| GVGIISEGNETVEDIAAR | |||
| EEAGGIAKPLEK | D1 DAR | ||
| LSPALSVILDYDTDVSLEK | |||
| IQPVTHSGQHST, YWAISSPFQYER |
Band number refers to location of band on Coomassie-stained gel in Fig. 1
Verification and Characterization of DAR Interactions with NKAα1—To verify the mass spectroscopy results, the DAR-NKA interactions were investigated by co-immunoprecipitation followed by Western blot analysis. For these experiments, HEK293T cells (which endogenously express the NKAα1 but do not natively express the D1 or D2 DARs) were transiently transfected with the FLAG tag alone, the FLAG-D1 receptor, or the FLAG-D1 receptor along with the NKAα1. Probing the Western blots with an anti-D1 receptor antibody showed that the D1 receptor was present only in cells that were transfected with the FLAG-D1 expression construct (Fig. 2A, left panel). Interestingly, overexpression of NKAα1 caused an apparent decrease in D1 receptor number (Fig. 2A, left panel, ∼50 kDa in size). The Western blot in Fig. 2A was stripped and re-probed with an antibody against NKAα1 (Fig. 2A, right panel, migrating at ∼95 kDa in size), revealing that endogenous NKAα1 only immunoprecipitates with the D1 receptor and not with the FLAG tag alone. Furthermore, overexpression of NKAα1 caused a proportionate increase in the amount of NKA interacting with the D1 receptor, despite the fact that less D1 receptor was present in these samples.
FIGURE 2.
Co-immunoprecipitation of D1 DAR with the NKAα1 subunit from transfected HEK293T cells. For all blots, proteins were extracted, immunoprecipitated (IP) with anti-FLAG agarose, electrophoresed, and immunoblotted (IB) as described under “Experimental Procedures.” A, HEK293T cells were transfected with 15 μg of FLAG tag alone, FLAG-D1 receptor, or FLAG-D1 receptor with NKAα1 subunit. Left panel, the blot was probed with a monoclonal anti-D1 antibody and visualized using ECL after incubation with an HRP-conjugated anti-rat secondary antibody. Right panel, the blot in left panel was stripped of all antibodies, re-probed with a monoclonal anti-NKAα1 antibody, and visualized by ECL after incubation with an HRP-conjugated anti-mouse secondary antibody. The arrow indicates the position of the NKAα1 subunit migrating at ∼95 kDa. B, HEK293T cells were transfected with 15 μg of FLAG tag, FLAG-D1, or FLAG-D1 receptor lacking the carboxyl terminus (FLAG-D1-T0) in the presence or absence of transiently co-expressed NKAα1 subunit (15 μg). The Western blot was probed with a monoclonal anti-NKAα1 antibody and visualized by ECL after incubation with an HRP-conjugated anti-mouse secondary antibody. The arrow indicates the position of the NKAα1 subunit migrating at ∼95 kDa.
The carboxyl terminus of the D1 receptor is a common site for protein interactions (30–32). A D1 receptor construct truncated at amino acid 347 to lack the carboxyl terminus (T0) was used to determine whether this region might be the location of the NKA-D1 receptor interaction. Because the D1T0 receptor construct does not express as well as the wild-type receptor, all co-immunoprecipitated samples had to be normalized for D1 receptor expression by radioligand binding. After normalizing for expression levels, the samples were resolved by SDS-PAGE, and resultant Western blots were probed with anti-NKAα1 antibody (Fig. 2B). The NKAα1-D1 interaction remained intact despite the removal of the D1 receptor carboxyl terminus, indicating that this is not the region of interaction between these two proteins.
To verify the original mass spectroscopy findings of the D2-NKA interactions, similar co-immunoprecipitation experiments were also conducted with the D2 receptor. In Fig. 3, HEK293T cells expressing the FLAG tag, FLAG-D2L alone, or FLAG-D2L along with NKAα1 were solubilized, immunoprecipitated with anti-FLAG-agarose, and resolved by SDS-PAGE. The Western blots from these experiments were probed with antibodies against the D2 receptor (Fig. 3, left panel, migrating as a glycosylated protein ∼50–65 kDa in size), then stripped, and re-probed with anti-NKAα1 antibodies (Fig. 3, right panel, ∼95 kDa in size). Similar to the D1 receptor experiments, there was a decrease in D2L receptor numbers when NKAα1 was overexpressed in these cells. There was also an increase in NKA-D2L association when the NKAα1 subunit was overexpressed, despite the apparent decrease in D2L receptor protein levels. In addition, there was no NKA immunoreactivity in the FLAG tag lane, indicating that the presence of NKA is receptor-specific in these experiments. Reverse co-immunoprecipitation assays were also performed from HEK293T cells. In these experiments, cells transfected with empty vector, FLAG-D2L, or FLAG-D2L along with NKAα1 were solubilized, and antibodies against the NKAα1 were used to immunoprecipitate the NKA-D2L complex. The NKAα1 and D2L proteins were both present in Western blots from these experiments (data not shown).
FIGURE 3.
Co-immunoprecipitation of D2L DAR with the NKAα1 subunit from transfected HEK293T cells. Proteins were extracted, immunoprecipitated (IP) with anti-FLAG agarose, electrophoresed, and immunoblotted (IB) as described under “Experimental Procedures.” HEK293T cells were transfected with 15 μg each of either FLAG tag alone, FLAG-D2L receptor, or FLAG-D2L receptor with NKAα1 subunit. Left panel, the blot was probed with a polyclonal anti-D2S/L antibody and visualized using ECL after incubation with an HRP-conjugated anti-rabbit secondary antibody. Right panel, the blot in left panel was stripped of all antibodies, re-probed with a monoclonal anti-NKAα1 antibody, and visualized by ECL after incubation with an HRP-conjugated anti-mouse secondary antibody. The arrow indicates the position of the NKAα1 subunit migrating at ∼95 kDa.
Of note, no visible change in endogenous NKAα1 expression was observed when the FLAG-D2L receptor was overexpressed (supplemental Fig. 1). In these experiments, cells were transfected with empty vector, FLAG-D2L alone, NKAα1 alone, or FLAG-D2L along with NKAa1. Cells were solubilized, and total lysates were resolved by SDS-PAGE, and Western blots were probed with antibodies against the NKAα1 or D2L proteins. Importantly, D2L DAR transfection produced no change in the expression levels of either endogenous or overexpressed NKAα1 (supplemental Fig. 1).
Endogenous proteins from brain tissue were also investigated to ensure that the interaction between D2 DAR and NKAα1 was not unique to HEK293T cells. In these experiments, bovine striatum was used to enrich the D2 receptor. The striatum was solubilized and immunoprecipitated with either anti-D2 antibody or species-matched, pre-immune serum using the Catch and Release kit as described under “Experimental Procedures.” The resultant Western blot was probed with anti-D2 antibody and revealed a less glycosylated form of the receptor migrating at ∼52 kDa (Fig. 4, left panel). The blot was stripped of all antibodies and re-probed with antibody against NKAα1 (Fig. 4, right panel, migrating at ∼95 kDa). In both Western blots, the proteins were not detected when control, pre-immune serum was used for the immunoprecipitation. These data indicate that the D2 receptor and NKAα1 subunit interact in a physiological setting with native levels of protein.
FIGURE 4.
Co-immunoprecipitation of the NKAα1 subunit with the D2 DAR from bovine striatum. Proteins were extracted from bovine striatum with 1% CHAPS in Tris-HCl buffer, immunoprecipitated (IP) via the Catch and Release system (Pierce) with the anti-D2S/L antibody, electrophoresed, and immunoblotted (IB) as described under “Experimental Procedures.” Left panel, the blot was probed with a polyclonal anti-D2S/L antibody and visualized using ECL after incubation with an HRP-conjugated anti-rabbit secondary antibody. Right panel, the blot in left panel was stripped of all antibodies, re-probed with a monoclonal anti-NKAα1 antibody, and visualized by ECL after incubation with an HRP-conjugated anti-mouse secondary antibody.
Confocal microscopy was employed to investigate D2 DAR and NKAα1 co-localization. In these experiments, HEK293T cells were co-transfected with fluorescent-tagged rat D2L-RFP and rat NKAα1-GFP, enabling subcellular visualization of the proteins. These experiments revealed that, in HEK293T cells, both the D2L-RFP (Fig. 5A) and the NKAα1-GFP (Fig. 5B) are predominantly expressed at the plasma membrane, and are similarly distributed when either fluorescent-tagged protein is expressed alone (supplemental Fig. 2). Of note, when the images are merged, it is clear that the two proteins co-localize at the cell surface (Fig. 5C). These data further support the finding ofaD2L-NKAα1 protein complex.
FIGURE 5.
Co-expression of the D2L-RFP with the NKAα1-eGFP. HEK293T cells were transfected with both 1 μg of D2L-RFP and 0.5 μg of NKAα1-eGFP as described under “Experimental Procedures.” After 24 h, cells were seeded on 35-mm glass-bottom culture dishes and visualized 48 h later. A, visualization of cells at a single line excitation of 543 nm, detecting NKAα1-eGFP. B, visualization of cells at a single line excitation of 488 nm, detecting D2L-RFP. C, overlay of A and B. Regions of yellow at the plasma membrane indicate protein co-localization. Images are representative of four separate experiments.
Overexpression of NKAα1 Decreases D1 and D2 Receptor Function and Density—To investigate the impact of NKAα1 on DAR function and expression, HEK293T cells were transiently transfected with D2L receptor cDNA and evaluated for cAMP levels in the presence of increasing concentrations of dopamine (Fig. 6A, solid line). In parallel experiments, cells expressing both the D2L receptor and the NKAα1 subunit were treated with various concentrations of dopamine, and inhibition of cAMP was evaluated (Fig. 6A, dashed line). Overexpression of the NKAα1 subunit caused a marked inhibition of D2L DAR function. The maximal dopamine-induced inhibition of cAMP decreased by ∼30% in the presence of NKAα1, and the EC50 was significantly shifted to lower affinity. A similar decrease in efficacy and potency was observed for the D1 receptor following overexpression of the NKAα1 subunit (Fig. 6B). The maximum dopamine-stimulated increase in cAMP was diminished by ∼20%, and the EC50 was significantly shifted to a lower affinity.
FIGURE 6.
DAR function following NKAα1 overexpression. HEK293T cells were co-transfected with 15 μg of DAR (D2L (A) or D1 (B)) and 7.5 μg of either empty pcDNA vector or NKAα1. Cells were washed 24 h later and re-plated into 24-well plates at a density of 200,000 cells per well; assays were conducted 48 h post-transfection. Cells were stimulated with forskolin and dopamine (A) or dopamine alone (B), and cAMP accumulation was measured as described under “Experimental Procedures.” Data were normalized for individual experiments as a percentage of the Emax value for DAR alone and are expressed as mean ± S.E. of four independent experiments, each conducted in triplicate; all statistical analyses were performed using Student's t test. For D2L alone (A), raw values for Emax (maximal dopamine-induced inhibition) ranged from 4.0 to 8.2 pmol of cAMP/200,000 cells; normalized Emax (% inhibition) values were 49.2 ± 1.1% for D2L alone and 20.2 ± 0.5% for D2L + NKA (***, p < 0.001). EC50 values were 5.0 nm for D2L alone and 14.7 nm for D2L + NKA (p < 0.01). For D1 alone (B), raw values for Emax ranged from 22.0 to 49.4 pmol of cAMP/200,000 cells; normalized Emax values were 99.7 ± 0.8% for D1 alone and 81.3 ± 0.9% for D1 + NKA (***, p < 0.001). EC50 values were 13.5 nm for D1 alone and 40.0 nm for D1 + NKA (p < 0.001).
Because receptor expression can impact function, the influence of NKAα1 on DAR expression was investigated. For these experiments, total receptor density was examined in transiently transfected HEK293T cells. For both the D1 and D2L receptors, overexpression of the NKAα1 subunit caused a significant decrease in total receptor density (Fig. 7) with no significant change in DAR affinity for radioligand. Total D2L density was reduced by ∼80% (Fig. 7A), and D1 density was decreased by ∼60% (Fig. 7B) when NKAα1 was co-expressed. As a control experiment, we co-transfected a plasmid expressing RFP, which is not predicted to interact with the DARs. Co-expression of RFP did not significantly alter the expression density of the receptors as determined using saturation radioligand binding assays for both D1 and D2 DARs (data not shown). These results indicate that the diminished DAR number following NKA expression is specific to the NKA.
FIGURE 7.
Total DAR density after NKAα1 overexpression. HEK293T cells were co-transfected with 15 μg of DAR (D2L (A) or D1 (B)) and 7.5 μg of either empty pcDNA vector or NKAα1. Cells were washed 24 h later and remained in the plate until the assay was conducted, 48 h post-transfection. Radioligand binding assays were performed using [3H]methylspiperone (D2L, A) or [3H]SCH23390 (D1, B) as described under “Experimental Procedures.” Data were normalized for individual experiments as a percentage of the Bmax value for DAR alone and are expressed as mean ± S.E. of four independent experiments, each conducted in triplicate; all statistical analyses were performed using Student's t test. For D2L alone (A), raw values for Bmax ranged from 7.2 to 13.1 fmol/mg protein; normalized Bmax for D2L + NKA was 20.3 ± 0.4% (***, p < 0.001). KD values were 23 ± 1 pm for D2L alone and 13 ± 2 pm for D2L + NKA. For D1 alone (B), raw values for Emax ranged from 4.1 to 11.2 fmol/mg protein; normalized Bmax for D1 + NKA was 37.2 ± 0.4% (***, p < 0.001). KD values were 126 ± 7 pm for D1 alone and 104 ± 5 pm for D1 + NKA.
Inhibition of NKA Function Enhances DAR Activity without Altering DAR Density—The cardiotonic steroid ouabain is known to inhibit NKA function (33). Ouabain was used to determine whether the activity or the presence of NKA was responsible for the observed decrease in DAR expression and function. An acute 15-min treatment with ouabain successfully inhibits NKA activity (22) without decreasing the expression of the protein. This short term ouabain treatment was used in all experiments to investigate the impact of NKA activity on DAR function and binding parameters. A relatively high concentration of ouabain (5 mm) was employed in these experiments for several reasons. First, the rat NKAα1, which is more resistant to ouabain (34), was used in confocal microscopy analysis (see below). Because ouabain experiments with this NKAα1-GFP construct require a higher dose of drug, the same ouabain concentration was employed for all assays to minimize confounding experimental variables. In addition, lower concentrations of ouabain (1 and 10 μm) were also employed in DAR functional assays (data not shown), and we found that the concentration of ouabain did not significantly impact the observed D1 or D2 DAR functional results.
In functional assays with the D2L receptor, ouabain treatment of cells overexpressing the NKA restored dopamine-induced cAMP inhibition to levels comparable with D2L alone (Fig. 8A). Moreover, inhibition of endogenous NKA with ouabain enhanced D2L function, causing a greater decrease in cAMP levels as compared with vehicle-treated D2L (∼60% decrease in cAMP with ouabain treatment versus ∼40% with vehicle; Fig. 8A). Similar results were obtained when ouabain concentrations of 1 or 10 μm were used (data not shown), indicating that the potential toxicity of 5 mm ouabain treatment is not responsible for the observed decrease in cAMP levels. To ascertain whether the NKA had a similar influence on D1 receptor function, cells expressing both the D1 and NKA were treated with ouabain. In these experiments, inhibition of overexpressed NKA caused ∼15% increase in maximal D1 response as compared with cells that were not treated with ouabain (Fig. 8B). Likewise, inhibiting the endogenous NKA with ouabain also enhanced D1 function by ∼15% (Fig. 8B); similar results were obtained when 10 μm ouabain was used (data not shown). Complete Emax and EC50 data for D1 and D2 receptors are presented in Table 3. These results for both D1 and D2 receptors suggest that endogenous NKA in HEK293T cells is able to regulate transfected DAR function. Interestingly, although ouabain treatment was able to surmount the NKA-induced decrease in Emax, it did not alter the rightward shift in EC50 that was observed for both D1 and D2 receptors after NKA overexpression (Table 3).
FIGURE 8.
DAR function after ouabain inhibition of NKAα1. HEK293T cells were co-transfected with 15 μg of DAR (D2L (A) or D1 (B)) and 7.5 μg of either empty pcDNA vector or NKAα1. Cells were washed 24 h later and re-plated into 24-well plates at a density of 200,000 cells per well; assays were conducted 48 h post-transfection. Five min prior to agonist treatment, cells were placed into media with (test) or without (control) ouabain. Cells were then stimulated with forskolin and dopamine (A) or dopamine alone (B) for 10 min in the presence or absence of ouabain (OUA), and cAMP accumulation was measured as described under “Experimental Procedures.” Data were normalized for individual experiments as a percentage of the Emax value for DAR alone and are expressed as mean ± S.E. of five independent experiments, each conducted in triplicate; all statistical analyses were performed using one-way ANOVA with Bonferroni's post test. For D2L alone (A), raw values for Emax (maximal dopamine-induced inhibition) ranged from 6.6 to 10.1 pmol of cAMP/200,000 cells. For D1 alone (B), raw values for Emax ranged from 37.9 to 68.4 pmol of cAMP/200,000 cells. Complete Emax and EC50 data are summarized in Table 3.
TABLE 3.
D1 and D2 DAR regulation of cAMP production following NKA expression and ouabain treatment cAMP accumulation assays were performed as described in Fig. 7. D2 DAR data are reported as percent dopamine-induced inhibition following forskolin stimulation of cAMP accumulation. C.I. means confidence interval. All statistical analyses were determined by one-way ANOVA with Bonferroni's post test.
|
D2 DAR
|
D1 DAR
|
|||
|---|---|---|---|---|
| max ± S.E. | EC50 (C.I.) | max ± S.E. | EC50 (C.I.) | |
| % | nm | % | nm | |
| Control | 37.7 ± 1.1 | 5.6 (3.5–8.9) | 99.8 ± 1.1 | 165 (139–197) |
| Ouabain/control | 56.9 ± 0.9a | 8.7 (6.9–11.0) | 115.1 ± 1.1a | 155 (135–178) |
| +NKA | 18.4 ± 1.2a | 9.8 (3.9–24.8)b | 62.6 ± 1.0a | 361 (284–459)a |
| Ouabain/+NKA | 40.7 ± 1.1c | 23.9 (16.2–35.2)b | 76.0 ± 1.7a,c | 392 (286–538)a |
Values are p < 0.001 as compared with control
Values are p < 0.01 as compared with control
Values are p < 0.001 as compared with +NKA
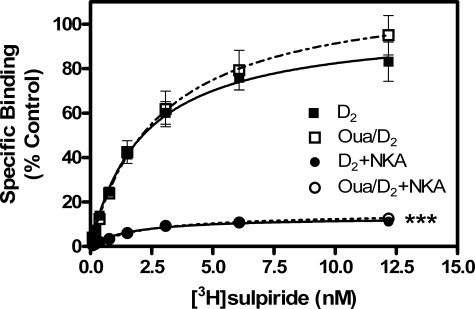
Total receptor binding assays were conducted in the presence and absence of ouabain to determine whether alterations in DAR density were responsible for the ouabain-induced increase in DAR function. Ouabain did not significantly increase D1 or D2 receptor density (Table 4); however, the treatment with ouabain was brief (15 min), and it is questionable that a significant increase in DAR number would occur in such a short time. Within this time frame, however, it might be possible to increase the surface expression of any intracellularly stored receptors, thus increasing the functional receptor pool. To investigate this possibility, the hydrophilic ligand [3H]sulpiride was used to determine the number of D2L receptors on the cell surface using intact cell binding assays (Fig. 9). No significant increase in cell-surface D2L expression was observed after a 15-min ouabain treatment for cells with either endogenous or overexpressed NKA (Fig. 9; summarized data in Table 4). These data imply that ouabain treatment does not increase the number of functional receptors at the cell surface.
TABLE 4.
Effect of ouabain on D1 and D2 DAR ligand binding parameters Radioligand binding assays for both D1 and D2 DARs were performed as described below. The maximum binding capacities (Bmax) were normalized to control values and are reported as percentages. All statistical analyses were performed by one-way analysis of variance with Bonferroni's post test.
|
D2
DARa
|
D1
DARb
|
|||
|---|---|---|---|---|
| % Bmax ± S.E. | KD ± S.E. | % Bmax ± S.E. | KD ± S.E. | |
| Control | 99.9 ± 0.8 | 23 ± 1 pm | 99.9 ± 1.3 | 113 ± 6 pm |
| Ouabain/control | 100.1 ± 0.6 | 51 ± 1 pmc | 99.9 ± 3.0 | 217 ± 21 pmc |
| +NKA | 15.3 ± 0.3d | 13 ± 2 pmc | 34.7 ± 0.4d | 88 ± 4 pm |
| Ouabain/+NKA | 25.4 ± 0.4d | 27 ± 2 pme | 48.2 ± 0.3d | 353 ± 7 pmc,e |
| Controlf | 100.0 ± 1.1 | 2.14 ± 0.07 nm | NDg | ND |
| Ouabain/controlf | 116.8 ± 1.0 | 2.80 ± 0.06 nm | ND | ND |
| +NKAf | 13.2 ± 0.2d | 1.61 ± 0.07 nmc | ND | ND |
| Ouabain/+NKAf | 15.1 ± 0.2d | 2.26 ± 0.10 nme | ND | ND |
D2 DAR binding in membrane preparations was determined with [3H]methylspiperone as described in Fig. 6
D1 DAR binding in membrane preparations was determined with [3H]SCH23390 as described in Fig. 6
Values are p < 0.01 as compared with control
Values are p < 0.001 as compared with control
Values are p < 0.01 as compared with +NKA
D2 DAR surface binding was determined with [3H]sulpiride using intact cell binding assays as described in Fig. 8
ND means not determined
FIGURE 9.
Cell-surface D2L DAR density following ouabain inhibition of NKAα1. HEK293T cells were co-transfected with 15 μg of D2L and 7.5 μg of either empty pcDNA vector or NKAα1. Cells were washed 24 h later and replated in 24-well plates at a density of 200,000 cells/well; assays were conducted 48 h post-transfection. Radioligand binding assays were performed by incubating cells concomitantly with [3H]sulpiride in the presence of vehicle or ouabain (OUA), as described under “Experimental Procedures.” Data were normalized for individual experiments as a percentage of the Bmax value for D2L alone and are expressed as mean ± S.E. of three independent experiments, each conducted in triplicate. All statistical analyses were performed using one-way ANOVA with Bonferroni's post-test; ***, p < 0.001 as compared with control. For D2L alone, raw values for Bmax ranged from 33.0 to 42.41 fmol/100,000 cells. Complete Bmax and KD data are reported in Table 4.
The possibility that either ouabain or dopamine could effectively alter the interaction between NKA and DAR was investigated by immunoprecipitating the DAR after drug treatment and probing Western blots with antibodies against the NKAα1 or the appropriate DAR. These experiments showed no apparent change in the relative amount of NKA, endogenous or overexpressed, interacting with the DAR after either drug treatment (data not shown). In addition, confocal microscopy was used to investigate the DAR-NKA interaction after ouabain or dopamine treatment. HEK293T cells were co-transfected with rat D2L-RFP and NKAα1-GFP, and images were obtained before and after a 15-min treatment with 5 mm ouabain (supplemental Fig. 3) or before and after a 10-min treatment with 10 μm dopamine (supplemental Fig. 4). Before ouabain treatment, both proteins clearly co-localize at the plasma membrane (supplemental Fig. 3A). Fifteen minutes after ouabain treatment, there was no change in D2 DAR or NKA distribution (supplemental Fig. 3B), indicating that the observed increase in DAR function is not caused by either a decrease in surface expression of NKA or an increase in surface expression of D2 DAR. Similarly, 10 min of dopamine treatment does not induce any observable changes in either NKA or D2L DAR distribution (supplemental Fig. 4, A and B), further indicating that dopamine treatment does not disrupt the DAR-NKA interaction. Taken together, these data imply that inhibiting NKA activity or stimulating the DARs does not disrupt the DAR-NKA interactions. More importantly these results indicate that NKA association with the D1 and D2 DARs leads to decreased signaling through a novel mechanism that is independent of receptor density or trafficking.
siRNA Inhibition of NKA Enhances DAR Activity—Because ouabain can have toxic effects in some cells, and because ouabain treatment only examines short term inhibition of the NKA, siRNA inhibition of NKAα1 was also utilized. HEK293T cells were co-transfected with FLAG-D2L and empty vector, FLAG-D2L and scrambled siRNA, or FLAG-D2L and anti-NKAα1 siRNA. After 48 or 72 h the FLAG-D2L was immunoprecipitated with anti-FLAG agarose and subjected to SDS-PAGE separation, and resulting Western blots were probed with antibody against the NKAα1. A 48-h treatment of cells with anti-NKAα1 siRNA essentially abolished the NKAα1-D2L interaction (Fig. 10). Because 48 h of treatment with siRNA was more effective at eliminating the NKA-DAR interaction than 72 h of treatment (Fig. 10), the 48-h time point was used for all subsequent experiments. The amount of total NKAα1 in whole cell lysates after siRNA treatment was also investigated. In these experiments, cells were transfected as above, and after 48 h, cell lysates were prepared and analyzed by SDS-PAGE followed by Western blotting. Probing Western blots for the NKAα1 subunit revealed an approximate 50% decrease in total levels of NKAα1 after inhibition with siRNA as compared with cells transfected with scrambled siRNA or empty vector (data not shown). This partial knockdown likely allows for better cell survival, as the NKA is critical for maintaining cellular ionic equilibrium. More importantly, the interaction between the NKA and the D2L DAR was essentially eliminated by siRNA inhibition (Fig. 10), thus facilitating specific investigation of the role of the NKA in the DAR signalplex without disrupting the overall cellular mechanics.
FIGURE 10.
Co-immunoprecipitation of NKAα1 with D2L DAR following siRNA treatment. HEK293T cells transiently expressing 5 μg of FLAG-D2L receptor were co-transfected with 3 μg of empty pcDNA vector, scrambled siRNA, or anti-NKAα1 siRNA for 48 or 72 h as indicated. Proteins were extracted, immunoprecipitated (IP) with anti-FLAG agarose, electrophoresed, and immunoblotted (IB) as described under “Experimental Procedures.” The blot was probed with a monoclonal anti-NKAα1 antibody and visualized by ECL after incubation with an HRP-conjugated anti-mouse secondary antibody. The arrow indicates the position of the NKAα1 subunit migrating at ∼95 kDa.
To determine the long term impact of NKA inhibition on DAR function, HEK293T cells were transfected with D2L receptor alone or D2L receptor with anti-NKAα1 siRNA and assessed for inhibition of cAMP after 48 h as described under “Experimental Procedures.” Inhibiting the NKAα1 with siRNA resulted in a 20% increase in dopamine-stimulated reduction of cAMP (Fig. 11A). The impact of siRNA-mediated NKAα1 inhibition on the D1 receptor was even more pronounced-an 80% increase in D1 function was observed after siRNA treatment (Fig. 11B). Total receptor binding experiments were also conducted to assess DAR densities after siRNA treatment. Although there was no significant change in D1 DAR density after NKA inhibition by siRNA (∼15% increase in D1 DAR Bmax as compared with scrambled siRNA; data not shown), it is noteworthy that the D2 DAR density was significantly decreased after anti-NKAα1 siRNA treatment (∼60% decrease as compared with scrambled siRNA; data not shown). The reason for these disparate results between D1 and D2 receptor densities is unclear, but it is quite remarkable that, despite the pronounced decrease in D2 density, there is still an increase in D2 DAR functional response following siRNA inhibition of NKAα1 expression. Because the NKAα1 protein is still present in the cells, but diminished in the DAR signalplex, these data further implicate a role for DAR-NKA protein interactions rather than signaling to mediate the observed functional results.
FIGURE 11.
DAR function following siRNA inhibition of NKAα1. HEK293T cells were co-transfected with 5 μg of DAR cDNA (D2L (A) or D1 (B)) and 3 μg of either scrambled siRNA vector or anti-NKAα1 siRNA. Cells were washed 24 h later and re-plated into 24-well plates at a density of 200,000 cells per well. Assays were conducted 48 h post-transfection, and cAMP accumulation was measured as described under “Experimental Procedures.” Data were normalized for individual experiments as a percentage of the Emax value for DAR alone and are expressed as mean ± S.E. of four independent experiments conducted in triplicate; curves represent single experiments. All statistical analyses were performed on pooled data using one-way ANOVA with Bonferroni's post-test. For D2L alone (A), raw values for Emax (maximal dopamine-induced inhibition) ranged from 5.3 to 7.5 pmol of cAMP/200,000 cells; normalized Emax (% inhibition) values were 35.4 ± 1.7% for D2L alone and 50.1 ± 2.9% for D2L + NKAα1 siRNA (*, p < 0.05). EC50 values were 2.2 nm for D2L alone and 8.1 nm for D2L+ NKAα1 siRNA. For D1 alone (B), raw values for Emax ranged from 11.3 to 32.4 pmol of cAMP/200,000 cells; normalized Emax values were 99.8 ± 2.0% for D1 alone and 209.0 ± 1.9% for D1 + NKAα1 siRNA (***, p < 0.001). EC50 values were 95.5 nm for D1 alone and 173.3 nm for D1 + NKAα1 siRNA (p < 0.05).
NKA Activity Is Modulated by DAR Expression—Endogenous NKA activity in HEK293T cells was investigated after expression of D1 and D2 receptors to determine the impact of the DARs on NKA function. Interestingly, expressing the D2 receptor alone caused a significant decrease (∼50%) in NKA function when compared with untransfected cells as measured by rubidium-86 ion (86Rb+) uptake (Fig. 12). Notably, as mentioned previously, total lysate and co-immunoprecipitation experiments demonstrated that there was no change in NKAα1 protein expression after transfection of the D2 receptor (supplemental Fig. 1), indicating that a decrease in NKA protein number does not mediate this decrease in function. Treatment of D2-expressing cells with 10 μm dopamine had no significant impact on 86Rb+ uptake as compared with either untransfected cells or vehicle-treated D2 cells (Fig. 12). Because the D2 receptor exhibits weak ligand-independent signaling (35, 36), constitutive activity was investigated as a possible mechanism for the D2-mediated decrease in NKA activity. Cells expressing the D2 receptor were treated with pertussis toxin to block the downstream signaling cascade, and NKA activity was measured. A 16-h pertussis toxin treatment completely eliminates D2L-mediated cAMP regulation in the HEK293 cells (data not shown) but had no impact on NKA activity (Fig. 12), indicating that constitutive activity of the D2 receptor was not responsible for the observed decrease in D2 function. These data reveal that the D2 receptor protein can decrease NKA activity in a ligand- and signal-independent manner, further suggesting a functional role for the putative DAR-NKA complex.
FIGURE 12.
NKA function following D2L DAR transfection. HEK293T cells were transfected with 7.5 μg of either D2L cDNA or empty pcDNA vector (UTs). Cells were washed 24 h later, and for cells involving pertussis toxin treatment (PTX), 125 ng/ml PTX was added to media 16 h prior to experimentation. 48 h post-transfection, cells were removed into assay buffer and 86Rb+ uptake, in the presence or absence of dopamine (DA), was measured as described under “Experimental Procedures.” Data were normalized for individual experiments as a percentage of the Emax value for UT cells alone and are expressed as mean ± S.E. of five independent experiments, each conducted in duplicate. All statistical analyses were performed using one-way ANOVA with Bonferroni's post-test; *, p < 0.05; **, p < 0.01. UT raw Emax values ranged from 109 to 219 pmol/mg protein/min.
Expression of the D1 receptor caused a similar decrease in NKA function (∼40%) as compared with untransfected cells (Fig. 13). Conversely, NKA-mediated 86Rb+ uptake was significantly increased in cells expressing the D1 receptor when treated with dopamine (Fig. 13). To see if this effect was second messenger-mediated, we attempted to inhibit adenylyl cyclase or PKA activities; however, all tested inhibitors independently altered NKA activity. In contrast, treatment with dibutyryl-cAMP, a cAMP analog and activator of PKA, did not alter NKA activity indicating that the Gαs signaling cascade downstream of D1 DAR activation is not solely responsible for the increase in NKA function (data not shown). Based upon these data, it cannot be determined whether the dopamine-stimulated increase in NKA activity (Fig. 13) is mediated by the D1 signaling cascade or by alterations in protein-protein interactions induced by dopamine binding to the receptor.
FIGURE 13.
NKA function following D1 DAR transfection. HEK293T cells were transfected with 5 μg of either D1 cDNA or empty pcDNA vector (UTs). Cells were washed 24 h later. 48 h post-transfection, cells were removed into assay buffer, and 86Rb+ uptake, in the presence or absence of dopamine (DA), was measured as described under “Experimental Procedures.” Data were normalized for individual experiments as a percentage of the Emax value for UT cells alone and are expressed as mean ± S.E. of five independent experiments, each conducted in duplicate. All statistical analyses were performed using one-way ANOVA with Bonferroni's post-test; *, p < 0.05; **, p < 0.01. UT raw Emax values ranged from 106 to 174 pmol/mg protein/min.
DISCUSSION
In this study, we used mass spectroscopy analysis to identify the NKAα1 subunit as an interacting protein in both the D1 and D2 DAR signalplexes. Although previous investigations have shown that dopamine can signal through its receptors to regulate NKA activity (15–17, 20–23), this is the first demonstration that the two proteins exist in a complex. This novel discovery of a DAR-NKA interaction was confirmed by co-immunoprecipitation and Western analyses in HEK293T cells, and was further substantiated by verifying the interaction in brain tissue between native DARs and NKA. Previous studies have shown that alterations in NKA function can occur via insertion or removal of NKA at the plasma membrane (15, 17, 19–23). However, confocal microscopy revealed that, in this system, both proteins predominantly co-localized at the plasma membrane rather than existing in separate vesicular pools implicating a mechanism apart from protein trafficking or endosomal recycling pathways as the mechanism for functional alterations in both proteins. At present, the precise location of the interaction between the NKA and DARs is unclear. Complete truncation of the carboxyl terminus of the D1 receptor, a common site for D1 protein-protein interactions (30–32), did not diminish the D1-NKA association. As both receptor and NKA proteins have multiple transmembrane-spanning domains, it is quite possible that their interactions occur within the plasma membrane regions. Alternatively, other nonmembranous regions of the proteins may mediate their interactions. These possibilities are currently under investigation.
Functional studies of the DAR-NKA interactions in HEK293T cells revealed diminished DAR signaling in the presence of overexpressed NKA with a concomitant decrease in total DAR expression. Inhibiting the overexpressed NKA with the cardiotonic steroid ouabain restored DAR function. Interestingly, ouabain inhibition of endogenous NKA also enhanced DAR signaling, indicating that the transfected DARs can be regulated by endogenous levels of NKA. Ouabain treatment did not increase total or cell-surface DAR expression, demonstrating that the amplified DAR signal was not caused by an increase in the number of functional DARs. Moreover, confocal microscopy and co-immunoprecipitation assays indicated that ouabain treatment did not disrupt the DAR-NKA interaction or cause endocytosis or membrane insertion of either protein. These data indicate that ouabain treatment impacts the DARs already at the plasma membrane, possibly by inducing compensatory conformational changes within the receptor structure upon ouabain binding to NKA.
To ensure that ouabain did not produce global cellular alterations as the mechanism for enhancing DAR function, siRNA treatment was employed as an alternate means to inhibit the NKA. Anti-NKAα1 siRNA decreased total cellular NKAα1 levels by ∼50% but reduced the amount of NKA interacting with the D2 DAR below detectable limits. This allowed examination of the role of NKA in the DAR signalplex over a longer period of time while still maintaining normal ionic balance in the cells. Transfection with anti-NKAα1 siRNA did not change the total D1 receptor density, but D1 receptor-stimulated cAMP accumulation approximately doubled after the siRNA inhibition of NKAα1. Interestingly, NKAα1 siRNA treatment dramatically and significantly diminished total D2 receptor density, but despite the reduction in D2 receptor number, there was a significant increase in D2 DAR function. Although the reason for the decrease in D2 receptor number is unknown, it is clear that loss of the NKA-DAR interaction can increase the D2 DAR signal, even when D2 DAR numbers are reduced. These data provide further evidence that endogenous NKA interactions can modulate DAR function. Of note, these data show that the NKA-DAR interaction, and not just the presence of the NKA protein in the cell, is critical for DAR functional regulation. More importantly, these data provide the first evidence for NKA control of DAR function and support a protein interaction mechanism, rather than downstream signaling molecules, for the novel NKA modulation of DARs.
Not only did NKA modify the DAR signal, but reciprocal modulation of NKA by DARs also occurred. Transfecting either the D1 or D2 DAR into HEK293T cells caused a marked decrease in NKAα1 activity without diminishing the NKA protein levels. The ability of DARs to diminish NKA function in the absence of ligand and without altering NKA numbers further supports the importance of the interacting complex. Because the D2 receptor exhibits constitutive activity (35, 36), pertussis toxin was used to inhibit the Gi/o signaling cascade and thus eliminate ligand-independent G-protein signaling as a potential mechanism for modulation of NKA activity. Pertussis toxin had no impact on D2 DAR modulation of NKA activity, indicating that the D2 protein itself, and not a second messenger, was responsible for the diminished NKA function. Dopamine binding to the D2 receptor did not further modify NKA activity, but dopamine stimulation of D1 receptors did significantly increase NKA function. It is possible that signaling downstream of the D1 receptor was responsible for amplifying NKA activity. To examine this possibility, PKA and adenylyl cyclase inhibitors were used to block the Gs signaling cascade. However, all tested inhibitors altered NKA function independent of the D1 DAR. Consequently, it remains unclear whether the dopamine-stimulated increase in NKA activity is mediated by the D1 signaling cascade or by alterations in protein-protein interactions induced by dopamine binding to the receptor. It is clear from our data that neither constitutive activity of DARs nor alterations in NKA number following DAR expression are responsible for the observed decrease in NKA activity. Moreover, confocal microscopy and co-immunoprecipitation assays indicated that dopamine treatment does not disrupt the DAR-NKA interaction or cause endocytosis or membrane insertion of either protein. This is significant in that prior studies have shown that dopaminergic control of NKA function occurs via NKA insertion or removal at the plasma membrane in a tissue-specific manner (15, 17, 19, 20–23). As such, it is likely that conformational changes occurring within the DAR-NKA protein complex mediate the observed decrease in NKA function. In addition, D1 receptor conformational changes induced upon dopamine binding may cause compensatory conformational changes in the NKA protein that further alter NKA activity.
Taken together, these data suggest a mechanism for reciprocal modulation of function between DARs and the NKA. The presented data show that NKA, when interacting with the DAR, decreases the DAR function. Similarly, expression of DARs decreases NKA activity without decreasing NKA expression. These data suggest a model wherein the co-existence of the two proteins in the signalplex causes a reciprocal dampening of each others' function (Fig. 14A). This could be caused by structural hindrance between the two proteins, resulting in diminished G-protein coupling to the receptor and ATP hydrolysis by the NKA. Within the signalplex, a conformational change in one protein likely induces a compensatory change in neighboring proteins. For the D1 receptor, the dopamine-binding event may induce conformational changes in the NKA promoting ATP hydrolysis and ion transport across the membrane (Fig. 14B). It is also possible that second messengers within the D1 signaling cascade and changes in the protein-protein interactions induced by dopamine binding both play a role in enhancing NKA function. Finally, inhibition of NKA within the DAR signalplex, using either cardiotonic steroids or siRNA, enhances DAR function. Because NKA activity is already reduced while interacting with the DARs, it is likely that a mechanism secondary to NKA inhibition is underlying the observed increase in DAR activity. As such, we hypothesize that ouabain binding to the NKA induces conformational changes in the proteins, thus allowing better coupling of the DAR to G-proteins (Fig. 14C). Although this model depicts a direct interaction between just the DAR and NKA, the possibility of an additional linker or tethering protein holding the DAR and NKA in close physical proximity cannot be eliminated at this time. Regardless, the apparent significance of this DAR-NKA complex and the observed reciprocal functional modulation of each protein is quite noteworthy.
FIGURE 14.
Model of DAR-NKA interactions as illustrated for the D1 DAR. A, close proximity of NKA and DAR, via protein-protein interactions, causes conformational hindrance of G-protein coupling to the receptor. This in turn diminished the downstream signaling cascade following ligand stimulation. B, binding of dopamine (DA) to the D1 DAR results in conformational changes to the DAR that cause compensatory conformational changes in the NKA. These changes to the pump allow enhanced transport of Na+ and K+ across the plasma membrane. C, ouabain binding to the NKA results in conformational changes to the sodium pump, enabling enhanced coupling between DAR and G-proteins. This causes an increase in the downstream signaling cascade upon dopamine stimulation.
In summary, interaction between the D1 or D2 DARs and NKA results in a reciprocal modulation of function between the two proteins, both in the presence and absence of ligands. This is the first demonstration of an interaction between these proteins, and this complex reveals the possibility of new mechanisms for functional regulation of both DARs and NKA. Physiologically, this interaction likely provides strict and immediate control of the cellular response to external stimuli by limiting the functional capacity of each protein when they are in complex. Although in complex with one another, inhibition of the NKA by endogenous cardiotonic steroids may enhance the native DAR signal, and dopamine signaling at the D1 receptor can activate the NKA. Within the brain, this NKA activation may be necessary for restoring resting membrane potential of neurons, and the amplified DAR signal may be critical for periods of enhanced responsiveness to dopaminergic cues. Further understanding the significance of this interaction in neuronal and other physiological environments will provide insight into both the DAR signalplex and native cellular control mechanisms.
Supplementary Material
Acknowledgments
We thank Dr. Alejandro Bertorello for advice and for the generous gift of the NKAα1-eGFP construct, Dr. Vanitha Ramakrishnanfor the HEK293tsa201 cells, and Jill Neiman, Steven Quinn, and Stephanie Gallanie for excellent technical assistance.
This work was supported, in whole or in part, by National Institutes of Health grant (Intramural Research Program of NINDS). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
Footnotes
The abbreviations used are: DAR, dopamine receptor; NKA, Na+,K+-ATPase; siRNA, small interfering RNA; DRIP, dopamine receptor-interacting protein; EBSS, Earle's balanced salt solution; MS, mass spectrometry; PKA, cAMP-dependent protein kinase; ANOVA, analysis of variance; eGFP, enhanced green fluorescent protein; RFP, red fluorescent protein; HRP, horseradish peroxidase; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid; BisTris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
References
- 1.El-Ghundi, M., O'Dowd, B. F., and George, S. R. (2007) Rev. Neurosci. 18 37-66 [DOI] [PubMed] [Google Scholar]
- 2.Goldman-Rakic, P. S. (1998) Adv. Pharmacol. 42 707-711 [DOI] [PubMed] [Google Scholar]
- 3.Missale, C., Nash, S. R., Robinson, S. W., Jaber, M., and Caron, M. G. (1998) Physiol. Rev. 78 189-225 [DOI] [PubMed] [Google Scholar]
- 4.Schultz, W. (2002) Neuron 36 241-263 [DOI] [PubMed] [Google Scholar]
- 5.Sealfon, S. C., and Olanow, C. W. (2000) Trends Neurosci. 23 S34-S40 [DOI] [PubMed] [Google Scholar]
- 6.Jose, P. A., Eisner, G. M., and Felder, R. A. (2003) Nephron Physiol. 95 19-27 [DOI] [PubMed] [Google Scholar]
- 7.Tonini, M., Cipollina, L., Poluzzi, E., Crema, F., Corazza, G. R., and De Ponti, F. (2004) Aliment. Pharmacol. Ther. 19 379-390 [DOI] [PubMed] [Google Scholar]
- 8.Zeng, C., Yang, Z., Asico, L. D., and Jose, P. A. (2007) Cardiovasc. Hematol. Agents Med. Chem. 5 241-248 [DOI] [PubMed] [Google Scholar]
- 9.Bockaert, J., Roussignol, G., Becamel, C., Gavarini, S., Joubert, L., Dumuis, A., Fagni, L., and Marin, P. (2004) Biochem. Soc. Trans. 32 851-855 [DOI] [PubMed] [Google Scholar]
- 10.Kabbani, N., and Levenson, R. (2007) Eur. J. Pharmacol. 572 83-93 [DOI] [PubMed] [Google Scholar]
- 11.Neve, K. A. (2005) Mol. Pharmacol. 68 275-278 [DOI] [PubMed] [Google Scholar]
- 12.Bergson, C., Levenson, R., Goldman-Rakic, P. S., and Lidow, M. S. (2003) Trends Pharmacol. Sci. 24 486-492 [DOI] [PubMed] [Google Scholar]
- 13.Moseley, A. E., Williams, M. T., Schaefer, T. L., Bohanan, C. S., Neumann, J. C., Behbehani, M. M., Vorhees, C. V., and Lingrel, J. B (2007) J. Neurosci. 27 616-626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dobretsov, M., and Stimers, J. R. (2005) Front. Biosci. 10 2373-2396 [DOI] [PubMed] [Google Scholar]
- 15.Barnard, M. L., Olivera, W. G., Rutschman, D. M., Bertorello, A. M., Katz, A. I., and Sznajder, J. I. (1997) Am. J. Respir. Crit. Care Med. 156 709-714 [DOI] [PubMed] [Google Scholar]
- 16.Bertorello, A. M., Hopfield, J. F., Aperia, A., and Greengard, P. (1990) Nature 347 386-388 [DOI] [PubMed] [Google Scholar]
- 17.Brismar, H., Asghar, M., Carey, R. M., Greengard, P., and Aperia, A. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 5573-5578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishi, A., Fisone, G., Snyder, G. L., Dulubova, I., Aperia, A., Nairn, A. C., and Greengard, P. (1999) J. Neurochem. 73 1492-1501 [DOI] [PubMed] [Google Scholar]
- 19.Efendiev, R., Yudowski, G. A., Zwiller, J., Leibiger, B., Katz, A. I., Berggren, P. O., Pedemonte, C. H., Leibiger, I. B., and Bertorello, A. M. (2002) J. Biol. Chem. 277 44108-44114 [DOI] [PubMed] [Google Scholar]
- 20.Ridge, K. M., Dada, L., Lecuona, E., Bertorello, A. M., Katz, A. I., Mochly-Rosen, D., and Sznajder, J. I. (2002) Mol. Biol. Cell 13 1381-1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bertorello, A. M., and Sznajder, J. I. (2005) Am. J. Respir. Cell Mol. Biol. 33 432-437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogimoto, G., Yudowski, G. A., Barker, C. J., Kohler, M., Katz, A. I., Feraille, E., Pedemonte, C. H., Berggren, P. O., and Bertorello, A. M. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 3242-3247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teixeira, V. L., Katz, A. I., Pedemonte, C. H., and Bertorello, A. M. (2003) Ann. N. Y. Acad. Sci. 986 587-594 [DOI] [PubMed] [Google Scholar]
- 24.Heinzel, S. S., Krysan, P. J., Calos, M. P., and DuBridge, R. B. (1988) J. Virol. 62 3738-3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Namkung, Y., and Sibley, D. R. (2004) J. Biol. Chem. 279 49533-49541 [DOI] [PubMed] [Google Scholar]
- 26.Monsma, F. J., Jr., Mahan, L. C., McVittie, L. D., Gerfen, C. R., and Sibley, D. R. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 6723-6727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gardner, B., Liu, Z. F., Jiang, D., and Sibley, D. R. (2001) Mol. Pharmacol. 59 310-321 [DOI] [PubMed] [Google Scholar]
- 28.Kim, O. J., Gardner, B. R., Williams, D. B., Marinec, P. S., Cabrera, D. M., Peters, J. D., Mak, C. C., Kim, K. M., and Sibley, D. R. (2004) J. Biol. Chem. 279 7999-8010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Free, R. B., Hazelwood, L. A., Cabrera, D. M., Spalding, H. N., Namkung, Y., Rankin, M. L., and Sibley, D. R. (2007) J. Biol. Chem. 282 21285-21300 [DOI] [PubMed] [Google Scholar]
- 30.Pei, L., Lee, F. J., Moszczynska, A., Vukusic, B., and Liu, F. (2004) J. Neurosci. 24 1149-1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woods, A. S., Ciruela, F., Fuxe, K., Agnati, L. F., Lluis, C., Franco, R., and Ferre, S. (2005) J. Mol. Neurosci. 26 125-132 [DOI] [PubMed] [Google Scholar]
- 32.Zhang, J., Vinuela, A., Neely, M. H., Hallett, P. J., Grant, S. G., Miller, G. M., Isacson, O., Caron, M. G., and Yao, W. D. (2007) J. Biol. Chem. 282 15778-15789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Therien, A. G., and Blostein, R. (2000) Am. J. Physiol. 279 C541-C566 [DOI] [PubMed] [Google Scholar]
- 34.Wasserstrom, J. A., and Aistrup, G. L. (2005) Am. J. Physiol. 289 H1781-H1793 [DOI] [PubMed] [Google Scholar]
- 35.Heusler, P., Newman-Tancredi, A., Castro-Fernandez, A., and Cussac, D. (2007) Neuropharmacology 52 1106-1113 [DOI] [PubMed] [Google Scholar]
- 36.Strange, P. G. (1999) Clin. Exp. Pharmacol. Physiol. 26 S3-S9 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.