Abstract
We report the isolation of a novel vitamin K-dependent protein from the calcified cartilage of Adriatic sturgeon (Acipenser nacarii). This 10.2-kDa secreted protein contains 16 γ-carboxyglutamic acid (Gla) residues in its 74-residue sequence, the highest Gla percent of any known protein, and we have therefore termed it Gla-rich protein (GRP). GRP has a high charge density (36 negative + 16 positive = 20 net negative) yet is insoluble at neutral pH. GRP has orthologs in all taxonomic groups of vertebrates, and a paralog (GRP2) in bony fish; no GRP homolog was found in invertebrates. There is no significant sequence homology between GRP and the Gla-containing region of any presently known vitamin K-dependent protein. Forty-seven GRP sequences were obtained by a combination of cDNA cloning and comparative genomics: all 47 have a propeptide that contains a γ-carboxylase recognition site and a mature protein with 14 highly conserved Glu residues, each of them being γ-carboxylated in sturgeon. The protein sequence of GRP is also highly conserved, with 78% identity between sturgeon and human GRP. Analysis of the corresponding gene structures suggests a highly constrained organization, particularly for exon 4, which encodes the core Gla domain. GRP mRNA is found in virtually all rat and sturgeon tissues examined, with the highest expression in cartilage. Cells expressing GRP include chondrocytes, chondroblasts, osteoblasts, and osteocytes. Because of its potential to bind calcium through Gla residues, we suggest that GRP may regulate calcium in the extracellular environment.
Vitamin K-dependent proteins are characterized by the presence of several γ-carboxyglutamic acid residues (Gla)3 resulting from vitamin K-dependent post-translation modifications of specific glutamates (Glu) by the γ-glutamyl carboxylase (1, 2). The identification in 1974 of the new amino acid Gla in prothrombin (3) opened the way for subsequent identifications of other Gla containing proteins in blood, all involved in coagulation (factors VII, IX, and X, and proteins C, S, and Z) and containing from 9 to 13 Gla residues (4). Lack of adequate γ-carboxylation was found to prevent Gla-mediated calcium binding and thus inhibit clot formation, highlighting the importance of Gla for normal coagulation process (5). Finding high levels of Gla in non-hepatic tissues such as bone led to the discovery of a different group of Gla-containing proteins not involved in hemostasis, e.g. osteocalcin (OC), characterized in 1976 (6, 7), and matrix Gla protein (MGP), first described in 1983 (8). Both proteins contain 3 to 5 Gla residues, appear to play a role in tissue mineralization, and form a distinct group within the VKD protein family (9). Later, additional VKD proteins were identified and found to be involved in diverse biological functions such as growth control and apoptosis (growth arrest-specific protein 6, Gas6) (10), and signal transduction (proline-rich Gla proteins 1, 2, 3, and 4, PRGP1-4) (11, 12). Although VKD proteins were initially identified in vertebrates, the presence of a γ-glutamyl carboxylase in Drosophila melanogaster and the identification of several of its substrates in marine snails from the genus Conus (13-15), together with the ubiquity of γ-glutamyl carboxylase expression in mammalian tissues (16), suggests a wider prevalence of γ-carboxylation, with a broad range of biological roles. We hereby report the purification and characterization of a new Gla-containing protein from the calcified cartilage of sturgeon, an ancient bony fish with a cartilaginous endoskeleton and in which bone is found essentially in the ganoid plaques forming the exogenous skeleton. An in silico approach, coupled with a cloning strategy, identified orthologs of this protein in all vertebrate groups, including humans. Its unprecedented high number of Gla residues and high evolutionary conservation suggests a fundamental function.
EXPERIMENTAL PROCEDURES
Biological Material—Specimens of Acipencer naccarii (Adriatic sturgeon) were kindly supplied by aquaculture Rio Frio (Granada, Spain), and kept until used at 20-22 °C in a closed circuit equipped with a biological filter and a natural photoperiod. Fish were fed once a day with commercial pellets. Frozen branchial arches from adult sturgeon specimens were also obtained from aquaculture Rio Frio. Samples of Rattus norvegicus (Norway rat) were obtained from Prof. José Belo from the University of Algarve.
GRP Extraction and Purification—In a typical purification, non-collagenous proteins were extracted from 150 g of ground branchial arches using a 10-fold excess of 10% formic acid for 4 h at 4 °C as described (17). The extracted proteins were separated from the insoluble collagenous matrix by filtration through filter paper and next dialyzed at 4 °C against 50 mm HCl using 3500 molecular weight tubing (Spectra-Por 3, Spectrum, Gardena, CA) with 4 changes of medium over 2 days to remove all dissolved mineral. The dialyzed extract was freezedried, dissolved in 6 m guanidine HCl, 0.1 m Tris, pH 9, and redialyzed against 5 mm ammonium bicarbonate to precipitate GRP. A portion of this solution was dialyzed against 50 mm HCl, dried, and aliquots of the resulting protein precipitate were analyzed by SDS-PAGE. The protein profile was revealed by staining either with Coomassie Brilliant Blue (Bio-Rad), or 4-diazobenzenesulfonic acid (Gla-specific stain; 8.5 mm 4-diazobenzene sulfonic acid (Sigma), as described (18). The crude precipitate was further purified by reverse phase-high performance liquid chromatography (RP-HPLC) to obtain pure GRP.
Reverse-phase HPLC—The crude protein precipitate was dissolved in 0.1 m Tris, 6 m guanidine HCl, pH 9, and aliquots were injected onto a Vydac C18 reverse-phase HPLC column (4.6 mm inner diameter × 25-cm length) equilibrated in 0.1% trifluoroacetic acid in water and at a flow rate of 1 ml/min (initial conditions). The HPLC was run for 7 min at initial conditions, and then proteins were eluted from the column with a 1.5-h linear gradient to 0.1% trifluoroacetic acid in 60% acetonitrile. Fractions of 1 ml were collected and their absorbance determined at 220 nm.
Sephadex G-75—Fractions from reverse-phase HPLC purification containing GRP were freeze-dried, re-dissolved in 0.5 ml of 50 mm HCl, and purified over a 25-ml Sephadex G-75 column equilibrated in 50 mm HCl. Fractions of 0.5 ml were collected and absorbance determined at 220 nm.
Amino Acid Analysis—Protein samples were dried and subjected to acid and alkaline hydrolysis as described (7, 8). Resulting acid hydrolysate was dried, whereas alkaline hydrolysate was titrated with 70% HClO4 to pH 7 (10). Both hydrolysates were then analyzed using a Biochrom 30-amino acid analyzer at the University of California, San Diego, Biochemical Genetics Research Facility.
N-terminal Protein Sequencing—Automatic Edman degradations were performed using an Applied Biosystems 494 sequencer equipped with a model 140C on-line HPLC system and employing the standard program supplied by the manufacturer. Phenylthiohydantoin-derivatives were separated using a 2.1 mm × 22 cm C-18 reverse-phase HPLC column (Applied Biosystems, Foster City, CA) and gradient conditions recommended by the manufacturer. They were detected by their absorbance at 269 nm.
RNA and DNA Preparation—Total RNA was extracted from sturgeon adult tissues (including bony and cartilaginous tissues, and major soft tissues), gilthead seabream (gills), European seabass (gills), and Norway rat (outer ear, heart, and femur) as described by Chomczynski and Sacchi (19). RNA integrity was checked by agarose-formaldehyde gel electrophoresis, and concentration was determined by spectrophotometry at 260 nm (GeneQuant, GE Healthcare). Sturgeon genomic DNA was extracted from muscular tissue according to a previously described procedure (20).
cDNA Cloning—Based on the N-terminal sequence of the sturgeon protein, a degenerated forward primer NGP_GN1F was designed and used in combination with universal adapter primer to amplify by PCR the 3′ ends of sturgeon, seabream, and seabass GRP cDNAs from reverse-transcribed RNA (using Moloney murine leukemia virus-reverse transcriptase, Invitrogen). The 5′ ends of those cDNAs were amplified by RACE PCR using specific reverse primers AnGRP1R, SaGRP1R, or DlGRP1R, and species-specific Marathon cDNA libraries (Clontech, Mountain View, CA) prepared according to the manufacturer's instructions from 1 μg of purified poly(A+) RNA (from a mixture of total RNA derived from soft and calcified tissues). The rat GRP cDNA was amplified by reverse transcriptase-PCR from total RNA of outer ear using specific primer RnGRP_ORF1F (designed according to EST sequences retrieved from GenBank™ sequence data base) and universal adapter primer. The rat MGP and OC cDNAs were amplified by reverse transcriptase-PCR from total RNA of heart and femur, respectively, using specific primers RnMGP_F/Rn-MGP_R and RnOC_F/RnOC_R, designed according to MGP and OC sequences retrieved from the GenBank™ sequence data base (accession numbers NM 012862 and NM 013414, respectively). Sequences of all PCR primers used in this study are presented in supplemental Table S1. All PCR products were cloned into pCRIITOPO (Invitrogen) and sequenced on both strands (Macrogen, South Korea).
Gene Cloning—Two GenomeWalker libraries (StuI and PvuII) were constructed using the Universal GenomeWalker kit (Clontech, Mountain View, CA), according to the manufacturer's recommendations. Sturgeon GRP gene was amplified using as template GenomeWalker libraries and genomic DNA, and primers constructed based first on cDNA sequence and later also on intronic sequences as they became available. Genomic DNA was used to obtain the following partial fragments: exon 1 to 3 (partial) with primers GWAnGRP1F (exon 1) and GWAnGRP1R (exon 3); exon 3 with primers AnGRP_ GBFI (intron 1) and GRPIntIIIR (intron 3); exon 4 to 5 with primers AnGRP_B1R (exon 5) and GRPIntIIIF (intron 3). Exon 4 (partial) and intron 3 (partial) were obtained with primers GWAnGRP2R and AP1 with the StuI library.
Measurement of Relative Gene Expression by Quantitative Real-time PCR—One microgram of total RNA was treated with RQ1 RNase-free DNase (Promega, Madison, WI) and reverse transcribed at 37 °C with Moloney murine leukemia virus-reverse transcriptase using specific reverse primers AnGRP1R, AnGAPDH_RT1R, and AnHPRTI_RT1R (supplemental Table S1). Quantitative real-time PCR (qPCR) was performed with an iCycler iQ apparatus (Bio-Rad), using primer sets AnGRP1F/AnGRP1R to amplify sturgeon GRP, AnGAPDH_RT1F/AnGAPDH_RT1R to amplify sturgeon glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and AnHPRTI_RT1F/AnHPRTI_RT1R to amplify sturgeon hypoxanthine phosphoribosyltransferase 1 (HPRT1). PCR, set up in duplicates, were as follows: 2 μl of cDNA (from reverse transcriptase diluted 1:10), 8 μl of primer mixture (each primer at 0.2 μm final concentration), and 10 μl of Absolute QPCR SYBR Green Fluorescein mixture (ABgene, Epsom, UK). PCR were submitted to an initial denaturation step at 95 °C for 15 min and 55 cycles of amplification (one cycle is 30 s at 95 °C and 30 s at 68 °C). Fluorescence was measured at the end of each extension cycle in the FAM-490 channel and melting profiles of each reaction were performed to check for unspecific product amplification. Levels of gene expression were calculated using the comparative method (ΔΔCt) and normalized using gene expression levels of GAPDH or HPRT1 housekeeping genes. Gene expression in heart was set to 1 and used as reference for relative expression in other tissues. qPCR was performed in quadruplicates and a normalized standard deviation was calculated. Sequences of all qPCR primers used in this study are presented in supplemental Table S1.
In Situ Hybridization—Samples were collected in freshly prepared sterile 4% paraformaldehyde solution, at 4 °C, dehydrated with increasing methanol concentrations, embedded in paraffin or decalcified for 1 week in sterile-buffered EDTA (0.3 m EDTA, 0.15 m NaCl, 0.1 m Tris-HCl, pH 7.6), sectioned (6-8 μm thick), and finally collected in 3-aminopropyltriethoxysilane (Sigma) coated slides. A 422-bp fragment of sturgeon GRP cDNA (spanning from nucleotide 217 to 639) and a 417-bp fragment of rat GRP cDNA (spanning from nucleotide 417 to the 3′ end) cloned in pCRII-TOPO were either linearized with ApaI and transcribed with SP6 RNA polymerase to generate an antisense riboprobe, or linearized with KpnI and transcribed with T7 RNA polymerase to generate a sense riboprobe. A 480-bp fragment of rat MGP cDNA (spanning from nucleotide 7 to 487) and a 481-bp fragment of rat OC cDNA (spanning from nucleotide 7 to 488) cloned in pCRII-TOPO were linearized with ApaI and KpnI, respectively, and transcribed with SP6 RNA polymerase to generate antisense riboprobes. Probes were then labeled with digoxigenin using the RNA labeling kit (Roche Applied Sciences) according to the manufacturer's instructions. Slides were hybridized with riboprobes as previously described (17, 20). Briefly, sections were digested with 40 μg/ml proteinase K (Sigma) in 1× phosphate-buffered saline containing 0.1% of Tween 20 (Sigma) for 30 min and then hybridized at 68 °C overnight in a humidified chamber. After hybridization, sections were washed and the signal revealed with the alkaline phosphatase-coupled antidigoxigenin-AP antibody (Roche) and nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate substrate solution (Sigma) as described (17, 20). Negative controls for GRP mRNA detection were performed with sense probes.
Morphometric Analysis—A total of 200 to 500 cells from each category (immature chondrocytes, i.e. cells without vacuoles, mature chondrocytes and chordoblasts) were counted from each of 3 consecutive sections of sturgeon vertebra, and used to estimate the percentage of cells expressing GRP, within the same defined area. For rat morphometry analysis, the chondrocytes were divided into 2 groups according to their developmental stage: immature chondrocytes, which included immature and proliferating chondrocytes; and mature chondrocytes, which included mature, columnar, and hypertrophic chondrocytes. A total of 500 cells of each defined group were counted from each of four consecutive sections of rat ribs, and used to estimate the ratio of cells expressing GRP as percentage of total cells of each group, counted in the same defined area. The ratio of rat osteocytes expressing GRP was obtained from 4 consecutive sections of tail, using the same procedure.
Sequence Reconstruction, Alignment, and Analysis—GenBank EST data base and Trace archives, as well as Ensembl genomes (release 45) were searched using BLAST (www.ncbi.nlm.nih.gov) for sequences showing similarities to sturgeon transcript. Species-specific sequences were first clustered, and elements of each cluster were assembled using the ContigExpress module from Vector NTI version 9 (Invitrogen) to generate, after manual correction, highly accurate consensus sequences. Virtual transcripts and genes were deduced from the joined consensus sequence using stringent overlap criteria. Virtual gene structure and splicing sites were predicted using comparative methods and GenScan. Separate alignments of GRP mature and precursor peptides were created using M-Coffee multiple sequence alignment software (21) with parameters set to default. Manual adjustments were made in a few cases to improve alignments. Sequence logos presented were created using WebLogo (22). The sequence logos are presented as graphical displays, where the height of each letter is made proportional to its frequency. This shows the conserved residues as larger characters. Putative signal peptides were identified in protein sequences using the SignalP (23).
Phylogenetic Analysis—Neighbor-joining tree was built from M-Coffee alignments of GRP mature peptides using MEGA version 3.0 (24). The PAM (percent accepted mutation) data matrix was chosen, and the rate of change was taken as site-independent (the use of a γ-distributed variable rate of change among sites was tried and produced worse results in all cases). The phylogenetic tree was generated using MEGA, where the internal branch labels, which are an estimate of branch assignment reliability, are branch support values.
RESULTS
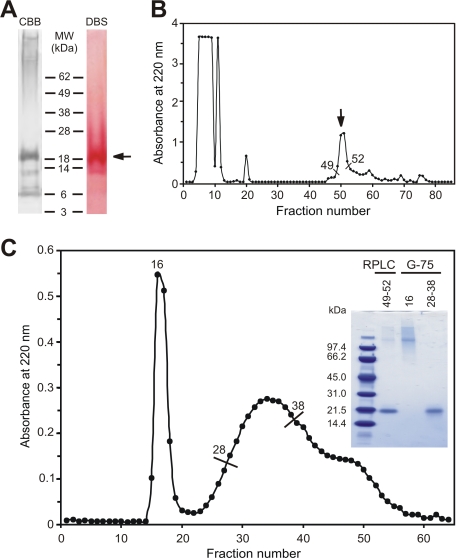
Isolation, Purification, and Characterization of Adriatic Sturgeon GRP—Matrix proteins from the calcified cartilage of Adriatic sturgeon were acid extracted from branchial arches of adult specimens as described (17), dialyzed against ammonium bicarbonate, and the resulting insoluble precipitate analyzed by SDS-PAGE, in reducing conditions. A protein with an apparent molecular mass of 21.5 kDa and a strong positive signal with the Gla-specific 4-diazobenzenesulfonic acid staining method was detected (Fig. 1A). Following transfer to a polyvinylidene difluoride membrane and protein sequence analysis, the first 20 aa were identified as STKSKDXVNAXNRQRLAADX (no phenylthiohydantoin-derivatives were detected for residues at positions 7, 11, and 20). Comparison to sequences available in GenBank databases using BLAST resource at NCBI identified various sequences sharing similarities with N-terminal of sturgeon GRP. Interestingly, all predicted open reading frames were unannotated, suggesting the discovery of a novel protein. The presence of a positive Gla-specific staining, combined with blanks obtained for 3 positions within the N-terminal sequence, later identified as Glu residues following cDNA cloning, provided additional evidence toward the possibility that this was a new Gla-containing protein, an hypothesis subsequently confirmed and thus it was named Gla-rich protein (GRP). To further purify sturgeon GRP, the insoluble material obtained after dialysis against ammonium bicarbonate was collected, dissolved in 0.1 m Tris, 6 m guanidine HCl, pH 9, and subjected to RP-HPLC using a C18 column (Fig. 1B). Fractions 49-52 were pooled and analyzed by SDS-PAGE. This peak contained GRP, but was found to be contaminated with high-molecular weight proteins (Fig. 1C, inset first lane) and was further purified by gel filtration using a Sephadex G-75 column. The resulting chromatogram showed a clear separation of the two main components (Fig. 1C). Aliquots of these two peaks were analyzed by SDS-PAGE, confirming the total separation of GRP, migrating as a 21.5-kDa entity (fractions 28-38; Fig. 1C, inset third lane), from the high molecular weight components (fractions 15-18; Fig. 1C, inset second lane).
FIGURE 1.
Isolation (A) and purification (B and C) of Adriatic sturgeon GRP. A, SDS-PAGE analysis of crude precipitate after acid extraction of branchial arches. The gel was stained with Coommassie Brilliant Blue (CBB) and 4-diazobenzenesulfonic acid (DBS). Arrow indicates the position of the GRP band for which the N-terminal sequence was determined. B, RP-HPLC separation of proteins in crude precipitate (see “experimental Procedures” for details). Fractions of 1 ml were collected and absorbance was determined at 220 nm. Arrow indicates the GRP elution peak. C, purification of GRP by Sephadex G-75 chromatography (see “Experimental Procedures” for details). Fractions of 0.5 ml were collected and absorbance was determined at 220 nm. The excluded and included volumes correspond to fractions 16 and 48 respectively. Four μg of RP-HPLC fractions 49-52 and 2 μg of Sephadex G-75 fractions 16 and 28-38 were analyzed by SDS-PAGE and stained with Coomassie Brilliant Blue (inset in panel C).
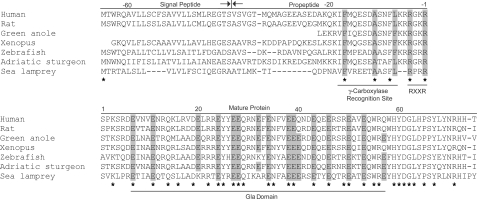
Sturgeon GRP Gene Organization, Tissue Distribution, and Sites of Expression—The full-length cDNA of sturgeon GRP was obtained through a combination of reverse transcriptase and RACE PCR amplifications. The longest cDNA spanned 1060 bp and contained an open reading frame of 420-bp encoding a polypeptide of 139 residues and 5′- and 3′-untranslated regions of 17 and 623 bp, respectively. The poly(A) tail was inserted 12 bp after two overlapping consensus polyadenylation signals (supplemental Fig. S1). Analysis of the deduced amino acid sequence revealed (i) a signal peptide of 26 aa, as predicted by SignalP, (ii) a propeptide of 39 aa containing a putative γ-glutamyl carboxylase (GGCX) recognition site (identified after alignment with propeptides of other VKD proteins), an AXXF motif and a RXXR furin-like cleavage site, and (iii) a mature protein of 74 aa with an estimated molecular mass of 9519 Da (considering only non-γ-carboxylated residues) (supplemental Figs. S1 and S3). The presence of 16 Glu residues in the deduced mature protein and the detection of 16.7 Gla residues by amino acid analysis in acid and alkaline hydrolysates of the purified protein strongly suggested that all Glu residues are γ-carboxylated in sturgeon GRP (supplemental Table S2), increasing its molecular mass to 10,207 Da. The overall composition of the purified protein confirmed the sequence deduced from the cDNA (supplemental Fig. 1) and the first 20 residues of the deduced mature protein were identical to those determined by N-terminal protein sequence analysis, confirming that the cloned cDNA encodes the purified GRP protein.
The sturgeon GRP gene was cloned through a combination of gene walking and genomic PCR strategies. It is organized in 5 coding exons and 4 introns (supplemental Fig. S1) all of phase 1 as defined by Patthy (25). The mature protein is encoded by exons 3, 4, and 5, all containing Gla residues, although 10 of them (from a total of 16) are present in exon 4, forming the putative functional core of the protein.
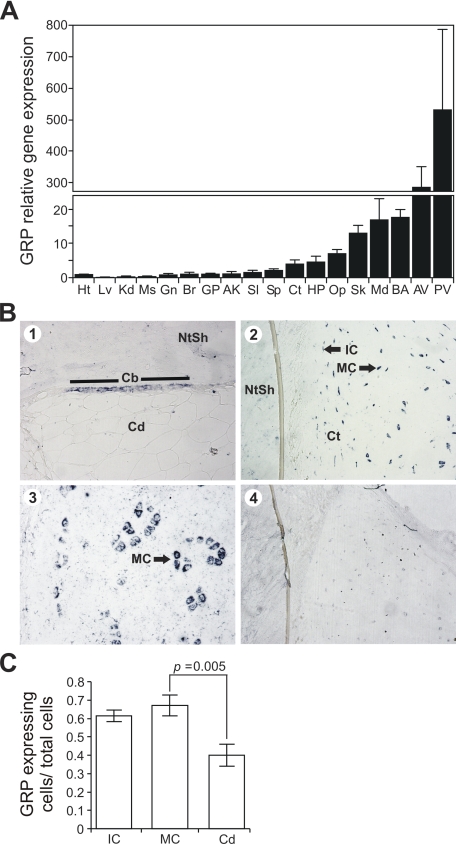
Levels of GRP gene expression were determined in a variety of adult sturgeon tissues (including calcified and non-calcified soft tissues) by real-time qPCR and normalized using HPRT1 as housekeeping gene (Fig. 2A). GRP gene expression was detected in all 18 tissues analyzed, but the highest levels were observed in cartilaginous tissues (skull, mandibula, branchial arches, anterior vertebra, and posterior vertebra), with posterior vertebra exhibiting the highest level. As expected from the high quantity of purified protein in branchial arches, high levels of gene expression were also observed in this tissue. Similar results were obtained when a different housekeeping gene, GAPDH, was used to normalize GRP gene expression (results not shown).
FIGURE 2.
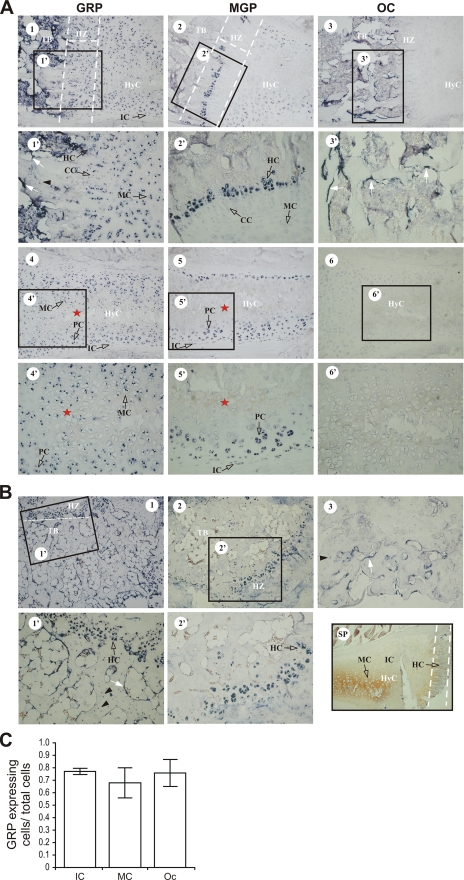
GRP tissue distribution (A), sites of gene expression (B), and corresponding quantification of GRP expression levels per cell type(C) in Adriatic sturgeon. A, levels of GRP gene expression measured by real-time qPCR in adult tissues related to levels in heart (Ht). Lv, liver; Kd, kidney; Ms, muscle; Gn, gonads; Br, brain; GP, ganoid plate; AK, anterior kidney; Sl, spleen; Sp, spine; Ct, cleithrum; HP, head plate; Op, operculum; Sk, skull; Md, mandibula; BA, branchial arches; AV, anterior vertebra; PV, posterior vertebra. B, sites of gene expression determined by in situ hybridization in vertebra (panels 1 and 2) and mandibula (panel 3). Cb, chordoblast layer of the notochord; Cd, chordocytes; NtSh, notochord sheath; Ct, cartilage; IC, immature chondrocytes; MC, mature chondrocytes. Hybridization with sense probe is presented in panel 4. Panels 1, 2, 3, and 4, magnification ×20. C, the percentage of IM, MC, and chordoblasts (Cd) expressing GRP was determined by dividing the number of cells of the indicated type expressing GRP by the total number of cells of that type detected in 3 consecutive vertebra sections. p was calculated using the unilateral Student's t test.
Sites of GRP gene expression were determined at single cell resolution by in situ hybridization of sections from a variety of adult sturgeon tissues. Fig. 2B shows that GRP mRNA was detected in mature and immature chondrocytes of vertebra and mandibula (Fig. 2B, panels 2 and 3), and in the chordoblast layer of the notochord in vertebra sections (Fig. 2B, panel 1), indicating that only cartilaginous tissues express significant levels of GRP in sturgeon. The cells found in the cartilaginous matrix of sturgeon vertebra are primarily vacuolated cells, identified as mature chondrocytes, and are surrounded by a thin layer of non-vacuolated cells, identified as immature chondrocytes and are clearly distinguishable in sections stained with hematoxylin-eosin (results not shown). Morphometric analysis performed for the different types of cells (Fig. 2C) shows that similar percentages of immature chondrocytes and mature chondrocytes (MC) express GRP, and that a statistically lower percentage of chordoblasts (Cd) express GRP. These results, coupled with the demonstration that cartilaginous tissues have the highest expression of GRP (shown in Fig. 2A) indicate that chondrocytes are the primary cells producing GRP in sturgeon.
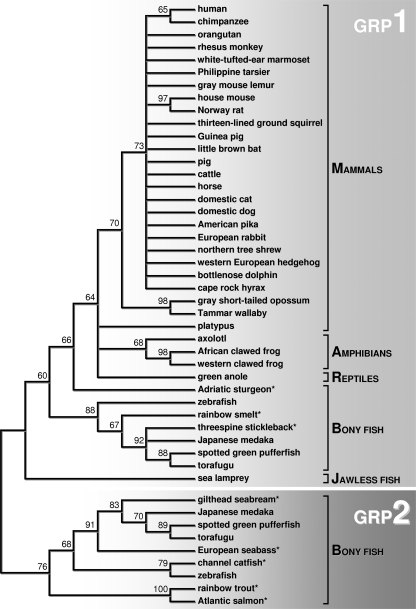
GRP Sequence Collection and Taxonomic Distribution—The GenBank and Ensembl databases were searched for GRP-related sequences using sturgeon GRP as query and BLAST facilities at NCBI. Resulting identified proteins were considered to be orthologs to GRP if they showed sequence similarity to the sturgeon sequence, and if this similarity extended throughout the entire protein. Positive search results, all unannotated sequences, were collected and species-specific sequences clustered using ContigExpress. A total of 52 sequences were collected and/or cloned and identified as GRP orthologs (48 sequences, either complete or partial, were reconstructed from EST, WGS, or chromosome sequences, and 4 cDNAs were cloned using a mixture of reverse transcriptase and RACE PCR (supplemental Fig. S2)). These sequences spanned 48 different species representing most classes of vertebrates (Fig. 3 and supplemental Figs. S3 and S4) including mammals, reptiles, amphibians, bony fish, and jawless fish. No evidence was found for the presence of any GRP sequences in invertebrates. Surprisingly, no GRP sequences were identified in chicken or other avian species, possibly due to lack of complete genomic information available from any avian organism. A GRP homolog was identified in one reptile, confirming its presence in the class Sauropsida, which includes most reptiles, birds, and dinosaurs. The identification of an ortholog in sea lamprey, the most ancestral species exhibiting GRP, together with its strong association to cartilaginous tissues, suggests that GRP may have appeared concomitantly with cartilage during jawless fish evolution.
FIGURE 3.
Alignment of GRP pre-propeptide of 7 species representative of vertebrates showing the conservation of residues at particular positions and highlighting functionally important domains and motifs. Stars indicate identical residues. Numbering is according to the first residue of the mature protein. Highly conserved residues in important motifs or in Gla domain are highlighted in gray (see supplemental Fig. S4). Arrows indicate cleavage position between the signal peptide and propeptide. RXXR, furin-like proteolytic cleavage site.
Through comparative genomics, a single GRP was identified per species, with the exception of bony fish (supplemental Fig. S4). In the four bony fish species for which nearly complete genome information is available (zebrafish, torafugu, spotted green pufferfish, and Japanese medaka) two distinct isoforms, encoded by two different genes, were identified and named GRP1 and GRP2. We believe that our inability to identify a second GRP isoform in all fish species analyzed is due to incomplete genome sequence information available. The presence of a second isoform in bony fish is no longer surprising. The existence of 2 paralogs in teleost fish, whereas only one ortholog is present in tetrapods, has been previously reported for other genes (hox genes, fzd8, sox11, f9, f7, OC, among others) (26, 27) and is associated with a fish-specific whole genome duplication event that has reportedly occurred in the teleost fish lineage after divergence from tetrapods, around 450 million years ago, and which likely affected most bony fish (27, 28). On the other hand, and despite our numerous efforts to PCR amplify a GRP2-like gene in sturgeon, we could only obtain the GRP1 isoform, supporting the most accepted theory that the fish-specific genome duplication event took place after the split of Acipenseriformes (sturgeons) and Semionotiformes (gars) from the lineage leading to teleost fish (28). In addition, a pseudogene (frameshift in exon 3 resulting in a truncated mature protein with a weak similarity to GRP) and an alternatively spliced transcript (skipping of exon 2) were identified in the threespine stickleback and mouse, respectively (supplemental Fig. S2).
GRP Phylogenetic Analysis—From the complete set of GRP sequences, a total of 47 mature proteins were deduced considering proteolytic cleavage at the conserved furin-like site. These proteins were aligned using M-Coffee (supplemental Fig. S5) and their relationship analyzed through a neighbor-joining phylogenetic tree using MEGA (Fig. 4). In agreement with the generally accepted taxonomy of vertebrates, GRP sequences were clustered into major taxonomic groups (i.e. amphibians, reptiles, mammals, bony fish, and jawless fish). Bony fish sequences clustered in separate groups confirmed the presence of 2 GRP isoforms. From the 47 initial mature proteins, 38 clustered as GRP1 and 9 clustered as GRP2, the bony fish-specific isoform.
FIGURE 4.
Phylogenetic analysis of the mature GRP peptides. Neighbor-joining tree constructed with MEGA3 demonstrating the taxon-specific clustering of mature GRP peptides and illustrating the bony fish-specific gene duplication event. Internal numbers are supporting values for the corresponding branch points (supporting values below 60% were omitted). *, fish species where only one GRP isoform was found, either GRP1 or GRP2.
GRP Conserved Features—GRP1 sequences (32 complete and 6 partial but containing the mature protein) were aligned using M-Coffee (supplemental Fig. S6A) then displayed as logos (supplemental Fig. S6B) using the WebLogo facilities. Sequence logos revealed 26 highly conserved residues (those residues having a bit score of 3.5 or greater), from which 21 are located within the mature protein and include 12 Glu residues. Additional conserved features were also identified, namely: (i) a transmembrane signal peptide of 26 or 27 aa; (ii) a propeptide of 38 or 39 aa containing a γ-carboxylase recognition site (GGCX) (targeting the VKD proteins to the γ-glutamyl carboxylase), an AXXF motif (first observed in MGP, where it serves as a site of proteolytic cleavage), and a furin-like cleavage site (likely used to cleave the propeptide at the RXXR polybasic cleavage site); and (iii) a mature protein ranging from 67 to 75 aa depending on species (Fig. 3 and supplemental Fig. S4). Although the N terminus of the precursor protein (residues -64 to -18 including signal peptide and the initial part of the propeptide) was found to be weakly conserved (14 identical residues between sturgeon and human), the remaining part of the protein (residues -17 to 74 including the γ-carboxylase recognition site, cleavage sites involved in the processing of the precursor protein, and the mature protein) exhibited a high degree of sequence identity (68 identical residues between sturgeon and human), indicating their involvement in the maintenance of important features for protein function and/or structure and thus conserved throughout vertebrate evolution. All 14 conserved Glu residues are sites of γ-carboxylation in sturgeon. No evidence for any additional conserved signatures of post-translational modification (e.g. phosphorylation and glycosylation) was found. Conserved features similar to those in GRP1 were observed in the GRP2 (supplemental Fig. S4), suggesting similar function and structure for both isoforms of GRP in fish.
The structure of the GRP1 gene was determined from 39 genes of species representing most classes of vertebrates (supplemental Fig. S7). For each gene, splicing sites and coding regions were predicted using either the Spidey mRNA to genomic alignment tool at NCBI (when both cDNA and gene are known for a species or in a closely related species) or by interspecies comparative sequence analysis. All genes shared the same simple gene organization with 5 coding exons (size of exon 4 > exon 5 > exon 3 > exon 2 > exon 1) and 4 introns. The phase of intron insertion (phase 1) was also rigorously conserved in all genes. Interestingly, the size of exon 4, which encodes the protein region with more Gla residues (9 or 10 depending on species), was rigorously maintained (99 bp) in all GRP1 (even in lamprey, the most ancestral species where this gene was identified and with the least conserved exon sizes) and GRP2 genes identified. Both fish isoforms exhibited a similar gene structure (i.e. in number of exons and phase of intron insertion) although the coding sequence was generally shorter in GRP2, mainly because of short exons 2 and 5. Altogether, these results suggest a constrained structure for GRP genes, in particular for the region encoding the core Gla domain (exon 4). Given the strict conservation of this domain throughout evolution, it further reinforces the fact that the function of GRP must be important and dependent on its Gla residues.
Sites of GRP Gene Expression in Rat—Given the high degree of conservation observed between fish and mammalian GRP at both protein and gene levels, we asked if sites of rat GRP gene expression were comparable with those identified in sturgeon. Sections of adult rat rib and tail (Fig. 5, A and B, respectively), and vertebra and femur (supplemental Fig. S8) were analyzed by in situ hybridization. Strong GRP mRNA expression was detected in cartilage chondrocyte cells from all phases of differentiation, i.e. in immature (IC), proliferating (PC), mature (MC), columnar (CC), and hypertrophic chondrocytes (HC) from rat ribs (Fig. 5A, 1, 1′, 4, and 4′), tail (Fig. 5B, 1 and 1′), vertebra (supplemental Fig. S8, panels A1, A1′, and A2), and femur (supplemental Fig. S8, panels B1 and B1′). In the hyaline cartilage from rib, GRP mRNA co-localized with MGP mRNA in IC, PC, and late HC in the hypertrophic zone of the articular region, although it was also present in HC of the central zone of hyaline cartilage, which did not stain for MGP (Fig. 5A, 4, 4′, 5, and 5′). In addition, CC were found to be positive for GRP mRNA but negative for MGP mRNA (Fig. 5A, 1′ and 2′). In vertebra, GRP mRNA was also detected in the chondrocytes of fibrocartilage (FC) (supplemental Fig. S8A2). Comparison between localization of GRP and MGP mRNAs clearly showed that whereas MGP expression was confined to immature, proliferating (Fig. 5A, 5 and 5′), and late hypertrophic chondrocytes (Fig. 5, A, 2, 2′, and B, 2, 2′, and supplemental Fig. S8, A4 and A4′), GRP mRNA was widely distributed in all cartilaginous cells. In addition, GRP mRNA was also detected in trabecular bone from rat rib, tail, vertebra, and femur, being specifically expressed in osteocytes and osteoblasts (Fig. 5, A, 1 and 1′, and B, 1 and 1′, and supplemental Fig. S8, A1, A1′, B1, and B2). Hybridization of consecutive sections with osteocalcin mRNA, a known marker for osteoblasts, demonstrated that the level of GRP expression in osteoblasts was comparable with the expression of OC (Fig. 5, A, 1′ and 3′, and B, 1′ and 3, and supplemental Fig. S8, A1′, A3′, B2, and B3′). In contrast, OC expression in osteocytes was barely detected (Fig. 5B, 3 and supplemental Fig. S8, A3′ and B3′), whereas GRP was highly expressed (Fig. 5B, 1 and 1′ and supplemental Fig. S8, A1′ and B2). Morphometric analysis was performed in rib and tail sections to evaluate the number and type of cells expressing GRP (Fig. 5C), and confirmed the qualitative in situ hybridization results. The percentage of immature and mature chondrocytes and osteocytes that express GRP is not significantly different. A morphometric analysis was not possible to perform for osteoblasts because it was not possible to individualize the GRP-positive osteoblasts present within each section, due to its continuous band-like shape. Nevertheless, a qualitative comparison between OC and GRP signal intensities was possible and showed a similar intensity of these two signals in osteoblasts, confirming the presence of GRP in these cells. These results demonstrate that GRP is not only localized to chondrocytes in sturgeon (a fish with an external bony skeleton) and rats, but is also expressed in bone cells of rats and therefore suggests a more widespread role for the protein throughout skeletal formation.
FIGURE 5.
GRP, MGP, and OC gene expression in rat cartilage and bone, determined by in situ hybridization in sections of rib (A) and tail (B), and corresponding quantification of GRP expression levels per cell type, performed through morphometric analysis (C). A, co-localization of GRP, MGP, and Oc in consecutive sections of rib. CC, columnar chondrocytes; HyC, hyaline cartilage; HZ, hypertrophic zone; TB, trabecular bone. White arrow and black arrowhead represents osteoblasts and osteocytes, respectively. Red asterisk is located within the hyaline cartilage central zone, which contain HC. B, co-localization of GRP, MGP, and Oc in consecutive sections of tail, as described for panel A. Hybridization with GRP sense probe is presented in the SP panel (×20). Magnification for panels A1-6 and panels B1 and B2, ×10. Magnification for panels A 1′, 2′, 3′, 4′, 5′, 6′, and panel B 1′, 2′, and 3, × 20. C, the percentage of immature and proliferating chondrocytes (IM), mature, columnar, and hypertrophic chondrocytes (MC), and osteocytes (Oc) expressing GRP was determined by dividing the number of cells of the indicated type expressing GRP by the total number of cells of that type detected in 4 consecutive sections of rib (IC and MC) and tail (Oc).
DISCUSSION
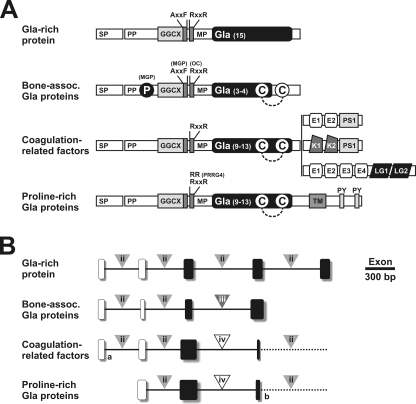
In the present work we report the purification and molecular characterization of a new member of the VKD protein family containing an unprecedented high density of Gla residues (22%) and thus named GRP. This protein, identified in the process of purification of Gla-containing proteins from the endogenous mineralized cartilaginous skeleton of the sturgeon A. naccarii, is a small secreted protein with a calculated molecular mass of 10.2 kDa (including Gla residues). The presence of 16 Gla residues, assigned through comparison between amino acid composition of the pure protein (indicating 16.7 Gla residues), and the number of Glu residues deduced from its cDNA, identifies GRP as the protein with the highest ratio of Gla residues/size identified to date, and emphasizes its uniqueness. The remarkably high degree of conservation among orthologs of GRP (identified in 48 different vertebrate species from most classes within this phyla and absent in invertebrates), as well as the presence of highly conserved features specific to all VKD proteins (a propeptide domain containing a γ-glutamyl carboxylase recognition site, an AXXF motif, a furin-like cleavage site, and a proven Gla-containing mature protein) strongly suggests that this is a new vertebrate-specific γ-carboxylated protein. The putative γ-carboxylase recognition site (GGCX) found in the GRP propeptide contains three highly conserved residues (e.g. Phe -17, Ala -11, and Leu -7) (Fig. 3 and supplemental Fig. S4) at positions shown by site-directed mutagenesis and kinetic studies to be important for carboxylase affinity (29-31). The conservation of these 3 residues and the high degree of γ-carboxylation observed in the sturgeon protein suggests that the affinity of carboxylase for GRP may be comparable with that observed for coagulation-related VKD proteins. Interestingly, the residue at position -7 is not totally conserved (Leu or Phe), and its pattern of distribution reveals that Leu is predominant in mammalian proteins, whereas Phe is predominant in fish, amphibians, and reptiles (with the exception of sturgeon and lamprey). Whether this variation results in a difference in carboxylase binding activity, and consequent change in degree of γ-carboxylation, remains to be clarified. A conserved AXXF motif, previously identified in MGP as a proteolytic cleavage site (9), was identified in all GRP propeptides. The AXXF motif is closely followed by a furin-like cleavage site (RXXR) located at the end of the propeptide, whose functionality was confirmed by N-terminal sequence analysis of the intact sturgeon protein. The most remarkable feature of this protein resides in the high number of Gla residues found in its mature form. This unique feature appears to have been conserved over more than 450 million years of evolution, with an impressive degree of conservation both in number and position of corresponding Glu residues within the protein structure (Fig. 3, and supplemental Figs. S4 and S6B), as well as in the putative γ-carboxylase binding site, thus strongly suggesting that this protein must be γ-carboxylated in all phyla. Despite including features found in other VKD proteins, GRP also shows significant differences in protein structure (Fig. 6A) and gene organization (Fig. 6B and supplemental Fig. S9), particularly in the vicinity of its Gla domain. Indeed, in all known VKD proteins, the Gla domain is followed, in the mature protein, by (i) an epidermal growth factor domain, (ii) a kringle domain, (iii) a transmembrane and proline-rich cytoplasmic region, or (iv) a disulfide bond (11). In contrast, the Gla domain of GRP spreads along the entire mature protein.
FIGURE 6.
Structural organization of the Gla domain of human VKD proteins at protein (A) and gene (B) levels. VKD protein superfamily is composed of 4 families, i.e. the Gla-rich protein family, the bone-associated Gla protein family (containing osteocalcin and matrix Gla protein), the coagulation factor family (containing factors 7, 9, and 10, prothrombin, proteins C, Z, and S, and gas6), and the proline-rich Gla protein family (PRGP1, -2, -3, and -4). A, SP, signal peptide; PP, propeptide; MM, mature peptide; GGCX, γ-glutamyl carboxylase recognition site; AXXF, conserved motif and proteolytic cleavage site in MGP; RXXR, proteolytic cleavage site, absent in MGP and PRGP4; P, serine phosphorylation; C, conserved cysteine residues; E, epidermal growth factor-like domain; PS, peptidase S1 domain; K, kringle domain; LG, laminin-G like domain; TM, transmembrane-like domain; PY, potential WW domain-binding motifs PPXY. Dashed line indicates disulfide bond. B, gene structure of human VKD proteins (limited to coding sequences and Gla domain; complete gene structure is presented in supplemental Fig. S8). Boxes represent coding exons; black boxes represent exons coding for the Gla domain; triangles indicate the phase of intron insertion; dash line represents the continuation of the gene. a indicates extra coding exon in coagulation factor 7 and protein Z, and b indicates fused exon in PRGP1 and -3 (see supplemental Fig. S9).
Two recent reports published during the development of our work identified a cartilage-associated secreted protein named unique cartilage matrix-associated protein, which appears to be identical to GRP (32, 33). The presence of both processed and unprocessed forms of unique cartilage matrix-associated protein in the extracellular matrix (33) indicates that cleavage may occur intracellularly, by a furin-like enzyme, or in the extracellular compartment, and is in agreement with our findings. However, those authors failed to identify the existence of γ-carboxylation in their protein, possibly due to incomplete or even complete absence of γ-carboxylation when the protein is synthesized in vitro. The molecular mass determination of the overexpressed protein, by matrix-assisted laser desorption ionization time-of-flight mass spectrometry, may have contributed to this failure because decarboxylation occurs during ionization and consequently Gla residues are converted to Glu (34). The production and secretion of recombinant VKD proteins has been previously shown to be complex, often hampered by the limited carboxylation capacity observed in the in vitro cell systems used (35) or by limitations in any of the intermediate phases of this process. Indeed, several protein factors are involved in the synthesis of functional VKD proteins: the γ-glutamyl carboxylase (36), the vitamin K oxidoreductase, a redox protein that regenerates vitamin K activity, a vitamin K co-factor, and a propeptide processing enzyme (36). Studies performed with Gla-containing coagulation proteins indicate that overall efficiency is dependent on the stoichiometry of all the components involved in this complex process (37). Our results show that, in sturgeon, GRP mRNA only associates with cartilaginous tissues, being expressed in chordoblasts as well as immature and mature chondrocytes. A similar cartilage specificity has been recently proposed for its mouse ortholog (unique cartilage matrix-associated protein) (32, 33), with a particular emphasis in resting and proliferating chondrocytes during development, and undergoing an accentuated postnatal decrease. However, our results in rat do not confirm that unique localization in mammals. Although we show a strong GRP expression in adult rat cartilage, associated to immature, mature, and hypertrophic chondrocytes, a result clearly in agreement with the immunolocalization shown by Surmann-Schmitt et al. (33), we also found a clear positive signal for GRP mRNA in trabecular bone, both in osteoblasts and osteocytes. This finding argues against the concept that this protein is cartilage specific in mammals, as claimed by those authors. Furthermore, our data are corroborated by available in silico information based on tissue-specific EST data and indicating expression of the human GRP ortholog in brain, connective tissue, ear, and uterus. Altogether, available data provide clear evidence against both the concept of its cartilage specificity and the adequacy of the protein name (unique cartilage matrix-associated protein) suggested by those authors.
The metal binding properties of Gla residues within VKD proteins have been associated with binding of calcium ions or calcium crystals, either through Ca2+ coordination in the Ca2+-dependent binding of coagulation factors to anionic phospho-lipids membrane surfaces (38), or through binding to hydroxyapatite crystals, the major mineral component present in mineralized extracellular matrix (39). This was extensively shown for MGP and OC, two VKD proteins also known to be associated with mineralized tissues, regulation of mineralization process, and vascular ectopic calcification. The selective extraction of GRP during demineralization of the sturgeon extracellular matrix shows that its Gla domain is capable of binding to calcium crystals. Considering our results in rat adult tissues and data reported for mouse cartilage, we propose that mammalian GRP is capable of binding to anionic proteoglycans highly abundant in cartilage matrix, probably through a calcium bridge with the Gla domain, mimicking the previously described function of the Gla domain in coagulation factors.
Cartilage plays a multiplicity of roles in multicellular organisms and is crucial for skeletal development and maintenance. However, disruption of genes known to be involved in cartilage homeostasis often leads to severe skeletal disorders. Of the ∼370 distinguishable skeletal dysplasias already reported, mutations in 115 genes were identified and found to be associated with about 150 disorders (40). The identification of GRP, a protein with (i) an unprecedented high Gla content allowing extracellular matrix binding and (ii) a strong expression in cartilage but also in other tissues including bone, may prove useful to further elucidate the molecular mechanisms responsible for some of these skeletal dysplasias. Indeed, the enormous potential of this protein for calcium binding makes it a choice candidate to play a role as physiological calcium modulator (41), and it would be highly relevant to investigate its possible involvement in cartilage development and maintenance as well as in those effects observed in extra-hepatic tissues upon treatment with various vitamin K antagonists, such as warfarin. These are widely used as anti-coagulants and known to induce side effects including accelerated bone loss and low bone mass, as well as arterial and heart valve calcification, a complex set of phenotypes that has not been entirely explained following genetic studies involving depletion of genes encoding the other already known Gla proteins (2). It is thus conceivable that some of these effects could be caused by impairment of GRP function, not previously addressed because GRP remained unidentified until today. Uncovering its structure should open new perspectives toward unveiling the complex mechanisms underlying in particular bone and cartilage formation and maintenance.
Supplementary Material
Acknowledgments
We thank Dr. P. Gavaia (CCMAR) for help in the course of in situ hybridization, S. Cavaco (CCMAR) for technical assistance in protein extraction, and Drs. A. Domezain (Aquaculture Rio Frio, Granada, Spain), J. Fuentes (CCMAR), and J. Belo (CBME/UALG, Faro, Portugal) for providing part of the biological material used in this study.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™ and UniProtKB databases with accession numbers EU022751 (sturgeon GRP cDNA), EU022752 (seabream GRP cDNA), EU022753 (seabass GRP cDNA), EU022754 (rat GRP cDNA), EF413586 (sturgeon GAPDH), EF413585 (sturgeon HPRT1), P85209 (sturgeon GRP N-terminal sequence), and EU482149 (sturgeon GRP gene).
This work was supported, in whole or in part, by National Institutes of Health Grant HL58090 (to P. A. P.) and a grant from the Centre of Marine Sciences (to M. L. C., V. L., and D. C. S.) and Project POCTI/MAR/57921/2004 from the Portuguese Science and Technology Foundation (including funds from FEDER and OE) (to D. C. S. and M. L. C.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S9 and Tables S1 and S2.
Footnotes
The abbreviations used are: Gla, γ-carboxyglutamic acid; GRP, Gla-rich protein; MGP, matrix Gla protein; OC, osteocalcin; HPRT1, hypoxanthine phosphoribosyltransferase 1; IC, immature chondrocytes; MC, mature chondrocytes; HC, hypertrophic chondrocytes; PC, proliferating chondrocytes; FC, fibrocartilage; RP-HPLC, reverse phase-high performance liquid chromatography; RACE, rapid amplification of cDNA ends; aa, amino acid; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; qPCR, quantitative PCR.
References
- 1.Vermeer, C. (1990) Biochem. J. 266 625-636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cranenburg, E. C., Schurgers, L. J., and Vermeer, C. (2007) Thromb. Haemostasis 98 120-125 [PubMed] [Google Scholar]
- 3.Stenflo, J., Fernlund, P., Egan, W., and Roepstorff, P. (1974) Proc. Natl. Acad. Sci. U. S. A. 71 2730-2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suttie, J. W. (1985) Annu. Rev. Biochem. 54 459-477 [DOI] [PubMed] [Google Scholar]
- 5.Zhang, L., and Castellino, F. J. (1993) J. Biol. Chem. 268 12040-12045 [PubMed] [Google Scholar]
- 6.Hauschka, P. V., Lian, J. B., and Gallop, P. M. (1975) Proc. Natl. Acad. Sci. U. S. A. 72 3925-3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Price, P. A., Otsuka, A. A., Poser, J. W., Kristaponis, J., and Raman, N. (1976) Proc. Natl. Acad. Sci. U. S. A. 73 1447-1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Price, P. A., Urist, M. R., and Otawara, Y. (1983) Biochem. Biophys. Res. Commun. 117 765-771 [DOI] [PubMed] [Google Scholar]
- 9.Laizé, V., Martel, P., Viegas, C. S. B., Price, P. A., and Cancela, M. L. (2005) J. Biol. Chem. 280 26659-26668 [DOI] [PubMed] [Google Scholar]
- 10.Manfioletti, G., Brancolini, C., Avanzi, G., and Schneider, C. (1993) Mol. Cell. Biol. 13 4976-4985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kulman, J. D., Harris, J. E., Haldeman, B. A., and Davie, E. W. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 9058-9062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kulman, J. D., Harris, J. E., Xie, L., and Davie, E. W. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 1370-1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walker, C. S., Shetty, R. P., Clark, K., Kazuko, S. G., Letsou, A., Olivera, B. M., and Bandyopadhyay P. K. (2001) J. Biol. Chem. 276 7769-7774 [DOI] [PubMed] [Google Scholar]
- 14.Brown, M. A., Hambe, B., Furie, B., Furie, B. C., Stenflo, J., and Stenberg, L. M. (2002) Toxicon 40 447-453 [DOI] [PubMed] [Google Scholar]
- 15.Hansson, K., Thamlitz, A. M., Furie, B., Furie, B. C., and Stenflo, J. (2006) Biochemistry 45 12828-12839 [DOI] [PubMed] [Google Scholar]
- 16.de Boer-van den Berg, M. A., Verstijnen, C. P., and Vermeer, C. (1986) J. Investig. Dermatol. 87 377-380 [DOI] [PubMed] [Google Scholar]
- 17.Simes, D. C., Williamson, M. K., Ortiz-Delgado, J. B., Viegas, C. S., Price, P. A., and Cancela, M. L. (2003) J. Bone Miner. Res. 18 244-259 [DOI] [PubMed] [Google Scholar]
- 18.Jie, K. S., Gijsbers, B. L., and Vermeer, C. (1995) Anal. Biochem. 224 163-165 [DOI] [PubMed] [Google Scholar]
- 19.Chomczynski, P., and Sacchi, N. (1987) Anal. Biochem. 162 156-159 [DOI] [PubMed] [Google Scholar]
- 20.Pinto, J. P., Ohresser, M. C., and Cancela, M. L. (2001) Gene (Amst.) 270 77-91 [DOI] [PubMed] [Google Scholar]
- 21.Wallace, I. M., O'Sullivan, O., Higgins, D. G., and Notredame, C. (2006) Nucleic Acids Res. 34 1692-1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneider, T. D., and Stephens, R. M. (1990) Nucleic Acids Res. 18 6097-6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. (1997) Protein Eng. 1 1-6 [DOI] [PubMed] [Google Scholar]
- 24.Kumar, S., Tamura, K., and Nei, M. (2004) Brief Bioinform. 5 150-163 [DOI] [PubMed] [Google Scholar]
- 25.Patthy, L. (1987) FEBS Lett. 214 1-7 [DOI] [PubMed] [Google Scholar]
- 26.Taylor, J. S., Braasch, I., Frickey, T., Meyer, A., and Van de Peer, Y. (2003) Genome Res. 13 382-390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laizé, V., Viegas, C. S. B., Price, P. A., and Cancela, M. L. (2006) J. Biol. Chem. 281 15037-15043 [DOI] [PubMed] [Google Scholar]
- 28.Roest Crollius, H., and Weissenbach, J. (2005) Genome Res. 15 1675-1682 [DOI] [PubMed] [Google Scholar]
- 29.Ratcliffe, J. V., Furie, B., and Furie, B. C. (1993) J. Biol. Chem. 268 24339-24345 [PubMed] [Google Scholar]
- 30.Stanley, T. B., Jin, D. Y., Lin, P. J., and Stafford, D. W. (1999) J. Biol. Chem. 274 16940-16944 [DOI] [PubMed] [Google Scholar]
- 31.Lin, P. J., Jin, D. Y., Tie, J. K., Presnell, S. R., Straight, D. L., and Stafford, D. W. (2002) J. Biol. Chem. 277 28584-28591 [DOI] [PubMed] [Google Scholar]
- 32.Tagariello, A., Luther, J., Streiter, M., Didt-Koziel, L., Wuelling, M., Surmann-Schmitt, C., Stock, M., Adam, N., Vortkamp, A., and Winterpacht, A. (2008) Matrix Biol. 27 3-11 [DOI] [PubMed] [Google Scholar]
- 33.Surmann-Schmitt, C., Dietz, U., Kireva, T., Adam, N., Park, J., Tagariello, A., Onnerfjord, P., Heinegård, D., Schlötzer-Schrehardt, U., Deutzmann, R., von der Mark, K., and Stock, M. (2008) J. Biol. Chem. 283 7082-7093 [DOI] [PubMed] [Google Scholar]
- 34.Blostein, M. D., Rigby, A. C., Jacobs, M., Furie, B., and Furie, B. C. (2000) J. Biol. Chem. 275 38120-38126 [DOI] [PubMed] [Google Scholar]
- 35.Lingenfelter, S. E., and Berkner, K. L. (1996) Biochemistry 35 8234-8243 [DOI] [PubMed] [Google Scholar]
- 36.Rehemtulla, A., Roth, D. A., Wasley, L. C., Kuliopulos, A., Walsh, C. T., Furie, B., Furie, B. C., and Kaufman, R. J. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 4611-4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hallgren, K. W., Qian, W., Yakubenko, A. V., Runge, K. W., and Berkner, K. L. (2006) Biochemistry 45 5587-5598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sperling, R., Furie, B. C., Blumenstein, M., Keyt, B., and Furie, B. (1978) J. Biol. Chem. 253 3898-3906 [PubMed] [Google Scholar]
- 39.Murshed, M., Schinke, T., McKee, M. D., and Karsenty, G. (2004) J. Cell Biol. 165 625-630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Funari, V. A., Day, A., Krakow, D., Cohn, Z. A., Chen, Z., Nelson, S. F., and Cohn, D. H. (2007) BMC Genomics 8 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simes, D. C., Viegas, C. S. B., and Cancela, M. L. (August 27, 2008) U. S. Patent 61/136.315
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.