Abstract
In mammals, the mechanisms regulating the number of active copies of the ∼200 ribosomal RNA (rRNA) genes transcribed by RNA polymerase I are unclear. We demonstrate that depletion of the transcription factor upstream binding factor (UBF) leads to the stable and reversible methylation-independent silencing of rRNA genes by promoting histone H1–induced assembly of transcriptionally inactive chromatin. Chromatin remodeling is abrogated by the mutation of an extracellular signal-regulated kinase site within the high mobility group box 1 domain of UBF1, which is required for its ability to bend and loop DNA in vitro. Surprisingly, rRNA gene silencing does not reduce net rRNA synthesis as transcription from remaining active genes is increased. We also show that the active rRNA gene pool is not static but decreases during differentiation, correlating with diminished UBF expression. Thus, UBF1 levels regulate active rRNA gene chromatin during growth and differentiation.
Introduction
Ribosome biogenesis plays an essential role in growth control, and transcription of the 45S precursor of the ribosomal RNA (rRNA) by RNA polymerase I (Pol I) is limiting for proliferation in most cellular systems (Jorgensen and Tyers, 2004). In addition, through its control of nucleolar formation, rRNA gene transcription indirectly regulates several other essential processes, including titration of oncogenes and tumor suppressors, cellular response to stress, and aging (for reviews see Moss, 2004; Mayer and Grummt, 2005).
A typical human cell contains ∼200 copies of the rRNA genes arranged in tandemly repeated arrays located in nucleolar organizing regions (NORs). Remarkably, despite rRNA gene transcription being limiting for growth, >50% of the rRNA genes are believed to be transcriptionally silent at any one time (for review see Grummt and Pikaard, 2003). The epigenetic mechanisms controlling the activity status of individual ribosomal genes and the reasons why a majority is silenced in higher eukaryotes remain unresolved questions (Huang et al., 2006).
One factor whose function has been linked to regulation of the rRNA gene locus is upstream binding factor (UBF). UBF belongs to the sequence-nonspecific class of high mobility group (HMG) proteins and appears to function exclusively in Pol I transcription (for review see Moss et al., 2007). UBF consists of two polypeptides (UBF1 and 2), which form hetero- and homodimers and arise from alternative splicing of a single transcript (O'Mahony and Rothblum, 1991). Although UBF1 supports robust rRNA gene transcription, UBF2 is fivefold less active (Hannan et al., 1996), but the underlying mechanisms that confer poor transcriptional activity to UBF2 are unclear. In one model, UBF acts through its multiple HMG boxes to induce looping of DNA. This creates the enhancesome, a nucleosome-like structure thought to be responsible for the ability of UBF to modulate rRNA gene transcription (Stefanovsky et al., 2001a,b). Early studies suggested that UBF1 acts predominantly at the promoter in the recruitment of SL1 (selectivity factor 1) and Pol I and in the formation of the preinitiation complex (Smith et al., 1990; McStay et al., 1991; Jantzen et al., 1992). More recently, additional roles have been ascribed to UBF1, including regulation of promoter escape (Panov et al., 2006) and transcription elongation (Moss et al., 2006). Importantly, the association of UBF1 with rRNA genes in vivo is not restricted to the promoter but extends across the entire transcribed portion and to a lesser extent to the intergenic spacer (IGS; Fig. 1, C and D; O'Sullivan et al., 2002). Indeed, consistent with its ability to modify DNA conformation, the inclusion of rRNA gene sequences with high affinity for UBF into ectopic sites on human chromosomes results in the formation of NOR-like structures indicative of “open” chromatin (Mais et al., 2005). In addition, targeting UBF1 to regions of heterochromatin is sufficient to induce large-scale chromatin decondensation (Chen et al., 2004). Together, these data suggest that UBF1 binding throughout the rRNA gene repeat might contribute to the formation of the active chromatin state of rRNA genes. However, direct experiments to demonstrate this function on endogenous rRNA genes and information on the relative contribution of the two UBF isoforms in chromatin remodeling are lacking.
Figure 1.
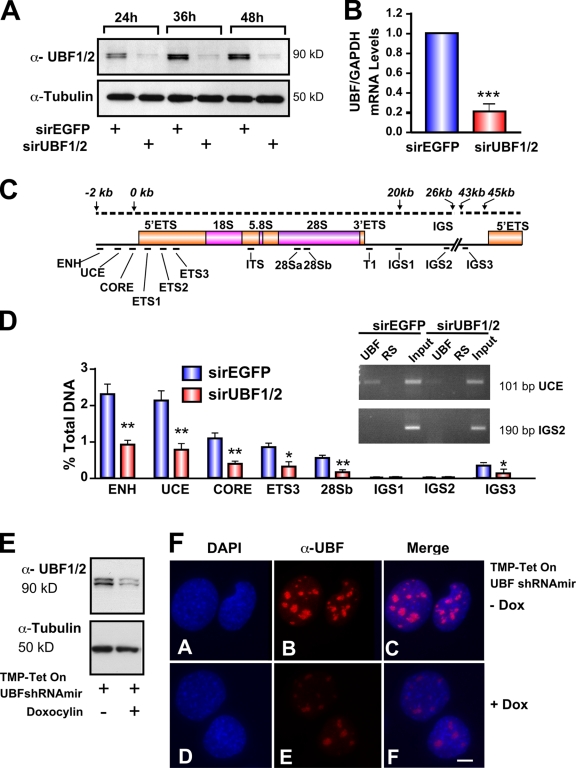
Silencing of UBF1/2 by siRNA. (A) NIH3T3 cells were transfected with siRNA-EGFP or -UBF1/2, and protein samples were harvested as indicated and analyzed by Western blotting. (B) RNA samples were harvested, and UBF and GAPDH mRNA levels were examined by reverse transcription qRT-PCR. n = 3; ***, P < 0.001. (C) Schematic of a murine rRNA gene and the positions of qRT-PCR amplicons. (D) qChIP analysis of UBF binding to the rRNA gene. The percentage of DNA immunoprecipitated with anti-UBF or rabbit serum (RS) antibodies was calculated relative to the unprecipitated input control. The percentage of DNA of rabbit serum controls was subtracted from corresponding UBF samples. n = 6; *, P < 0.05; **, P < 0.01. A representative ethidium bromide gel shows the amount of UCE and IGS2 products amplified after 22 PCR cycles. (E) NIH3T3 cells stably transduced with tetracycline-inducible TMP-UBF shRNAmir were cultured in the presence or absence of doxocyclin and analyzed by Western blotting. (F) Samples in E were analyzed by immunofluorescence for UBF1/2. ENH, enhancer; ITS, internal transcribed spacer; T, terminator region. Mean ± SEM (error bars). Bar, 2 μm.
In this study, we provide strong evidence that UBF1, but not UBF2, regulates the open chromatin structure found in active rRNA genes by preventing linker histone H1–induced assembly of transcriptionally inactive chromatin. Long-term rRNA gene silencing in response to UBF depletion is stably propagated through the cell cycle and through many generations and is not associated with heterochromatic marks related to nucleolar remodeling complex (NoRC)–dependent remodeling, including DNA methylation. Restoring UBF levels rescues the number of active genes. Thus, in contrast to epigenetically silenced rRNA genes, which are methylated, silencing of rRNA genes through UBF depletion is reversible. We also demonstrate that, contrary to accepted dogma, the pool of active ribosomal genes is not static but decreases during differentiation and that this decrease correlates with diminished UBF levels in the absence of changes in ribosomal DNA (rDNA) methylation. Together, these data suggest that modulation of UBF levels might be an important determinant of the relative proportion of active and silent rRNA genes during development.
Results
UBF1 depletion silences rRNA genes
To determine whether UBF is necessary for maintenance of the open chromatin structure found in active NORs, we transfected siRNAs targeting conserved regions of both isoforms of UBF (UBF1/2; Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200805146/DC1) into NIH3T3 cells and examined the effects of UBF knockdown on ribosomal gene chromatin (r-chromatin). UBF knockdown reduced UBF1/2 mRNA and protein levels by 80% compared with the control siRNA oligonucleotide duplex against EGFP (siRNA-EGFP; Fig. 1, A and B).
Quantitative chromatin immunoprecipitation (ChIP [qChIP]) analysis was used to examine the effect of UBF1/2 depletion on the level of UBF associated with the rRNA genes. Consistent with recent experiments (O'Sullivan et al., 2002), we found that UBF1/2 is enriched not only at the proximal promoter but also across the transcribed portion of the 45S rRNA gene (Fig. 1, C and D). Depletion of UBF1/2 reduced the levels of UBF1/2 associated with the rRNA genes by a mean of 60% (Fig. 1 D). Immunofluorescence microscopy of NIH3T3 fibroblasts in interphase demonstrated a significant reduction in both the number of UBF-positive nucleoli and the intensity of UBF immunostaining (Fig. 1, E and F) after inducible UBF1/2 knockdown using retroviral delivery of a tetracycline-inducible UBF1/2 short hairpin RNA (shRNA; Dickins et al., 2007).
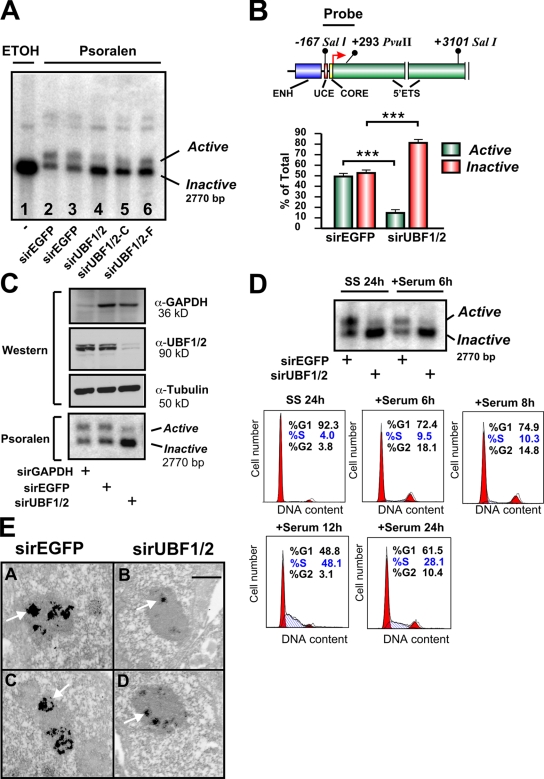
Next, we directly examined the effect of UBF depletion on the relative proportion of active and inactive (silent) ribosomal genes using psoralen, a DNA cross-linking agent. Active rRNA genes have an open chromatin structure that is accessible to psoralen and associated with nascent rRNA transcripts. Conversely, the silent genes are inaccessible to psoralen and associated with regularly spaced nucleosomes. After psoralen cross-linking, the active and inactive rRNA genes can be distinguished with Southern blotting by their differing rates of migration (Conconi et al., 1989; Dammann et al., 1993). Strikingly, psoralen analysis of the rRNA genes demonstrated a 70% reduction in the number of active genes with a reciprocal increase in the fraction of silenced genes (47.4 and 52.6% to 18.8 and 81.2%) after UBF knockdown in NIH3T3 cells (Fig. 2, A and B). The change in the proportion of active genes was not a result of off-target effects, as silencing of UBF1/2 using siRNAs to different regions of the UBF1/2 coding sequence (Fig. S1, A and B) also reduced the number of active ribosomal genes (Fig. 2 A, lanes 4–6), whereas siRNA targeting EGFP or glyceraldehyde 3-phosphate dehydrogenase (GAPDH) had no effect on the ratio of active to inactive genes (Fig. 2, A and C). UBF1/2 knockdown also increased the percentage of silenced rRNA genes in cells arrested in G0–G1 phase after serum starvation (SS; Fig. 2 D, SS 24 h) and also in cells in early G1 (Fig. 2 D, +Serum 6 h). Thus, silencing of rRNA genes (rDNA) after depletion of UBF is not dependent on growth factors or chromatin remodeling during S phase. We further examined the effect of UBF depletion using argyrophilic NOR (AgNOR) staining of nucleoli from interphase NIH3T3 cells. Nucleolar fibrillar centers contain undercondensed and thus transcriptionally competent rRNA genes. Like mitotic NORs, they can be silver stained because of their association with the Pol I transcription machinery (Roussel and Hernandez-Verdun, 1994; Weisenberger and Scheer, 1995; Heliot et al., 1997). AgNOR staining of the nucleoli demonstrated an ∼60–70% decrease in the overall intensity of silver staining and number of fibrillar centers after UBF1/2 knockdown (Fig. 2 E). The strong correlation between decreased AgNOR staining and UBF depletion is consistent with experiments demonstrating that UBF is responsible for recruiting a majority of the Pol I transcription machinery to the rDNA repeats (Mais et al., 2005). Together, these data provide strong evidence that the association of UBF with rRNA genes is necessary for maintenance of the open chromatin structure found in active NORs.
Figure 2.
UBF1/2 depletion decreases the number of active ribosomal genes. (A) NIH3T3 cells transfected with siRNA-EGFP (n = 2) or three independent siRNAs targeting UBF1/2. Nuclei were extracted and irradiated in the presence of ethanol (ETOH; lane 1) or psoralen (lanes 2–6). Genomic DNA was isolated and analyzed by Southern blotting for rRNA genes. (B) A map of the murine rRNA gene promoter and the probe used for Southern blotting (top). The proportion of active versus inactive rDNA from experiments in A was quantitated (bottom). ENH, enhancer. n = 5; mean ± SEM (error bars); ***, P < 0.001. (C) NIH3T3 cells were transfected with siRNA-GAPDH, -EGFP, or -UBF1/2 and analyzed by Western blotting (top) and psoralen cross-linking experiments (bottom). (D) NIH3T3 cells transfected with siRNA-EGFP or -UBF1/2 were serum deprived for 24 h (SS) and serum refed for 6 h (+Serum). The psoralen cross-linking assay was performed as in A (top). Cell cycle analysis of NIH3T3 cells serum starved for 24 h and serum refed for the indicated times. Fixed cells were stained with propidium iodide and analyzed by flow cytometry. The percentages of cells in G0–G1, S, and G2–M phases were determined using Modfit 3.0 software. (E) NIH3T3 were transfected with siRNA-EGFP (A and C) and -UBF1/2 (B and D), and AgNOR staining was performed. Arrows indicate stained interphase NORs. Bar, 1 μm.
UBF2 does not function in r-chromatin remodeling
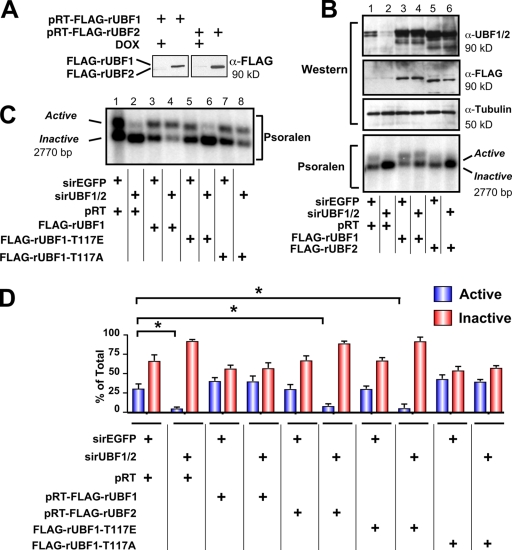
UBF2, the naturally occurring splice variant of UBF1, is a poor activator of rDNA transcription compared with UBF1 (O'Mahony et al., 1992; O'Neill et al., 1993; Kuhn et al., 1994; Hannan et al., 1999). However, the relative ability of UBF1 and UBF2 to regulate r-chromatin in vivo has not been investigated. To address this, we performed replacement experiments with rat UBF1 and UBF2, which contain nucleic acid sequence differences in the region targeted by the murine-specific siRNA-UBF1/2 (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200805146/DC1). Immortalized mouse embryonic fibroblast (MEF) cell lines (NIH3T3 MEFs) expressing tetracycline-inducible Flag epitope–tagged rat UBF1 or rat UBF2 were generated (Fig. 3 A). Western analysis demonstrated that the rat UBF1 and 2 transgenes were expressed approximately two- to threefold above endogenous levels and were resistant to the siRNA-UBF1/2 (Fig. 3 B). Knockdown of UBF1/2 reduced the percentage of active and inactive genes from 31.7 and 68.3% to 5.3 and 94.7%, respectively, after 48 h (Fig. 3, B and D). Expression of rat UBF1 in these cells was able to prevent the loss of active genes (41.3 and 58.7%), whereas rat UBF2 could not (8.5 and 91.5%; Fig. 3, B and D). Thus, UBF2 does not function in r-chromatin remodeling in vivo in the absence of UBF1.
Figure 3.
Differential regulation of active r-chromatin by UBF1 and UBF2. (A) RevTet-Off–inducible FLAG-rUBF1 and -rUBF2 NIH3T3 MEF cell lines were stimulated by doxocyclin for 48 h and analyzed by Western blotting. (B) RevTet-Off–inducible RNAi-resistant UBF1, UBF2, and pRT (empty vector) NIH3T3 MEF cell lines were transfected with siRNA-EGFP or -UBF1/2 and analyzed by Western blotting (top). Nuclei from corresponding samples were analyzed by psoralen cross-linking (bottom). (C) Nuclei from pRT and RNAi-resistant wild-type UBF1, UBF1-T117E, and UBF1-T117A–overexpressing NIH3T3 MEF cell lines were collected 48 h after transfection with siRNA-EGFP or -UBF1/2 and analyzed by psoralen cross-linking. (D) The results (n = 3) in B and C were quantitated. Mean ± SEM (error bars); *, P < 0.05.
The ability of UBF1 to regulate r-chromatin is abrogated by mutation of an extracellular signal-regulated kinase site within the HMG box 1 domain
To examine in more detail the structural requirements for UBF1 to regulate r-chromatin, we performed UBF replacement experiments as described in the previous section with structure/function mutants that affect the ability of UBF1 to bend and loop DNA in vitro and to activate transcription (Stefanovsky et al., 2001a). Phosphorylation of T117 and T201 in HMG boxes 1 and 2 of UBF1 reduces their ability to bend DNA, leading to a cooperative unfolding of the enhancesome structure, as does substitution of these residues with glutamic acid (UBF-threonin to glutamic acid mutation; Stefanovsky et al., 2001b, 2006b). The T117A UBF1 mutant that can still bend DNA and form the enhancesome was similar to wild-type UBF in its ability to remodel chromatin (Fig. 3, C and D). However, the T117E UBF1 mutant was unable to prevent the loss of inactive ribosomal genes in UBF1/2-depleted fibroblasts (Fig. 3, C and D). Thus, mutations that block the ability of UBF to bend DNA and form the enhancesome in vitro also significantly reduce the ability of UBF1 to remodel r-chromatin into an active configuration in vivo.
rRNA gene silencing in response to UBF depletion does not require NoRC-mediated chromatin-remodeling events
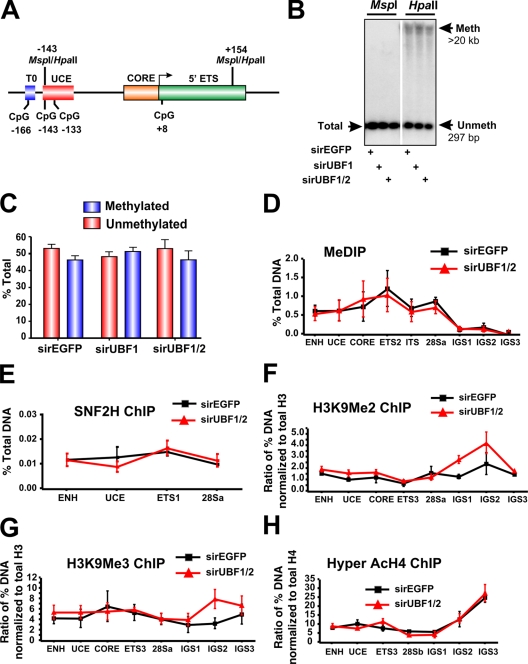
The chromatin-remodeling complex NoRC recruits DNA methyltransferases and histone deacetylases to the rRNA gene promoter–proximal terminator, contributing to the formation of a closed nucleosomal structure (Santoro and Grummt, 2005). In particular, NoRC-induced DNA methylation of a CpG dinucleotide at −133 in the core region of the rRNA gene promoter has been implicated in silencing of murine rRNA genes (Santoro and Grummt, 2001) and also has been shown to reduce UBF binding to the rRNA gene promoter. We examined the methylation status of the CpG dinucleotide at −133 in response to acute UBF1/2 depletion by Southern analysis of genomic DNA digested with the enzymes HpaII and MspI. These enzymes demonstrate differential sensitivity to methylation (Fig. 4 A; Santoro and Grummt, 2001), and the analysis revealed that ∼48% of the ribosomal genes were unmethylated at the CpG dinucleotide −133 in exponentially growing NIH3T3 cells (Fig. 4, B and C), which is in accordance with the aforementioned psoralen data and previous experiments (Santoro and Grummt, 2001). This percentage was not affected by depletion of UBF1/2 or UBF1 only (Fig. 4, B and C; and Fig. S1). We extended this analysis across the entire rRNA gene using methylated DNA immunoprecipitation (MeDIP; Fig. 4 D) but failed to observe a significant change in CpG methylation at any of the amplicons upon UBF depletion. Moreover, ChIP analysis demonstrated that enrichment of SNF2H, a component of the NoRC complex (Strohner et al., 2001; Santoro et al., 2002; Percipalle et al., 2006), was not significantly altered at the rDNA promoter and transcribed region in response to UBF depletion (Fig. 4 E). In addition, the levels of histone marks associated with NoRC-dependent gene silencing such as H3K9Me2 and H3K9Me3 (Fig. 4, F–H) were unchanged after UBF depletion. Thus, the loss of UBF is sufficient to induce a closed r-chromatin state without the need for increased rDNA methylation and other NoRC-mediated chromatin-remodeling events.
Figure 4.
DNA methylation and other heterochromatic markers are unaffected in response to UBF1/2 depletion. (A) Schematic of the murine rRNA gene promoter and 5′ ETS region with the position of restriction enzyme sites indicated. (B) Southern blot analysis of rDNA using methylation-sensitive restriction enzymes. Genomic DNA from an NIH3T3 cell transfected with siRNA-EGFP, -UBF1, or -UBF1/2 was digested with HpaII or MspI, subjected to electrophoresis, and hybridized to the probe depicted in Fig. 2 A. The white line indicates that intervening lanes have been spliced out. (C) The results (n = 2) in B were quantitated, and HpaII-sensitive (unmethylated) bands were quantitated as a proportion of the total rDNA (MspII band), and the difference was designated as methylated. (D) Analysis of DNA methylation across the rDNA by MeDIP in siRNA-EGFP or -UBF1/2 cells (n = 3). Samples were analyzed by qRT-PCR as described in Fig. 1 D. (E–H) Loss of UBF1/2 does not alter SNF2H binding or histone modifications associated with NoRC silencing of rDNA. qChIP analysis of the rRNA genes in siRNA-EGFP– or -UBF1/2–transfected NIH3T3 cells using antibodies against SNF2H (E), H3K9Me2 (F), H3K9Me3 (G), or hyperacetylated H4 (H). Samples were analyzed by qRT-PCR as described in Fig. 1 D. qChIPs (F–H) were normalized to total histone H3 or H4 loading (Fig. 6, A and B) and expressed as a ratio of the percentage of DNA (n = 3). ENH, enhancer; ITS, internal transcribed spacer; T, terminator region. Mean ± SEM (error bars).
Silencing of rDNA in response to UBF depletion is reversible
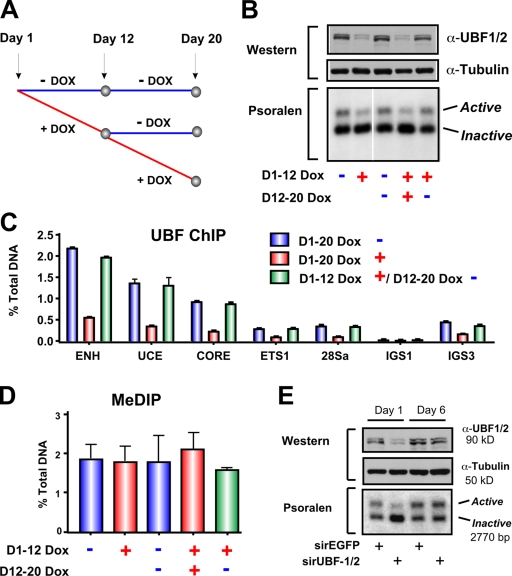
Next, we examined whether the newly silenced repeats would eventually become methylated and permanently silenced after long-term UBF depletion or, alternatively, whether they would remain unmethylated and thus could be returned to an open chromatin configuration by restoration of UBF levels. Long-term tetracycline-inducible knockdown of UBF (12 d; Fig. 5, A and B) led to a sustained reduction in the number of active rRNA genes (Fig. 5 B, first and second lanes) and reduced loading of UBF on the rDNA repeats (Fig. 5 C). This was not accompanied by increased methylation at the rRNA gene promoter (Fig. 5 D). Removal of tetracycline after 12 d of knockdown led to a recovery of UBF expression (Fig. 5 B, top) and its occupancy of the rRNA genes (Fig. 5 C), which correlated with the restoration of the number of active genes back to wild-type levels (Fig. 5 B, compare the third, fourth, and fifth lanes). CpG methylation at the rDNA promoter was again unchanged (Fig. 5 D). We also performed recovery experiments by transient knockdown of UBF using siRNA oligonucleotides (Fig. 1, A and B) and followed the recovery of UBF expression and active rRNA genes with time as the siRNA oligonucleotides were depleted. Loss of UBF siRNA led to a recovery of UBF to control levels and restoration of the active gene number (Fig. 5 E). Thus, silencing of rRNA genes in response to UBF depletion is stable and reversible.
Figure 5.
Restoration of UBF levels after short- or long-term depletion rescues the number of active rRNA genes. (A) Schematic of experimental timeline. NIH3T3 cells stably transduced with tetracycline-inducible TMP-UBF shRNAmir were cultured in the presence or absence of doxocyclin (DOX) for 12 d. Doxocyclin-treated cells were either maintained in doxocyclin-supplemented media or grown without doxocyclin for a further 8 d. (B) Cells in A were analyzed by Western blotting (top) and psoralen cross-linking experiments (bottom). The white line indicates that intervening lanes have been spliced out. (C) qChIP analysis of UBF binding to the rDNA in cells in A. UBF enrichment was determined as in Fig. 1 D. Mean ± SEM (error bars) of samples in duplicates. (D) MeDIP analysis of rDNA promoter methylation in cells in A. Samples were analyzed by qRT-PCR using the core primers as described in Fig. 1 D. Mean ± SEM (error bars; n = 2). (E) siRNA-EGFP– or -UBF1/2–transfected NIH3T3 cells from Fig. 2 C were maintained in culture for a further 7 d to allow restoration of UBF1/2 protein levels and were harvested for Western blotting (top) and psoralen cross-linking (bottom). ENH, enhancer.
rRNA gene silencing leads to increased association of linker histone H1 with r-chromatin
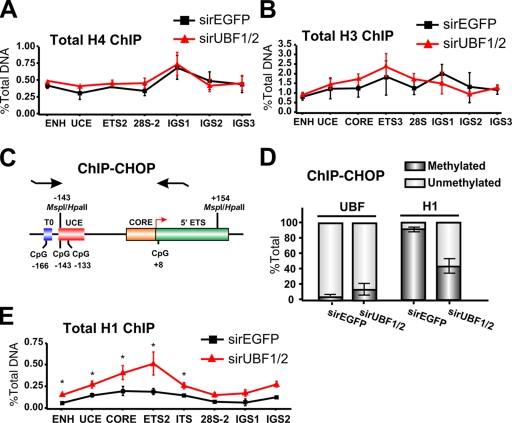
Next, we examined the mechanism by which the UBF–DNA complexes might promote decondensation of the rRNA genes. One possibility is that association of UBF with the rDNA leads to ejection of the core histone octamers, thus promoting an open r-chromatin structure. However, qChIP analysis failed to demonstrate any significant changes in the relative amount of total H3 and H4 occupancy at the promoter and transcribed portion of the rRNA genes after silencing induced by UBF1/2 knockdown (Fig. 6, A and B).
Figure 6.
Loss of UBF1/2 leads to an increase in total levels of histone H1 associated with rRNA genes. (A and B) Depletion of UBF does not alter the total levels of core histones associated with rDNA. qChIP analysis of the rRNA genes in siRNA-EGFP– or -UBF1/2–transfected NIH3T3 cells using antibodies against total histone H4 (A) or total histone H3 (B). Samples were analyzed by qRT-PCR as described in Fig. 1 D (n = 3). (C) Schematic of the ChIP-CHOP assay representing the murine rDNA promoter and the 5′ ETS region as in Fig. 4 A with the position of restriction enzyme sites and primers used for qRT-PCR indicated. (D) ChIP-CHOP assay of UBF or histone H1 ChIPs from siRNA-EGFP– or -UBF1/2–transfected NIH3T3 cells. Samples were either mock digested or digested with HpaII. The relative level of HpaII-resistant methylated rDNA was determined by qRT-PCR using the core primers, and the difference was designated as unmethylated rDNA (n = 4). (E) qChIP analysis of the rRNA genes in siRNA-EGFP– or -UBF1/2–transfected NIH3T3 cells using antibodies to total histone H1. Chromatin samples were analyzed by qRT-PCR as described in Fig. 1 D (n = 3; *, P < 0.05). Mean ± SEM (error bars).
Alternatively, experiments performed with cell-free systems demonstrate that UBF out competes linker histone H1 for binding to a nucleosome core (Kermekchiev et al., 1997), suggesting that UBF might form active r-chromatin by preventing the formation of higher order repressive chromatin structures regulated by histone H1. This model predicts that the association of UBF and histone H1 with r-chromatin in vivo would be mutually exclusive and that UBF knockdown would increase histone H1 association with the pool of rRNA genes that are undergoing silencing. To monitor relative UBF and histone H1 occupancy at silenced (methylated) and active (unmethylated) rRNA genes, we used the ChIP-CHOP assay (Lawrence et al., 2004). In this assay, immunoprecipitated DNA from UBF and histone H1 ChIP experiments was digested with HpaII before quantitative real-time PCR (qRT-PCR) using the core primers (Fig. 1 C) that span the −133 CpG dinucleotide in the rRNA gene promoter (Fig. 6 C). The percentage of unmethylated versus methylated DNA was determined and used to distinguish between active and silent rRNA genes. In wild-type cells, the active repeats are unmethylated and sensitive to HpaII cleavage, whereas the silent ones are methylated and resistant to HpaII digestion (Santoro and Grummt, 2001).
Using ChIP-CHOP, we found that immunoprecipitated UBF was almost exclusively associated with active, unmethylated rRNA gene promoters (>90%; Fig. 6 D). In contrast, immunoprecipitated histone H1 was almost entirely associated with the silenced, methylated rRNA gene promoters (>90%; Fig. 6 D). Thus, the rRNA genes are either associated with UBF or histone H1; they are mutually exclusive. UBF1/2 knockdown led to a twofold increase in the amount of H1 at the promoter and transcribed portion of the total pool of rRNA genes (Fig. 6 E). ChIP-CHOP demonstrated that this enrichment occurred on the unmethylated fraction of genes (Fig. 6, D and E). These would, presumably, correspond to the fraction of newly silenced genes depleted of UBF. Together, these results suggest a model in which direct association of UBF with the unmethylated rRNA genes prevents the assembly of transcriptionally inactive higher order chromatin structures catalyzed by linker histone H1.
UBF1 overexpression does not increase UBF loading on the rRNA genes
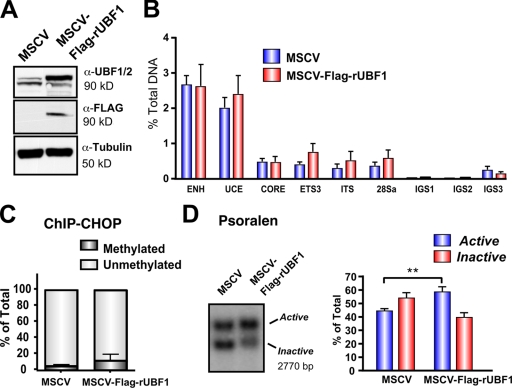
Our aforementioned data and previously published experiments (Santoro and Grummt, 2001) demonstrate that UBF does not bind to methylated rRNA genes. When considered with the UBF rescue experiments in Fig. 5, the data suggest a model whereby methylated rRNA genes are permanently silenced because of their inability to load UBF, which is required for rDNA activation. Consistent with this model, three- to fourfold overexpression of Flag-tagged UBF1 (Fig. 7 A) failed to significantly increase the amount of UBF1 at the promoter, transcribed, and IGS regions of the rDNA (Fig. 7 B). In addition, ChIP-CHOP experiments revealed that in control as well as UBF1-overexpressing cells, immunoprecipitated UBF was almost exclusively associated with unmethylated rDNA promoters (Fig. 7 C). Thus, UBF cannot be loaded onto the methylated pool of rRNA genes. Surprisingly, despite the inability to increase the net loading of UBF onto the rRNA genes, overexpression of UBF1 induced a modest but statistically significant increase in the proportion of active genes from 45.8 to 60.4% (Fig. 7 D). One explanation for the increase in active gene number without more UBF loading is that overexpression of UBF1 increases the preponderance of UBF1–UBF1 homodimers, which our data (Fig. 3) indicate would be more active in r-chromatin remodeling than UBF1–UBF2 or UBF2–UBF2 dimers.
Figure 7.
Overexpression of UBF1 is not sufficient to activate silenced rDNA. (A) Western blot analysis of NIH3T3 cell lines expressing rUBF1-MSCV or empty vector MSCV. (B) qChIP analysis of UBF binding to rDNA in cell lines in A. Samples were analyzed by qRT-PCR as described in Fig. 1 D (n = 3). (C) ChIP-CHOP assay of ChIP samples in B was performed as in Fig. 6 D (n = 3). (D) Nuclei from cells listed in A analyzed by psoralen cross-linking (left). The proportion of active versus inactive rDNA was quantitated (right; n = 5; **, P < 0.01). ENH, enhancer; ITS, internal transcribed spacer. Mean ± SEM (error bars).
Increased rRNA gene silencing leads to increased transcription from the remaining active rRNA genes
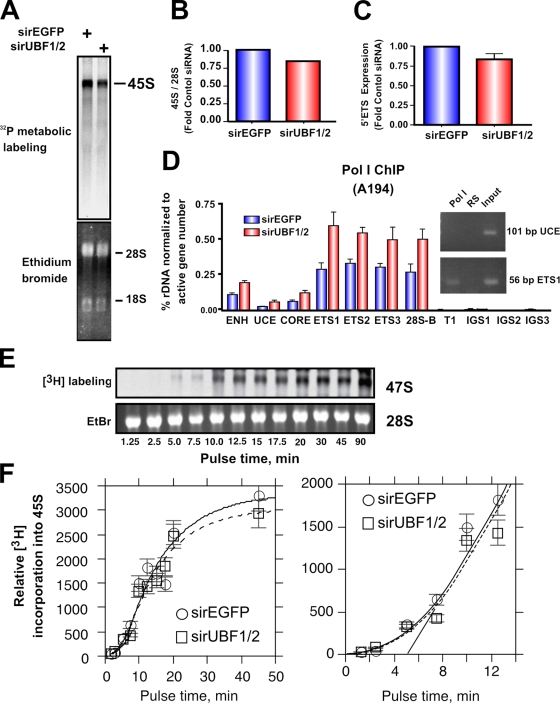
Unexpectedly, rRNA synthesis rates as measured by metabolic labeling (Fig. 8, A and B) or levels of the 5′ external transcribed spacer (ETS; Fig. 8 C) were reduced by only ∼15% in response to depletion of UBF, which is fourfold less than the decrease in number of active genes (∼70% decrease; Fig. 2, A and B). This suggests that the rate of transcription on the remaining active genes was increased. Consistent with this, ChIP analysis using antibodies to the largest subunit of Pol I (RPA194) demonstrated that UBF1/2 knockdown led to a twofold increase in Pol I loading on the remaining 15% of active rRNA genes (Fig. 8 D). This would maintain a nearly constant transcriptional output. We also examined Pol I transcription elongation rates, which are regulated by UBF and are limiting for rRNA gene transcription (Stefanovsky et al., 2006a). In vivo elongation rates were determined as we previously described (Stefanovsky et al., 2006a) using [3H]uridine pulse labeling (Fig. 8, E and F). If slowing of elongation in response to UBF depletion occurs, it would lead to an observable lag before the linear phase of label incorporation is reached. However, if the elongation rates are similar, no lag should be apparent. When 45S rRNA labeling was followed with time in NIH3T3 cells, no difference in the initial incorporation curves in the control siRNA-EGFP– and -UBF1/2–transfected cells was observed (Fig. 8 F), suggesting that UBF depletion did not affect elongation rates. Thus, increased Pol I loading per gene in the absence of appreciable changes in elongation suggests that initiation rates must have increased on the genes remaining active after UBF knockdown.
Figure 8.
UBF1/2 depletion causes a modest decrease in net rDNA transcription. (A) NIH3T3 cells transfected with siRNA-EGFP or -UBF1/2 and incubated in phosphate-free DME for 2 h and in phosphate-free DME/FBS containing 0.125 mCi/ml [32P]orthophosphate for 30 min. 32P-labeled cellular RNAs were resolved on 1.2% MOPS-formaldehyde gels and exposed on a PhosphoImaging screen. Total levels of 28S and 18S rRNAs were detected by ethidium bromide staining. (B) 45S rRNA levels in A were quantitated and normalized to corresponding total 28S levels. (C) Total RNA was extracted from siRNA-EGFP– or -UBF1/2–transfected NIH3T3 cells and normalized to an equal number of cells for each sample, and 45S rRNA precursor levels were determined by reverse transcription qRT-PCR using primers to the 5′ ETS (n = 3). (D) qChIP analysis of Pol I (A194 subunit) binding to the rDNA. Pol I enrichment was calculated as described in Fig. 1 D and normalized to the number of active rRNA genes as determined by psoralen cross-linking experiments in Fig. 2 B (n = 3). A representative ethidium bromide gel showing the amount of UCE and ETS1 products amplified after 22 PCR cycles. (E) UBF depletion does not affect Pol I elongation rates in NIH3T3 cells. NIH3T3 cells were transfected with siRNA-EGFP or -UBF1/2 and labeled with 10 μCi [3H]uridine for the indicated times. 3H-labeled cellular RNAs were extracted and resolved on 1% formaldehyde gels, transferred to membrane, and exposed to x-ray films. Total levels of 28S rRNAs were detected by ethidium bromide (EtBr) staining. (F) Duplicate analyses of 3H-labeled 45S rRNA in E were quantitated and normalized to corresponding total 28S levels (EtBr). The curves fitted to the data were calculated as previously shown (Stefanovsky et al., 2006a). The mean per gene elongation time was estimated to be 5 min by extrapolation of the linear phase of incorporation onto the time axis. ENH, enhancer; ITS, internal transcribed spacer; T, terminator region. Mean ± SEM (error bars).
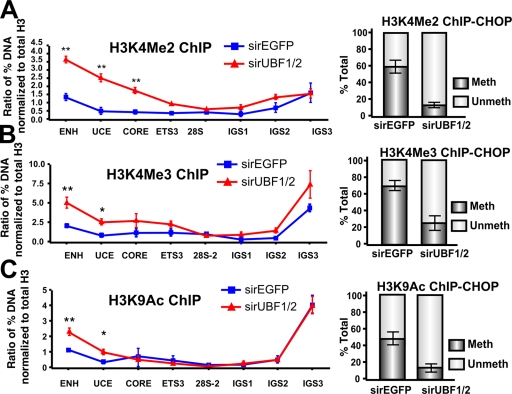
Intriguingly, the increased rate of transcription on the surviving active genes correlated with a 2.5–4-fold and a 1.5–2.5-fold increase in the euchromatic marks H3K4Me2 and H3K4Me3, respectively, across the promoter (Fig. 9, A and B; left). Similarly, we observed a twofold increase in acetylated H3K9, a marker of gene activation, at the enhancer and upstream control element (UCE; Fig. 9 C, left) of the rRNA genes. ChIP-CHOP demonstrated that in each case the increase in active chromatin marks occurred on an unmethylated fraction of rRNA genes (Fig. 9, A–C; right).
Figure 9.
UBF1/2 depletion correlates with an increase in euchromatic histone modifications at rDNA. (A–C) qChIP analysis of the rDNA in siRNA-EGFP– or -UBF1/2–transfected NIH3T3 cells using antibodies against H3K4Me2 (A), H3K4Me3 (B), and acetylated H3K9 (C). ChIP samples were analyzed by qRT-PCR as described in Fig. 1 D, normalized to total H3 loading (Fig. 6 A), and expressed as a ratio of the percentage of DNA (n = 3; *, P < 0.05; **, P < 0.01). ChIP-CHOP assays performed as in Fig. 6 D are represented graphically on the right of corresponding ChIPs (n = 3). Mean ± SEM (error bars).
Granulocyte differentiation is characterized by increased rRNA gene silencing, which correlates with decreased UBF levels but not rDNA methylation
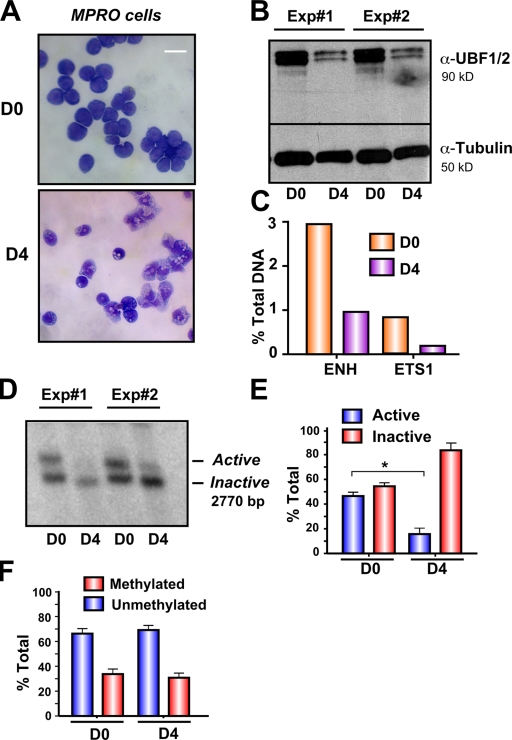
The prevailing model is that the relative amounts of active and inactive ribosomal genes are stably maintained and are not regulated in higher eukaryotic cells (Conconi et al., 1989; Stefanovsky and Moss, 2006). We have previously shown that down-regulation of rRNA gene transcription associated with terminal differentiation of the murine promyelocytic (MPRO) cell line correlates with decreased UBF1/2 expression and the amount of UBF1/2 associated with the rRNA genes (Poortinga et al., 2004). In light of our observations that UBF1 is necessary to maintain active r-chromatin, we examined whether myeloid differentiation might also be associated with a decrease in the number of active genes. Induction of the terminal differentiation of the MPRO cell line (Fig. 10 A; D0 = undifferentiated and D4 = differentiated) led to a 90% decrease in UBF1/2 expression and reduced UBF1/2 enrichment at the rRNA gene promoter and transcribed region (Fig. 10, B and C), as we have previously shown (Poortinga et al., 2004). Psoralen analysis demonstrated that the reduction in rRNA gene transcription during differentiation (Poortinga et al., 2004) correlated with a significant reduction in the number of active genes (43.7 ± 2.8% active in day 0 compared with 19.4 ± 6% active in day 4; Fig. 10, D and E). This occurred in the absence of changes in rRNA gene promoter methylation (Fig. 10 F). Thus, the pool of active ribosomal genes is not static but decreases during terminal differentiation of granulocytes most likely as a result of decreased UBF1 expression.
Figure 10.
Loss of UBF1/2 expression correlates with rDNA silencing during granulocyte differentiation. (A) Phase-contrast microscopy of uninduced granulocytic MPRO cells (D0) and differentiated granulocytes (D4) stained with May-Grunwald–Giemsa. (B) Western blots of UBF1/2 and tubulin in day 0 and day 4 cells (n = 2). (C) One representative qChIP analysis of previously published data (Poortinga et al., 2004) showing UBF binding to the rDNA in day 0 and day 4 cells. UBF enrichment was determined as in Fig. 1 D. ENH, enhancer. (D) Nuclei from day 0 and day 4 cells (n = 2) were analyzed by psoralen cross-linking assay. (E) The results (n = 3) similar to D were quantitated (*, P < 0.05). (F) Genomic DNA from day 0 and day 4 cells was extracted, digested with HpaII or MspI, and analyzed by Southern blotting of rDNA. The relative amounts of methylated and unmethylated rRNA genes are calculated as in Fig. 4 C and represented graphically (n = 3). Mean ± SEM (error bars). Bar, 30 μm.
Discussion
Despite previous investigation, the specific mechanisms that remodel the r-chromatin from an inactive condensed state into an open structure and the ensuing steps that result in the initiation of Pol I transcription remain unclear. Studies in higher eukaryotes have implicated rRNA gene promoter methylation and the posttranslational modification of histones in regulating the epigenetic silencing switch of rRNA gene clusters (Hirschler-Laszkiewicz et al., 2001; Santoro and Grummt, 2001; Lawrence et al., 2004). However, these findings are difficult to reconcile with two recent studies that indirectly implicate UBF in regulating the open chromatin state of active ribosomal genes in mammalian cells (Chen et al., 2004; Mais et al., 2005). In this study, we show that depletion of UBF1/2 leads to an increase in the number of rRNA genes in an inactive condensed state. rRNA genes inactivated in response to UBF depletion have characteristics of epigenetically silenced rDNA repeats, as they are psoralen inaccessible and stably propagated though the cell cycle and through many cell generations. However, in contrast to classical epigenetic silencing of NORs, rRNA gene inactivation in response to UBF depletion is not associated with CpG methylation and thus is reversible; restoring UBF levels restores the wild-type ratio of active to inactive genes. Thus, we term this form of rRNA gene inactivation methylation-independent silencing or pseudosilencing to distinguish it from epigenetic silencing characteristic of imprinting. These data provide strong evidence that UBF binding to rRNA repeats is necessary for maintenance of the open chromatin structure found in active NORs.
Through its HMG boxes, UBF has been shown to bend ∼140 bp of DNA into a near 360° loop. This protein DNA structure, coined the enhancesome, is central to the ability of UBF to regulate Pol I transcription at the level of elongation (Stefanovsky et al., 2006a). Our data demonstrate that mutations that block the ability of UBF1 to bend DNA and form the enhancesome in vitro also severely reduce the ability of UBF to define a unique psoralen-accessible chromatin structure across the rRNA gene in vivo, suggesting that these two processes are linked. Specifically, a UBF1 mutant (UBF-T117E) that mimics extracellular signal-regulated kinase phosphorylation at threonine 117 and is thus defective in the ability to form a functional enhancesome (Stefanovsky et al., 2001a,2001b, 2006b) was unable to prevent the loss of active genes. In contrast, the equivalent nonphosphorylatable mutant (UBF-T117A) that retains its ability to form the enhancesome in vitro was able to remodel r-chromatin to the same extent as recombinant wild-type UBF.
Interestingly, although the T117A mutant is able to function in enhancesome formation, it, like the T117E mutant, is severely compromised in its ability to regulate transcription by Pol I compared with wild-type UBF1 (Stefanovsky et al., 2001b). However, we do not think this demonstrates that transcription and chromatin remodeling can be separated. Rather, we conclude that psoralen cross-linking measures the number of actively transcribed genes but does not differentiate between highly transcribed genes (i.e., rescue with wild-type UBF1) and poorly transcribed genes (i.e., rescue with T117A). This is consistent with the observation that the number of active genes, as determined by psoralen cross-linking, does not vary between exponentially growing cells and serum-starved cells, although clearly the rate of transcription on the latter pool of rRNA genes is considerably repressed (Stefanovsky and Moss, 2006).
Our data also demonstrate that UBF1 and its natural splice variant UBF2 possess distinct abilities to remodel r-chromatin. UBF1 was able to efficiently replace endogenous UBF1/2 to maintain rRNA genes in an open chromatin state. In marked contrast, UBF2 exhibited little, if any, chromatin remodeling capability. UBF2 contains a 27–amino acid deletion in HMG box 2, which reduces the DNA-binding capacity of this domain (Stefanovsky and Moss, 2008). Thus, reminiscent of the aforementioned UBF1 mutants, UBF2 may function poorly in remodeling r-chromatin as a result of an inability to bend and loop DNA as efficiently as UBF1 (Stefanovsky and Moss, 2008). Moreover, these data also suggest that in vivo variations in UBF2 levels might function to fine tune the number of active rRNA genes by forming less active UBF2–UBF2 homodimers or UBF1–UBF2 heterodimers.
NoRC activity is not required for sustained silencing of ribosomal genes in response to UBF depletion
Importantly, our data demonstrate that ribosomal genes pseudosilenced in response to UBF depletion do not require increased CpG methylation or histone deacetylation across the rDNA repeats. These data are consistent with a model in which rRNA genes loaded with UBF are open; rRNA genes devoid of UBF are closed regardless of their CpG methylation status. In this model, the chromatin-remodeling complex NoRC lies upstream of UBF in the rRNA gene–silencing program, most likely by controlling the local r-chromatin landscape, including CpG methylation, to regulate the overall level of UBF binding to the rRNA genes (Santoro and Grummt, 2001).
Our data also demonstrate that rRNA genes pseudosilenced by UBF depletion do not become CpG methylated even after many generations, and, thus, the silencing is stable and can be reversed by restoring UBF levels. This suggests that UBF does not normally function to prevent “constitutive” NoRC-mediated methylation of rRNA genes, which is consistent with a model in which epigenetic silencing of genes through CpG methylation and UBF exclusion only occurs during defined stages during development.
Our data also allow us to make some conclusions about the mechanism by which UBF facilitates decondensation and formation of an active chromatin environment at rRNA genes. First, it is apparent that the UBF–DNA complexes and the core histones can coexist on rRNA genes. We found no evidence that UBF depletion was associated with increases in the total level of the core histones associated with the rRNA genes, which would be expected if UBF chromatin remodeling functioned to eject nucleosomes from DNA. These findings do not necessarily imply that the histones associated with active rRNA genes are nucleosomal. Indeed, much earlier experiments suggested that actively transcribed ribosomal genes remain associated with core histones, but in an unfolded “half-histone” state (Prior et al., 1983).
Second, rRNA gene silencing through loss of UBF leads to significant increases in the level of linker histone H1 associated with the new fraction of silenced genes. Thus, the combined data argue strongly that UBF contributes to the open r-chromatin structure by preventing the assembly of transcriptionally inactive higher order chromatin structures catalyzed by linker histone H1 binding.
Regulation of rRNA synthesis at the level of rate per gene rather than number of active genes
Another important finding of our study is that the reduction in the number of inactive genes through the depletion of UBF does not lead to a commensurate decrease in net cellular rRNA gene transcription rates. This was because the density of Pol I loading on the remaining active genes was increased, thus maintaining rRNA synthesis rates per cell. Such a phenomenon has been observed in yeast where artificially reducing the rRNA gene copy number from 143 to 42 does not affect growth rates because the mean number of polymerases per rRNA gene increases to maintain transcription output (French et al., 2003). Interestingly, upon UBF knockdown, a pool of unmethylated rRNA genes exhibited increases in markers of gene activation (acetylated H3K9 and di- and trimethylated H3K4) at their promoter regions (Fig. 9, A–C). It is probable that these changes are functionally associated with the increased Pol I loading.
UBF regulates the active ribosomal gene pool during differentiation
The prevailing view is that the relative amounts of active and inactive ribosomal genes are stably maintained as a result of CpG methylation of a fixed number of rRNA repeats (Conconi et al., 1989; Stefanovsky and Moss, 2006). In contrast, our experiments demonstrate that the number of silenced genes increases markedly during granulocyte differentiation. Furthermore, the silencing correlates with a reduction in UBF levels and UBF loading on the rDNA repeat but not promoter methylation of rRNA genes, suggesting that the loss of UBF is causative in the rRNA gene inactivation. Furthermore, as we and others have shown that reduced UBF expression is common during the terminal differentiation of many cell types (Larson et al., 1993; Datta et al., 1997; Alzuherri and White, 1999; Poortinga et al., 2004; Li et al., 2006; Liu et al., 2007), it is likely that down-regulation of UBF is a widespread mechanism for the silencing of active rRNA genes during development.
The level of rRNA gene silencing during granulocyte differentiation is similar to that observed during UBF depletion in NIH3T3 cells. However, UBF depletion did not change rRNA gene transcription rates significantly. Thus, although rRNA gene silencing may be required for the down-regulation of Pol I transcription during differentiation, it is not sufficient to regulate this process. Interestingly, our preliminary experiments show that during granulocyte differentiation, in addition to UBF, the expression of a majority of the other components of the Pol I complex is down-regulated (unpublished data). We propose that this results in limiting amounts of the Pol I complex, which prevents increased loading of Pol I on the remaining active rRNA genes after UBF depletion. This would ensure that rRNA gene silencing during differentiation leads to decreased rRNA synthesis rates. Future studies are needed to determine whether coordinate regulation of Pol I transcription factors and modulation of the number of active rRNA genes is a general mechanism to effect long-term changes in rRNA gene transcription rates during differentiation.
Materials and methods
Antibodies
Anti–trimethyl H3K9, anti–dimethyl H3K9, anti–trimethyl H3K4, anti–dimethyl H3K4, anti-H4, and anti-SNF2H antibodies were obtained from Abcam. Anti–hyperacetylated H4, anti–acetyl H3K9, anti-H3, anti-H2A, and anti-H1 (AE-4) antibodies were obtained from Millipore. Anti–5-methyl cytosine antibody (anti-5mC) was obtained from Megabase Research Products. Antibodies to α-tubulin and GAPDH were obtained from Sigma-Aldrich and Abcam, respectively. In-house rabbit anti–Pol I, -UBF1/2, and –rabbit sera were used for Western and ChIP assays (Poortinga et al., 2004).
RNAi
siRNAs encoding the sequences of EGFP or UBF were synthesized by Sigma-Aldrich. A complementary sequence of each oligonucleotide was designed to produce a two-nucleotide overhang at both of the 3′ ends of the duplex. The oligonucleotide RNA sequences and positions on UBF1/2 are marked in Fig. S1. ON-TARGETplus siCONTROL GAPDH siRNA was purchased from Thermo Fisher Scientific. Short hairpin–targeting UBF1/2 sequences (Fig. S1) were subcloned into the tetracycline-regulated retroviral vector TMP containing the entire micro-RNA cassette.
Cell culture, transfection, and retroviral infection
NIH3T3 cells were cultured in DME with 10% FBS at 37°C. Lipofectamine reagent (Invitrogen) was used to transfect siRNA at 25 nM according to the manufacturer's protocol, and cells were harvested 48 h after transfection unless otherwise specified. For inducible RNAi targeting UBF, NIH3T3 cells were stably cotransduced with the pRevTet-On tetracycline transactivator (Clontech Laboratories, Inc.) and TMP-UBF shRNA–micro RNA#1 (shRNAmir#1) retroviral vectors. Single clones were isolated and selected with 1 μg/ml doxocyclin to induce UBF1/2 knockdown. To establish cell lines overexpressing rattus UBF 1 (rUBF1), NIH3T3 cells were stably transduced with rUBF1–murine stem cell virus (MSCV)–GFP and empty MSCV-GFP retroviral vectors. Cells were sorted for high GFP expression using flow cytometry. The tetracycline-regulated rat UBF1, UBF2, rUBF1-T117E, and rUBF1-T117A NIH3T3 MEF cell lines were established using a RevTet-Off expression system (Clontech Laboratories, Inc.). NIH3T3 MEF cells expressing the RevTet-Off–responsive elements were infected with pRevTRE (pRT)-UBF recombinant viruses. The MPRO granulocyte cell line was generated from whole bone marrow of mice and maintained in culture as described previously (Walkley et al., 2004). Differentiation of MPRO cells into mature granulocytes was induced by stimulation with 10−6 M of the retinoid agonist AGN 194204 for 4 d (McArthur et al., 2002).
ChIP
ChIP was performed as described previously (Poortinga et al., 2004; Walkley et al., 2004). Cross-linking was achieved with 0.6% formaldehyde and assays performed using 106 cells per immunoprecipitation. For all ChIPs, 4 μg of purified antibody or 8 μl of sera was used per immunoprecipitation. Samples were analyzed in triplicate using the SYBR green dye on the ABI Prism 7000 (Applied Biosystems). To calculate the percentage of total DNA bound, unprecipitated input samples from each condition were used as reference for all qRT-PCR reactions. Primer sequences are listed in Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200805146/DC1). ChIP-CHOP assays were performed by digesting DNA with HpaII before qRT-PCR. The relative level of HpaII resistance was calculated after normalization to mock-digested DNA.
Immunofluorescence and microscopy
Cells were fixed in 3% paraformaldehyde for 10 min at room temperature, permeabilized with ice-cold 0.05% Triton X-100 in PBS for 15 min, and blocked with 5% skim milk powder and 0.5% chicken serum in PBS for 30 min. Anti-UBF sera was used at 1:500 dilution and detected with Alexa Fluor 594 anti–rabbit secondary antibody (Invitrogen) at 1:1,000 dilution. DNA was counterstained with DAPI in ProLong Gold antifade reagent (Invitrogen). Images were acquired on a microscope (BX-51; Olympus) equipped with a camera (RT model 25.4; SPOT) using the UPlanAPO 60× NA 1.2 water immersion objective. Images were acquired using Advanced software (version 4.6.4.3; SPOT). All UBF images were taken at a 1-s exposure with a gain (excitation power) of two. Settings for adjusting the image after acquisition (i.e., γ adjust and background subtract settings) were identical for all images.
RNA extraction and expression analysis
Cells were lysed in 4 M guanidine thiocyanate, 25 mM sodium citrate, pH 7.0, 0.5% sarcosyl, and 0.1 M β-mercaptoethanol, and RNA was extracted according to standard methods. To estimate RNA recovery rate, a 32P-labeled RNA probe was mixed with RNA lysates before extraction. RNA amounts were quantitated and normalized to equal numbers of cells. First-strand cDNA was synthesized using a random hexamer primer and avian myeloblastosis virus reverse transcription (Promega) according to the manufacturer's protocol. qRT-PCR was performed as described in the ChIP section. Mouse 5′ ETS primer sequences were previously published (Poortinga et al. 2004).
Sliver staining
AgNOR staining was performed as described previously (Ploton et al., 1984), and nucleoli were visualized using transmission EM.
Psoralen cross-linking assay
Cells were lysed in 10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, and 0.5% NP-40, and nuclei were pelleted, resuspended in 50 mM Tris-HCl, pH 8.3, 40% glycerol, 5 mM MgCl2, and 0.1 mM EDTA, and irradiated in the presence of 4,5,8′-trimethylpsoralen (Sigma-Aldrich) with a 366-nm UV light box at a distance of 6 cm (Conconi et al., 1989). 200 μg/ml psoralen was added at 1:20 dilution every 4 min for a total irradiation time of 20 min. Genomic DNA was isolated, digested with SalI, and separated on a 0.9% agarose gel, and alkaline Southern blotting was performed. To reverse psoralen cross-linking, filters were treated with 254-nm UV rays at 1,875 × 100 μJ/cm2 using a UV cross-linker (Stratalinker 2400; Agilent Technologies). The membrane was then hybridized to a purified 32P (Amersham)-labeled rDNA as depicted in Fig. 2 A, visualized by scanning on a PhosphoImager (GE Healthcare), and quantitated using ImageQuant (TLv2005.04; GE Healthcare).
DNA methylation and MeDIP
Genomic DNA was digested with the methylation-sensitive enzyme HpaII or MspI (Santoro and Grummt, 2001). Southern blotting was performed using the aforementioned probe. MeDIP was performed by incubating 4 μg of heat-denatured sonicated, genomic DNA with 4 μg anti-5mC antibody for 2 h at 4°C in 0.14 M NaCl, 16.7 mM Tris, pH 8.0, and 0.05% Triton X-100. DNA–antibody complexes were incubated with protein A–Sepharose beads (Millipore) for 2 h at 4°C, and the precipitates were eluted in 50 mM Tris, pH 8.0, 10 mM EDTA, and 0.5% SDS. DNA was purified and analyzed by qRT-PCR, and the percentage of bound DNA was calculated after normalization to 20 ng of input DNA.
Online supplemental material
Figs. S1 and S2 describe siRNA oligonucleotide sequences and positions within the murine UBF1/2 coding regions. Table S1 includes ChIP RT-PCR primer sequences. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200805146/DC1.
Supplementary Material
Acknowledgments
We thank Sarah Ellis for performing the EM experiments and Anna Jenkins for her contributions.
This work was supported by grants from the National Health and Medical Research Council (NHMRC) of Australia and the Cancer Council Victoria to R.D. Hannan, R.B. Pearson, and G.A. McArthur and by a National Institutes of Health grant (5R01HL077814 USA) to L. Rothblum. R.D. Hannan, R.B. Pearson, and G.A. McArthur were supported by NHMRC fellowships.
E. Robb's present address is Mental Health Research Institute, Parkville, Victoria 3052, Australia.
Abbreviations used in this paper: AgNOR, argyrophilic NOR; ChIP, chromatin immunoprecipitation; ETS, external transcribed spacer; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HMG, high mobility group; IGS, intergenic spacer; MeDIP, methylated DNA immunoprecipitation; MEF, mouse embryonic fibroblast; MPRO, murine promyelocytic; MSCV, murine stem cell virus; NOR, nucleolar organizing region; NoRC, nucleolar remodeling complex; Pol I, polymerase I; pRT, pRevTRE; qChIP, quantitative ChIP; qRT-PCR, quantitative real-time PCR; r-chromatin, ribosomal gene chromatin; rDNA, ribosomal DNA; rRNA, ribosomal RNA; rUBF, rattus UBF; shRNA, short hairpin RNA; shRNAmir, shRNA–micro RNA; SS, serum starvation; UBF, upstream binding factor; UCE, upstream control element.
References
- Alzuherri, H.M., and R.J. White. 1999. Regulation of RNA polymerase I transcription in response to F9 embryonal carcinoma stem cell differentiation. J. Biol. Chem. 274:4328–4334. [DOI] [PubMed] [Google Scholar]
- Chen, D., A.S. Belmont, and S. Huang. 2004. Upstream binding factor association induces large-scale chromatin decondensation. Proc. Natl. Acad. Sci. USA. 101:15106–15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conconi, A., R.M. Widmer, T. Koller, and J.M. Sogo. 1989. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell. 57:753–761. [DOI] [PubMed] [Google Scholar]
- Dammann, R., R. Lucchini, T. Koller, and J.M. Sogo. 1993. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 21:2331–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta, P.K., S. Budhiraja, R.R. Reichel, and S.T. Jacob. 1997. Regulation of ribosomal RNA gene transcription during retinoic acid-induced differentiation of mouse teratocarcinoma cells. Exp. Cell Res. 231:198–205. [DOI] [PubMed] [Google Scholar]
- Dickins, R.A., K. McJunkin, E. Hernando, P.K. Premsrirut, V. Krizhanovsky, D.J. Burgess, S.Y. Kim, C. Cordon-Cardo, L. Zender, G.J. Hannon, and S.W. Lowe. 2007. Tissue-specific and reversible RNA interference in transgenic mice. Nat. Genet. 39:914–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French, S.L., Y.N. Osheim, F. Cioci, M. Nomura, and A.L. Beyer. 2003. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol. Cell. Biol. 23:1558–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt, I., and C.S. Pikaard. 2003. Epigenetic silencing of RNA polymerase I transcription. Nat. Rev. Mol. Cell Biol. 4:641–649. [DOI] [PubMed] [Google Scholar]
- Hannan, R.D., V. Stefanovsky, L. Taylor, T. Moss, and L.I. Rothblum. 1996. Overexpression of the transcription factor UBF1 is sufficient to increase ribosomal DNA transcription in neonatal cardiomyocytes: implications for cardiac hypertrophy. Proc. Natl. Acad. Sci. USA. 93:8750–8755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan, R., V. Stefanovsky, T. Arino, L. Rothblum, and T. Moss. 1999. Cellular regulation of ribosomal DNA transcription: both rat and Xenopus UBF1 stimulate rDNA transcription in 3T3 fibroblasts. Nucleic Acids Res. 27:1205–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heliot, L., H. Kaplan, L. Lucas, C. Klein, A. Beorchia, M. Doco-Fenzy, M. Menager, M. Thiry, M.F. O'Donohue, and D. Ploton. 1997. Electron tomography of metaphase nucleolar organizer regions: evidence for a twisted-loop organization. Mol. Biol. Cell. 8:2199–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschler-Laszkiewicz, I., A. Cavanaugh, Q. Hu, J. Catania, M.L. Avantaggiati, and L.I. Rothblum. 2001. The role of acetylation in rDNA transcription. Nucleic Acids Res. 29:4114–4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S., L.I. Rothblum, and D. Chen. 2006. Ribosomal chromatin organization. Biochem. Cell Biol. 84:444–449. [DOI] [PubMed] [Google Scholar]
- Jantzen, H.M., A.M. Chow, D.S. King, and R. Tjian. 1992. Multiple domains of the RNA polymerase I activator hUBF interact with the TATA-binding protein complex hSL1 to mediate transcription. Genes Dev. 6:1950–1963. [DOI] [PubMed] [Google Scholar]
- Jorgensen, P., and M. Tyers. 2004. How cells coordinate growth and division. Curr. Biol. 14:R1014–R1027. [DOI] [PubMed] [Google Scholar]
- Kermekchiev, M., J.L. Workman, and C.S. Pikaard. 1997. Nucleosome binding by the polymerase I transactivator upstream binding factor displaces linker histone H1. Mol. Cell. Biol. 17:5833–5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn, A., R. Voit, V. Stefanovsky, R. Evers, M. Bianchi, and I. Grummt. 1994. Functional differences between the two splice variants of the nucleolar transcription factor UBF: the second HMG box determines specificity of DNA binding and transcriptional activity. EMBO J. 13:416–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson, D.E., W. Xie, M. Glibetic, D. O'Mahony, B.H. Sells, and L.I. Rothblum. 1993. Coordinated decreases in rRNA gene transcription factors and rRNA synthesis during muscle cell differentiation. Proc. Natl. Acad. Sci. USA. 90:7933–7936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, R.J., K. Earley, O. Pontes, M. Silva, Z.J. Chen, N. Neves, W. Viegas, and C.S. Pikaard. 2004. A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol. Cell. 13:599–609. [DOI] [PubMed] [Google Scholar]
- Li, J., G. Langst, and I. Grummt. 2006. NoRC-dependent nucleosome positioning silences rRNA genes. EMBO J. 25:5735–5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, M., X. Tu, G. Ferrari-Amorotti, B. Calabretta, and R. Baserga. 2007. Downregulation of the upstream binding factor1 by glycogen synthase kinase3beta in myeloid cells induced to differentiate. J. Cell. Biochem. 100:1154–1169. [DOI] [PubMed] [Google Scholar]
- Mais, C., J.E. Wright, J.L. Prieto, S.L. Raggett, and B. McStay. 2005. UBF-binding site arrays form pseudo-NORs and sequester the RNA polymerase I transcription machinery. Genes Dev. 19:50–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, C., and I. Grummt. 2005. Cellular stress and nucleolar function. Cell Cycle. 4:1036–1038. [DOI] [PubMed] [Google Scholar]
- McArthur, G.A., K.P. Foley, M.L. Fero, C.R. Walkley, A.J. Deans, J.M. Roberts, and R.N. Eisenman. 2002. MAD1 and p27(KIP1) cooperate to promote terminal differentiation of granulocytes and to inhibit Myc expression and cyclin E-CDK2 activity. Mol. Cell. Biol. 22:3014–3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McStay, B., M.W. Frazier, and R.H. Reeder. 1991. xUBF contains a novel dimerization domain essential for RNA polymerase I transcription. Genes Dev. 5:1957–1968. [DOI] [PubMed] [Google Scholar]
- Moss, T. 2004. At the crossroads of growth control; making ribosomal RNA. Curr. Opin. Genet. Dev. 14:210–217. [DOI] [PubMed] [Google Scholar]
- Moss, T., V. Stefanovsky, F. Langlois, and T. Gagnon-Kugler. 2006. A new paradigm for the regulation of the mammalian ribosomal RNA genes. Biochem. Soc. Trans. 34:1079–1081. [DOI] [PubMed] [Google Scholar]
- Moss, T., F. Langlois, T. Gagnon-Kugler, and V. Stefanovsky. 2007. A housekeeper with power of attorney: the rRNA genes in ribosome biogenesis. Cell. Mol. Life Sci. 64:29–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Mahony, D.J., and L.I. Rothblum. 1991. Identification of two forms of the RNA polymerase I transcription factor UBF. Proc. Natl. Acad. Sci. USA. 88:3180–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Mahony, D.J., S.D. Smith, W. Xie, and L.I. Rothblum. 1992. Analysis of the phosphorylation, DNA-binding and dimerization properties of the RNA polymerase I transcription factors UBF1 and UBF2. Nucleic Acids Res. 20:1301–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill, T.E., J.G. Smith, and E.M. Bradbury. 1993. Histone octamer dissociation is not required for transcript elongation through arrays of nucleosome cores by phage T7 RNA polymerase in vitro. Proc. Natl. Acad. Sci. USA. 90:6203–6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan, A.C., G.J. Sullivan, and B. McStay. 2002. UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol. Cell. Biol. 22:657–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panov, K.I., J.K. Friedrich, J. Russell, and J.C. Zomerdijk. 2006. UBF activates RNA polymerase I transcription by stimulating promoter escape. EMBO J. 25:3310–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percipalle, P., N. Fomproix, E. Cavellan, R. Voit, G. Reimer, T. Kruger, J. Thyberg, U. Scheer, I. Grummt, and A.K. Farrants. 2006. The chromatin remodelling complex WSTF-SNF2h interacts with nuclear myosin 1 and has a role in RNA polymerase I transcription. EMBO Rep. 7:525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploton, D., M. Menager, and J.J. Adnet. 1984. Simultaneous high resolution localization of Ag-NOR proteins and nucleoproteins in interphasic and mitotic nuclei. Histochem. J. 16:897–906. [DOI] [PubMed] [Google Scholar]
- Poortinga, G., K.M. Hannan, H. Snelling, C.R. Walkley, A. Jenkins, K. Sharkey, M. Wall, Y. Brandenburger, M. Palatsides, R.B. Pearson, et al. 2004. MAD1 and c-MYC regulate UBF and rDNA transcription during granulocyte differentiation. EMBO J. 23:3325–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior, C.P., C.R. Cantor, E.M. Johnson, V.C. Littau, and V.G. Allfrey. 1983. Reversible changes in nucleosome structure and histone H3 accessibility in transcriptionally active and inactive states of rDNA chromatin. Cell. 34:1033–1042. [DOI] [PubMed] [Google Scholar]
- Roussel, P., and D. Hernandez-Verdun. 1994. Identification of Ag-NOR proteins, markers of proliferation related to ribosomal gene activity. Exp. Cell Res. 214:465–472. [DOI] [PubMed] [Google Scholar]
- Santoro, R., and I. Grummt. 2001. Molecular mechanisms mediating methylation-dependent silencing of ribosomal gene transcription. Mol. Cell. 8:719–725. [DOI] [PubMed] [Google Scholar]
- Santoro, R., and I. Grummt. 2005. Epigenetic mechanism of rRNA gene silencing: temporal order of NoRC-mediated histone modification, chromatin remodeling, and DNA methylation. Mol. Cell. Biol. 25:2539–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro, R., J. Li, and I. Grummt. 2002. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat. Genet. 32:393–396. [DOI] [PubMed] [Google Scholar]
- Smith, S.D., E. Oriahi, H.F. Yang-Yen, W.Q. Xie, C. Chen, and L.I. Rothblum. 1990. Interaction of RNA polymerase I transcription factors with a promoter in the nontranscribed spacer of rat ribosomal DNA. Nucleic Acids Res. 18:1677–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovsky, V., and T. Moss. 2006. Regulation of rRNA synthesis in human and mouse cells is not determined by changes in active gene count. Cell Cycle. 5:735–739. [DOI] [PubMed] [Google Scholar]
- Stefanovsky, V.Y., and T. Moss. 2008. The splice variants of UBF differentially regulate RNA polymerase I transcription elongation in response to ERK phosphorylation. Nucleic Acids Res. 36:5093–5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovsky, V.Y., G. Pelletier, D.P. Bazett-Jones, C. Crane-Robinson, and T. Moss. 2001. a. DNA looping in the RNA polymerase I enhancesome is the result of non-cooperative in-phase bending by two UBF molecules. Nucleic Acids Res. 29:3241–3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovsky, V.Y., G. Pelletier, R. Hannan, T. Gagnon-Kugler, L.I. Rothblum, and T. Moss. 2001. b. An immediate response of ribosomal transcription to growth factor stimulation in mammals is mediated by ERK phosphorylation of UBF. Mol. Cell. 8:1063–1073. [DOI] [PubMed] [Google Scholar]
- Stefanovsky, V., F. Langlois, T. Gagnon-Kugler, L.I. Rothblum, and T. Moss. 2006. a. Growth factor signaling regulates elongation of RNA polymerase I transcription in mammals via UBF phosphorylation and r-chromatin remodeling. Mol. Cell. 21:629–639. [DOI] [PubMed] [Google Scholar]
- Stefanovsky, V.Y., F. Langlois, D. Bazett-Jones, G. Pelletier, and T. Moss. 2006. b. ERK modulates DNA bending and enhancesome structure by phosphorylating HMG1-boxes 1 and 2 of the RNA polymerase I transcription factor UBF. Biochemistry. 45:3626–3634. [DOI] [PubMed] [Google Scholar]
- Strohner, R., A. Nemeth, P. Jansa, U. Hofmann-Rohrer, R. Santoro, G. Langst, and I. Grummt. 2001. NoRC–a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J. 20:4892–4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkley, C.R., L.E. Purton, H.J. Snelling, Y.D. Yuan, H. Nakajima, P. Chambon, R.A. Chandraratna, and G.A. McArthur. 2004. Identification of the molecular requirements for an RAR alpha-mediated cell cycle arrest during granulocytic differentiation. Blood. 103:1286–1295. [DOI] [PubMed] [Google Scholar]
- Weisenberger, D., and U. Scheer. 1995. A possible mechanism for the inhibition of ribosomal RNA gene transcription during mitosis. J. Cell Biol. 129:561–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.