Abstract
γ-Synuclein (Syn G) is highly expressed in retinal ganglion cells and the loss of these cells in glaucoma is associated with significant reduction of the intracellular Syn G level. However, a causative relationship between these two events has not been established. Here we show that the knockdown of Syn G results in a decreased viability of the immortalized retinal ganglion cells (RGC-5). The Syn G silencing reduces phosphorylation of serine 112 (Ser112) in Bad protein, a member of the Bcl-2 family that plays a critical role in apoptotic cell death signaling. Our gene expression analysis data suggests that changes in Bad phosphorylation status may be caused by a coordinated shift in activities of kinases controlling Bad phosphorylation and phosphatases catalyzing its dephosphorylation. Moreover, increased phosphorylation of Bad-sequestering protein 14-3-3 detected in these cells is also pro-apoptotic. These results suggest that the homeostatic level of Syn G in RGC-5 cells is required for transcriptional regulation of protein kinases and phosphatases, controlling phosphorylation of Bad and 14-3-3. Lowering Syn G causes Bad dephosphorylation, dissociation from phosphorylated 14-3-3, and translocation to mitochondria where it initiates apoptotic death cascade.
Glaucoma is a leading cause of irreversible world vision loss (1). This neuropathy is characterized by progressive damage of the optic nerve associated with a selective loss of the retinal ganglion cells (RGC)2 (2–6). The precise mechanisms involved in glaucoma pathogenesis have yet to be determined, but a better understanding of the factors involved in ganglion cell death is central to the development of treatment of this neuropathy (7–9). It has been established that in glaucoma RGCs die by apoptosis (10) and a variety of key events in apoptosis focus on mitochondria, including the participation of pro- and antiapoptotic Bcl-2 family proteins (11, 12). These results imply that dysregulation of molecular mechanisms controlling mitochondrial apoptotic signaling may be important for the progression of glaucoma. In glaucoma progression, considerable changes in the transcriptome occur in the optic nerve (13), whole retina (14, 15), RGC (16–19), trabecular meshwork cells (20, 21), and lymphocytes (22). However, the mechanisms controlling disease-induced changes in transcriptional regulation of mitochondrial apoptotic cascade in RGCs are not completely understood.
Syn G is one of the genes that is highly expressed in RGC (16, 23, 24) and down-regulated in the course of glaucomatous alterations (25, 26). The reduction of Syn G in RGC may have vital consequences for these cells, because Syn G is involved in cellular signaling and modulates the level of transcription of selected genes (27, 28). It is not clear whether the reduction of Syn G in RGC initiates the changes leading to glaucomatous alterations or it is just a result of the upstream biochemical processes that take place in glaucoma. The role of Syn G in the regulation of kinases and signaling pathways is well established (28–30), but the involvement of this mechanism in glaucoma progression is not studied.
In this study, we used siRNA knockdown of Syn G as an approach to mimic this protein decrease observed in the RGCs affected by glaucoma. Experimental silencing of Syn G in RGC-5 cells resulted in decreased cell viability and correlated with the reduction of Bad phosphorylation and the increase in 14-3-3 phosphorylation. Given the level of phosphorylation these proteins serve as an important survival/death checkpoint in RGC, potentially critical for their loss in glaucoma (31, 32), it is feasible to suggest that a decrease in Syn G causes this dys-regulation. The changes in Bad and 14-3-3 phosphorylation may be a result of misbalance in the expression of kinases and phosphatases in Syn G-silenced cells.
EXPERIMENTAL PROCEDURES
Cell Culture—The transformed rat RGC line, RGC-5 (33), was kindly provided by Dr. Raghu Krishnamoorthy (University of North Texas Health Science Center, Fort Worth, TX). Cells were maintained in growth medium containing low-glucose Dulbecco's modified Eagle's medium with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin (Sigma) in a humidified atmosphere of 95% air and 5% CO2 at 37 °C, as described previously (33, 34).
siRNA—The inhibition of Syn G expression by siRNA was carried out by vector-based RNA interference approaches. pSUPER.retro.neo+gfp was used as a vector (Oligoengine, Inc., Seattle, WA). This retroviral vector ensures efficient siRNA expression using H1 RNA polymerase III promoter, which drives the endogenous production of siRNA. For oligonucleotide design the software from Dharmacon and Whitehead Institute were used. The designed oligonucleotides correspond to different parts of the rat Syn G gene, including exons 3 and 4 (E3 and E4) and 3′-untranslated region (3′-UTR) (Table 1). As a control, scrambled (Scr) nucleotide sequence corresponding to E3 was used. The scrambled sequence has the same nucleotide composition as the input sequence and does not possess a significant homology to other genes, according to BLAST analysis. Each double strand oligo contained the BglII site on 5′- and 3′-end on the HindIII. The oligonucleotides for candidate siRNAs were analyzed by BLAST search to exclude considerable similarity to other genes. Oligonucleotides were inserted into pSUPER vector by ligation using BglII and HindIII sites as recommended by the manufacturer (Oligoengine, Inc.). Further steps (annealing, linearization, cloning the annealed oligonucleotides into the vector, and transformation of Escherichia coli) were performed according to the manufacturer's protocols. The correctness of all constructs was confirmed by sequencing. On the next step, pSUPER.retro.plasmids were transfected into 293T cells together with a packaging plasmid pCLEco (Imgenex, San Diego, CA) using FUGENE-HD transfection reagent (Roche). Transfection into 293T cells was performed following the manufacturer's protocol. The cells (105/ml) were split into 60-mm plates with Dulbecco's modified Eagle's medium and grown overnight. Before transfection the media was changed to a fresh one. DNA was incubated with FUGENE-HD (ratio 3:2, 30 min, room temperature) to form a complex. Each mixture contained 1.5 μg of pSUPER-plasmid DNA and 0.5 μg of pCLeo DNA. After incubation the mixture was added to cells in the media containing fetal bovine serum without antibiotics. In 48 h the fluorescence was analyzed using the Bio-Rad Microradians Plus confocal system coupled to a Nikon Eclipse inverted microscope TE300 (Melville, NY). The efficiency of transfection was ∼50%. The media from the 293T cells containing the virus together with 10 μg/ml of Polybrene (Millipore Corporation, Billerica, MA) were used for the infection of RGC-5. After 24 h of growth the efficiency of infection determined as described above was ∼30%. Because pSUPER plasmid contained green fluorescent protein encoding nucleotide sequence, we used flow cytometry (BD-FACSAria Cell Sorting System, BD Biosciences) to enrich the population of cell containing siRNA.
TABLE 1.
Sequences of oligonucleotides used for small interfering RNAs
| Oligo to | Sense strand | Antisense strand | mRNA target |
|---|---|---|---|
| Scrambled | GCTGGCGGACCGATAAAGTTT | ACTTTATCGGTCCGCCAGCTT | GCUGGCGGACCGAUAAAGU |
| Exon 4 | GGAGGCCAAAGAGCAAGAGTT | CTCTTGCTCTTTGGCCTCCTT | GGAGGCCAAAGAGCAAGAG |
| 3′-UTR | GCCCAATGCACAAGCCTATTT | ATAGGCTTGTGCATTGGGCTT | GCCCAAUGCACAAGCCUAU |
| Exon 3 | GGCCAATGCCGTGAGTGAATT | TTCACTCACGGCATTGGCCTT | GGCCAAUGCCGUGAGUGAA |
Cell Viability—Cell viability was assessed using the “Quick Cell Proliferation Assay Kit” (BioVision Research Products, Mountain View, CA). The cells were split into 96-well plates at a density of 5 × 104–105 cells/well in triplicate and incubated overnight. Then the media was changed to a fresh one containing a cytotoxic agent and incubated as indicated in the legend to Fig. 2. 10 μl of WST-1/Electro Coupling Solution was added to each well and the incubation continued for an additional 4 h at 37 °C. The formazan dye produced by viable cells was quantified by measuring the absorbance of the dye solution at 470 and 650 nm using a microtiter plate reader (BioTek Synergy HT-K24, Winooski, VT). Cell viability was assessed in the presence of the following toxic agents: etoposide and staurosporine (Sigma), 4-hydroxylnonenal (HNE, Cayman Chemical, Ann Arbor, MI), and hydrogen peroxide (Sigma).
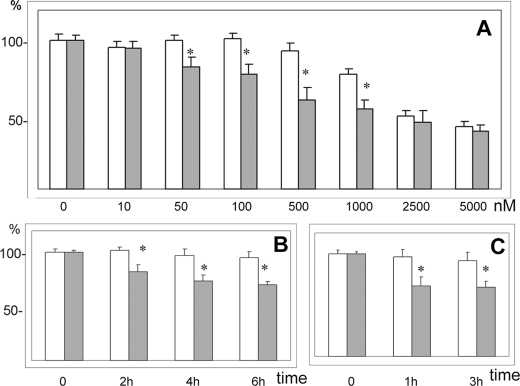
FIGURE 2.
Effect of Syn G silencing on RGC-5 viability was assessed in the presence of cytotoxic agents. The cells were incubated overnight, cytotoxic agent was added and cell viability was determined by the formazan method. White bars, control cells treated with scrambled siRNA; gray bars, cells treated with siRNA to E3. A, cell viability was assessed after a 15-h incubation in the presence of 10, 50, 100, and 500 nm and 1, 2.5, and 5 μm staurosporine. B, cells were incubated in the presence of 35 μm HNE for 2, 4, and 6 h. C, cells were incubated in the presence of 100 μm HNE for 1 and 3 h. A reduction of cell viability was observed in cells with Syn G knockdown in the presence of both cytotoxic substances. * indicates p < 0.05.
Preparation of Cell Lysates—Cells grown in 75-cm2 flask were rinsed with ice-cold phosphate-buffered saline (PBS) and suspended in 0.5 ml of boiling 1× cell lysis buffer (10 mm Tris-HCl, pH 7.4, 1 mm sodium orthovanadate, 1% SDS-Na). Then the cells were scraped off the flask, transferred to microcentrifuge tubes, heated in water bath at 95 °C for 30 s, and sonicated three times on ice for 5 s each. The samples were centrifuged for 10 min at 14,000 × g, at 4 °C, and the supernatant was transferred to a new tube. The amount of total protein in cell lysates was determined using BCA (Sigma) with bovine serum albumin as a standard.
Western Blotting—For the analysis of Syn G expression, the cells were disrupted by ultrasonic treatment and the extracts analyzed by Western blot in 12% polyacrylamide gels. 30 μg of total protein were loaded on each well. After electrophoresis, proteins were transferred onto Immobilon-FL transfer membrane with 0.45 μm pore size (Millipore, Chelmsford, MA). Nonspecific binding sites were blocked by immersing the membrane in Tris-buffered saline with 0.05% Tween 20 (TBS-T) and 10% nonfat dry milk for 1 h at room temperature on an orbital shaker. For quantitative imaging of films Kodak Image Station 440CF (Eastman Kodak Co., Rochester, NY) and Kodak Digital Science Image Analysis Software were used. Other details are described in our previous publications (26–28).
Antibodies for Western Blot and Immunoprecipitation—The following antibodies were used for Western blot: Syn G rabbit polyclonal Ab from ABCam (Ab 6169, Cambridge, MA), dilution 1:1,000; Bad, rabbit mAb 9628 (Cell signaling Technology, Danvers, MA), dilution 1:40; phospho-BAD-Ser112-7E11 mouse monoclonal antibody 9296 from Cell Signaling Technology, dilution 1:1,000; 14-3-3P affinity purified rabbit polyclonal antibody (Phosphosolutions, Aurora, CA), dilution 1:1,000; 14-3-3-β/ζ monoclonal antibody from QED Bioscience Inc. (San Diego, CA), dilution 1:500; Bcl-XL (13.6), rabbit polyclonal Ab (gift of Dr. L. H. Boise, University of Miami, FL), dilution 1:500; anti-cytochrome oxidase (Cox) subunit IV, monoclonal 20E8 (Invitrogen), dilution 1:5,000; α-tubulin, monoclonal antibody (clone DM1/a, ascites fluid, Sigma), dilution 1:10,000.
The following antibodies were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA): c-Jun (N), rabbit polyclonal Sc-45, dilution 1:400; JNK1 (F3), mouse monoclonal Sc-1648, dilution 1:250; Bad (K-17) Sc-942, dilution 1:400; Bax (YTH6A7), mouse monoclonal, Sc-80658, dilution 1:250; Bcl2, mouse monoclonal antibody Sc-7382, dilution 1:500.
Immunoprecipitation—200 μl of cell lysate in RIPA buffer (10 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.1% SDS-Na, 1% Triton, 1% deoxycholate) were mixed with 2 μg of Bad antibody 11E3 (rabbit monoclonal from Cell Signaling Technology) and incubated with gentle shaking overnight at 4 °C. Protein A-agarose beads (Sigma) (20 μl of 50% bead slurry) were added and incubated with gentle shaking for 2 h at 4 °C. The samples were centrifuged for 30 s at 4 °C and the pellets were washed five times with 500 μl of 1× cell lysis buffer. The pellets were resuspended in 20 μl of 3× SDS sample buffer, mixed by vortexing, and then centrifuged for 30 s. The samples were heated at 95–100 °C for 2–5 min and centrifuged for 1 min at 14,000 × g. 20 μl of the supernatants were loaded on a 12% SDS-PAGE gel for Western blot.
Isolation of Mitochondria-rich Fraction—A mitochondria-rich fraction was isolated as described previously (35) with slight modifications. Briefly, RGC-5 cells were harvested, washed with phosphate-buffered saline, and pelleted at 500 × g for 5 min at room temperature. Cell pellets were homogenized in isotonic buffer (5 mm HEPES, pH 7.2, 300 mm sucrose, 1 mm EDTA) supplemented with the protease inhibitor mixture diluted 1:7 (Roche). Homogenates were centrifuged at 500 × g for 5 min at 4 °C and the supernatant was centrifuged again at 500 × g for 5 min to remove all the remaining unbroken cells and nuclei. The resultant supernatant was centrifuged at 10,000 × g for 20 min at 4 °C to obtain the mitochondria-rich fraction and the supernatant was considered as the cytosolic fraction.
RNA Extraction and Quality Control—RNA was isolated from 4 × 106 cultured RGC-5 cells with RNeasy Protect Mini Kit (Qiagen, Valencia, CA) using spin columns and stored at -80 °C. The isolated RNA was quantified by spectrophotometric absorbance considering that 1 unit at 260 nm in 10 mm Tris-HCl, pH 7.5, corresponds to 44 μg of RNA/ml. Only samples with an A260/A280 ratio higher than 1.9 were used. The integrity of RNA was checked by denaturing 1.2% agarose gel electrophoresis in the presence of formaldehyde. Ribosomal RNAs appeared as sharp bands and the ratio of 28 S rRNA to 18 S rRNA quantified under UV light with Kodak Image Station 440CF (Eastman Kodak Co., Rochester, NY) was equal to 2:1. The RNA was converted into cDNA using the High Capacity cDNA Reverse Transcription Kit with random primers (Applied Biosystems, Foster City, CA) following the recommendations of the manufacturer and was treated by DNase. The concentrations of RNA in samples were equalized to 300 ng/μl. A minus reverse transcriptase (RT) control containing all the reaction components except the reverse transcriptase was included in all qRT-PCR experiments to test genomic DNA contamination.
RNA Amplification, Labeling, and Array Hybridization—Amplification was performed using the Amino Allyl Message-Amp™ Kit (Ambion) that is configured to incorporate the modified nucleotide 5-(3-aminoallyl)-UTP (aaUTP) into the aRNA during in vitro transcription, as described previously (17). Prior to amplification, the RNA quality was tested in each sample using the NanoDrop and Agilent 2100 Bioanalyzer. Each of the two compared samples of amplified RNAs from control and silenced RGC-5 cells was labeled with Cy-3 and Cy-5 dye (Amersham Biosciences). Once purified and fragmented, the dye-labeled amplified RNA was used for microarray hybridization. Equivalent amounts of the two compared experimental amplified RNAs were hybridized with the Agilent Rat Oligo Microarrays (Agilent, Santa Clara, CA) using dye-swap design according to the manufacturer's instructions.
Microarray Profiling—Agilents two-color Genomic Rat Oligo microarrays that represent ∼41,000 rat genes and transcripts were used to analyze changes in global gene expression induced by siRNA silencing of the Syn G gene in the RGC-5. Hybridization was carried out in SureHyb chambers. A total of four replicates (two biological and two technical) were performed using a dye-swap design to eliminate the “dye bias” error. Data analysis included filtration, normalization, and signal quality control, and was carried out at an Agilent-certified Microarray Facility. Real-time quantitative PCR and Western blot were used to validate changes in gene expression levels. Differentially expressed genes with changes exceeding 1.3-fold at the FDR < 1% were identified, clustered based on their functions, and analyzed for relative enrichment on generic pathway maps using Metacore Analytical software and data base (Genego Inc., St. Joseph, MI).
Microarray Data Processing and Analysis—For the purpose of gene expression comparison each array was normalized for signal intensities across the whole array and locally, using the Lowess normalization (36). We analyzed only those genes that passed several Agilent-recommended quality control criteria, as described previously (17). To determine differential gene expression, the “naïve RGC-5/reference RNA” ratios were divided by the ratios of “silenced RGC-5/reference RNA.” One class significance analysis of microarrays with FDR < 1% and -fold change of 2.0 was used to determine genes with consistent differences in expression (37, 38). Our final list of significantly changed genes was obtained by application of one class significance analysis of microarrays (FDR < 1%) to identify genes with consistent ratios of naïve RGC-5/silenced RGC-5.
qRT-PCR—Relative quantification was performed as described by the manufacturer (Applied Biosystems) with TaqMan reagents starter kit for a two-step qRT-PCR. The following probes (Applied Biosystems) specific for rat mRNAs were used to validate the gene array data: periostin, Rn00580566_m1; Kcnh2, Rn 00588515_m1; Sema 3A, Rn00436469_m1; Gap43, Rn00567901_m1; Fbn1, Rn00582774_m1; Wilms tumor 1 (Wt-1), Rn00580566_m1; Pim-3, Rn00575067_m1; MAPK-8, Rn01218952_m1; PI3K class 1, Rn00579366_m1; PP2A regulatory α-isoform, Rn00588874_m1; PP2A regulatory β-isoform, Rn00710561_m1; calmodulin 1, Rn00821407_g1; p70 S6 kinase 1, Rn00583148_m1. β-Actin (Rn00667869-m1) was used as an endogenous control. Amplification was performed on an Applied Biosystems 7300 instrument using the following conditions: 2 min at 50 °C (hold), 10 min at 95 °C (hold), 15 s at 95 °C, and 1 min at 60 °C (45 cycles). The samples were amplified in triplicate and three independent isolations of RNA/cDNA were used. In each experiment non-template controls (without cDNA) did not give amplification signals. Amplification products were confirmed by sequencing. The results of qRT-PCR were in agreement with the gene array data (supplemental data Table).
Statistical Analysis—At least three sets of transfection were performed for each experiment with siRNA. In experiments with cell viability a p < 0.05 was used to determine significant differences between the groups. Columns represent the mean of at least three individual experiments; bars are S.D. A paired Student's t test was used to assess significant differences between groups.
RESULTS
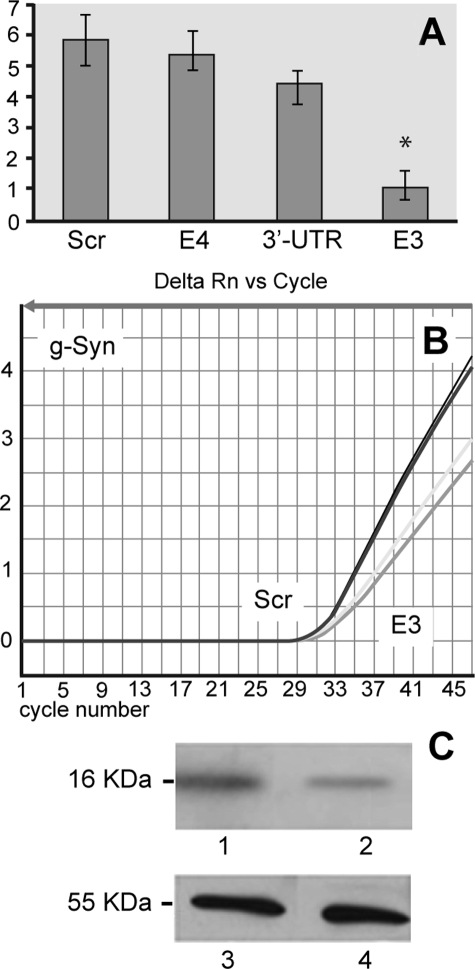
Syn G Silencing Reduces both Syn G mRNA and Protein Levels—To identify a specific target sequence that is efficient for silencing the Syn G gene, we generated three siRNA to the three unique regions, i.e. to exon 3 (E3), exon 4 (E4), and 3′-UTR (Table 1). The efficiency of each siRNA in silencing the Syn G gene was first assessed by analyzing the amount of corresponding mRNA using qRT-PCR. The results obtained showed that siRNA to E4 and 3′-UTR caused only moderate reduction of Syn G mRNA compared with control (11 and 23% reduction, respectively), whereas siRNA to E3 significantly inhibited Syn G expression (82% reduction) (Fig. 1A). Fig. 1B demonstrates a representative amplification plot with mRNA isolated from cells with knockdown Syn G (siRNA to E3) and scrambled control siRNA (Scr). In addition to assessing levels of the Syn G mRNA in the knockdown and control cells, the amounts of corresponding protein was compared in these cell lysates by Western blot. In good agreement with the gene expression data, Western blot results showed ∼85% reduction in the Syn G protein level in RGC-5 silenced with siRNA to E3 (Fig. 1C). Thus, in further experiments we used siRNA to E3, identified as the most effective target for siRNA-mediated silencing of the Syn G gene in RGC-5 stable clones.
FIGURE 1.
Effect of siRNAs on the expression of Syn G in RGC-5 cells. A, qRT-PCR analysis of Syn G expression. Gene expression plot is shown for scrambled siRNA (Scr), siRNA designed for exon 4 (E4), siRNA to 3′-UTR, and siRNA to exon 3 (E3). The combined results of relative quantification for three independent experiments demonstrate that siRNA to E3 has the highest inhibitory potential. Star shows statistically significant difference (p < 0.05). B, a representative amplification plot with cDNAs from two clones generated with scrambled siRNA (Scr, two left curves) and two clones generated with siRNA to exon 3 (E3, two right curves). C, effect of siRNA specific to E3 on the amount of Syn G analyzed by Western blot. Lane 1, scrambled siRNA; lane 2, siRNA to E3; lanes 3 and 4, samples identical to 1 and 2 were treated with an Ab to α-tubulin. These results show considerable (82–85%) reduction of both Syn G mRNA (A) and protein (C) in clones treated with siRNA to E3.
Effect of Syn G Silencing on Cell Viability—To determine whether down-regulation of Syn G affects cell viability we have subjected both RGC-5 cultures with Syn G knockdown and control cells to four cytotoxic compounds, i.e. etoposide, staurosporine, HNE, and hydrogen peroxide. As shown on Fig. 2, the viability of cells with Syn G silencing was decreased in the presence of staurosporine (A) and HNE (B and C). The maximal effect of Syn G knockdown on cell viability relative to “scrambled” control was 33% (500 nm staurosporine, 15 h, Fig. 2A) and 23% (35 μm HNE, 6 h, Fig. 2B). In the presence of etoposide and hydrogen peroxide no statistically significant changes in cell viability were observed (not shown).
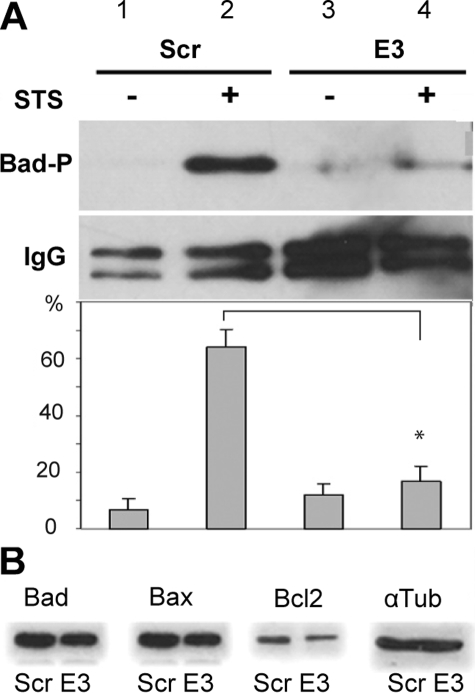
Effect of Syn G Silencing on Prosurvival and Proapoptotic Proteins in Syn G-silenced Cells—We did not find any statistically significant differences in the expression level of pro-apoptotic (Bad and Bax) and found only a slight reduction in the level of pro-survival proteins (Bcl-2) in the RGC-5 cells with silenced Syn G (Fig. 3A). To further elucidate a mechanism of the reduced cell viability of the Syn G-silenced cells, we then examined the phosphorylation status of Bad protein. We selected Bad, because the phosphorylation status of this pro-apoptotic member of the Bcl2 family plays a critical role in RGC survival (31, 32, 39, 40). As shown in Fig. 3B, in cells with Syn G knock-down, the amount of phosphorylated Bad (Bad-P) at Ser112 was reduced to 16% (B, lane 4) compared with the control RGC-5 cells treated with scrambled siRNA (B, lane 2).
FIGURE 3.
Effect of Syn G silencing on Bcl-2 family members. The amount of proapoptotic and antiapoptotic proteins was compared by immunoblot in cells with Syn G knockdown (E3) and control cells (scrambled, Scr). A, decrease in BAD phosphorylation in Syn G knockdown cells is shown. RGC-5 control cells (Scr) and cells with Syn G silencing (E3) were treated with staurosporine (0.1 μm, 12 h) (STS+) or 0.01% DMSO (STS-). Cell lysates were immunoprecipitated with Ab recognizing total endogenous Bad. The precipitates were dissolved in 2× SB buffer and used for Western blot with Ab specific to Bad-P (phosphor-Bad, Ser-112, mouse clone 7E11). In cells with Syn G knockdown (E3), the amount of Bad-P phosphorylated at Ser112 was reduced to 16% (B, lane 4) compared with the control RGC-5 cells treated with scrambled siRNA (B, lane 2). The amount of high chain IgG is shown to compare the loading. A bar chart shows quantification of Bad-P by densitometric analysis. The data are presented as percentage of each band relative to all four bands. The blots were repeated three times. The asterisk indicates that p < 0.01 when comparing the amount of Bad P in E3 (lane 4) and scrambled treated cells (lane 2). B, the amount of Bad, Bax, and Bcl-2 was assessed in cells with knockdown Syn G and scrambled controls. Only a small and statistically insignificant decrease in the level of Bcl-2 in γ-synuclein silenced cells compared to scrambled controls was found.
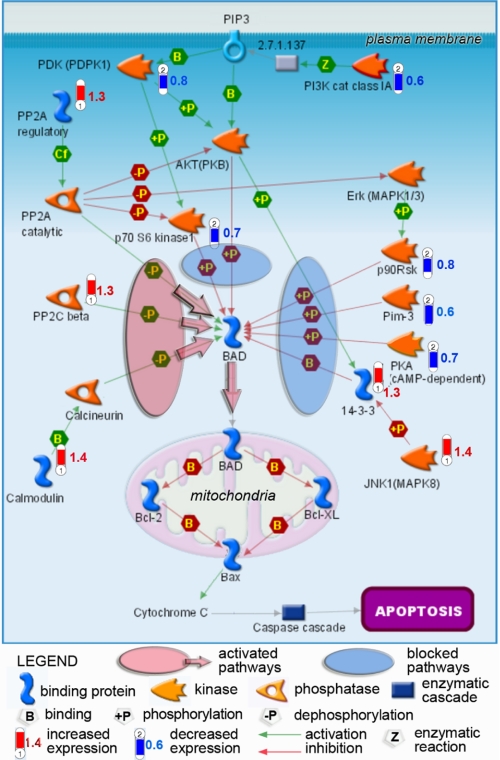
Expression of Protein Kinases and Phosphatases in Syn G Knockdown Cells—Given Bad activity is controlled by multiple phosphorylation/dephosphorylation pathways, we utilized gene arrays to determine whether Syn G silencing has any effect on the expression levels of corresponding kinases and phosphatases. Our pathway analysis has revealed coordinated shift in the activity of genes controlling phosphorylation/dephosphorylation of Ser112 in the Bad protein. As shown on Fig. 4, Syn G knockdown in RGC-5 cells resulted in decreased levels of genes encoding several kinases, including serine/threonine kinase PIM-3, p70 S6 kinase 1, p90 ribosomal S6 kinase (RSK), and protein kinase A. In addition, gene transcripts for two upstream activators of AKT (PKB) kinase, PDK (PDPK1) and PI3K Class 1A, were also reduced in the knockdowns. In synergy, the gene for PP2A kinase, the negative regulator of AKT (PKB) and p70 S6 kinase 1, were activated. At the same time, the expression levels of genes for enzymes catalyzing dephosphorylation of Ser112 in Bad, i.e. protein phosphatases 2A (PP2A) and PP2C, were increased. In accord, the gene for the upstream activator of the calcineurin phosphatase calmodulin was up-regulated. To validate the microarray results, we conducted qRT-PCR analysis of 12 genes including 7 genes encoding kinase, phosphatase, or their regulators. The results indicated overall agreement between the microarray data and the qRT-PCR results (supplemental data Table). Thus, as shown in Fig. 4, the concerted alterations of these enzymatic pathways at gene expression levels would most likely result in reduced phosphorylation of Bad at Ser112 and subsequent translocation of the activated protein into mitochondria.
FIGURE 4.
Regulatory map of Bad phosphorylation pathways with changes in transcriptional activities revealed in RGC-5 cells with Syn G knockdown. Transcriptional changes detected by microarray analysis and mapped by the Metacore software show coordinated down-regulation (blue-“thermometers”) of several kinases and their activators, paralleled by activation (red“thermometers”) of phosphatases targeting Ser112 in Bad protein. In synergy with increased phosphorylation of 14-3-3 leading to release of the bound Bad in the cytoplasm, these events will result in dephosphorylation-dependent translocation of activated Bad to mitochondria, which is confirmed by our Western blot data. Increase in non-phosphorylated BAD that heterodimerizes with Bcl-XL or Bcl-2 in mitochondria, causes cytochrome c release and activation of the apoptotic cascade, is in good agreement with the decreased cell survival. Numbers next to thermometers indicate -fold change in expression levels of corresponding genes in Syn G knockdown cells.
14-3-3 Phosphorylation in Cells with Syn G Knockdown—Because the pro-apoptotic activation of Bad depends both on its own phosphorylation status and on binding to 14-3-3 proteins, we examined the level of phosphorylation of 14-3-3 that regulates such binding (32, 41). We compared the effect of Syn G silencing on the stoichiometry of phosphorylated (14-3-3P) and non-phosphorylated 14-3-3. The cells were sensitized by the apoptosis inducer staurosporine, which affects signaling cascades and inhibits protein kinases (42).
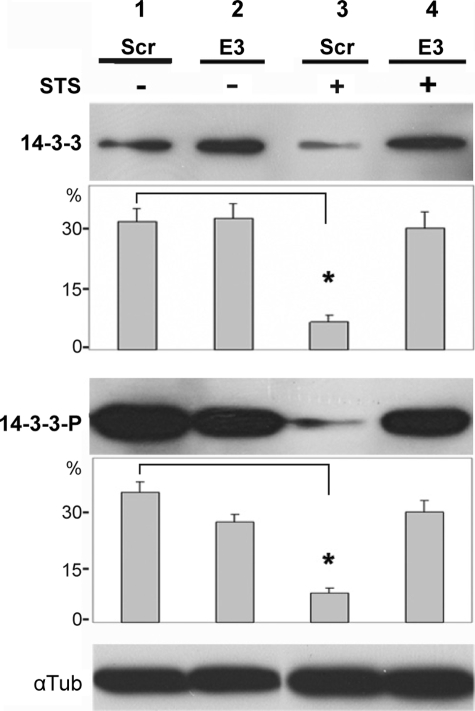
The Syn G knockdown suppressed a significant decrease in both the amount (Fig. 5, top panel, lanes 1 and 3) and phosphorylation status (Fig. 5, middle panel, lanes 1 and 3) of 14-3-3 observed in control RGC-5 cells in response to staurosporine. In Syn G knockdown cells both phosphorylated and non-phosphorylated 14-3-3 was up-regulated ∼4 times compared with corresponding control cells (6.8 versus 31.3% for 14-3-3, p < 0.01; 8.0 versus 35.1% for 14-3-3P, p < 0.01). This increase in 14-3-3 and 14-3-3P was revealed only in the presence of staurosporine (Fig. 5, A–D, lanes 3 and 4).
FIGURE 5.
Analysis of 14-3-3 and 14-3-3P in Syn G knockdown cells by immunoblot. To compare the levels of 14-3-3 (A and B) and 14-3-3P (C and D), 30 μg of cell lysates were separated on 12% PAGE, transferred to a membrane, and probed with Ab against total 14-3-3 (A) or 14-3-3P (C). Lysates from scrambled cells (Scr) and cells with Syn G knockdown (E3) were treated with STS (+) or incubated without STS (-). C shows the blot striped and probed with antibody against α-tubulin for loading control. B and D, quantification of 14-3-3 (A) and 14-3-3P (C), respectively, by densitometric analysis. The data are presented as percentage of each band relative to all four bands shown in A and C. The blots were repeated three times; stars show statistically significant differences (p < 0.05).
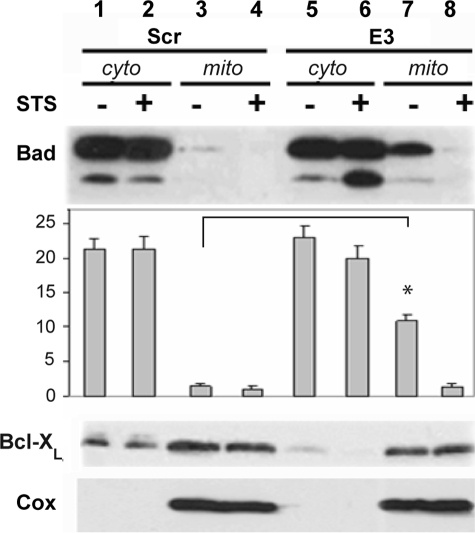
Bad Translocation to Mitochondria in Syn G Knockdown Cells—We further analyzed the localization of Bad in the cytoplasm and mitochondria of Syn G knockdown and control cells. No statistically significant differences in the amount of Bad in the cytosolic fraction have been detected between Syn G knock-down and control cells (Fig. 6, top panel, lanes 1, 2, 5, and 6). However, in the mitochondrial fraction of Syn G knockdown cells the amount of Bad was significantly increased relative to control cells. These differences have been observed only in the absence of staurosporine (1.5 versus 10.9%, p < 0.05, Fig. 6, top panel, lanes 3 and 7). Significantly, the translocation of Bad to mitochondria in Syn G knockdown cells was paralleled by similarly increased translocation of BcL-XL to this fraction from the cytoplasm (compare lanes 7 and 8 with lanes 5 and 6 in the middle panel of Fig. 6).
FIGURE 6.
Translocation of Bad and BcL-XL to mitochondrial fraction in Syn G knockdown cells. The amount of Bad and BcL-XL was assessed in cytoplasmic (cyto) and mitochondrial (mito) fractions by immunoblot. 35 μg of protein were loaded on 12% PAGE. A, lanes 1–4, lysates from control clone (scrambled, Scr); lanes 5–8, lysates from clone with Syn G knockdown (E3). Lanes 1, 2, 5, and 6, cytosolic fractions (cyto); lanes 3, 4, 7, and 8, mitochondrial fractions (mito). The blot was probed with K-17 Bad Ab Sc942 to determine the localization of Bad in cytoplasmic and mitochondrial fractions. Anti-rabbit horseradish peroxidase (Amersham Biosciences) was used as the secondary Ab at a 1:40,000 dilution. B, the blot was probed with BcL-XL Ab to determine the localization of BcL-XL in cytoplasmic and mitochondrial fractions. The secondary Ab was the same as in A. C, COX IV Ab was used to characterize the purity of isolated fractions. D, quantification of Bad by densitometric analysis. The data are presented as percentage of each band relative to all eight bands together shown in A. The blots were repeated three times; stars show statistically significant differences (p < 0.05).
DISCUSSION
High level of Syn G expression in RGC (16, 23, 24) and its down-regulation in the course of glaucomatous alterations (25, 26) has put forward questions concerning both normal function(s) of this protein and the molecular and cellular effects of a precipitous decline observed in glaucoma. In this study, we investigated how the decrease of Syn G in RGC affects cell viability, signaling pathways, and transcription of other genes. We utilized efficient silencing of the Syn G gene by siRNA designed to the third exon (E3) of the Syn G. Treatment with this siRNA decreases the levels of both specific mRNA and protein in RGC-5 cells. Knocking down endogenous Syn G expression in RGC-5 cells provides an in vitro model for precipitous down-regulation of the protein in glaucomatous neurons.
We detected a drop in RGC-5 viability following treatment with staurosporine or HNE in Syn G knockdown relative to that of control cells. Reduced viability of RGC-5 cells with the Syn G knockdown following treatment with HNE or staurosporine may be explained by a role of Syn G in transcriptional regulation of genes that modulate death/survival signaling (27–30). Alternatively, Syn G may influence cell viability via chaperone activity, as suggested previously for all three members of the synuclein family (43, 44). Chaperone activity is known to be implicated in protection from stressors and cytotoxic agents.
The absence of a very significant effect of Syn G knockdown on cell viability may be explained by incomplete knockdown of the protein in RGC-5 cells, so that the activity of the residual ∼20% of the protein might have been sufficient to support survival in the majority of cells. In addition, the partially compensatory effect by the two other members of the synuclein family, which are structurally similar (45), may provide an alternative explanation, as suggested earlier (46–48).
A relatively moderate effect of Syn G silencing on RGC-5 cell viability in our experiments is in good agreement with the data obtained using the Syn G knock-out mouse (47). The analysis of these knock-out mice showed insignificant changes in neuron survival in the brains of such animals.
The role of Syn G in regulation of kinases was established as an important regulatory mechanism leading to pathology, including tumor development via modulation of the JNK pathway (29, 30). However, the role of this mechanism in glaucoma progression has not been established yet. One of the well characterized pathways of RGC death involves activation of the Bcl-2 family of proapoptotic proteins, which includes Bad (32, 49). The phosphorylation status of Bad, a member of the Bcl-2 protein family, is an important checkpoint governing life or death decisions. A strong correlation has been established between reduced levels of phosphorylation of serine residues 112, 136, and 155 on BAD and apoptotic cell death (39, 50–53). Similarly, significant reduction in the amount of phosphorylated Ser112 on Bad in our knockdown experiments with RGC-5 cells correlated with increased sensitivity to staurosporine and HNE. These results may imply that Syn G is required for maintaining the homeostatic level of Bad phosphorylation to block apoptosis in RGCs, whereas removal of such blockade via pathological down-regulation of Syn G will likely facilitate apoptotic RGC death. Importantly, significant reduction in both Syn G gene expression and in Bad phosphorylation was recently reported in rodent models of intraocular pressure-induced glaucoma (25, 32).
Bad phosphorylation at Ser112 is controlled by a combined activity of several pathways such as p90 ribosomal S6 kinase (RSK2) (53, 54), protein kinase A (55), and proto-oncogenic kinase Pim-3 (52), all of which are reduced in cells with Syn G knockdown. Other down-regulated pathways include AKT and p70 S6 kinases, which have been implicated in phosphorylation of Bad at Ser136 (56). At the same time, our analysis revealed transcriptional activation of JNK1, which is known to suppress Ser112 phosphorylation at Bad, because increased JNK activity has been shown to correlate with a decrease in phosphorylation of this residue (54). Moreover, JNK has been implicated in Bad dephosphorylation at Ser136, phosphorylation of 14-3-3, and induces release of Bad from the cytoplasmic complex with 14-3-3 (57–59).
The competing process of Bad Ser112 dephosphorylation is catalyzed by a number of enzymes including PP2A (60), PP2C (61), and calcineurin (62) that showed transcriptional activation following Syn G knockdown (Fig. 4). The results of gene expression analysis showed that coordinated down-regulation of kinases phosphorylating Bad Ser112 is accompanied by transcriptional activation of phosphatases that dephosphorylate the same serine residue in Bad. Therefore, despite relatively mild changes in transcriptional regulation of enzymes catalyzing Bad phosphorylation/dephosphorylation at Ser112, their synergy suggested a coordinated decrease in Bad Ser112 phosphorylation as a result of Syn G knockdown. A smaller magnitude of these changes can also be attributed to incomplete silencing of the gene by siRNA targeting E3 in Syn G.
Previous reports have demonstrated that anti-apoptotic activity of 14-3-3 proteins block translocation of Bad to mitochondria via binding and sequestering it in the cytoplasm. When dephosphorylated and released from 14-3-3, Bad binds and inactivates anti-apoptotic Bcl-XL and Bcl-2 proteins in the mitochondria, triggering cytochrome c-mediated apoptotic signaling (53, 63). Both dephosphorylation of Bad and phosphorylation of 14-3-3 by JNK1 serve to inhibit binding with 14-3-3 proteins (53, 60, 64–67), Bad translocation to mitochondria, and RGC death (32). Interestingly, synucleins and 14-3-3 possess some structural similarity (68) and regulation of activity by phosphorylation (69, 70).
Earlier Syn G role in the modulation of kinases was established as an important regulatory mechanism leading to pathology (28–30). In particular, Pan and collaborators (30) showed that Syn G contributes to tumor development by protecting cancer cells through modulating JNK pathway. However, the role of Syn G in controlling this mechanism in glaucoma progression was not described.
Interesting results were obtained on the effect of Syn G knockdown on regulation of 14-3-3 activity in RGC-5 cells. In contrast to control, where both phosphorylated and non-phosphorylated 14-3-3 forms were considerably down-regulated in the presence of staurosporine, cells with Syn G knockdown preserved both forms of 14-3-3 virtually unaffected by staurosporine (Fig. 5). Staurosporine-induced down-regulation of 14-3-3 has been described previously (71), but a strong interference of Syn G silencing with this effect (Fig. 5) is a novel observation. Further experiments are necessary to elucidate the role and mechanism of Syn G regulation of 14-3-3 family members. Our transcriptomics-based pathway analysis in RGC-5 cells with Syn G knockdown suggests that transcriptional activation of both 14-3-3 and JNK1 (MAPK-8) kinase, which phosphorylates 14-3-3, may provide a plausible hypothesis for such regulation (Fig. 4). If validated, this regulatory mechanism might explain a decreased sequestering of Bad in the cytoplasm and translocation into mitochondria.
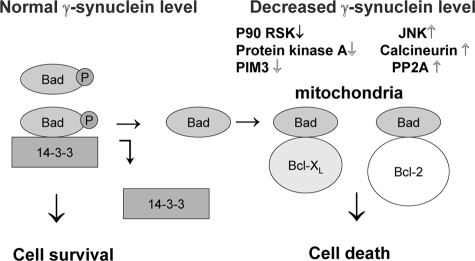
Thus, changing the intracellular concentration of Syn G may affect proapoptotic and prosurvival pathways in RGC in the following way (Fig. 7). At the normal Syn G concentration phosphorylated Bad is associated with 14-3-3 in the cytosol (left). Lowering of Syn G down-regulates kinases, up-regulates phosphatases, and causes Bad dephosphorylation. Dephosphorylated Bad then triggers the mitochondrial pathway of apoptosis through interaction with pro-survival Bcl-2 family members, such as BcL-XL and Bcl-2 (Fig. 7, right).
FIGURE 7.
Schematic model for Syn G effect on proapoptotic and prosurvival pathways in RGC. At the normal Syn G intracellular concentration phosphorylated Bad complexes with 14-3-3 in the cytosol (left). Reduction of the Syn G level results in Bad dephosphorylation. Dephosphorylated Bad then triggers the mitochondrial pathway of apoptosis through interaction with pro-survival Bcl-2 family members, such as Bcl-XL and Bcl-2 (right).
In conclusion, combined results of this and previous studies suggest that Syn G plays an essential regulatory role in neuronal homeostasis, resistance to stress, and neuroprotection. Both artificial silencing and pathological down-regulation of this protein in animal models of glaucoma correlate with increased susceptibility to cytotoxic agents, neurotoxicity, and RGC loss. These results correlate well with previous data suggesting a dual role of another member of the synuclein family, α-synuclein, in neuronal survival (72). Similar to our findings, the protective effect of α-synuclein against neurodegeneration is realized through regulation of PI3K/Akt and Bcl-2 pathways. Although it is well studied in cancer, the exact role of Syn G in progression of glaucoma and other retinal diseases has yet to be elucidated.
In the vast majority of late-stage breast and ovarian cancers Syn G is dramatically up-regulated and its overexpression can enhance tumorigenicity (30, 72–74). Thus, siRNA-based silencing of Syn G utilized in this study may prove efficient in cancer gene therapy.
Supplementary Material
Acknowledgments
We thank Dr. Raghu Krishnamoorthy (University of North Texas Health Science Center, Fort Worth, TX) for RGC-5 cells, Dr. L. H. Boise (University of Miami, FL), for antibody against Bcl-XL. We thank Julia Shestopalov for help with illustrations and the technical help of Kristin Cain is acknowledged.
This work was supported, in whole or in part, by National Institutes of Health Grants EY02687, EY017991, and Center Grant P30 EY014801. This work was also supported by a Veterans Affairs Merit Review grant, The Glaucoma Foundation, Midwest Biomedical Research Foundation (MBRF), and an unrestricted grant to the University of Miami Department of Ophthalmology from Research to Prevent Blindness. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains a supplemental Table.
Footnotes
The abbreviations used are: RGC, retinal ganglion cells; Syn G, γ-synuclein; siRNA, small interference RNA; UTR, untranslated region; E3, exon 3; E4, exon 4; Scr, scrambled; Wt-1, Wilms tumor 1; HNE, 4-hydroxynonenal; p70 S6 kinase 1, p70 ribosomal protein S6 kinase 1; p90 RSK, p90 ribosomal S6 kinase RSK; PI3K, phosphatidylinositol 3-kinase; PP2A, protein phosphatase 2A; PP2C, phosphatase 2C; Ab, antibody; qRT, quantitative real time; MAPK, mitogen-activated protein kinase; JNK, c-Jun NH2-terminal kinase.
References
- 1.Qigley, H. A. (1996) Br. J. Ophthalmol. 80 389-393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fortune, B., Bui, B. V., Morrison, J. C., Johnson, E. C., Dong, J., Cepurna, W. O., Jia, L., Barber, S., and Cioffi, G. A. (2004) Investig. Ophthalmol. Vis. Sci. 45 1854-1862 [DOI] [PubMed] [Google Scholar]
- 3.Gupta, N., and Yücel, Y. H. (2007) Curr. Opin. Ophthalmol. 18 110-114 [DOI] [PubMed] [Google Scholar]
- 4.Morgan, J. E., Uchida, H., and Caprioli, J. (2000) Br. J. Ophthalmol. 84 303-310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pease, M. E., McKinnon, S. J., Quigley, H. A., Kerrigan-Baumrind, L. A., and Zack, D. J. (2000) Investig. Ophthalmol. Vis. Sci. 41 764-774 [PubMed] [Google Scholar]
- 6.Tezel, G., and Wax, M. B. (2007) Glaucoma Chem. Immunol. Allergy 92 221-227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Valenzuela, E., Shareef, S., Walsh, J., and Sharma, S. C. (1995) Exp. Eye Res. 61 33-44 [DOI] [PubMed] [Google Scholar]
- 8.Levin, L. A., and Peeples, P. (2008) Am. J. Manag. Care 14 S11-S14 [PubMed] [Google Scholar]
- 9.Quigley, H., Nickells, R., Kerrigan, L. A., Pease, M. E., Thibault, D. J., and Zack, D. J. (1995) Investig. Ophthalmol. Vis. Sci. 36 774-786 [PubMed] [Google Scholar]
- 10.McKinnon, S. J., Lehman, D. M., Kerrigan-Baumrind, L. A., Merges, C. A., Pease, M. E., Kerrigan, D. F., Ransom, N. L., Tahzib, N. G., Reitsamer, H. A., Levkovitch-Verbin, H., Quigley, H. A., and Zack, D. J. (2002) Investig. Ophthalmol. Vis. Sci. 43 1077-1087 [PubMed] [Google Scholar]
- 11.Green, D. R., and Reed, J. C. (1998) Science 28 1309-1312 [DOI] [PubMed] [Google Scholar]
- 12.Nickells, R. W. (2007) Can. J. Ophthalmol. 42 78-87 [Google Scholar]
- 13.Johnson, E. C., Jia, L., Cepurna, W. O., Doser, T. A., and Morrison, J. C. (2007) Investig. Ophthalmol. Vis. Sci. 48 3161-3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piri, N., Kwong, J. M., Song, M., Elashoff, D., and Caprioli, J. (2006) Mol. Vis. 12 1660-1673 [PubMed] [Google Scholar]
- 15.Yang, Z., Quigley, H. A., Pease, M. E., Yang, Y., Qian, J., Valenta, D., and Zack, D. J. (2007) Investig. Ophthalmol. Vis. Sci. 48 5539-5548 [DOI] [PubMed] [Google Scholar]
- 16.Farkas, R. H., Qian, J., Goldberg, J. L., Quigley, H. A., and Zack, D. J. (2004) Investig. Ophthalmol. Vis. Sci. 45 2503-2513 [DOI] [PubMed] [Google Scholar]
- 17.Ivanov, D., Dvoriantchikova, G., Nathanson, L., McKinnon, S. J., and Shestopalov, V. I. (2006) FEBS Lett. 580 331-335 [DOI] [PubMed] [Google Scholar]
- 18.Khalyfa, A., Chlon, T., Qiang, H., Agarwal, N., and Cooper, N. G. (2007) Mol. Vis. 13 293-308 [PMC free article] [PubMed] [Google Scholar]
- 19.Tezel, G., and Yang, X. (2005) Exp. Eye Res. 81 207-217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diskin, S., Kumar, J., Cao, Z., Schuman, J. S., Gilmartin, T., Head, S. R., and Panjwani, N. (2006) Investig. Ophthalmol. Vis. Sci. 47 1491-1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liton, P. B., Luna, C., Challa, P., Epstein, D. L., and Gonzalez, P. (2006) Mol. Vis. 12 774-790 [PMC free article] [PubMed] [Google Scholar]
- 22.Golubnitschaja-Labudova, O., Liu, R., Decker, C., Zhu, P., Haefliger, I. O., and Flammer, J. (2000) Curr. Eye Res. 21 867-876 [DOI] [PubMed] [Google Scholar]
- 23.Surguchov, A., McMahon, B., Masliah, E., and Surgucheva, I. (2001) J. Neurosci. Res. 65 68-77 [DOI] [PubMed] [Google Scholar]
- 24.Surgucheva, I., Weissman, A., Goldberg, J. L., Shnyra, A., and Surguchov, A. (2008) Mol. Vis. 14 1540-1548 [PMC free article] [PubMed] [Google Scholar]
- 25.Soto, I., Oglesby, E., Buckingham, B. P., Son, J. L., Roberson, E. D., Steele, M. R., Inman, D. M., Vetter, M. L., Horner, P. J., and Marsh-Armstrong, N. (2008) J. Neurosci. 28 548-561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Surgucheva, I., McMahan, B., Ahmed, F., Tomarev, S., Wax, M. B., and Surguchov, A. (2002) J. Neurosci. Res. 68 97-106 [DOI] [PubMed] [Google Scholar]
- 27.Surgucheva, I., Sivak, J. M., Fini, M. E., Palazzo, R. E., and Surguchov, A. P. (2003) Arch. Biochem. Biophys. 410 167-176 [DOI] [PubMed] [Google Scholar]
- 28.Surguchov, A., Palazzo, R. E., and Surgucheva, I. (2001) Cell Motil. Cytoskeleton 49 218-228 [DOI] [PubMed] [Google Scholar]
- 29.Gupta, A., Inaba, S., Wong, O. K., Fang, G., and Liu, J. (2003) Oncogene 22 7593-7599 [DOI] [PubMed] [Google Scholar]
- 30.Pan, Z. Z., Bruening, W., Giasson, B. I., Lee, V. M., and Godwin, A. K. (2002) J. Biol. Chem. 277 35050-35060 [DOI] [PubMed] [Google Scholar]
- 31.Huang, W., Fileta, J. B., Dobberfuhl, A., Filippopolous, T., Guo, Y., Kwon, G., and Grosskreutz, C. L. (2005) Proc. Natl. Acad. Sci. U. S. A 102 12242-12247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang, X., Luo, C., Cai, J., Pierce, W. M., and Tezel, G. (2008) Investig. Ophthalmol. Vis. Sci. 49 2483-2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krishnamoorthy, R. R., Agarwal, P., Prasanna, G., Vopat, K., Lambert, W., Sheedlo, H. J., Pang, I. H., Shade, D., Wordinger, R. J., Yorio, T., Clark, A. F., and Agarwal, N. (2001) Brain Res. Mol. Brain Res. 86 1-12 [DOI] [PubMed] [Google Scholar]
- 34.Charles, I., Khalyfa, A., Kumar, D. M., Krishnamoorthy, R. R., Roque, R. S., Cooper, N., and Agarwal, N. (2005) Investig. Ophthalmol. Vis. Sci. 46 1330-1338 [DOI] [PubMed] [Google Scholar]
- 35.Morales, A. A., Gutman, D., Lee, K. P., and Boise, L. H. (2008) Blood 111 5152-5162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang, Y. H., Dudoit, S., Luu, P., Lin, D. M., Peng, V., Ngai, J., and Speed, T. P. (2002) Nucleic Acids Res. 30 15-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tusher, V. G., Tibshirani, R., and Chu, G. (2001) Proc. Natl. Acad. Sci. U. S. A 98 5116-5137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khatri, P., Draghici, S., Ostermeier, G. C., and Krawetz, S. A. (2002) Genomics 79 266-336 [DOI] [PubMed] [Google Scholar]
- 39.Kamada, H., Nito, C., Endo, H., and Chan, P. (2007) J. Cereb. Blood Flow Metab. 27 521-533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang, E., Zha, J., Jockel, J., Boise, L. H., Thompson, C. B., and Korsmeyer, S. J. (1995) Cell 80 285-291 [DOI] [PubMed] [Google Scholar]
- 41.Sunayama, J., Tsuruta, F., Masuyama, N., and Gotoh, Y. (2005) J. Cell Biol. 170 5-304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wan, X., Yokoyama, Y., Shinohara, A., Takahashi, Y., and Tamaya, T. (2002) Cell Death Differ. 9 414-420 [DOI] [PubMed] [Google Scholar]
- 43.Jiang, Y., Liu, Y. E., Goldberg, I. D., and Shi, Y. E. (2004) Cancer Res. 64 4539-4546 [DOI] [PubMed] [Google Scholar]
- 44.Souza, J. M., Giasson, B. I., Lee, V. M., and Ischiropoulos, H. (2000) FEBS Lett. 474 116-119 [DOI] [PubMed] [Google Scholar]
- 45.Surguchov, A. (2009) Intl. Rev. Cell Mol. Biol. 270 227-317 [Google Scholar]
- 46.Kuhn, M., Haebig, K., Bonin, M., Ninkina, N., Buchman, V. L., Poths, S., and Riess, O. (2007) Neurogenetics 8 71-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ninkina, N., Papachroni, K., Robertson, D. C., Schmidt, O., Delaney, L., O'Neill, F., Court, F., Rosenthal, A., Fleetwood-Walker, S. M., Davies, A. M., and Buchman, V. L. (2003) Mol. Cell. Biol. 23 8233-8245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Senior, S. L., Ninkina, N., Deacon, R., Bannerman, D., Buchman, V. L., Cragg, S. J., and Wade-Martins, R. (2008) Eur. J. Neurosci. 27 947-957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim, H. S., and Park, C. K. (2005) Brain Res. 1057 17-28 [DOI] [PubMed] [Google Scholar]
- 50.Bonni, A., Brunet, A., West, A. E., Datta, S. R., Takasu, M. A., and Greenberg, M. E. (1999) Science 286 1358-1362 [DOI] [PubMed] [Google Scholar]
- 51.del Peso, L., González-Garcia, M., Page, C., Herrera, R., and Nuñez, G. (1997) Science 278 687-689 [DOI] [PubMed] [Google Scholar]
- 52.Li, Y. Y., Popivanova, B. K., Nagai, Y., Ishikura, H., Fujii, C., and Mukaida, N. (2006) Cancer Res. 66 6741-6747 [DOI] [PubMed] [Google Scholar]
- 53.Ohi, N., Nishkawa, Y., Tokairin, T., Yamamoto, Y., Doi, Y., Omori, Y., and Enomoto, K. (2006) Am. J. Pathol. 168 1097-1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.She, Q. B., Ma, W. Y., Zhong, S., and Dong, Z. (2002) J. Biol. Chem. 277 24039-24048 [DOI] [PubMed] [Google Scholar]
- 55.Kaiser, R. A., Liang, Q., Bueno, O., Huang, Y., Lackey, T., Klevitsky, R., Hewett, T. E., and Molkentin, J. D. (2005) J. Biol. Chem. 280 32602-32608 [DOI] [PubMed] [Google Scholar]
- 56.Harada, H., Andersen, J. S., Mann, M., Terada, N., and Korsmeyer, S. J. (2001) Proc. Natl. Acad. Sci. U. S. A. 14 9666-9670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Han, J. Y., Jeong, E. Y., Kim, Y. S., Roh, G. S., Kim, H. J., Kang, S. S., Cho, G. J., and Choi, W. S. (2008) J. Neurosci. Res. 86 3221-3229 [DOI] [PubMed] [Google Scholar]
- 58.Hu, X. F., Li, J., Yang, E., Vandervalk, S., and Xing, P. X. (2007) Br. J. Cancer 96 918-927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoshida, K., Yamaguchi, T., Natsume, T., Kufe, D., and Miki, Y. (2005) Nat. Cell Biol. 7 278-285 [DOI] [PubMed] [Google Scholar]
- 60.Chiang, C. W., Kanies, C., Kim, K. W., Fang, W. B., Parkhurst, C., Xie, M., Henry, T., and Yang, E. (2003) Mol. Cell. Biol. 23 6350-6362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Klumpp, S., Selke, D., and Krieglstein, J. (2003) Neurochem. Int. 42 555-560 [DOI] [PubMed] [Google Scholar]
- 62.Wang, H. G., Pathan, N., Ethell, I. M., Krajewski, S., Yamaguchi, Y., Shibasaki, F., McKeon, F., Bobo, T., Franke, T. F., and Reed, J. C. (1999) Science 284 339-343 [DOI] [PubMed] [Google Scholar]
- 63.Springer, J. E., Azbill, R. D., Nottingham, S. A., and Kennedy, S. E. (2000) J. Neurosci. 20 7246-7251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Datta, S. R., Dudek, H., Tao, X., Masters, S., Fu, H., Gotoh, Y., and Greenberg, M. E. (1997) Cell 91 231-241 [DOI] [PubMed] [Google Scholar]
- 65.Hui, L., Rodrik, V., Pielak, R. M., Knirr, S., Zheng, Y., and Foster, D. A. (2005) J. Biol. Chem. 280 35829-35835 [DOI] [PubMed] [Google Scholar]
- 66.Scheid, M. P., Schubert, K. M., and Duronio, V. (1999) J. Biol. Chem. 274 31108-31113 [DOI] [PubMed] [Google Scholar]
- 67.Zha, J., Harada, H., Yang, E., Jockel, J., and Korsmeyer, S. J. (1996) Cell 87 619-628 [DOI] [PubMed] [Google Scholar]
- 68.Ostrerova-Golts, N., Petrucelli, L., Hardy, J., Lee, J. M., Farer, M., and Wolozin, B. (2000) J. Neurosci. 20 6048-6054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pronin, A. N., Morris, A. J., Surguchov, A., and Benovic, J. L. (2000) J. Biol. Chem. 275 26515-26522 [DOI] [PubMed] [Google Scholar]
- 70.Fujiwara, H., Hasegawa, M., Dohmae, N., Kawashima, A., Masliah, E., Goldberg, M. S., Shen, J., Takio, K., and Iwatsubo, T. (2002) Nat. Cell Biol. 4 160-164 [DOI] [PubMed] [Google Scholar]
- 71.Lee, S. W., Choi, S. M., Chang, Y. S., Kim, K. T., Kim, T. H., Park, H. T., Park, B. S., Sohn, Y. J., Park, S. K., Cho, S. H., Chung, W. T., and Yoo, Y. H. (2007) J. Ethnopharmacol. 111 213-218 [DOI] [PubMed] [Google Scholar]
- 72.Seo, J. H., Rah, J. C., Choi, S. H., Shin, J. K., Min, K., Kim, H. S., Park, C. H., Kim, S., Kim, E. M., Lee, S. H., Lee S., Suh, S. W., and Suh, Y. H. (2002) FASEB J. 16 1826-1828 [DOI] [PubMed] [Google Scholar]
- 73.Ahmad, M., Attoub, S., Singh, M. N., Martin, F. L., and El-Agnaf, O. M. (2007) FASEB J. 21 3419-3430 [DOI] [PubMed] [Google Scholar]
- 74.Jia, T., Liu, Y. E., Liu, J., and Shi, Y. E. (1999) Cancer Res. 59 742-747 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.